Abstract
Background:
Low-dose CT (LDCT) screening trials have shown that lung cancer early detection saves lives. However, a better stratification of the screening population is still needed. In this respect, we generated and prospectively validated a plasma miRNA signature classifier (MSC) able to categorize screening participants according to lung cancer risk. Here, we aimed to deeply characterize the peripheral immune profile and develop a diagnostic immune signature classifier to further implement blood testing in lung cancer screening.
Methods:
Peripheral blood mononuclear cell (PBMC) samples collected from 20 patients with LDCT-detected lung cancer and 20 matched cancer-free screening volunteers were analyzed by flow cytometry using multiplex panels characterizing both lymphoid and myeloid immune subsets. Data were validated in PBMC from 40 patients with lung cancer and 40 matched controls and in a lung cancer specificity set including 27 subjects with suspicious lung nodules. A qPCR-based gene expression signature was generated resembling selected immune subsets.
Results:
Monocytic myeloid-derived suppressor cell (MDSC), polymorphonuclear MDSC, intermediate monocytes and CD8+PD-1+ T cells distinguished patients with lung cancer from controls with AUCs values of 0.94/0.72/0.88 in the training, validation, and lung cancer specificity set, respectively. AUCs raised up to 1.00/0.84/0.92 in subgroup analysis considering only MSC-negative subjects. A 14-immune genes expression signature distinguished patients from controls with AUC values of 0.76 in the validation set and 0.83 in MSC-negative subjects.
Conclusions:
An immune-based classifier can enhance the accuracy of blood testing, thus supporting the contribution of systemic immunity to lung carcinogenesis.
Impact:
Implementing LDCT screening trials with minimally invasive blood tests could help reduce unnecessary procedures and optimize cost-effectiveness.
Introduction
Lung cancer represents one of the major burdens for patients and for the healthcare system with an estimated mortality rate of 22%, considering all cancer-related deaths (1). In the last decade, large randomized trials have reported that clear benefits in terms of both tumor specific and overall mortality can be achieved by promoting extensive lung cancer screening programs based on low-dose CT (LDCT; refs. 2–4). On the other hand, limitations of LDCT screening remain the overdiagnosis, the high number of false positive cases and the management of suspicious nodules (5). In this scenario, the identification of noninvasive biomarkers that could be employed either alone or in combination with LDCT for the early diagnosis of lung cancer is of extreme interest.
A continuous evolution of antitumor immunity, starting from preneoplasia to invasive non–small cell lung cancer (NSCLC), has evidenced a gradual loss of immune activating pathways with concomitant increase of immune suppressive pathways, leading to immune escape (6). Research efforts directed to the identification of new biomarkers based on the profiles of patient's immunity recently achieved remarkable findings. For instance, neutrophils are the most relevant immune cell type infiltrating lung cancer tissues and the presence of tumor-infiltrating lymphocytes in the lung tumor microenvironment (TME) was associated with better survival (6–8). Conversely, high numbers of T-regulatory cells (Treg) or pro-tumorigenic macrophages in the TME were correlated with worse survival (9, 10).
Concerning peripheral blood, higher levels of neutrophils-related molecules have been found in patients with early-stage NSCLC (11), while the absolute count of lymphocytes subpopulation or natural killer (NK) cells was associated with a better prognosis (12). It has also been demonstrated that the immunoprofiles and/or changes in the repertoires of the T-cell receptor of circulating T cells are able to predict response to anti–PD-1 and anti-CTLA4 therapies (13). Changes in the gene expression profiles of peripheral blood mononuclear cells (PBMC) were already described for early-stage pancreatic and renal cell carcinoma and could be potentially used as diagnostic cancer biomarkers (14, 15). Similarly, a PBMC and a whole blood gene expression classifier were described as able to distinguish nonmalignant from malignant lung nodules (16, 17).
In lung cancer screening settings, we developed and ultimately validated a plasma microRNA signature classifier (MSC) able to improve performance of LDCT alone (18–20). The results of the prospective BioMILD lung cancer screening trial on 4,119 high-risk individuals have shown that the MSC test at baseline defines individual lung cancer risk profiles independently from LDCT result and reduces unnecessary LDCT repeat (20). In details, MSC showed a major added value in LDCT+ participants, where it resulted in a positive predictive value (PPV) and a negative predictive value (NPV) of 18% and 94% in discriminating cancerous and noncancerous lung nodules (20). In the current study, we aimed to test whether differential frequencies of specific immune cell subsets in the peripheral blood of subjects enrolled in the BioMILD screening trial could further implement the performance of the MSC test.
By using multiplex flow cytometry–based approach, we first assessed whether the frequency of specific immune cell subsets detectable in the peripheral blood of subjects enrolled in the BioMILD screening trial could contribute to discriminate patients with screening-detected lung cancer from disease-free smokers. A set of immune-related genes was then selected on the basis of flow cytometry results to develop an easy-to-use and potentially clinical-grade test to improve early detection of patients with lung cancer and the accuracy of the MSC algorithm.
Materials and Methods
Patients’ selection and samples’ collection
Samples composing the training and validation sets were collected from heavy smokers enrolled in the BioMILD lung cancer screening trial ongoing in our institution. The training and validation sets consisted of 20 and 40 PBMC samples collected at the time of tumor pathological diagnosis and matched 1:1 for gender, age, pack-year, and MSC result with screening participants who did not develop lung cancer in the following 5 years. For the specificity set, 27 additional PBMC samples were selected from subject with positive CT findings identified in our institution. Of these, 19 had a diagnosis of lung cancer, 6 of benign disease, and 2 were diagnosed as primary pulmonary lymphomas (Supplementary Fig. S1). Peripheral blood samples (20 mL) were collected in K2EDTA tubes and centrifuged to collect the plasma as previously described (21). Viable PBMC were separated by density gradient medium centrifugation within 2 hours of blood collection, using Histopaque-1077 (Sigma Chemicals) and SepMate tubes (STEMCELL Technologies), following the manufacturer's instructions. Isolated PBMC were frozen in RPMI1640 (Lonza) containing 10% dimethylsulfoxide (DMSO, Sigma) and 30% FCS (Euroclone) and stored in liquid nitrogen to be then simultaneously tested by multicolor flow cytometry. The MSC test was prospectively performed on plasma samples as by standard protocol of the BioMILD screening trial (21).
Flow cytometry analysis
Multiplex flow cytometry was applied to samples of the training set composed by 20 patients with LDCT-detected lung cancer and 20 matched controls. Results were confirmed in a validation set composed by 40 patients and 40 matched controls and a third specificity set including 27 subjects with clinically detected LDCT suspicious nodules. Thawed PBMC were incubated with live/dead (Thermo Fisher Scientific) staining for 30 minutes on ice and washed, treated with Fc blocking reagent (Miltenyi Biotec; 10 minutes at room temperature), before incubating with the different mAbs for 30 minutes at 4°C. Thereafter, samples were washed, fixed, and acquired. The monoclonal fluorochrome-conjugated antibodies applied are listed in Supplementary Table S1 and allowed the analysis of immune subsets defined in Supplementary Table S2.
Samples were acquired by Cytoflex flow cytometer and data were analyzed with Kaluza software (all Beckman Coulter). Gates were set on the basis of internal references. The distinct cell subsets were quantified in terms of frequency within PBMC and parent populations. For quality control (QC) purpose, PBMC of the same cohort (training, validation, and specificity sets) were evaluated simultaneously, in separate experimental sessions, with samples randomized within the same session. Every experiment included PBMC from one or two healthy donors, stained with all single mAbs plus the mix, to set flow cytometer compensation. Daily QC included the use of Flow-Check Pro (Beckman, A63493) and CytoFLEX Daily QC Fluorospheres (Beckman, B53230), fluorescent microspheres for optical alignment and fluidics system verification. All used Abs were titrated to reach the optimal concentration to use in the antibody panel mixes. Single mAb lots were used within the same experimental session. To detect polymorphonuclear myeloid-derived suppressor cell (PMN-MDSC) we applied our verified procedure of reliable quantification in thawed PBMC, using the ‘doublet exclusion gate’, without the live/dead exclusion (22).
RT-qPCR
RNA was extracted from PBMC samples following the Maxwell RSC RNA Tissue Kit (Promega) protocol, eluted in 50 μL of buffer and stored at –80°C. Starting from 500 ng of eluted RNA, RT‐PCR was performed using TaqMan Universal Master Mix II (Thermo Fisher Scientific) and 96-well plates according to the manufacturer's instructions using QuantStudio Real-Time PCR System (Thermo Fisher Scientific). Details of the assays adopted are reported in Supplementary Table S3.
Raw data were extrapolated using automated background subtraction and Ct threshold using the QuantStudio Real-Time PCR Software Version 1.1 (Thermo Fisher Scientific). Ct values were normalized according to the –ΔΔCt method: HPRT was adopted as housekeeping gene and the minor expressor as calibrator. Data were further standardized to the unit variance for computational analysis.
For the analysis of plasma microRNAs, the MSC test was performed as previously described (21).
Computational and statistical analysis
All data were transformed to the respective log2 value to have better-behaved data for statistical and computational analysis. Flow cytometry data were obtained using two different lots of reagents: one for the training and specificity set and one for the validation set. To correct for the batch effect, flow cytometry data of the training and validation sets were standardized using the training set as reference. Indeed, given the high similarity of the two sets in terms of patients and controls characteristics (Table 1), data of the validation set were scaled to the mean and SD of the training set. Unsupervised clustering analyses were performed centering and scaling genes and using one minus correlation and average linkage. The differences between groups were assessed using the Student t test for continuous variables and χ2, or Fisher exact test (properly selected) for contingency tables. Pearson R and relative P value was adopted to correlate continuous data with a normal distribution. All tests were two-sided and a P value < 0.05 was taken as statistically significant.
Table 1.
Clinico-pathological characteristics of patients (pts) with lung cancer and controls in the training, validation and lung cancer specificity set. Chi-square or Fisher's exact test P values are properly reported.
| Training Set | Validation Set | Lung cancer Specificity Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 20 | 20 | 40 | 40 | 19 | 8 | |||||
| Lung cancer pts | Controls | P | Lung cancer pts | Controls | P | Lung cancer pts | non–Lung cancer pts | P | ||
| Gender | ||||||||||
| Female | 10 | 10 | 1.0000 | 15 | 15 | 1.0000 | 8 | 4 | 1.0000 | |
| Male | 10 | 10 | 25 | 25 | 11 | 4 | ||||
| Age | ||||||||||
| ≤60 | 9 | 8 | 0.7491 | 16 | 18 | 0.8213 | 4 | 3 | 0.6334 | |
| >60 | 11 | 12 | 24 | 22 | 15 | 5 | ||||
| Pack-year | ||||||||||
| <44 | 8 | 11 | 0.3422 | 16 | 21 | 0.2622 | 14 | 5 | 0.6578 | |
| ≥44 | 12 | 9 | 24 | 19 | 5 | 3 | ||||
| MSC | ||||||||||
| Positive | 14 | 14 | 1 | 25 | 25 | 1 | 9 | 3 | 0.6957 | |
| Negative | 6 | 6 | 15 | 15 | 10 | 5 | ||||
| Histology | ||||||||||
| ADC | 13 | / | 29 | / | 16 | / | ||||
| Other | 7 | / | 11 | / | 3 | / | ||||
| Stage | ||||||||||
| I | 14 | / | 25 | / | 9 | / | ||||
| II-IV | 6 | / | 15 | / | 10 | / | ||||
To identify immune cell subsets discriminating patients vs. controls in the training set, class comparison analyses were performed and features with P value < 0.05 and fold change > 1.75 were selected. The compound covariate predictor method was adopted to define the linear classifier in the training set. By recursive feature elimination, the minimum amount of immune subsets able to discriminate patients and controls with an AUC > 0.9 was established to define the flow cytometry–based immune signature classifier (ISC).
The area under the receiver operating characteristic (ROC) curve method was adopted to estimate the performance of the developed tests in the training, validation, and specificity sets. The performance of the ISC diagnostic test in terms of sensitivity (Se), specificity (Sp), NPV, and PPV, was evaluated by setting the threshold for positivity as the maximum Youden's index (J) value in the whole cohort, where J = sensitivity+specificity-1. For the simulation of the ISC application in the BioMILD lung cancer screening trial, we considered the performance of ISC in the validation and lung cancer specificity set combined.
For the RT-qPCR based ISC, an algorithm using normalized expression data of genes representative of specific immune subpopulations was defined as follows:
To maintain interdependence with flow cytometry data, a weight (Wi) corresponding to the difference between the mean values in the 3 higher expressors and in the 3 lowest expressors of the calibration set was given to each normalized gene expression value (Xi).
A first immune score for the single-cell subsets was calculated as the Wi Xi mean value of genes representative of each immune subset.
To define a scoring system where higher values correspond to patients, the final ISC was given by the sum of the specific immune scores considering the inverse values for the immune subsets lower in patients than in controls.
Analyses were performed using BRB-ArrayTools v4.6.1 developed by Dr. Richard Simon and Amy Peng Lam and R software version 3.5.1. Figures were obtained using Microsoft Office 2007 package and GraphPad Prism version 5.02 statistical software.
Analysis of public datasets
Two public datasets (GSE13255 and GSE108375) were downloaded from the public Gene Expression Omnibus (GEO) database on February 14, 2022 (16, 17). To select a population as similar as possible to that of the volunteers of the screening programs the following filters were applied. In the GSE13255 dataset, including patients with NSCLC and non-healthy controls, only current smokers older than 40 were filtered in. In the GSE108375 dataset, composed by patients with malignant or benign nodules, current smokers with a pack-years >20 and older than 50 were filtered in. To apply the ISC algorithm, normalized data were first calibrated on the minor expressor and standardized to the unit variance. The ISC algorithm was then applied as described above.
Data availability
The data generated in this study are available within the article and its supplementary data files. Expression profile data analyzed in this study were obtained from GEO at GSE108375 and GSE13255.
Results
Patients’ characteristics
To test our hypothesis we selected PBMC samples from the BioMILD lung cancer screening trial to compose the following cohorts: the training set comprised 20 patients with lung cancer and 20 matched controls, the validation set comprised 40 patients with lung cancer and 40 matched controls (Supplementary Fig. S1). In addition, 27 subjects with positive CT, including 19 with tumor and 8 with non-tumor lung nodules were analyzed in the specificity set (Supplementary Fig. S1). In all the 3 sets, no significant differences between patients and controls were observed in terms of gender, age, smoking habits (pack-year), and MSC risk level (Table 1). Adenocarcinoma was the main histology type and stage I tumors were 14 of 20 (70%) in the training set, 25 of 40 (63%) in the validation set and 9 of 19 (47%) in the specificity set (Table 1).
Peripheral immune cells distinguish patients from controls in the training set
PBMC samples from the training set were analyzed by flow cytometry using 4 multiplex panels (Fig. 1A; Supplementary Tables S1 and S2). Consequential gating strategies were applied to analyze samples of the training set with the 23 markers characterizing both lymphoid (Fig. 2A) and myeloid immune subsets, including immune suppressive components (Fig. 2B). At the univariate level, NK-like T CD56neg significantly differed according to gender (P = 0.0234), C-Mo (P = 0.0440), and PMN-MDSC (P = 0.0485) according to pack-year, while TCD8+PD-1+ (P = 0.0071) and MoCX3CR1+ (P = 0.0070) according to tumor stage (Supplementary Table S4).
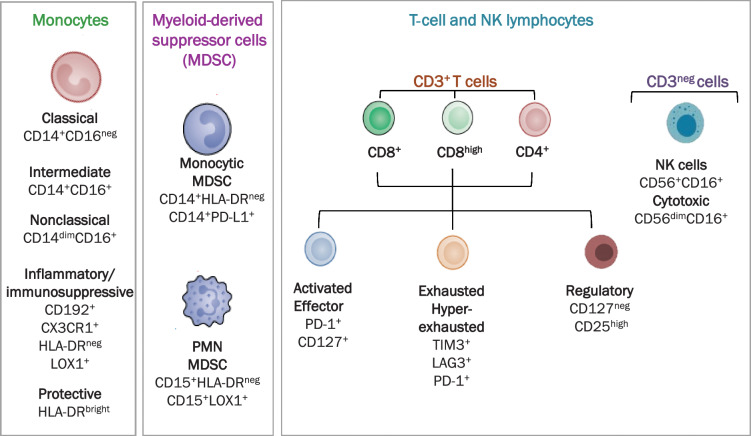
Figure 1.
High-dimensional flow cytometry analysis. Scheme of immune subpopulation analyzed in patients with lung cancer and controls.
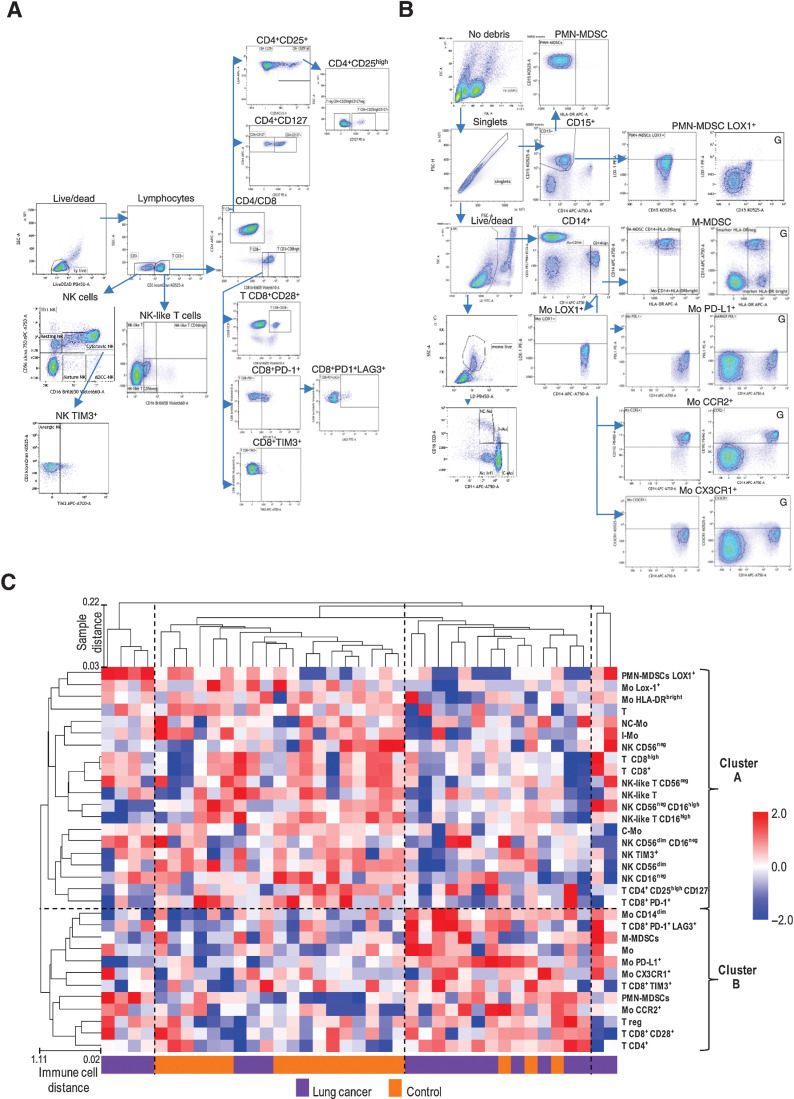
Figure 2.
The comprehensive peripheral immune cell profile of patients with screening-detected lung cancer and controls. Gating strategies of lymphoid populations (A) and myeloid populations (B). Plots marked with “G” are shown to indicate the setting of the gate. C, Unsupervised clustering considering all 32 subpopulation analyzed in the training set. Dashed lines separate the 4 and the 2 clusters of samples and immune cells at the larger distance (1-correlation), respectively.
Unsupervised clustering analysis of flow cytometry results identified 4 clusters of samples with the larger distance, as reported in Fig. 2C. Indeed, two small clusters were composed exclusively by 6 patients with lung cancer, whereas the two larger clusters in the middle included the majority of the controls (16/20) and 11 of the 14 remaining patients, respectively. When considering the immune cell tree, two main clusters with the largest distance can be identified, Cluster A and Cluster B including immune cell subsets with lower and higher frequency in PBMC samples of patients compared with controls, respectively. Among myeloid subsets, intermediate (CD14+CD16+), both classical (CD14+CD16neg) and nonclassical (CD14dimCD16+), Lox1+ and protective (HLA-DRbright) monocytes were found in Cluster A (lower frequency in patients PBMC with respect to controls). Conversely, monocytes with inflammatory (CD14dim), pro-angiogenic (CX3CR1+) and migration-prone (CCR2+) phenotypes, as well as M-MDSC and PMN-MDSC (both HLA-DRneg) compose Cluster B, i.e., (higher frequency in patients versus controls. When considering lymphoid markers, all NK and NK-like T were included in the Cluster A. This cluster also includes T lymphocytes, activated (CD3+CD8high) and cytotoxic (CD3+CD8+PD-1+) T cells, as well as CD4+ effector T cells (CD3+CD4+CD25highCD127+). Instead, Tregs (CD3+CD4+CD25highCD127neg), anergic (CD3+CD8+TIM3+), and exhausted T (CD3+CD8+PD-1+LAG3+) characterized by an immunosuppressive potential were all included in Cluster B.
Development of a phenotypic lung cancer ISC
By class comparison analysis, 8 immune subsets were found differentially expressed at the univariate level (P < 0.05) comparing patients with lung cancer and controls in the training set (Supplementary Table S5). According to the compound covariate predictor method, the 8 immune cell subsets discriminated patients and controls with an AUC of 0.95 (Supplementary Fig. S2A). By recursive feature elimination, 4 immune subsets [M-MDSC, PMN-MDSC, intermediate monocyte (I-Mo), and T CD8+PD-1+] maintained an AUC > 0.90 (Supplementary Fig. S2B), and were thus selected to compose a flow cytometry–based ISC.
PBMC samples of the validation and specificity sets were analyzed to confirm our findings. Considering the 4 selected immune subsets separately, they were concordantly found at higher (M-MDSC and PMN-MDSC) and lower levels (I-Mo and T CD8+PD-1+) in patients and controls of the validation sets and, except for the I-Mo subset, in the lung cancer specificity set (Fig. 3A).
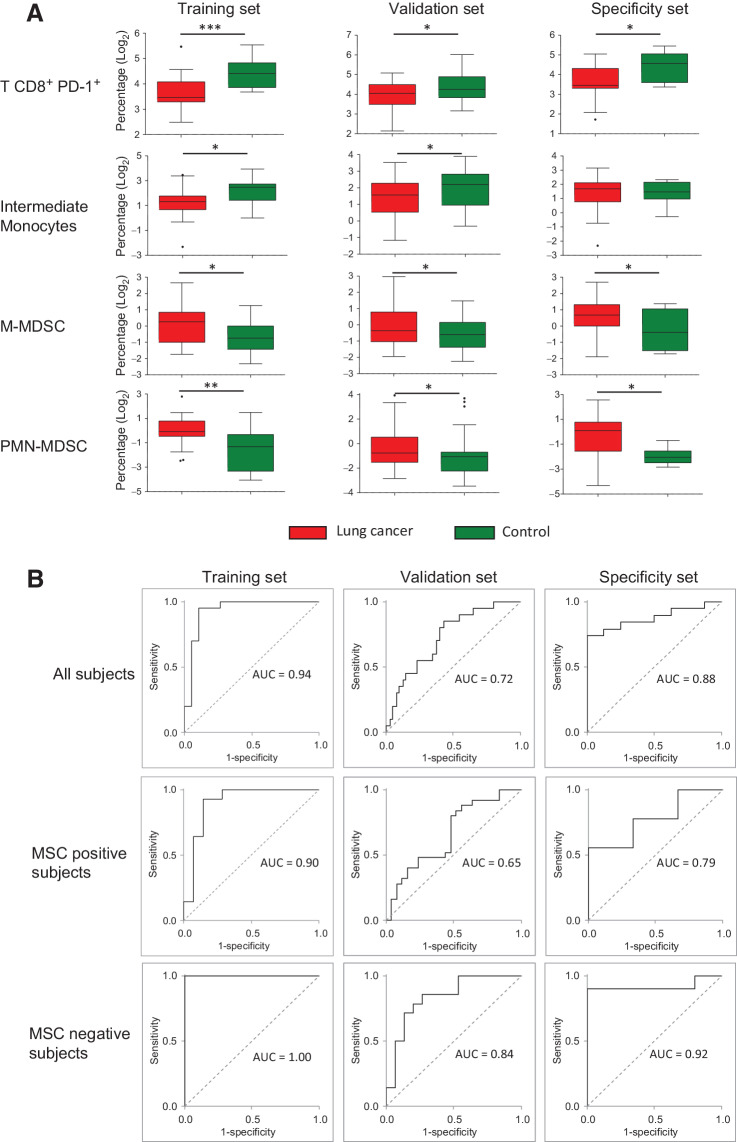
Figure 3.
Immune cell subsets discriminate patients with lung cancer and controls. A, Box plots reporting the levels of selected peripheral immune subsets in patients with lung cancer and controls measured by flow cytometry in the training, validation and specificity sets. Asterisks represent the level of significance by unpaired t test P value (*, < 0.05; **, < 0.01; ***, < 0.001). B, Performance of the ISC, estimated using the AUC-ROC method, in the training, validation and specificity sets, considering all the analyzed subjects or stratified according to MSC test results.
The performance of the ISC algorithm in discriminating patients and controls was further evaluated in the training, the validation and specificity sets. As reported in Fig. 3B, when considering all patients the AUCs were 0.94, 0.72 and 0.88, respectively. Subset analyses were also performed in MSC-positive and MSC-negative patients. Despite the ISC algorithm clearly discriminates patients and controls in both the subgroups, in MSC-negative patients a better performance of ISC classifier was observed with higher AUCs values in the training (AUC = 1.00), the validation (AUC = 0.84), and the specificity set (AUC = 0.92).
Clinical utility of ISC in lung cancer screening
The performance of the ISC in terms of Se, Sp, NPV, and PPV, was then estimated and the threshold for positivity defined as the maximum J value in the whole cohort. Results are reported in Supplementary Table S6: in both the training and validation sets, Se (95% and 80%, respectively) is higher than Sp (85% and 60%), favoring a higher NPV (94% and 75%) rather than the PPV (86% and 67%). Conversely, in the lung cancer specificity set, given the imbalance of cancerous compared with noncancerous lung nodules, the PPV was larger than the NPV (89% vs. 67%). Also in this analysis, when stratifying subjects according to MSC test results, higher values in terms of Se, Sp, NPV, and PPV were observed in the MSC-negative subgroup in all the 3 datasets (Supplementary Table S6).
To have a first insight if tumor characteristics such as histology, stage and nodule size could affect ISC outcome, we considered all 79 patients with lung cancer belonging to the 3 sets together. Regarding histology, our cohort is composed mainly of Adenocarcinoma (ADC; n = 58), but it also includes 8 squamous cell carcinoma (SCC), 7 small cell lung cancer (SCLC), and 6 carcinoma not otherwise specified (CaNOS). Indeed, no main differences in terms of Se were observed, being 49 of 58 (84%) ADC, 7/8 (88%) SSC, 6/7 (86%) SCLC, and 5/6 (83%) CaNOS positive to ISC. Similarly, stratifying by stage, 42/48 (87%) stage I and 25/31 (81%) stage > I were positive to ISC; while a positive ISC outcome was observed in 28/32 (87%) patients with lung cancer with indeterminate nodules (113–260 mm3) and 39/47 (83%) with nodules larger than 260 mm3.
To better have insights about the clinical utility of ISC in the setting of lung cancer screening, we took advantage of BioMILD data recently published to simulate its application (20). Focusing on the 655 LDCT+ BioMILD participants at the baseline, lung cancer was diagnosed within the first 2 years in 38 of the 209 (18%) MSC-positive and 28 of 446 (6%) MSC-negative participants. By performing the ISC test in MSC-negative subjects only, 24 of the 28 (86%) malignant nodules could be recovered by ISC at the cost of 105 of 418 (25%) non–lung cancer nodules classified as ISC positive. As a result, the NPV of the two tests combined would raise from 94% of the MSC alone to 99%, while maintaining 18% PPV.
Improvement of a molecular lung cancer ISC
To set-up a handy qPCR-based clinical grade assay, a panel of 26 genes representative of the 4 immune cell subsets discriminating patients with lung cancer from controls was first assessed in RNA extracted from PBMC samples of 12 screening participants characterized by extreme levels of at least one immune cell subset, according to flow cytometry data. The full list of genes can be found in Supplementary Table S3. Genes to be tested were selected on the basis of the phenotype (CD3D, CD8A, PDCD1, FUT4, CD14 and FCGR3A) and functional activity (CD274, GZMB, PRF1, IFNG, APBA2, S100A9, IL6, CCL2, TGFB1, SEMA4B, CCR5, FPR1 and HCAR2) of immune cell subpopulations as determined by flow cytometry analysis and published immune cell gene signatures (23–29). In addition, genes encoding markers specific for immune cell types (ARG1, CEACAM8, CCL13, S1PR3, CCL26 and GFRA2) were also evaluated (30).
Twenty of the 26 selected genes were detected by RT-qPCR in at least the 90% of samples. The expression value of each gene was then correlated with the amount of the represented immune cell subset considering the 3 highest and the 3 lowest flow cytometry values. According to our results, 14 genes showed a direct correlation with the represented immune population (Supplementary Fig. S3): 5 genes were representative of T cells CD8+PD-1+ (CD8A, GZMB, PRF1, PDCD1 and APBA2), 1 gene represented the I-Mo (GFRA2), 5 (CD274, S1PR3, SEMA4B, TGFβ1 and CD14) the M-MDSC and 3 (FUT4, FPR1 and HCAR2) the PMN-MDSC immune subsets.
The 14 genes were thus considered to define the final ISC as described in the materials and methods section. A molecular based immunoscore was first defined for each of the 4 immune subsets in samples of the validation set. Considering the whole series, a positive direct correlation was found comparing the flow cytometry data with the molecular immunoscore of each cell subset (Fig. 4A). Once the 4 immunoscores were then combined in the final molecular ISC, the performance reported an AUC of 0.75 in the entire validation set (Fig. 4B). Furthermore, consistent with flow cytometry data, the performance of the molecular ISC was lower in the subset of MSC-positive subjects (AUC = 0.70; Figure 4C), rather than in MSC-negative subjects (AUC = 0.83; Figure 4D). Even when the molecular ISC score was dichotomize into positive and negative result to evaluate Se, Sp, PPV, and NPV, results were similar to those obtained by flow cytometry (Supplementary Table S7).
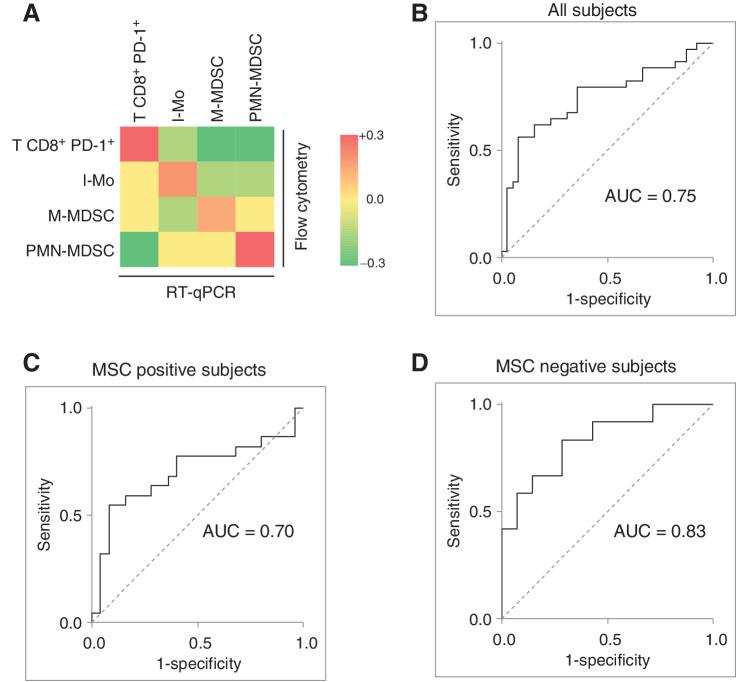
Figure 4.
Development of a RT-qPCR based lung cancer-related immune-score (ISC) for the analysis of PBMC samples. A, Heatmap reporting the correlations (Pearson R) between the gene expression values measured by RT-qPCR of 14 genes representative and the values of the 4 immune subset (T CD8+ PD-1+, I-Mo, M-MDSC, and PMN-MDSC) evaluated by flow cytometry on PBMC samples of the validation set. Genes are highlighted with the same color/s of the represented immune subset/s. ROC curves and respective AUCs evaluating the discriminatory capacity of the RT-qPCR based ISC considering the whole series (B) as well as the subsets composed by MSC-positive (C) and MSC-negative (D) subjects.
Microarray gene expression dataset obtained from PBMC (GSE13255) and whole blood (GSE108375) samples in public available datasets were further analyzed to validate the molecular ISC. After applying filtering parameters on age and smoking status to more closely mirror individuals eligible for lung cancer screening trials, 27 patients with NSCLC and 6 with nonmalignant lung disease were selected in the GSE13255 dataset, and 29 patients with malignant nodules and 39 with benign nodules were selected in the GSE108375 dataset (Supplementary Table S8). By the microarray platform, the CD274 gene was not detected in any of the two datasets, while TGFB was not detected in whole blood samples of the GSE108375 dataset. These genes were thus excluded from the classifier for this analysis. The AUC obtained by applying the molecular ISC was 0.69 and 0.63 in the GSE13255 and in the GSE108375, respectively (Supplementary Fig. S4).
Discussion
New advances in immune-oncology have drastically changed the management for the treatment of patients with lung cancer. Nevertheless, the role of adaptive and innate systemic immunity in lung carcinogenesis remains poorly understood. Studying peripheral blood immune cells could thus provide insights into the pathogenesis of lung tumors and allow the identification of novel biomarkers for early diagnosis and intervention. In the current study, by taking advantage of multicolor flow cytometry, immunologic markers related to myeloid lineage and T-cell anergy/exhaustion were included to capture early disease-related immunologic alterations. We observed differential frequencies of 4 specific immune cell subsets in the peripheral blood of patients with lung cancer compared with disease-free smokers in both the training and validation sets.
Notwithstanding the good performance, difficulties in bringing a flow cytometry–based assay into clinical practice, where PBMCs should remain viable from blood withdrawal until the analysis, might limit the utility of such a test in a general laboratory. Translation of the flow cytometry assay into a molecular test, which includes qPCR-based expression values of those genes most representatives of the 4 immune subsets, revealed to be successful with good correlation between the 2 assays and a significant diagnostic value.
The final molecular ISC was composed by 14 genes representative of activated T cells and of intermediate monocytes (i-Mo) as well as of genes characterizing M-MDSC and PMN-MDSC. Downregulation of T cells and of I-Mo genes, which are associated with cancer immunosurveillance with concomitant upregulation of MDSC in patients with lung cancer compared with controls are indicators of an immunosuppressive feature. The role of i-Mo in tumor development is still controversial. Indeed, these cells could play a role in the immune response against pathogens (31) but they are also able to secrete anti-inflammatory cytokines such as IL10 (32). In patients with lung cancer, a high number of circulating i-Mos was observed in the blood compared with controls (33). Regarding the role of circulating T cells, it has been shown that alterations in specific T and B cell subsets in the blood of patients with lung cancer were associated to prognosis (34).
The frequencies of circulating M-MDSC or PMN-MDSC differed in patients with lung cancer compared with healthy donors and were associated with the risk of recurrence in resected patients (35). The importance of PMN-MDSC in enhancing tumor growth and their immunosuppressive role in TME is well known (36). Furthermore, the ratio of circulating T cells and PMN-MDSC number was a predictive biomarker of response in patients treated with anti–PD-1 therapy (37).
Our findings in a cohort of patients with lung cancer detected within LDCT screening trial suggest that imbalance of specific blood cell subsets might represent an useful biomarker for screening and early diagnosis. So far, the characterization of peripheral blood immune cells and their gene expression content as diagnostic biomarker was assessed in few studies. Interestingly, a 29-gene signature in PBMC, able to discriminate patients with lung cancer from controls with nonmalignant nodules with 91% sensitivity and 80% specificity, was first identified (17). This signature, composed by genes related to immune response, NK function and apoptosis, was modulated in post-surgery patients (38). The same authors also reported that differential expression of 26 genes in PBMC was correlated with survival independently from tumor stage. Some of the genes associated to prognosis were linked to monocytes, myeloid cells and neutrophils (39).
In line with the hypothesis of the current study, our group has already reported that 24 circulating microRNAs, originating mostly from lung stromal and hematopoietic cells, compose a miRNA signature risk Classifier (MSC) able to predict tumor development in heavy smokers (18, 19, 40). Interestingly MSC also identifies a subgroup of patients who do not benefit from immunotherapy treatments (41). Full clinical validation of MSC as a companion diagnostic tool in the BioMILD prospective LDCT screening trial was recently published (20). The combination of LDCT and MSC biomarker showed clinical utility in targeting screening intervals on the basis of initial risk prediction and identified individuals with major differences in lung cancer risk despite similar age and tobacco exposure.
The main limitation of MSC is sensitivity to hemolysis, which leads to a negative MSC result. In addition, false negative results might hamper sensitivity of such biomarker. Thus implementing the MSC test by other complementary biomarkers is crucial to translate the test into clinical practice. Interestingly the ISC algorithm developed in the current study showed significantly higher AUCs values in MSC-negative subsets in the training (AUC = 1.00), validation (AUC = 0.84) and lung cancer specificity (AUC = 0.92) sets.
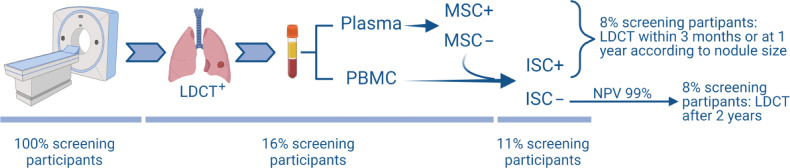
By simulating a chained-rule approach in the context of the BioMILD screening trial and considering the 655 LDCT+ participants, ISC was able to increase the NPV from the 94% of MSC alone to 99%. In accordance with current guidelines, additional radiological examinations are planned for LDCT+ participants after 3 months or one year according to nodule size, but only 10% actually have cancer (20). Indeed, with 99% NPV, the 48% (314/655) of LDCT+ screening participants who are both MSC and ISC negative could avoid additional unnecessary procedures for up to 2 years (Fig. 5). It should also be considered that both tests can be performed on a single 5mL blood sample: MSC using plasma and ISC using PBMC, thus optimizing time, resources and, more importantly, inducing less stress in screening participants.
Figure 5.
Lung cancer screening workflow integrating LDCT with blood-based tests such as the MSC and the ISC. Below are reported the expected percentages of participants who will pass through every step. Created with BioRender.com.
While the strength of this study relies in the direct validation of immune related biomarkers in the ‘intended to use’ context, given the availability of prospectively enrolled screening cohorts of smokers at risk of developing lung tumors, nonetheless we acknowledge a few limitations including the retrospective nature of the study and the limited number of samples analyzed. On the other hand, the two GEO datasets that we interrogated as external validation were unbalanced in term of cases:control ratio and did not include lung cancer screening series. Moreover, given the different technology adopted, some of the genes composing the ISC algorithm were not detected, thus leading to less optimal AUCs (0.69 and 0.63). Further analyses are needed to prospectively validate the ISC test in larger LDCT screening cohorts.
Altogether, these findings suggest that an immunosuppressive systemic immunity could concur to lung carcinogenesis. Hence, a peripheral myeloid/lymphoid molecular immunoscore can help the early detection of lung cancer and may implement the accuracy of other blood biomarkers such as miRNA-based classifier.
Supplementary Material
Acknowledgments
The study was supported by grants from the Italian Association for Cancer Research [AIRC 5xmille IG 12162; Investigator grant nos. 23244 to G. Sozzi, 25078 to V. Huber, Italian Ministry of Health (RF-2018-12367824 to G. Sozzi; GR-2016-02361849 to M. Boeri)].
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 1993
Footnotes
Note: Supplementary data for this article are available at Cancer Epidemiology, Biomarkers & Prevention Online (http://cebp.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
O. Fortunato: Conceptualization, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. V. Huber: Conceptualization, data curation, formal analysis, writing–original draft, writing–review and editing. M. Segale: Formal analysis, methodology, writing–review and editing. A. Cova: Formal analysis, investigation, methodology, writing–review and editing. V. Vallacchi: Formal analysis, investigation, methodology, writing–review and editing. P. Squarcina: Formal analysis, investigation, methodology. L. Rivoltini: Supervision, writing–original draft, writing–review and editing. P. Suatoni: Data curation, formal analysis, investigation, methodology, writing–review and editing. G. Sozzi: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing. U. Pastorino: Supervision, writing–original draft, writing–review and editing. M. Boeri: Conceptualization, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pastorino U, Silva M, Sestini S, Sabia F, Boeri M, Cantarutti A, et al. Prolonged lung cancer screening reduced 10-year mortality in the MILD trial. Ann Oncol 2019;30:1162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Koning HJ, van der Aalst CM, de Jong PA, Scholten ET, Nackaerts K, Heuvelmans MA, et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med 2020;382:503–13. [DOI] [PubMed] [Google Scholar]
- 5. Jonas DE, Reuland DS, Reddy SM, Nagle M, Clark SD, Weber RP, et al. Screening for lung cancer with low-dose computed tomography: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA 2021;325:971–87. [DOI] [PubMed] [Google Scholar]
- 6. Dejima H, Hu X, Chen R, Zhang J, Fujimoto J, Parra ER, et al. Immune evolution from preneoplasia to invasive lung adenocarcinomas and underlying molecular features. Nat Commun 2021;12:2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hiraoka K, Miyamoto M, Cho Y, Suzuoki M, Oshikiri T, Nakakubo Y, et al. Concurrent infiltration by CD8+ T cells and CD4+ T cells is a favourable prognostic factor in non–small cell lung carcinoma. Br J Cancer 2006;94:275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tassi E, Grazia G, Vegetti C, Bersani I, Bertolini G, Molla A, et al. Early effector T lymphocytes coexpress multiple inhibitory receptors in primary non–small cell lung cancer. Cancer Res 2017;77:851–61. [DOI] [PubMed] [Google Scholar]
- 9. Shimizu K, Nakata M, Hirami Y, Yukawa T, Maeda A, Tanemoto K. Tumor-infiltrating Foxp3+ regulatory T cells are correlated with cyclooxygenase-2 expression and are associated with recurrence in resected non–small cell lung cancer. J Thorac Oncol 2010;5:585–90. [DOI] [PubMed] [Google Scholar]
- 10. Dai F, Liu L, Che G, Yu N, Pu Q, Zhang S, et al. The number and microlocalization of tumor-associated immune cells are associated with patient's survival time in non–small cell lung cancer. BMC Cancer 2010;10:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ye X, Zhang N, Jin Y, Xu B, Guo C, Wang X, et al. Dramatically changed immune-related molecules as early diagnostic biomarkers of non–small cell lung cancer. FEBS J 2020;287:783–99. [DOI] [PubMed] [Google Scholar]
- 12. Xia Y, Li W, Li Y, Liu Y, Ye S, Liu A, et al. The clinical value of the changes of peripheral lymphocyte subsets absolute counts in patients with non–small cell lung cancer. Transl Oncol 2020;13:100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soyano AE, Dholaria B, Marin-Acevedo JA, Diehl N, Hodge D, Luo Y, et al. Peripheral blood biomarkers correlate with outcomes in advanced non–small cell lung cancer patients treated with anti–PD-1 antibodies. J Immunother Cancer 2018;6:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baine MJ, Chakraborty S, Smith LM, Mallya K, Sasson AR, Brand RE, et al. Transcriptional profiling of peripheral blood mononuclear cells in pancreatic cancer patients identifies novel genes with potential diagnostic utility. PLoS One 2011;6:e17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Twine NC, Stover JA, Marshall B, Dukart G, Hidalgo M, Stadler W, et al. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res 2003;63:6069–75. [PubMed] [Google Scholar]
- 16. Kossenkov AV, Qureshi R, Dawany NB, Wickramasinghe J, Liu Q, Majumdar RS, et al. A gene expression classifier from whole blood distinguishes benign from malignant lung nodules detected by low-dose CT. Cancer Res 2019;79:263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Showe MK, Vachani A, Kossenkov AV, Yousef M, Nichols C, Nikonova EV, et al. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non–small cell lung cancer from patients with nonmalignant lung disease. Cancer Res 2009;69:9202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA 2011;108:3713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sozzi G, Boeri M, Rossi M, Verri C, Suatoni P, Bravi F, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: A correlative MILD Trial Study. J Clin Oncol 2014;32:768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pastorino U, Boeri M, Sestini S, Sabia F, Milanese G, Silva M, et al. Baseline computed tomography screening and blood microRNA predict lung cancer risk and define adequate intervals in the BioMILD trial. Ann Oncol 2022;33:395–405. [DOI] [PubMed] [Google Scholar]
- 21. Mensah M, Borzi C, Verri C, Suatoni P, Conte D, Pastorino U, et al. MicroRNA based liquid biopsy: the experience of the plasma miRNA Signature Classifier (MSC) for lung cancer screening. J Vis Exp 2017;56326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huber V, Di GL, Lalli L, Giardiello D, Cova A, Squarcina P, et al. Back to simplicity: a four-marker blood cell score to quantify prognostically relevant myeloid cells in melanoma patients. J Immunother Cancer 2021;9:e001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, et al. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 2011;118:e16–31. [DOI] [PubMed] [Google Scholar]
- 24. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 25. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huber V, Vallacchi V, Fleming V, Hu X, Cova A, Dugo M, et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J Clin Invest 2018;128:5505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fridlender ZG, Sun J, Mishalian I, Singhal S, Cheng G, Kapoor V, et al. Transcriptomic analysis comparing tumor-associated neutrophils with granulocytic myeloid-derived suppressor cells and normal neutrophils. PLoS One 2012;7:e31524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwak T, Wang F, Deng H, Condamine T, Kumar V, Perego M, et al. Distinct populations of immune-suppressive macrophages differentiate from monocytic myeloid-derived suppressor cells in cancer. Cell Rep 2020;33:108571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kapellos TS, Bonaguro L, Gemünd I, Reusch N, Saglam A, Hinkley ER, et al. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front Immunol 2019;10:2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019;366:eaax9198. [DOI] [PubMed] [Google Scholar]
- 31. Fingerle G, Pforte A, Passlick B, Blumenstein M, Strobel M, Ziegler-Heitbrock HW. The novel subset of CD14+/CD16+ blood monocytes is expanded in sepsis patients. Blood 1993;82:3170–6. [PubMed] [Google Scholar]
- 32. Skrzeczyńska-Moncznik J, Bzowska M, Loseke S, Grage-Griebenow E, Zembala M, Pryjma J. Peripheral blood CD14high CD16+ monocytes are main producers of IL-10. Scand J Immunol 2008;67:152–9. [DOI] [PubMed] [Google Scholar]
- 33. Kwiecień I, Rutkowska E, Polubiec-Kownacka M, Raniszewska A, Rzepecki P, Domagała-Kulawik J. Blood monocyte subsets with activation markers in relation with macrophages in non–small cell lung cancer. Cancers (Basel) 2020;12:2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y, Zhou N, Zhu R, Li X, Sun Z, Gao Y, et al. Circulating activated immune cells as a potential blood biomarkers of non–small cell lung cancer occurrence and progression. BMC Pulm Med 2021;21:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamauchi Y, Safi S, Blattner C, Rathinasamy A, Umansky L, Juenger S, et al. Circulating and tumor myeloid-derived suppressor cells in resectable non–small cell lung cancer. Am J Respir Crit Care Med 2018;198:777–87. [DOI] [PubMed] [Google Scholar]
- 36. Yang Z, Guo J, Weng L, Tang W, Jin S, Ma W. Myeloid-derived suppressor cells-new and exciting players in lung cancer. J Hematol Oncol 2020;13:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim HR, Park SM, Seo SU, Jung I, Yoon HI, Gabrilovich DI, et al. The ratio of peripheral regulatory T cells to Lox-1(+) polymorphonuclear myeloid-derived suppressor cells predicts the early response to anti–PD-1 therapy in patients with non–small cell lung cancer. Am J Respir Crit Care Med 2019;199:243–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kossenkov AV, Vachani A, Chang C, Nichols C, Billouin S, Horng W, et al. Resection of non–small cell lung cancers reverses tumor-induced gene expression changes in the peripheral immune system. Clin Cancer Res 2011;17:5867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kossenkov AV, Dawany N, Evans TL, Kucharczuk JC, Albelda SM, Showe LC, et al. Peripheral immune cell gene expression predicts survival of patients with non–small cell lung cancer. PLoS One 2012;7:e34392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fortunato O, Borzi C, Milione M, Centonze G, Conte D, Boeri M, et al. Circulating mir-320a promotes immunosuppressive macrophages M2 phenotype associated with lung cancer risk. Int J Cancer 2019;144:2746–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boeri M, Milione M, Proto C, Signorelli D, Lo RG, Galeone C, et al. Circulating miRNAs and PD-L1 tumor expression are associated with survival in advanced NSCLC patients treated with immunotherapy: a prospective study. Clin Cancer Res 2019;25:2166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Expression profile data analyzed in this study were obtained from GEO at GSE108375 and GSE13255.