Bone marrow T-cell clonal expansion, immune phenotypes, and specificities at the single-cell level were determined in treatment-naïve multiple myeloma. Dominant T-cell clones rarely recognize antigens presented on myeloma cells, are not neoantigen-specific, and show low immune checkpoint expression.
Abstract
Multiple myeloma is a hematologic malignancy of monoclonal plasma cells that accumulate in the bone marrow. Despite their clinical and pathophysiologic relevance, the roles of bone marrow–infiltrating T cells in treatment-naïve patients are incompletely understood. We investigated whether clonally expanded T cells (i) were detectable in multiple myeloma bone marrow, (ii) showed characteristic immune phenotypes, and (iii) whether dominant clones recognized antigens selectively presented on multiple myeloma cells. Single-cell index sorting and T-cell receptor (TCR) αβ sequencing of bone marrow T cells from 13 treatment-naïve patients showed dominant clonal expansion within CD8+ cytolytic effector compartments, and only a minority of expanded T-cell clones expressed the classic immune-checkpoint molecules PD-1, CTLA-4, or TIM-3. To identify their molecular targets, TCRs of 68 dominant bone marrow clones from five selected patients were reexpressed and incubated with multiple myeloma and non–multiple myeloma cells from corresponding patients. Only 1 of 68 TCRs recognized antigen presented on multiple myeloma cells. This TCR was HLA-C–restricted, self-peptide–specific and could be activated by multiple myeloma cells of multiple patients. The remaining dominant T-cell clones did not recognize multiple myeloma cells and were, in part, specific for antigens associated with chronic viral infections. In conclusion, we showed that dominant bone marrow T-cell clones in treatment-naïve patients rarely recognize antigens presented on multiple myeloma cells and exhibit low expression of classic immune-checkpoint molecules. Our data provide experimental context for experiences from clinical immune-checkpoint inhibition trials and will inform future T cell–dependent therapeutic strategies.
Introduction
Multiple myeloma is a hematologic malignancy characterized by the accumulation of monoclonal plasma cells in the bone marrow. Immune cells constituting the bone marrow microenvironment have a major role in disease pathophysiology (1–3), and immunomodulatory agents, as well as monoclonal antibodies, are part of effective treatment regimens (4, 5).
Clonal T-cell expansion is an antigen-driven process, and the roles of clonally expanded multiple myeloma-infiltrating T cells are incompletely understood. In solid tumors, neoantigens can drive specific clonal T-cell expansion, resulting in functionally exhausted T-cell compartments (6–9). The peripheral blood of heavily pretreated multiple myeloma patients can contain variable amounts of neoantigen-specific T cells (10); however, their occurrence in the bone marrow of treatment-naïve patients, degrees of clonal expansion, impact on multiple myeloma pathophysiology, and clinical significance remain unclear. Tumor mutational burden, which correlates with the presence of tumor-specific T cells and response to immune-checkpoint inhibition in solid tumors, can be considered intermediate in multiple myeloma (11) and increases with the number of previous treatment regimens. T cells can recognize multiple myeloma–associated antigens but acquire features of functional inhibition during disease progression (12, 13). However, immune-checkpoint blockade targeting PD-1/PD-L1 interactions to reverse functional T-cell inhibition has been ineffective as a monotherapy in multiple myeloma (14) and has led to increased severe adverse events without improving clinical responses in combination therapies (15, 16). Furthermore, multiple myeloma cells can reduce antigen presentation (17), and regulatory T cells (Treg) may contribute to the impairment of effective anti-myeloma T-cell responses (18).
Hypothesizing that T-cell expansion can be driven by chronically persisting multiple myeloma-associated antigens, we studied differentiation states and specificities of dominant bone marrow T-cell clones in treatment-naïve patients at the single-cell level. We investigated whether (i) clonal expansion was restricted to bone marrow T cells, (ii) clonal T-cell expansion associated with characteristic immune phenotypes, and (iii) dominant bone marrow T-cell clones recognized antigens selectively presented on multiple myeloma cells.
Materials and Methods
Patient and healthy donor samples
The study was approved by the local institutional review board (Ethikkommission der Charité, protocol EA2/096/15 to LH). All patients and one healthy peripheral blood donor were accrued at Charité–Universitätsmedizin Berlin, provided written informed consent, and all research was conducted in accordance with the Declaration of Helsinki. All samples were acquired between August 2015 and October 2019. None of the participants had clinically apparent signs of infection at the time of sample collection. Bone marrow mononuclear cells and peripheral blood mononuclear cells (PBMC), if available, were isolated using Ficoll-Paque PLUS (GE Healthcare) according to the manufacturer's instructions and were cryopreserved in RPMI-1640 (Thermo Fisher Scientific) containing 50% fetal bovine serum (FBS, Thermo Fisher Scientific) and 10% dimethyl sulfoxide (DMSO, Carl Roth GmbH) before further processing. For cryopreservation, vials were stored in liquid nitrogen. Patient characteristics and genetic features are listed in Table 1 and Supplementary Table S1, respectively.
Table 1.
Patient characteristics.
| Patient | Age (years) | Sex | BM infiltration (%) | ISS stage | Disease type | CMV IgG | EBV VCA- IgG | HLA-class I |
|---|---|---|---|---|---|---|---|---|
| MM195 | 47 | Female | 60–70 | II | IgG κ | Neg | Pos | A*01:01; A*02:01 |
| B*27:05; B*44:02 | ||||||||
| C*02:02; C*05:01 | ||||||||
| MM104 | 69 | Female | 30–40 | III | λ light chain | Pos | Pos | A*03:01; A*24:02 |
| B*51:01; B*55:01 | ||||||||
| C*01:02; C*03:03 | ||||||||
| MM078 | 41 | Male | 60 | II | IgG κ | Pos | Pos | A*02:01; A*23:01 |
| B*15:03; B*15:03 | ||||||||
| C*02:10; C*02:10 | ||||||||
| MM165 | 66 | Female | 90 | III | κ light chain | Neg | Pos | A*01:01; A*02:01 |
| B*51:01; B*08:01 | ||||||||
| C*02:02; C*07:01 | ||||||||
| MM061 | 64 | Male | 30 | III | λ light chain | Neg | Neg | A*26:01; A*32:01 |
| B*44:02; B*38:01 | ||||||||
| C*05:01; C*12:03 | ||||||||
| MM112 | 40 | Female | 95 | III | IgA λ | Neg | Pos | A*03:01; A*24:02 |
| B*07:02; B*51:01 | ||||||||
| C*01:02; C*07:02 | ||||||||
| MM204 | 61 | Female | 70 | III | λ light chain | Neg | Pos | A*02:01; A*29:02 |
| B*08:01; B*44:02 | ||||||||
| C*05:01; C*07:18 | ||||||||
| MM008 | 86 | Male | 70 | II | IgG κ | N.d. | N.d. | A*01:01; A*03:01 |
| B*07:02; B*08:01 | ||||||||
| C*07:01; C*07:02 | ||||||||
| MM003 | 71 | Male | 60 | II | IgG κ | Pos | N.d. | A*01:01; A*30:02 |
| B*08:01; B*35:03 | ||||||||
| C*04:01; C*07:01 | ||||||||
| MM157 | 83 | Male | 80 | II | IgG κ | Neg | Pos | A*03:01; A*11:01 |
| B*07:02; B*44:02 | ||||||||
| C*05:01; C*07:02 | ||||||||
| MM050 | 85 | Female | 50 | III | IgG λ | Pos | Pos | A*03:01; A*03:01 |
| B*07:02; B*51:01 | ||||||||
| C*01:02; C*07:02 | ||||||||
| MM139 | 73 | Female | 70 | II | IgG κ | Pos | Pos | A*01:01; A*24:02 |
| B*15:17; B*35:01 | ||||||||
| C*04:29; C*07:01 | ||||||||
| MM160 | 68 | Female | 25 | III | IgA κ | Pos | Pos | A*02:01; A*23:01 |
| B*44:03; B*51:01 | ||||||||
| C*15:02; C*04:09N |
Note: Bone marrow plasma cell infiltration was determined by histology. Genetic characteristics can be found in Supplementary Table S1.
Abbreviations: BM, bone marrow; CMV, cytomegalovirus; EBV, Epstein–Barr virus; ISS, international staging system; n.d., not determined; VCA, virus capsid antigen.
Fluorescence-activated cell sorting
Cells isolated from bone marrow, peripheral blood, and cell lines were stained with multiparameter panels according to the manufacturer's instructions. Briefly, up to 5 × 106 cells were washed in pure phosphate-buffered saline (PBS, Thermo Fisher Scientific), centrifuged at 300 × g, supernatant was decanted, and cells were stained with the indicated antibodies at 4°C for 30 minutes protected from light. After staining, cells were washed with PBS containing 2% FBS (Thermo Fisher Scientific) and resuspended in 100 μL PBS containing 2% FBS for subsequent analysis or sorting. Index sorting for single-cell sequencing was done using a FACSAria Fusion high-speed cell sorter (BD Biosciences) equipped with a 70-μm nozzle as previously described (19, 20). Antibodies used for index sorting are listed in Supplementary Table S2. Flow cytometry for cell analysis was performed using LSRFortessa (BD Biosciences), Navios (Beckman Coulter), or Aurora (Cytek) instruments.
T-cell receptor αβ, cytokine, and transcription factor sequencing
Single-cell nucleic acid amplification, library preparation, sequencing, and data processing were done as described previously (19–22). For peripheral blood bulk T-cell receptor (TCR)β repertoire sequencing, we used the T-cell receptor beta Deep immunoSEQ assay (Adaptive Biotechnologies) with 10 μg PBMC DNA per patient as input.
Single-cell gene expression to determine cell type–associated AP5M1 expression
Single-cell gene expression, VDJ immunoglobulin sequencing, and feature barcoding of bone marrow mononuclear cells and PBMCs to illustrate cell type–associated AP5M1 expression were done using the Chromium Single-Cell Immune Profiling platform, Chromium Next GEM Single-Cell 5′ Kit v2, Chromium Single-Cell Human BCR Amplification Kit, and 5′ Feature Barcode Kit (all 10X Genomics) according to the manufacturer's instructions. For feature barcoding, cells were stained with the following antibodies according to the manufacturer's instructions: CD38 (RRID: AB_2800758), CD138 (RRID: AB_2810567), CD45 (RRID: AB_2800762), CD56 (RRID: AB_2801024), CD19 (RRID: AB_2800741), CD20 (RRID: AB_2800743), CD3 (RRID: AB_2800723), CD14 (RRID: AB_2810576), TIM-3 (RRID: AB_2800925), BTLA (RRID: AB_2800919), PD-1 (RRID: AB_2800862), CD39 (RRID: AB_2800853), CD8 (RRID: AB_2800922), CXCR4 (RRID: AB_2800791), CD57 (RRID: AB_2801030; all BioLegend). Sequencing was done on a NovaSeq 6000 system (Illumina) with the following specifications: Single-cell gene expression was sequenced 27 × 97 bp paired end, feature barcodes and BCR sequences were sequenced 27 × 92 bp paired end. Numbers of sequenced cells are indicated in the respective figure descriptions. Cellranger 5.0.1, Loupe Browser 5.0, and Loupe VDJ Browser 4.0 (all 10X Genomics) were used for data processing and illustration according to the manufacturer's instructions.
Identification of somatic mutations
DNA and RNA were isolated from study patients’ multiple myeloma bone marrow cells using the AllPrep DNA/RNA micro kit or QIAamp DNA mini kit (both Qiagen). PBMC DNA of the respective patients was used as germline control. In cases where peripheral blood was not available as a germline control (patients MM003 and MM165), bone marrow was CD38 depleted by magnetic-activated cell separation (MACS), as described below, and subsequently CD38−CD138−CD19− cells were sorted via FACS as specified.
For DNA library preparations, we used SureSelect Human All Exome V7 kits (Agilent), for RNA library preparation, TruSeq Stranded mRNA kits (Illumina) were used. DNA was sequenced 100 bp paired end or 150 bp paired end, RNA was sequenced 75 bp paired end, all on a HiSeq 4000 (Illumina). Sequencing data were mapped against the human genome release GRCh37 with decoy regions (hs37d5). BWA (23) was used to align exome data, STAR (24) to align mRNA data, applying the GENECODE release 19 as annotations. Somatic single-nucleotide variants were detected using MuTect (25). The median sequencing depth of exome data were 215× for 13 samples sequenced with 100 cycles, and 629× for 5 samples sequenced with 150 cycles. The median number of reads for expression data was 3.33 × 108 reads/sample. Presentation of expressed mutations was predicted using NetMHC 4.0 and NetMHCpan 4.0 (26). Peptides with affinity < 1,000 nmol/L and/or %rank ≤ 2 were considered presented.
MACS
Multiple myeloma cells from patient bone marrow were magnetically enriched using the Plasma Cell Isolation Kit II, human (Miltenyi Biotec) according to the manufacturer's instructions with the following modification: only CD38 MicroBeads from the Plasma Cell Isolation Kit II, human (Miltenyi Biotec) were used, and positive selection was applied. For enrichment of CD8+ TCR-transduced T cells, we performed depletion of CD4+ T cells immediately before coincubation experiments, using the CD4 MicroBeads, human kit (Miltenyi Biotec).
Cell lines
Multiple myeloma cell lines U266B1 (RRID: CVCL_0566), JJN-3 (RRID: CVCL_2078), L-363 (RRID: CVCL_1357), NCI-H929 (RRID: CVCL_1600), and KMS-12-PE (RRID: CVCL_1333), as well as HEK293T (RRID: CVCL_0063), were provided by the Blankenstein Laboratory. 58α−β− T hybridoma cells were created by the Dornmair laboratory (27) and kindly provided in 2019. For all experiments, cryopreserved vials of the respective cell lines were thawed and cultivated at 37°C and 5% CO2 for 7 to 14 days before further use. All cell lines were authenticated by microscopy and flow cytometry, and the absence of Mycoplasma was confirmed by LookOut Mycoplasma PCR-Detektions-Kit (Sigma-Aldrich). All cell lines were cultured in RPMI-1640 containing 10% FBS, 10,000 U/mL penicillin, and 10 mg/mL streptomycin (all Thermo Fisher Scientific).
TCR expression on human peripheral blood lymphocytes
TCR gene transfer for recombinant TCR expression was performed as previously described (28) with minor modifications: All TCRαβ chains (Supplementary Table S3) were linked via a self-cleaving peptide (P2A, amino acid sequence: ATNFSLLKQAGDVEENPGP), and human TCR constant regions were replaced by their murine counterparts as described previously (29). All TCR DNA sequences (Supplementary Table S3) were codon-optimized for mammalian expression and synthesized (GeneArt, Life Technologies).
TCR expression on 58α−β− reporter cells
All TCR DNA sequences (Supplementary Table S3) were synthesized (GeneArt, Life Technologies). TCRs were expressed on 58α−β− T hybridoma cells, which also expressed human CD8αβ chains and green fluorescent protein (GFP) under the control of nuclear factor of activated T cells (NFAT), as described (21, 27). These cells are referred to as “58α−β−” reporter cells. Recombinant TCR expression was confirmed by CD3 staining (Brilliant Violet 421 anti-mouse CD3, BioLegend, RRID: AB_10900227) according to the manufacturer's instructions.
T-cell stimulation, incubation with potential target cells, and target peptide identification
60,000 TCR-recombinant 58α−β− cells were stimulated with plate-bound anti-mouse CD3 (clone 17A2, BioLegend, RRID: AB_2810314) for 16 hours at 37°C and 5% CO2 as a positive control. Mouse IL2 secretion was measured using the Mouse IL2 Duo Set ELISA Kit (R&D Systems) according to the manufacturer's instructions. Activation of TCR-transduced human PBL was detected using the Human IFNγ ELISA Set BD OptEIA (BD Biosciences, RRID: AB_2869029) according to the manufacturer's instructions. ELISA plates were analyzed using the Infinite M Plex (Tecan) plate reader, and Magellan (Tecan) data analysis software version 7.0. Coincubation details are listed in Supplementary Table S4.
Potential target peptides (predicted from the somatic mutation data as indicated) were synthesized (JPT Peptide Technologies GmbH or GenScript Biotech Corporation) and loaded onto L-363 as antigen-presenting cells at 5 μmol/L concentration. For each coincubation with TCR-recombinant T cells, 100,000 L-363 were used. For target peptide screening and coincubation with multiple myeloma cells, TCRs specific for cytomegalovirus-derived peptides (TPR, amino acid sequence: TPRVTGGGAM; or VTE, amino acid sequence: VTEHDTLLY), and an Epstein–Barr virus-derived peptide (GLC, amino acid sequence: GLCTLVAML) were used as positive controls. The sequences of the TPR-specific TCR are derived from MM008 15E10, and the VTE-specific TCR from MM139 10F7 (Supplementary Table S3); the GLC-specific TCR was previously published (30). CDR3 amino acid sequences of all TCRs detailed in Supplementary Table S3 were screened for potential target epitopes using VDJdb (https://vdjdb.cdr3.net).
All coincubations of TCR-recombinant and bone marrow cells (Supplementary Table S4) were performed in 96-well plates in 150 μL RPMI-1640 containing 10% FBS, 10,000 U/mL penicillin, and 10 mg/mL streptomycin (all Thermo Fisher Scientific) for 16 hours at 37°C and 5% CO2.
For target peptide screening, we generated pools of eight Micro-Scale Peptides (JPT Peptide Technologies GmbH) and added a cytomegalovirus pp65-derived control peptide (TPRVTGGGAM, called TPR, presented on HLA-B*07:02, GenScript Biotech Corporation). All peptide pools (final concentration of 1–5 μmol/L per peptide) were incubated with 60,000 reporter cells expressing the TCR in question or a control peptide-specific TCR for 16 hours at 37°C and 5% CO2. In case of T-cell activation determined by GFP fluorescence, each peptide in the respective pool was tested individually and activating peptides were synthesized with high purity (GenScript Biotech Corporation) for verification. For HLA-blocking, purified antibodies against HLA-A2 (clone BB7.2, BioLegend, RRID: AB_1659243), HLA-BC (clone B1.23.2, Thermo Fisher Scientific, RRID: AB_2866114), or HLA-class I (clone W6/32, BioLegend, RRID: AB_2561492) were titrated and used at the indicated concentrations. Ultra-LEAF Purified Mouse IgG2a κ isotype control (clone MOPC-173, BioLegend, RRID: AB_11148947) or Purified Mouse IgG1 κ isotype control (clone MOPC-21, BioLegend, RRID: AB_2891079) were used as isotype controls at the indicated concentrations.
HLA knockout using CRISPR-Cas9
The following guide RNAs (5′ → 3′) were used for HLA knockout: ACAGCGACGCCGCGAGCCAG (HLA-A), CGTACTGGTCATGCCCGCGG (HLA-B), and GACACAGAAGTACAAGCGCC (HLA-C). All guide RNAs were coded within pSpCas9 plasmids (GenScript). JJN-3 and U266B1 cells were transfected by electroporation (Gene Pulser Xcell, Bio-Rad), and HEK293T were transfected with FuGENE HD Transfection Reagent (Promega). HLA-knockout cell lines were expanded for 4 to 6 weeks from single cells sorted on a FACSAria Fusion high-speed cell sorter (BD Biosciences) equipped with a 70-μm nozzle. Cell culture conditions are specified in the cell line section. HLA knockout was confirmed by flow cytometry using antibodies against HLA-B7 (clone BB7.1, BioLegend, RRID: AB_2650776), HLA-C (clone DT9, BD Biosciences, RRID: AB_2739715), or HLA-class I (clone W6/32, BioLegend, RRID: AB_493669) according to the manufacturer's instructions.
Recombinant HLA expression
HLA coding sequences (IPD accession numbers HLA00401, HLA00404, HLA00413, HLA00420, HLA01451, HLA00427, HLA00433, HLA00434, HLA01672, HLA00446, HLA00455, and HLA00467) were downloaded from IPD-IMGT/HLA (https://www.ebi.ac.uk/ipd/imgt/hla/), synthesized (Thermo Fisher Scientific), cloned into pHSE3’ as previously described (27), and nucleofected (SF Cell Line 4D-Nucleofector X Kit L, Lonza) into 250,000 JJN-3 cells. HLA expression was confirmed by flow cytometry as detailed above.
U266B1 gene expression and peptidome data
Multiple myeloma U266B1 gene-expression data are publicly available (LL-100; ref. 31) and were used. Published U266B1 peptidome data were provided by the authors (32) and combined with a data set we generated as previously described (33) with minor modifications. Briefly, 1×108 U266B1 cells were lysed in 200 μL lysis buffer containing 1% cholamidopropyl dimethylammonio propanesulfate (CHAPS), 200 mmol/L Tris-HCl, 150 mmol/L NaCl, and 1× Halt Protease Inhibitor Cocktail (all Thermo Fisher Scientific) for 60 minutes at 4°C. The lysate was centrifuged at 13,000 × g for 10 minutes, and the supernatant was incubated with HLA-class I–specific antibody (clone W6/32, BioLegend, RRID: AB_2561492) at 60 μg/mL for 60 minutes at 4°C. Recombinant (r)Protein A Sepharose Fast Flow beads (Sigma) were added at 100 μL beads/mL lysate and incubated for another 5 hours. Peptides were then eluted with 50 μL 10% acetic acid (Sigma) and desalted on C18 STAGE tips (Thermo Fisher Scientific) as described previously (34). Peptides were taken up in 0.1% formic acid, 3% acetonitrile (both Thermo Fisher Scientific), and acquired on an EASY-nLC 1200 system (Thermo Fisher Scientific) coupled to a Q Exactive HF-X orbitrap mass spectrometer (Thermo Fisher Scientific). Samples were separated on a 110-minute gradient ramping from 5% to 55% acetonitrile using an in-house prepared nano-LC column (0.074 mm × 250 mm, 1.9 μm Reprosil C18, Dr. A. Maisch HPLC GmbH) and a flow rate of 250 nL/minute. MS acquisition was operated in positive mode at an MS1 resolution of 60,000 and a scan range from 350 to 1,800 m/z. For data-dependent MS2 acquisition, the top 20 peaks were selected for MS2 with a resolution of 45,000, a maximum injection time of 250 ms and an isolation window of 1.3 m/z. Automatic gain control (AGC) target was set to 1×105 and dynamic exclusion was specified to 30 seconds. Raw files were analyzed in MaxQuant (version 1.6.2.6; ref. 35) applying an unspecific search against a human Uniprot database (2017), which allowed a peptide length of 7 to 14 amino acids.
Data availability statement
Detailed TCR sequence and mutation data can be found in the supplementary data. Whole-exome and RNA-sequencing raw data are publicly available at sequence read archive (https://www.ncbi.nlm.nih.gov/sra under BioProject accession number PRJNA862671).
Statistical analysis
Statistics were calculated using R (36) version 4.0.3. Applied tests and P value thresholds are indicated in all figure descriptions.
Results
Multiple myeloma bone marrow is infiltrated with clonally expanded CD8+ cytolytic effector T cells
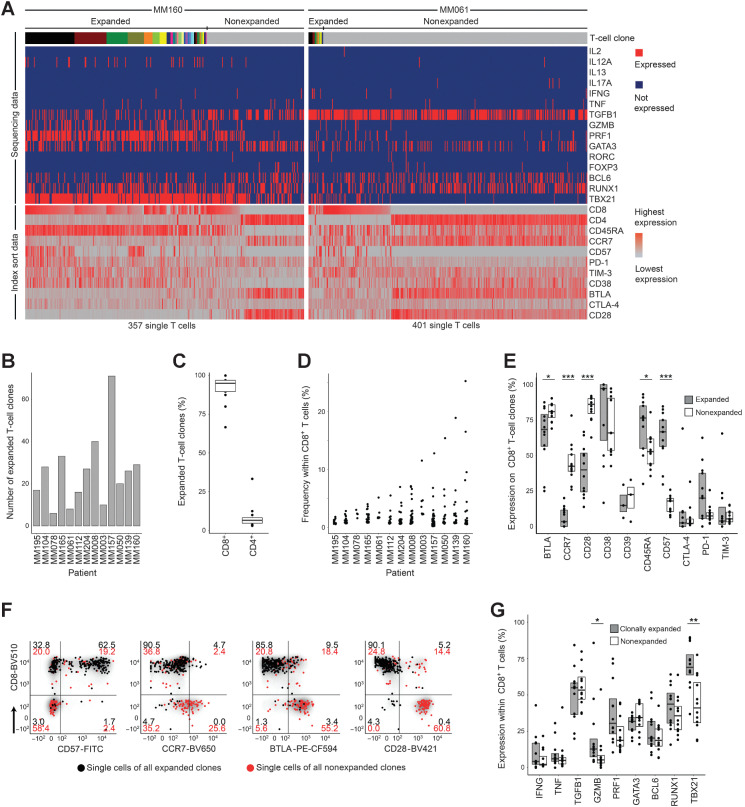
We hypothesized that multiple myeloma could drive specific clonal T-cell expansion, and we determined 13-dimensional immune phenotypes and TCRαβ sequences of single bone marrow T cells from 13 treatment-naïve patients (Table 1; Supplementary Table S1). A total of 6,744 bone marrow T cells (368-1,288 T cells per patient) were FACS index sorted (gating in Supplementary Fig. S1, antibody panel in Supplementary Table S2). TCR sequences were determined in 5,695 cells (84% of sorted cells), and clones were considered expanded if identical TCRαβ complementarity determining region 3 (CDR3) amino acid sequences were detected in at least two cells within a patient. Clones were defined as positive for a fluorescent parameter if the majority of cells were positive. The number of expanded T-cell clones varied across patients (range, 6-71 expanded clones per patient), with detailed data of two patients are shown as representative examples for relatively strong and weak clonal expansion in Fig. 1A, and the number of expanded T-cell clones within all patients was determined (Fig. 1B). Clonal expansion predominantly occurred within CD8+ compartments (Fig. 1C), and size of most expanded clones per patient ranged from 2% to 25% of CD8+ T cells (Fig. 1D). Compared with nonexpanded T cells, expanded clones were more frequently positive for CD57, while being less frequently positive for CCR7 and CD28 (Fig. 1E). Significant differentially expressed markers between expanded and nonexpanded T-cell clones are shown for patient MM160 as an example in Fig. 1F (all patients in Supplementary Fig. S2). Regarding transcription factor and cytokine expression, TBX21 and GZMB were more frequently expressed in clonally expanded compared with nonexpanded T cells (Fig. 1G, all parameters in Supplementary Fig. S3). Expression of the immune-checkpoint molecules PD-1, CTLA-4, and TIM-3 varied across patients but was not significantly different between expanded and nonexpanded T-cell clones and was generally low on CD8+ T-cell clones (Fig. 1E). In contrast, BTLA was expressed on more than 50% of T cells, irrespective of clonal expansion (Fig. 1E). Data on clone-associated immune phenotypes were consistent across technical replicates and not affected by total numbers of sequenced cells per patient (Supplementary Fig. S4). Taken together, immune phenotypes of expanded bone marrow T-cell clones were significantly different from those of nonexpanded clones and characteristic of CD8+ cytolytic effector differentiation.
Figure 1.
Clonal expansion–associated phenotypes of bone marrow T cells. A, Parallel determination of TCRαβ sequences, transcription factors, and cytokine gene expression from amplified cDNA of single FACS-sorted T cells. Single cells are arranged in columns with each column representing one cell. The top bar indicates TCR sequences; adjacent cells with the same color in the top bar indicate identical TCRαβ CDR3 amino acid sequences. Clonal expansion was defined as the detection of at least two cells with identical TCRαβ CDR3 sequences. The upper part of the heat map indicates single-cell gene expression determined by targeted panel sequencing. The lower part of the heat map visualizes corresponding FACS index sort data. The heat maps show data from patients MM160 and MM061 as examples for strong and weak clonal T-cell expansion, respectively. B, Absolute numbers of expanded bone marrow T-cell clones per patient (n = 13). Patients were ordered by size of the most expanded clone per patient as shown in D. C, Percentages of expanded T-cell clones expressing CD8 or CD4. Data points indicate individual patients. D, Frequencies of expanded CD8+ T-cell clones within CD8+ T cells. Each data point represents one unique T-cell clone. E, Immune phenotypes of CD8+ expanded and nonexpanded T-cell clones determined by FACS index sorting. Data from all 13 patients are summarized as box plots, and data points indicate individual patients. BTLA and CD28 were stained in n = 12; CD38, CTLA-4, and TIM-3 in 11; and CD39 in 3 (out of 13 patients). A clone was determined positive for the indicated parameter if the majority of cells belonging to this clone were positive. F, Gating example for patient MM160. Each data point represents one T-cell belonging to an expanded (black) or nonexpanded (red) clone. Numbers within gates indicate percentages. G, Single-cell gene expression of selected cytokines and transcription factors that were detected in more than 5% of expanded or nonexpanded CD8+ T cells. Data points represent individual patients. All panels show data from 13 patients, unless otherwise stated. Single-cell FACS and sequencing for each patient were done once resulting in 13 experiments. Box plots represent 25th to 75th percentiles; lines within boxes indicate medians. Statistical significance was calculated using the two-sided Wilcoxon test; *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Dominant bone marrow T-cell clones are present within peripheral blood TCRβ repertoires
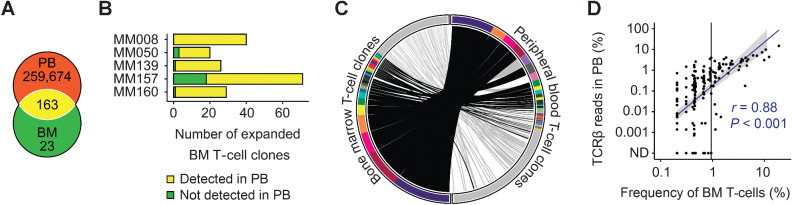
To investigate whether expansion was a feature of selected bone marrow T-cell clones, we determined the corresponding peripheral blood TCRβ repertoires from five patients with substantial clonal T-cell expansion in the bone marrow. We detected a total of 259,837 peripheral blood T-cell clones (on average 51,967 clones per patient). The most expanded peripheral blood clone of each patient accounted for on average 8.0% of reads (range, 4.0%–15.0%). 163 of 186 expanded bone marrow T-cell clones were also detectable in peripheral blood (Fig. 2A). On average, 90% of expanded bone marrow T-cell clones from each patient overlapped with peripheral blood (Fig. 2B), and clonal overlap within patient MM160 is detailed as an example (Fig. 2C). Clones with frequencies above 0.95% of total bone marrow T cells were always detectable and proportionally represented within peripheral blood TCRβ repertoires (Fig. 2D). Whether detection of dominant bone marrow T-cell clones in peripheral blood was due to hemodilution of bone marrow aspirate specimens or indicated physiologic circulation within peripheral blood cannot be concluded from our data. However, high frequencies of usually bone marrow–resident multiple myeloma plasma cells (on average 29.1%, range, 20.8%–50.4% of total cells) within the aspirate specimens confirmed high representation of cells of the bone marrow immune microenvironment (Supplementary Fig. S5).
Figure 2.
Circulation of expanded bone marrow T-cell clones in peripheral blood. A, Overlap of matched expanded bone marrow and total peripheral blood T-cell clones from 5 patients. Numbers indicate total numbers of T-cell clones. B, Total numbers of expanded bone marrow clones that were also detectable in peripheral blood for each individual patient. C, Left: single-cell TCRαβ sequencing data of bone marrow T cells from patient MM160. Cells that belonged to the same T-cell clone were arranged next to each other and represented by identical colors. Clones are ordered by size, and nonexpanded T cells with unique TCRαβ sequences are represented in gray. Right: corresponding peripheral blood TCRβ repertoire sequencing data. T-cell clones are ordered by TCRβ read number. Clones with a frequency below 0.3% are represented in gray. Links connect bone marrow T-cell clones to peripheral blood T-cell clones with identical CDR3β amino acid sequences (black links: expanded bone marrow T-cell clones, gray links: nonexpanded bone marrow T-cell clones). D, Spearman correlation of clonal T-cell expansion in bone marrow and peripheral blood for all 5 sample pairs combined. Bone marrow T-cell clones with a frequency above 0.95% (vertical black line) were always detectable in corresponding peripheral blood. The gray area indicates 95% confidence interval for an assumed linear correlation (blue line). Sequencing of bone marrow and peripheral blood T cells was done once for each patient resulting in 5 experiments. BM, bone marrow; PB, peripheral blood; ND, not detected.
Only 1 of 68 expanded bone marrow T-cell clones recognize antigen presented on multiple myeloma cells
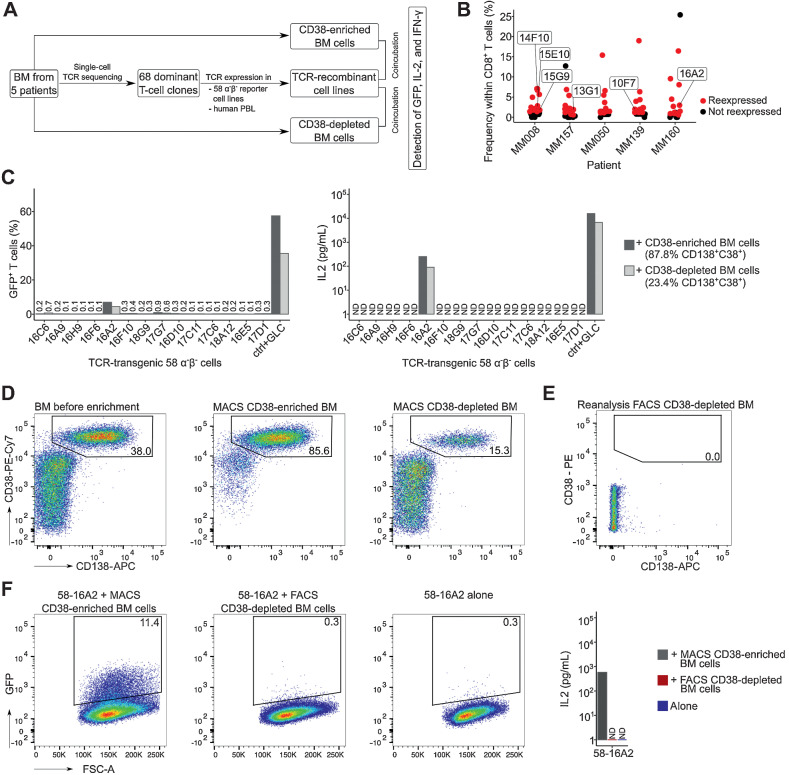
To determine whether the expansion of dominant bone marrow T-cell clones was driven by antigens selectively presented on multiple myeloma cells, we tested selected TCRs for recognition of CD38-enriched multiple myeloma and CD38-depleted nonmyeloma cells (Fig. 3A). Based on their frequencies, we chose a total of 68 dominant bone marrow T-cell clones from five patients with substantial clonal T-cell expansion (Fig. 3B; Supplementary Table S3). The selected clones accounted for, on average, 3.4% (range, 0.6%–19%) of CD8+ T cells. TCRs were expressed on 58α−β− reporter T cells and named “58-name of the TCR.” GFP expression and IL2 production indicated T-cell activation. When stimulated with plate-bound anti-CD3 (positive control), all TCR-recombinant cell lines produced GFP and IL2 (Supplementary Fig. S6). Upon incubation with their corresponding CD38-enriched multiple myeloma cells, three TCR-transgenic cell lines (58-14F10 and 58-15G9 from MM008, and 58-16A2 from MM160; Supplementary Table S3) produced GFP above background (58-16A2 in Fig. 3C, gating and all patients in Supplementary Fig. S7, coculture conditions in Supplementary Table S4). Significant IL2 production was observed only from 58-16A2. Activation of 58-16A2 with CD38-depleted nonmyeloma cells was due to the remaining 23.4% CD38++CD138+ multiple myeloma cells, and potentially other B-lineage cells, after magnetic cell separation (Supplementary Table S5), which was confirmed by repeating the enrichment (Fig. 3D) and eliminating the remaining multiple myeloma/B-lineage cells from MACS CD38-depleted bone marrow by subsequent FACS (Fig. 3E, gating in Supplementary Fig. S8). The resulting population was named “FACS CD38-depleted BM,” and neither GFP nor IL2 production could be detected upon incubation with 58-16A2; only CD38-enriched multiple myeloma cells (85.6% CD38++CD138+) activated 58-16A2 (Fig. 3F). The two other TCR-recombinant cell lines that potentially recognized multiple myeloma cells (58-15G9 and 58-14F10 from MM008) showed very low GFP expression (<1.2%) and did not produce IL2 upon incubation with CD38-enriched cells (Supplementary Fig. S7). Therefore, we could not confirm that these two T-cell clones were multiple myeloma reactive.
Figure 3.
Recognition of multiple myeloma cells by dominant bone marrow T-cell clones. A, Experimental setup: From five patients with substantial bone marrow T-cell expansion, 68 clones were selected for TCR expression in 58α−β− reporter T cells. TCR-recombinant cells were incubated with respective CD38-enriched multiple myeloma or CD38-depleted nonmyeloma cells for 16 hours at 37°C in 5% CO2 to determine antigen recognition. B, Patients and T-cell clones that were selected for expression (red) based on their frequencies. Data points indicate T-cell clones. Clones of mentioned in the manuscript are labeled. C, Incubation of TCR-recombinant 58α−β− reporter T cells with enriched multiple myeloma and nonmyeloma cells from patient MM160. 58α−β− cells expressing an HLA-A*02:01–restricted TCR specific for the EBV-derived peptide GLCTLVAML (GLC) were incubated for 16 hours at 37°C in 5% CO2 with myeloma and nonmyeloma cells from the same patient loaded with the target peptide as control (ctrl). GFP and murine IL2 were measured as readouts for T-cell activation. D, MACS CD38 enrichment and depletion of bone marrow from patient MM160. Cells were pregated on mononuclear cells. To remove remaining multiple myeloma cells (CD38++CD138+) from MACS CD38-depleted cells, (E) we performed subsequent FACS (CD19−CD38−CD138−, sorting gates in Supplementary Fig. S8), resulting in 0.0% remaining CD38++CD138+ multiple myeloma cells within the FACS CD38-depleted BM population. F, Repeated incubation of 58-16A2 with CD38-enriched and FACS CD38-depleted bone marrow from patient MM160 or 58-16A2 alone. Cells were pregated on single live murine CD3+ T cells after exclusion of human bone marrow cells by scatter characteristics, CD38, CD138, and CD45 staining. For each patient, incubations of reporter cells with CD38-enriched or depleted bone marrow cells were done in one experiment resulting in 5 experiments, unless stated otherwise. BM, bone marrow; PBL, peripheral blood lymphocytes; n.d., not detectable.
In contrast to the dominant immune phenotypes of expanded bone marrow T cells outlined above (CD57+PD-1−TIM-3−), 16A2 was CD57−TIM-3+ and 15G9 was PD-1+ (Supplementary Fig. S9). TCRβ CDR3 sequences of all three clones (16A2 from MM160; 15G9 and 14F10 from MM008) were detectable in peripheral blood TCRβ repertoires. To determine whether nonreactivity of the majority of reexpressed dominant TCRs with bone marrow cells was due to the sensitivity of 58α−β− reporter cell lines, a total of 21 TCRs from patients MM050 and MM139 were also expressed in healthy donor peripheral blood lymphocytes. None of the TCR-transduced lymphocytes recognized antigens selectively presented on multiple myeloma cells, confirming our results obtained with 58α−β− reporter cell lines (Supplementary Fig. S10).
Multiple myeloma–reactive expanded bone marrow T-cell clones recognize self-antigens
The identification of one expanded T-cell clone (16A2) that recognized CD38-enriched multiple myeloma cells and two additional clones (15G9 and 14F10) with potentially similar cell type specificities raised the question of which antigens caused their expansion in vivo. Epitopes derived from chronically persistent viruses and neoantigens have been shown to be drivers of clonal T-cell expansion (10, 37). Indeed, TCRα or β chains from 6 of the 68 selected dominant T-cell clones had been deposited in VDJdb and reported to be specific for cytomegalovirus (CMV)- or influenza A–derived epitopes; however, paired TCRαβ information was not publicly available (38). We identified the corresponding TCR chains and confirmed the specificity of three TCRs (15E10 from MM008, 10F7 from MM139, and 13G1 from MM157) for CMV epitopes (Supplementary Table S6; Supplementary Fig. S11). The epitope specificities of the remaining three TCRs (18A12 from MM160, 12D1 and 13E11 from MM157), suggested by VDJdb (32), could not be confirmed and remain unknown.
Regarding neoantigens, we determined somatic nonsynonymous mutations in multiple myeloma cells from nine selected patients that were representative of strong and weak clonal T-cell expansion. Neoantigen load and genes harboring mutations were representative of previously published data on treatment-naïve multiple myeloma (10), and potential neoantigen presentation was predicted bioinformatically (Supplementary Fig. S12; Supplementary Table S7). Multiple myeloma cells from patients MM008 and MM160 contained 10 and 7 possibly presented somatic mutations, respectively, resulting in a total of 22 potential neoepitopes (Supplementary Table S8, and Supplementary Appendix for detailed mutation data). However, neither the multiple myeloma-reactive TCR 16A2 nor the two potentially multiple myeloma-reactive TCRs 15G9 and 14F10 recognized these neoepitopes (Supplementary Fig. S13), suggesting that they may be specific for self-antigens or epitopes from persistent pathogens.
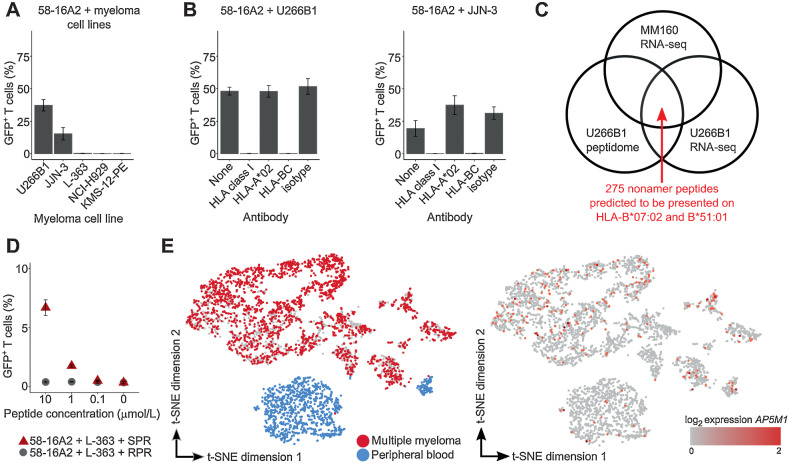
Considering self-antigens as targets, we hypothesized that TCRs 15G9, 14F10, and 16A2 could recognize broadly available multiple myeloma cell lines. Of the five multiple myeloma cell lines, 58-16A2 was activated by U266B1 and JJN-3 (Fig. 4A). 58-15G9 and 58-14F10 were not activated by any of the tested cell lines (Supplementary Fig. S14) and excluded from further analysis.
Figure 4.
TCR 16A2 recognizes the AP5M1-derived peptide SPRPPLISV (SPR). A and B, 3 × 104 58-16A2 were incubated with 5 × 104 cells of different multiple myeloma cell lines for 16 hours at 37°C 5% CO2. Blocking antibodies against HLA-class I, HLA-A*02, or HLA-BC were used at 20 μg/mL, 20 μg/mL, or 40 μg/mL, respectively. C, Potential target peptides of 16A2 were selected from a total of 3,686 nonamer peptides of the U266B1 peptidome applying the following criteria: (i) RNA expression within multiple myeloma cells of MM160 and U266B1, and (ii) presentation on HLA-B*07:02 and HLA-B*51:01. Peptides with predicted HLA binding affinity ≤ 700 nmol/L (NetMHC) were considered presented. D, L-363 cells that did not activate 58-16A2 without additional peptide loading were used as antigen-presenting cells. RPR (amino acid sequence: RPRPPVLSV) was used as a negative control peptide representative for all other 274 nonamer peptides that did not activate 58-16A2. E, Combined single-cell gene expression of CD38-enriched multiple myeloma bone marrow and peripheral blood cells from patient MM160. Each data point represents 1 of 4,430 single cells. Left: Multiple myeloma cells were identified by monoclonal immunoglobulin light chain CDR3 sequences. Right: AP5M1 expression was generally low and not restricted to multiple myeloma cells. A, B, and D indicate mean ± standard deviation. Data are representative of 3 experiments unless otherwise stated.
Surprisingly, no HLA-class I allele was shared between patient MM160, U266B1, and JJN-3 (Table 2). Nevertheless, blocking antibodies against HLA class I and HLA-BC completely inhibited 58-16A2 activation by U266B1 and JJN-3 (Fig. 4B), suggesting that the target antigen might be presented on the HLA-B*07 supertype, including HLA-B*07:02 and B*51:01, at least one of which was present in patient MM160, U266B1, and JJN-3.
Table 2.
HLA types of patient MM160 and multiple myeloma cell lines.
| HLA-class I | MM160 | U266B1 | JJN-3 | L-363 | NCI-H929 | KMS-12-PE |
|---|---|---|---|---|---|---|
| A | 02:01; 23:01 | 02:01; 03:01 | 03:01; 33:01 | 02:01; 31:01 | 03:01; 24:02 | 26:01; 33:03 |
| B | 44:03; 51:01 | 07:02; 40:01 | 07:02; 14:02 | 07:02; 40:01 | 07:02; 18:01 | 35:01; 44:03 |
| C | 15:02; 04:09N | 03:04; 07:02 | 07:02; 08:02 | 03:04; 07:02 | 07:02; 07:02 | 03:03; 14:03 |
Note: HLA type of MM160 was determined by sequencing. All cell line HLA types were downloaded from the TRON Cell Line Portal (http://celllines.tron-mainz.de/).
To identify potential target epitopes of TCR 16A2, we used peptidome data of U266B1 in combination with gene-expression data from MM160 and U266B1 applying the following criteria: the potential target epitope should be (i) part of the HLA class I peptidome of U266B1, (ii) presented on the HLA-B*07 supertype, and (iii) expressed in MM160 and U266B1 (schematic in Fig. 4C). A total of 275 nonamer peptides met these criteria (Supplementary Table S9), were synthetized, loaded on L-363 as HLA-B*07:02-expressing antigen-presenting cells, and tested for activation of 58-16A2. Only SPRPPLISV (SPR), which is part of Adapter Related Protein Complex 5 Subunit Mu 1 (AP5M1), was recognized by 58-16A2 (Fig. 4D). Although 58-16A2 was selectively activated by multiple myeloma/B-lineage cells, AP5M1 expression, determined by single-cell gene expression in patient MM160 and one additional patient as an example, was generally low and not restricted to plasma cell compartments (Fig. 4E; Supplementary Fig. S15).
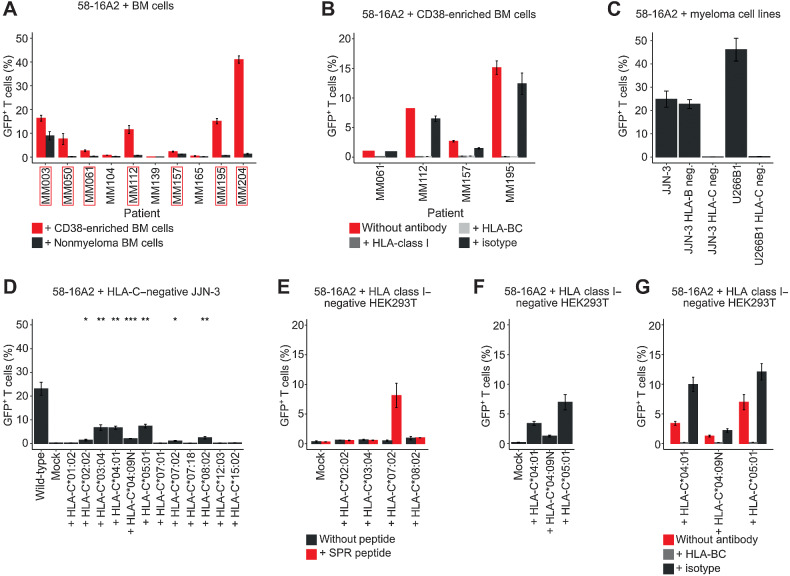
TCR 16A2 recognizes multiple myeloma cells from additional patients across a variety of HLA-C alleles
Having identified SPR as a potential self-antigen–derived target epitope, we investigated whether TCR 16A2 could recognize multiple myeloma cells in patients other than MM160. Multiple myeloma cells from 7 of 10 additionally tested patients activated 58-16A2 (MM003, MM050, MM061, MM112, MM157, MM195, and MM204; Fig. 5A). Nonmyeloma cells did not activate 58-16A2, except for patient MM003, of whom we did not have enough bone marrow to remove remaining multiple myeloma/B-lineage cells from the CD38-depleted population by FACS. 58-16A2 activation did not follow the expected HLA-B*07 supertype pattern (Table 1) but could be blocked by antibodies against HLA-class I and HLA-BC as shown for four patients as examples (Fig. 5B), leading us to question the assumed HLA-B*07 supertype restriction.
Figure 5.
16A2 is a degenerate TCR that recognizes multiple myeloma cells of different patients in the context of a variety of HLA-C alleles. A, 58-16A2 were incubated with CD38-enriched multiple myeloma or MACS CD38-depleted nonmyeloma cells. Axis descriptions of patients who activated 58-16A2 are highlighted by red rectangles. “Nonmyeloma BM cells” represent CD38-depleted bone marrow (BM). In patients MM157 and MM195, remaining multiple myeloma/B-lineage cells were removed from MACS CD38-depleted bone marrow by additional FACS sorting (sorting criteria: CD38−CD138−CD19−, gating in Supplementary Fig. S8). B, 58-16A2 were incubated for 16 hours at 37°C in 5% CO2 with multiple myeloma cells from four patients in the presence of blocking antibodies against HLA-class I or HLA-BC. C, 58-16A2 were incubated with JJN-3 or U266B1 cells for 16 hours at 37°C in 5% CO2. HLA-B or HLA-C were deleted using CRISPR-Cas9. D, 58-16A2 were incubated for 16 hours at 37°C in 5% CO2 with HLA-C–negative JJN-3 transiently transfected with selected HLA-C alleles. Wild-type, unmanipulated JJN-3 expressing all endogenous HLA alleles; mock, nucleofection of HLA-C–negative JJN-3 without HLA-containing plasmid. Statistical significance between mock and indicated conditions was determined by paired Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. E–G, Selected HLA-C alleles were expressed in HLA-class I–negative HEK293T cells and incubated with 58-16A2 for 16 hours at 37°C in 5% CO2. Mock, transfection without HLA-containing plasmid. E, HLA-recombinant HEK293T were also loaded with SPR. G, Blocking antibodies against HLA-BC or an isotype antibody were added before coincubation. Bar charts indicate mean ± standard deviation. Data are representative of 3 experiments.
To systematically determine the HLA-restriction of TCR 16A2, we incubated 58-16A2 with U266B1 and JJN-3 after CRISPR-Cas9–mediated deletion of HLA-B or HLA-C. Only HLA-C, but not HLA-B, deletion abolished activation of 58-16A2 (Fig. 5C), excluding the previously assumed HLA-B restriction of TCR 16A2. Subsequently, we transiently reexpressed all HLA-C alleles from all patients and cell lines that activated 58-16A2 in HLA-C-negative JJN-3. We included the null allele HLA-C*04:09N because it was present in patient MM160. On average, 17.3% of JJN-3 were transiently transfected with the respective HLA-C alleles (Supplementary Fig. S16), and activation of 58-16A2 required expression of at least one of the following HLA-C alleles: 02:02, 03:04, 04:01, 04:09N, 05:01, 07:02, or 08:02, indicated by significant GFP expression compared with mock-transfected HLA-C–negative JJN-3 (Fig. 5D). Accordingly, each of the eight patients whose multiple myeloma cells were recognized by 58-16A2 expressed at least one of these HLA-C alleles (Table 1). Because we identified an array of HLA-C alleles that could present antigens to 58-16A2, we asked, whether (i) activation was due to allo-reactivity and (ii) TCR 16A2 could recognize the previously identified target peptide, SPR, when presented on HLA-C. All HLA-C alleles that led to activation of 58-16A2 when expressed on JJN-3 were individually expressed in HLA class I–negative HEK293T (Supplementary Fig. S17 for HLA class I knockout) and incubated with 58-16A2. Only HLA-C*07:02, which was not expressed in patient MM160, presented SPR and activated 58-16A2 (Fig. 5E). Expression of HLA-C*02:02, 03:04, or 08:02 did not result in activation of 58-16A2 with or without SPR peptide. Expression of HLA-C*04:01, 04:09N, and 05:01 resulted in activation of 58-16A2 irrespective of SPR (Fig. 5F), suggesting allo- and/or self-peptide reactivity. Again, 58-16A2 activation in context of HLA-C*04:01, 04:09N, and 05:01 could be blocked with antibodies against HLA-BC (Fig. 5G). In summary, 16A2 is a degenerate TCR that was activated by multiple myeloma cells of multiple patients in context of a variety of HLA-C alleles. None of the dominant TCRs that recognized multiple myeloma cells could be confirmed to be neoantigen specific.
Discussion
Major findings of our study included (i) clonal expansion occurred within cytolytic effector T-cell compartments, (ii) dominant T-cell clones rarely recognized multiple myeloma cells, (iii) multiple myeloma–reactive TCRs of dominant T-cell clones were not neoantigen-specific, and (iv) 16A2 is a self-reactive, degenerate TCR that recognizes antigens presented on multiple HLA-C alleles. Clonal T-cell expansion predominantly occurred within CD8+ T cells, which is in line with findings on tumor-infiltrating T cells in other malignancies (21, 39). We confirmed that bone marrow T cells in multiple myeloma showed exhausted and cytotoxic phenotypes (40–42). We also determined the effector memory RA differentiation (CD45RA+CCR7−) of clonally expanded bone marrow T-cell compartments, which has been associated with potent effector functions (43).
Data on immune-checkpoint molecule expression in multiple myeloma are controversial (42, 44). We showed that PD-1, TIM-3, and CTLA-4 were expressed on a minority of expanded T-cell clones, and only 1 of 68 dominant clones reproducibly recognized multiple myeloma cells, providing experimental context for results of clinical trials applying immune-checkpoint blockade in multiple myeloma (13–16, 44, 45). However, TIM-3 expression on one multiple myeloma–reactive dominant T-cell clone suggests that immune-checkpoint modulation could be therapeutically relevant in carefully selected conditions. Future T cell–based immunotherapy approaches will potentially target low-abundance clones and include strategies in addition to disinhibition of preexisting neoantigen-specific T cells.
Almost all dominant bone marrow T-cell clones were detectable in peripheral blood. Parallel aspiration of peripheral blood during bone marrow acquisition or circulation of bone marrow T cells in peripheral blood could be a possible explanation. Our study focused on expanded bone marrow T-cell clones, irrespective of overlap with peripheral blood as (i) hematopoiesis takes place in the bone marrow, (ii) circulation in peripheral blood does not exclude TCR specificity for antigens presented in the bone marrow, (iii) neoantigen-specific T-cell clones can be present in bone marrow and peripheral blood alike (10), and (iv) frequencies of multiple myeloma cells within the investigated bone marrow specimens indicated high representation of cells of the bone marrow microenvironment.
Besides potential specificity against multiple myeloma cells, bone marrow is a site of T- and B-cell memory (46), and major proportions of expanded bone marrow and circulating T-cell clones can be specific for persistent pathogens (47, 48). Most patients in our study had been in contact with Epstein–Barr virus (EBV) and CMV, and we confirmed that three expanded bone marrow T-cell clones were CMV specific. Considering data from other studies and concepts of immunologic memory (9, 46), we expected the number of virus-specific TCRs in our data set to be considerably higher.
None of the dominant T-cell clones that recognized multiple myeloma cells could be confirmed to be neoantigen specific. Neoantigen-specific T cells have mostly been identified in malignancies with high mutational burden (e.g., malignant melanoma; refs. 6, 9), and it is possible that higher neoantigen loads in pretreated multiple myeloma could result in higher frequencies of neoantigen-specific T cells among dominant clones. We deliberately studied treatment-naïve patients to avoid therapy-induced effects on T-cell phenotypes and TCR repertoires. The number of somatic mutations within our treatment-naïve cohort was relatively low, representative of data from larger studies (10, 49–51), and could be a reason for the absence of neoantigen-specific T cells within dominant clones. We assume multiple myeloma-reactive T cells to be part of less expanded bone marrow clones; however, their frequencies cannot be concluded from our study, which was specifically designed to define phenotypes and TCR sequences of dominant clones at the single-cell level. The presence of multiple myeloma-reactive T cells within less expanded compartments is further suggested by therapeutic effects of adoptively transferred, previously in vitro activated and expanded, bone marrow-infiltrating lymphocytes (52). In patients refractory to multiple lines of therapy, neoantigen-specific T cells have been shown to become detectable in bone marrow and peripheral blood (10, 53). Furthermore, immunoediting is likely to affect frequencies of neoantigen-specific T-cell clones (54). Potentially immunogenic mutations and multiple myeloma-specific T-cell clones should be studied in parallel at different stages of disease development including monoclonal gammopathy of undetermined significance, smoldering myeloma, treatment-naïve and pretreated multiple myeloma in future studies.
One expanded T-cell clone (16A2) could be confirmed as multiple myeloma–reactive and was studied in detail. Whether the target antigen of TCR 16A2 was specifically presented on multiple myeloma cells cannot be concluded from our data. Especially in cases where we depleted the remaining multiple myeloma cells by FACS, we eliminated the entire B-lineage compartment. CD19 was included as a sorting parameter because multiple myeloma cells can be CD19+ even if the majority are CD19− (55, 56). However, TCR 16A2 could not be activated by cells other than B-lineage cells.
Using HLA knockout and recombinant HLA expression, we excluded the initially assumed HLA-B*07 supertype restriction (57) of TCR 16A2 and identified the following activation characteristics: (i) peptide-specific activation by SPR presented on HLA-C*07:02; (ii) activation by unknown target peptides presented on HLA-C*02:02, 03:04, 08:02; and (iii) alloreactivity against HLA-C*04:01 and 05:02. Activation by SPR required high peptide concentrations and presentation on HLA-C*07:02, which was not present in patient MM160, questioning its role as a relevant target epitope in vivo. HLA-C*04:09N was the only HLA-C allele in patient MM160 that led to 58-16A2 activation when expressed on HLA-C-negative JJN-3 cells. HLA-C*04:09N is identical to HLA-C*04:01, except for a frameshift mutation in its cytoplasmic domain, which has been assumed to lead to intracellular, but not cell surface, HLA expression (58). However, HLA-C*04:09N expression in HLA-C–negative JJN-3 and HLA class I–negative HEK293T cells resulted in 58-16A2 activation, which could be blocked by HLA-BC–specific antibodies, suggesting TCR–HLA–peptide interaction at the cell surface. It is possible that HLA-C*04:09N can become accessible for antigen presentation in situations of high cellular turnover and/or on selected cell types, including multiple myeloma plasma cells, which have extraordinarily high secretory activity. Independent of the role of HLA-C*04:09N, recognition of multiple antigens in context of a variety of HLA-C alleles characterizes 16A2 as a profoundly degenerate TCR that, in patient MM160, has potentially been functionally inhibited by TIM-3–expression in vivo.
In conclusion, our study provides insights into immune phenotypes and targets of clonally expanded bone marrow T cells in treatment-naïve multiple myeloma. Dominant clones rarely recognized multiple myeloma cells and, in most cases, were not inhibited by expression of classic immune-checkpoint molecules.
Supplementary Material
Acknowledgments
We thank Kirstin Rautenberg at MDC Berlin for expert help with FACS sorting, Josefin Garmshausen for technical assistance, and Vivien Boldt for providing fluorescence in situ hybridization information. This work was supported by Deutsche Krebshilfe e.V. (70113355; L. Hansmann), Berliner Krebsgesellschaft e.V. (HAFF202013 MM; L. Hansmann), the German Cancer Consortium (DKTK; L. Hansmann), the European Union (ERC Advanced Grant 882963; T. Blankenstein), a research fellowship from the German Research Foundation (DFG, PE 3127/1-1; L. Penter), and DFG EXC 2145 (SyNergy) ID 390857198 (K. Dornmair). L. Penter is a scholar of the American Society of Hematology.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
L. Bullinger reports personal fees from AbbVie, Amgen, Astellas, Bristol Myers Squibb, Celgene, Daiichi Sankyo, Gilead, Hexal, Janssen, Jazz Pharmaceuticals, Menarini, Novartis, Pfizer, Sanofi, and Seattle Genetics and grants from Bayer and Jazz Pharmaceuticals outside the submitted work. A. Moosmann reports grants from DZIF–German Center for Infection Research and grants from Wilhelm Sander-Stiftung during the conduct of the study; grants from BioSynGen Pte. Ltd. outside the submitted work. L. Hansmann reports grants from Deutsche Krebshilfe e.V. and Berliner Krebsgesellschaft during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
C. Welters: Conceptualization, investigation, visualization, methodology, writing–original draft, writing–review and editing. M.F. Lammoglia Cobo: Formal analysis, investigation. C. Stein: Software, formal analysis, visualization. M.-T. Hsu: Investigation, methodology. A. Ben Hamza: Investigation, visualization. L. Penter: Software, visualization. X. Chen: Investigation, methodology. C. Buccitelli: Software, formal analysis, methodology. O. Popp: Software, formal analysis, methodology. P. Mertins: Software, formal analysis, methodology. K. Dietze: Investigation, methodology. L. Bullinger: Resources, formal analysis. A. Moosmann: Resources, formal analysis. E. Blanc: Data curation, software, formal analysis, methodology. D. Beule: Data curation, software, formal analysis, methodology. A. Gerbitz: Formal analysis. J. Strobel: Formal analysis, investigation, methodology. H. Hackstein: Formal analysis, investigation, methodology. H. Rahn: Investigation, methodology. K. Dornmair: Resources, investigation, methodology, writing–review and editing. T. Blankenstein: Resources, formal analysis, supervision, methodology, writing–review and editing. L. Hansmann: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Mitsiades CS, Mitsiades NS, Richardson PG, Munshi NC, Anderson KC. Multiple myeloma: a prototypic disease model for the characterization and therapeutic targeting of interactions between tumor cells and their local microenvironment. J Cell Biochem 2007;101:950–68. [DOI] [PubMed] [Google Scholar]
- 2. Hansmann L, Blum L, Ju CH, Liedtke M, Robinson WH, Davis MM. Mass cytometry analysis shows that a novel memory phenotype B cell is expanded in multiple myeloma. Cancer Immunol Res 2015;3:650–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mitsiades CS, Mitsiades N, Munshi NC, Anderson KC. Focus on multiple myeloma. Cancer Cell 2004;6:439–44. [DOI] [PubMed] [Google Scholar]
- 4. Cavo M, Pantani L, Pezzi A, Petrucci MT, Patriarca F, Di Raimondo F, et al. Bortezomib-thalidomide-dexamethasone (VTD) is superior to bortezomib-cyclophosphamide-dexamethasone (VCD) as induction therapy prior to autologous stem cell transplantation in multiple myeloma. Leukemia 2015;29:2429–31. [DOI] [PubMed] [Google Scholar]
- 5. Moreau P, Attal M, Hulin C, Arnulf B, Belhadj K, Benboubker L, et al. Bortezomib, thalidomide, and dexamethasone with or without daratumumab before and after autologous stem-cell transplantation for newly diagnosed multiple myeloma (CASSIOPEIA): a randomised, open-label, phase 3 study. Lancet 2019;394:29–38. [DOI] [PubMed] [Google Scholar]
- 6. Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 2016;22:433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gros A, Tran E, Parkhurst MR, Ilyas S, Pasetto A, Groh EM, et al. Recognition of human gastrointestinal cancer neoantigens by circulating PD-1+ lymphocytes. J Clin Invest 2019;129:4992–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malekzadeh P, Yossef R, Cafri G, Paria BC, Lowery FJ, Jafferji M, et al. Antigen experienced T cells from peripheral blood recognize p53 neoantigens. Clin Cancer Res 2020;26:1267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oliveira G, Stromhaug K, Klaeger S, Kula T, Frederick DT, Le PM, et al. Phenotype, specificity and avidity of antitumour CD8(+) T cells in melanoma. Nature 2021;596:119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perumal D, Imai N, Lagana A, Finnigan J, Melnekoff D, Leshchenko VV, et al. Mutation-derived neoantigen-specific T-cell responses in multiple myeloma. Clin Cancer Res 2020;26:450–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dhodapkar MV, Krasovsky J, Osman K, Geller MD. Vigorous premalignancy-specific effector T cell response in the bone marrow of patients with monoclonal gammopathy. J Exp Med 2003;198:1753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suen H, Brown R, Yang S, Weatherburn C, Ho PJ, Woodland N, et al. Multiple myeloma causes clonal T-cell immunosenescence: identification of potential novel targets for promoting tumour immunity and implications for checkpoint blockade. Leukemia 2016;30:1716–24. [DOI] [PubMed] [Google Scholar]
- 14. Lesokhin AM, Ansell SM, Armand P, Scott EC, Halwani A, Gutierrez M, et al. Nivolumab in patients with relapsed or refractory hematologic malignancy: preliminary results of a phase Ib study. J Clin Oncol 2016;34:2698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mateos MV, Blacklock H, Schjesvold F, Oriol A, Simpson D, George A, et al. Pembrolizumab plus pomalidomide and dexamethasone for patients with relapsed or refractory multiple myeloma (KEYNOTE-183): a randomised, open-label, phase 3 trial. Lancet Haematol 2019;6:e459–e69. [DOI] [PubMed] [Google Scholar]
- 16. Usmani SZ, Schjesvold F, Oriol A, Karlin L, Cavo M, Rifkin RM, et al. Pembrolizumab plus lenalidomide and dexamethasone for patients with treatment-naive multiple myeloma (KEYNOTE-185): a randomised, open-label, phase 3 trial. Lancet Haematol 2019;6:e448-e58. [DOI] [PubMed] [Google Scholar]
- 17. Racanelli V, Leone P, Frassanito MA, Brunetti C, Perosa F, Ferrone S, et al. Alterations in the antigen processing-presenting machinery of transformed plasma cells are associated with reduced recognition by CD8+ T cells and characterize the progression of MGUS to multiple myeloma. Blood 2010;115:1185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giannopoulos K, Kaminska W, Hus I, Dmoszynska A. The frequency of T regulatory cells modulates the survival of multiple myeloma patients: detailed characterisation of immune status in multiple myeloma. Br J Cancer 2012;106:546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol 2014;32:684–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Penter L, Dietze K, Bullinger L, Westermann J, Rahn HP, Hansmann L. FACS single cell index sorting is highly reliable and determines immune phenotypes of clonally expanded T cells. Eur J Immunol 2018;48:1248–50. [DOI] [PubMed] [Google Scholar]
- 21. Penter L, Dietze K, Ritter J, Lammoglia Cobo MF, Garmshausen J, Aigner F, et al. Localization-associated immune phenotypes of clonally expanded tumor-infiltrating T cells and distribution of their target antigens in rectal cancer. Oncoimmunology 2019;8:e1586409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ballhausen A, Ben Hamza A, Welters C, Dietze K, Bullinger L, Rahn HP, et al. Immune phenotypes and checkpoint molecule expression of clonally expanded lymph node-infiltrating T cells in classical Hodgkin lymphoma. Cancer Immunol Immunother 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, e-prints p.arXiv:1303.3997. [Google Scholar]
- 24. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016;32:511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siewert K, Malotka J, Kawakami N, Wekerle H, Hohlfeld R, Dornmair K. Unbiased identification of target antigens of CD8+ T cells with combinatorial libraries coding for short peptides. Nat Med 2012;18:824–8. [DOI] [PubMed] [Google Scholar]
- 28. Engels B, Cam H, Schuler T, Indraccolo S, Gladow M, Baum C, et al. Retroviral vectors for high-level transgene expression in T lymphocytes. Hum Gene Ther 2003;14:1155–68. [DOI] [PubMed] [Google Scholar]
- 29. Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res 2006;66:8878–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lammoglia Cobo MF, Welters C, Rosenberger L, Leisegang M, Dietze K, Pircher C, et al. Rapid single-cell identification of Epstein-Barr virus-specific T-cell receptors for cellular therapy. Cytotherapy 2022;24:818–26. [DOI] [PubMed] [Google Scholar]
- 31. Quentmeier H, Pommerenke C, Dirks WG, Eberth S, Koeppel M, MacLeod RAF, et al. The LL-100 panel: 100 cell lines for blood cancer studies. Sci Rep 2019;9:8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walz S, Stickel JS, Kowalewski DJ, Schuster H, Weisel K, Backert L, et al. The antigenic landscape of multiple myeloma: mass spectrometry (re)defines targets for T-cell-based immunotherapy. Blood 2015;126:1203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khodadoust MS, Olsson N, Wagar LE, Haabeth OA, Chen B, Swaminathan K, et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 2017;543:723–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2007;2:1896–906. [DOI] [PubMed] [Google Scholar]
- 35. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- 36. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 37. Messaoudi I, Lemaoult J, Guevara-Patino JA, Metzner BM, Nikolich-Zugich J. Age-related CD8 T cell clonal expansions constrict CD8 T cell repertoire and have the potential to impair immune defense. J Exp Med 2004;200:1347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res 2020;48:D1057-D62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lucca LE, Axisa PP, Lu B, Harnett B, Jessel S, Zhang L, et al. Circulating clonally expanded T cells reflect functions of tumor-infiltrating T cells. J Exp Med 2021;218:e20200921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paiva B, Azpilikueta A, Puig N, Ocio EM, Sharma R, Oyajobi BO, et al. PD-L1/PD-1 presence in the tumor microenvironment and activity of PD-1 blockade in multiple myeloma. Leukemia 2015;29:2110–3. [DOI] [PubMed] [Google Scholar]
- 41. Raitakari M, Brown RD, Sze D, Yuen E, Barrow L, Nelson M, et al. T-cell expansions in patients with multiple myeloma have a phenotype of cytotoxic T cells. Br J Haematol 2000;110:203–9. [DOI] [PubMed] [Google Scholar]
- 42. Zelle-Rieser C, Thangavadivel S, Biedermann R, Brunner A, Stoitzner P, Willenbacher E, et al. T cells in multiple myeloma display features of exhaustion and senescence at the tumor site. J Hematol Oncol 2016;9:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol 2005;175:5895–903. [DOI] [PubMed] [Google Scholar]
- 44. Suen H, Brown R, Yang S, Ho PJ, Gibson J, Joshua D. The failure of immune checkpoint blockade in multiple myeloma with PD-1 inhibitors in a phase 1 study. Leukemia 2015;29:1621–2. [DOI] [PubMed] [Google Scholar]
- 45. Oliva S, Troia R, D'Agostino M, Boccadoro M, Gay F. Promises and pitfalls in the use of PD-1/PD-L1 inhibitors in multiple myeloma. Front Immunol 2018;9:2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang HD, Radbruch A. Maintenance of quiescent immune memory in the bone marrow. Eur J Immunol 2021;51:1592–601. [DOI] [PubMed] [Google Scholar]
- 47. Hislop AD, Annels NE, Gudgeon NH, Leese AM, Rickinson AB. Epitope-specific evolution of human CD8(+) T cell responses from primary to persistent phases of Epstein-Barr virus infection. J Exp Med 2002;195:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol 2007;25:587–617. [DOI] [PubMed] [Google Scholar]
- 49. Kortum KM, Langer C, Monge J, Bruins L, Zhu YX, Shi CX, et al. Longitudinal analysis of 25 sequential sample-pairs using a custom multiple myeloma mutation sequencing panel (M(3)P). Ann Hematol 2015;94:1205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leich E, Weissbach S, Klein HU, Grieb T, Pischimarov J, Stuhmer T, et al. Multiple myeloma is affected by multiple and heterogeneous somatic mutations in adhesion- and receptor tyrosine kinase signaling molecules. Blood Cancer J 2013;3:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies FE, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018;132:587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Noonan KA, Huff CA, Davis J, Lemas MV, Fiorino S, Bitzan J, et al. Adoptive transfer of activated marrow-infiltrating lymphocytes induces measurable antitumor immunity in the bone marrow in multiple myeloma. Sci Transl Med 2015;7:288ra78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noonan K, Matsui W, Serafini P, Carbley R, Tan G, Khalili J, et al. Activated marrow-infiltrating lymphocytes effectively target plasma cells and their clonogenic precursors. Cancer Res 2005;65:2026–34. [DOI] [PubMed] [Google Scholar]
- 54. Nakamura K, Smyth MJ, Martinet L. Cancer immunoediting and immune dysregulation in multiple myeloma. Blood 2020;136:2731–40. [DOI] [PubMed] [Google Scholar]
- 55. Hansmann L, Han A, Penter L, Liedtke M, Davis MM. Clonal expansion and interrelatedness of distinct B-lineage compartments in multiple myeloma bone marrow. Cancer Immunol Res 2017;5:744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nerreter T, Letschert S, Gotz R, Doose S, Danhof S, Einsele H, et al. Super-resolution microscopy reveals ultra-low CD19 expression on myeloma cells that triggers elimination by CD19 CAR-T. Nat Commun 2019;10:3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol 2008;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Balas A, Santos S, Aviles MJ, Garcia-Sanchez F, Lillo R, Alvarez A, et al. Elongation of the cytoplasmic domain, due to a point deletion at exon 7, results in an HLA-C null allele, Cw*0409 N. Tissue Antigens 2002;59:95–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed TCR sequence and mutation data can be found in the supplementary data. Whole-exome and RNA-sequencing raw data are publicly available at sequence read archive (https://www.ncbi.nlm.nih.gov/sra under BioProject accession number PRJNA862671).