The tumor microenvironment (TME) of clear cell ovarian cancer is characterized. Immune infiltration and collagen deposition differ based on disease stage, TME localization, and ARID1A status, highlighting the necessity of evaluating multiple features when determining treatment strategy.
Abstract
Some patients with advanced clear-cell ovarian cancer (CCOC) respond to immunotherapy; however, little is known about the tumor microenvironment (TME) of this relatively rare disease. Here, we describe a comprehensive quantitative and topographical analysis of biopsies from 45 patients, 9 with Federation Internationale des Gynaecologistes et Obstetristes (FIGO) stage I/II (early CCOC) and 36 with FIGO stage III/IV (advanced CCOC). We investigated 14 immune cell phenotype markers, PD-1 and ligands, and collagen structure and texture. We interrogated a microarray data set from a second cohort of 29 patients and compared the TMEs of ARID1A-wildtype (ARID1Awt) versus ARID1A-mutant (ARID1Amut) disease. We found significant variations in immune cell frequency and phenotype, checkpoint expression, and collagen matrix between the malignant cell area (MCA), leading edge (LE), and stroma. The MCA had the largest population of CD138+ plasma cells, the LE had more CD20+ B cells and T cells, whereas the stroma had more mast cells and αSMA+ fibroblasts. PD-L2 was expressed predominantly on malignant cells and was the dominant PD-1 ligand. Compared with early CCOC, advanced-stage disease had significantly more fibroblasts and a more complex collagen matrix, with microarray analysis indicating “TGFβ remodeling of the extracellular matrix” as the most significantly enriched pathway. Data showed significant differences in immune cell populations, collagen matrix, and cytokine expression between ARID1Awt and ARID1Amut CCOC, which may reflect different paths of tumorigenesis and the relationship to endometriosis. Increased infiltration of CD8+ T cells within the MCA and CD4+ T cells at the LE and stroma significantly associated with decreased overall survival.
Introduction
Advanced clear-cell ovarian cancer (CCOC) has low response rates to current treatments, especially in the recurrent setting (1). Some patients with recurrent, heavily pretreated CCOC may benefit from treatment with programmed cell death protein 1 (PD-1) checkpoint inhibition (2, 3), with a number of clinical trials currently evaluating the response to this immunotherapy. Data on the tumor microenvironment (TME) of CCOC are limited, making it difficult to develop hypotheses as to why some patients respond to checkpoint blockade while others do not.
ARID1A (AT-rich interaction domain 1A, BAF250a) is mutated in up to 67% of CCOC cases (4). In the preclinical setting, ARID1A has been linked to effective functioning of the mismatch repair protein MSH2 due to its role in facilitating the interaction with chromatin during DNA replication and repair (5), high expression of programmed death-ligand 1 (PD-L1; ref. 6), and increased IL6 production (7), suggesting a number of potential mechanisms by which loss of ARID1A function can impact the TME.
In this study, we performed a quantitative, qualitative, and topographical analysis of the TME of advanced CCOC. We then compared the TME of advanced CCOC with that of early-stage disease to understand the changes that occurred, which may facilitate immune evasion and metastatic dissemination. Finally, we explored the impact of ARID1A mutation on the TME of advanced CCOC.
Materials and Methods
Biobank samples
Formalin-fixed, paraffin-embedded (FFPE) samples, which were surplus to diagnostic and therapeutic requirements, were collected from 49 patients with CCOC under the Barts Gynae Tissue Bank HTA license number 12199 (REC nos.: 10/H0304/14 and 15/EE/0131). Studies were reviewed by Barts Gynae Tissue Bank Institutional Review Board and were conducted in accordance with the Declaration of Helsinki and the International Ethical Guidelines for Biomedical Research Involving Human Subjects. Samples were sourced from patients who had given written consent from Barts Cancer Institute (n = 15), University College London Hospital (n = 14), and the Northern Ireland Biobank (n = 20). All specimens were reviewed locally by a pathologist to ensure clear-cell morphology and tumor burden; four samples with low tumor burden were excluded from subsequent analysis. Federation Internationale des Gynaecologistes et Obstetristes (FIGO) stages I–II were termed early-stage CCOC (n = 9), and FIGO stages III–IV were advanced-stage CCOC (n = 36); clinical information and overall survival (OS) data were available for 38 patients. Because of a limited number of FFPE slides available per patient, we randomly allocated samples into cohorts to allow immunohistological profiling of each immune marker (Supplementary Table S1).
IHC
IHC was carried out on 4-μm-thick FFPE tissue sections. Antibodies and their respective concentrations, incubation times, antigen retrievals, and control tissues are shown in Supplementary Table S2. The ARID1A antibody, ab182561, which has 100% concordance between ARID1A mutational status and protein expression (8), was used to identify tumors which were ARID1A-wildtype (ARID1Awt) or ARID1A-mutant (ARID1Amut). Slides were deparaffinized in xylene (Thermo Fisher Scientific, 10588070) and rehydrated with successive mixtures of ethanol (Thermo Fisher Scientific, E/0650DF/17) of decreasing concentration (100%, 95%, 90%, 70%, and 50%), each for 2 minutes, followed by a wash in water. Where antigen retrieval was used, the slides were placed in an antigen retrieval solution in a water bath at the target temperature for the specified duration. A pH9 antigen retrieval solution was created with 1 mL Antigen Retrieval Buffer 100x Tris-EDTA buffer (Abcam, ab93684) mixed with 100 mL distilled water. A pH6 antigen retrieval solution was achieved by mixing 2.5 mL of citrate-based antigen unmasking solution (Vector Labs, H-3300) with 246.5 mL of distilled water. When removed from the water bath, the slides were placed under cold water for 5 minutes, followed by three washes with PBS and then were placed in a hydrogen peroxidase (Thermo Fisher Scientific, 10687022) block. The slides were then washed once in PBST (PBS with Tween 20; Sigma-Aldrich, P7949) and twice in PBS before both an avidin and biotin block (Vector Labs, SP-2001) are applied, each for 10 minutes. After this, 200 μL/slide of PBS + 2.5% BSA (Merck, A2058) + 2.5% goat serum (Gibco, 16210064) was applied for 45 minutes. Primary antibody was then applied. After incubation, the slides were washed once with PBST and twice more with PBS alone. The appropriate secondary antibody was then applied for 45 minutes. All secondary antibodies were diluted to 1:200 concentration and matched to the appropriate primary host species using either biotinylated rabbit anti-goat IgG (Vector; BA-5000), Biotinylated Goat Anti-Rabbit IgG (Vector; BA-1000) or Biotinylated Goat Anti-Mouse IgG (Vector; BA-900). Slides were washed once with PBST and twice with PBS before 200 μL/slide of ultrasensitive ABC staining kit solution (Thermo Fisher Scientific, 32050) was applied for 30 minutes. After three washes with PBS, 200 μL/slide of DAB+ substrate/chromagen (DAKO, K3468) was applied. Counterstain was with 100% Gills I hematoxylin (GHS116-500ML) before dehydration with successive mixtures of ethanol of increasing concentration. All slides were scanned a Pannoramic 250 High Throughput Scanner.
Hematoxylin and eosin staining
Hematoxylin and eosin (H&E) staining performed using the Lecia Autostainer XL. FFPE sections were deparaffinized in xylene (Thermo Fisher Scientific, X/0200/17) then rehydrated with successive mixtures of ethanol (Thermo Fisher Scientific, M/455/17) of decreasing concentrations (100%, 90%, 70%) followed by wash in water. Slides were submerged in 1% acid alcohol (Lecia, 3803651E was), followed by eosin (Leica, 3801601E) and then hematoxylin (Leica, 3801542E) before dehydration with successive mixtures of ethanol of increasing concentration (70%, 90%, and 100%) and finally xylene. All slides were scanned a Pannoramic 250 High Throughput Scanner.
Regions of interest and immune marker detection
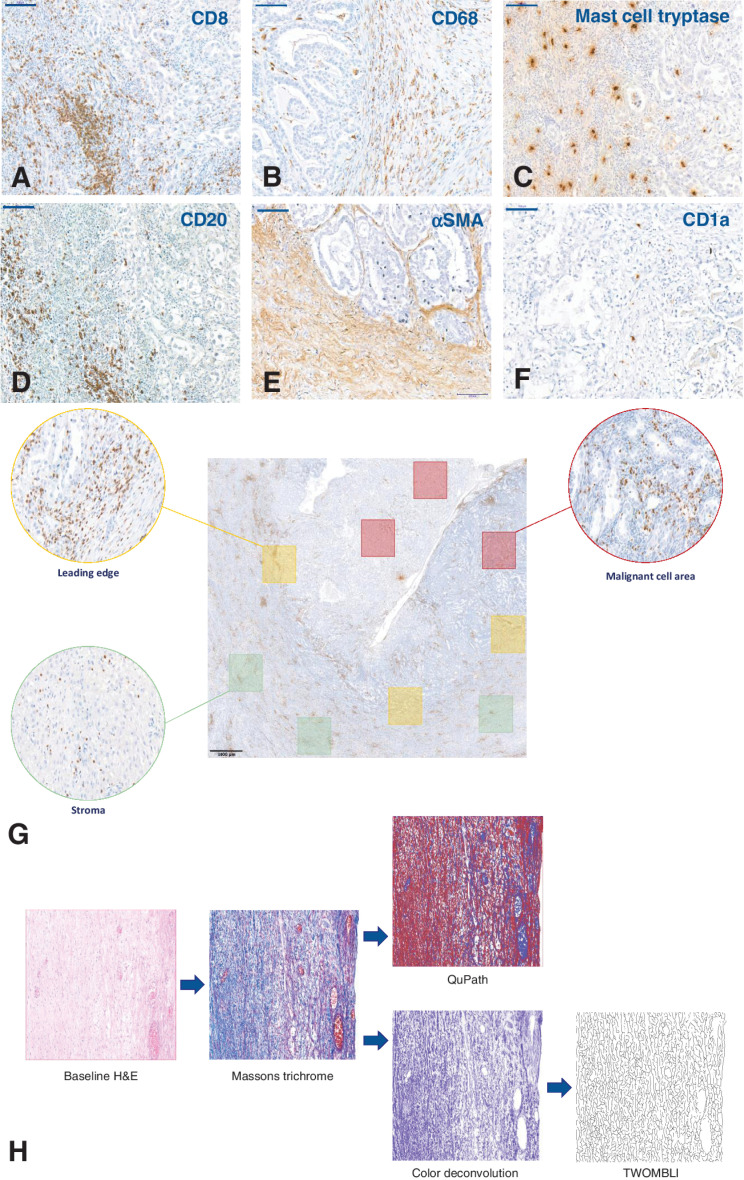
The baseline density, location, and proximity of tumor-infiltrating lymphocytes all significantly associate with response to checkpoint blockade across a number of tumor types (9); therefore, we wanted to evaluate the spatial composition of the TME based on immune markers. For IHC samples, the malignant cell area (MCA) was defined as the region that contained predominantly malignant cells, the stromal area contained no malignant cells, and the leading edge (LE) contained equal amounts of both malignant cells and stroma. Three 1×1 mm squares within each of these regions of interest (ROI) were identified, and the median result selected as the value for that region. To remove any potential selection bias, each ROI was selected from the baseline H&E slide and translated onto consecutive slides using QuPath V0.1.2, a software which has been shown to be functional and reproducible when quantifying immune infiltrates (10). Cellular stains were quantified using positive cell detection. For cytoplasmic stains, which were irregular and made individual cells hard to distinguish, quantification was by positive area detection.
Masson's trichrome staining
FFPE slides were deparaffinized in xylene (Thermo Fisher Scientific, 10588070) and rehydrated with successive mixtures of ethanol (Thermo Fisher Scientific, E/0650DF/17) of decreasing concentration (100%, 95%, 90%, 70%, and 50%, each for 2 minutes), followed by a wash in water. Slides were then submerged in Bouin's solution (Sigma-Aldrich; HT10132) for 1 hour at 60°C. Working Weigert's Iron Hematoxylin Solution (Sigma-Aldrich, HT1079-1SET) was applied for 5 minutes, and slides washed again in water. Slides were then processed using a Trichrome Stain Masson's Kit (Sigma-Aldrich HT15-1KT) according to the manufacturer's instructions. All slides were scanned a Pannoramic 250 High Throughput Scanner.
Collagen quantification
Collagen structure and texture are altered in a number of malignancies, with features such as collagen thickness being associated with survival (11). Textural analysis of the stained collagen (via Masson's trichrome) was carried out using Haralick features (12) on QuPath V0.1.2, and structure was analyzed using TWOMBLI (13), a plugin for FIJI (www.imagej.net). Images of Masson's trichrome within each ROI were either directly input into QuPath or underwent a deconvolution step before analysis by TWOMBLI. Deconvolution allowed the isolation of blue collagen from the image and was performed using the H PAS setting on Image J. Branch points and endpoints were standardized by dividing by total length; the average fiber thickness was found by dividing the high-density matrix by the total fiber length.
Microarrays, data sets, and bioinformatic analysis
A microarray data set of ovarian cancer patient samples, referred to as “the AOCS data set,” was obtained from a previous study (14), and contained 29 samples of CCOC paired with clinical data and the expression matrix of 19,701 microarray probes. This data set was generated using the Affymetrix U133 Plus 2.0 array. It was supplemented in parts with the GSE44104 (15) and GSE7305 (16) data sets.
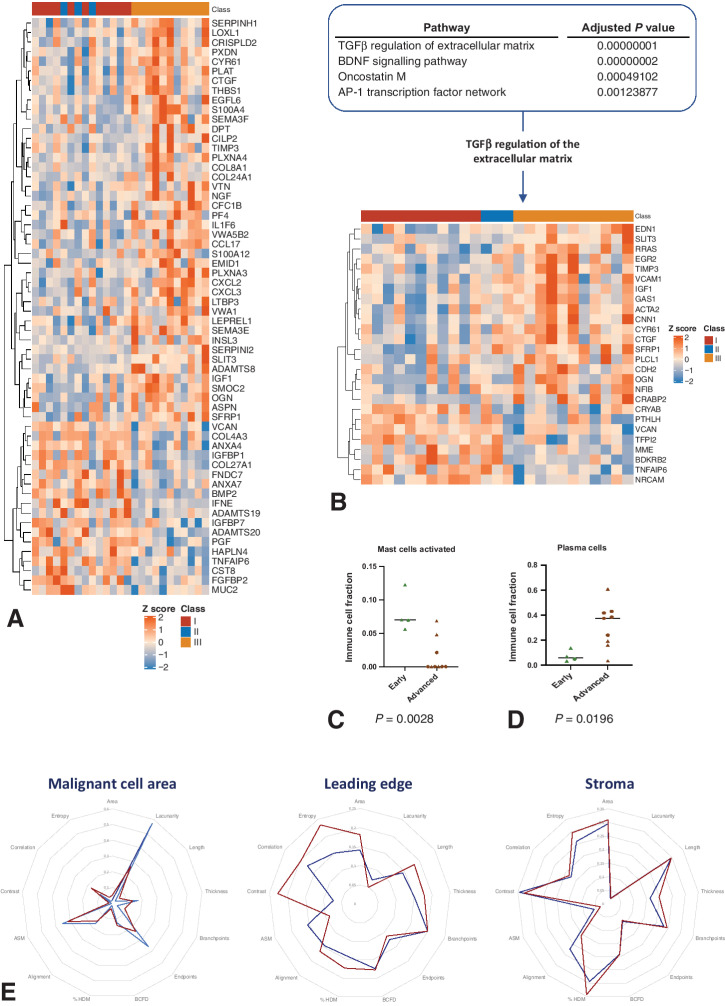
CIBERSORT (17) analysis was run on the AOCS and GSE44104 data sets, using the L22 signature matrix and P < 0.05 as a goodness-of-fit cutoff, to assess significantly enriched immune cell subsets for 4 patients with early-stage and 9 patients with advanced-stage CCOC; from the ACOS data set, 3 were early-stage and 4 advanced-stage; from the GSE44104 data set, 1 was early-stage and 5 were advanced-stage. Differential gene expression analysis (DGEA) for late (stage III) versus early (stage I/II) CCOC, excluding four early CCOC that had recurrent disease, was performed on the AOCS data set using the linear model in limma (18), resulting in 181 genes with P < 0.05. MSigDB matrisome genes (NABA_MATRISOME; gene set enrichment analysis, GSEA M5889; ref. 19) were identified in the differentially expressed genes (DEG). Pathway enrichment analysis was performed on the 181 DEGs using the Enrichr platform (20) and the BioPlanet database (21). DGEA was also performed on the AOCS data set for ARID1Awt versus ARID1Amut samples, using a linear model and taking into account disease stage with an additive ARID1A status+stage design. The ranked t-statistic of all genes was then used as input for GSEA preranked on the GenePattern platform (22) using Gene Ontology Biological Processes and Canonical Pathways (www.genepattern.org). Significantly enriched pathways were identified with a FDR q < 0.05. The endometriosis gene signature was generated from the GSE7305 data set (16) and Ahn and colleagues (23). First, the DEGs in the GSE7305 Affymetrix human U133 plus 2.0 array data set comparing ovarian endometriosis with normal ovary were extracted using GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r), with P < 0.001 and log2 fold change > |1|. This gene list was combined with the gene list of significant DEGs between ectopic endometriosis versus control endometrium reported by Ahn and colleagues (23). Heat maps were generated with R package ComplexHeat map, and all bioinformatic analyses were performed using R version 4.0.4. The heat map in Fig. 3B illustrates all differentially expressed matrisome genes between early- and advanced-stage CCOC of the AOCS data set with a threshold of P ≤ 0.05.
Figure 3.
Differences in the TME between early- and advanced-stage CCOC. A, Differentially expressed matrisome genes between early- and advanced-stage CCOC were determined for the AOCS data set (P ≤ 0.05). Heat map shows row z-scores of normalized gene expression. Genes (rows) are clustered with euclidean distance and the complete clustering method. Samples (columns) are annotated according to FIGO stage; FIGO stage I shown in red; FIGO II sown in blue; FIGO III shown in gold. B, Significantly enriched pathways in early- versus advanced-stage CCOC were determined for the AOCS data set. Top: ranking of the significantly enriched pathways in early- versus advanced-stage CCOC (Padj < 0.05 and combined score > 50). C and D, CIBERSORT analysis from early-stage (n = 4) and advanced-stage (n = 9) CCOC from the AOCS and GSE44104 (9) data sets (ACOS data set: 3 early-stage, 4 advanced-stage; GSE44104 data set: 1 early-stage, 5 were advanced-stage). Error bars indicate median. C, Activated mast cells (P = 0.0028). D, Plasma cells (P = 0.00196). ACOS data: triangles; GSE44104: circles. Error bars indicate median. E, Radar chart of TWOMBLI and Haralick analysis features across the MCA, LE, and stroma of early- and advanced-stage CCOC. Collagen was identified in FFPE samples using Masson's trichrome and quantified using TWOMBLI and Haralick features, with results normalized to allow comparison among the three ROI. Red: advanced-stage CCOC; blue: early-stage CCOC.
Statistical analysis
For all calculations, P <0.05 was considered significant. For comparisons across the MCA, LE, and stroma of paired samples, a Friedman test with a post hoc Dunn multiple comparisons test was used. When comparing metrics between ARID1Awt and ARID1Amut samples for markers quantified by the number of positive cells, a χ2 with Yates’ continuity correction was used; for immune markers quantified by positive area, detection analysis was performed with a generalized linear model with quasi-binomial distribution; for all other metrics, a Mann–Whitney test was used. Correlations were analyzed by nonparametric Spearman correlation. OS was assessed using Cox proportional hazards analysis and Kaplan–Meier analysis. Statistical analysis was performed on Prism version 8.4.3 and R version 4.0.4.
Data availability
The data generated in this study are available within the article and its Supplementary Data. The “the AOCS data set” was obtained from a previous study (14); restrictions apply to the availability of these data, which were used under license for this study. Data are available from the authors upon reasonable request with the permission of the original study authors. Additional data analyzed in this study were obtained from the Gene Expression Omnibus under accession numbers, GSE44104 (15) and GSE7305 (16).
Results
TME of advanced CCOC
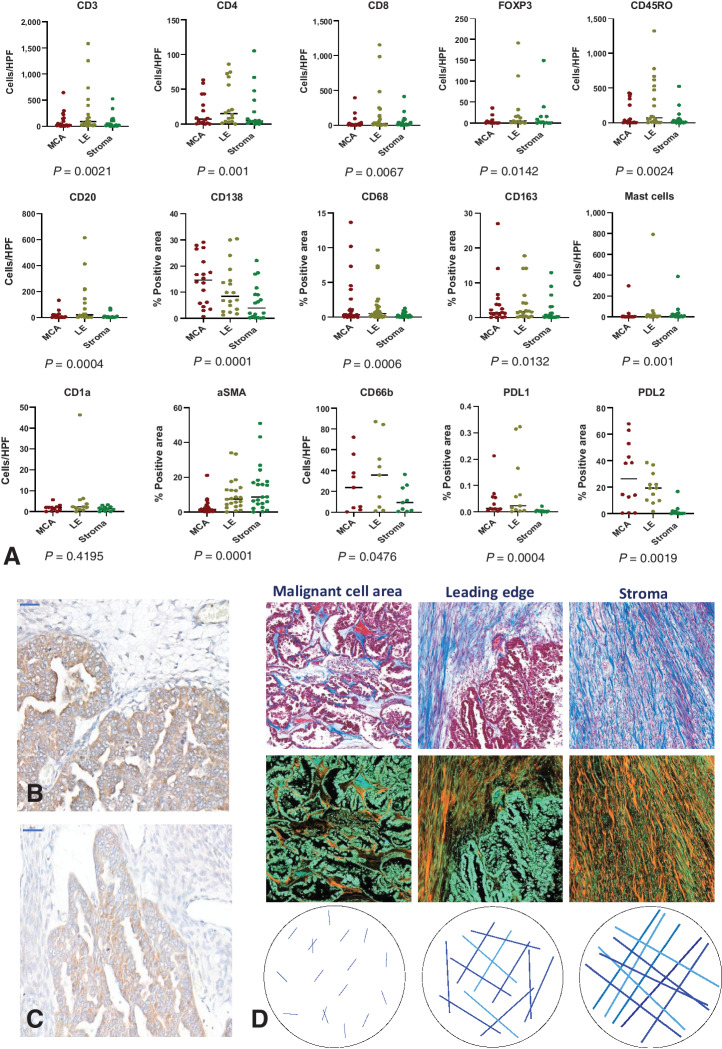
We first conducted a quantitative, qualitative, and topographical analysis of 14 immune cell phenotype markers using IHC on FFPE samples from 36 cases of treatment-naïve advanced CCOC. We also conducted structural and textural analysis of collagen using Masson's trichrome staining (representative staining and spatial analysis shown in Fig. 1). Except for CD1a+ dendritic cells (DC), significant variability in the immune cell markers across the MCA, LE, and stroma of advanced CCOC biopsies was observed (Fig. 2A). The MCA had the largest population of CD138+ plasma cells. The LE had significantly more CD20+, CD8+, CD4+, and CD45RO+ lymphocytic cells than any other region. Within the stroma, there were more mast cells and αSMA+ fibroblasts than any other region.
Figure 1.
Examples of IHC identification of immune markers in CCOC and subsequent quantification methods. A–F, Representative IHC images used to identify immune cell populations in advanced CCOC. CD8 (A), CD68 (B), mast cell tryptase (C), CD20 (D), αSMA (E), CD1a (F). All images taken at 20× magnification; scale bar: 100 μm. G, Topographical quantification. For spatial analysis of the TME, quantification occurred within ROI. Red boxes: MCA, defined as a region that contained predominantly malignant cells; Green boxes: stromal area, defined as areas containing no malignant cells; Yellow boxes: the LE, defined as areas containing equal amounts of both malignant cells and stromal cells. Three 1 × 1 mm squares within each ROI were identified, with each slide having nine individually quantified areas. To remove any potential selection bias, each ROI was selected from the baseline H&E slide and translated onto consecutive slides. The immune cells shown in this example are CD8+ cells; scale bar: 1,000 μm. H, Image processing pipeline for collagen analysis. Textural analysis of stained collagen was carried out using Haralick features on QuPath, and structure was analyzed using TWOMBLI. Representative images of Masson's trichrome within each ROI were either directly input into QuPath or underwent a deconvolution step on ImageJ before analysis by TWOMBLI.
Figure 2.
Variation in immune cell markers and the collagen matrix throughout the TME of advanced CCOC, with IHC identification of PDL2. A, Scatter plots showing median values for the indicated immune markers across the MCA, LE, and stroma of paired samples. Immune markers were assessed using IHC and quantified by either positive cells per high powered field (HPF) or percent positive area. The distribution of markers across the three ROI was analyzed in paired samples using a Friedman test with a post hoc Dunn multiple comparisons test; P values shown, error bars indicate medians. CD3, n = 21; CD8, n = 22; CD4, n = 16; FOXP3, n = 13; CD45RO, n = 20; CD20, n = 18; CD138, n = 18; CD68, n = 29, CD163, n = 18; mast cells, n = 18; CD1a, n = 12, αSMA, n = 34; CD66b, n = 9; PD-L1, n = 12; PD-L2, n = 12. B and C, PDL2 was evaluated using IHC on 12 samples and was confirmed using two different antibodies; scale bar: 50 μm. D, Masson's trichrome was used to identify collagen (blue in the top row of images) across the MCA, LE, and stroma of 36 samples. Images were inverted to better visualize collagen content (orange in the bottom row).
We also evaluated sections for PD-1 and its ligands. Significantly more PD-L2 positivity than either PD-1 or PD-L1 (P = <0.0001) was observed. PD-L2 was identified with two different antibodies and predominantly localized to malignant cells (Fig. 2B and C), with the largest area of positive cells being within the MCA. The only significant correlation between PD-1 and its ligands was between PD-1 and PD-L2 within the LE (P = 0.003; r = 0.477), suggesting that PD-L2 may be the dominant ligand in the TME of advanced CCOC.
Following analysis of the immune infiltrate, we investigated the extracellular matrix (ECM) compartment of CCOC tumors, focusing on the collagen matrix. From the MCA through to the LE and into the stroma, we found an increase in the area of collagen defined by Masson's trichrome positivity (P = <0.0001; Fig. 2D; Supplementary Fig. S1). The decrease in fiber endpoints moving from the MCA to the stroma, alongside an increase in fiber length and thickness (all P = <0.0001), indicated fewer, but longer and thicker, fibers in the stroma compared with the LE and MCA. In parallel, we observed an increase in branchpoints (P = <0.0001), indicating that these collagen fibers were overlapping more frequently in the stroma compared with the LE or MCA. The decrease in lacunarity (P = 0.0118) alongside an increase in box counting fractal dimension (BCFD; P = <0.0001) moving from the MCA into the stroma, indicated that the collagen was closer together and more spatially complex in the stroma. No difference in fiber alignment among the three areas of the biopsies was seen (P = 0.083), indicating that at no point were the fibers more likely to be orientated in a similar direction in the TME. The converse decrease in angular second moment (ASM) and increase in entropy (both P = <0.0001) moving from the MCA through the LE and into the stroma showed that in the stroma, collagen had a more heterogeneous and complex texture. The increasing contrast from the MCA through to the stroma (P = <0.0001) showed that the fibers were smoother in the MCA compared with the rougher fibers of the stroma. The correlation, which infers information on the directional distribution of texture change, was highest at the LE (P = <0.0001) which may be due to it being a transition point between the collagen of the MCA and that of the stroma.
In summary, our data showed that the collagen matrix changes from the MCA through the LE and into the stroma, with fewer but longer and thicker collagen fibers that overlap more frequently as part of a more spatially complex and textually diverse matrix. We also found significant variability in immune cell phenotype across the three ROI, suggesting that it is important to consider topographical location when interpreting data to avoid, for example, comparing the MCA of one sample with the LE of another, given the significant intratumoral difference between these regions in terms of immune marker populations and ECM structure.
Early versus advanced CCOC
We next aimed to understand what changed within the TME of CCOC between early and advanced disease that may allow for immune evasion and metastatic dissemination. This was achieved by comparing nine FFPE samples from early-stage disease with the advanced-stage cohort, as well as through interrogation of the AOCS transcriptional data set annotated for stage (14). Quantitative IHC results for T cells (CD3+), B cells (CD20+), mast cells (mast cell tryptase+), macrophages (CD68+), and fibroblasts (αSMA+) in nine FFPE samples of early-stage CCOC were compared with the advanced-stage cohort. Across the MCA, LE, and stroma, the only significant difference was an increase in αSMA+ fibroblasts within the stroma of advanced-stage CCOC compared with early-stage disease (P = 0.024; Supplementary Table S3). Annotation of ECM-related gene transcripts from the AOCS data set demonstrated separation between early- and advanced-stage CCOC (Fig. 3A), with Enrichr analysis identifying TGFβ regulation of the ECM to be the most significantly enriched pathway in advanced CCOC compared with early-stage disease (P = <0.001; Fig. 3B).
CIBERSORT analysis of transcriptional data from four early-stage CCOC and nine advanced-stage CCOC from the AOCS and GSE44104 (15) data sets identified two immune cell populations that were significantly different between early- and advanced-stage CCOC, with an increase in plasma cells in advanced-stage disease (P = 0.00196) and a decrease in activated mast cells (P = 0.0028); this is consistent with how these populations respond to increased TGFβ expression (refs. 24, 25; Fig. 3C and D). ACTA2 expression, a gene correlating to cancer-associated fibroblasts (CAF; ref. 26), was significantly upregulated in advanced-stage disease (P = 0.0152) within the ACOS data set compared with early-stage CCOC. ACTA2 had significant positive correlations with TGFB1 (P = <0.001; r = 0.64), TGFBI (P = 0.002; r = 0.5985), TGFBR1 (P = 0.011; r = 0.4985), and TGFB1I1 (P = <0.001; r = 0.7692), suggesting a link between CAFs and TGFβ signaling in CCOC.
The collagen analysis included nine early-stage and 36 advanced-stage samples. The differences were most noticeable within the MCA, and stromal collagen was similar in early and advanced disease (Fig. 3E). In the MCA, advanced-stage tumors had more collagen (P = 0.0134), consisting of longer (P = 0.0159) fibers which overlapped more frequently (P = 0.0014) and had fewer endpoints (P = 0.0058) compared with early-stage disease. These fibers were closer together (P = 0.0146) and formed a more diffuse matrix, with fibers orientated in many different directions (P = 0.0222). The texture of collagen in advanced-stage disease was more heterogeneous (P = 0.0213), complex (P = 0.018), and rough (P = 0.018), with the variations of texture occurring in numerous directions (P = 0.0053). At the LE, the fibers had similar appearance; however, advanced-stage LE continued to have more collagen (P = 0.0079) containing longer fibers (P = 0.0087) that were closer together (P = 0.0138) and part of a more diffuse matrix (P = 0.0366). In summary, compared with early CCOC, the collagen fibers of advanced disease formed a more diffuse and complex matrix within the MCA and LE, with TGFβ-mediated regulation of the ECM being the most enriched pathway as CCOC progressed. In advanced disease, a significant increase in stromal αSMA, with ACTA2 having significant positive correlations with TGFβ signaling, suggests a link between CAFs and TGFβ expression.
Impact of ARID1A
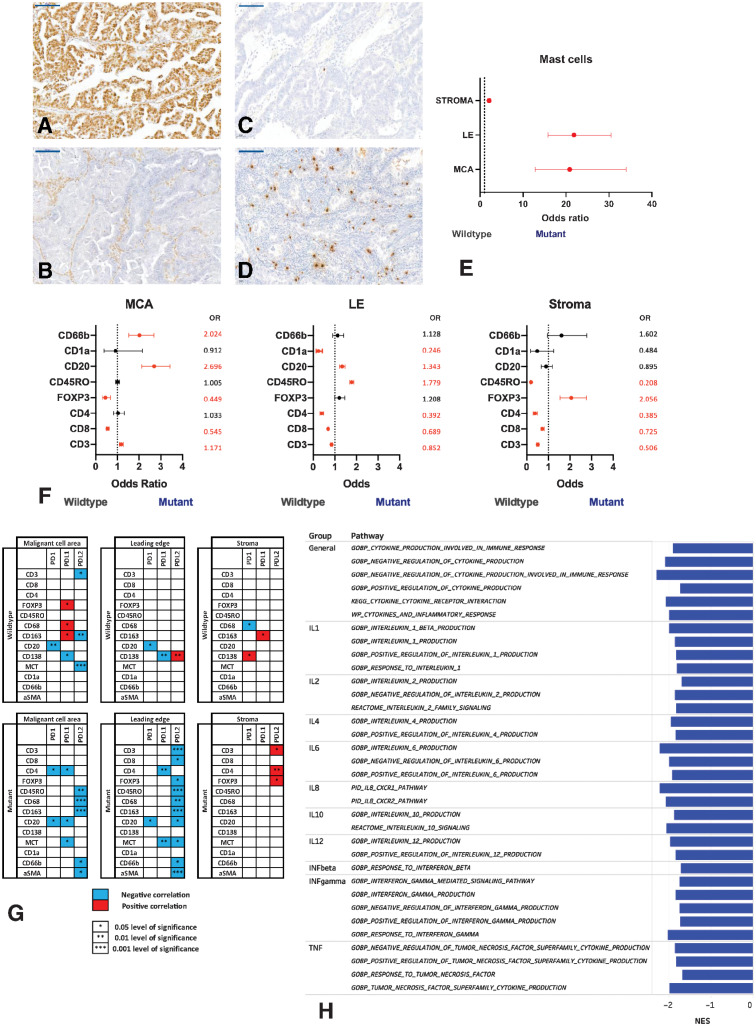
We next sought to determine the impact of ARID1A mutation on the TME of CCOC. ARID1A status was determined using IHC (Fig. 4A and B). Of the 36 FFPE samples of advanced CCOC, 21 were classified as ARID1Awt, 14 were ARID1Amut, and one sample had mixed expression and was excluded from subsequent analysis. For the 14 immune markers quantified, we included at least five samples in each group (Supplementary Table S1). In the AOCS transcriptional data set, ARID1A status was known for 22 of the 29 CCOC samples; of these seven were ARID1Awt and 15 ARID1Amut.
Figure 4.
The immune microenvironment of ARID1A-wildtype (ARID1Awt) and -mutant (ARID1Amut) CCOC. A and B. Representative immunohistological images of ARID1A in advanced CCOC. Anti-ARID1A was used to evaluate protein expression in biopsies. Representative ARID1Awt sample (A) and representative ARID1Amut sample (B). All images taken at 20× magnification; scale bar: 100 μm. C–E, Mast cells in ARID1Awt and ARID1Amut CCOC were assessed. Mast cells were identified using anti-mast cell tryptase. Representative images of mast cells in anARID1Awt CCOC biopsy (C); in an ARID1Amut CCOC biopsy (D). E, Mast cells in the MCA, LE, and stroma of ARID1Amut CCOC biopsies compared with ARID1Awt tumors. Mast cells were quantified by positive cells per high powered field and significance determined using χ2. The OR is plotted; if <1, the immune population was higher in ARID1Awt tumors; >1, the population was higher in ARID1Amut tumors. All results were statistically significant. F, Comparison of ARID1Awt and ARID1Amut tumors for immune markers quantified by number of cells per high powered field. χ2 was used to determine significance. Immune markers were quantified using positive cells per high powered field via IHC across the MCA, LE, and stroma. The OR is plotted, with significant values indicated in red and nonsignificant ratios in black; if <1, the immune population was higher in ARID1Awt tumors; if >1, the population was higher in ARID1Amut tumors. G, Associations between the PD-1 axis and immune cell populations across the TME of ARID1Awt and ARID1Amut advanced CCOC was determined. Shown are associations for PD-1, PD-L1, and PD-L2 with immune cell markers across the MCA, LE, and stroma. Significance is indicated with a star, with positive associations indicated in red and negative associations in blue. H, Cytokine pathways in ARID1Awt and ARID1Amut CCOC. DEGs from the AOCS data set were ranked and pathway enrichment performed. A FDR q < 0.05 was considered significant. The normal enrichment score (NES) is plotted for pathways enriched in ARID1Amut tumors.
In the FFPE cohort, we found significant differences between ARID1Awt and ARID1Amut CCOC for all immune markers, with the exception of CD68+ macrophages. Across all three ROIs, ARID1Amut CCOC had significantly more mast cells (Fig. 4C–E). Within the MCA, ARID1Amut tumors had greater immune cell diversity compared with ARID1Awt tumors, including significantly more mast cells, CD3+, CD20+ CD138+, CD66b+ cells, and αSMA+ fibroblasts. In comparison, ARID1Awt biopsies had significantly larger populations of CD8+ and CD4+ T cells. At the LE, ARID1Amut biopsies continued to have significantly more immune cells across a number of lineages, including mast cells, CD20+, CD138+, and CD45RO+ cells. At the LE of ARID1Awt, the increase in immune cell populations was limited to some T-cell lineages (CD3+, CD8+, CD4+) and CD1a+ DCs. Although ARID1Amut CCOC had larger populations of FOXP3+ cells and mast cells in the stroma, in ARID1Awt tumors, this region contained significantly more fibroblasts and CD3+, CD8+, CD4+, and CD45RO+ T cells (Fig. 4F).
We observed no significant differences in PD-1 or PD-L1 expression between the two groups; however, ARID1Awt biopsies had more PD-L2 expression within the MCA (P = 0.0004) and LE (P = 0.038) compared with ARID1Amut biopsies. When we explored how the PD-1 checkpoint correlated with surrounding immune cells, we again found significant differences between ARID1Awt and ARID1Amut groups (Fig. 4G). ARID1Amut tumors had more significant correlations between immune cells and the PD-1 axis than ARID1Awt biopsies (26 associations vs. 15, respectively), and within the MCA and LE, these were universally negative. PD-L1 was the ligand with the most immune correlations in ARID1Awt tumors, whereas in ARID1Amut tumors, PD-L2 had the most associations.
Enrichment of DEGs from the AOCS data set found 643 unique pathways enriched between ARID1Awt and ARID1Amut CCOC (FDR q = <0.05). These included 38 general leukocyte and lymphocyte pathways that were enriched in ARID1Amut CCOC compared with ARID1Awt. Other pathways enriched in ARID1Amut compared with ARID1Awt biopsies involved T cells, B cells, macrophages, mast cells, natural killer cells, neutrophils, eosinophils, and fibroblasts and included activation, proliferation, differentiation, regulation, and migration of immune cells. This analysis also demonstrated a number of cytokine-related pathways that were significantly different between ARID1Awt and ARID1Amut CCOC (Fig. 4H) and 11 collagen-specific pathways (Supplementary Table S4).
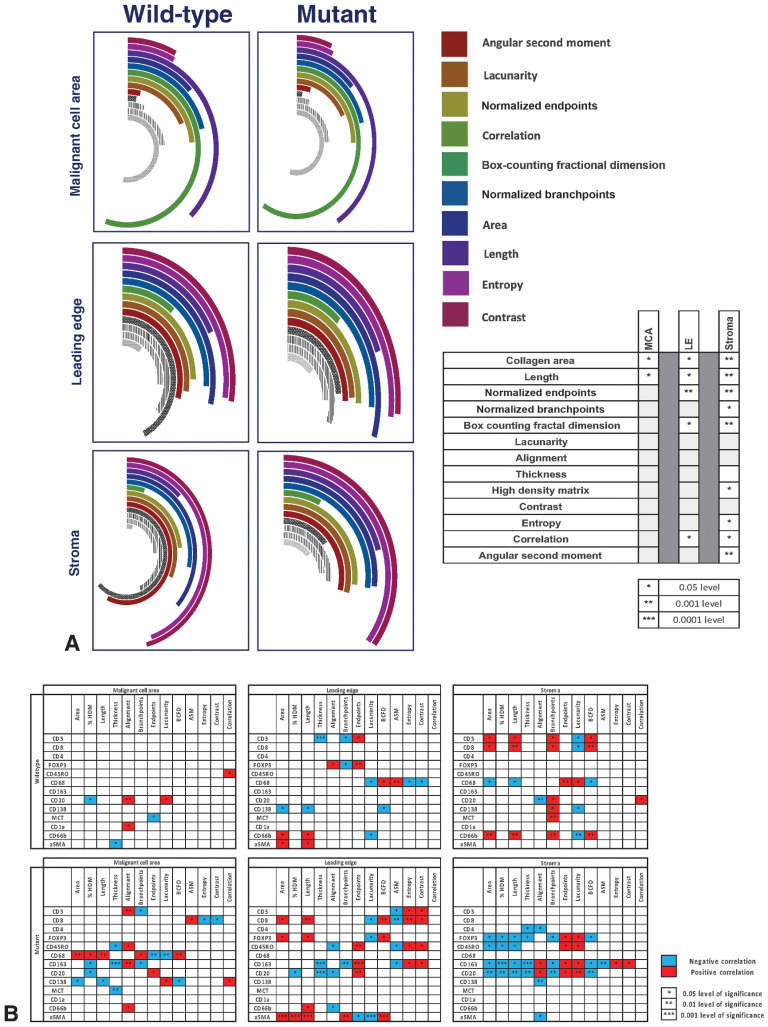
Analysis of collagen from FFPE samples demonstrated that throughout the TME, ARID1Awt tumors had more collagen than ARID1Amut tumors (Fig. 5A). Within the LE and stroma, ARID1Awt tumors had fewer collagen endpoints, indicating fewer, but longer, fibers that were part of a more diffuse matrix, as indicated by the higher BCFD. The differences were most pronounced within the stroma, where the collagen fibers of ARID1Awt tumors overlapped more frequently as part of a high-density matrix.
Figure 5.
The collagen matrix of ARID1A–wild-type (ARID1Awt) and -mutant (ARID1Amut)CCOC and relationship with immune infiltrates. A, Collagen across the TME of ARID1Awt (n = 21) and ARID1Amut (n = 14) advanced CCOC FFPE samples was identified via Masson's trichrome and underwent structural and textural analysis with TWOMBLI and Haralick features. The radial bar chart displays variability in the structure and texture of collagen across the across the MCA, LE, and stroma. All results are normalized to allow direct comparison between both groups and across the different ROI. Significant values, calculated with Mann–Whitney test, are indicated in the adjoining table. B, Correlations between the collagen matrix and immune cell markers for ARID1Awt and ARID1Amut CCOC, including across the MCA, LE, and stroma. Significance is indicated by stars, with a positive association shown in red and a negative association in blue.
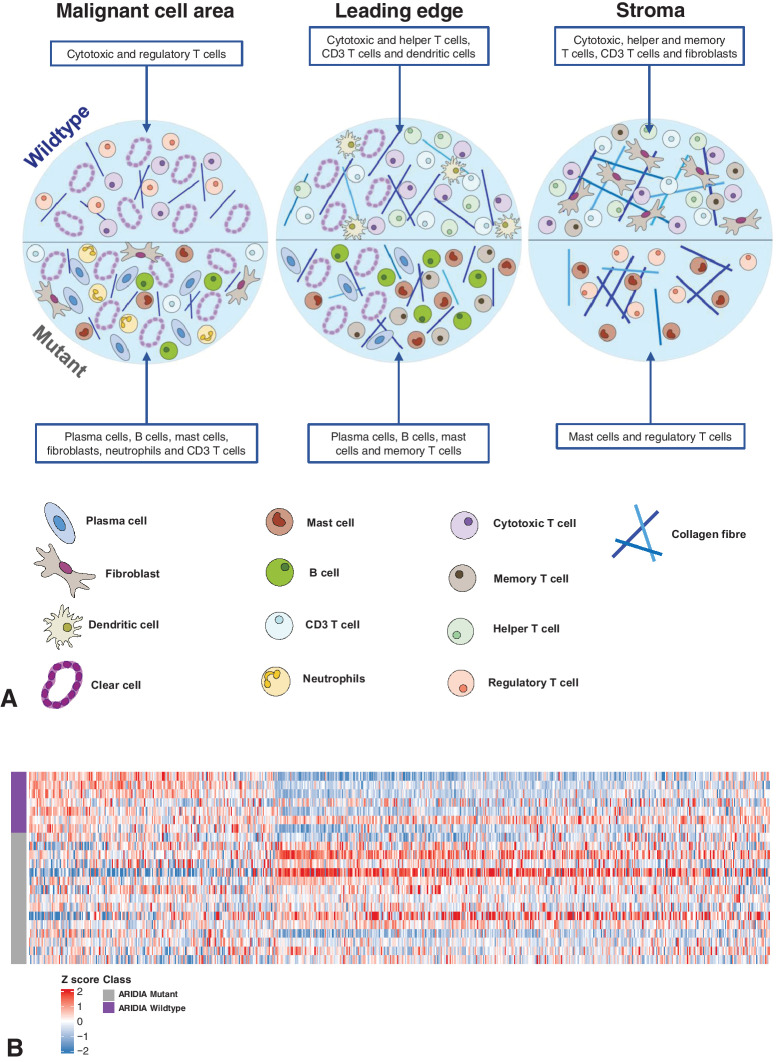
Given the significant differences in both the collagen matrix and immune cell populations, between ARID1Awt and ARID1Amut CCOC, we wanted to investigate whether ECM metrics inhibited or promoted different immune phenotypes (Fig. 5B). We found that ARID1Awt tumors had half the number of significant associations between ECM-metrics and immune markers compared with ARID1Amut tumors (52 vs. 103). In ARID1Awt tumors, the stroma had the most immune-collagen associations (26 associations), with the number of significant associations decreasing through the LE (19 associations) and into the MCA (7 associations). In comparison, ARID1Amut CCOC showed little regional variation in the number of significant immune-collagen associations (27 in MCA; 38 for both LE and stoma). In ARID1Awt tumors, the decreasing number of associations between immune cells and the collagen ECM moving from the stroma into the LE, and eventually the MCA, suggests that the malignant core of these tumors developed within a relatively immune-privileged environment. This may be a consequence of cancer arising within a structure with a preexisting physical barrier, such as an adenofibroma (27, 28), or due to the successful remodeling of the stromal ECM by malignant cells. In contrast, in ARID1Amut tumors, there were more associations between the collagen ECM and immune cells, with little variation in the number of associations across the TME. This pattern makes immune exclusion less likely to be part of tumorigenesis and points toward immune modulation of an inflammatory precursor lesion, such as endometriosis (29). This theory was further supported by our observation that ARID1Amut tumors had more significant correlations between immune cells and the PD-1 axis than what was observed in ARID1Awt biopsies; within the MCA and LE, these interactions were universally negative. In summary, analysis of both RNA and protein revealed that ARID1Awt and ARID1Amut CCOC tumors had significant differences in immune cell populations, cytokine and checkpoint expression, and collagen structure and texture (Fig. 6A).
Figure 6.
Diagrammatic summary of the differences in the TME of ARID1A-wildtype (ARID1Awt) and -mutant (ARID1Amut) advanced CCOC and relationship to endometriosis. A, Schematic highlighting the immune cell populations significantly different between ARID1Awt and ARID1Amut CCOC across the MCA, LE, and stroma. Top half illustrates the composition of ARID1Awt tumors and the bottom half illustrates the composition of ARID1Amut tumors. B, Heat map showing expression of endometriosis-associated genes in ARID1Awt and ARID1Amut CCOC. Two data sets were used (10, 23) to assess significant DEGs between ectopic endometriosis and normal endometrium. A total of 641 DEGs in the AOCS data set are shown in the heat map. Red: upregulation and blue: downregulation in ARID1Awt tumors. N = 7 ARID1Awt (purple) and N = 15 ARID1Amut (gray).
Relationship to endometriosis
The links between endometriosis and CCOC are well established, with the loss of ARID1A function thought to be an early event in the transformation from endometriosis into CCOC (29). However, not all CCOCs associate with endometriosis, with some cases arising from clear-cell adenofibromas in the absence of endometriosis (27, 28). Because our findings suggested that ARID1Awt CCOC may develop within a preexisting physical barrier and that ARID1Amut may develop via immune modulation of an inflammatory precursor lesion, we hypothesized that ARID1A status could identify differing tumorigenesis paths due to different premalignant microenvironments, rather than solely the action or inaction of a single gene. To investigate this hypothesis, we interrogated the AOCS data set with an endometriosis gene signature derived from two data sets (refs. 16, 23; see Materials and Methods). This demonstrated a separation between the ARID1Awt and ARID1Amut groups (Fig. 6B), indicating differences in how these two groups relate to this precursor lesion. Given the already established links between ARID1A loss and the development of CCOC from endometriosis (29), alongside our results suggesting that immune-modulation plays a significant role in the tumorigenesis of ARID1Amut disease, we hypothesize that ARID1Amut identifies CCOC arising from endometriosis and that ARID1Awt identifies CCOC from the relatively immune-privileged environment of an adenofibroma.
Survival analysis
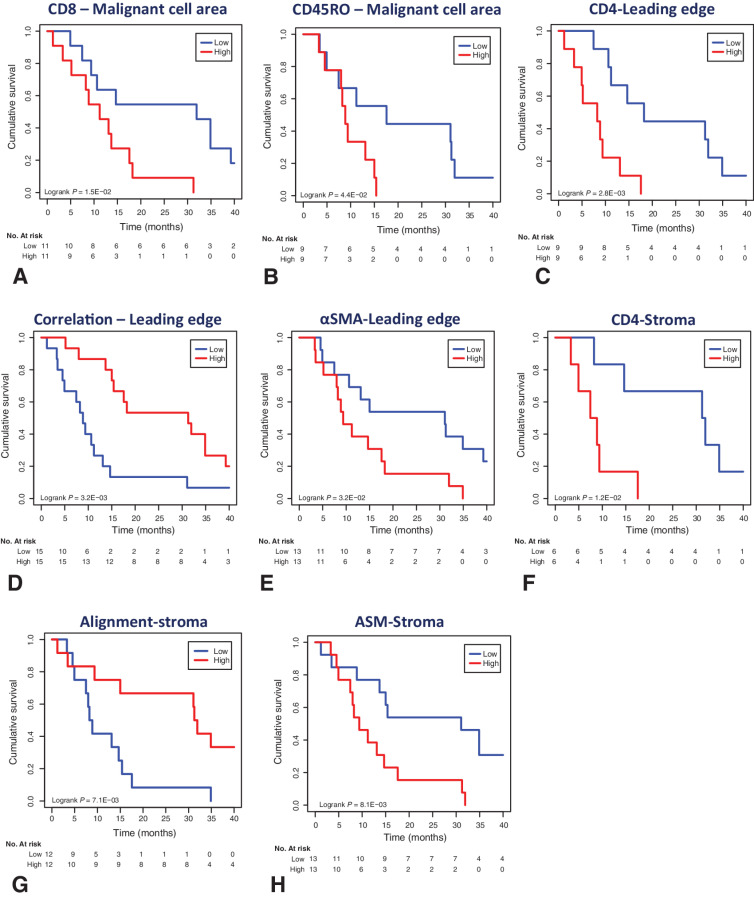
For 38 patients where clinical information and OS data were available, we evaluated the prognostic effects of each TME metric by Cox proportional hazards analysis and Kaplan–Meier analysis using the median values as thresholds. Eight metrics showed a significant correlation with OS following univariate analysis (Fig. 7A–H). When we performed multivariate analysis, taking into account FIGO stage, ARID1A status, Aletti score, and whether adjuvant chemotherapy was received or not, five metrics remained significant (Supplementary Table S5). A higher number of CD8+ T cells in the MCA, a higher ASM in the stroma, and a higher number of CD4+ T cells in both LE and stoma significantly associated with poorer OS, whereas a higher correlation at the LE and higher alignment in the stroma associated with an improved OS.
Figure 7.
TME metrics which collate with OS in advanced CCOC. A–H, Kaplan–Meier curves of OS using the indicated eight TME metrics that were significant factors following univariate analysis (Supplementary Table S5). N = 38 patients from the FFPE cohort with clinical information and OS data were stratified into high and low groups for the indicated metrics using the median values as thresholds. Kaplan–Meier analysis was performed using the median values as thresholds. ASM, angular second moment.
Discussion
To our knowledge, this is the most comprehensive profiling of the TME of advanced CCOC that has been performed to date. We demonstrated that the TME of advanced CCOC is complex and diverse; with the exception of CD1a+ DCs, all fourteen immune phenotype markers profiled were present and displayed quantitative, qualitative, and topographical variation.
A high CD8+ T-cell infiltrate within the MCA and high CD4+ T-cell infiltrate at the LE and stroma significantly associated with a reduced OS following multivariate analysis. Increased infiltration of CD8+ T cells is typically associated with an improved overall in survival in malignancy, however a similar link to poor prognosis is observed in renal cell carcinoma (30). The mechanism for this is unclear; however, it may be due to a lack of cancer antigen specificity, as reflected by a high T-cell receptor diversity and low clonality (31). Similarly, the link between an increased CD4+ infiltrate in the LE and stroma and decreased survival may reflect Th cells lacking the ability to detect cancer antigens. We found very few CD1a+ cells in our immune phenotype profiling suggesting few DCs, and this, along with high expression of PDL2 on malignant cells, may contribute to the finding. Although both CD8+ and CD4+ T cells are linked with a poorer outcome, ultimately these patients may be the ones most likely to benefit from checkpoint inhibition, given their primed TME.
Similar to other studies, our analysis found low expression of PD-1 and PD-L1 throughout the TME (32, 33); however substantial PD-L2 expression, which primarily localized to the malignant cells, was seen, providing a mechanism by which CCOC may be able to evade T-cell immune detection. High PD-L2 expression also suggests that CCOC may benefit more from therapies targeting PD-1 rather than PD-L1.
We recognize that the use of single-plex IHC markers is a limitation of this study, making it difficult to attribute individual markers to specific cell types without the presence of confirmatory markers on the same cells. The single-plex technique was selected to allow this work to be easily reproducible without the need for special equipment, and we made efforts to address the limitations associated with this technique by using consecutive slides and digital image overlays.
The collagen ECM displayed structural properties that varied across the MCA, LE, and stroma. A collagen stroma that contains highly aligned collagen fibers, along with a general lack of heterogeneity of collagen texture within the stroma and at the LE, associated with a poor prognosis. TGFβ signaling can induce the expression of ECM proteins and is a major driver in the transformation of the TME from early-stage to advanced-stage CCOC. TGFβ can be released by cancer cells, stromal fibroblasts, and other cells within the TME, and our current data suggest a link between stromal CAFs and TGFβ, which may play a role in the change in the collagen matrix between early- and advanced-stage CCOC.
Other clear-cell gynecologic malignancies, such as uterine and cervical, are morphologically and immunophenotypically similar to CCOC, despite emerging from different tissues (34). Similarly, we hypothesize that although the malignant cells within ARID1Awt and ARID1Amut CCOC may appear histologically alike, these two groups represent distinct disease entities. CCOC has two differentiation paths—one that involves the highly inflammatory microenvironment of endometriosis (29) and one that involves clear-cell adenofibromas (27, 28). It is difficult to imagine that malignant cells generated within these two very different microenvironments would somehow coalesce into one uniform disease entity. Because loss of ARID1A function is thought to be an early event in malignant transformation of endometriosis into CCOC (29), we suggest that ARID1Amut identifies CCOC emerging from endometriosis, whereas ARID1Awt CCOC does not.
ARID1Amut tumors had significantly more mast cells, CD20+ B cells, and CD138+ plasma cells compared with ARID1Awt, with endometriosis also having large populations of these cell types (35, 36). Endometriosis modulates the phenotype and frequency of leukocytes (37), with a reduction in infiltration and proliferation of T cells alongside a reduction in CD8+ and CD4+ T-cell activity (38), similar to ARID1Amut CCOC. Endometriosis exhibits upregulation of both PD-1 and PD-L1 (39), which may explain why in ARID1Amut CCOC, the PD-1 axis has more immune associations that were universally negative within the MCA and LE. We found significant enrichment of many cytokines known to be increased in endometriosis, including IL2, IL4, IL6, IL8, IL10, IL12, and TNF (40–43), in ARID1Amut CCOC.
ARID1Awt and ARID1Amut tumors had significant differences in the collagen content and structure. Matrix metallopeptidases (MMP) mediate endometrial breakdown at menstruation, and the regulation of MMPs and their inhibitors allows for ectopic endometriosis to invade other tissues by remodeling the ECM (44). We hypothesize that CCOC emerging from endometriosis has a very different collagen matrix to CCOC that does not arise from endometriosis, due to the actions of MMPs and related molecules. If ARID1Awt CCOC represents disease that has developed within the relatively immune-privileged environment of an adenofibroma, this may explain why most immune-collagen associations are within the stroma and decrease moving through the LE and into the MCA, whereas ARID1Amut CCOC shows little change in the number of immune-collagen associations throughout the TME.
Despite the differences between ARID1Awt and ARID1Amut CCOC, they shared some features. No significant difference in TGFβ pathway enrichment was found, and we hypothesize that TGFβ signaling may be important to the development of a clear-cell pathology, given its high expression in clear-cell renal cell carcinoma (45), where higher signaling promotes the invasive potential of renal clear cells (46). TGFβ is elevated in endometriosis (47), but it is also upregulated in response to hypoxia, as is observed in clear-cell renal cell carcinoma (48), which may be a mechanism for the development of CCOC within an adenofibroma.
These data do not provide a clear indication that one group of CCOC is more or less likely to respond to immunotherapy. A definitive answer to the role of ARID1A as a biomarker for response in CCOC will be obtained from the results of ongoing clinical trials. Although the results of these clinical trials are not yet available, we hypothesize that on balance, ARID1Amut CCOC will be more likely to respond to checkpoint inhibition, given that these tumors have more negative associations between the PD-1 axis and immune cells within the MCA and LE. Interruption of this axis is more likely to generate an anticancer immune response compared with ARID1Awt disease, where the number of PD-1–immune associations are fewer and not universally negative. The TME of ARID1Amut CCOC also contained other positive predictors of immune response, including increased populations of B cells and plasma cells [both of which are linked to checkpoint response (49)] and an increase in IFNγ, IL6, and IL10, a combined profile that correlates with a higher response rate in melanoma following PD-1 blockade (50). We believe the summation of these TME differences means that we cannot continue to treat this disease as a single entity, particularly when considering response to checkpoint blockade.
Supplementary Material
Acknowledgments
This work was also supported by a European Society for Medical Oncology Translational Fellowship (to M.-J. Devlin), the Medical Research Council Clinical Research Training Fellowship (MR/S00629×1, to M.-J. Devlin), Cancer Research UK Centre Grant (C355/A25137), CRUK Programme Grants (A16354, A25714, to F.R. Balkwill), CRUK Centre of Excellence Award to Barts Cancer Centre (C16420/A18066, to E. Maniati). The Northern Ireland biobank has received funds from HSC Research and Development Division of the Public Health Agency of Northern Ireland.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
M.-J. Devlin reports grants from Medical Research Council and European Society for Medical Oncology during the conduct of the study. R. Miller reports other support from GSK, AZD, Clovis oncology, and Ellipses outside the submitted work. R. Kristeleit reports personal fees and nonfinancial support from GSK, InCyte, Regeneron, and Shattuck Labs; grants, personal fees, and nonfinancial support from MSD and Clovis outside the submitted work. D.D. Bowtell reports grants from Genentech Roche, AstraZeneca, Beigene, and personal fees from Exo Therapeutics outside the submitted work. F.R. Balkwill reports personal fees from iOmx Therapeutics and GlaxoSmithKlein outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
M.-J. Devlin: Conceptualization, resources, data curation, software, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. R. Miller: Conceptualization, supervision, methodology, writing–review and editing. F. Laforets: Software, methodology, writing–review and editing. P. Kotantaki: Software, methodology, writing–review and editing. D.W. Garsed: Resources, data curation, writing–review and editing. R. Kristeleit: Resources, writing–review and editing. D.D. Bowtell: Resources, writing–review and editing. J. McDermott: Resources, formal analysis, methodology, writing–review and editing. E. Maniati: Resources, data curation, formal analysis, methodology, writing–original draft, writing–review and editing. F.R. Balkwill: Conceptualization, resources, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing.
References
- 1. Crotzer DR, Sun CC, Coleman RL, Wolf JK, Levenback CF, Gershenson DM. Lack of effective systemic therapy for recurrent clear cell carcinoma of the ovary. Gynecol Oncol 2007;105:404–8. [DOI] [PubMed] [Google Scholar]
- 2. Matulonis UA, Shapira-Frommer R, Santin AD, Lisyanskaya AS, Pignata S, Vergote I, et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann Oncol 2019;30:1080–7. [DOI] [PubMed] [Google Scholar]
- 3. Hamanishi J, Mandai M, Ikeda T, Minami M, Kawaguchi A, Murayama T, et al. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J Clin Oncol 2015;33:4015–22. [DOI] [PubMed] [Google Scholar]
- 4. Shibuya Y, Tokunaga H, Saito S, Shimokawa K, Katsuoka F, Bin L, et al. Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosomes Cancer 2018;57:51–60. [DOI] [PubMed] [Google Scholar]
- 5. Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med 2018;24:556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fukumoto T, Fatkhutdinov N, Zundell JA, Tcyganov EN, Nacarelli T, Karakashev S, et al. HDAC6 inhibition synergizes with Anti-PD-L1 therapy in ARID1A-inactivated ovarian cancer. Cancer Res 2019;79:5482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandler RL, Damrauer JS, Raab JR, Schisler JC, Wilkerson MD, Didion JP, et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat Commun 2015;6:6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khalique S, Naidoo K, Attygalle AD, Kriplani D, Daley F, Lowe A, et al. Optimised ARID1A immunohistochemistry is an accurate predictor of ARID1A mutational status in gynaecological cancers. J Pathol Clin Res 2018;4:154–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loughrey MB, Bankhead P, Coleman HG, Hagan RS, Craig S, McCorry AMB, et al. Validation of the systematic scoring of immunohistochemically stained tumour tissue microarrays using QuPath digital image analysis. Histopathology 2018;73:327–38. [DOI] [PubMed] [Google Scholar]
- 11. Zhou Z-H, Ji C-D, Xiao H-L, Zhao H-B, Cui Y-H, Bian X-W. Reorganized collagen in the tumor microenvironment of gastric cancer and its association with prognosis. J Cancer 2017;8:1466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haralick RM, Dinstein I, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern 1973;3:610–21. [Google Scholar]
- 13. Wershof E, Park D, Barry DJ, Jenkins RP, Rullan A, Wilkins A, et al. A FIJI macro for quantifying pattern in extracellular matrix. Life Sci Alliance 2021;4:e202000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anglesio MS, George J, Kulbe H, Friedlander M, Rischin D, Lemech C, et al. IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin Cancer Res 2011;17:2538–48. [DOI] [PubMed] [Google Scholar]
- 15. Wu Y-H, Chang T-H, Huang Y-F, Chen C-C, Chou C-Y. COL11A1 confers chemoresistance on ovarian cancer cells through the activation of Akt/c/EBPβ pathway and PDK1 stabilization. Oncotarget 2015;6:23748–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hever A, Roth RB, Hevezi P, Marin ME, Acosta JA, Acosta H, et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proc Natl Acad Sci U S A 2007;104:12451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics 2012;11:M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang R, Grishagin I, Wang Y, Zhao T, Greene J, Obenauer JC, et al. The NCATS BioPlanet - an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front Pharmacol 2019;10:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet 2006;38:500–1. [DOI] [PubMed] [Google Scholar]
- 23. Ahn SH, Khalaj K, Young SL, Lessey BA, Koti M, Tayade C. Immune-inflammation gene signatures in endometriosis patients. Fertil Steril 2016;106:1420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gomez G, Ramirez CD, Rivera J, Patel M, Norozian F, Wright HV, et al. TGF-beta 1 inhibits mast cell Fc epsilon RI expression. J Immunol 2005;174:5987–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lebman DA, Edmiston JS. The role of TGF-beta in growth, differentiation, and maturation of B lymphocytes. Microbes Infect 1999;1:1297–304. [DOI] [PubMed] [Google Scholar]
- 26. Chakravarthy A, Khan L, Bensler NP, Bose P, De Carvalho DD. TGF-β-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat Commun 2018;9:4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao C, Wu LSF, Barner R. Pathogenesis of ovarian clear cell adenofibroma, atypical proliferative (borderline) tumor, and carcinoma: clinicopathologic features of tumors with endometriosis or adenofibromatous components support two related pathways of tumor development. J Cancer 2011;2:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamamoto S, Tsuda H, Takano M, Hase K, Tamai S, Matsubara O. Clear-cell adenofibroma can be a clonal precursor for clear-cell adenocarcinoma of the ovary: a possible alternative ovarian clear-cell carcinogenic pathway. J Pathol 2008;216:103–10. [DOI] [PubMed] [Google Scholar]
- 29. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 2010;363:1532–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Remark R, Alifano M, Cremer I, Lupo A, Dieu-Nosjean M-C, Riquet M, et al. Characteristics and clinical impacts of the immune environments in colorectal and renal cell carcinoma lung metastases: influence of tumor origin. Clin Cancer Res 2013;19:4079–91. [DOI] [PubMed] [Google Scholar]
- 31. Braun DA, Street K, Burke KP, Cookmeyer DL, Denize T, Pedersen CB, et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell 2021;39:632–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Webb JR, Milne K, Kroeger DR, Nelson BH. PD-L1 expression is associated with tumor-infiltrating T cells and favorable prognosis in high-grade serous ovarian cancer. Gynecol Oncol 2016;141:293–302. [DOI] [PubMed] [Google Scholar]
- 33. Willis BC, Sloan EA, Atkins KA, Stoler MH, Mills AM. Mismatch repair status and PD-L1 expression in clear cell carcinomas of the ovary and endometrium. Mod Pathol 2017;30:1622–32. [DOI] [PubMed] [Google Scholar]
- 34. Ju B, Wang J, Yang B, Sun L, Guo Y, Hao Q, et al. Morphologic and immunohistochemical study of clear cell carcinoma of the uterine endometrium and cervix in comparison to ovarian clear cell carcinoma. Int J Gynecol Pathol 2018;37:388–96. [DOI] [PubMed] [Google Scholar]
- 35. Anaf V, Chapron C, El Nakadi I, De Moor V, Simonart T, Noël J-C. Pain, mast cells, and nerves in peritoneal, ovarian, and deep infiltrating endometriosis. Fertil Steril 2006;86:1336–43. [DOI] [PubMed] [Google Scholar]
- 36. Riccio LGC, Baracat EC, Chapron C, Batteux F, Abrão MS. The role of the B lymphocytes in endometriosis: a systematic review. J Reprod Immunol 2017;123:29–34. [DOI] [PubMed] [Google Scholar]
- 37. Jones RK, Bulmer JN, Searle RF. Immunohistochemical characterization of stromal leukocytes in ovarian endometriosis: comparison of eutopic and ectopic endometrium with normal endometrium. Fertil Steril 1996;66:81–9. [DOI] [PubMed] [Google Scholar]
- 38. Osuga Y, Koga K, Hirota Y, Hirata T, Yoshino O, Taketani Y. Lymphocytes in endometriosis. Am J Reprod Immunol 2011;65:1–10. [DOI] [PubMed] [Google Scholar]
- 39. Wu L, Lv C, Su Y, Li C, Zhang H, Zhao X, et al. Expression of programmed death-1 (PD-1) and its ligand PD-L1 is upregulated in endometriosis and promoted by 17beta-estradiol. Gynecol Endocrinol 2019;35:251–6. [DOI] [PubMed] [Google Scholar]
- 40. Qiu X-M, Lai Z-Z, Ha S-Y, Yang H-L, Liu L-B, Wang Y, et al. IL-2 and IL-27 synergistically promote growth and invasion of endometriotic stromal cells by maintaining the balance of IFN-γ and IL-10 in endometriosis. Reproduction 2020;159:251–60. [DOI] [PubMed] [Google Scholar]
- 41. Malutan AM, Drugan C, Drugan T, Ciortea R, Mihu D. The association between interleukin-4 -590C/T genetic polymorphism, IL-4 serum level, and advanced endometriosis. Cent Eur J Immunol 2016;41:176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kocbek V, Vouk K, Bersinger NA, Mueller MD., Lanišnik Rižner T. Panels of cytokines and other secretory proteins as potential biomarkers of ovarian endometriosis. J Mol Diagn 2015;17:325–34. [DOI] [PubMed] [Google Scholar]
- 43. Galo S, Zúbor P, Szunyogh N, Kajo K, Macháleková K, Biringer K, et al. [TNF-alpha serum levels in women with endometriosis: prospective clinical study]. Ces Gynekol 2005;70:286–90. [PubMed] [Google Scholar]
- 44. Li Y, Wang L, Tian F, Xiao F, Xu D, Zhao Y. Expression of MMPs/TIMP in endometriosis. Fertil Steril 2002;77:S32.11849634 [Google Scholar]
- 45. Mallikarjuna P, Sitaram RT, Landström M, Ljungberg B. VHL status regulates transforming growth factor-β signaling pathways in renal cell carcinoma. Oncotarget 2018;9:16297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sitaram RT, Mallikarjuna P, Landström M, Ljungberg B. Transforming growth factor-β promotes aggressiveness and invasion of clear cell renal cell carcinoma. Oncotarget 2016;7:35917–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Omwandho COA, Konrad L, Halis G, Oehmke F, Tinneberg H-R. Role of TGF-betas in normal human endometrium and endometriosis. Hum Reprod 2010;25:101–9. [DOI] [PubMed] [Google Scholar]
- 48. Mallikarjuna P, Raviprakash TS, Aripaka K, Ljungberg B, Landström M. Interactions between TGF-β type I receptor and hypoxia-inducible factor-α mediates a synergistic crosstalk leading to poor prognosis for patients with clear cell renal cell carcinoma. Cell Cycle 2019;18:2141–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Griss J, Bauer W, Wagner C, Simon M, Chen M, Grabmeier-Pfistershammer K, et al. B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma. Nat Commun 2019;10:4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamazaki N, Kiyohara Y, Uhara H, Iizuka H, Uehara J, Otsuka F, et al. Cytokine biomarkers to predict antitumor responses to nivolumab suggested in a phase 2 study for advanced melanoma. Cancer Sci 2017;108:1022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data. The “the AOCS data set” was obtained from a previous study (14); restrictions apply to the availability of these data, which were used under license for this study. Data are available from the authors upon reasonable request with the permission of the original study authors. Additional data analyzed in this study were obtained from the Gene Expression Omnibus under accession numbers, GSE44104 (15) and GSE7305 (16).