Abstract
ADAMTS family members control mammalian development and disease, primarily through their function as proteases, by regulation of extracellular matrix composition. Until recently, ADAMTS6 was known as one of the orphan proteinases of the nineteen-member family with a relatively unknown expression pattern and function. Emerging focus on this enzyme has started to uncover these unknowns and revealed a vast importance and requirement of ADAMTS6 in cardiovascular and musculoskeletal development. In addition, ADAMTS6 has been linked to numerous disease settings including several types of cancer. This review summarizes the necessity of ADAMTS6 during development, its role in disease and requirement for essential prospective studies to fully realize its biological implications and potential for therapeutic intervention.
Keywords: ADAMTS6, matrix metalloprotease, extracellular matrix, cardiovascular, musculoskeletal, cancer
Introduction
A disintegrin-like and metalloprotease with thrombospondin type 1 motif (ADAMTS) genes encode secreted proteases that have demonstrated critical roles during adult tissue homeostasis and disease acting within the extracellular matrix (ECM) (Mead and Apte, 2018). Their roles during development are well addressed with many ADAMTS gene knockouts resulting in embryonic lethality or causative in disease (Dubail and Apte, 2015). The functional subdivisions or clades of the 19 family members is based upon common affinities towards preferred substrates and evolutionary homology (Brunet et al., 2015). They share similar domain organization with a common structure, yet distinct modules. They all have a propeptide region, an N-terminal protease domain, variable number of thrombospondin type 1 repeats (TSRs) with differing C-terminal domains. The biological functions of ADAMTS proteases are largely determined by their substrates and the subsequent actions or necessity of the substrates in biological processes. While many ADAMTS family members have been extensively studied in vitro and in vivo with identified substrates and mapped cleavage sites, such as in the case of multiple ADAMTS proteases cleaving the proteoglycans aggrecan and versican, other family members are not that well defined in terms of expression profile, necessity during tissue development and homeostasis or substrate identification (Verma and Dalal, 2011; Nandadasa et al., 2014; Santamaria, 2020; Martin et al., 2021).
ADAMTS6 maps to human chromosome 6 and mouse chromosome 13 and was first identified to be expressed in human placental tissue and the domain structures were subsequently identified (Hurskainen et al., 1999). Like other ADAMTS protease family members, ADAMTS6 is secreted as a zymogen and subsequently processed, and thus activated, by furin (Turpeinen et al., 2011; Cain et al., 2016). Protein O-fucosyltransferase-2 (POFUT2) is an enzyme that adds O-linked fucose to the TSRs, which are numerous in ADAMTS6, in the endoplasmic reticulum to promote trafficking of substrates that include ADAMTS proteases (Neupane et al., 2022). TSR1-3 were modified with the O-fucose disaccharide while TSR1 was additionally modified with C-mannose (Neupane et al., 2022). Notably, inactivation of POFUT2 resulted in decreased ADAMTS6 secretion. Knockout and conditional knockout of Pofut2 resulted in shortening of limbs and accumulation of fibrillin-2 microfibrils, which mirrored Adamts6-deficient mice (this will be discussed below) (Mead et al., 2022; Neupane et al., 2022). It was further suggested that ADAMTS6 does not undergo autocatalysis, as evident by lack of additional bands in western blot analysis and comparable band intensity to catalytically inactivated ADAMTS6 (Mead et al., 2022).
Adamts6 is expressed in numerous vertebrates in a variety of tissues. In zebrafish, it is enhanced 200-500-fold after 8 h post fertilization and expressed in all organs surveyed including high expression in the liver, kidney and skeletal muscle as assessed by quantitative real-time PCR (qRT-PCR) (Brunet et al., 2015). Expression of Adamts6 mRNA was shown by RNAscope in situ hybridization in murine tissues including cardiac outflow tract, heart valves, myocardium, cartilage, tendon, and skeletal muscle amongst other tissues (Prins et al., 2018; Mead et al., 2022). In human tissue, ADAMTS6 is expressed in retinal pigment epithelial cells (RPE) as assayed by RT-PCR and northern blot analysis as well as adult chondrocytes (Bevitt et al., 2003; Kevorkian et al., 2004; Davidson et al., 2006).
It is also expressed in a variety of tumors from different origins and associated with other pathological situations as described in detail below. However, the role of ADAMTS6 in these and other yet undescribed tissues and disease settings are not fully uncovered. ADAMTS6 has long been considered an “orphan protein” with very little data on its expression profile, function, and substrate targets. Very recent studies have started to uncover these unknowns and provide a strong case for the necessity of ADAMTS6 in ECM maintenance during development and its implications during disease processes. The main processes that ADAMTS6 has been identified to participate in will be discussed in the following sections, spanning embryology through cancer and other disease settings.
ADAMTS6 in cardiovascular development and disease
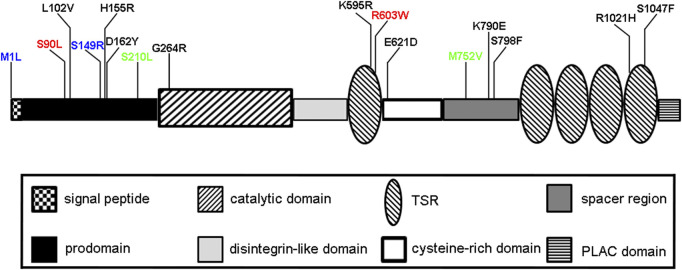
Adamts6 is expressed throughout the developing heart with strong staining noted in the outflow tract, valves and myocardium (Prins et al., 2018). As part of the “bench to bassinet program”, an ethylnitrosourea (ENU) mutagenesis forward screen uncovered two point mutations in Adamts6 that result in late embryonic death likely due to defective cardiovascular development (Li et al., 2015). One mutation effected the start codon located at position 1 (p.Met1Ile) while the other was a Ser-Arg mutation at position 149 (p.Ser149Arg) in the prodomain, both resulting in lack of ADAMTS6 protein (Figure 1). A subsequent study showed that the p.Ser149Arg mutation, with a conserved serine residue across numerous species, resulted in no secretion in the medium in a cell culture assay effectively resulting in a knockout (Prins et al., 2018). These Adamts6 S149R/S149R mice present with a myriad of congenital heart defects including double outlet right ventricle, persistent truncus arteriosus, defective atrial septation and ventricular hypertrophy (Prins et al., 2018). However, the cardiovascular targets of ADAMTS6 and the underlying mechanisms of its necessity during outflow tract development remain unknown and warrants further investigation.
FIGURE 1.
Schematic of ADAMTS6 domain structure and allelic mutations. Mutations shown in blue (M1L, S149R) are recessive mutations identified in mice that cause prenatal/neonatal lethality (Li et al., 2015). Two rare non-synonymous variants rs61736454 (p.Ser90Leu) and rs114007286 (p.Arg603Trp) are shown in red, predicted damaging variants in cardiac QRS interval are shown in black and benign-predicted variants (S210L, M752V) are shown in green (Prins et al., 2018).
Cardiac conduction is controlled by electrical impulses in a network of specialized cardiomyocytes that regulate ventricular contraction. If this is not properly regulated, cardiac conduction disorders, such as an arrhythmia, may occur. The duration of ventricular depolarization is noted by the QRS interval on an electrocardiogram. Prolonged QRS intervals are an independent predictor of mortality and commonly associated with left side heart failure (Desai et al., 2006; Badheka et al., 2013; Mozos and Caraba, 2015). A genome-wide association study uncovered rare, non-synonymous ADAMTS6 variants that are associated with altered cardiac ventricular conduction, namely a prolonged QRS interval (Figure 1) (Prins et al., 2018). These predicted pathogenic mutations had reduced secretion from the cells into the medium as compared to predicted benign variants, resulting is diminished available ADAMTS6 protein and thus proteolytic activity. To this end, Adamts6 S149R/S149R mice, which is considered an Adamts6-deficient mouse, had reduced connexin 43 staining in embryonic myocardium. Adamts6 S149R/+ juvenile and adult myocardium also had reduced connexin 43 staining as compared to control, indicative of reduced gap junctions, and is one likely explanation for defective cardiac contraction (Prins et al., 2018). As is with the outflow tract, the role of ADAMTS6 in myocardium, and in particular the cardiac conduction system, is not fully understood and might be aided by a conditional loss of function approach in mice.
ADAMTS6 in musculoskeletal tissue development and disease
The role of ADAMTS6 in musculoskeletal tissue is starting to come into focus. ADAMTS6 is highly expressed in both normal and osteoarthritic cartilage and synovium as identified from qRT-PCR analysis (Kevorkian et al., 2004; Davidson et al., 2006). Its expression is upregulated in primary annulus cells grown from degenerated human lumbar discs as shown by microarray analysis (Gruber et al., 2010). ADAMTS6 expression was downregulated, as shown by RNA-seq and validated by qRT-PCR, in adipose-derived stem cells undergoing osteogenesis and a genome wide association study (GWAS) identified ADAMTS6 as a promising, but not yet genome-wide significant, susceptible loci for osteosarcoma (Savage et al., 2013; Shaik et al., 2019).
A recent study illustrated the role of ADAMTS6 in musculoskeletal tissue utilizing a mouse model. RNAscope in situ hybridization showed Adamts6 was detected not only in resting and proliferative chondrocytes, but also in the perichondrium, which lines the developing cartilage anlagen (Mead et al., 2022). Further expression was demonstrated throughout the embryonic musculoskeletal system including in the skeletal muscle, ligaments and tendons with strong expression noted in the joint regions. Adamts6-deficient mice die at birth due to defective cardiovascular development, however, they also present with appendicular skeletal abnormalities including club foot-like phenotype as well as a myriad of axial skeleton manifestations including shorter ribs, defective sternum ossification as well as facial deformities including micrognathia and cleft palate (Mead et al., 2022). Adamts6 −/− embryos are runted and the forelimb and hindlimb bones are all shortened and widened as compared to control. The distal hindlimb tibia and fibula bones are additionally bent, with noted severe reduction of ossification in the fibula, which manifests in a club limb-like phenotype. There is an apparent delay of endochondral ossification and mineralization from embryonic day (E) 14.5 to E18.5 in the hindlimb bones with the fibula being the most drastic example. Adamts6 −/− growth plates are disorganized and have a reduction of key chondrogenic markers, including aggrecan, cartilage link protein and Sox9 with no change in chondrocyte proliferation or cell death (Mead et al., 2022). Staining of Col X, a marker of hypertrophic chondrocytes, is enlarged and points to an expanded hypertrophic chondrocyte zone, possibly at the expense of other chondrocyte zones or bone formation.
Due to the malformation in the distal hindlimb, the Adamts6 −/− knee joint was a point of further investigation. Disorganized and increased collagen staining is apparent in Adamts6 −/− perichondrium and adjacent mesenchyme with increased staining of MAGP1 and fibrillin-2 (Mead et al., 2022). Overexpression of ADAMTS6 resulted in no apparent fibrillin-1, fibrillin-2 or fibronectin microfibrils in an in vitro model. Conversely, in the absence of Adamts6 or in the presence of catalytically inactive ADAMTS6 (loss of protease activity), there were abundant microfibrils present, which points to a direct role of ADAMTS6 in microfibril formation or turnover. Therefore, these proteins were further explored as possible substrates. Direct binding of ADAMTS6 to fibrillin-1, fibrillin-2 and fibronectin was noted in Biacore assays and proteomic assays revealed that they were all indeed substrates through identification of specific cleavage sites (Mead et al., 2022). Reduced BMP, but not TGFβ signaling, was shown in Adamts6 −/− hindlimb tissue as compared to control, pointing to a potential regulation of this major signaling pathway.
Due to an increase of fibrillin-2 staining in Adamts6 −/− hindlimbs, absence of fibrillin-2 microfibrils in the presence of active ADAMTS6, direct binding of ADAMTS6 to fibrillin-2 and a specific cleavage site identified in fibrillin-2 due to ADAMTS6 activity, the relationship of ADAMTS6 and fibrillin-2 was further explored in vivo. Fbn2 haploinsufficiency on the Adamts6 −/− background (Adamts6 −/−;Fbn2 +/−) resulted in genetic reversal of limb abnormalities exhibited in Adamts6 −/− mice with restoration of chondrocyte growth plate markers (Mead et al., 2022). Reversal of axial skeleton abnormalities were also evident. Notably, reduction of Fbn1 did not affect Adamts6 −/− skeletal defects pointing to a direct role of ADAMTS6 cleavage of fibrillin-2 in the musculoskeletal tissues as necessary for proper skeletal formation.
Beyond its recently discovered roles during musculoskeletal development and associated malformations in its absence, it is important to study the role of ADAMTS6 in postnatal musculoskeletal tissues. This can be achieved in a murine conditional loss of function approach in numerous musculoskeletal tissue-specific lines. ADAMTS6 has already been implicated in osteoarthritis and it could prove fruitful to study this in an osteoarthritis mouse model, for example.
ADAMTS6 relationship to ADAMTS10
ADAMTS6 is a sister protease of ADAMTS10, based on the ADAMTS phylogenetic tree, with identical gene domain structures, high amino similarity of both the full length and catalytic domain (60% identical amino acid residues) (Apte, 2004; Somerville et al., 2004; Karoulias et al., 2020; Rose et al., 2021). Their relationship was explored in in vitro studies, utilizing human retinal pigmented epithelial cells. Knockdown of ADAMTS10 resulted in an increase of ADAMTS6 expression and overexpression of ADAMTS10 resulted in decreased ADAMTS6 expression, strongly suggesting that ADAMTS10 negatively regulates ADAMTS6 and that ADAMTS6 can compensate for loss of ADAMTS10 (Cain et al., 2016). Overexpression of ADAMTS6 resulted in increased ADAMTS10 while knockdown of ADAMTS6 resulted in decreased ADAMTS10, suggestive of an ADAMTS6 positive regulation of ADAMTS10. Their relationship was further explored in vivo using Adamts6 −/− and Adamts10 −/− embryonic limb, heart, and lung samples (Mead et al., 2022). In this study, transcriptional adaptation was apparent in that Adamts6 was upregulated in Adamts10 −/− samples while Adamts10 was upregulated in Adamts6 −/− tissues. The difference between the two studies in term of mutual regulation could be an in vitro versus an in vivo difference or cell type specific differences in how ADAMTS6 and ADAMTS10 interact. The finding of cooperation was demonstrated in double-knockout mice (Adamts6 −/−;Adamts10 −/−) which had additional musculoskeletal phenotypes compared to Adamts6 −/− alone consisting of forelimb buckling in a similar manner to Adamts6 −/− hindlimbs and subcutaneous hemorrhage, neither of which appear in Adamts10 −/− mice (Mead et al., 2022). It would be of interest to investigate Adamts6 −/−;Adamts10 −/− embryonic hearts to see if the Adamts6 −/− congenital malformations worsen or new phenotypes arise as a result of concurrent loss of Adamts10, and therefore loss of compensation, in Adamts6 −/− hearts.
Emerging roles of ADAMTS6 in cancer
ADAMTS6, like many other ADAMTS family members, has a pro- or anti-tumor role in various types of cancer, spanning several organ systems. In some instances, ADAMTS6 was shown to be either upregulated or downregulated in the same cancer type based on microarrays, clinical samples, and in vitro assays. White the overall majority of studies show that increased expression of ADAMTS6 is pro-cancer and correlates with a negative outcome, there are a few exceptions noted.
A survey of ADAMTS family members showed that ADAMTS6 expression is upregulated in breast carcinoma tissue samples with highest expression in myoepithelial cells (Porter et al., 2004). In agreement, a further study showed that elevated levels of ADAMTS6 shown by RNA-seq and validated by qRT-PCR, via NFATC2 upregulation, was demonstrated in a metastasized breast cancer cell line (Xu et al., 2021). Knockdown of ADAMTS6 in this cancer cell line resulted in delayed wound healing and decreased cell migration. Furthermore, elevated levels of ADAMTS6 mRNA associated with poor prognosis in breast cancer patients in overall survival (Xu et al., 2021). In a contrasting study, increased level of ADAMTS6 was associated with breast cancer tumor suppression in numerous breast cancer cells lines (Xie et al., 2016). In the cancer cell lines surveyed, ADAMTS6 protein and mRNA levels were downregulated. Artificially enhanced ADAMTS6 correlated with suppression of breast cancer cell migration, invasion and tumorigenesis and associated with disease-free survival. This favorable finding was in part mediated through regulation of the ERK signaling pathway through decreased phosphorylation and ADAMTS6 was shown to bind a microRNA, which reduced its expression (Xie et al., 2016).
ADAMTS6 has been linked to non-small cell lung cancer (NSCLC) through regulation of anterior gradient 2 (AGR2), an endoplasmic reticulum protein family member (Luu et al., 2020). In this cancer, tumor cell-acquired drug resistance is problematic, especially in patients who initially respond favorably to epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKI) and subsequently acquire resistance to treatment (Pao et al., 2005). AGR2 is upregulated in drug-resistant NSCLC tumor cells and corelated with cancer cell proliferation, migration, and invasion. Knockdown of AGR2 resulted in increased expression of ADAMTS6 as well as decreased VEGFA, a marker of vascular endothelium and surrogate marker of angiogenesis (Luu et al., 2020). Importantly, knockdown of ADAMTS6 resulted in upregulation of AGR2 mRNA and protein and provides the evidence for a negative feedback loop in these NSCLC cells. The relationship of ADAMTS6 and NSCLC was further delineated by another group, focusing on epigenetic regulators of histone methylation. Chromatin immunoprecipitation and qRT-PCR analysis revealed that ADAMTS6 expression is over 60-fold upregulated during epithelial to mesenchymal transition (EMT) and associated with loss of H3K27me3 on its promoter (Lachat et al., 2020). Increased ADAMTS6 staining was more evident in EMT+ tumors than in EMT- tumors while knockdown of ADAMTS6 decreased cell invasion (Lachat et al., 2020). Therefore, the authors concluded that increased ADAMTS6 expression mediated by H3K27 demethylation favored invasion during EMT.
Bulk RNA-Seq identified ADAMTS6 as one of the most differentially expressed genes in a survey of data sets of human colorectal cancer as compared to normal tissue (Xiao et al., 2015). Further qRT-PCR validation revealed that ADAMTS6 is significantly downregulated in colon cancer while significantly upregulated in rectal cancer (Xiao et al., 2015). This dichotomy points to differing regulatory mechanisms and signaling pathways at work in colon and rectal cancer (Li et al., 2012). A subsequent study, utilizing RNA-Seq data and qRT-PCR analysis, identified ADAMTS6 as highly expressed in colon cancer tissues and cell lines and was associated with cancer progression and predicted worse prognosis in colon cancer patients (Wang et al., 2020). Knockdown of ADAMTS6 in a colon cancer cell line resulted in repression of colon cancer cell growth, invasion, and migration while overexpression led to the opposite. This was in part mediated by the regulation of E-cadherin, N-cadherin, Vimentin, Snail p-AKT and p-p65 and point to the regulation of EMT and AKT/NFκB signaling pathways, respectively (Wang et al., 2020). Therefore, ADAMTS6 might act as a catalyst for EMT in colon cancer progression thereby enhancing colon cancer cell growth, proliferation, migration, and invasion. Further studies are warranted to fully understand the role of ADAMTS6 in colon cancer cell biology.
A study utilizing The Cancer Genome Atlas (TCGA) and RNA-seq data revealed that high ADAMTS6 expression in gastric cancer patients associated with poor clinical outcome and reliably predicted overall survival (Zhu et al., 2021). Consistent with this finding, immunohistochemistry and qRT-PCR analysis showed increased ADAMTS6 protein staining and mRNA expression, respectively, in gastric cancer samples. Furthermore, ADAMTS6 expression is correlated with tumor stage and histological grade and shown to promote the development and occurrence of stomach cancer (Zhu et al., 2021). Gene set enrichment analysis showed that high ADAMTS6 mRNA levels led to elevated VEGF and TNF pathway expression amongst other cancer-related pathways. Therefore, elevated levels of ADAMTS6 in gastric cancer is a tumor promoter and mediates occurrence, tumor cell proliferation, invasion and metastasis that leads to increased mortality.
Several other cancers have been linked to ADAMTS6. ADAMTS6 was significantly downregulated in placental site trophoblastic tumor (PSTT), a rare type of gestational trophoblastic neoplasms through interrogation of microarray data and confirmed by qRT-PCR analysis (Gan et al., 2022). This finding was further validated via immunohistochemistry with diffuse ADAMTS6 positive staining in normal villi while reduced staining was apparent in PSTT cells (Gan et al., 2022). Increased staining of ADAMTS6 is an indicator of poor prognosis in patients with esophageal squamous cell carcinoma (Liu et al., 2018). Shown to be abundantly apparent in the cytoplasm and nucleus, ADAMTS6 showed strong staining in the cancer cell invasive front. While it was not shown to be correlated with the patient’s age, sex or tumor size, ADAMTS6 staining was corelated with clinical stage, metastasis, recurrence and poor overall survival (Liu et al., 2018). A further study utilizing qRT-PCR analysis showed increased expression of ADAMTS6 in aggressive and aggressive-invasive prolactin pituitary tumors, as compared to non-invasive tumors, and further showed that this increase in expression corelated with tumor recurrence and progression (Wierinckx et al., 2007; Raverot et al., 2010).
With the dual role of ADAMTS6 as either cancer tumor suppressor and pro-tumoral agent, analysis and correlation studies are of key importance to decipher the precise role of ADAMTS6 in cancer development. The actions of ADAMTS6 may depend upon several factors including the type of cells present as well as the interactions with other ECM components. Further studies that add to current in vitro data, such as inhibiting or enhancing ADAMTS6 in tumor mouse models is warranted to study these cancers in depth to reveal therapeutic potential.
ADAMTS6 in disease
Along with ARID1B, disruption of ADAMTS6 is associated with developmental delay, speech impairment, dysmorphic features and intellectual learning disability (Malli et al., 2014). The genomic translocation breakpoint identified that a sequence from intron 11 of ADAMTS6 fused to ARID1B intron 2. Whether this genomic translocation is a result of either ADAMTS6 or ARID1B or both being disrupted is unknown. While a few patients with developmental delay have been identified with large deletions in the ADAMTS6 locus, ARID1B is a known regulator of neural development and has previously been associated with intellectual disability (Firth et al., 2009; Halgren et al., 2012; Hoyer et al., 2012; Santen et al., 2012; Tsurusaki et al., 2012). This warrants further investigation.
Recently, ADAMTS6 has been linked to many microRNAs (miR) in several disease settings. ADAMTS6 was down regulated by miR-210-3p in a screen of preeclampsia biomarkers utilizing the Gene Expression Omnibus database (Jiang et al., 2020). It was further shown that Inc-CTD2383M3.1 acted as a competing endogenous RNA by competitively binding to miR-210-3p, thus alleviating suppression of ADAMTS6 (Jiang et al., 2020). A further study, utilizing bioinformatics, showed that ADAMTS6 is a potential hub for miRNAs of oral fluids related to orthodontics (Kapoor et al., 2021). Bioinformatics, RNAseq and qRT-PCR analysis, along with in vitro studies, showed that circular (circ)ADAMTS6 RNA, a type of noncoding RNA, was downregulated after treatment of IL-1β (Fu et al., 2021). circADAMTS6, originated from the ADAMTS6 gene and located in cytoplasm, is a 669 nucleotide circRNA that was formed from joining the 5′ splice site of exon 21 to the 3’ splice site of exon 24 (Fu et al., 2021). Overexpression of circADAMTS6 inhibited human chondrocyte apoptosis while overexpression of miR-431-5p had an opposing effect. Suggested binding of circADAMTS6 to miR-431-5p was shown through RNA-FISH and luciferase assays showing both located in direct contact in the cytoplasm, although definitive studies determining their relationship need to be explored further. Another connection of circADAMTS6 to a miRNA was explored in a follow up osteoarthritis study. qRT-PCR analysis showed that circADAMTS6 expression is reduced in osteoarthritic cartilage as compared to normal control cartilage (Shen et al., 2022). Knockdown of circADAMTS6 reduced chondrocyte proliferation and increased apoptosis and elevated inflammatory response in control human cartilage, which mirrored IL-1β stimulation in normal cartilage. Overexpression of circADAMTS6 with concurrent inhibition of miR-324-5p enhanced human chondrocyte proliferation while suppressing chondrocyte apoptosis, inflammatory response and the PI3K/AKT/mTOR pathway (Shen et al., 2022). Taken together, circADAMTS6 negates IL-1β-induced chondrocyte apoptosis and warrants further studies in the treatment of osteoarthritis. Up regulation of another circADAMTS6 was associated with poor overall survival in patients with esophageal squamous cell carcinoma (ESCC) (Bu et al., 2022). Conversely, knockdown of circADAMTS6 inhibited ESCC cell proliferation, migration and invasion in xenograft tumors. Thus, as with the role of ADAMTS6 in cancer, the role of circADAMTS6 in disease is tissue specific.
Additional disease settings involving ADAMTS6 have been reported, but have limited data and require follow up studies. ADAMTS6 was upregulated, as assessed by microarray and confirmed by qRT-PCR analysis, in kidney epithelial cells in an in vitro model of Fabry disease, an X-linked lysosomal disorder (Shin et al., 2015). ADAMTS6 is highly expressed in osteoarthritic cartilage and synovium as identified from qRT-PCR analysis (Kevorkian et al., 2004; Davidson et al., 2006). Two GWAS identified ADAMTS6 as a susceptible locus for inguinal hernia (Jorgenson et al., 2015; Fadista et al., 2022). Adamts6 expression, as determined by RNA-seq and qRT-PCR analysis, was noted in mouse connective tissue equivalent to human transversalis fascia, which when weaken or defective can lead to an inguinal hernia (Jorgenson et al., 2015). And finally, GWAS located single nucleotide polymorphisms (SNPs) in the region of ADAMTS6 associated with changes in central corneal thickness in the eye and may lead to disease if not properly maintained (Lu et al., 2013).
ADAMTS6 substrates
Like other family members, ADAMTS6 participates in ECM remodeling and thus targets will likely be found in this area. Evidence that fibrillin-1, fibrillin-2 and fibronectin were potential substrates was shown through increased immunofluorescence staining in Adamts6 −/− hindlimbs, as compared to control, with no change in RNA levels (Mead et al., 2022). They were therefore subjected to BIAcore analysis and shown to all bind to ADAMTS6 and followed up by targeted proteomic searches in which the substrate of interest was overexpressed in the presence or absence of active ADAMTS6, which identified specific cleavage sites in each. Cleavage of fibrillin-2 was verified through western blot analysis that showed cleavage products when the C-terminal half of fibrillin-2 was co-cultured with ADAMTS6 overexpressing cells, but not in the presence of inactivate ADAMTS6 (Mead et al., 2022). In a series of experiments utilizing retinal epithelial cells, Biacore analysis revealed that ADAMTS6 binds to heparin, syndecan-4, fibrillin-1 and LTBP-1 (Cain et al., 2016). ADAMTS6 was shown to cleave LTBP-1 in a purified protein to protein assay to show direct interaction, but the precise cleavage site is unknown (Cain et al., 2016). The cleavage of LTBP-1 by ADAMTS6 suggested a potential role in regulation of TGFβ signaling. While TGFβ signaling, as measured by pSMAD1/5/8 staining, was unchanged in Adamts6-deficient embryonic hindlimbs, another tissue and setting could uncover a yet unknown role and function (Mead et al., 2022). Western blot analysis also showed cleavage of syndecan-4 by ADAMTS6, but no cleavage site was determined (Cain et al., 2016).
While these few ADAMTS6 substrates have been suggested and fibrillin-2 verified through cleavage site identification and genetic interaction, additional focus on substrate verification and identification is warranted. ADAMTS6 is expressed in numerous cell types and tissues during development and disease and its roles in these settings will not be fully realized until additional tissue-specific substrates are identified. Thus, an unbiased proteomic search in select ECM-enriched populations is necessary to uncover additional substrates. Once these cleavage sites are verified and shown to be critical, utilizing in vitro and in vivo means, only then can the role of ADAMTS6 in these settings be realized which is necessary to reveal new avenues of research towards therapeutic intervention.
Conclusion and perspectives in ADAMTS6 functional biology
This review summarizes the known ADAMTS6 connections to development and diseases known currently. This will undoubtedly expand in the near future and beyond as this once “orphan protein” starts to be recognized and more studies are undertaken, such as whole genome sequencing of rare disorders in humans and preclinical models as well as proteomic screenings. ADAMTS6 participates in a myriad of processes including cardiac and musculoskeletal development as well as cancer and disease and interacts with many ECM molecules and signaling pathways as a result. Greater understanding of the ADAMTS6 regulatory network is vital to predict undesired outcomes when attempting to modify consequences of disease.
Adamts6-deficent mice have made a considerable impact on the ECM field and have led to the discovery of its necessity in cardiovascular and musculoskeletal biology (Li et al., 2015; Prins et al., 2018; Mead et al., 2022). However, the mice die at birth and thus preclude investigation into any postnatal studies or reveal further complications from absence of Adamts6 in disease settings. Adamts6 is expressed in differing cells lineages and it is difficult at present to discern cell type specific functions. Therefore, a conditional genetic approach is warranted in future in vivo studies. The full picture of the role of ADAMTS6 can only begin to be realized once the substrates are identified and verified. This will allow for delineation of molecular pathway and mechanistic insight into its role in the ECM during tissue development, homeostasis, and disease settings. While a few substrates have been suggested and some verified, an unbiased, proteomics approach is warranted to discern the full landscape. Only then will it be readily suitable to generate targeted approaches for therapeutic applications.
Fibrillin microfibrils are a necessary component of the ECM that provides mechanical stability to tissues and regulates several signaling pathways such as TGFβ and BMP (Ramirez and Rifkin, 2009). That ADAMTS6 regulates fibrillin microfibrils in development warrants further investigation in disease settings. ADAMTS6 was shown to bind to and cleave both fibrillin-1 and fibrillin-2, yet only fibrillin-2 increased abundance was apparent in Adamts6 −/− musculoskeletal tissues during development (Mead et al., 2022). Whether that holds true in other tissues during development or in adult tissue homeostasis or disease remains to be explored. Fibrillin microfibrils can form homodimers or heterodimers and it is possible that the dominance of fbn1 postnatal expression as compared to fbn2, which is virtually absent starting in early postnatal tissues, is a result of abundance fibrillin-1 transcription masking a fibrillin-2 microfibril core (Zhang et al., 1995; Lin et al., 2002; Charbonneau et al., 2003; Marson et al., 2005). Therefore, it is possible that ADAMTS6 targets fibrillin-1 predominantly in postnatal tissues but may also target fibrillin-2 if unmasked. This will be necessary to explore and understand if targeting one versus the other is preferred in a disease setting.
A further function of ADAMTS6 was uncovered in focal adhesion biology. Through a series of experiments utilizing a human retinal epithelial cell line, it was shown that ADAMTS6 negatively regulates focal adhesions (Cain et al., 2016). Addition of the C-terminal half of ADAMTS6 stimulated adhesions and was bound to heparin and syndecan-4, both focal adhesion components (Woods et al., 2000). Overexpression of full length, active ADAMTS6 resulted in lack of focal adhesions while ADAMTS6 knockdown or mutations in the ADAMTS6 catalytic domain or in the furin cleavage site (both mutations rendering ADAMTS6 inactive) resulted in an over-abundance of focal adhesions (Cain et al., 2016). Additionally, heparin sulfate, which is necessary for induction of focal adhesions (Gopal et al., 2010), was depleted in the presence of ADAMTS6 and abundant in ADAMTS6 knockdown cells as determined by a prominent cell surface glycocalyx (Cain et al., 2016). The lack of focal adhesions in the presence of ADAMTS6 influenced microfibril deposition. Overexpression of ADAMTS6 resulted in lack of microfibril and fibronectin deposition, in agreement with a subsequent study utilizing mouse embryonic fibroblasts (Cain et al., 2016; Mead et al., 2022).
ADAMTS6 was shown to have a role in gap junctions in ventricular myocardium. Adamts6-deficient embryonic myocardium had a reduction of Cx43 staining, a predominant myocardial gap junction protein (Thomas et al., 1998; Gutstein et al., 2001; Prins et al., 2018). This was also evident in adult Adamts6 +/− myocardium without a change in Gja1 mRNA levels, which suggested post-transcriptional regulation (Prins et al., 2018). These findings point to the possible function of ADAMTS6 in prolonged QRS interval resulting from impaired myocardial connectivity.
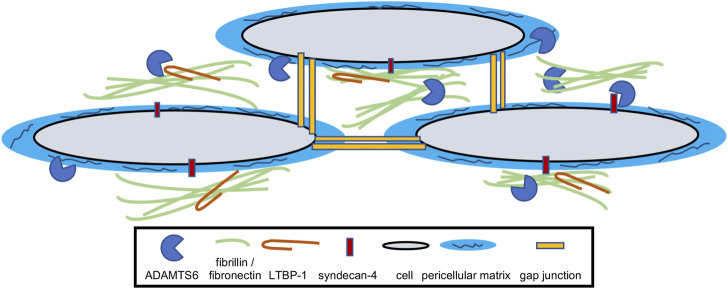
In summary, ADAMTS6 is a major component of the ECM and regulates several biological processes during development and disease (Figure 2). Further studies on patient diseases and biological samples will uncover additional regulatory network roles of ADAMTS6, which will generate supplementary scientific avenues and tools to reveal therapeutic potential. Understanding the full landscape of ADAMTS6 targets is necessary to unlock its potential in congenital malformations and disease.
FIGURE 2.
Model for the function of ADAMTS6. ADAMTS6 locates and functions within the extracellular matrix and has been shown to cleave fibrillin-1, fibrillin-2, fibronectin, LTBP-1 and syndecan-4. It directly or indirectly effects cellular gap junctions as well as pericellular matrix/glycocalyx quantity.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by funding from the National Institutes of Health (RO1HL156987).
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Apte S. S. (2004). A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motifs: The ADAMTS family. Int. J. Biochem. Cell Biol. 36, 981–985. [DOI] [PubMed] [Google Scholar]
- Badheka A. O., Singh V., Patel N. J., Deshmukh A., Shah N., Chothani A., et al. (2013). QRS duration on electrocardiography and cardiovascular mortality (from the national Health and nutrition examination survey-III). Am. J. Cardiol. 112, 671–677. [DOI] [PubMed] [Google Scholar]
- Bevitt D. J., Mohamed J., Catterall J. B., Li Z., Arris C. E., Hiscott P., et al. (2003). Expression of ADAMTS metalloproteinases in the retinal pigment epithelium derived cell line ARPE-19: Transcriptional regulation by TNFalpha. Biochim. Biophys. Acta 1626, 83–91. 10.1016/s0167-4781(03)00047-2 [DOI] [PubMed] [Google Scholar]
- Brunet F. G., Fraser F. W., Binder M. J., Smith A. D., Kintakas C., Dancevic C. M., et al. (2015). The evolutionary conservation of the A Disintegrin-like and Metalloproteinase domain with Thrombospondin-1 motif metzincins across vertebrate species and their expression in teleost zebrafish. BMC Evol. Biol. 15, 22. 10.1186/s12862-015-0281-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu J., Gu L., Liu X., Nan X., Zhang X., Meng L., et al. (2022). The circRNA circADAMTS6 promotes progression of ESCC and correlates with prognosis. Sci. Rep. 12, 13757. 10.1038/s41598-022-17450-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain S. A., Mularczyk E. J., Singh M., Massam-Wu T., Kielty C. M. (2016). ADAMTS-10 and -6 differentially regulate cell-cell junctions and focal adhesions. Sci. Rep. 6, 35956. 10.1038/srep35956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau N. L., Dzamba B. J., Ono R. N., Keene D. R., Corson G. M., Reinhardt D. P., et al. (2003). Fibrillins can co-assemble in fibrils, but fibrillin fibril composition displays cell-specific differences. J. Biol. Chem. 278, 2740–2749. 10.1074/jbc.M209201200 [DOI] [PubMed] [Google Scholar]
- Davidson R. K., Waters J. G., Kevorkian L., Darrah C., Cooper A., Donell S. T., et al. (2006). Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res. Ther. 8, R124. 10.1186/ar2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A. D., Yaw T. S., Yamazaki T., Kaykha A., Chun S., Froelicher V. F. (2006). Prognostic significance of quantitative QRS duration. Am. J. Med. 119, 600–606. 10.1016/j.amjmed.2005.08.028 [DOI] [PubMed] [Google Scholar]
- Dubail J., Apte S. S. (2015). Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol. 44-46, 24–37. 10.1016/j.matbio.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Fadista J., Skotte L., Karjalainen J., Abner E., Sorensen E., Ullum H., et al. (2022). Comprehensive genome-wide association study of different forms of hernia identifies more than 80 associated loci. Nat. Commun. 13, 3200. 10.1038/s41467-022-30921-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth H. V., Richards S. M., Bevan A. P., Clayton S., Corpas M., Rajan D., et al. (2009). DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533. 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Li L., Wang B., Wu J., Li H., Han Y., et al. (2021). CircADAMTS6/miR-431-5p axis regulate interleukin-1β induced chondrocyte apoptosis. J. Gene Med. 23, e3304. 10.1002/jgm.3304 [DOI] [PubMed] [Google Scholar]
- Gan J., Chen Z., Feng X., Wei Z., Zhang S., Du Y., et al. (2022). Expression profiling of lncRNAs and mRNAs in placental site trophoblastic tumor (PSTT) by microarray. Int. J. Med. Sci. 19, 1–12. 10.7150/ijms.65002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal S., Bober A., Whiteford J. R., Multhaupt H. A., Yoneda A., Couchman J. R. (2010). Heparan sulfate chain valency controls syndecan-4 function in cell adhesion. J. Biol. Chem. 285, 14247–14258. 10.1074/jbc.M109.056945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H. E., Hoelscher G. L., Hanley E. N., Jr (2010). Annulus cells from more degenerated human discs show modified gene expression in 3D culture compared with expression in cells from healthier discs. Spine J. 10, 721–727. 10.1016/j.spinee.2010.05.014 [DOI] [PubMed] [Google Scholar]
- Gutstein D. E., Morley G. E., Tamaddon H., Vaidya D., Schneider M. D., Chen J., et al. (2001). Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circ. Res. 88, 333–339. 10.1161/01.res.88.3.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren C., Kjaergaard S., Bak M., Hansen C., El-Schich Z., Anderson C. M., et al. (2012). Corpus callosum abnormalities, intellectual disability, speech impairment, and autism in patients with haploinsufficiency of ARID1B. Clin. Genet. 82, 248–255. 10.1111/j.1399-0004.2011.01755.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer J., Ekici A. B., Endele S., Popp B., Zweier C., Wiesener A., et al. (2012). Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am. J. Hum. Genet. 90, 565–572. 10.1016/j.ajhg.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurskainen T. L., Hirohata S., Seldin M. F., Apte S. S. (1999). ADAM-TS5, ADAM-TS6, and ADAM-TS7, novel members of a new family of zinc metalloproteases. General features and genomic distribution of the ADAM-TS family. J. Biol. Chem. 274, 25555–25563. 10.1074/jbc.274.36.25555 [DOI] [PubMed] [Google Scholar]
- Jiang R., Wang T., Zhou F., Yao Y., He J., Xu D. (2020). Bioinformatics-based identification of miRNA-lncRNA-and mRNA-associated ceRNA networks and potential biomarkers for preeclampsia. Med. Baltim. 99, e22985. 10.1097/MD.0000000000022985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E., Makki N., Shen L., Chen D. C., Tian C., Eckalbar W. L., et al. (2015). A genome-wide association study identifies four novel susceptibility loci underlying inguinal hernia. Nat. Commun. 6, 10130. 10.1038/ncomms10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P., Chowdhry A., Bagga D. K., Bhargava D., Aishwarya S. (2021). MicroRNAs in oral fluids (saliva and gingival crevicular fluid) as biomarkers in orthodontics: Systematic review and integrated bioinformatic analysis. Prog. Orthod. 22, 31. 10.1186/s40510-021-00377-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoulias S. Z., Taye N., Stanley S., Hubmacher D. (2020). The ADAMTS/fibrillin connection: Insights into the biological functions of ADAMTS10 and ADAMTS17 and their respective sister proteases. Biomolecules 10, E596. 10.3390/biom10040596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevorkian L., Young D. A., Darrah C., Donell S. T., Shepstone L., Porter S., et al. (2004). Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 50, 131–141. 10.1002/art.11433 [DOI] [PubMed] [Google Scholar]
- Lachat C., Bruyere D., Etcheverry A., Aubry M., Mosser J., Warda W., et al. (2020). EZH2 and KDM6B expressions are associated with specific epigenetic signatures during EMT in non small cell lung carcinomas. Cancers (Basel) 12, E3649. 10.3390/cancers12123649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. N., Zhao L., Wu J., Wu B., Yang H., Zhang H. H., et al. (2012). Differences in gene expression profiles and carcinogenesis pathways between colon and rectal cancer. J. Dig. Dis. 13, 24–32. 10.1111/j.1751-2980.2011.00551.x [DOI] [PubMed] [Google Scholar]
- Li Y., Klena N. T., Gabriel G. C., Liu X., Kim A. J., Lemke K., et al. (2015). Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521, 520–524. 10.1038/nature14269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Tiedemann K., Vollbrandt T., Peters H., Batge B., Brinckmann J., et al. (2002). Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J. Biol. Chem. 277, 50795–50804. 10.1074/jbc.M210611200 [DOI] [PubMed] [Google Scholar]
- Liu L., Yang Z., Ni W., Xuan Y. (2018). ADAMTS-6 is a predictor of poor prognosis in patients with esophageal squamous cell carcinoma. Exp. Mol. Pathol. 104, 134–139. 10.1016/j.yexmp.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Lu Y., Vitart V., Burdon K. P., Khor C. C., Bykhovskaya Y., Mirshahi A., et al. (2013). Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat. Genet. 45, 155–163. 10.1038/ng.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu T. T., Bach D. H., Kim D., Hu R., Park H. J., Lee S. K. (2020). Overexpression of AGR2 is associated with drug resistance in mutant non-small cell lung cancers. Anticancer Res. 40, 1855–1866. 10.21873/anticanres.14139 [DOI] [PubMed] [Google Scholar]
- Malli T., Duba H. C., Erdel M., Marschon R., Kranewitter W., Deutschbauer S., et al. (2014). Disruption of the ARID1B and ADAMTS6 loci due to a t(5;6)(q12.3;q25.3) in a patient with developmental delay. Am. J. Med. Genet. A 164A, 3126–3131. 10.1002/ajmg.a.36738 [DOI] [PubMed] [Google Scholar]
- Marson A., Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K., et al. (2005). Homotypic fibrillin-1 interactions in microfibril assembly. J. Biol. Chem. 280, 5013–5021. 10.1074/jbc.M409029200 [DOI] [PubMed] [Google Scholar]
- Martin D. R., Santamaria S., Koch C. D., Ahnstrom J., Apte S. S. (2021). Identification of novel ADAMTS1, ADAMTS4 and ADAMTS5 cleavage sites in versican using a label-free quantitative proteomics approach. J. Proteomics 249, 104358. 10.1016/j.jprot.2021.104358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead T. J., Apte S. S. (2018). ADAMTS proteins in human disorders. Matrix Biol. 71-72, 225–239. 10.1016/j.matbio.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead T. J., Martin D. R., Wang L. W., Cain S. A., Gulec C., Cahill E., et al. (2022). Proteolysis of fibrillin-2 microfibrils is essential for normal skeletal development. Elife 11, e71142. 10.7554/eLife.71142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozos I., Caraba A. (2015). Electrocardiographic predictors of cardiovascular mortality. Dis. Markers 2015, 727401. 10.1155/2015/727401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandadasa S., Foulcer S., Apte S. S. (2014). The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol. 35, 34–41. 10.1016/j.matbio.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane S., Berardinelli S. J., Cameron D. C., Grady R. C., Komatsu D. E., Percival C. J., et al. (2022). O-fucosylation of thrombospondin type 1 repeats is essential for ECM remodeling and signaling during bone development. Matrix Biol. 107, 77–96. 10.1016/j.matbio.2022.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., et al. (2005). Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73. 10.1371/journal.pmed.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S., Scott S. D., Sassoon E. M., Williams M. R., Jones J. L., Girling A. C., et al. (2004). Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin. Cancer Res. 10, 2429–2440. 10.1158/1078-0432.ccr-0398-3 [DOI] [PubMed] [Google Scholar]
- Prins B. P., Mead T. J., Brody J. A., Sveinbjornsson G., Ntalla I., Bihlmeyer N. A., et al. (2018). Exome-chip meta-analysis identifies novel loci associated with cardiac conduction, including ADAMTS6. Genome Biol. 19, 87. 10.1186/s13059-018-1457-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F., Rifkin D. B. (2009). Extracellular microfibrils: Contextual platforms for TGFbeta and BMP signaling. Curr. Opin. Cell Biol. 21, 616–622. 10.1016/j.ceb.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raverot G., Wierinckx A., Dantony E., Auger C., Chapas G., Villeneuve L., et al. (2010). Prognostic factors in prolactin pituitary tumors: Clinical, histological, and molecular data from a series of 94 patients with a long postoperative follow-up. J. Clin. Endocrinol. Metab. 95, 1708–1716. 10.1210/jc.2009-1191 [DOI] [PubMed] [Google Scholar]
- Rose K. W. J., Taye N., Karoulias S. Z., Hubmacher D. (2021). Regulation of ADAMTS proteases. Front. Mol. Biosci. 8, 701959. 10.3389/fmolb.2021.701959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria S. (2020). ADAMTS-5: A difficult teenager turning 20. Int. J. Exp. Pathol. 101, 4–20. 10.1111/iep.12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen G. W., Aten E., Sun Y., Almomani R., Gilissen C., Nielsen M., et al. (2012). Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat. Genet. 44, 379–380. 10.1038/ng.2217 [DOI] [PubMed] [Google Scholar]
- Savage S. A., Mirabello L., Wang Z., Gastier-Foster J. M., Gorlick R., Khanna C., et al. (2013). Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat. Genet. 45, 799–803. 10.1038/ng.2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik S., Martin E. C., Hayes D. J., Gimble J. M., Devireddy R. V. (2019). Transcriptomic profiling of adipose derived stem cells undergoing osteogenesis by RNA-seq. Sci. Rep. 9, 11800. 10.1038/s41598-019-48089-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Ji C., Lin J., Yang H. (2022). Regulation of circADAMTS6-miR-324-5p-PIK3R3 ceRNA pathway may be a novel mechanism of IL-1β-induced osteoarthritic chondrocytes. J. Bone Min. Metab. 40, 389–401. 10.1007/s00774-021-01308-0 [DOI] [PubMed] [Google Scholar]
- Shin Y. J., Jeon Y. J., Jung N., Park J. W., Park H. Y., Jung S. C. (2015). Substrate-specific gene expression profiles in different kidney cell types are associated with Fabry disease. Mol. Med. Rep. 12, 5049–5057. 10.3892/mmr.2015.4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville R. P., Jungers K. A., Apte S. S. (2004). Discovery and characterization of a novel, widely expressed metalloprotease, ADAMTS10, and its proteolytic activation. J. Biol. Chem. 279, 51208–51217. 10.1074/jbc.M409036200 [DOI] [PubMed] [Google Scholar]
- Thomas S. A., Schuessler R. B., Berul C. I., Beardslee M. A., Beyer E. C., Mendelsohn M. E., et al. (1998). Disparate effects of deficient expression of connexin43 on atrial and ventricular conduction: Evidence for chamber-specific molecular determinants of conduction. Circulation 97, 686–691. 10.1161/01.cir.97.7.686 [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y., Okamoto N., Ohashi H., Kosho T., Imai Y., Hibi-Ko Y., et al. (2012). Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 44, 376–378. 10.1038/ng.2219 [DOI] [PubMed] [Google Scholar]
- Turpeinen H., Kukkurainen S., Pulkkinen K., Kauppila T., Ojala K., Hytonen V. P., et al. (2011). Identification of proprotein convertase substrates using genome-wide expression correlation analysis. BMC Genomics 12, 618. 10.1186/1471-2164-12-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P., Dalal K. (2011). ADAMTS-4 and ADAMTS-5: Key enzymes in osteoarthritis. J. Cell. Biochem. 112, 3507–3514. 10.1002/jcb.23298 [DOI] [PubMed] [Google Scholar]
- Wang Y. P., Zhao Y. J., Kong X. L. (2020). A metalloproteinase of the disintegrin and metalloproteinases and the ThromboSpondin motifs 6 as a novel marker for colon cancer: Functional experiments. Genet. Mol. Biol. 43, e20190266. 10.1590/1678-4685-GMB-2019-0266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierinckx A., Auger C., Devauchelle P., Reynaud A., Chevallier P., Jan M., et al. (2007). A diagnostic marker set for invasion, proliferation, and aggressiveness of prolactin pituitary tumors. Endocr. Relat. Cancer 14, 887–900. 10.1677/ERC-07-0062 [DOI] [PubMed] [Google Scholar]
- Woods A., Longley R. L., Tumova S., Couchman J. R. (2000). Syndecan-4 binding to the high affinity heparin-binding domain of fibronectin drives focal adhesion formation in fibroblasts. Arch. Biochem. Biophys. 374, 66–72. 10.1006/abbi.1999.1607 [DOI] [PubMed] [Google Scholar]
- Xiao W. H., Qu X. L., Li X. M., Sun Y. L., Zhao H. X., Wang S., et al. (2015). Identification of commonly dysregulated genes in colorectal cancer by integrating analysis of RNA-Seq data and qRT-PCR validation. Cancer Gene Ther. 22, 278–284. 10.1038/cgt.2015.20 [DOI] [PubMed] [Google Scholar]
- Xie Y., Gou Q., Xie K., Wang Z., Wang Y., Zheng H. (2016). ADAMTS6 suppresses tumor progression via the ERK signaling pathway and serves as a prognostic marker in human breast cancer. Oncotarget 7, 61273–61283. 10.18632/oncotarget.11341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Li N., Wang Y., Yu J., Mi J. (2021). Calcium channel TRPV6 promotes breast cancer metastasis by NFATC2IP. Cancer Lett. 519, 150–160. 10.1016/j.canlet.2021.07.017 [DOI] [PubMed] [Google Scholar]
- Zhang H., Hu W., Ramirez F. (1995). Developmental expression of fibrillin genes suggests heterogeneity of extracellular microfibrils. J. Cell Biol. 129, 1165–1176. 10.1083/jcb.129.4.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. Z., Liu Y., Liao X. W., Luo S. S. (2021). Identified a disintegrin and metalloproteinase with thrombospondin motifs 6 serve as a novel gastric cancer prognostic biomarker by bioinformatics analysis. Biosci. Rep. 41, BSR20204359. 10.1042/BSR20204359 [DOI] [PMC free article] [PubMed] [Google Scholar]