Small-molecule inhibition of ENL leads to suppression of oncogenic gene expression and blocks the progression of MLL-rearranged and NPM1-mutated acute myeloid leukemia (AML), demonstrating ENL as a promising therapeutic target for AML subsets.
Abstract
The chromatin reader eleven–nineteen leukemia (ENL) has been identified as a critical dependency in acute myeloid leukemia (AML), but its therapeutic potential remains unclear. We describe a potent and orally bioavailable small-molecule inhibitor of ENL, TDI-11055, which displaces ENL from chromatin by blocking its YEATS domain interaction with acylated histones. Cell lines and primary patient samples carrying MLL rearrangements or NPM1 mutations are responsive to TDI-11055. A CRISPR-Cas9–mediated mutagenesis screen uncovers an ENL mutation that confers resistance to TDI-11055, validating the compound's on-target activity. TDI-11055 treatment rapidly decreases chromatin occupancy of ENL-associated complexes and impairs transcription elongation, leading to suppression of key oncogenic gene expression programs and induction of differentiation. In vivo treatment with TDI-11055 blocks disease progression in cell line– and patient-derived xenograft models of MLL-rearranged and NPM1-mutated AML. Our results establish ENL displacement from chromatin as a promising epigenetic therapy for molecularly defined AML subsets and support the clinical translation of this approach.
Significance:
AML is a poor-prognosis disease for which new therapeutic approaches are desperately needed. We developed an orally bioavailable inhibitor of ENL, demonstrated its potent efficacy in MLL-rearranged and NPM1-mutated AML, and determined its mechanisms of action. These biological and chemical insights will facilitate both basic research and clinical translation.
This article is highlighted in the In This Issue feature, p. 2483
INTRODUCTION
Acute myeloid leukemia (AML) is one of the deadliest hematologic malignancies with an overall 5-year survival rate of 26% (1, 2). Though genetically heterogeneous, AML is characterized by and highly “addicted” to dysregulated transcriptional programs that ultimately result in uncontrolled proliferation and malignant self-renewal of myeloid progenitor cells. Accordingly, genetic alterations in regulators of gene expression are drastically overrepresented in AML (3). However, direct therapeutic intervention remains challenging for many of these oncogenic drivers. As such, pharmacologically tractable proteins that themselves are not necessarily mutated but essential for sustaining dysregulated transcription programs in AML have been increasingly recognized as attractive therapeutic targets.
Previously, we and others identified the eleven–nineteen leukemia protein (ENL; encoded by the MLLT1 gene) as a critical dependency in AML (4, 5). ENL belongs to a protein family that contains a well-conserved YEATS domain, which can recognize histone lysine acylation, including acetylation and crotonylation (6–13). The human genome encodes four YEATS domain–containing proteins: ENL, AF9, GAS41, and YEATS2. These proteins have been found in nuclear complexes with a variety of molecular functions spanning chromatin remodeling, histone modification, and transcription (7, 14), and they have been increasingly implicated in cancer. In leukemia, ENL and its paralog, AF9, are frequently fused with the mixed lineage leukemia protein (MLL1, also known as KMT2A) as a result of chromosomal translocations. The resultant MLL fusions, often missing the ENL/AF9 YEATS domains, drive the onset of distinct subsets of AML and acute lymphoblastic leukemia (ALL), both of which are associated with poor prognosis (15, 16). In addition to participating in oncogenic fusions, the wild-type ENL is co-opted to support oncogenic gene expression programs important for the maintenance of AML, in particular with those harboring MLL fusions (4, 5). It has been shown that genetic depletion of ENL, or mutational disruption of its YEATS domain–mediated interaction with acylated histones, leads to growth impairment of MLL-rearranged (MLL-r) leukemia in cellular and animal studies (4, 5). Furthermore, recurrent mutations in the ENL YEATS domain have been discovered in Wilms tumor and are associated with a high risk of relapse (17, 18). These mutations, as shown in our recent study (19), can confer ENL-increased transcriptional activity and drive hyperactivation of developmentally important genes, notably the HOXA cluster, and such a function requires the acyl-lysine binding activity of ENL. Together, these studies establish important roles for ENL in human cancer and nominate its acyl-binding YEATS domain as a critical functional domain and potential therapeutic target.
Chemical probes of chromatin readers serve as valuable tools for investigating regulatory mechanisms and functional roles of their target proteins in normal and disease states and could have significant clinical potential, as evidenced by the success of inhibitors against bromodomain and extraterminal (BET) proteins (20–24). Previous structural studies suggest that YEATS domains are pharmacologically tractable, as they contain both a deep pocket to accommodate their acyl-lysine substrate and nearby regions that could potentially complement the placement of other functional groups of an inhibitor (4, 6, 8, 10). Indeed, recent reports of a few peptide-mimic and small-molecule chemical probes have convincingly supported the tractability of ENL/AF9 YEATS domains for drug discovery (25–31). However, only two reported chemical probes of ENL/AF9 have been evaluated for their antiproliferative effects in AML cell lines so far (30, 31). In each case, the probe exhibited either moderate cellular activity (30) or poor pharmacologic properties (31) that limited further use for in vivo efficacy studies. Therefore, inhibitors with improved in vivo bioavailability will be needed to fully explore the translational potential of ENL inhibition in cancer. Furthermore, how specific perturbation of the ENL YEATS–acylation interaction by a direct-acting inhibitor remodels chromatin to affect oncogenic programs in AML remains to be defined.
In this study, we develop the potent and orally bioavailable ENL/AF9 YEATS domain inhibitor TDI-11055 through the structure-based optimization of a reported chemical probe, and we use TDI-11055 to explore the therapeutic potential and mechanisms of ENL inhibition in AML. TDI-11055 potently inhibits the proliferation and colony-forming ability of human leukemia cell lines and primary AML patient cells harboring MLL1 translocations and NPM1 mutations. The on-target activity of TDI-11055 was extensively validated through a CRISPR-Cas9–mediated mutagenesis screen in AML cells. Oral treatment with TDI-11055 in mice blocks disease progression in cell line– and patient-derived xenograft (PDX) models of MLL-r and NPM1-mutated AML. Mechanistically, we show that displacement of ENL from chromatin by TDI-11055 leads to a rapid decrease in the recruitment of select ENL-associated complexes and Pol II elongation. These chromatin changes occur prominently at top ENL-occupied loci, which enrich for key leukemogenic genes (including MYC and HOXA9), thus leading to the suppression of oncogenic gene expression programs and induction of cellular differentiation. These findings establish the displacement of ENL from chromatin as a promising epigenetic therapy for molecularly defined AML subsets and support future translation of this approach to the clinical setting.
RESULTS
A Potent, Selective, and Orally Bioavailable Inhibitor of the ENL/AF9 YEATS Domains
The goal of our study was to develop a potent and in vivo bioavailable inhibitor that blocks the reader function of ENL as a tool to explore the therapeutic potential and mechanisms of ENL targeting in cancer models. At the beginning of this study, SGC-iMLLT was the first and most potent small-molecule chemical probe reported for the ENL/AF9 YEATS domains (Supplementary Fig. S1A; ref. 25). However, SGC-iMLLT has not been functionally characterized in AML beyond proximal target engagement and was noted to have metabolic instability and correspondingly a modest half-life in primary human hepatocytes (25). Consistent with these findings, our pharmacokinetic (PK) characterization in mice revealed a short half-life for SGC-iMLLT that limits its further use for in vivo efficacy studies (t1/2 = 0.83 hours at oral dose 100 mg/kg; Supplementary Fig. S1B). As such, we performed a structure-guided optimization of SGC-iMLLT with the goal of improving its drug-like properties to enable in vivo utility while maintaining its favorable potency and selectivity profiles. These efforts resulted in the development of TDI-11055 (Fig. 1A), which was obtained by (i) removing one of the benzimidazole nitrogen atoms due to its ability to tautomerize or protonate and (ii) introducing a new nitrogen atom into the ring system to predispose the molecule to adopt the conformation in which it binds to the ENL YEATS domain. Time-resolved fluorescence energy transfer (TR-FRET) assays were used to measure the inhibitory effects of TDI-11055 on the interactions between different YEATS domains and acylated histone peptides (Fig. 1B). As designed, TDI-11055 retained potency and selectivity for the YEATS domains of ENL (IC50 = 0.05 μmol/L) and AF9 (IC50 = 0.07 μmol/L) but showed no inhibition of the YEATS domains of GAS41 and YEATS2 (IC50 >100 μmol/L; Fig. 1B; Supplementary Fig. S1C). The direct binding of TDI-11055 to the ENL YEATS domain was further confirmed by isothermal titration calorimetry (ITC; Fig. 1C, Kd = 119 nmol/L, N = 1). To evaluate the target engagement of TDI-11055 in a cellular context, we performed standard cellular thermal shift assays (32) in the human leukemia cell line MOLM-13. TDI-11055 binds to and stabilizes endogenously expressed ENL but not GAS41 or YEATS2 in cells (Fig. 1D). The docking model of TDI-11055, which was based on the experimental crystal structure of ENL YEATS in complex with SGC-iMLLT (25), suggested that TDI-11055 binds directly to the acyl-binding site in ENL and engages with key acyl-recognizing residues that we have previously identified (ref. 4; Fig. 1E). In support of this model, we found that although TDI-11055 can bind to and stabilize wild-type ENL proteins in cellular thermal shift assays, its stabilization effect on an ENL mutant (Y78A) that lacks the ability to bind to acetylated histone (4) was markedly reduced (Fig. 1F).
Figure 1.
A potent, selective, and orally bioavailable inhibitor of the ENL/AF9 YEATS domains. A, Chemical structure for TDI-11055. B, Left, schematic depiction of the TR-FRET assay used to quantify the ability of a compound to disrupt YEATS domain and acylated histone H3 peptide interaction. Right, dose-dependent inhibition of the TR-FRET signal by TDI-11055. The IC50 shown represents the mean of four independent experiments. C, ITC experiment demonstrating direct binding of TDI-11055 to ENL YEATS with 1:1 stoichiometry. One of two independent replicates is shown. D, Top, immunoblots showing endogenous levels of ENL, GAS41, and YEATS2 protein after heat treatment in MOLM-13 leukemia cells at increasing temperatures. Bottom, quantification of immunoblot signals. One of two independent experiments is shown.E, Docking studies of TDI-11055 bound to ENL YEATS from cocrystal structures of SGC-iMLLT [Protein Data Bank (PDB): 6HT1] and H3K27ac (PDB: 5J9S). Ribbon representations indicate the protein, and stick representations show key residues involved in ENL and TDI-11055 interactions. TDI-11055 is depicted as sticks with colors corresponding to the atom type (green, carbon; blue, nitrogen; red, oxygen). F, Left, immunoblots showing levels of ectopically expressed FLAG-tagged wild-type (WT) or Y78A-mutant ENL proteins in HEK293 cells after heat treatment at increasing temperatures. Right, quantification of immunoblot signals. One of two independent experiments is shown. G, Rank-ordered heat map of FLAG-ENL ChIP-seq signals at ENL-bound peaks in MOLM-13 cells treated with DMSO or TDI-11055 (5 μmol/L) for 24 hours. H3K27ac and H3K9ac (GSE80779) ChIP-seq signals at ENL-bound peaks in untreated cells are also shown to demonstrate ENL localization to acetylated chromatin. See Supplementary Table S1. H, The genome browser view of indicated ChIP-seq signals at key ENL-bound genes (MYC, HOXA9/10) in parental [H3K27ac (gray), H3K9ac (black)] as well as DMSO (blue)- or TDI-11055 (red)–treated (FLAG-ENL) MOLM-13 cells. I, PK studies in mice (n = 3) performed for TDI-11055 and SGC-iMLLT, demonstrating unbound plasma concentration of indicated compounds after an oral dose of 30, 50, and 100 mg/kg.
Previously, structure-based mutagenesis helped establish the acyl-lysine binding activity of the YEATS domain as an important contributor to ENL chromatin localization (4, 5). To test whether TDI-11055 acts as a chromatin-competitive antagonist of ENL in cells, we treated MOLM-13 cells ectopically expressing a FLAG-tagged ENL at levels equivalent to the endogenous ENL proteins (Supplementary Fig. S1D) with DMSO or TDI-11055 for 24 hours and then examined ENL genomic occupancy by chromatin-immunoprecipitation followed by sequencing (ChIP-seq) using an anti-FLAG antibody. ENL predominantly localized to the transcription start site (TSS) and gene body of genes that are highly enriched for H3K9ac and H3K27ac (Fig. 1G), consistent with previous reports (4, 5). TDI-11055 treatment led to a substantial displacement of ENL from target genes, including well-established leukemogenic genes in AML such as MYC and the HOXA cluster (Fig. 1H; Supplementary Table S1). Because TDI-11055 treatment did not decrease the bulk levels of ENL proteins in cells (Supplementary Fig. S1E), these results establish TDI-11055 as a validated chemical tool for efficiently and specifically perturbing the chromatin reader function of ENL in living cells.
Next, we evaluated the potential utility of TDI-11055 as a candidate for in vivo studies by assessing its drug-like properties using in vitro and subsequently in vivo profiling. TDI-11055 exhibited moderate/high permeability in a Caco2 cell monolayer with minimal efflux and high solubility (Supplementary Fig. S1F), properties that forecast sufficient oral absorption in vivo. Upon oral administration at doses of 30 to 100 mg/kg (Supplementary Fig. S1G), TDI-11055 was well tolerated with high oral bioavailability (Fpo >100%). Notably, the half-life for TDI-11055, particularly at higher oral doses, was much improved over that of SGC-iMLLT and reached 5 hours, thus providing the opportunity to deliver high and sustained concentrations of the unbound drug via oral administration (Fig. 1I). Taken together, our characterization indicates that TDI-11055 is a potent, selective, and orally bioavailable ENL/AF9 YEATS inhibitor with a profile appropriate for in vivo studies. Therefore, we used it for subsequent studies in AML.
TDI-11055 Inhibits the Growth of MLL-r and NPM1-Mutated Leukemia Cells In Vitro
To study the antileukemic potential of TDI-11055, we first assessed alterations in the proliferation of a panel of human leukemia cell lines after treatment with TDI-11055. We tested four MLL-r leukemia cell lines that have been previously shown to be sensitive to genetic depletion of ENL: MV4;11 (MLL–AF4), MOLM-13 (MLL–AF9), OCI-AML2 (MLL–AF6), and ML2 (MLL–AF6; refs. 4, 5). Exposure to TDI-11055 led to a profound reduction in cell proliferation in a concentration-dependent manner in each of these lines (Fig. 2A). By contrast, TDI-11055 exerted minimal effects on the growth of HL60, K562, U937, and JURKAT (Fig. 2B), all of which are non–MLL-r leukemia cell lines and largely insensitive to genetic loss of ENL (4, 5). We noticed that TDI-11055 also had a strong inhibitory effect on the growth of OCI-AML3 (Fig. 2A), a cell line that harbors a genetic mutation in nucleophosmin (NPM1)—a gene that is mutated in >30% of AML (33–35). Because OCI-AML3 had not been previously tested for its reliance on ENL, we used CRISPR-Cas9 editing to disrupt ENL and performed growth competition assays. We found that the genetic perturbation of ENL inhibited the growth of OCI-AML3 cells to a degree similar to the ENL-dependent MLL-r cell line MV4;11 (Supplementary Fig. S2A and S2B). Further analysis showed that growth inhibition by TDI-11055 in MV4;11 and OCI-AML3 cells was accompanied by a decrease in cell-cycle progression (Supplementary Fig. S2C and S2D) and, to a lesser extent, an increase in apoptosis (Supplementary Fig. S2E and S2F).
Figure 2.
TDI-11055 suppresses the cellular growth of MLL-r and NPM1-mutated leukemia cells through on-target inhibition of ENL. A and B, Dose–response viability curves (8 days) for TDI-11055 in human leukemia cell lines with high (A) and low (B) dependence on ENL defined by genetic studies. n = 3. NPM1 mut, NPM1-mutated. C–F, Representative images (left) and quantification (right) of colonies formed by indicated human leukemia cell lines treated with DMSO or TDI-11055 (TDI; 1 and 10 μmol/L). Error bars represent mean ± SEM (n = 3). One of two independent experiment batches is shown. G, Schematic of CRISPR-Cas9 mutagenesis scanning workflow. MOI, multiplicity of infection; sgRNA, single-guide RNA; T_final, end time point; T_initial, start time point. H, Violin plots showing the log2 (fold-change) sgRNA enrichment (T_final over T_initial) in MV4;11 cells. Neg, negative controls, sgRNAs targeting known nonessential genes; Pos, positive controls, sgRNAs targeting known essential genes. Black lines denote median. See Supplementary Table S2. I, Scatter plot showing log2 (fold-change sgRNA enrichment) in MV4;11 in TDI-11055 vs. DMSO comparison. sgRNAs are positioned along the x axis by the ENL coding sequence. Data represent average across three replicate transductions. Regions 1 and 2 (R1 and R2) highlighted in blue contain 4 or more enriched sgRNAs. See Supplementary Table S3. sgENL326 shows the strongest enrichment in TDI-11055 treatment condition over DMSO among all sgRNAs.J, Schematic for a proliferation competition assay used in K. K, Left and center, plots showing the relative fitness of indicated sgRNA+ cells under DMSO (left) or TDI-11055 (TDI; 5 μmol/L, center) treatment conditions; right, plots showing the log2 fold-change enrichment (TDI-11055 vs. DMSO) of indicated sgRNA+ cells. An sgRNA targeting Rosa26 (black) or RPA3 (gray) serves as a negative or a positive control, respectively. Among the 10 ENL sgRNAs tested, sgENL326 (red)- and sgENL323 (orange)-induced mutant(s) conferred relative growth advantage under TDI-11055 treatment. L, Pie charts showing the relative abundance of ENL mutations induced by sgENL326 in MV4;11 cells at indicated time points. Each slice represents a single mutant ENL allele. See Supplementary Table S5. WT, wild-type. M, Schematic for a proliferation competition assay used in N. N, Left and center: plots showing the relative fitness of cells transduced with indicated mCherry-ENL transgenes over nontransduced parental cells under DMSO (left) or TDI-11055 (5 μmol/L, center) treatment. Right, plots showing the fold change in fitness (TDI-11055 vs. DMSO) for cells expressing indicated mCherry-ENL transgenes.
Next, we tested the effects of TDI-11055 on the clonogenic potential of leukemia cells, an in vitro surrogate assay for leukemia stem cell activity. TDI-11055 treatment strongly inhibited the clonogenic potential of ENL-dependent cells (MV4;11 and OCI-AML3) when compared with DMSO control, which was reflected by a marked decrease in colony number and size in a concentration-dependent manner (Fig. 2C and D). In contrast, the colony formation by leukemia cells with low ENL dependence (HL60 and K562) was only modestly affected by TDI-11055 (Fig. 2E and F). Taken together, these results indicate that TDI-11055 treatment recapitulates the genetic disruption of ENL and exhibits preferential inhibitory activity against MLL-r and NPM1-mutated leukemia.
A CRISPR-Cas9–mediated Mutagenesis Screen Identifies a Drug-Resistant ENL Mutant Allele
Many well-characterized small-molecule inhibitors suffer from off-target effects in cells despite exhibiting specific effects in biochemical assays and cellular phenotypes (36). To rigorously validate that the antiproliferative effects of TDI-11055 in AML cells are indeed due to inhibition of ENL, we performed a CRISPR-Cas9–mediated mutagenesis screen to systematically alter ENL protein sequence in situ. This strategy allowed us to ask if any genetic mutation(s) in ENL could alter cellular response to TDI-11055, a strategy previously used to successfully identify physiologic targets of anticancer agents and generate inhibitor-resistant mutants for mechanistic studies (37–40). Specifically, we designed a pool of all possible NGG Protospacer Adjacent Motif (PAM)-restricted single-guide RNAs (sgRNA) targeting the YEATS domains of ENL and AF9 (Supplementary Table S2). We transduced these sgRNAs into Cas9-expressing MV4;11 cells and treated the cells with DMSO or TDI-11055 for 4 weeks. Genomic DNA from cells at the initial and final time points of the experiments was subjected to deep sequencing to determine sgRNA distribution (Fig. 2G). Consistent with the essential nature of ENL in MV4;11 cells, most sgRNAs targeting ENL were depleted in both the DMSO and TDI-11055 treatment conditions. In contrast, sgRNAs targeting AF9 were largely not depleted (Fig. 2H; Supplementary Table S2), reinforcing previous findings that AF9 is dispensable for the growth of MV4;11 cells (4, 5) and indicating that the effects of TDI-11055 in this context could not be attributed to its inhibition of AF9.
We then examined the differential enrichment of ENL sgRNAs in DMSO- and TDI-11055–treated groups. We reasoned that if a given ENL sgRNA is preferentially enriched under TDI-11055 exposure, it may produce mutant ENL allele(s) that retain, at least partially, ENL's functionality while conferring relative resistance to TDI-11055. There were 19 ENL sgRNAs exhibiting >2-fold enrichment in the TDI-11055–treated group when compared with the control group (Fig. 2I; Supplementary Tables S3 and S4), and noticeably, four or more of these sgRNA targeted two hotspot regions within the ENL YEATS domain (Fig. 2I; R1 and R2). We then evaluated whether individual sgRNAs enriched in these two hotspot regions could render cells less sensitive to TDI-11055 using a growth competition assay (Fig. 2J). Cells harboring each of the ENL sgRNAs tested were initially depleted in both the DMSO and TDI-11055 treatment conditions (Fig. 2K; Supplementary Table S4), suggesting that these sgRNAs predominantly generated loss-of-function mutations in ENL. Under continuous exposure to TDI-11055, cells transduced with ENL sgRNA #326 and, to a lesser extent, sgRNA #323, gained a competitive growth advantage versus nontransduced cells (Fig. 2K), indicating the emergence and expansion of drug-resistant clone(s). The drug-resistant phenotypes induced by sgENL326 were also observed in NPM1-mutated OCI-AML3 cells (Supplementary Fig. S3A and S3B).
Deep sequencing of ENL in sgENL326-positive cells before and after TDI-11055 treatment revealed a strong enrichment for an in-frame deletion mutation, p.108_111delLEGN, after prolonged drug treatment (Fig. 2L; Supplementary Table S5). The same mutation was also enriched, albeit to a lesser extent, in sgENL323-positive cells after drug treatment (Supplementary Fig. S3C; Supplementary Table S5). The deletion encompasses the predicted Cas9 cutting site induced by sgENL326 and sgENL323 (Supplementary Fig. S3D and S3E) and is located in the loop region adjacent to the acyl-binding pocket in ENL (Supplementary Fig. S3F). To directly test if the ENL deletion mutant confers a growth advantage under drug selection, we transduced MV4;11 cells with mCherry-linked wild-type or mutant ENL transgenes and monitored their growth relative to nontransduced cells over time under DMSO and TDI-11055 treatment conditions (Fig. 2M). Cells ectopically expressing the wild-type ENL transgene slightly outgrew nontransduced cells under continuous exposure to TDI-11055, possibly due to higher levels of total ENL proteins; such a growth advantage was more pronounced in cells ectopically expressing the mutant ENL transgene (Fig. 2N). We found that purified ENL YEATS domains harboring the deletion exhibited a lower binding affinity with TDI-11055 when compared with the wild type [Kd = 0.51 μmol/L (deletion) vs. 0.15 μmol/L (wild type); Supplementary Fig. S3G], revealing a potential mechanism by which the mutation confers relative resistance to TDI-11055–induced growth inhibition that warrants future investigation. Collectively, our results provide compelling “genetic proof” that the antigrowth activity of TDI-11055 is largely attributed to its inhibition of ENL, further validating the on-target activity of the compound.
Displacement of ENL from Chromatin by TDI-11055 Suppresses Key Oncogenic Gene Expression Programs in MLL-r and NPM1-Mutated AML
We next sought to investigate the molecular mechanism by which TDI-11055 affects AML. We performed RNA sequencing (RNA-seq) analysis of MV4;11 cells treated with DMSO or TDI-11055 for 24 hours, a time point before any noticeable phenotypes were observed. There were significantly more genes downregulated (923) than upregulated (219) upon TDI-11055 treatment (1.5-fold change, P < 0.05; Fig. 3A; Supplementary Table S6), consistent with observations obtained from genetic depletion of ENL in the same cell line (5). To assess the extent to which ENL-dependent genes were perturbed by TDI-11055, we performed gene set enrichment analysis (GSEA). We found that genes that were previously shown to be downregulated by dTAG-mediated degradation of ENL were significantly downregulated by TDI-11055; by contrast, genes that were upregulated upon dTAG-mediated degradation of ENL were upregulated by TDI-11055 (Fig. 3B; Supplementary Table S7). To further evaluate the specificity of TDI-11055–induced transcriptional changes, we profiled ENL-dependent gene sets along with >6,000 gene sets from the Molecular Signatures Database (MSigDB; refs. 41, 42). Remarkably, ENL-dependent gene sets were the topmost enriched upon TDI-11055 treatment (Fig. 3C; Supplementary Table S7). These data not only strongly support the on-target activity of TDI-11055 in gene regulation but also indicate that specific perturbation of its reader function is sufficient for disrupting ENL-dependent transcriptional programs in AML cells. Among the genes with significantly downregulated expression upon TDI-11055 treatment, we found several key oncogenes in AML, including MYC, HOXA9/10, and MYB (Fig. 3A). The expression of these oncogenes was decreased by TDI-11055 in a concentration-dependent manner, starting at a concentration as low as 0.1 μmol/L (Fig. 3D). In terms of affected pathways, GSEA revealed that MYC target gene signatures were among the most repressed among >6,000 gene sets (Fig. 3E; Supplementary Table S7), in line with MYC being a top gene suppressed by TDI-11055 (Fig. 3A). Furthermore, TDI-11055 treatment led to the loss of a leukemia stem cell (LSC) signature (43) along with the gain of a myeloid lineage differentiation signature in MV4;11 cells (Fig. 3E; Supplementary Table S7), consistent with the well-established roles of key ENL target genes, including MYC and HOXA9, in regulating LSC function.
Figure 3.
Inhibition of ENL recruitment to chromatin suppresses key oncogenic gene expression programs in MLL-r and NPM1-mutated AML. A, Volcano plot of RNA-seq data [with External RNA Controls Consortium (ERCC) RNA spike-in] obtained from MV4;11 cells treated with DMSO or 5 μmol/L TDI-11055 (TDI) for 24 hours. Key differentially expressed genes are shown in red. Data represent mean across two replicates. See Supplementary Table S6. B, GSEA plots evaluating gene expression changes in MV4;11 cells treated with TDI-11055 (5 μmol/L for 24 hours), with genes downregulated (DN; left) or upregulated (UP; right) upon dTAG-mediated degradation of ENL. FDR, false discovery rate; NES, normalized enrichment score. See Supplementary Table S7. C, Unbiased GSEA using all signatures from MSigDB v7.2 C2 together with ENL genetic signatures for RNA-seq data presented in A. Each gene set is represented as a single dot. ENL signatures are indicated in red with numeral rank from the topmost enriched gene set. D, RT-qPCR analysis showing mRNA expression levels (normalized to B2M) of selected genes in MV4;11 cells upon treatment with different concentrations of TDI-11055 (0.01, 0.1, 1 μmol/L) for 72 hours. Error bars represent mean ± SEM (n = 3). E, GSEA plots evaluating gene expression changes in MV4;11 cells treated with 5 μmol/L TDI-11055 for 24 hours with genes associated with MYC target genes (left), LSCs (center), and monocyte differentiation (right). DMAP, differentiation map. F, Volcano plot of RNA-seq data (with ERCC RNA spike-in) obtained from OCI-AML3 cells treated with DMSO or 1 μmol/L TDI-11055 for 24 hours. Key differentially expressed genes are in red. Data represent mean across two replicates. See Supplementary Table S8. G, GSEA plots evaluating gene expression changes in OCI-AML3 cells treated with 1 μmol/L TDI-11055 for 24 hours with genes associated with MYC target genes (left), LSCs (center), and monocyte differentiation (right). NPM1 mut, NPM1-mutated. H and I, Pearson correlation of downregulated genes that were induced by TDI-11055 in either MV4;11 (H) or OCI-AML3 (I) cells and leukemia stem cell genes in the TCGA and TARGET datasets. Each dot represents one sample from the TCGA and TARGET datasets. r, Pearson correlation coefficient. P values by the two-tailed Pearson correlation test.
We next asked whether TDI-11055 suppressed the growth of ENL-dependent NPM1-mutated AML cells by regulating similar transcriptional changes as in MLL-r leukemia cells. We first examined the expression of MYC, MYB, and the HOXA cluster in OCI-AML3 cells and observed downregulated expression in response to TDI-11055 as seen in MV4;11 cells (Supplementary Fig. S4A). To fully assess transcriptional changes induced by TDI-11055, we performed RNA-seq for OCI-AML3 cells treated with DMSO or TDI-11055 for 24 hours (Fig. 3F; Supplementary Table S8). Although transcriptional changes induced by TDI-11055 in OCI-AML3 cells were less profound than those in MV4;11 cells (Fig. 3F), we found that top ENL-dependent genes in MV4;11 cells were suppressed by TDI-11055 in OCI-AML3 cells and vice versa (Supplementary Fig. S4B and S4C; Supplementary Tables S6 and S8). Moreover, similar to MV4;11 cells, OCI-AML3 cells exhibited downregulated expression of MYC target genes and of an LSC gene signature along with upregulated expression of genes associated with myeloid differentiation upon TDI-11055 treatment (Fig. 3G; Supplementary Table S7).
To explore the clinical relevance of ENL-regulated genes identified in cell line systems, we examined the relationship between the expression levels of genes that are downregulated by TDI-11055, identified in either MV4;11 or OCI-AML3 cells, and the expression levels of LSC genes (43) in primary AML samples from The Cancer Genome Atlas (TCGA; ref. 44) and the Therapeutically Applicable Research to Generate Effective Treatments (TARGET; ref. 45) datasets. This analysis revealed a strong positive correlation between ENL-dependent genes and LSC genes in patient samples (Fig. 3H and I). Collectively, these results suggest that specific inhibition of the reader function of ENL suppresses highly similar and clinically relevant oncogenic gene expression programs in MLL-r and NPM1-mutated leukemia.
TDI-11055 Induces Rapid Changes in Transcription Elongation at Top ENL-Bound Genes
ENL has been reported to interact with multiple chromatin-associated proteins, including the super elongation complex (SEC/P-TEFb), DOT1L, and RNA polymerase II–associated factor 1 (PAF1), all of which are involved in transcription regulation and implicated in AML (46–50). However, the precise contribution of these various binding partners to the function of ENL in AML remains incompletely understood. Previous work showed that genetic depletion of ENL results in decreased SEC/P-TEFb occupancy at a subset of ENL target genes (4, 5). It has remained unclear whether ENL also plays a role in the recruitment of other partners such as DOT1L and PAF1 and, importantly, whether isolated inhibition of its reader function is sufficient to recapitulate ENL depletion in chromatin regulation.
We treated MV4;11 cells ectopically expressing an HA-tagged ENL at levels equivalent to the endogenous ENL proteins with DMSO or TDI-11055 for 30 minutes and then determined ENL genomic occupancy by ChIP-seq using an anti-HA antibody. Such a short-term treatment with TDI-11055 was sufficient to globally displace ENL from chromatin (Fig. 4A; Supplementary Table S9). Next, we performed ChIP-seq for AFF1 (a scaffold subunit of SEC/P-TEFb), DOT1L, and PAF1 in MV4;11 cells. In the control cells, we found that AFF1, DOT1L, and PAF1 localized to largely similar genes across the genome as ENL (Supplementary Fig. S5A). To further investigate the relationship between each of these proteins and ENL, we grouped genes in the genome into strongly ENL-bound (top), weakly ENL-bound (low), as well as non–ENL-bound (non) genes based on HA-ENL ChIP-seq signals (Fig. 4B; Supplementary Table S10). Metagene profiles showed that AFF1, DOT1L, and PAF1 ChIP-seq signals positively correlated with the levels of ENL occupancy (Supplementary Fig. S5B; Supplementary Tables S11–S13). After 30 minutes of TDI-11055 treatment, there was an evident reduction of AFF1 (Fig. 4C–E) and DOT1L (Fig. 4C, F, and G) occupancy at ENL-bound genes. Notably, the fold-change decrease in AFF1 and DOT1L occupancy significantly correlated with the original ENL occupancy at target genes, with top ENL-bound genes (e.g., ZEB2, MYC, and HOXA9/10) demonstrating the most pronounced decrease after TDI-11055 treatment (Fig. 4C–G; Supplementary Tables S14 and S15). Similar results were observed after 24 hours of TDI-11055 treatment (Supplementary Fig. S5C and S5D; Supplementary Tables S11 and S12). In sharp contrast to AFF1 and DOT1L, PAF1 ChIP-seq signals were largely unaffected after 24 hours of TDI-11055 treatment (Supplementary Fig. S5E; Supplementary Table S13). Together, these results reveal a direct contribution of the reader function of ENL to the recruitment of select ENL-associated proteins (SEC/P-TEFb and DOT1L, but not PAF1) to at least a subset of ENL target genes in AML cells.
Figure 4.
TDI-11055 treatment induces rapid changes in transcription elongation at top ENL-bound genes. A, Rank-ordered heat map of HA-ENL ChIP-seq signals at ENL-bound peaks in MV4;11 cells treated with DMSO or TDI-11055 (5 μmol/L) for 0.5 hours. See Supplementary Table S9. RPM, reads per million. B, HA-ENL–bound genes in MV4;11 cells. HA-ENL ChIP-seq signals at the TSS ± 3 kb region of individual ENL-bound genes were plotted against their rank among all ENL-bound genes. Red depicts top ENL-bound genes (n = 165), and blue depicts low ENL-bound genes (n = 1,367). See Supplementary Table S10. C, The genome browser view of HA-ENL, AFF1, DOT1L, and Pol II S2P ChIP-seq signals at select ENL target genes under DMSO and TDI-11055 (TDI) treatment conditions in MV4;11 cells. D and E, Average occupancies (D) and quantification (E) of AFF1 on top, low, and non–ENL-bound genes along the transcription unit in DMSO and TDI-11055 (5 μmol/L for 0.5 hours) treatment conditions. See Supplementary Table S14. FC, fold change. F and G, Average occupancies (F) and quantification (G) of DOT1L on top, low, and non–ENL-bound genes along the transcription unit in DMSO and TDI-11055 (5 μmol/L for 0.5 hours) treatment conditions. See Supplementary Table S15. H–J, Average occupancies (H) and quantification (I and J) of Pol II S2P on top, low, and non–ENL-bound genes along the transcription unit in DMSO and TDI-11055 [5 μmol/L for 2 hours (I) or 24 hours (J)] treatment conditions. TES, transcription end site. In E, G, I, J, black solid lines denote median, and black dash lines denote quartiles. P values by the Welch two-tailed t test. See Supplementary Tables S16 and S17.
We then asked if the decreased occupancy of SEC/P-TEFb and DOT1L at ENL target genes mediates transcriptional changes induced by TDI-11055. We first examined serine 2 phosphorylation on Pol II (Pol II S2P), which is catalyzed by the CDK9 subunit in SEC/P-TEFb and a key indicator of productive transcription elongation (51). We observed a decrease of Pol II S2P ChIP-seq signals at top ENL-bound genes following 2 hours of TDI-11055 treatment (Fig. 4C, H, and I), and such a decrease became more pronounced after 24 hours of treatment (Fig. 4J). As in the case of AFF1, the fold-change decrease in Pol II S2P occupancy was most evident at top ENL-bound genes (Fig. 4I and J; Supplementary Tables S16 and S17). In contrast to Pol II S2P, we did not observe a significant decrease in total Pol II occupancy at ENL target genes. Instead, there was a slight but noticeable increase in total Pol II ChIP-seq signals at the promoter-proximal region of ENL target genes (Supplementary Fig. S5F; Supplementary Table S18), suggesting a potential defect in the release of Pol II from promoter-proximal pausing into productive elongation. We calculated the degree of Pol II pausing based on the ratio of promoter to gene body Pol II density (52) and found that TDI-11055 treatment increased the pausing index of Pol II (Supplementary Fig. S5G). Finally, the decrease in Pol II S2P occupancy was significantly associated with reduced gene expression (Supplementary Fig. S5H). Together, these findings bolster the reported function of ENL in SEC/P-TEFb recruitment and transcription elongation revealed by genetic studies (4, 5) and provide direct experimental evidence that such a function is critically dependent on the reader activity of ENL.
Unlike SEC/P-TEFb, the regulation of DOT1L chromatin localization by ENL had not been previously determined. Given that TDI-11055 treatment led to a rapid decrease in DOT1L occupancy at a subset of ENL target genes, we wanted to confirm that this is indeed due to ENL displacement using a genetic approach. We ectopically expressed an ENL–FKBP12(F36V) transgene in Cas9-positive MV4;11 cells and then infected these cells with an sgRNA that targets only the endogenous ENL to reduce the levels of endogenous proteins (Supplementary Fig. S5I). We observed a substantial decrease of ENL–FKBP12 proteins upon dTAG13 treatment for 6 or 24 hours (Supplementary Fig. S5J). Despite some residual levels of endogenous ENL proteins, the occupancy of DOT1L at several top ENL targets (e.g., HOXA9/10 and MYC) was evidently decreased upon dTAG13 treatment (Supplementary Fig. S5K). Thus, our results from both chemical and genetic studies support a role of ENL in DOT1L localization to a subset of target genes in AML cells that we have tested. DOT1L catalyzes methylation at histone H3 lysine 79—a chromatin mark generally associated with active genes—and inhibition of the DOT1L enzymatic activity has been shown to suppress the expression of key oncogenes (e.g., HOXA9/10) important for MLL-r and NPM1-mutated leukemia (53–55). We found that the reduced DOT1L occupancy (Supplementary Fig. S5D) led to only a very modest decrease in the H3K79me2 ChIP-seq signal at top ENL target genes following 24 hours of TDI-11055 treatment (Supplementary Fig. S6A–S6C; Supplementary Table S19), possibly due to the slow turnover of H3K79 methylation as previously noted (53, 54). Moreover, TDI-11055 decreased the expression of HOXA9/10 after 2 hours of treatment (Supplementary Fig. S6D), whereas the potent DOT1L inhibitor EPZ-5676 suppressed expression of the same genes at much later time points (Supplementary Fig. S6E), consistent with the reported slow kinetics of DOT1L inhibitor effects on gene expression (53–55). Thus, our results suggest that rapid transcriptional changes induced by TDI-11055 at early time points (e.g., within hours) are likely attributed to a decrease in SEC/P-TEFb–mediated Pol II elongation rather than a decrease in DOT1L-mediated H3K79 methylation. Nevertheless, we do not rule out the possibility that the rapid decrease in DOT1L occupancy in response to ENL inhibition might contribute to gene regulation in an H3K79 methylation–independent manner (56) or that changes in DOT1L-mediated H3K79 methylation might contribute to antileukemia effects of ENL inhibition at later time points.
TDI-11055 Impairs the Clonogenic Potential and Induces Differentiation of MLL-r and NPM1-Mutated Primary AML Patient Samples
Next, we explored the activity of ENL YEATS inhibition in primary AML patient samples. First, we tested the effects of TDI-11055 on the clonogenic potential of primary AML patient samples harboring different MLL1 translocations (MLL–AF4; MLL–AF9; MLL–ENL). TDI-11055 treatment strongly inhibited the colony formation of all five MLL-r leukemia patient samples tested in a concentration-dependent manner (Fig. 5A; Supplementary Table S20). Next, we assessed the activity of TDI-11055 in primary AML samples harboring NPM1 mutations and found a similar reduction in clonogenic potential (Fig. 5B; Supplementary Table S20). Notably, the effect of TDI-11055 in NPM1-mutated AML samples did not seem to be dependent on the type of secondary mutations, as the samples tested harbored diverse combinations of mutations and were all sensitive to TDI-11055 treatment. By contrast, treatment with TDI-11055 at the same concentrations had no effects on the colony formation of normal human hematopoietic progenitor cells (CD34+ umbilical cord blood cells; Fig. 5C) or normal mouse hematopoietic stem and progenitor–enriched Lin−Sca-1+c-Kit+ (LSK) populations (Fig. 5D). In addition to suppressing colony formation, TDI-11055 treatment induced differentiation phenotypes in primary AML samples, as reflected by the morphologic changes (Fig. 5E and F) and increased surface expression of the myeloid differentiation marker CD11b (Supplementary Fig. S7A and S7B) in TDI-11055–treated samples. Furthermore, the expression of key ENL target genes, including MYC, HOXA9, and MYB, was significantly downregulated upon TDI-11055 treatment in primary AML samples with MLL1 translocations or NPM1 mutations (Fig. 5G and H).
Figure 5.
TDI-11055 impairs the clonogenic potential and induces differentiation of MLL-r and NPM1-mutated primary AML samples. A and B, Quantification of colonies formed by five different MLL-r (A) and five different NPM1-mutated (B) primary AML samples under DMSO or TDI-11055 (1 and 10 μmol/L) treatment conditions. Error bars represent mean ± SEM (n = 3). See Supplementary Table S20 for more information about the primary patient samples. C and D, Quantification of colonies formed by normal human hematopoietic progenitor cells (CD34+ cord blood cells; C) and normal mouse hematopoietic stem and progenitor–enriched Lin−Sca-1+c-Kit+ (LSK) cells (D) under DMSO or TDI-11055 (1 and 10 μmol/L) treatment conditions. Error bars represent mean ± SEM (n = 3).E and F, Wright–Giemsa-stained cytospins for four different MLL-r (E) and four different NPM1-mutated (F) primary AML samples under DMSO or TDI-11055 (TDI; 1 and 10 μmol/L) treatment conditions. G and H, RT-qPCR analysis showing mRNA expression levels of select ENL-regulated genes in MLL-r (G) and NPM1-mutated (H) primary AML samples treated with DMSO or TDI-11055 (1 μmol/L). Error bars represent mean ± SEM (n = 3).
TDI-11055 Blocks Disease Progression in Models of MLL-r Leukemia
Given the strong inhibitory effects of TDI-11055 on AML cell lines and primary patient samples in vitro, we next wanted to explore the therapeutic potential of ENL inhibition in vivo using mouse models, a critical step toward clinical translation. To determine the optimal dosing strategies for in vivo studies, we integrated the unbound plasma concentration versus time profile following oral administration of TDI-11055 at different dose levels (Fig. 1I) with the in vitro potency against MV4;11 (IC50 = 0.27 μmol/L; Fig. 2A) to predict the magnitude and duration of target coverage achievable in vivo. Based on this analysis, we chose 100 and 200 mg/kg doses combined with once (q.d.) or twice (b.i.d.) daily dosing regimens for subsequent studies, as they had the potential to achieve high and sustained levels of target coverage in vivo.
We first carried out PK/pharmacodynamic (PD) studies to assess the effect of TDI-11055 on tumor growth in a mouse xenograft model using MV4;11 cells subcutaneously implanted into BALB/c nude mice (Fig. 6A). After tumors reached approximately 300 to 400 mm3, treatment with TDI-11055 [100 or 200 mg/kg, per os (p.o.), b.i.d.] or vehicle was initiated and continued for 8 consecutive days. Serial sampling of blood from TDI-11055–treated, tumor-bearing mice throughout the course of the treatment revealed sustained unbound drug concentrations above the in vitro IC50 for MV4;11 cell proliferation (0.27 μmol/L; Supplementary Fig. S8A and S8B), supporting our modeling. Dosing with TDI-11055 twice daily resulted in a significant inhibition of tumor growth over 8 days of treatment (Fig. 6B) with no overt effect on body weight (Supplementary Fig. S8C). Gene expression analysis of tumors at the end of treatment revealed a substantial reduction in the expression of key ENL target genes (HOXA9, MYC, MEIS1, and MYB) in tumors in the TDI-11055–treated group compared with vehicle controls (Fig. 6C). Furthermore, the degree of inhibition on tumor growth and target gene expression correlated with the levels of TDI-11055 in the blood (Supplementary Fig. S8A and S8B) and in tumor samples (Supplementary Fig. S8D), indicating a positive relationship between drug exposure and response in vivo.
Figure 6.
In vivo activity of TDI-11055 in xenograft models of MLL-r and NPM1-mutated leukemia. A, Schematic of tumor growth in a mouse xenograft model with MV4;11 cells and treatment workflow. Treatment with vehicle or TDI-11055 (100 or 200 mg/kg, p.o., b.i.d.) was initiated 20 days after transplantation and lasted for a total of 8 days. subQ, subcutaneous. B, Tumor volume over time in vehicle- or TDI-11055 (TDI)–treated mice in the MV4;11 subcutaneous transplantation model. Error bars represent mean ± SEM (n = 5). P < 0.05 using unpaired, two-tailed Student t test. C, RT-qPCR analysis showing mRNA expression levels of indicated genes in tumors from mice treated with vehicle or TDI-11055 for 8 days. Error bars represent mean ± SEM (n = 5 tumors from n = 5 mice). D, Schematic of the MV4;11 xenotransplantation model and treatment workflow. Treatment with vehicle or TDI-11055 (200 mg/kg, p.o., q.d.) was initiated 21 days after transplantation and lasted for 28 days. E, Flow-cytometric quantification of human CD45+ cells in the peripheral blood harvested at indicated time points from mice transplanted with MV4;11 cells. Note that mice in the vehicle-treated group developed terminal disease before reaching the end of the 28-day treatment (day 49 after transplantation). Bars represent the median (n = 5). F, Kaplan–Meier survival curves of vehicle (n = 5)- or TDI-11055 (n = 5)–treated mice in the MV4;11-disseminated model. P value using the log-rank test.G, Schematic of the MLL-r PDX-2263 xenotransplantation model and treatment workflow. Treatment with vehicle or TDI-11055 (200 mg/kg, p.o., q.d. or 200 mg/kg, p.o., b.i.d., with 5 days on and 2 days off) was initiated 7 days after transplantation and lasted for a total of 28 days. H, Flow-cytometric quantification of human CD45+ cells in the peripheral blood harvested at indicated time points from the MLL-r PDX-2263 model. As five of seven mice in the control group died before completing the treatment, their percentage hCD45 values shown were at the time of death rather than at end of the treatment (day 35 after transplantation). I, Kaplan–Meier survival curves of mice treated with vehicle (n = 9) or TDI-11055 (TDI; n = 9, 200 mg/kg, p.o., q.d.) in the MLL-r PDX-2263 model. P value using the log-rank test. J, Flow-cytometric quantification of human CD45+ cells in the peripheral blood harvested at indicated time points from the MLL-r PDX-6315 model. As two of 10 mice in each of the control and TDI-11055 treatment groups died before completing the treatment, their percentage hCD45 values shown were at the time of death rather than at experimental endpoint (44 days after transplantation; 8 days after completing 28-day treatment). K and L, Flow-cytometric quantification of human CD45+ cells in the bone marrow (K) and spleen (L) harvested 8 days after completing the treatment in the MLL-r PDX-6315 model. Bars represent the median. P value using unpaired, two-tailed Student t test. For J, K, and L, treatment with vehicle (n = 10) or TDI-11055 (n = 10, 200 mg/kg, p.o., q.d.) was initiated 8 days after transplantation and lasted 28 days. M, Kaplan–Meier survival curves of vehicle (n = 9)- or TDI-11055 (n = 10)–treated mice in the OCI-AML3 model. Treatment with vehicle or TDI-11055 (200 mg/kg, p.o., q.d.) was initiated 7 days after transplantation and lasted for 28 days. P value using the log-rank test. NPM1 mut, NPM1-mutated. N, Flow-cytometric quantification of human CD45+ cells in the bone marrow harvested 1 week after completing the treatment in the NPM1-mutated PDX-3055 model. Treatment with vehicle (n = 7) or TDI-11055 (n = 8, 200 mg/kg, p.o., q.d.) was initiated 14 days after transplantation and lasted 28 days. Bars represent the median. P value using an unpaired two-tailed Student t test.
We next tested the effects of TDI-11055 in a disseminated human MLL-r leukemia model. MV4;11 cells were injected into NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice via the tail vein, and treatment with TDI-11055 was initiated 21 days later to allow efficient engraftment of leukemic cells (Fig. 6D). Based on the PK/PD studies (Fig. 6A–C), we selected a 200 mg/kg q.d. regimen and treated the mice for 28 consecutive days without observing significant body weight loss (Supplementary Fig. S8E). TDI-11055 treatment resulted in lower leukemia burden in mice after 14 days of treatment (day 35 after transplantation) as assessed by the percentage of cells expressing human CD45 in the peripheral blood (Fig. 6E). All mice in the control group succumbed to the disease before reaching the end of treatment (day 49 after transplantation) and showed a high percentage of human leukemia cells in the peripheral blood (Fig. 6E), the bone marrow, and the spleen at necropsy (Supplementary Fig. S8F). By contrast, most mice in the TDI-11055–treated group remained alive and had low or nondetectable levels (0%–1.6%) of human leukemia cells in peripheral blood by the end of drug treatment (Fig. 6E; day 49 after transplantation). Finally, the overall survival of mice treated with TDI-11055 was significantly improved when compared with mice in the control group (Fig. 6F; P = 0. 0018), with a 70% increase in median survival (37 vs. 63 days).
Next, we sought to assess the activity of TDI-11055 in PDX models, which maintain the histopathologic features and genetic profiles of the original human primary tumor samples and thus represent highly clinically relevant preclinical models. We derived a PDX model from a pediatric patient with MLL–AF4 B-ALL who experienced multiple relapses and eventual lineage switch to AML. Patient leukemia cells were first expanded in NSG primary recipients, and then human leukemia cells were harvested from the bone marrow and transplanted into NSG secondary recipient mice via tail-vein injection. Treatment with vehicle or TDI-11055 was initiated 7 days after transplantation and continued for 28 days (Fig. 6G). Because of the aggressive and treatment-refractory nature of this PDX model, we started with two different dosing regimens (Fig. 6G): 200 mg/kg q.d. for 28 consecutive days, and 200 mg/kg b.i.d. with a 5 day on and 2 day off cycle for a total of 28 days. Both dosing regimens were well tolerated without causing overt toxicity and significant body weight loss (Supplementary Fig. S8G). Leukemia developed rapidly in most vehicle-treated mice, as manifested by a substantial increase in the percentage of human CD45+ cells in peripheral blood over time (Fig. 6H). Before reaching the end of the treatment (day 35 after transplantation), most mice (five of seven) in the control group developed terminal disease, as reflected by high levels of leukemic blasts in the bone marrow and spleen at necropsy (Supplementary Fig. S8H). In contrast, all mice in the two TDI-11055–treated groups had low or nondetectable levels of leukemia cells in the peripheral blood at the end of treatment (Fig. 6H). As there were no noticeable differences in the leukemia burden between the two treatment regimens, we chose the once-daily regimen to assess the survival benefit of TDI-11055 treatment in a separate batch of experiments with a larger cohort of mice. TDI-11055–treated mice had substantially prolonged survival (Fig. 6I; P = 0.0003) with a 53% increase in median survival over the vehicle-treated mice (32 vs. 49 days). We further assessed the activity of TDI-11055 in a second MLL-r PDX model derived from an AML patient sample harboring an MLL–ENL fusion (57). Treatment of mice with TDI-11055 (200 mg/kg, p.o., q.d.) was initiated 8 days after transplantation and continued for 28 days. Although the percentage of human CD45+ cells in the peripheral blood increased over time in the vehicle controls, it remained largely unchanged in the TDI-11055–treated group during the treatment period (Fig. 6J), suggesting a significant block in disease progression. After 4 weeks of treatment, the leukemia burden was substantially reduced in both the bone marrow and spleen in the TDI-11055–treated mice when compared with vehicle controls (Fig. 6K and L). In summary, our results show that TDI-11055 administration as a single agent for 28 days was able to significantly block disease progression in models of aggressive MLL-r leukemia.
TDI-11055 Blocks Disease Progression in Models of NPM1-Mutated Leukemia
Next, we explored the therapeutic effects of TDI-11055 in disseminated models of NPM1-mutated AML. We first utilized the OCI-AML3 (with NPM1 and DNMT3A mutations) xenotransplantation model, which represents an aggressive model of NPM1-mutated AML in which terminal disease develops within 3 weeks after transplantation. Treatment with vehicle or TDI-11055 (200 mg/kg, p.o., q.d.) was initiated 7 days after transplantation and continued for 28 consecutive days. As expected, the vehicle-treated mice quickly developed terminal leukemia with a median survival of 23 days. TDI-11055 treatment resulted in a significant increase in survival (Fig. 6M; P = 0.0065). We further tested the activity of TDI-11055 in a PDX model of NPM1-mutated leukemia (3055, with NPM1 mutation and FLT3-ITD; ref. 58). Treatment with vehicle or TDI-11055 (200 mg/kg, p.o., q.d.) was initiated 14 days after transplantation and continued for 28 consecutive days. Because of the relatively long latency of leukemia development in this model (58), treatment efficacy was assessed by comparing the leukemia burden, defined by the percentage of bone marrow cells expressing human CD45, 1 week after completing the treatment. We found that TDI-11055–treated mice had a significantly lower leukemia burden than the vehicle controls (Fig. 6N).
Collectively, our results from numerous AML models provide the first demonstration of in vivo efficacy of a YEATS domain inhibitor against cancer. TDI-11055 demonstrates pronounced in vivo activity, manifested by reduced leukemia growth and prolonged survival in xenograft models of advanced MLL-r and NPM1-mutated leukemia. This correlates with markedly downregulated expression of key ENL target genes including MYC and HOXA9, which might represent valid biomarkers for assessing the effectiveness of ENL inhibitors in future clinical studies.
The Impact of TDI-11055 on Normal Hematopoiesis
Having established the antileukemia effect of TDI-11055 both in vitro and in vivo, we sought to assess whether TDI-11055 affects normal hematopoiesis in mice. We treated healthy C57BL/6 mice with vehicle or TDI-11055 using the same regimen applied to the leukemia models (200 mg/kg, p.o., q.d.) for 28 consecutive days (Fig. 7A). There was no overt toxicity and no evident loss of body weight or spleen weight following TDI-11055 treatment (Fig. 7B and C). Complete blood count analyses (Fig. 7D) showed no significant changes in levels of red blood cells (RBC), hematocrit (HCT), and platelets. The levels of white blood cells (WBC) and hemoglobin (HGB) were slightly decreased in the TDI-11055–treated group compared with vehicle controls, but these levels remained within the normal range (59, 60). Next, we analyzed bone marrow samples isolated from the mice and observed a modest decrease in the number of total bone marrow cells in TDI-11055–treated mice (Fig. 7E). We further profiled different hematopoietic cell compartments using flow-cytometric analysis (Supplementary Table S21). Interestingly, TDI-11055 treatment led to an increase in both relative and absolute numbers of stem cell–enriched LSK cells (Fig. 7F; Supplementary Fig. S9A and S9B). Among LSK cells, we observed a modest decrease in the absolute number of phenotypically defined long-term hematopoietic stem cells (LT-HSC, Lin−Sca-1+c-Kit+CD150+CD48−), whereas multipotent progenitors (MPP; Lin−Sca-1+c-Kit+CD150−CD48−) and downstream hematopoietic progenitor cell subsets were increased (HPC-1, Lin−Sca-1+c-Kit+CD150−CD48+ and HPC-2, Lin−Sca-1+ c-Kit+CD150+CD48+; Fig. 7F; Supplementary Fig. S9A and S9B; ref. 61). TDI-11055 treatment induced minimal changes in the myeloerythroid progenitor–enriched LKS− population (Lin−c-Kit+Sca-1−) and the granulocyte–macrophage progenitor–enriched population (GMP; Lin−Sca-1−c-Kit+CD41−CD150−CD16/32+; refs. 61–63; Fig. 7F; Supplementary Fig. S9C and S9D). Furthermore, analysis of differentiated cell populations in the bone marrow revealed a modest decrease in the absolute number of myeloid cells (Mac1+Gr1+) in the TDI-11055–treated mice, accompanied by an increase in the relative and absolute numbers of CD4+ or CD8+ T cells (Fig. 7G; Supplementary Fig. S9E). We also observed a decrease in developing B cells (B220+CD19+CD93+) in the bone marrow of TDI-11055–treated mice, whereas the pool of mature recirculating B cells (B220+CD19+CD93−) was less affected (Fig. 7G; Supplementary Fig. S9E; ref. 64).
Figure 7.
The impact of TDI-11055 on normal hematopoiesis. A, Schematic of TDI-11055 treatment workflow in normal C57BL/6 mice. B, Body weight variance over time for vehicle- or TDI-11055 (TDI)–treated mice. Error bars represent mean ± SEM (n = 6). C, Representative images (left) and weight quantification (right) of spleen harvested from mice at the end of 28-day treatment with vehicle or TDI-11055. Error bars represent mean ± SEM (n = 6). n.s., not signficant. D, Complete blood count analyses [white blood cell (WBC), red blood cell (RBC), hemoglobin (HGB), hematocrit (HCT) and platelets] of peripheral blood samples harvested from mice at the end of 28-day treatment with vehicle or TDI-11055. Bars represent the median (n = 6). E–G, Number of total cells (E), immature cells [LSK, long-term hematopoietic stem cells (LT-HSC), Lin−c-Kit+Sca-1− (LKS−), multipotent progenitor (MPP), and granulocyte–macrophage progenitor (GMP); F], and differentiated cells (Mac1+Gr1+ myeloid, CD4+ T, CD8+ T, B220+CD19+CD93− B, and B220+CD19+CD93+ B; G) in bone marrow samples harvested from mice at the end of 28-day treatment with vehicle or TDI-11055. Bars represent the median (n = 6). BM, bone marrow. H, Complete blood count analyses (WBC, RBC, HGB, HCT, platelets) of peripheral blood samples harvested from mice 80 days after completing treatment with vehicle or TDI-11055. Bars represent the median (n = 3). I–K, Number of total cells (I), immature cells (LSK, LT-HSC, MPP, LKS−, and GMP; J), and differentiated cells (Mac1+Gr1+ myeloid, CD4+T, CD8+ T, B220+CD19+CD93− B, and B220+CD19+CD93+ B; K) in bone marrow samples harvested from mice 80 days after completing treatment with vehicle or TDI-11055. Bars represent the median (n = 3). C–K,P values by an unpaired, two-tailed Student t test.
To ask whether the changes in different hematopoietic cell compartments induced by TDI-11055 treatment were reversible, we monitored a small cohort of mice for an additional 80 days after completing the treatment and performed an analysis of peripheral blood and bone marrow samples. Notably, no significant differences were observed between TDI-11055–treated mice compared with the vehicle controls in complete blood counts (Fig. 7H) and total bone marrow cell numbers (Fig. 7I). Furthermore, TDI-11055–induced changes in different hematopoietic cell compartments (Fig. 7J and K; Supplementary Fig. S10A–S10E) largely disappeared.
Together, these results show that TDI-11055 treatment caused modest and well-tolerated changes in specific hematopoietic compartments, which resolved after cessation of treatment. Note, our biochemical studies show that TDI-11055 exhibits similar activity against the YEATS domains of ENL and its paralog, AF9 (Fig. 1B). Previous studies have established an important role of AF9 in the expansion and transplantation potential of human hematopoietic stem/progenitor cells (65), whereas the function of ENL in normal hematopoiesis remains undetermined. Thus, the effects induced by TDI-11055 in normal hematopoiesis could be due to the suppression of AF9 function in hematopoietic stem/progenitor cells or the combinatorial inhibition of AF9 and ENL. Nevertheless, the transient and well-tolerated effects of TDI-11055 in normal steady-state hematopoiesis, together with its potent effects in AML models, indicate a promising therapeutic window for this ENL inhibitor.
DISCUSSION
Cancer cells are characterized by dysregulated transcriptional programs, and these programs can render cancer cells highly dependent on certain regulators of gene expression. Recognition of modified histones by reader proteins constitutes a key mechanism underlying transcriptional control; therefore, targeting such pathways holds clinical promise (66, 67). In recent years, the YEATS domain–containing proteins have emerged as novel acyl-lysine readers with important roles in cancer and, as such, attractive drug targets (7). In particular, the identification of the ENL YEATS–histone acylation interaction as a critical dependency in MLL-r leukemia (4, 5) has motivated ongoing early drug development. Although a few small-molecule inhibitors of ENL have been reported so far (25–27, 30, 31), pharmacologic evaluation of ENL as a therapeutic target remained inadequately explored due to the current lack of an in vivo bioavailable inhibitor. Furthermore, the direct chromatin changes induced by specific inhibition of the ENL YEATS–acylation interaction and how they lead to suppression of the malignant state remained undefined.
In this study, we describe TDI-11055 as a potent and the first orally bioavailable small-molecule inhibitor of the ENL/AF9 YEATS domains. TDI-11055 retains the potency and selective profile of SGC-iMLLT and, through structure-based chemical design, exhibits markedly improved PK properties that enable in vivo utility. We show that TDI-11055 inhibits the proliferation and colony-forming potential of human leukemia cell lines bearing MLL1 translocations or NPM1 mutations, and the antiproliferative activity of TDI-11055 phenocopies genetic perturbation of ENL. A particularly powerful demonstration of the on-target activity of TDI-11055 is the finding that a specific deletion mutation in the ENL YEATS domain, discovered through a CRISPR-Cas9–mediated mutagenesis screen, can render cells less sensitive to the compound. Importantly, the development of TDI-11055 has allowed us for the first time to explore the therapeutic potential of ENL inhibition in human primary AML patient samples in vitro and in vivo. TDI-11055 is very effective in decreasing the clonogenic potential and inducing differentiation in primary MLL-r or NPM1-mutated AML patient samples, and, notably, these effects do not seem to depend on specific MLL1 fusion partners or secondary mutations co-occurring with NPM1 mutation. Furthermore, administration of TDI-11055 as a single agent in vivo blocks disease progression and prolongs survival in both MLL-r and NPM1-mutated leukemia models, including PDXs with a high clinical relevance. Importantly, treatment with TDI-11055 has a transient and well-tolerated impact on normal hematopoiesis in mice without causing other overt toxicity, indicating the existence of a good therapeutic window for ENL YEATS inhibition. Collectively, our results provide crucial preclinical evidence for pharmacologic inhibition of ENL in molecularly defined AML subsets, supporting rapid translation of this approach to the clinical setting.
The development and rigorous validation of TDI-11055 has allowed us to interrogate the precise function of the ENL YEATS–histone acylation interaction on chromatin. Although previous studies using genetic depletion have established a contribution of ENL to the recruitment of SEC/P-TEFb and Pol II elongation (4, 5), our inhibitor studies have now provided direct experimental evidence that this function of ENL occurs in a YEATS domain–dependent manner. Remarkably, 30 minutes of TDI-11055 treatment is sufficient to globally displace ENL from chromatin. This in turn leads to a rapid decrease in the occupancy of SEC/P-TEFb and Pol II S2P at a subset of ENL-bound genes preceding decreased gene expression, indicating that TDI-11055 suppresses gene expression, at least in part, through the regulation of the SEC/P-TEFb/Pol II S2P axis. In addition to SEC/P-TEFb, ENL has been shown to interact with other chromatin-associated proteins, including DOT1L and PAF1, yet the contribution of ENL to the recruitment of these proteins had not been previously determined. Using both genetic and chemical approaches, we provide evidence for a direct role of ENL and its YEATS domain in DOT1L chromatin localization to at least a subset of target genes in AML, revealing an additional mechanism underlying DOT1L regulation (68). It is worth pointing out that the rapid transcriptional changes induced by TDI-11055 in early time points (within hours) are more likely attributed to a decrease in SEC/P-TEFb–mediated Pol II elongation rather than a decrease in DOT1L-mediated H3K79 methylation, as only a very modest decrease in H3K79 methylation is observed at top ENL target genes after 24 hours of TDI-11055 treatment. Nevertheless, it will be interesting for future studies to explore whether the decrease in DOT1L occupancy in response to ENL inhibition could contribute to gene regulation in an H3K79 methylation–independent manner (56) and whether changes in DOT1L-mediated H3K79 methylation contribute to the efficacy of ENL inhibitors in AML at later time points. Finally, in contrast to SEC/P-TEFb and DOT1L, the chromatin occupancy of PAF1, another ENL-interacting partner that also plays a critical role in transcription elongation (69), is largely unaffected by TDI-11055, suggesting that ENL acts downstream of PAF1 (47) or independently of PAF1 in the context that we have tested. Our results thus reveal immediate consequences of ENL YEATS inhibition on chromatin and offer molecular insights that support the effectiveness of YEATS inhibitors in disrupting ENL-mediated function.
Notably, TDI-11055–induced chromatin changes occur most prominently at top ENL-bound genes, which account for a very small fraction (<10%) of all ENL-bound genes. These genes, as previously noted (5), strongly enrich for key AML oncogenes, including MYC and HOXA9/10. The expression of these oncogenes is sensitive to TDI-11055 treatment in both MLL-r and NPM1-mutated leukemia cell lines and primary patient samples, indicating their expression levels as potential PD biomarkers for the clinical translation of ENL YEATS inhibitors. These findings, together with results from genetic studies (4, 5), reveal that ENL exerts its regulatory function at highly selective loci. Such a property may underlie the context-specific roles of ENL as reflected by the fact that ENL gene inactivation is not detrimental to the growth of most cancer cell lines in the Dependency Map database (70, 71). This is distinct from BRD4, a well-studied histone acetylation reader, which binds to a broad range of acetylated enhancers and protomers and is a commonly essential gene across diverse cell types (70, 71). Future studies are warranted to investigate mechanisms underlying the chromatin distribution of ENL in detail and to identify additional biological contexts in which ENL plays a critical role to fully explore the therapeutic potential of ENL targeting.
The findings that MLL-r and NPM1-mutated leukemia cells are sensitive to genetic and pharmacologic inhibition of ENL provide a blueprint for future clinical investigation. MLL-r and NPM1-mutated leukemia are both characterized by high expression of the HOXA cluster. They also exhibit dependence on a shared subset of chromatin regulators, notably DOT1L and Menin as identified in previous studies (53–55, 58, 72–77), as well as ENL as shown in the current study. These observations support the notion that distinct AML genetic drivers can co-opt similar chromatin machinery to sustain core oncogenic gene expression programs important for disease maintenance. Indeed, TDI-11055 treatment induces gene expression changes in a common subset of genes in MLL-r and NPM1-mutated leukemia cells that we have tested. Future studies will be needed to explore the cross-talk between ENL and other chromatin regulators required for MLL-r and NPM1-mutated leukemia, including DOT1L and Menin, whose inhibitors are currently under clinical evaluation. Such studies will enable a better understanding of the regulatory circuits governing leukemic gene expression programs and have the potential to nominate more effective treatment strategies that might overcome specific mechanisms of resistance to individual epigenetics-targeted agents. Furthermore, although our findings suggest that MLL-r and NPM1-mutated AML may be the most sensitive to ENL inhibition, it will be important for future studies to test the responsiveness of different molecular subtypes of AML more broadly to ENL inhibition.
In summary, our study establishes TDI-11055 as a comprehensively validated ENL inhibitor and provides the first demonstration of in vivo activity and therapeutic efficacy of a YEATS domain inhibitor against cancer. The biological and chemical insights discovered herein offer a solid foundation for further evaluation of ENL inhibition as a therapeutic strategy against subsets of AML and pave the way toward meaningful biology endpoints. We envision that validated chemical probes of YEATS domain–containing proteins, such as TDI-11055 described herein, will serve both as powerful tools for studying this newly identified group of chromatin readers in broad biological contexts and as starting points for further development of better and more potent inhibitors for clinical evaluation.
METHODS
Compound Synthesis
For the chemical synthesis of TDI-11055, see Supplementary Methods.
Molecular Modeling
The binding mode of TDI-11055 was predicted by docking the ligand to the SGC-iMLLT binding site of the experimental crystal structure (Protein Data Bank: 6HT1) using Schrödinger's Glide SP program. The protein structure was prepared using the Protein Preparation Wizard in Maestro with default settings, keeping the buried water molecule that forms water-mediated interactions with the ligand. The ligand structure was prepared using LigPrep in Maestro, selecting for tautomers that have the amide-NH and a neutral state of azaindole. Free-energy calculations were performed with Schrödinger's FEP+ (78). Before prospective application, we performed a validation using 18 compounds spanning 3 orders of magnitude in binding affinity. The mean unsigned error was 0.87 kcal/mol, meaning that predictions are on average within 10-fold of the experiment. The R2 correlation was 0.51.
Caco2 Cell Permeability
The apical-to-basolateral (A-B) and basolateral-to-apical (B-A) transport of 5 μmol/L test compounds in HBSS (10 mmol/L HEPES, pH 7.4) was measured across Caco2 cell monolayers (ATCC HTB-37). Duplicate incubations were performed at approximately 37°C for 120 minutes, with functionality of the test system confirmed using 5 μmol/L propranolol and digoxin as control compounds. Aliquots (50 μL) from both apical and basolateral wells were transferred into two fresh 96-well plates and quenched with acetonitrile solution containing analytical internal standards. Samples were vortexed mixed and centrifuged, and a 100 μL aliquot of the resulting supernatant was mixed with an equal volume of ultra-pure water prior to analysis by ultra-performance liquid chromatography–mass spectrometry (UPLC-MS/MS). The concentrations of the test compound and control compounds in the incubation medium of donor and receiver compartments at the beginning and the end of the incubation period were used to calculate the apparent permeability (Papp) from the A-B and B-A directions. The efflux ratio was expressed as PappB -A/PappA-B. The integrity of the cell monolayers after 2 hours of incubation was confirmed using the marker reagent Lucifer yellow (Sigma-Aldrich, #L0259).
LogD Determination
LogD7.4, which is a partition coefficient between 1-octanol and aqueous buffer pH 7.4, of the compounds was measured on the chromatographic procedure whose condition was developed based on a published method (79). TDI-11055 (15 μL of 10 mmol/L DMSO stock solution), 500 μL of 1-octanol saturated PBS (pH 7.4), and 500 μL of PBS-saturated 1-octanol were added to glass vials. Solutions were shaken at 25°C at 1,100 rpm for 1 hour. Following separation of the octanol and buffer layers, aliquots of 5 μL were taken from the octanol phase and added to 495 μL of acetonitrile. Following mixing, a 50 μL aliquot was further diluted with 450 μL of acetonitrile. For the buffer phase, 50 μL aliquots were taken and diluted with 450 μL of acetonitrile. Aliquots of each final dilution (100 μL) were transferred to a fresh 96-well plate and mixed with an equal volume of water prior to analysis by UPLC-MS/MS.
TR-FRET Assay
An optimized TR-FRET approach was used to determine to what degree the compounds inhibit interaction between indicated YEATS domain proteins and histone H3 acyl peptides. The general protocol was as follows: 4 μL compound or DMSO control (5%) was preincubated for 15 minutes with 4 μL of 5 × 6HIS-tagged YEATS domain protein: ENL (625 or 50 nmol/L), AF9 (75 nmol/L), GAS41 (200 nmol/L), and YEATS2 (200 nmol/L). This was followed by the addition of 4 μL 5× biotinylated peptide (see the list below) and incubated for 30 minutes at room temperature in the appropriate assay buffer in a 384-well plate. Eight microliters of a 2.5× mix of 37.5 nmol/L anti-6HIS ULight (PerkinElmer) and 1.25 nmol/L streptavidin Europium-chelate were added, and plates were incubated for 60 minutes at room temperature under subdued lighting. Signals (615 nm and 665 nm) were sequentially measured on a PerkinElmer 2104 EnVision (320-nm excitation, 615-nm emission filter ± 4-nm bandwidth, and 665-nm emission filter ± 3.5-nm bandwidth). TR-FRET signal was calculated as follows: TR-FRET signal = (665 nm signal/615 nm signal) × 10,000. For dose–response curves, data were plotted using 4-parameter logistic sigmoidal curve fitting via GraphPad Prism (v9). IC50 apparent was calculated for compounds when possible or was noted as not determinable (ND). Peptides used for TF-FRET assays were as follows: ENL: 25 nmol/L H3K9cr (aa 1–20); AF9: 15 nmol/L H3K9cr (aa 1–20); GAS41: 50 nmol/L H3K9,14,18cr (aa 1–20); and YEATS2: 10 nmol/L H3.3K27cr (aa 15–34). Buffer conditions were as follows: ENL-125 nmol/L: 50 mmol/L Tris pH 8 + 150 mmol/L NaCl + 0.5% Casein + 0.1% NP-40; ENL-10 nmol/L: 20 mmol/L Tris pH 7.5 + 125 mmol/L NaCl + 0.01% BSA + 0.01% NP-40; AF9: 50 mmol/L Tris pH 8 + 150 mmol/L NaCl + 0.5% Casein + 0.1% NP-40; GAS41: 20 mmol/L Tris pH 7.5 + 50 mmol/L NaCl + 0.01% BSA + 0.01% NP-40; and YEATS2: 20 mmol/L Tris pH 7.5 + 25 mmol/L NaCl + 0.01% BSA + 0.01% NP-40.
ITC Titration
All ITC titrations were performed at 25°C using a Nano ITC (TA Instruments). Protein was thawed in room temperature water and transferred to ice as soon as thawed. The protein sample was dialyzed overnight in ITC buffer (20 mmol/L HEPES 7.5; 200 mmol/L KCl; 1 mmol/L TCE; 2% DMSO, added before titration; 0.01% Tween 20, added before titration), kept on ice afterward, and used within 2 days after dialysis. Compounds were dissolved in DMSO based on provided molecular weight and weight information. The compound was further diluted to 50× assay concentration in DMSO prior to dilution into ITC buffer just prior to titration. Both titrand and titrant were diluted in dialysis buffer to the desired concentration. Solvent and detergent concentrations were matched between protein and compound solutions. The recombinant YEATS proteins were extensively dialyzed against ITC buffer. All samples were centrifuged at 13,000 rpm at 25°C for ∼10 to 15 minutes before titration. ITC cell was rinsed with buffer, and then protein solution at test concentration was added to the cell. Contents were pipetted up/down several times to mix with any trace buffer in the cell. A small volume of protein solution was removed from the cell for concentration recheck using NanoDrop. The compound was added to the main solution in 20 increments with 250-second intervals between injections. Usually, compound at 150 μmol/L was added to protein solution at 20 to 25 μmol/L, and H3 peptides at 2.0 to 2.2 mmol/L were titrated into proteins at 0.15 mmol/L. Data were analyzed using TA NanoAnalyze software. First injection data were always excluded from the analysis.
Cellular Thermal Shift Assay
A cellular thermal shift assay was performed following a previously reported protocol (32). Briefly, 2 × 107 cells were treated with DMSO or 5 μmol/L TDI-11055 and incubated in a 37°C incubator for 1 hour. After treatment, cells were collected, washed once with PBS, and resuspended in 500 μL PBS with a protease inhibitor cocktail (Roche, complete tablets, EDTA-free). Cell suspension was then aliquoted equally into eight PCR tubes, heated at indicated temperatures for 3 minutes using a thermocycler, followed by cooling at 25°C for another 3 minutes. The samples were lysed by adding 30 μL lysis buffer (50 mmol/L Tris, pH 7.4, 250 mmol/L NaCl, 5 mmol/L EDTA, 50 mmol/L NaF, 1 mmol/L Na3VO4, 1% NP-40, protease inhibitor cocktail) and treated with 3 repeated freeze–thaw cycles using liquid nitrogen. The lysate was collected by centrifugation at 15,000 × g for 20 minutes at 4°C using a benchtop centrifuge. The supernatants were transferred to a new tube and analyzed by immunoblotting.
Plasmids
The 3xFLAG-HA-tagged wild-type human ENL construct was generated previously (4). HA-tagged wild-type human ENL was cloned into a LentiV_Neo vector (Addgene 108101, a gift from Junwei Shi, University of Pennsylvania, Philadelphia, PA). Neomycin was replaced by mCherry using the in-fusion cloning method (Clontech, #638909). Mutations in ENL were introduced using a site-mutagenesis approach. Constitutive Cas9 vectors (LentiV_Cas9_Puro vector, ref. 80; LentiCas9_Blast vector, Addgene 52962, ref. 81; a gift from Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA) were used to generate Cas9-expressing cells. sgRNAs were cloned into the LRG2.1T (82). All sgRNA sequences used are provided in Supplementary Tables S2–S4.
Antibodies
All antibodies used are provided in Supplementary Table S21.
Cell Culture and Virus Transduction
Human leukemia cell lines (MV4;11, MOLM-13, ML2, OCI-AML2, OCI-AML3, HL60, K562, U937, and Jurkat) were maintained in RPMI 1640 (Fisher Scientific, #MT10040CV) supplemented with 10% FBS (HyClone, #SH30910.03) and 100 U/mL penicillin/streptomycin (Fisher Scientific, #15-140-122). Human HEK293 cells (ATCC, #CRL-1573) were grown in EMEM (Fisher Scientific, #MT10009CV) with 10% FBS and 100 U/mL penicillin–streptomycin. HEK293T cells were maintained in DMEM (Fisher Scientific, #MT10013CV) supplemented with 10% FBS and 100 U/mL penicillin/streptomycin. All cell lines were Mycoplasma-negative and were tested for authentication by short tandem repeat profiling. Lentivirus packaging was performed in HEK293T cells using PEI MAX reagent [DNA (μg):PEI (μL) = 1:4; Fisher Scientific, #NC1038561] in accordance with the manufacturer's instructions. Medium containing virus was filtered and stored at −80°C. Leukemia cells were spin infected at 2,500 rpm at 33°C for 60 minutes with 10 μg/mL polybrene (EMD Millipore, #TR-1003-G). HEK293 cells were infected by incubation with viral supernatants for 24 hours in the presence of 10 μg/mL polybrene. Transduced cell populations were usually selected or analyzed 72 hours after infection.
Colony Formation of Human Leukemia Cells
For leukemia cell lines, colony-forming assays were performed in Methocult (STEMCELL Technologies, #H4435). The indicated number of cells were resuspended in 100 μL PBS and added to 900 μL MethoCult. After 7 days, the number of colonies was determined by counting. For primary cells from patients, the indicated number of cells were resuspended in 500 μL 20% FBS IMDM containing DMSO or TDI-11055 and added to 1.5 mL human methylcellulose-enriched media (R&D Systems, #HSC005). The number of colonies was determined after incubation in a 37°C incubator for ∼2 weeks.
Flow-Cytometric Analysis of Different Hematopoietic Cell Compartments in the Bone Marrow
Vehicle- or TDI-11055–treated C57BL/6J mice were sacrificed, and bone marrow cells were extracted from femurs, coxae, and vertebral columns. Bone marrow cells were stained with murine c-Kit, Sca-1, lineage antibodies, and Zombie Aqua fixable viability kit (BioLegend, #423102). Cells were run through the FACSAria Flow Cytometer (BD) to perform hematopoietic cell compartment analysis.
LSK Cell Sorting and Colony Formation
Eight-week-old C57BL/6J mice were sacrificed, and bone marrow cells were extracted from femurs, coxae, and vertebral columns. c-Kit–positive cells were enriched using the EasySep Mouse CD117 (cKIT) positive selection kit (STEMCELL Technologies, #18757). Then, c-Kit–enriched cells were stained with murine c-Kit, Sca-1, lineage antibodies, and Zombie Aqua fixable viability kit (BioLegend, #423102). Cells were sorted using a FACSAria Flow Cytometer (BD) to obtain the LSK cells. For the colony formation assay, 2,000 LSK cells were resuspended in 100 μL IMDM and add to 1.5 mL methylcellulose media (STEMCELL Technologies, #M3234) supplemented with 10 μg/mL IL3, 20 μg/mL SCF, and 10 μg/mL IL6 (PeproTech, #231-13, 250-03, 216-16). Colonies were counted after incubation in a 37°C incubator for 7 days.
Cell Viability Assays
Cell viability assays for TDI-11055 treatment were performed using CellTiter-Glo Luminescent Cell Viability Assay (Promega, #G7572) according to the manufacturer's guidelines. Dose–response curves were generated using GraphPad Prism (v9) with a three-parameter log(inhibitor) versus response fitting. IC50 apparent was calculated for compounds when possible or was noted as ND.
Competitive Proliferation Assays Using sgRNAs
Cas9-positive leukemia cells were transduced with lentivirus containing indicated sgRNAs or cDNA constructs and analyzed for GFP or mCherry expression 5 days after infection. The percentage of sgRNA-expressing cells (GFP+) or cDNA construct expressing cells (mCherry+) was measured over time using flow cytometry and normalized to the starting time point.
Cell-Cycle and Apoptosis Analyses
Cell-cycle analysis for DMSO- or TDI-11055–treated cells was performed using the APC BrdU Flow Kit (BD, #552598) according to the manufacturer's guidelines. Data were analyzed by FlowJo software (v10). For apoptosis assays, DMSO- or TDI-11055–treated cells were resuspended in 100 μL Annexin V Binding buffer (BioLegend, #422201). Five microliters of Annexin V-PE (BioLegend, #640908) and 5 μL of 7-AAD viability staining solution (BioLegend, #420403) were added to the buffer. Cells were gently mixed and incubated at room temperature in the dark for 15 minutes. Cells were then washed once with Annexin V binding buffer and analyzed using the Attune Flow Cytometer (Thermo Fisher).
Cytospin for Primary AML Patient Samples
Primary AML patient cells (40,000) were resuspended in 200 μL PBS. Slides and filters were placed into appropriate slots with the cardboard filters facing the center of the cytospin. Cells were added to the cytofunnel and spun at 700 rpm for 10 minutes. Filters were removed from the slide without disturbing the cells. Slides were stained with the Wright–Giemsa Stain Kit (Jorgensen Labs, #J0322A). Images were captured using a Zeiss Widefield Microscope.
CRISPR-Cas9–mediated Saturated Mutagenesis Screen of ENL and AF9 YEATS Domains
sgRNA sequences targeting the YEAST domains of human ENL (NM_005934.3; total 88 sgRNAs) and AF9 (NM_004529.3; total 66 sgRNAs) were designed using the Genetic Perturbation Platform (Broad Institute; ref. 83). Briefly, sgRNA oligonucleotides were synthesized via microarray (CustomArray) and cloned into the ipUSEPR lentiviral sgRNA vector (a gift from Scott A. Armstrong, Dana-Farber Cancer Institute, Boston, MA) that coexpresses a puromycin-resistant gene [puroR] and a red fluorescent protein [tagRFP]. The CRISPR library was packaged by HEK293T cells (ATCC) cotransfected with psPAX2 (Addgene) and pMD2.G (Addgene) to produce lentiviral particles. The lentiviral library was pretitrated to obtain ∼20% multiplicity of infection (monitored by flow cytometry for tagRFP expression) in the MV4;11-Cas9+ cells. Each screen culture was calculated to maintain at least 1,000× of the number of constructs in each library. The infected cultures were selected using puromycin (1 μg/mL; InvivoGen, #ant-pr-1) and treated with DMSO or 1 μmol/L TDI-11055 up to 32 days. At each designated time point, cells from the cultures that covered at least 1,000× of the number of the constructs in the library were collected. For sequencing sgRNAs, the genomic DNA of the screened cells was PCR-amplified using primers DCF01 5′-CTTGTGGAAAGGACGAAACACCG-3′ and DCR03 5′-CCTAGGAACAGCGGTTTAAAAAAGC-3′ and subjected to single-end, 75-bp (SE75) high-throughput sequencing using a NextSeq 550 (Illumina). To quantify sgRNA reads in the library, the 20-nucleotide sequences that matched the sgRNA backbone structure (5′ prime CACCG and 3′ prime GTTT) were extracted from raw fastq reads. Extracted reads were then mapped to the sgRNA library sequences using Bowtie2 (84). The frequency for individual sgRNAs was calculated by the read counts of each sgRNA divided by the total read counts mapped to the library. The CRISPR score was defined by the log fold change of the frequency of individual sgRNAs between the initial (day 0) and final (day 32) screened samples using the edgeR R package (85).
Animal Experiments
All animal experiments related to leukemia cells engraftment study were approved by the Institutional Animal Care and Use Committee (IACUC) of the Center for Childhood Cancer Research, Children's Hospital of Philadelphia. All animal experiments related to PK/PD study were approved by the IACUC of Pharmaron following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care.
PK Analysis in Mice.
Six- to 8-week-old, female BALB/c mice were used for the PK analysis. Mice were treated with different dosages of TDI-11055 or SGC-iMLLT via oral gavage (30–200 mg/kg). The dose formulation used for oral dosing was 5% DMSO:95% of 10% w/v Kleptose HPB (KLEPTOSE, #346112), administered at a dose level of 10 mL/kg. Serial blood samples were collected up to 24 hours after the start of dosing via direct vein puncture of the dorsal metatarsal vein. At the end of the study, the mice were euthanized. Blood was centrifuged to yield plasma, and then plasma samples (10 μL) were extracted using protein precipitation with 200 μL of acetonitrile containing an analytical internal standard. An aliquot of the supernatant was analyzed by reverse-phase LC-MS/MS. Samples were assayed against calibration standards prepared in control plasma, and the assay was qualified through inclusion of quality control samples. PK parameters were obtained from the plasma concentration–time profiles using noncompartmental analysis with Phoenix (WinNonlin) PK software version 6.1.0 (Pharsight). Unbound concentration and AUC in plasma were calculated as total plasma concentration (or AUC) × unbound fraction in plasma.
Plasma Protein Binding.
Equilibrium dialysis methods were used to determine the unbound fraction in mouse plasma that was obtained frozen from BioIVT. Duplicate plasma samples containing 1 μmol/L of test compounds or PBS, pH 7.4, were placed in separate wells of a 96-well equilibrium dialysis plate (HTDialysis). The dialysis plate was placed in an incubator at 37°C with 5% CO2 at approximately 100 rpm for 6 hours. After incubation, samples of plasma and buffer were matrix matched, quenched with acetonitrile (containing analytical internal standards), and centrifuged. Samples of the supernatant were analyzed for the peak area ratio by UPLC-MS/MS. The unbound fraction was determined using standard equations.
Blood:Plasma Ratio.
Whole blood from mice was obtained from Pharmaron laboratories. Blood was collected using K2EDA as the anticoagulant. Solutions of the test compound were prepared in solvent and added to blood from each species, in duplicate, to give a final concentration of 5 μmol/L. All of the blood samples were incubated at 37°C for 60 minutes. After the incubation, aliquots from two of these blood samples were combined and centrifuged for 10 minutes at 10,000 × g (37°C) to provide the reference blood sample (REF BL). Simultaneously, plasma was prepared from initial blood samples by centrifugation, and an aliquot of plasma was transferred into new tubes to provide the plasma sample (PL). The reference blood and plasma samples were matrix matched to an equivalent volume, mixed, and quenched using acetonitrile (containing analytical internal standards). Following further mixing and centrifugation, an aliquot of each supernatant was analyzed for peak area ratio (I) in the reference blood or plasma samples by UPLC-MS/MS. The blood:plasma ratio was determined as IREF BL/IPL.
PKPD and Gene Expression Analysis of MV4;11 Cells.
Female Balb/c nude mice (6–8 weeks, Beijing Anikeeper Biotech. Ltd) were inoculated subcutaneously on the right flank with MV4;11 tumor cells (1 × 107) in 0.1 mL of 1:1 v/v of IMDM:Matrigel for tumor development. Compound treatment was initiated when the mean tumor volume reached about 300 to 400 mm3. Mice were randomly assigned to respective treatment groups such that the average starting tumor size and body weight were the same for each treatment group. Mice were dosed with vehicle (5% DMSO:95% of 10% w/v Kleptose HPB, pH 4.0) or compound at 100 or 200 mg/kg p.o., b.i.d. at 12 hours intervals. For PD readouts (tumor volume and gene analysis), five animals per group were used. Animals were checked daily and body weights measured every 2 days throughout the study. Tumor volume was estimated using the formula: TV = a × b2/2 throughout the study, where “a” and “b” are the long and short diameters of a tumor, respectively. Individual tumor samples for gene analysis were taken at 2 hours after dose 8 days of dosing, and the following genes were analyzed: MYC (Hs00153408_m1, Thermo Fisher), HOXA9 (Hs00365956_m1, Thermo Fisher), MEIS1 (Hs00180020_m1, Thermo Fisher), and MYB (see Supplementary Table S4).
In Vivo Efficacy Studies in Disseminated Aml Models.
To prepare TDI-11055 for in vivo treatment, the compound was resuspended in DMSO at 400 mg/mL and diluted with 10% hydroxypropyl β-cyclodextrin (HPB; KLEPTOSE, #346112). The solution was adjusted for pH to 4.0 with 2 M HCl, sonicated at 4°C for 2 minutes to totally dissolve TDI-11055, and further diluted 1:20 with 10% HPB to a final concentration of 20 mg/mL. A vehicle solution was prepared with the same protocol but without the addition of TDI-11055. Cells were transplanted by tail-vein injection into 6- to 8-week-old, female NSG or NSGS recipient mice (cell number: MV4;11, 2 million; OCI-AML3, 200 K; MLL-r PDX-2263, 10 K; MLL-r PDX-6315, 1 million; and NPM1-mutated PDX-3055, 4 million). For the MLL-r PDX-2263 xenotransplantation model, mice were treated with vehicle or 200 mg/kg TDI-11055 q.d. for 28 consecutive days, or 200 mg/kg b.i.d. with a 5 day on and 2 day off cycle for a total of 28 days via oral gavage starting 7 days after transplantation. For all other xenotransplantation models, mice were treated with vehicle or 200 mg/kg TDI-11055 once daily via oral gavage starting from indicated time points after transplantation (MV4;11, 21 days; OCI-AML3, 7 days; MLL-r PDX-6315, 8 days; and NPM1-mutated PDX-3055, 14 days) and for 28 consecutive days. Animals were monitored daily, and body weights were measured every 2 days throughout the treatment period. To assess leukemia progression, peripheral blood was extracted from mice via retro-orbital bleeding, stained for human CD45%, and subjected to flow cytometry analysis. Bone marrow, spleen, and peripheral blood cells were extracted from mice at the experimental endpoint, stained for human CD45%, and subjected to flow cytometry analysis. Kaplan–Meier survival curves were created using GraphPad Prism (v9) software.
RT-qPCR Analyses
Total RNA was isolated using the RNeasy kit (Qiagen, #74106) and reverse transcribed with the high-capacity cDNA reverse transcription kit (Thermo Fisher, #4368814) in accordance with the manufacturer's instructions. Quantitative PCR was performed using the SYBR Green PCR Master Mix (Fisher Scientific, #A25778) with the ViiA 7 Real-time PCR System (Thermo Fisher). qPCR primers used are provided in Supplementary Table S4.
Rna-seq with External RNA Controls Consortium Rna Spike-in Mix Normalization
The cell number was counted, and 0.5 million live cells were collected in each tube. One microliter of a 1:10 dilution of External RNA Controls Consortium (ERCC) spike-in mix (Invitrogen, #4456740) was added into each tube. Total RNA was extracted using the RNeasy plus mini kit (Qiagen, #74136). Total RNA (1,000 ng) was used for library preparation using the polyA mRNA magnetic isolation module (NEB, #E7490) and the RNA library prep kit (NEB, #7770) following the manufacturer's instructions. RNA samples were sequenced using Illumina NextSeq 500. Sequencing data were analyzed with spike-in as normalization. Raw reads were mapped to the human reference genome (hg19) and transcriptome using HISAT2 v 2.2.1 with default parameters. Unmapped reads from hg19 alignment were mapped to the spike-in ERCC control transcriptome using Kallisto v 0.46.2 (86) as spike-in control. Lowly expressed genes with raw counts less than 10 in any individual sample were filtered before downstream analysis. The size factor of Spike-In control was estimated using estimateSizeFactors from R Bioconductor DESeq2 v1.28.1 package (87). Then the size factor was assigned to the RNA-seq result using the assignment function sizeFactors. Differentially expressed genes between conditions were statistically determined using R Bioconductor DESeq2 v1.28.1 package (87). Gene-specific normalization factors for each sample were provided using pregenerated sizeFactors from spike-in control, which preempts sizeFactors in DESeq2 calculations. Genes with adjusted P < 0.05 and fold change ≥1.5 were reported as differential genes. Volcano plots were generated directly in R.
GSEA
GSEA was performed using GSEA v4.1.0 software with 1,000 gene set permutations. Gene sets used were manually curated from published datasets or our own dataset. A detailed description of GSEA methodology and interpretation can be found at http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html. Gene rank lists were generated by ordering the expression fold change (TDI-11055 vs. DMSO) from largest to smallest. The enrichment score (ES) reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes. The normalized enrichment score (NES) is the ES with normalization across analyzed gene sets. The false discovery rate q value (FDR q-val) is the estimated probability that a gene set with a given NES represents a false-positive finding. All gene sets used in this study are provided in Supplementary Table S7.
ChIP-Seq and ChIP-qPCR
In general, cells were collected, washed, and cross-linked with 1% formaldehyde in PBS for 10 minutes and stopped with 125 mmol/L glycine for 5 minutes. For FLAG-ENL, HA-ENL, and H3K79me2 ChIP, cells were first fixed at room temperature. For AFF4, DOT1L, PAF1, RNA Pol II, and Pol II S2P, cells were fixed at 37°C for 10 minutes. Fixed cells were resuspended and sonicated in RIPA 0.3 buffer [0.1% SDS, 1% Triton X-100, 10 mmol/L Tris-HCl (pH 7.4), 1 mmol/L EDTA (pH 8.0), 0.1% NaDOC, 0.3 M NaCl, 0.25% sarkosyl, 1 mmol/L DTT, and protease inhibitors] using a Covaris Ultrasonicator. Antibody (10 μg) was preincubated with 75 μL Protein A Dynabeads at 4°C for >4 hours. The beads were washed 3 times with PBS plus 0.01% Tween 20, and added to and incubated with the samples overnight at 4°C. The immunoprecipitates were washed twice with low-salt wash buffer [50 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, and 0.1% SDS], twice with high-salt wash buffer [50 mmol/L Tris (pH 8.0), 500 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, and 0.1% SDS], twice with LiCl wash buffer [50 mmol/L Tris (pH 8.0), 150 mmol/L LiCl, 1 mmol/L EDTA, 1% NP-40, 0.5% Na-Deoxycholate, and 0.1% SDS], and once with TE buffer (1 mmol/L EDTA and 10 mmol/L Tris-HCl (pH 8.0)] plus 50 mmol/L NaCl. Bound DNA was eluted using 200 μL ChIP elution buffer [1% SDS, 50 mmol/L Tris-HCl (pH 8.0), 10 mmol/L EDTA, and 200 mmol/L NaCl], reverse cross-linked, and purified using a PCR purification kit (Qiagen, #28106) in a final volume of 50 μL. ChIP DNA (5 μL) was diluted 50-fold using water for qRT-PCR using the ViiA 7 Real-time PCR System (Thermo Fisher) and the Power SYBR Green PCR Master Mix (Fisher Scientific, #A25778).
ChIP-seq sample libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit (NEB, #E7645L) following the manufacturer's instructions. Then samples were sequenced using Illumina NextSeq 500 or NextSeq 2000. Raw reads were aligned to the UCSC hg-19 genome sequence build from the Bsgenome.Hsapiens.UCSC.hg19 Bioconductor package using the Rsubread v 2.26 package's align function (88). Aligned BAM files were sorted using the function sortBam from Rsubread v 2.2.6. Secondary alignments, unmapped reads, and duplicate reads in the sorted BAM files were filtered out using sambamba v 0.7.1. The filtered and sorted BAM files were then indexed using the function indexBam from Rsubread v 2.2.6. The genome-wide ChIP-seq profiles were generated using MACS2 v 2.2.7.1 (89) with the following command (macs2 callpeak -t BAMFILE -c INPUTFORTHEBAMFILE -n NAMING -f BAM -g hs -p 1e-8 –nomodel –keep-dup all). Peaks were annotated to corresponding genes using function annotatePeaks.pl from HOMER v 4.10 (90). To make the heat maps, filtered, sorted, and indexed BAM files were converted to bigwig files using bamCoverage from deeptools v3.5.0 (91) and then normalized by counts per million. Heat maps were generated using computeMatrix from deeptools v 3.5.0 and plot heat map from deeptools v3.5.0. Metagene profiles of promoter and gene body regions from ChIP-seq data were generated using ngs.plot.r v2.41.4 (92).
To compare the chromatin occupancy of each protein under different treatment conditions, the count of ChIP-seq alignments was extracted for indicated genomic regions [TSS ± 3 kb for HA-ENL, AFF1, DOT1L, and H3K79me2; gene body (TSS + 300 bp to transcription end site, or TES) for Pol II S2P and PAF1; promoter-proximal region (TSS ± 300 bp) and gene body (TSS + 300 bp to TES) for total Pol II] using package bedtools multicov v2.29.2 (93). Total read counts for individual samples were obtained by samtools function view v1.9 (94). RPM values were obtained by normalizing read counts to total read counts. Box plots showing log2 fold changes (TDI-11055 vs. DMSO) in chromatin occupancy of indicated proteins at different subsets of genes were made in Prism V9. To define top, low, and non–ENL-bound genes, HA-ENL ChIP-seq data were used for ENL peak calling. The closest gene for each peak was defined as an ENL-bound gene. ENL-bound genes were then divided into top- and low-bound groups based on HA-ENL ChIP-seq signals at TSS ± 3 kb regions. Non-ENL–bound genes were defined by subtracting all ENL-bound genes from the genome (hg19, https://www.gencodegenes.org/human/release_36lift37.html).
Clinical Patient Data Analysis
Pearson correlation of TDI-downregulated gene set enrichment scores and LSC gene set enrichment scores was calculated by gene set variation analysis (v1.40.1 in R). Each dot represents one sample from the TCGA-LAML (44) or TARGET LAML (45) datasets.
Statistical Analyses
No statistical methods were used to predetermine the sample size. Experimental data are presented as mean ± SEM unless stated otherwise. Statistical significance was calculated by a two-tailed, unpaired t test on two experimental conditions, with P < 0.05 considered statistically significant unless stated otherwise. Statistical significance levels are denoted as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Data Availability
The ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus database under accession numbers GSE185093 and GSE185091, respectively. All other raw data generated or analyzed during this study are included in this article (and its Supplementary Information files). Codes used for data analysis are available upon request.
Supplementary Material
Acknowledgments
The authors thank members of the Wan laboratory for technical support and scientific input throughout the study; Ronen Marmorstein, M. Andres Blanco, and Krista Budinich for critical reading of the manuscript; Leigh Baxt, Qiao Liu, Jingqi Huang, Tan Pang, Yi Wang, and Shaofeng Zhang for technical assistance; Oleg Fedorov for sharing compounds; and the Stem Cell and Xenograft Core (SCXC) facility at the University of Pennsylvania for primary human AML cells. The chemical development of the compound was initiated when L.Wan was a postdoctoral fellow in the Allis laboratory at The Rockefeller University. The authors are grateful for the generous support from C. David Allis, The Rockefeller University, the Leukemia & Lymphoma Society, and the St. Jude Children's Research Hospital Collaborative Initiative on Chromatin Regulation in Pediatric Cancer during the initial stages of the work. The authors also gratefully acknowledge the support generously provided by the Tri-Institutional Therapeutics Discovery Institute (TDI), a 501(c)(3) organization. TDI receives financial support from Takeda Pharmaceutical Company, TDI's parent institutes (Memorial Sloan Kettering Cancer Center, The Rockefeller University, and Weill Cornell Medicine), and a generous contribution from Mr. Lewis Sanders, and other philanthropic sources. The research was supported by an NIH Director's New Innovator Award 1DP2HG012443 (to L. Wan), an NIH Pathway to Independence Award R00CA226399 (to L. Wan), a Pew-Stewart Scholar Award (to L. Wan), a V Foundation Scholar Award (to L. Wan), an American Society of Hematology Scholar Award (to L. Wan), a Leukemia Research Foundation grant (to L. Wan), SU2C and Cancer Research UK (#RT617 to C.-W. Chen), and the Biff Ruttenberg Foundation (to M. Carroll).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
M. Michino reports a patent for WO2021127166A1 issued. A.W. Stamford reports other support from the Tri-Institutional Therapeutics Discovery Institute, Bridge Medicines, and Deerfield Management outside the submitted work, as well as a patent for U.S. No. 62/949,160 pending. N. Liverton reports a patent for AU2020407589A1 pending and licensed to Bridge Medicines. L.M. Renzetti reports personal fees from Bridge Medicines during the conduct of the study; personal fees from Bridge Medicines outside the submitted work; a patent for inhibitors of ENL/AF9 YEATS pending; and is an employee of Bridge Medicines. Bridge Medicines provided financial support for the studies, and L.M. Renzetti provided intellectual input and commentary on their design and the interpretation of results. Bridge Medicines has submitted a patent application for inhibitors of ENL/AF9 YEATS. S. Kargman is an employee of the Tri-Institutonal Therapeutics Discovery Institute and Bridge Medicines. Bridge Medicines and the Tri-Institutional Therapeutics Discovery Institute funded a portion of this work and have patents pending. Bridge Medicines is a licensee of the Tri-Institutional Therapeutics Discovery Institute patent. Employees of the Tri-Institutional Therapeutics Discovery Institute may benefit from the further development and licensure of molecules described herein. P.T. Meinke reports a patent for AU2020407589A1 pending and licensed to Bridge Medicines. M.A. Foley reports other support from Takeda during the conduct of the study; other support from the Tri-Institutional Therapeutics Discovery Institute outside the submitted work; and is an employee of Deerfield Management. M. Carroll reports personal fees from Janssen Pharmaceuticals and Cartography Biosciences outside the submitted work. C.-W. Chen reports grants from the NIH/NCI, Stand Up To Cancer (SU2C), the Leukemia & Lymphoma Society, the American Society of Hematology, Alex's Lemonade Stand Foundation, and Circle of Service Foundation during the conduct of the study. I. Maillard reports grants from the National Institute of Allergy and Infectious Diseases during the conduct of the study, as well as grants from Regeneron, Genentech, and the Leukemia & Lymphoma Society and personal fees from Garuda Therapeutics outside the submitted work. D.J. Huggins reports a patent for WO 2021/127166 A1 issued and with royalties paid and a patent for U.S. 2013/0018073 A1 issued and with royalties paid. K.M. Bernt reports grants from Syndax outside the submitted work. L. Wan reports grants from the NIH, the V Foundation, The Pew Charitable Trusts and The Alexander and Margaret Stewart Trust, the American Society of Hematology, and the Leukemia Research Foundation during the conduct of the study; personal fees from Bridge Medicines and Panorama Medicine outside the submitted work; and a patent for U.S. No. 62/949,160 pending. No disclosures were reported by the other authors.
Authors’ Contributions
Y. Liu: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. Q. Li: Formal analysis, investigation, visualization, methodology, writing–review and editing. F. Alikarami: Data curation, investigation. D.R. Barrett: Data curation, investigation. L. Mahdavi: Data curation, investigation. H. Li: Data curation, formal analysis, investigation, visualization, methodology. S. Tang: Data curation, investigation, writing–review and editing. T.A. Khan: Investigation, visualization, methodology, writing–review and editing. M. Michino: Investigation, visualization, methodology, writing–review and editing. C. Hill: Data curation, investigation. L. Song: Data curation, investigation. L. Yang: Investigation. Y. Li: Data curation, investigation, methodology. S.P. Pokharel: Data curation, investigation. A.W. Stamford: Investigation. N. Liverton: Investigation, writing–review and editing. L.M. Renzetti: Resources, supervision. S. Taylor: Formal analysis, investigation, methodology, writing–original draft. G.F. Watt: Investigation. T. Ladduwahetty: Resources, investigation. S. Kargman: Resources, investigation, writing–review and editing. P.T. Meinke: Resources, supervision, writing–review and editing. M.A. Foley: Resources, supervision. J. Shi: Resources. H. Li: Investigation. M. Carroll: Resources. C.-W. Chen: Investigation, methodology. A. Gardini: Investigation. I. Maillard: Investigation, writing–review and editing. D.J. Huggins: Investigation, methodology, writing–original draft, writing–review and editing. K.M. Bernt: Supervision, investigation, methodology, writing–review and editing. L. Wan: Conceptualization, data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. Longo DL, Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 3. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wan L, Wen H, Li Y, Lyu J, Xi Y, Hoshii T, et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017;543:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo HS, et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 2017;543:270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hsu C-C, Shi J, Yuan C, Zhao D, Jiang S, Lyu J, et al. Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer. Gene Dev 2018;32:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao D, Li Y, Xiong X, Chen Z, Li H. YEATS domain—a histone acylation reader in health and disease. J Mol Biol 2017;429:1994–2002. [DOI] [PubMed] [Google Scholar]
- 8. Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 2014;159:558–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao D, Guan H, Zhao S, Mi W, Wen H, Li Y, et al. YEATS2 is a selective histone crotonylation reader. Cell Res 2016;26:629–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mi W, Guan H, Lyu J, Zhao D, Xi Y, Jiang S, et al. YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat Commun 2017;8:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol 2016;12:396–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsu C-C, Zhao D, Shi J, Peng D, Guan H, Li Y, et al. Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity. Cell Discov 2018;4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol Cell 2016;62:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem Cell Biol 2009;87:65–75. [DOI] [PubMed] [Google Scholar]
- 15. Winters AC, Bernt KM. MLL-rearranged leukemias—an update on science and clinical approaches. Front Pediatr 2017;5:691–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 2007;7:823–33. [DOI] [PubMed] [Google Scholar]
- 17. Gadd S, Huff V, Walz AL, Ooms AHAG, Armstrong AE, Gerhard DS, et al. A Children's Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet 2017;49:1487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gadd S, Arold ST, Radhakrishnan A, Gerhard DS, Jennings L, Huff V, et al. MLLT1 YEATS domain mutations in clinically distinctive favourable histology Wilms tumours. Nat Commun 2015;6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 2019;577:121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146:904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011;478:529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung C, et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010;468:1119–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moustakim M, Christott T, Monteiro OP, Bennett J, Giroud C, Ward J, et al. Discovery of an MLLT1/3 YEATS domain chemical probe. Angew Chem Int Ed 2018;57:16302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christott T, Bennett J, Coxon C, Monteiro O, Giroud C, Beke V, et al. Discovery of a selective inhibitor for the YEATS domains of ENL/AF9. Slas Discov Adv Sci Drug Discov 2019;24:133–41. [DOI] [PubMed] [Google Scholar]
- 27. Asiaban JN, Milosevich N, Chen E, Bishop TR, Wang J, Zhang Y, et al. Cell-based ligand discovery for the ENL YEATS domain. Acs Chem Biol 2020;15:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Y, Chen G, Li X-M, Liu S, Tian G, Li Y, et al. Selective targeting of AF9 YEATS domain by cyclopeptide inhibitors with preorganized conformation. J Am Chem Soc 2020;142:21450–9. [DOI] [PubMed] [Google Scholar]
- 29. Li X, Li X-M, Jiang Y, Liu Z, Cui Y, Fung KY, et al. Structure-guided development of YEATS domain inhibitors by targeting π-π-π stacking. Nat Chem Biol 2018;14:1140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma XR, Xu L, Xu S, Klein BJ, Wang H, Das S, et al. Discovery of selective small-molecule inhibitors for the ENL YEATS domain. J Med Chem 2021;64:10997–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garnar-Wortzel L, Bishop TR, Kitamura S, Milosevich N, Asiaban JN, Zhang X, et al. Chemical inhibition of ENL/AF9 YEATS domains in acute leukemia. ACS Central Sci 2021;7:815–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundbäck T, Nordlund P, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 2014;9:2100–22. [DOI] [PubMed] [Google Scholar]
- 33. Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008;358:1909–18. [DOI] [PubMed] [Google Scholar]
- 34. Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood 2020;136:1707–21. [DOI] [PubMed] [Google Scholar]
- 35. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 2005;352:254–66. [DOI] [PubMed] [Google Scholar]
- 36. Lin A, Giuliano CJ, Palladino A, John KM, Abramowicz C, Yuan ML, et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci Transl Med 2019;11:eaaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neggers JE, Kwanten B, Dierckx T, Noguchi H, Voet A, Bral L, et al. Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes. Nat Commun 2018;2377:47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ipsaro JJ, Shen C, Arai E, Xu Y, Kinney JB, Joshua-Tor L, et al. Rapid generation of drug-resistance alleles at endogenous loci using CRISPR-Cas9 indel mutagenesis. PLoS One 2017;12:e0172177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Donovan KF, Hegde M, Sullender M, Vaimberg EW, Johannessen CM, Root DE, et al. Creation of novel protein variants with CRISPR/Cas9-mediated mutagenesis: turning a screening by-product into a discovery tool. PLoS One 2017;12:e0170445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vinyard ME, Su C, Siegenfeld AP, Waterbury AL, Freedy AM, Gosavi PM, et al. CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML. Nat Chem Biol 2019;15:529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. P Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Somervaille TCP, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell 2009;4:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cancer Genome Atlas Research Network; Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bolouri H, Farrar JE, Triche T, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018;24:103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 2010;37:429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, et al. Human polymerase-associated factor complex (PAFc) connects the super elongation complex (SEC) to RNA polymerase II on chromatin. Proc Nat Acad Sci U S A 2011;108:E636–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 2010;17:198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mueller D, Bach C, Zeisig D, Garcia-Cuellar M-P, Monroe S, Sreekumar A, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 2007;110:4445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KSJ, Allis CD, et al. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Nat Acad Sci U S A 2011;108:15751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 2006;23:297–305. [DOI] [PubMed] [Google Scholar]
- 52. Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, et al. RNA polymerase is poised for activation across the genome. Nat Genet 2007;39:1507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 2013;122:1017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent Small-molecule DOT1L inhibitor. Cancer Cell 2011;20:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kühn MWM, Song E, Feng Z, Sinha A, Chen CW, Deshpande AJ, et al. Targeting chromatin regulators inhibits leukemogenic gene expression in NPM1 mutant leukemia. Cancer Discov 2016;6:1166–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cao K, Ugarenko M, Ozark PA, Wang J, Marshall SA, Rendleman EJ, et al. DOT1L-controlled cell-fate determination and transcription elongation are independent of H3K79 methylation. Proc National Acad Sci U S A 2020;117:27365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miao H, Kim E, Chen D, Purohit T, Kempinska K, Ropa J, et al. Combinatorial treatment with menin and FLT3 inhibitors induces complete remission in AML models with activating FLT3 mutations. Blood 2020;136:2958–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Klossowski S, Miao H, Kempinska K, Wu T, Purohit T, Kim E, et al. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J Clin Invest 2019;130:981–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. O'Connell KE, Mikkola AM, Stepanek AM, Vernet A, Hall CD, Sun CC, et al. Practical murine hematopathology: a comparative review and implications for research. Comparative Med 2015;65:96–113. [PMC free article] [PubMed] [Google Scholar]
- 60. Santos EW, de Oliveira DC, Hastreiter A, da Silva GB, de Oliveira Beltran JS, Tsujita M, et al. Hematological and biochemical reference values for C57BL/6, Swiss Webster and BALB/c mice. Braz J Vet Res Animal Sci 2015;53:138–45. [Google Scholar]
- 61. Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell 2013;13:102–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pronk CJH, Rossi DJ, Månsson R, Attema JL, Norddahl GL, Chan CKF, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 2007;1:428–42. [DOI] [PubMed] [Google Scholar]
- 63. Cheng H, Zheng Z, Cheng T. New paradigms on hematopoietic stem cell differentiation. Protein Cell 2020;11:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev Immunol 2005;5:497–508. [DOI] [PubMed] [Google Scholar]
- 65. Calvanese V, Nguyen AT, Bolan TJ, Vavilina A, Su T, Lee LK, et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature 2019;576:281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Arrowsmith CH, Schapira M. Targeting non-bromodomain chromatin readers. Nat Struct Mol Biol 2019;26:863–9. [DOI] [PubMed] [Google Scholar]
- 67. Dawson MA, Kouzarides T. Cancer Epigenetics: from mechanism to therapy. Cell 2012;150:12–27. [DOI] [PubMed] [Google Scholar]
- 68. Wood K, Tellier M, Murphy S. DOT1L and H3K79 methylation in transcription and genomic stability. Biomol 2018;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 2010;140:491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun 2018;9:4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Krivtsov AV, Evans K, Gadrey JY, Eschle BK, Hatton C, Uckelmann HJ, et al. A Menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell 2019;36:660–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol 2012;8:277–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 2015;27:589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 2011;20:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yokoyama A, Somervaille TCP, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The Menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 2005;123:207–18. [DOI] [PubMed] [Google Scholar]
- 77. Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 2008;14:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang L, Wu Y, Deng Y, Kim B, Pierce L, Krilov G, et al. Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J Am Chem Soc 2015;137:2695–703. [DOI] [PubMed] [Google Scholar]
- 79. Nakashima S, Yamamoto K, Arai Y, Ikeda Y. Impact of physicochemical profiling for rational approach on drug discovery. Chem Pharm Bulletin 2013;61:1228–38. [DOI] [PubMed] [Google Scholar]
- 80. Tarumoto Y, Lu B, Somerville TDD, Huang YH, Milazzo JP, Wu XS, et al. LKB1, salt-inducible kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol Cell 2018;69:1017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Grevet JD, Lan X, Hamagami N, Edwards CR, Sankaranarayanan L, Ji X, et al. Domain-focused CRISPR screen identifies HRI as a fetal hemoglobin regulator in human erythroid cells. Science 2018;361:285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 87. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 2019;47:gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 2014;42:W187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shen L, Shao N, Liu X, Nestler E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 2014;15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus database under accession numbers GSE185093 and GSE185091, respectively. All other raw data generated or analyzed during this study are included in this article (and its Supplementary Information files). Codes used for data analysis are available upon request.



![Figure 1. A potent, selective, and orally bioavailable inhibitor of the ENL/AF9 YEATS domains. A, Chemical structure for TDI-11055. B, Left, schematic depiction of the TR-FRET assay used to quantify the ability of a compound to disrupt YEATS domain and acylated histone H3 peptide interaction. Right, dose-dependent inhibition of the TR-FRET signal by TDI-11055. The IC50 shown represents the mean of four independent experiments. C, ITC experiment demonstrating direct binding of TDI-11055 to ENL YEATS with 1:1 stoichiometry. One of two independent replicates is shown. D, Top, immunoblots showing endogenous levels of ENL, GAS41, and YEATS2 protein after heat treatment in MOLM-13 leukemia cells at increasing temperatures. Bottom, quantification of immunoblot signals. One of two independent experiments is shown.E, Docking studies of TDI-11055 bound to ENL YEATS from cocrystal structures of SGC-iMLLT [Protein Data Bank (PDB): 6HT1] and H3K27ac (PDB: 5J9S). Ribbon representations indicate the protein, and stick representations show key residues involved in ENL and TDI-11055 interactions. TDI-11055 is depicted as sticks with colors corresponding to the atom type (green, carbon; blue, nitrogen; red, oxygen). F, Left, immunoblots showing levels of ectopically expressed FLAG-tagged wild-type (WT) or Y78A-mutant ENL proteins in HEK293 cells after heat treatment at increasing temperatures. Right, quantification of immunoblot signals. One of two independent experiments is shown. G, Rank-ordered heat map of FLAG-ENL ChIP-seq signals at ENL-bound peaks in MOLM-13 cells treated with DMSO or TDI-11055 (5 μmol/L) for 24 hours. H3K27ac and H3K9ac (GSE80779) ChIP-seq signals at ENL-bound peaks in untreated cells are also shown to demonstrate ENL localization to acetylated chromatin. See Supplementary Table S1. H, The genome browser view of indicated ChIP-seq signals at key ENL-bound genes (MYC, HOXA9/10) in parental [H3K27ac (gray), H3K9ac (black)] as well as DMSO (blue)- or TDI-11055 (red)–treated (FLAG-ENL) MOLM-13 cells. I, PK studies in mice (n = 3) performed for TDI-11055 and SGC-iMLLT, demonstrating unbound plasma concentration of indicated compounds after an oral dose of 30, 50, and 100 mg/kg.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b374/9627135/19daef3cb0e8/2684fig1.jpg)
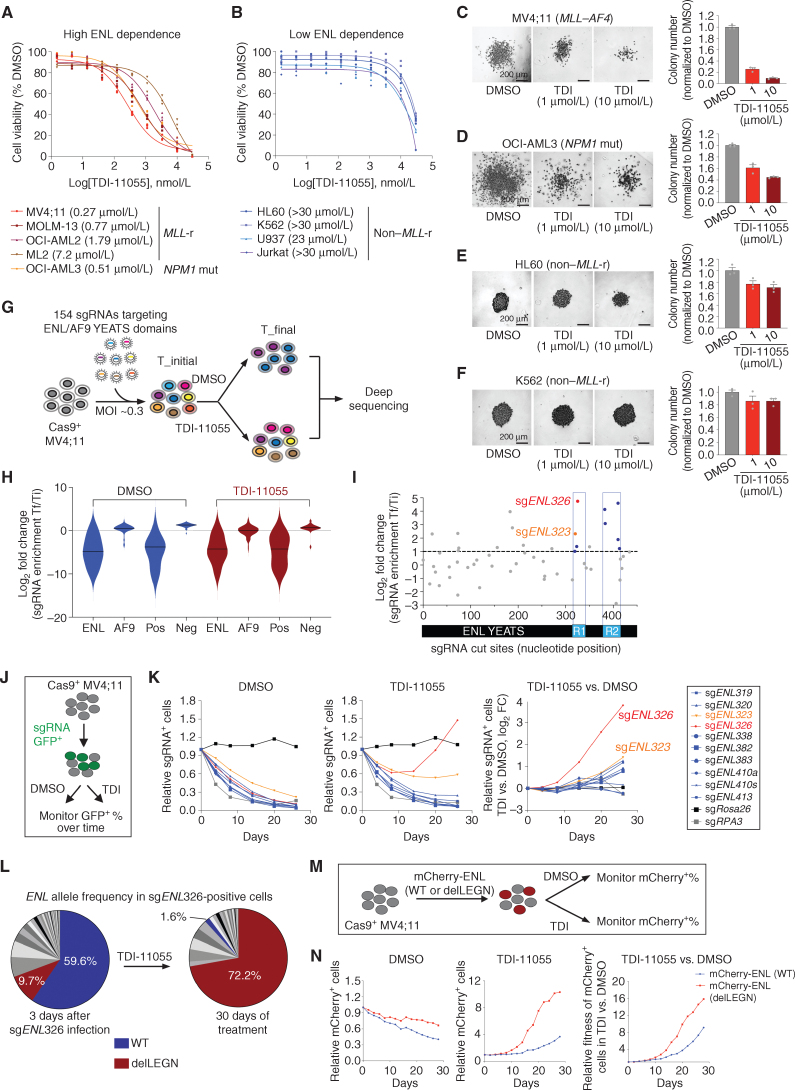
![Figure 3. Inhibition of ENL recruitment to chromatin suppresses key oncogenic gene expression programs in MLL-r and NPM1-mutated AML. A, Volcano plot of RNA-seq data [with External RNA Controls Consortium (ERCC) RNA spike-in] obtained from MV4;11 cells treated with DMSO or 5 μmol/L TDI-11055 (TDI) for 24 hours. Key differentially expressed genes are shown in red. Data represent mean across two replicates. See Supplementary Table S6. B, GSEA plots evaluating gene expression changes in MV4;11 cells treated with TDI-11055 (5 μmol/L for 24 hours), with genes downregulated (DN; left) or upregulated (UP; right) upon dTAG-mediated degradation of ENL. FDR, false discovery rate; NES, normalized enrichment score. See Supplementary Table S7. C, Unbiased GSEA using all signatures from MSigDB v7.2 C2 together with ENL genetic signatures for RNA-seq data presented in A. Each gene set is represented as a single dot. ENL signatures are indicated in red with numeral rank from the topmost enriched gene set. D, RT-qPCR analysis showing mRNA expression levels (normalized to B2M) of selected genes in MV4;11 cells upon treatment with different concentrations of TDI-11055 (0.01, 0.1, 1 μmol/L) for 72 hours. Error bars represent mean ± SEM (n = 3). E, GSEA plots evaluating gene expression changes in MV4;11 cells treated with 5 μmol/L TDI-11055 for 24 hours with genes associated with MYC target genes (left), LSCs (center), and monocyte differentiation (right). DMAP, differentiation map. F, Volcano plot of RNA-seq data (with ERCC RNA spike-in) obtained from OCI-AML3 cells treated with DMSO or 1 μmol/L TDI-11055 for 24 hours. Key differentially expressed genes are in red. Data represent mean across two replicates. See Supplementary Table S8. G, GSEA plots evaluating gene expression changes in OCI-AML3 cells treated with 1 μmol/L TDI-11055 for 24 hours with genes associated with MYC target genes (left), LSCs (center), and monocyte differentiation (right). NPM1 mut, NPM1-mutated. H and I, Pearson correlation of downregulated genes that were induced by TDI-11055 in either MV4;11 (H) or OCI-AML3 (I) cells and leukemia stem cell genes in the TCGA and TARGET datasets. Each dot represents one sample from the TCGA and TARGET datasets. r, Pearson correlation coefficient. P values by the two-tailed Pearson correlation test.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b374/9627135/77e73bdb3cac/2684fig3.jpg)
![Figure 4. TDI-11055 treatment induces rapid changes in transcription elongation at top ENL-bound genes. A, Rank-ordered heat map of HA-ENL ChIP-seq signals at ENL-bound peaks in MV4;11 cells treated with DMSO or TDI-11055 (5 μmol/L) for 0.5 hours. See Supplementary Table S9. RPM, reads per million. B, HA-ENL–bound genes in MV4;11 cells. HA-ENL ChIP-seq signals at the TSS ± 3 kb region of individual ENL-bound genes were plotted against their rank among all ENL-bound genes. Red depicts top ENL-bound genes (n = 165), and blue depicts low ENL-bound genes (n = 1,367). See Supplementary Table S10. C, The genome browser view of HA-ENL, AFF1, DOT1L, and Pol II S2P ChIP-seq signals at select ENL target genes under DMSO and TDI-11055 (TDI) treatment conditions in MV4;11 cells. D and E, Average occupancies (D) and quantification (E) of AFF1 on top, low, and non–ENL-bound genes along the transcription unit in DMSO and TDI-11055 (5 μmol/L for 0.5 hours) treatment conditions. See Supplementary Table S14. FC, fold change. F and G, Average occupancies (F) and quantification (G) of DOT1L on top, low, and non–ENL-bound genes along the transcription unit in DMSO and TDI-11055 (5 μmol/L for 0.5 hours) treatment conditions. See Supplementary Table S15. H–J, Average occupancies (H) and quantification (I and J) of Pol II S2P on top, low, and non–ENL-bound genes along the transcription unit in DMSO and TDI-11055 [5 μmol/L for 2 hours (I) or 24 hours (J)] treatment conditions. TES, transcription end site. In E, G, I, J, black solid lines denote median, and black dash lines denote quartiles. P values by the Welch two-tailed t test. See Supplementary Tables S16 and S17.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b374/9627135/d16c30a7fa82/2684fig4.jpg)
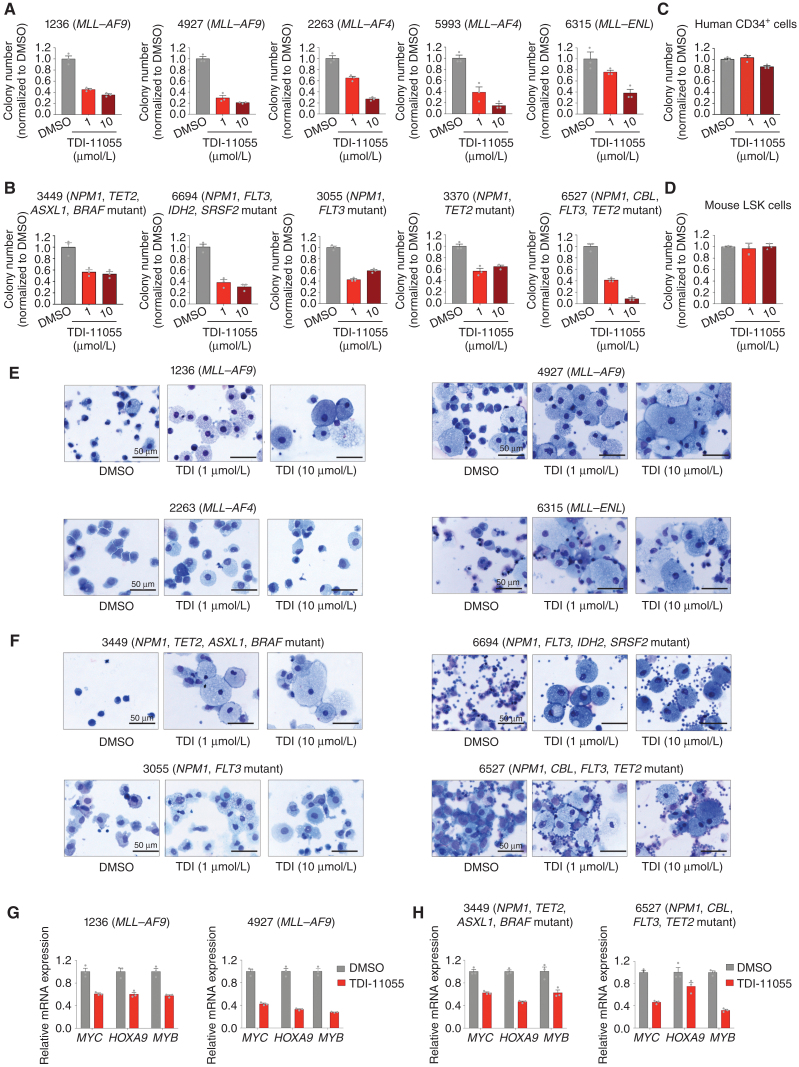
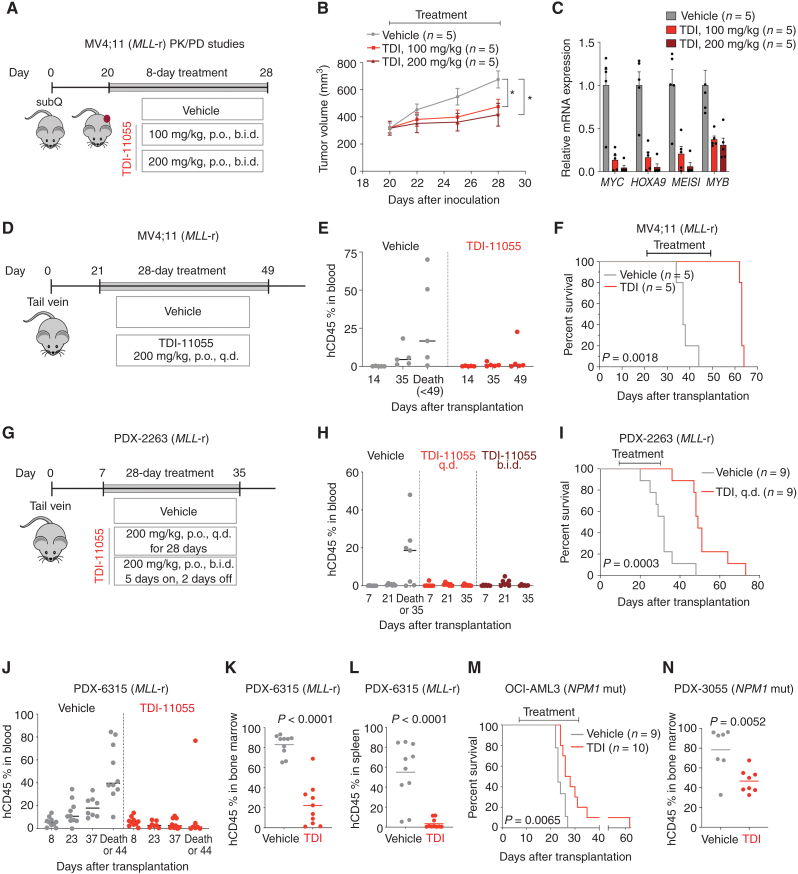
![Figure 7. The impact of TDI-11055 on normal hematopoiesis. A, Schematic of TDI-11055 treatment workflow in normal C57BL/6 mice. B, Body weight variance over time for vehicle- or TDI-11055 (TDI)–treated mice. Error bars represent mean ± SEM (n = 6). C, Representative images (left) and weight quantification (right) of spleen harvested from mice at the end of 28-day treatment with vehicle or TDI-11055. Error bars represent mean ± SEM (n = 6). n.s., not signficant. D, Complete blood count analyses [white blood cell (WBC), red blood cell (RBC), hemoglobin (HGB), hematocrit (HCT) and platelets] of peripheral blood samples harvested from mice at the end of 28-day treatment with vehicle or TDI-11055. Bars represent the median (n = 6). E–G, Number of total cells (E), immature cells [LSK, long-term hematopoietic stem cells (LT-HSC), Lin−c-Kit+Sca-1− (LKS−), multipotent progenitor (MPP), and granulocyte–macrophage progenitor (GMP); F], and differentiated cells (Mac1+Gr1+ myeloid, CD4+ T, CD8+ T, B220+CD19+CD93− B, and B220+CD19+CD93+ B; G) in bone marrow samples harvested from mice at the end of 28-day treatment with vehicle or TDI-11055. Bars represent the median (n = 6). BM, bone marrow. H, Complete blood count analyses (WBC, RBC, HGB, HCT, platelets) of peripheral blood samples harvested from mice 80 days after completing treatment with vehicle or TDI-11055. Bars represent the median (n = 3). I–K, Number of total cells (I), immature cells (LSK, LT-HSC, MPP, LKS−, and GMP; J), and differentiated cells (Mac1+Gr1+ myeloid, CD4+T, CD8+ T, B220+CD19+CD93− B, and B220+CD19+CD93+ B; K) in bone marrow samples harvested from mice 80 days after completing treatment with vehicle or TDI-11055. Bars represent the median (n = 3). C–K, P values by an unpaired, two-tailed Student t test.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b374/9627135/81ce346ef8cd/2684fig7.jpg)