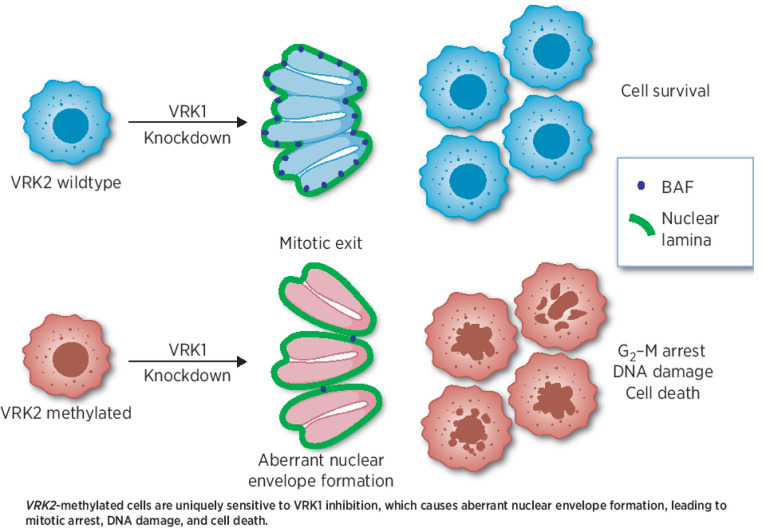
A paralog synthetic–lethal interaction between VRK1 and VRK2 sensitizes VRK2-methylated glioblastoma to perturbation of VRK1 kinase activity, supporting VRK1 as a drug discovery target in this disease.
Abstract
Synthetic lethality is a genetic interaction that results in cell death when two genetic deficiencies co-occur but not when either deficiency occurs alone, which can be co-opted for cancer therapeutics. Pairs of paralog genes are among the most straightforward potential synthetic–lethal interactions by virtue of their redundant functions. Here, we demonstrate a paralog-based synthetic lethality by targeting vaccinia-related kinase 1 (VRK1) in glioblastoma (GBM) deficient of VRK2, which is silenced by promoter methylation in approximately two thirds of GBM. Genetic knockdown of VRK1 in VRK2-null or VRK2-methylated cells resulted in decreased activity of the downstream substrate barrier to autointegration factor (BAF), a regulator of post-mitotic nuclear envelope formation. Reduced BAF activity following VRK1 knockdown caused nuclear lobulation, blebbing, and micronucleation, which subsequently resulted in G2–M arrest and DNA damage. The VRK1–VRK2 synthetic–lethal interaction was dependent on VRK1 kinase activity and was rescued by ectopic expression of VRK2. In VRK2-methylated GBM cell line–derived xenograft and patient-derived xenograft models, knockdown of VRK1 led to robust tumor growth inhibition. These results indicate that inhibiting VRK1 kinase activity could be a viable therapeutic strategy in VRK2-methylated GBM.
Significance:
A paralog synthetic–lethal interaction between VRK1 and VRK2 sensitizes VRK2-methylated glioblastoma to perturbation of VRK1 kinase activity, supporting VRK1 as a drug discovery target in this disease.
Graphical Abstract
Introduction
Recent years have witnessed a profound interest in targeting vulnerabilities in cancer stemming from synthetic–lethal interactions—an approach to cancer treatment that specifically targets cancer cells while sparing normal healthy cells, thus increasing the therapeutic index of the therapeutic agent (1–3). The success of PARP-1 inhibitors that are synthetic–lethal with BRCA1 and BRCA2 mutations as well as other “BRCA-like” defects in homologous recombination demonstrated the potential of this therapeutic approach, and a subsequent large-scale cancer dependency map (DepMap) resulted in discovery of additional novel synthetic–lethal relationships (4–6). Using our proprietary bioinformatics pipeline, referred to as Tango Cancer Dependency Map, we analyzed public functional genomics data, including the Cancer Cell Line Encyclopedia (CCLE) and identified one such novel paralog synthetic lethality, wherein vaccinia-related kinase (VRK2)–methylated GBM cell lines were sensitive to loss of VRK1.
VRKs are a family of serine/threonine kinases that play a role in regulating transcription factors, chromatin remodeling, nuclear envelope formation, and cell-cycle progression (7). There are three members of the VRK family—VRK1, VRK2, and a pseudokinase VRK3. VRK1 is found in both the nucleus and cytosol, and VRK2 localizes to the endomembrane of the endoplasmic reticulum and nuclear envelope (7). Functionally, VRK1 phosphorylates multiple substrates involved in both cell-cycle progression and cell-cycle arrest. Specifically, in response to mitogenic stimuli, VRK1 phosphorylates histones H3 and H2AX to facilitate chromatin remodeling, transcription factors ATF2, CREB, Sox2, and farnesoid X nuclear receptor HR1H4 to promote cell-cycle progression, and barrier to autointegration factor (BAF) to regulate nuclear envelope formation (8, 9). In response to stress signals, such as DNA damage, VRK1 phosphorylates p53, c-Jun, and 53BP1 to initiate cell-cycle arrest for DNA damage repair (8, 9). The functional role of VRK2 is less clear, however, it has been reported to downregulate apoptosis via direct interaction with antiapoptotic protein Bcl-xL and by downregulating proapoptotic Bax (10). p53 and BAF have also been reported as substrates for VRK2 (11, 12). In addition, cells with low expression of VRK2 have shown enhanced sensitivity to chemotherapeutics (10).
GBM is the most common primary malignant brain tumor and is uniformly fatal due to minimal success with current and novel therapies (13). Approximately 90% of grade 4 gliomas are IDH–wild-type, and these patients have lower overall survival (OS) compared with IDH-mutant (14). O6-methylguanine-DNA methyltransferase (MGMT) promoter status is also used to stratify GBMs. Patients with methylated (or partially methylated) MGMT promoters are more likely to respond to the standard-of-care chemotherapeutic temozolomide, in contrast with those with unmethylated MGMT who are unlikely to benefit from the chemotherapeutic (15). In addition to temozolomide, the current standard of care includes surgical resection when possible and adjuvant radiotherapy; however, these treatments are associated with a median survival of only 15 months and a 5-year survival rate of 6.8% (13, 16, 17).
Here, we use CRISPR-based viability studies and cDNA rescue experiments to validate the paralog synthetic lethality between VRK1 and VRK2. Specifically, we show that in tumor cell lines with high VRK2 promoter methylation, and thus low VRK2 expression, knockdown of VRK1 induces cell death via G2–M arrest and DNA damage. We demonstrate that the kinase function of VRK1 is required for the synthetic lethality that posits that VRK1-targeting can be used as an approach to treat VRK2-methylated GBM.
Materials and Methods
Cell culture
Original cell lines were acquired from the ATCC, ECACC, and JCRB. HAP1 isogenic cell lines were purchased from Horizon Discovery. All cell line stocks were routinely tested for Mycoplasma. HAP1, LN229, YKG1, KNS60, U118MG, H4, LN18, T98G, YH13, KS1, KALS1, and SW1088 were maintained in DMEM supplemented with 10% FBS and 1% sodium pyruvate. U251MG cell lines were maintained in EMEM supplemented with 10% FBS. All cell lines were maintained in a cell culture incubator at 37°C, 95% humidified air, and 5% CO2 atmosphere. GBM patient-derived xenograft (PDX) models were passaged in immune-deficient NSG mice, dissociated, and then injected into the flank of female NSG mice. Xenograft tumors were dissociated using a papain dissociation kit to obtain cancer stem cell (CSC) and non-CSC tumor fractions, cultured in supplemented neurobasal medium, and then sorted on the basis of CD133 status. CD122+ (CSC) cells were maintained in supplemented neurobasal medium, and CD133− (non-CSC) cells were maintained in DMEM supplemented with 10% FBS and 1% pen/strep. For detailed information see Supplementary Methods.
DNA constructs and cell line engineering
We used a dual vector lentiviral system for both CRISPR–Cas9 and tetracycline-inducible CRISPR–dCas9–KRAB cloning. Lentivirus was generated by transiently transfecting Lenti-X 293T cells with lentiviral packaging mix, lipofectamine 3000 transfection reagent, and constructs diluted in Opti-MEM. Virus was collected from the supernatant 48 hours after transfection and filtered with a 0.45-μm filter. Cells were infected with lentivirus and polybrene and selected in medium containing puromycin, blasticidin, or geneticin antibiotic as determined by the construct. For shRNA lentivirus generation for use with PDX models, DNA was transfected into 293T cells using calcium phosphate and then concentrated using polyethylene glycol. GBM PDX models were plated on Geltrex plates for infection with shRNA at MOI approximately 1, then selected in medium containing puromycin two days later. For detailed information see Supplementary Methods.
Colony forming assays
Cas9-expressing cells were seeded in tissue culture plates such that cells would reach 80%–90% confluency at endpoint. The next day, cells were infected with lentivirus containing CRISPR guides and polybrene transfection reagent, and the following day, selected with puromycin. Cells were left to grow for 7–14 days and stained with crystal violet. For inducible CRISPR-dCas9-KRAB experiments, doxycycline was added to medium the day after seeding, and medium containing doxycycline was refreshed every 3–4 days during the 14-day growth period.
Immunoblotting
Cells were rinsed in cold PBS and lysed in RIPA buffer supplemented with protease and phosphatase inhibitors and universal nuclease. Lysates were cleared of insoluble material by centrifuging at 20,000 × g for 10 minutes at 4°C and protein concentration was determined with the BCA Protein Assay. For immunoblotting 20–40 μg of protein in equal volumes were heated in LDS-sample buffer containing DTT for 5 minutes at 95°C. Samples were centrifuged at 20,000 × g, separated by SDS-PAGE electrophoresis in 4% to 12% Bis-Tris gels, and transferred to nitrocellulose membranes. For detailed information see Supplementary Methods.
GBM PDM growth assays
GBM PDX cells were plated on Geltrex 96-well plates at 2,000 cells per well in triplicate. Doxycycline at 1 μg/mL was added to induce shRNA expression in VRK1 shRNA transduced GBM PDX models. Images were taken every 8 hours for 10 days using the Incucyte.
Murine xenograft studies
All protocols were approved by the Institutional Animal Care and Use Committee of Pharmaron (Beijing, China) or Cleveland Clinic (Cleveland, Ohio) following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care. U251MG VRK2 low and VRK2 high cell lines were inoculated subcutaneously into 6- to 8-week-old female NOG mice (10 million cells/animal with 50% high density Matrigel in EMEM) and allowed to form palpable tumors. Once the tumors reached approximately 150 mm3, the mice were assigned to treatment groups with similar mean tumor volumes and treated with either saline or doxycycline (25 mg/kg) QD by oral gavage. Tumors were measured twice weekly throughout treatment. For intracerebral PDX implantation, mice were anesthetized, fit to a stereotaxic apparatus, and cells (30,000 cells/animal in DMEM/F12) were slowly injected 0.5-mm rostral and 1.8-mm lateral to the Bregma. Animals were monitored over time for changes in body mass and the presentation of the set of neurological and behavioral symptoms associated with end-stage brain cancer. For detailed methodology, see Supplementary Methods.
Cell-cycle analysis
Cells were treated with doxycycline at the indicated concentrations and times, trypsinized, washed in PBS, fixed in 70% ethanol, and stained with propidium iodide/RNase staining buffer. Individual cells were characterized for forward and side scatter and DNA content was determined in 10,000 cells as measured by flow cytometry (excitation at 488 nm, emission measured using 600-nm bandpass filter) with an Attune cytometer or Novocyte cytometer. Histograms and cell counts were generated using FlowJo X software.
High-content immunofluorescence imaging
Cells were cultured in CellCarrier-96 Ultra microplates in the presence or absence of 1 μg/mL doxycycline for 5 days. Cells were fixed, blocked, and incubated with primary and secondary antibodies as indicated. Plates were imaged using Phenix high-content imaging system and analysis was performed using the Harmony software. Briefly, nuclei were identified using the “find nuclei” function, and nuclei located at the periphery were removed using the “remove border objects” feature. Nuclear envelope roundness was quantified using Alexa-488 signal, and based on roundness, abnormal nuclear envelope positive cells were scored by the software. For detailed information, see Supplementary Methods.
Phospho- and total proteomics analysis
Cells were treated with and without doxycycline for the indicated times, washed twice in ice-cold PBS before harvesting. Proteins were digested using LysC/Trypsin and labeled for multiplexing using TMT labeling. Phospho proteins were collected using pSTY enrichment and fractionation and flow through from the pSTY enrichment was used for total protein-level analysis. All mass spectra were acquired on an Orbitrap Fusion Lumos coupled to an EASY nanoLC-1200 liquid chromatography system. Heatmaps were generated on normalized scaled signal data using Morpheus Software. Differential analysis between treatment groups was performed using an empirical Bayes method as in ref. 18 and gene set enrichment analysis was calculated on proteins that were altered greater than ±2 log fold-change. For detailed information, see Supplementary Methods.
Data availability
Mass spectrometry raw files are available at UCSD MassIVE repository ID # MSV000089783. All data were generated by the authors and are available upon request.
Results
VRK1 is a potential synthetic–lethal target in VRK2-methylated cancer
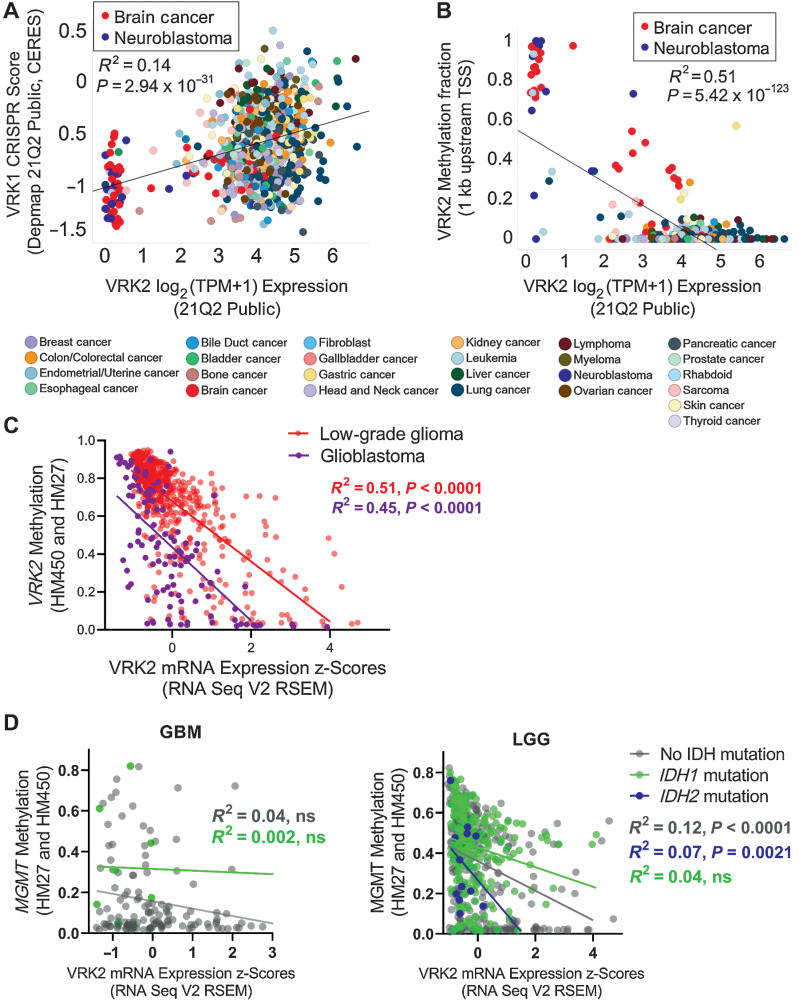
To discover novel synthetic–lethal interactions, we analyzed the large-scale cancer dependency database Achilles, where 808 cancer cell lines were screened with a genome-wide CRISPR Cas9 library to uncover genes essential for cell proliferation (19, 20). We identified a subset of cell lines with low VRK2 expression that were sensitive to VRK1 knockdown (Fig. 1A). Low VRK2 expression was primarily found in brain cancer and neuroblastoma cell lines, suggesting a neural lineage expression pattern. Genotype-Tissue Expression (GTEx) data reveal that normal human brain tissue has lower expression of VRK2 transcripts than other tissues, further suggesting a lineage-specific context (Supplementary Fig. S1A and S1B). Unlike VRK2, VRK1 is consistently expressed across all normal tissues (Supplementary Fig. S1C and S1D). Further analysis of CCLE data demonstrated that decreased expression of VRK2 strongly associated with increased promoter methylation (Fig. 1B). VRK2 is rarely deleted or mutated in cancer cell lines or human tumors (Supplementary Fig. S1E), suggesting that the VRK2 methylation is the most likely cause of low VRK2 expression. To determine whether VRK2-methylation occurs in primary human tumors, we analyzed data from The Cancer Genome Atlas low-grade glioma (LGG) and GBM datasets (21, 22). Approximately two thirds of LGG and GBM have low VRK2 expression that correlates with VRK2 promoter methylation, demonstrating that the “VRK2-low” epigenetic context is present in human brain cancer (Fig. 1C). The current prognostic markers for LGG and GBM are MGMT promoter methylation and IDH1/2 mutation, so we queried whether VRK2-low status co-occurred with either of these markers. We do not observe strong correlations between VRK2 expression and MGMT methylation status or IDH mutation in GBM and LGG, suggesting that these epigenetic and genetic events occur independently (Fig. 1D; Supplementary Fig. S1F).
Figure 1.
VRK2-methylated glioblastoma and neuroblastoma cell lines are sensitive to VRK1 loss. A, Scatter plot depicting VRK2 expression and VRK1 CRISPR knockdown sensitivity score in 783 cancer cell lines. Color coded by primary lineage. B, Scatter plot depicting VRK2 expression and VRK2 promoter methylation in 902 cancer cell lines. Color coded by primary lineage. C, Scatter plot depicting VRK2 expression and VRK2 methylation for 530 low grade glioma (LGG; peach) and 116 glioblastoma (GBM; purple) tumors. D, Scatter plots with VRK2 expression and MGMT methylation for GBM and LGG tumors as in (C), color coded by IDH mutation status.
VRK1–VRK2 synthetic lethality in vitro and in vivo in GBM cell lines
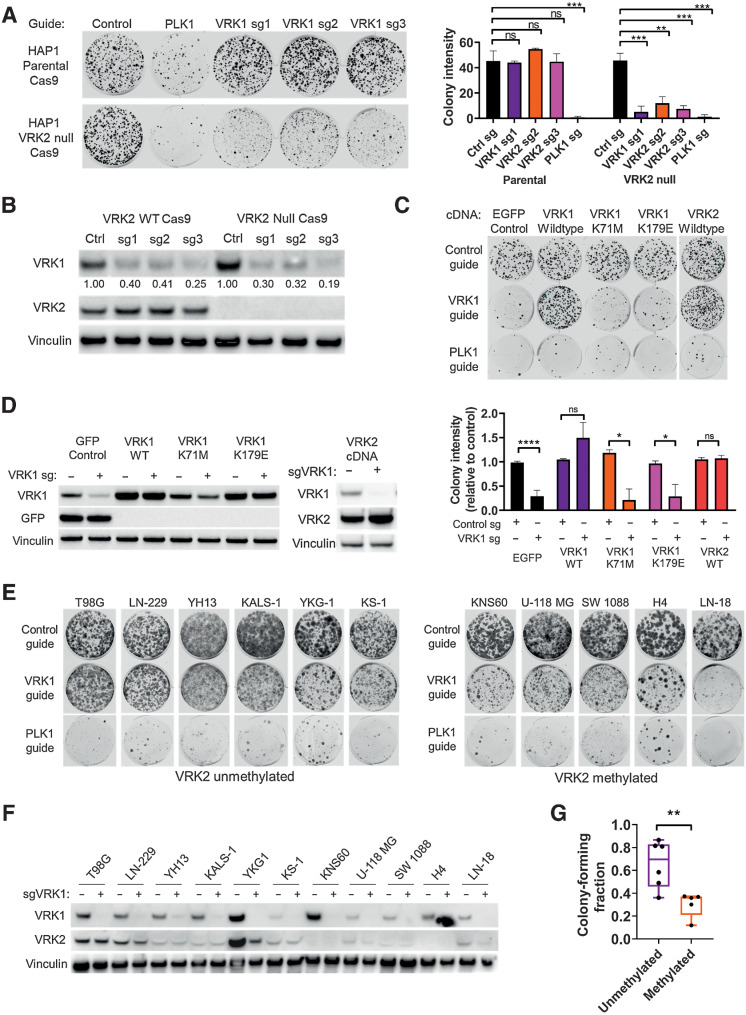
To validate the synthetic–lethal relationship between VRK1 and VRK2, we obtained a VRK2–wild-type and -null isogenic cell line pair derived from the HAP1 model. We generated Cas9 derivatives of these cell lines and knocked out VRK1 using three different sgRNAs. Knockout of VRK1 was lethal in the VRK2-null cell line in a 14-day colony forming assay, whereas the HAP1 VRK2 wild-type cells continued to proliferate (Fig. 2A). To control for Cas9 efficiency, we used sgRNA to the pan-lethal gene PLK1 and observed cell death in both cell lines. Immunoblots for VRK1 and VRK2 demonstrated 60%–80% knockdown with all three VRK1 guides and confirmed VRK2 expression levels in the cell lines (Fig. 2B). On-target knockdown was confirmed by rescuing the lethal phenotype in the HAP1 VRK2-null cells by expressing a CRISPR edit-resistant VRK1 cDNA (Fig. 2C). To determine whether the VRK1 kinase activity is important for the synthetic–lethal interaction, we engineered VRK1-kinase dead and reduced activity mutations. Lysine 71 is the catalytic lysine in the VRK1 active site and the K71M mutation eliminated VRK1 kinase activity as determined by in vitro phosphorylation activity on a VRK1 substrate (Supplementary Fig. S2A). Edit-resistant VRK1 kinase dead (K71M) mutant and a previously published reduced activity mutant (K179E; ref. 23) failed to rescue the antiproliferative phenotype, indicating that the kinase activity of VRK1 is required for the synthetic–lethal interaction. Re-expression of VRK2 in the VRK2-null cells also rescued the lethality confirming that VRK2 loss in the VRK2-null cell line is the cause of the antiproliferative phenotype. Immunoblots for VRK1 and VRK2 confirm the expression of the respective cDNA constructs (Fig. 2D). To further evaluate the synthetic–lethal interaction in VRK2-methylated GBM cell lines, we tested the effects of VRK1 knockdown in a panel of VRK2-methylated and -unmethylated GBM cell lines (Supplementary Fig. S2B). VRK2-methylated cell lines were more sensitive to VRK1 knockdown than VRK2-unmethylated cell lines in a 14-day colony forming assay (Fig. 2E and F). The data were quantified by normalizing colony intensity compared with intron cutting controls and were controlled for Cas9 efficiency by subtracting the colony intensity of PLK1 pan-lethal controls (Fig. 2G). To understand whether VRK1 knockdown affected other cancer lineages, we tested VRK1 CRISPR knockout in two non-GBM, VRK2-high expressing cell lines, RKO and SNU-398, and observed no proliferative defects in colony forming assay (Supplementary Fig. S2B and S2C). Because TP53 is mutated in 28% of glioblastoma (22), we queried whether VRK1 knockdown correlated with TP53 mutation status. We observed no correlation between TP53 mutation status and VRK1 CRISPR score in 79 CCLE cell lines of the central nervous system (Supplementary Fig. S2B and S2D). To understand the potential toxicity effects of VRK1 knockdown in normal neural cells, we knocked down VRK1 using siRNA in rat primary astrocytes and observed no proliferative defects, suggesting minimal VRK1 dependence in normal cells (Supplementary Fig. S2E).
Figure 2.
VRK1 is synthetic–lethal in VRK2-null and VRK2-methylated cell lines. A, Fourteen-day colony forming assay in HAP1 parental and VRK2-null cells with CRISPR knockdown for intron-cutting negative controls, positive control PLK1, and three VRK1 guides (left) and quantification of colonies (right). **, P < 0.01; ***, P < 0.001, one-way ANOVA and Dunnett post hoc multiple comparison test. B, Immunoblots from A at 3 days. Quantification of VRK1 bands, normalized to vinculin and relative to intron-cutting controls, are indicated below the blot. C, Fourteen-day colony forming assays of VRK1 knockdown in HAP1 VRK2-null cell line with ectopic expression of the indicated cDNA constructs (top) and quantification of colonies (bottom). *, P < 0.05; ****, P < 0.0001, two-tailed t test. D, Immunoblots from C at 3 days. E, Fourteen-day colony forming assays of VRK1 knockdown in a panel of VRK2-unmethylated and -methylated GBM cell lines. F, Immunoblots from E at 3 days depicting VRK1 and VRK2 protein expression. G, Quantification of colony forming intensities of E from two biological replicates, corrected for Cas9 cutting efficiency. **, P < 0.001, two-tailed t test. n.s., not significant.
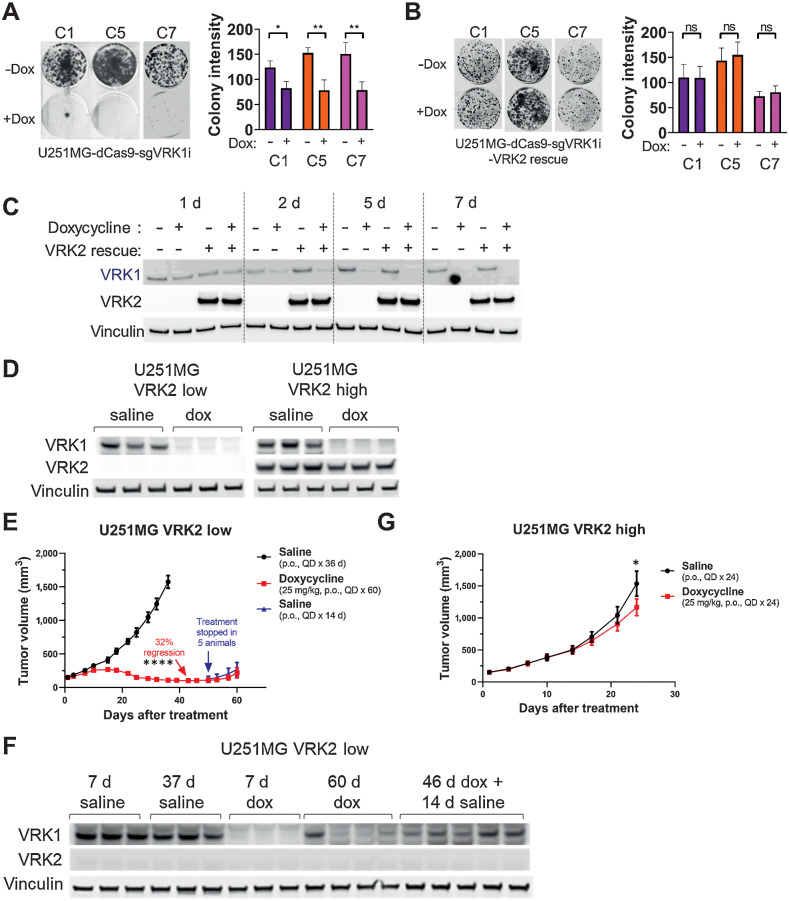
To extend our in vitro findings to the in vivo setting, we first established doxycycline-inducible VRK1 knockdown in the VRK2-methylated U251MG GBM model. Clonal U251MG cells expressing ubiquitous dCas9–KRAB and doxycycline-inducible CRISPR guides for the VRK1 promoter (henceforth referred to as “VRK2-low”) were sensitive to knockdown of VRK1 in a 14-day colony forming assay (Fig. 3A). We observed a modest, but significant increase in apoptosis at the 7-day timepoint with a luminescence-based Annexin V assay (Supplementary Fig. S3A). We generated “VRK2-high” derivatives of the clonal cell lines by re-expressing ubiquitous VRK2 cDNA and observed a full rescue of the antiproliferative and apoptosis phenotypes (Fig. 3B; Supplementary Fig. S3A). Protein levels of VRK1 and VRK2 were verified by immunoblotting (Fig. 3C). Similarly, doxycycline-inducible knockdown of VRK1 in a VRK2-unmethylated LN229 GBM cell line showed no proliferative defect in vitro (Supplementary Fig. S3B and S3C). To evaluate the synthetic–lethal interaction in vivo, mice harboring established, subcutaneous xenografts of U251MG VRK2-low and VRK2-high derivative cell lines were treated with doxycycline or saline solution via oral gavage. A subset of tumors from all arms of the study were collected at seven days after treatment and immunoblots demonstrated successful doxycycline-induced VRK1 knockdown in both models and sustained VRK2 expression in the VRK2-high tumors (Fig. 3D). Tumors in the VRK2-low derivative models regressed after 36 days on treatment compared with the vehicle-treated arm. Doxycycline treatment was stopped in five mice and continued in four mice at day 50, at which time, the tumors started to regrow in both arms (Fig. 3E). Immunoblots of these tumors at endpoint (day 60) reveal re-expression of VRK1 protein, suggesting that selection pressure to maintain VRK1 expression is required for survival in vivo in the absence of VRK2 expression (Fig. 3F). Tumors in the VRK2-high group continued to grow in the presence of doxycycline compared with the saline arm (Fig. 3G), suggesting that VRK2 is a key predictor of response to VRK1 inhibition in this model.
Figure 3.
The VRK1–VRK2 synthetic lethality is maintained in vivo. A, Fourteen-day colony forming assays in U251MG VRK2-low cell line in the absence or presence of 1 μg/mL doxycycline to induce VRK1 knockdown (left) and quantification (right). *, P < 0.05; **, P < 0.001, two-tailed t test. B, Assay similar to A in the U251MG VRK2-high cell line. C, Immunoblots from A to B at 1, 2, 5, and 7 days after doxycycline treatment. D, Immunoblots from U251MG VRK2-low and VRK2-high xenografts in tumor-bearing mice treated with or without 25-mg/kg doxycycline QD for 7 days. E, Tumor growth curves in mice bearing established 150 mm3 U251MG VRK2-low xenografts were treated with the indicated treatments. Data are presented as mean tumor volume ± SEM with 9 mice/data point up to day 42. ****, P < 0.0001 (two-tailed t test at day 42). F, Seven-day and endpoint immunoblots tumors from E. G, Tumor growth curves as in E with U251MG VRK2-high xenografts. Data are presented as mean tumor volume ± SEM with 9 mice/data point. *, P = 0.032 (two-tailed t test at day 24). ns, not significant.
VRK1 regulates BAF activity to maintain nuclear envelope integrity
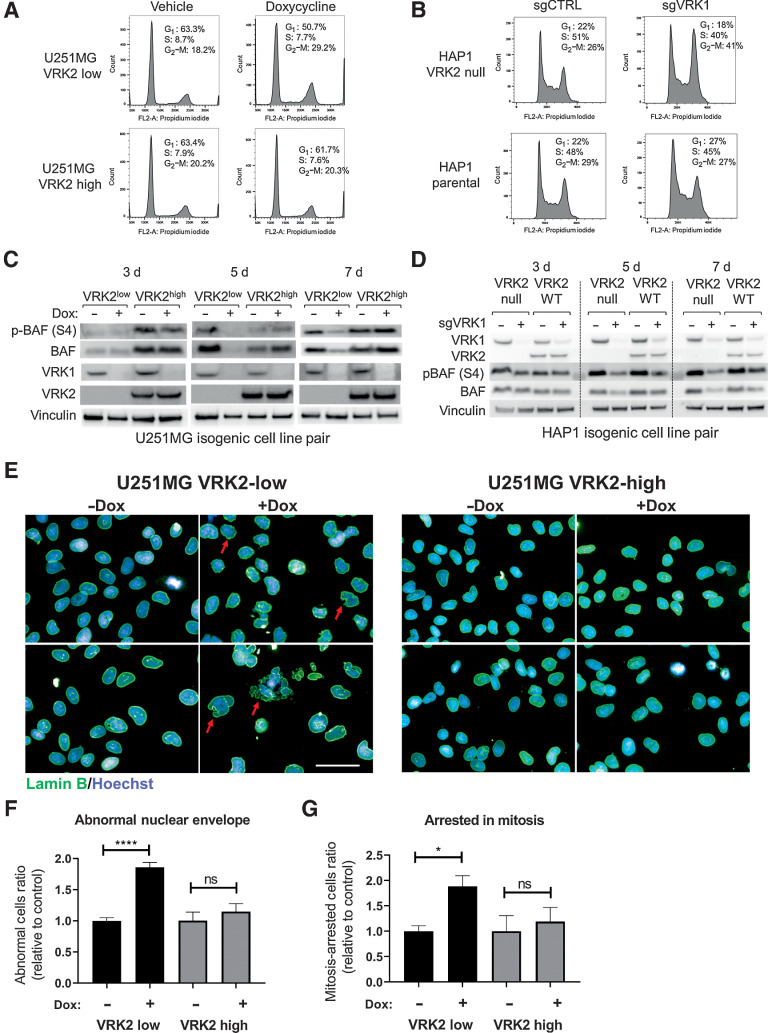
Because VRK1 plays an important role in cell-cycle progression (9), we queried whether VRK1 knockdown led to aberrant cell cycling in the VRK2-low context. Doxycycline-induced VRK1 knockdown for seven days provoked a G2–M arrest in the U251MG VRK2-low cell line as demonstrated by flow cytometry of propidium iodide–stained cells (Fig. 4A; Supplementary Fig. S4A). The VRK2-high U251MG derivative continued to cycle, suggesting that the mechanism of VRK1–VRK2 synthetic lethality involved a G2–M arrest. A similar G2–M arrest was also observed in the HAP1 VRK2-null, but not HAP1 VRK2–wild-type cells upon VRK1 knockdown (Fig. 4B; Supplementary Fig. S4B). To query whether VRK1 knockdown altered the ability of cells to migrate, we performed a wound healing assay in the U251MG VRK2-low and VRK2-high cell lines and observed no increase in cellular migration (Supplementary Fig. S4C). Because VRK1 phosphorylates several substrates involved in mitosis, we profiled the phosphorylation status of the reported substrates histone H3 (T3, S10), p53 (T18), and BAF (S4). We were unable to detect any phosphorylation of p53 at Thr18 with several commercially available antibodies (data not shown). Consistent with a recent study (24), we did not observe significant alterations in the phosphorylation of Histone H3 at Thr3 and Ser10 (Supplementary Fig. S4D). As there is no commercially available antibody for the detection of phosphorylated BAF, we generated a custom polyclonal phospho-BAF (S4) antibody. Lambda phosphatase-treatment of HAP1 lysates demonstrated that the antibody recognized a mixture of phospho-BAF and total-BAF proteins (Supplementary Fig. S4E). Phosphorylation of BAF (S4) was depleted upon VRK1 knockdown (Fig. 4C; Supplementary Fig. S4D) and restored upon VRK2 re-expression (Fig. 4C). A similar alteration in phosphorylation of BAF (S4) was observed in the HAP1 VRK2-null cell line with VRK1 knockout (Fig. 4D), and in the U251MG VRK2-low xenografts in vivo (Supplementary Fig. S4F). Consistent with previously published data, total BAF was also depleted upon VRK1 knockdown in both cell line models (25, 26). BAF, encoded by the BANF1 gene, is a highly conserved chromatin binding factor that regulates post-mitotic nuclear envelope formation by linking nuclear envelope proteins to the DNA (27). Depletion of BAF results in aberrant nuclear envelope formation, nuclear blebbing, and multinucleation (28). Knockdown of VRK1 for five days led to abnormal nuclear envelope, nuclear lobulation, multinucleation, and arrested mitotic spindles as determined by lamin B immunofluorescent staining (Fig. 4E–G). This phenotype was not due to a doxycycline effect because a nontargeting control guide did not produce nuclear envelope defects in the presence of doxycycline (Supplementary Fig. S4G). Re-expression of VRK2 rescued the abnormal nuclear envelope phenotype (Fig. 4E–G). Importantly, VRK1 knockdown in the VRK2-high LN229 cell line did not result in proliferation defects, mitotic arrest, or nuclear envelope defects (Supplementary Figs. S3B and S4H and S4I). Taken together, these data suggest that the synthetic lethality depends, in part, on the VRK1 and VRK2 substrate BAF.
Figure 4.
VRK1 knockdown results in G2–M arrest, BAF deregulation, and nuclear lamina defects. A, Cell-cycle distributions were determined by flow cytometric analyses in U251MG VRK2-low and VRK-high cells in the presence or absence of 1 μg/mL doxycycline for 7 days. B, Cell-cycle distributions were determined by flow cytometric analyses in HAP1 VRK2-null and parental cells 5 days after VRK1 CRISPR knockdown. C, Immunoblots of U251MG VRK2-low and VRK2-high cells treated with or without doxycycline for 3, 5, and 7 days. D, Immunoblots of HAP1 VRK2-null and parental cells knocked out with VRK1 or intron-cutting control guides for 3, 5, and 7 days. E, U251MG VRK2-low and VRK2-high cells were treated with or without doxycycline for 5 days and immunostained for lamin B (green) and Hoechst (blue) and imaged by high-content imaging. Scale bar, 50 μm. F, Quantification of abnormal nuclear envelope from E. G, Quantification of cells arrested in mitosis from E. *, P < 0.01; ****, P < 0.0001; ns, not significant (two-tailed t test).
The VRK1–VRK2 synthetic lethality activates the DNA damage response pathway
To further understand the mechanism underlying the synthetic lethality, we performed global phospho- and total proteomic profiling in the U251MG VRK2-low and VRK2-high cell lines at 5 and 7 days after doxycycline treatment. Global changes in the proteome and phosphoproteome were more marked at 7 than 5 days, and thus, we performed differential analysis between VRK2-low and VRK2-high cell lines in the presence of doxycycline at the 7-day timepoint in both datasets (Fig. 5A; Supplementary Table S1). Consistent with our previous findings, differential analysis of proteomic profiling revealed notable gene set enrichment analysis enrichment of proteins involved in G2–M arrest such as PLK1, AURKA, AURKB, BIRC5, CDC6, and CCNB1 at seven days after doxycycline treatment (Fig. 5B and C; Supplementary Table S2). The changes in G2–M proteins were observed as early as 5 days (Supplementary Fig. S5) and at the later 7-day timepoint, we also observed an accumulation of proteins involved in DNA repair such as RAD51, PCNA, RPA3, and members of the RFC complex. The phosphoproteomics data (Fig. 5D) also indicate a G2–M arrest due increased phosphorylation of CDK1 (CDC2) at Tyr15 that inhibits the progression of the cell cycle. We additionally observed increased phosphorylation of CDC20 at Ser51 that is reported to accumulate during mitosis (29). Consistent with the nuclear envelope defect in VRK2-low cells upon VRK1 knockdown, we observed an accumulation of phosphorylated lamin A/C and TMPO (Fig. 5D) that signal nuclear envelope breakdown (30). Phosphorylated BAF was not detected in this dataset likely due to the small-molecular weight and low abundance of the protein. The G2–M and DNA damage response (DDR) proteins were not altered in the VRK2-high cell line, suggesting that these processes are involved in the VRK1–VRK2 synthetic–lethal interaction. A subset of G2–M and DDR markers were validated by immunoblotting (Fig. 5E). These data suggest that the synthetic–lethal mechanism of VRK1 perturbation in a VRK2-low GBM cell line is G2–M cell-cycle arrest and subsequent DNA damage.
Figure 5.
Phospho- and total proteomics reveals DNA repair pathways are activated upon VRK1 knockdown in VRK2-low cells. A, Heat maps showing total proteomics (left) and phosphoproteomics (right) in VRK2-low and VRK2-high U251MG cells at 5 and 7days after doxycycline. B, Gene set enrichment analysis of 7-day total proteomics data in the doxycycline conditions. C, Volcano plots of differential expression analysis of total proteomic data. The x-axis represents log2-fold change and the y-axis represents the FDR [−log10(q value)]; black circles, proteins with greater than ± 2 log-fold change. D, Volcano plots of differential expression analysis of phosphoproteomic data. The x-axis represents log2-fold change and the y-axis represents the FDR [−log10(q value)]; black circles, proteins with greater than ± 2 log-fold change. E, Immunoblots of select proteins from U251MG VRK2-low and VRK2-high cells treated with or without doxycycline for 7 days.
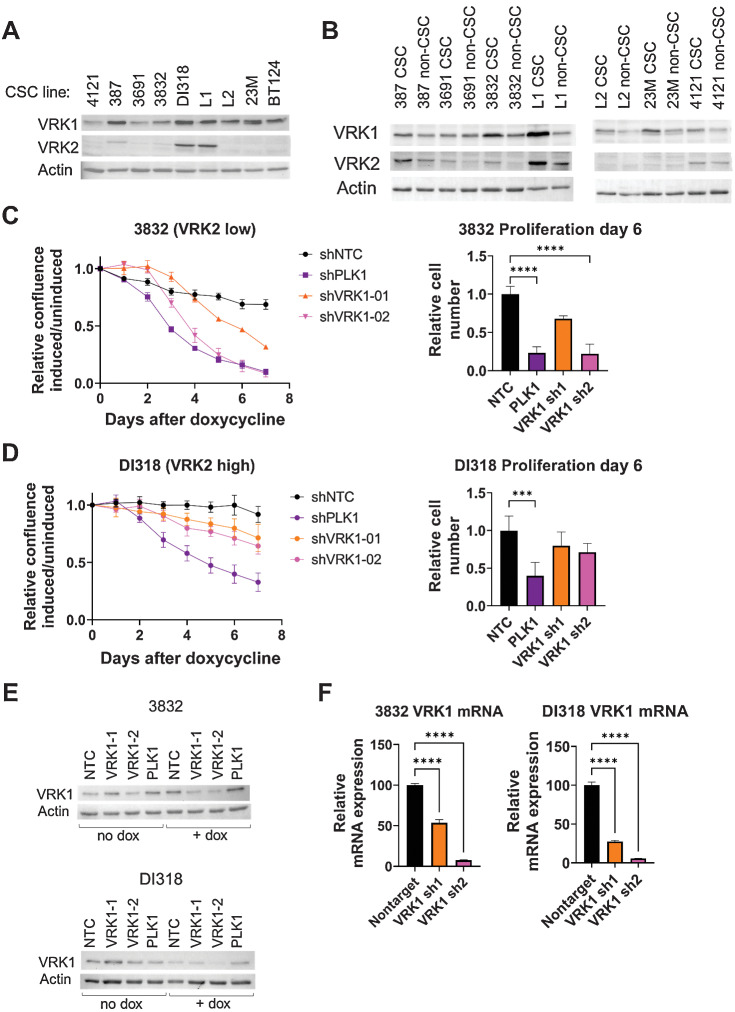
VRK2-methylated patient-derived GBM models are sensitive to VRK1 knockdown
Because commercially available cell line models of GBM are not representative of the heterogeneous nature of the disease (31), we evaluated the VRK2 expression levels in patient-derived GBM models. We performed immunoblotting for VRK2 in a panel of PDX GBM models and observed low VRK2-expression in seven out of nine models (Fig. 6A; Supplementary Fig. S6A). Measurement of VRK2 mRNA transcript levels by quantitative RT-PCR corroborated the immunoblot results (Supplementary Fig. S6B). Because GBM contains self-renewing, tumorigenic CSCs that contribute to tumor initiation and therapeutic resistance (32), we asked whether VRK2 expression differs between differentiating and nondifferentiating media conditions. Each of the models were cultured either in CSC (nonadherent, serum-free) or in non-CSC (adherent, with serum) promoting conditions as previously described (33). VRK2 protein levels measured by immunoblotting remained unchanged in both media conditions, suggesting stability of the promoter methylation regardless of differentiation status (Fig. 6B). In a subset of these models, we queried the methylation status of the VRK2 promoter using bisulfite-treatment of genomic DNA followed by PCR amplification and Sanger sequencing. We observed no notable difference in the CpG methylation status in the different media conditions (Supplementary Fig. S6C). These data demonstrate that the VRK2-low context is maintained in patient-derived GBM models.
Figure 6.
VRK2-methylated context is maintained in patient-relevant glioblastoma models. A, Immunoblots from 9 GBM CSC models. B, Select CSC models were cultured in CSC or non-CSC medium and immunoblotted for VRK1 and VRK2 protein. C, Seven-day growth curves of the 3832 model with shRNA knockdown induced by doxycycline of the indicated genes (left), and data from day 6 quantified (right). ****, P < 0.001, one-way ANOVA and Dunnett post hoc multiple comparison test. D, Data as in C for the DI318 model. E and F, Immunoblots (E) and mRNA levels (F) for VRK1 at 3 days after doxycycline.
To assess the effects of VRK1 knockdown in PDX models in vitro, we picked the 3832 (VRK2-low) and DI318 (VRK2-high) models and derived stable cells lines with doxycycline-inducible shRNA's targeting VRK1, PLK1 (pan-lethal control), and nontargeting control. VRK1 knockdown in the VRK2 low 3832 model resulted in robust antiproliferative effects, with the more efficient hairpin (shVRK1–2) achieving effects similar to PLK1 knockdown (Fig. 6C). We observed a modest, but not significant, antiproliferative effect in the VRK2-high model (Fig. 6D). Immunoblots and qPCR for VRK1 at 3-days after doxycycline induction demonstrate that the knockdown efficiency correlates with the antiproliferative effects observed in the 3832 model (Fig. 6E and F).
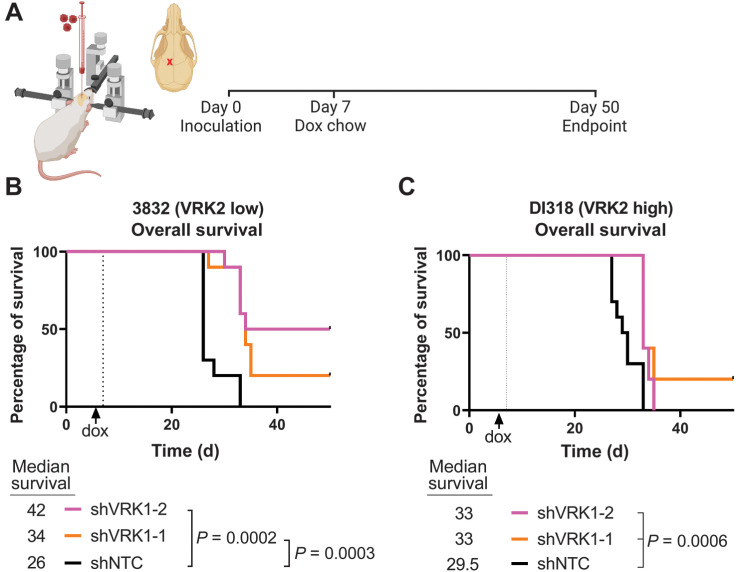
To extend these findings to the in vivo orthotopic setting, we performed intracerebral injections of the 3832 (VRK2-low) and DI318 (VRK2-high) shRNA-derived PDX models in mice. Tumors were established for 7 days before VRK1 knockdown with doxycycline (Fig. 7A). Animals tolerated the doxycycline chow well and were observed daily for signs of neurobehavioral symptoms associated with end-stage brain cancer. OS improved significantly in the 3832 VRK2-low model upon VRK1 knockdown, with 20% to 50% of the animals surviving past study endpoint in the shVRK1 groups (Fig. 7B). Corroborating the in vitro data, a modest improvement in survival (0%–20%) was observed in the animals implanted with the DI318 VRK2-high shVRK1 groups (Fig. 7C). These data suggest that the VRK2-low 3832 PDX model is more sensitive to VRK1-knockdown compared with the VRK2-high DI318 PDX model in vitro and in vivo in the brain microenvironment.
Figure 7.
VRK2-methylated patient-derived intracranial xenografts are sensitive to VRK1 knockdown. A, Schematic of experimental setup (created in BioRender.com). B and C, Kaplan–Meier curves depicting OS in mice with intracranial xenografts of 3832 and DI318 shRNA derivative models (10 mice/group); median survival (days) indicated below graphs; P values calculated with the log-rank test.
Discussion
Recent data from genome-scale CRISPR-Cas9 screening across hundreds of cancer cell lines have resulted in nomination of multiple novel targets for potential therapeutic development (34–36). In this study, we identified VRK1 as a paralog synthetic–lethal target in VRK2-methylated GBM and neuroblastoma cell lines. Using CRISPR- and shRNA-based genetic tools, we demonstrate that knockdown of VRK1 in VRK2-null and VRK2-low expressing GBM cell lines is lethal, and results in defective nuclear envelope formation, G2–M arrest, and subsequent DNA damage. The synthetic–lethal interaction is recapitulated in vivo in a VRK2-methylated U251MG GBM xenograft model, where VRK1 knockdown leads to tumor regressions. Xenografts from an isogenic VRK2-high U251MG cell line are insensitive to VRK1 knockdown, suggesting that the sensitivity depends solely on VRK2 expression. The sensitivity to VRK1 knockdown is phenocopied in a VRK2-low PDX model implanted intracranially in mice, wherein we observe significantly improved OS. Importantly, our study demonstrates that the enzymatic activity of VRK1 is required for the VRK1–VRK2 synthetic lethality, which provides a path for small molecule drug discovery. Patient data indicate that the VRK2-methylated/VRK2-low context is present and may be common in LGG and GBM tumors. Taken together, these findings suggest that VRK1 is a promising synthetic–lethal drug target in VRK2-methylated brain tumors, an aggressive indication with few therapeutic strategies currently available.
Past studies have identified synthetic–lethal relationships among paralog genes such as SMARCA2-SMARCA4, ARID1A-ARID1B, and CREBBP-EP300 (37). The advantage of a synthetic–lethal therapeutic approach for cancer is the inherent large therapeutic index that maximizes antitumor efficacy while minimizing dose-limiting on-target toxicities (1). However, creating a paralog-selective inhibitor with these qualities hinges on developing a highly selective inhibitor that spares the nontarget paralog despite nearly identical protein sequence homologies. Through mutant and wild-type cDNA rescue experiments, we show that the kinase function of VRK1 is essential in the synthetic–lethal interaction. We believe that selectively targeting VRK1 over VRK2 may be possible as the kinase domains of VRK1 and VRK2 have approximately 80% protein sequence identity. Recent structural biology analysis revealed differential mechanisms for stabilization of an ATP-competitive inhibitor in the binding pocket of VRK1 compared with VRK2 (38, 39), further supporting the possibility for development of a paralog-selective VRK1 kinase inhibitor.
VRK1 is one of the most abundant nuclear kinases in human cells and its overexpression is associated with poor prognosis in many solid tumors, including GBM and neuroblastoma (40–43). We have shown for the first time that tumors with the epigenetic context of VRK2-methylation may benefit from selective inhibition or degradation of VRK1. Publicly accessible cancer cell line data suggest that the VRK2-methylated context is restricted to cancer stemming from the neural lineage, that is, GBM and neuroblastoma. Healthy human tissue expression data from the GTEx project further corroborate the lineage effect with lower VRK2 expression in neural-derived tissues compared with other tissues (Supplementary Fig. S1A). Additional evidence of a lineage effect comes from genetics on people with germline mutation of VRK1, which results in a neurological disease that manifests as prenatal microcephaly with pontocerebellar hypoplasia (44–46). Paralleling the human disease, mice with partial Vrk1 knockdown have reduced brain weight and mild motor dysfunction (47). We posit the susceptibility of the developing brain to loss of VRK1 may be due to a naturally occurring synthetic–lethal interaction stemming from reduced VRK2 expression in the neural cells compared with other lineages. As it pertains to drug discovery, on-target toxicity of a VRK1 kinase inhibitor may be limited to the neural lineage, that is, the central and peripheral nervous systems. Given the mitotic mechanism of synthetic lethality demonstrated in this report, and the lack of proliferative defects when VRK1 is knocked down in normal neural cells (Supplementary Fig. S2E), the toxicity of a potential VRK1 inhibitor will likely be minimal. These assumptions would need to be tested in drug toxicity studies once a selective VRK1 therapeutic agent is available.
We demonstrate that knockdown of VRK1 in VRK2-null and VRK2-low expressing GBM cell lines is lethal, and results in defective nuclear envelope formation, G2–M arrest, and subsequent DNA damage. Previous studies have identified VRK1 as a regulator of cell cycle with roles in G1–S progression (48, 49) and mitosis (50–52). In our studies, VRK1 knockdown in the VRK2-low context results in G2–M arrest (Fig. 4A and B; Supplementary Fig. S4A and S4B). Given the localization of VRK2 to nuclear envelope and surrounding endo-membranes (53) and the nuclear envelope phenotype we observe (Fig. 4E–G), it is plausible that the redundancy in VRK1 and VRK2 lies in their role governing nuclear envelope formation. Although many substrates have been reported for VRK1 (54), our results demonstrate that BAF may be critical in the VRK1–VRK2 synthetic–lethal interaction. We observe that knockdown of VRK1 leads to downregulation of BAF activity and results in a nuclear envelope defect that phenocopies BAF depletion (28, 55–58). It is interesting to note that we and others observe decreases in both phosphorylated and total BAF, which may be due to protein instability in the absence of phosphorylation, a hypothesis that will require further testing (25, 26). In addition to post-mitotic nuclear envelope assembly, BAF is involved in regulation of the DDR and intrinsic immunity (27). Inherited germline BANF1 mutations, which results in instability of the BAF protein, cause Nestor-Guillermo Progeria Syndrome, a premature aging disease characterized by genome instability and accumulation of DNA damage (59, 60). On the basis of this study and previously published data, we postulate that the increased DDR observed is due to genomic instability arising from the defective post-mitotic nuclear envelope. Interestingly, BAF also regulates cell-intrinsic immunity during mitosis by preventing cytosolic cGAS activation on genomic self-DNA (61). BAF dynamically outcompetes cGAS for DNA binding and prohibits the formation of DNA-cGAS complexes that are essential for inducing the type I interferon response. We did not observe cGAS activation or type I IFN response in our proteomics dataset, suggesting that the pathway is not involved in the VRK1–VRK2 synthetic lethality in the U251MG cell line. Taken together, our study suggests that BAF plays a role in the VRK1–VRK2 synthetic–lethal interaction.
Despite aggressive treatments for newly diagnosed GBM, almost all patients relapse with more aggressive tumors with minimal treatment options within 1 to 2 years. To date, efforts to develop treatments based on genetic alterations such as EGFR amplification, CDKN2A loss, TERT promoter mutation or PTEN mutation have been unsuccessful (62). Our findings indicate that VRK1 is a potential target for synthetic–lethal therapy in VRK2-methylated GBM, an aggressive indication with few therapeutic options. Given the epigenetic mechanism of VRK2 loss, it is plausible that demethylation of the VRK2 promoter will be a resistance mechanism to a potential VRK1 therapy. We did not observe any re-expression of VRK2 in the U251MG VRK2-methylated xenografts that recurred in vivo (Fig. 3E–G), suggesting stability of VRK2 promoter methylation. Nonetheless, it will be important to consider a combination strategy to overcome such resistance mechanisms. Interestingly, a recent study demonstrated that knockdown of VRK1 in vitro synergizes with temozolomide treatment by augmenting the DDR (63). Together, these data suggest that a VRK1 inhibitor may be used as a single agent or in combination with the current standard of care to augment therapeutic response.
The results of this study uncover a novel paralog synthetic–lethal interaction between VRK1 and VRK2 in GBM. We demonstrate that knockdown of VRK1 is lethal in VRK2-methylated GBM cell lines in vitro and in vivo, and the kinase activity of VRK1 is important in the interaction. The mechanism underpinning the lethality is BAF deregulation, resulting in aberrant nuclear envelope formation, G2–M arrest, and subsequent DNA damage. These findings support the significant therapeutic potential of a VRK1 inhibitor in VRK2-methylated GBM.
Supplementary Material
Acknowledgments
The authors thank Dr. D. Whittington for structural analysis of VRK1 and Dr. M. “Masha” Alimova for assistance with high-content imaging analysis. They acknowledge their contract research partners for their contributions: Dr. X. Wang and team at Chempartner Shanghai and Dr. T. Pang and team at Pharmaron Beijing. This work was supported by Tango Therapeutics.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
J.A. Shields reports personal fees and other support from Tango Therapeutics outside the submitted work. S.R. Meier reports personal fees and other support from Tango Therapeutics outside the submitted work. M. Bandi reports personal fees and other support from Tango Therapeutics outside the submitted work. M.D. Ferdinez reports personal fees from Tango Therapeutics Inc. and other support from Tango Therapeutics outside the submitted work. J.L. Engel reports personal fees and other support from Tango Therapeutics outside the submitted work. B. Shen reports personal fees and other support from Tango Therapeutics Inc. outside the submitted work. W. Zhang reports personal fees and other support from Tango Therapeutics outside the submitted work. S.-C. Zhao reports personal fees and other support from TNGX outside the submitted work. A. Bejnood reports personal fees and other support from Tango Therapeutics outside the submitted work. M. Zhang reports personal fees and other support from Tango Therapeutics outside the submitted work. R. Tjin Tham Sjin reports personal fees and other support from Tango Therapeutics outside the submitted work; as well as reports employment and shareholder of Tango Therapeutics. E. Wilker reports other support from Tango Therapeutics outside the submitted work. J.N. Andersen reports other support from Tango Therapeutics outside the submitted work. Y. Chen reports personal fees and other support from Tango Therapeutics outside the submitted work. B. Weber reports other support from Tango Therapeutics outside the submitted work. A. Huang reports personal fees and other support from Tango Therapeutics outside the submitted work. N. Emmanuel reports personal fees and other support from Tango Therapeutics Inc. outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
J.A. Shields: Conceptualization, data curation, formal analysis, supervision, visualization, methodology, writing–original draft, writing–review and editing. S.R. Meier: Data curation, software, formal analysis, methodology, writing–original draft. M. Bandi: Data curation, software, formal analysis. E.E. Mulkearns-Hubert: Data curation, formal analysis, visualization, methodology. N. Hajdari: Data curation, formal analysis. M.D. Ferdinez: Data curation, formal analysis. J.L. Engel: Data curation, formal analysis, visualization, methodology. D.J. Silver: Data curation, formal analysis. B. Shen: Data curation, methodology. W. Zhang: Data curation, formal analysis. C.G. Hubert: Formal analysis. K. Mitchell: Data curation, formal analysis. S. Shakya: Data curation. S.-C. Zhao: Data curation, formal analysis, methodology. A. Bejnood: Formal analysis. M. Zhang: Data curation, formal analysis, methodology. R. Tjin Tham Sjin: Formal analysis, supervision, methodology. E. Wilker: Formal analysis, supervision, methodology. J.D. Lathia: Supervision, visualization, methodology, writing–review and editing. J.N. Andersen: Resources, supervision, validation, visualization, writing–review and editing. Y. Chen: Resources, supervision, validation, investigation, methodology, writing–review and editing. F. Li: Data curation, formal analysis, supervision, investigation, visualization, methodology, writing–review and editing. B. Weber: Resources, data curation, formal analysis, supervision, visualization, writing–review and editing. A. Huang: Conceptualization, resources, supervision, visualization, writing–review and editing. N. Emmanuel: Conceptualization, data curation, formal analysis, supervision, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. Huang A, Garraway LA, Ashworth A, Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat Rev Drug Discov 2020;19:23–38. [DOI] [PubMed] [Google Scholar]
- 2. Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science 1997;278:1064–8. [DOI] [PubMed] [Google Scholar]
- 3. Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 2005;5:689–98. [DOI] [PubMed] [Google Scholar]
- 4. Neggers JE, Paolella BR, Asfaw A, Rothberg MV, Skipper TA, Yang A, et al. Synthetic lethal interaction between the ESCRT paralog enzymes VPS4A and VPS4B in cancers harboring loss of chromosome 18q or 16q. Cell Rep 2020;33:108493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan EM, Shibue T, McFarland J, Gaeta B, McPartlan JS, Ghandi M, et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 2019;568:551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pacini C, Dempster JM, Boyle I, Gonçalves E, Najgebauer H, Karakoc E, et al. Integrated cross-study datasets of genetic dependencies in cancer. Nat Commun 2021;12:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klerkx EPF, Lazo PA, Askjaer P. Review Emerging biological functions of the Vaccinia-Related Kinase (VRK) family. Histol Histopathol 2009;24:749–59 [DOI] [PubMed] [Google Scholar]
- 8. Martín-Doncel E, Rojas AM, Cantarero L, Lazo PA. VRK1 functional insufficiency due to alterations in protein stability or kinase activity of human VRK1 pathogenic variants implicated in neuromotor syndromes. Sci Rep 2019;9:13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valbuena A, Sanz-García M, López-Sánchez I, Vega FM, Lazo PA. Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell Signal 2011;23:1267–72. [DOI] [PubMed] [Google Scholar]
- 10. Monsalve DM, Merced T, Fernández IF, Blanco S, Vázquez-Cedeira M, Lazo PA. Human VRK2 modulates apoptosis by interaction with Bcl-xL and regulation of BAX gene expression. Cell Death Dis 2013;4:e513–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blanco S, Klimcakova L, Vega FM, Lazo PA. The subcellular localization of vaccinia-related kinase-2 (VRK2) isoforms determines their different effect on p53 stability in tumour cell lines. Febs J 2006;273:2487–504. [DOI] [PubMed] [Google Scholar]
- 12. Nichols RJ, Wiebe MS, Traktman P. The Vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell 2006;17:2451–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Louis DN, Perry A, Reifenberger G, von DA, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016;131:803–20. [DOI] [PubMed] [Google Scholar]
- 15. Kamson DO, Grossman SA. The role of temozolomide in patients with newly diagnosed Wild-type IDH, unmethylated MGMTp glioblastoma during the COVID-19 Pandemic. JAMA Oncol 2021;7:675–6. [DOI] [PubMed] [Google Scholar]
- 16. Finch A, Solomou G, Wykes V, Pohl U, Bardella C, Watts C. Advances in research of adult gliomas. Int J Mol Sci 2021;22:924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stupp R, Hegi ME, Mason WP, den BMJv, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009;10:459–66. [DOI] [PubMed] [Google Scholar]
- 18. Kammers K, Cole RN, Tiengwe C, Ruczinski I. Detecting significant changes in protein abundance. Eupa Open Proteom 2015;7:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dempster JM, Rossen J, Kazachkova M, Pan J, Kugener G, Root DE, et al. Extracting biological insights from the project achilles genome-scale CRISPR screens in cancer cell lines. bioRxiv 2019;720243. doi: https://doi.org/10.1101/720243. [Google Scholar]
- 21. Network CGAR, Brat DJ, Verhaak RGW, Aldape KD, Yung WKA, Salama SR, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New Engl J Med 2015;372:2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vega FM, Sevilla A, Lazo PA. p53 Stabilization and Accumulation induced by human vaccinia-related kinase 1. Mol Cell Biol 2004;24:10366–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cartwright TN, Harris RJ, Meyer SK, Mon AM, Watson NA, Tan C, et al. Dissecting the roles of Haspin and VRK1 in histone H3 phosphorylation during mitosis. Sci Rep 2022;12:11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ren Z, Geng J, Xiong C, Li X, Li Y, Li J, et al. Downregulation of VRK1 reduces the expression of BANF1 and suppresses the proliferative and migratory activity of esophageal cancer cells. Oncol Lett 2020;20:1163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Molitor TP, Traktman P. Depletion of the protein kinase VRK1 disrupts nuclear envelope morphology and leads to BAF retention on mitotic chromosomes. Mol Biol Cell 2014;25:891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jamin A, Wiebe MS. barrier to autointegration factor (BANF1): interwoven roles in nuclear structure, genome integrity, innate immunity, stress responses and progeria. Curr Opin Cell Biol 2015;34:61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Samwer M, Schneider MWG, Hoefler R, Schmalhorst PS, Jude JG, Zuber J, et al. DNA Cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 2017;170:956–72.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kapanidou M, Curtis NL, Bolanos-Garcia VM. Cdc20: at the crossroads between chromosome segregation and mitotic exit. Trends Biochem Sci 2017;42:193–205. [DOI] [PubMed] [Google Scholar]
- 30. Heald R, McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 1990;61:579–89. [DOI] [PubMed] [Google Scholar]
- 31. Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006;9:391–403. [DOI] [PubMed] [Google Scholar]
- 32. Lathia JD, Mack SC,EE Mulkearns-Hubert, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Gene Dev 2015;29:1203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lathia JD, Gallagher J, Myers JT, Li M, Vasanji A, McLendon RE, et al. Direct in vivo evidence for tumor propagation by glioblastoma cancer stem cells. PLoS ONE 2011;6:e24807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adane B, Alexe G, Seong BKA, Lu D, Hwang EE, Hnisz D, et al. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma. Cancer Cell 2021;39:827–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gillani R, Seong BKA, Crowdis J, Conway JR, Dharia NV, Alimohamed S, et al. Gene fusions create partner and collateral dependencies essential to cancer cell survival. Cancer Res 2021;81:3971–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Malone CF, Dharia NV, Kugener G, Forman AB, Rothberg MV, Abdusamad M, et al. Selective modulation of a Pan-essential protein as a therapeutic strategy in cancer. Cancer Discov 2021;11:2282–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, et al. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov 2016;6:430–45. [DOI] [PubMed] [Google Scholar]
- 38. Couñago RM, Allerston CK, Savitsky P, Azevedo H, Godoi PH, Wells CI, et al. Structural characterization of human vaccinia-related kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations. Sci Rep-uk 2017;7:7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Serafim RAM, Gama FH de S, Dutra LA, Reis CVD, Vasconcelos SNS, Santiago A da S, et al. Development of Pyridine-based Inhibitors for the human vaccinia-related kinases 1 and 2. Acs Med Chem Lett 2019;10:1266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santos CR, Rodríguez-Pinilla M, Vega FM, Rodríguez-Peralto JL, Blanco S, Sevilla A, et al. VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol Cancer Res 2006;4:177–85. [DOI] [PubMed] [Google Scholar]
- 41. Ben Z, Gong L, Qiu Y. High expression of VRK1 is related to poor prognosis in glioma. Pathology Res Pract 2018;214:112–8. [DOI] [PubMed] [Google Scholar]
- 42. Li J, Wang T, Pei L, Jing J, Hu W, Sun T, et al. Expression of VRK1 and the downstream gene BANF1 in esophageal cancer. Biomed Pharmacother 2017;89:1086–91. [DOI] [PubMed] [Google Scholar]
- 43. Colmenero-Repiso A, Gómez-Muñoz MA, Rodríguez-Prieto I, Amador-Álvarez A, Henrich K-O, Pascual-Vaca D, et al. Identification of VRK1 as a new neuroblastoma tumor progression marker regulating cell proliferation. Cancers 2020;12:3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Renbaum P, Kellerman E, Jaron R, Geiger D, Segel R, Lee M, et al. Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the VRK1 Gene. Am J Hum Genetics 2009;85:281–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gonzaga-Jauregui C, Lotze T, Jamal L, Penney S, Campbell IM, Pehlivan D, et al. Mutations in VRK1 associated with complex motor and sensory axonal neuropathy plus microcephaly. JAMA Neurol 2013;70:1491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stoll M, Teoh H, Lee J, Reddel S, Zhu Y, Buckley M, et al. Novel motor phenotypes in patients with VRK1 mutations without pontocerebellar hypoplasia. Neurology 2016;87:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vinograd-Byk H, Renbaum P, Levy-Lahad E. Vrk1 partial knockdown in mice results in reduced brain weight and mild motor dysfunction, and indicates neuronal VRK1 target pathways. Sci Rep 2018;8:11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Valbuena A, López-Sánchez I, Lazo PA. Human VRK1 is an early response gene and its loss causes a block in cell-cycle progression. PLoS ONE 2007;3:e1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kang T-H, Park D-Y, Kim W, Kim K-T. VRK1 phosphorylates CREB and mediates CCND1 expression. J Cell Sci 2008;121:3035–41. [DOI] [PubMed] [Google Scholar]
- 50. Kang T-H, Park D-Y, Choi YH, Kim K-J, Yoon HS, Kim K-T. Mitotic histone H3 phosphorylation by Vaccinia-related kinase 1 in mammalian cells. Mol Cell Biol 2007;27:8533–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jeong M-W, Kang T-H, Kim W, Choi YH, Kim K-T. Mitogen-activated protein kinase phosphatase 2 regulates histone H3 phosphorylation via interaction with vaccinia-related kinase 1. Mol Biol Cell 2013;24:373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moura DS, Campillo-Marcos I, Vázquez-Cedeira M, Lazo PA. VRK1 and AURKB form a complex that cross inhibit their kinase activity and the phosphorylation of histone H3 in the progression of mitosis. Cell Mol Life Sci Cmls 2017;75:2591–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kc B, May DG, Benson BV, Kim DI, Shivega WG, Ali MH, et al. VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Mol Biol Cell 2017;28:2241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Campillo-Marcos I, García-González R, Navarro-Carrasco E, Lazo PA. The human VRK1 chromatin kinase in cancer biology. Cancer Lett 2021;503:117–28. [DOI] [PubMed] [Google Scholar]
- 55. Furukawa K, Sugiyama S, Osouda S, Goto H, Inagaki M, Horigome T, et al. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J Cell Sci 2003;116:3811–23. [DOI] [PubMed] [Google Scholar]
- 56. Gorjánácz M, Klerkx EP, Galy V, Santarella R, López-Iglesias C, Askjaer P, et al. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. Embo J 2007;26:132–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. P Natl Acad Sci Usa. 2005;102:3290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc National Acad Sci 2000;97:8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Puente XS, Quesada V, Osorio FG, Cabanillas R, Cadiñanos J, Fraile JM, et al. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am J Hum Genet 2011;88:650–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Burla R, Torre ML, Merigliano C, Vernì F, Saggio I. Genomic instability and DNA replication defects in progeroid syndromes. Nucl Austin Tex 2018;9:368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guey B, Wischnewski M, Decout A, Makasheva K, Kaynak M, Sakar MS, et al. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 2020;369:823–8. [DOI] [PubMed] [Google Scholar]
- 62. Rhun EL, Preusser M, Roth P, Reardon DA, den BMv, Wen P, et al. Molecular targeted therapy of glioblastoma. Cancer Treat Rev 2019;80:101896. [DOI] [PubMed] [Google Scholar]
- 63. Navarro-Carrasco E, Lazo PA. VRK1 Depletion Facilitates the synthetic lethality of temozolomide and olaparib in glioblastoma cells. Front Cell Dev Biol 2021;9:683038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry raw files are available at UCSD MassIVE repository ID # MSV000089783. All data were generated by the authors and are available upon request.



![Figure 5. Phospho- and total-proteomics reveals DNA repair pathways are activated upon VRK1 knockdown in VRK2-low cells. A, Heat maps showing total-proteomics (left) and phospho–proteomics (right) in VRK2-low and VRK2-high U251MG cells at 5 and 7days after doxycycline. B, Gene Set Enrichment Analysis (GSEA) of 7-day total proteomics data in the doxycycline conditions. C, Volcano plots differential expression analysis of total proteomic data; The x-axis represents log2 fold change and the y-axis represents the FDR [−log10(q value)]; black circles—proteins with greater than ± 2 log-fold change. D, Volcano plots differential expression analysis of phospho–proteomic data; The x-axis represents log2 fold change and the y-axis represents the FDR [−log10(q value)]; black circles—proteins with greater than ± 2 log-fold change. E, Immunoblots of select proteins from U251MG VRK2-low and VRK2-high cells treated with or without doxycycline for 7 days](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c488/9627132/e9384e61be20/4044fig5.jpg)