Abstract
Rapid and precise serum cytokine quantification provides immense clinical significance in monitoring the immune status of patients in rapidly evolving infectious/inflammatory disorders, examplified by the ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. However, real-time information on predictive cytokine biomarkers to guide targetable immune pathways in pathogenic inflammation is critically lacking, because of the insufficient detection range and detection limit in current label-free cytokine immunoassays. In this work, we report a highly sensitive localized surface plasmon resonance imaging (LSPRi) immunoassay for label-free Interleukin 6 (IL-6) detection utilizing rationally designed peptide aptamers as the capture interface. Benefiting from its characteristically smaller dimension and direct functionalization on the sensing surface via Au–S bonding, the peptide-aptamer-based LSPRi immunoassay achieved enhanced label-free serum IL-6 detection with a record-breaking limit of detection down to 4.6 pg/mL, and a wide dynamic range of ~6 orders of magnitude (values from 4.6 to 1 × 106 pg/mL were observed). The immunoassay was validated in vitro for label-free analysis of SARS-CoV-2 induced inflammation, and further applied in rapid quantification of serum IL-6 profiles in COVID-19 patients. Our peptide aptamer LSPRi immunoassay demonstrates great potency in label-free cytokine detection with unprecedented sensing capability to provide accurate and timely interpretation of the inflammatory status and disease progression, and determination of prognosis.
Keywords: Antibody-derived peptide aptamer (ADPA), Interluekin-6 (IL-6), localized surface plasmon resonance (LSPR), nanoplasmonic immunoassay, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
Graphical Abstract
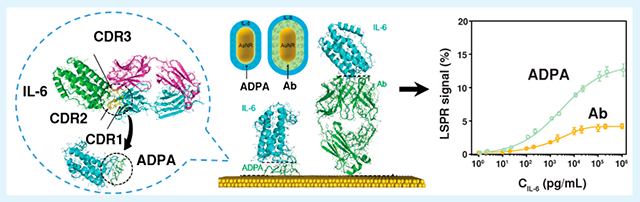
INTRODUCTION
Cytokines are low-molecular-weight signaling proteins that play central roles in dynamic regulation of the innate and adaptive immunity, and in the process of inflammation.1 Rapid, precise, and sensitive quantification of cytokines offers valuable clinical and biological information in monitoring the immune status of patients in cancer, autoimmune, and infectious diseases.2 The newly emerged coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces a hyper-inflammatory response, the major cause of disease severity and death.3 Recent studies have suggested that elevated serum levels of inflammatory cytokines such as interleukin 6 (IL-6) have been observed in patients, and could be strong indicators of COVID severity and good therapeutic targets for effective treatment via the right timing and drug dosage.4–6 These have necessitated the profiling of cytokine biomarkers in a real-time manner to guide immunomodulatory therapeutic interventions to mitigate the pathogenic inflammation.7,8
However, accurate and real-time detection of cytokines remains challenging, because of their low trace amount, highly dynamic secretion, and short half-lives.9,10 The commonly used enzyme-linked immunosorbent assay (ELISA) requires labor-intensive sample handling, labeling, and washing processes, falling short of meeting the urgent demands for sensitive, dynamic cytokine monitoring. In contrast, label-free biosensing circumvents the need for secondary tagging by providing direct physical signals upon target binding, rendering them a consolidated approach for high-speed diagnosis.11–13 Among the various transduction mechanisms of optical,14,15 electrical,16,17 and mechanical18,19 sensors, nanoplasmonic biosensors have shown great promise in the compact, simple, and rapid detection of protein biomarkers.20–23 By utilizing the localized surface plasmon resonance (LSPR), nanoplasmonic biosensing allows remote transduction of biomolecular binding in a highly localized environment (~20–60 nm) around the plasmonic nanoparticle (NP) surface,20,24 presenting excellent sensing performance to detect small target analytes such as cytokines. Despite the great potential, the limit of detection (LOD) of label-free LSPR immunoassay is reaching its theoretical limit of ~10 pg/mL due to fundamental constraints in the quality (Q)-factor and sensing volume of the plasmonic NPs, and the binding affinity and mass ratio of the antibody–antigen pairs.25 As the serum cytokine levels span across a board range with lower boundaries at a few pg/mL (i.e., 1.6–4823 pg/mL of IL-6 in COVID-19 patients),3,23 the current label-free LSPR biosensors cannot afford the rapid cytokine detection with desirable LOD and dynamic range for precise immune monitoring of patients.
Since LSPR features an enhanced electromagnetic field in the vicinity of NP surface and decays exponentially into the medium,12,26 ideal LSPR biosensing craves a thin recognition layer with analyte binding events occurring in close proximity of the sensing surface. While antibodies have been predominantly used as the recognition elements, these Y-shaped proteins with considerably large dimensions (~150 kDa) display clear disadvantages in probing nanosized cytokines (6–70 kDa) in a label-free LSPR sensing scheme. This opens opportunities to explore novel probes with much smaller sizes and larger analyte-probe mass ratios to surpass the theoretical limit of LSPR biosensing. Recent studies in nanobody,27 a single domain of camelid heavy-chain antibodies with a molecular weight of 15 kDa, have demonstrated high binding affinity to target proteins and evolved as a new class of therapeutic and diagnostic probes. Peptide aptamers derived from directed evolution have emerged as another alternative, in which a shorter amino acid sequence (~3–5 kDa) with high affinity against the selected target is embedded in an inert peptide scaffold.28,29 Nonetheless, the conventional approach in generating nanobodies and peptide aptamers involves cumbersome processes for the construction of phage libraries, affinity selection, and purification, which greatly hinders the scalable production of these small-sized probe elements and their practical application in sensitive and timely biomarker analysis.30
Herein, we report a LSPR imaging (LSPRi) immunoassay enabled by rationally designed antibody-derived peptide aptamers (ADPAs) for label-free detection of ultralow concentration of IL-6 in serum. The ADPAs were constructed by joining three complementarity-determining region (CDR) loops of a camelid anti-IL-6 antibody with minimalist linkers that could connect various N- and C-termini of loops in the structure without imposing structural strains (Chart 1a). Compared to its antibody counterpart, the ADPA exhibits a much smaller dimension (1.5 nm) and lower mass ratio to cytokines whiling maintaining high specificity and reasonable affinity, serving as an “ideal” biosensing interface for enhanced nanoplasmonic cytokine detection (Chart 1b). By integrating with a simple microfluidic device and a dark field imaging system, our ADPA-LSPRi immunoassay achieved label-free detection of IL-6 with a total assay time of 35 min and a LOD down to 4.6 pg/mL, using a minimal sample of 3 μL (Chart 1c). The immunoassay was validated by physiological samples extracted from cocultivated epithelial cells, macrophages, and SARS-CoV-2 S-protein, and further applied for rapid analysis of IL-6 profile in 16 blood samples from COVID-19 patients under different treatment conditions.
Chart 1.
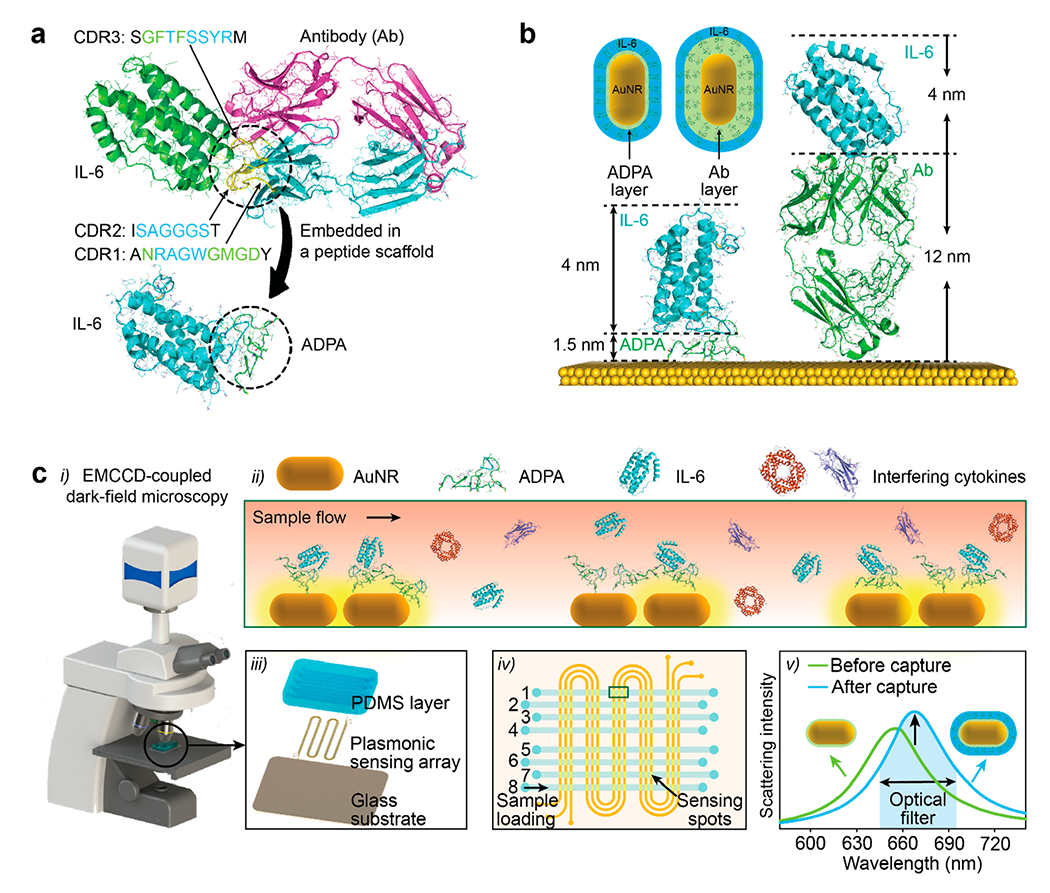
Schematic Illustration of ADPA Design and ADPA-LSPRi Immunoassay: (a) The Solved Antibody-Antigen Complex Structure,a (b) Schematic Structures of IL-6 Captured by Antibody (Left) and ADPA (Right) on a Bare Gold Surface,b and (c) The Illustration of APDA-LSPRi Immunoassay Platformc
aHere, IL-6 (olive)was recognized by three CDR loops from heavy chain on the antibody (cyan). bThe dimensions of the antibody, ADPA, and IL-6 were calculated to be 12.0, 1.5 and 4.0 nm, respectively. cCit-AuNRs were firstly patterned on a glass substrate, followed by the direct functionalization of ADPA. As-fabricated LSPRi chip was mounted under an optical dark-field microscope. IL-6 in samples captured by the ADPA-functioned AuNRs generated an increase in scattering light intensity, which can be monitored by a high-resolution EMCCD in real time.
RESULTS AND DISCUSSION
Rational Design of Antibody-Derived Peptide Aptamers.
IL-6 is a critical pro-inflammatory pleiotropic cytokine involved in a variety of biological processes, such as the acute inflammatory response. Therefore, anti-IL-6 antibodies have been extensively developed as effective theragnostic agents for the detection and suppression of IL-6 activity to alleviate inflammation. Based on the camelid monoclonal anti-IL-6 antibody 61H7 with well-identified CDR structure,31,32 we explored a strategy for the rational development of anti-IL-6 ADPAs. In the crystallography structure of the antigen–antibody complex (Chart 1a, PDB ID 4o9h), all three CDR loops of 61H7 heavy chain are in contact with IL-6, contributing to the binding specificity and affinity. Therefore, we designed ADPA sequences by connecting three CDR loops (CDR1–3) of 61H7 heavy chain with linkers (see the Experimental Section). Briefly, minimalist linkers were designed to ensure that all three CDRs in the ADPA can adopt similar conformations upon binding IL-6 as in the original antibody–i.e., not too short to impose structural strains between neighboring loops in the bound state and also not too long and flexible with increased entropy loss upon binding the target. Cystines were introduced to the termini of loops as the anchors for direct surface functionalization on the gold nanoparticles (AuNPs, 40 nm × 80 nm) through Au–S bonding. The shape and size of our sensor AuNRs have been specifically selected to yield optimal sensing performance33 with small LOD values. The engineered nanoparticles are highly sensitive to the local refractive index with a relatively large sensing volume, displaying a notable scattering intensity change upon analyte surface binding.34,35 Two schemes of peptide aptamers (denoted as “Scheme 1” and “Scheme 2”) were designed with distinct ordering of the CDR loops (Figure 1b). Given the close proximity, only one cysteine was included at the C-terminus of CDR1 before the second linker. Because the second linker features close proximity between the CDR2 C-terminus and the N-terminus of either CDR1 or CDR3 and also a sharp turn of the backbone, we adopted the sequence of Gly-Proline-Gly known to favor turn structure in proteins.36 For the first linker, we used the flexible and hydrophilic (Gly-Ser)n tandem repeats that are commonly used in protein engineering.37,38 The repeat number n was chosen to be the minimum length that can physically connect the C-terminus of CDR2 or CDR3 with the N-terminus of CDR2 without introducing structural strains in the bound state of ADPA with IL-6 and had the value of 2 and 4 for Scheme 1 and Scheme 2, respectively (e.g., the structure of Scheme 1 ADPA is depicted in Figure S1 in the Supporting Information). Atomistic discrete molecular dynamics (DMD) simulations39–41 were applied to assess the possible binding between the designed ADPA and IL-6 on the surface of AuNP (see Figure 1a, as well as Movie 1 in the Supporting Information). Our DMD simulations confirmed that the designed sequence shown in Figure 1a could adopt the “native-like” conformation compatible for target binding and cysteines could hop on the surface, to allow peptide conformational sampling (Movie 1 in the Supporting Information). Compared to the Fab of 61H7, the ADPA displayed further reduced molecule weight (~3 kDa) and mass/size ratio to the bound IL-6, as clearly shown in the generated simulation figures (see Chart 1b and Figure 1a).
Figure 1.
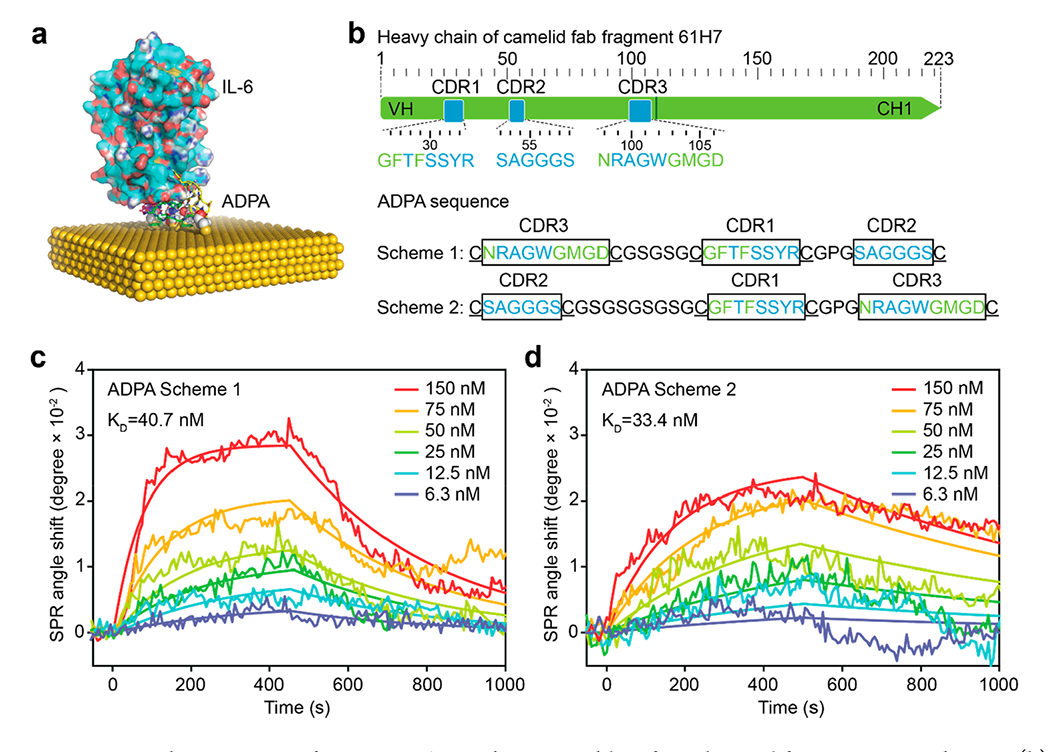
(a) A representative snapshot structure of ADPA-IL-6 complex on a gold surface obtained from DMD simulations. (b) Three CDR loops were adopted from a full sequence of anti-IL-6 antibody (PDB ID 4o9h). Two ADPA schemes were designed by connecting three CDR loops with spacers and cystine residues were introduced for site-specific functionalization on gold. (c, d) SPR kinetics of interaction between two ADPA schemes and human IL-6.
SPR Characterization of Peptide Aptamer.
The affinities of anti-IL-6 ADPAs were analyzed by surface plasmon resonance (SPR). A series of human IL-6 standards with concentrations of 6.3–150 nM were injected into an SPR sensor chip functionalized with Scheme 1 or Scheme 2 at a surface coverage of 83.3 pg/mm2. By fitting the sensorgram with a one-to-one binding model, Scheme 1 displayed an association constant (ka) of 6.9 × 104 M−1·s−1, a dissociation constant (kd) of 2.8 × 10−3 s−1, and an equilibrium dissociation constant (KD) of 40.7 nM (see Figure 1c and Table 1), while, for Scheme 2, the measured values were ka = 3.3 × 104 M−1 s−1, kd = 1.1 × 10−3 s−1, and KD = 33.4 nM, respectively (see Figure 1d and Table 1). The two schemes showed similar affinity, while Scheme 1 displayed a faster binding rate and dissociation rate with IL-6. Compared to the antibody (with estimated values of ka = 2.0–3.2 × 105 M−1 s−1 and kd = 0.6 × 10−4 s−1), both ADPAs exhibited lower ka and higher kd, indicating a slower initial binding and faster disassociation behavior. This reduced affinity resulted from the fact that the light chain in contact with IL-6 (see Figure S1a in the Supporting Information) also contributed to the ultrahigh affinity of the original antibody. In addition, the weaker affinity might also be due to the structure alteration of the framework region (FR), which serves as a scaffold of the CDRs and a stabilizer of the antigen-binding site. The mutations of FRs loop have been reported to be associated with the affinity variation of antibodies.42 In addition, the amino acid–gold interactions possibly resulted in undesired interaction between CDRs and the gold surface, followed by unexpected folding of the CDRs. For instance, Arg, Asn in CDRs can be adsorbed on gold through Au–N interaction, while the carboxylic group on the side chain of Asp allows its proximity to the gold surface.43 Despite a slightly reduced affinity than the corresponding antibody, our ADPAs still showed a comparable affinity level with some other anti-IL-6 antibodies, such as siltuximab (11.7 nM), an FDA-approved anti-IL-6 monoclonal antibody.44 Thereby, both ADPAs were used for the following investigations on their immobilization optimization for enhanced plasmonic sensing.
Table 1.
Properties and Kinetic Constants of Peptide Aptamers
| molecular weight (kDa) | isoelectric point | Ka(M−1 s−1) | Kd (s−1) | KD (M) | |
|---|---|---|---|---|---|
| Scheme 1 | 3.40 | 7.54 | 6.93 × 104 | 2.82 × 10−3 | 4.07 × 10−8 |
| Scheme 2 | 3.69 | 7.54 | 3.29 × 104 | 1.10 × 10−3 | 3.34 × 10−8 |
Optimized Immobilization of ADPA on Gold Nanorods (AuNRs) for Enhanced Plasmonic Sensing.
Based on the Mie theory, the highly localized electric field of LSPR is sensitive to subtle refractive index variations surrounding the AuNP, while the strength of the electric field decays exponentially with the distance away from the nanoparticle/medium interface, as shown below:45
| (1) |
where R is the plasmonic response (e.g., wavelength shift, intensity change), m is the refractive-index sensitivity from the plasmonic nanoparticle, Δη is the refractive index difference induced from adsorbed layer, ld is the decay length, and d1 and d2 are the thicknesses of recognition layer and analyte layer, respectively. Accordingly, R increases with thinner recognition layers and/or thicker analyte layers (suggesting smaller probes), larger analyte-to-probe mass ratios, and more available binding sites for enhanced plasmonic sensing. As a result, the tiny-sized anti-IL-6 ADPA with a much smaller MW, relative to its antibody counterpart, allows the binding of IL-6 molecules in much closer proximity to the AuNP surface and a relatively thicker analyte binding layer (larger d2/d1 ratio; see Figures 2a and 2d), leading to higher IL-6 sensitivity. Moreover, the conventional approach to assemble the biomolecular probes on the AuNP sensing surface is mainly through 1-ethyl-3-[3-(dimethylamino)propyl] carbodiimide/N-hydroxysuccinimide (EDC/NHS) coupling chemistry (see Figure S2 in the Supporting Information). Such non-site-specific labeling often results in random orientations of immobilized antibodies, which could reduce the density of available antigen binding sites and thereby compromise the sensitivity of the immunoassay.46,47 In the computationally designed anti-IL-6 ADPA, we intentionally introduced cystine residues in the FRs, enabling site-specific direct functionalization of ADPA via Au–S bonding for optimized sensing performance. In addition, the ADPAs possess net positive charges in physiological conditions (pH 7.4) that are naturally in favor of interacting with negatively charged citrate gold nanorods (Cit-AuNR).48–50 Hence, Cit-AuNR was selected in place of commonly used hexadecyltrimethylammonium bromide capped gold nanorods (CTAB-AuNR) as the sensing NPs, due to easier ligand exchange.51,52
Figure 2.
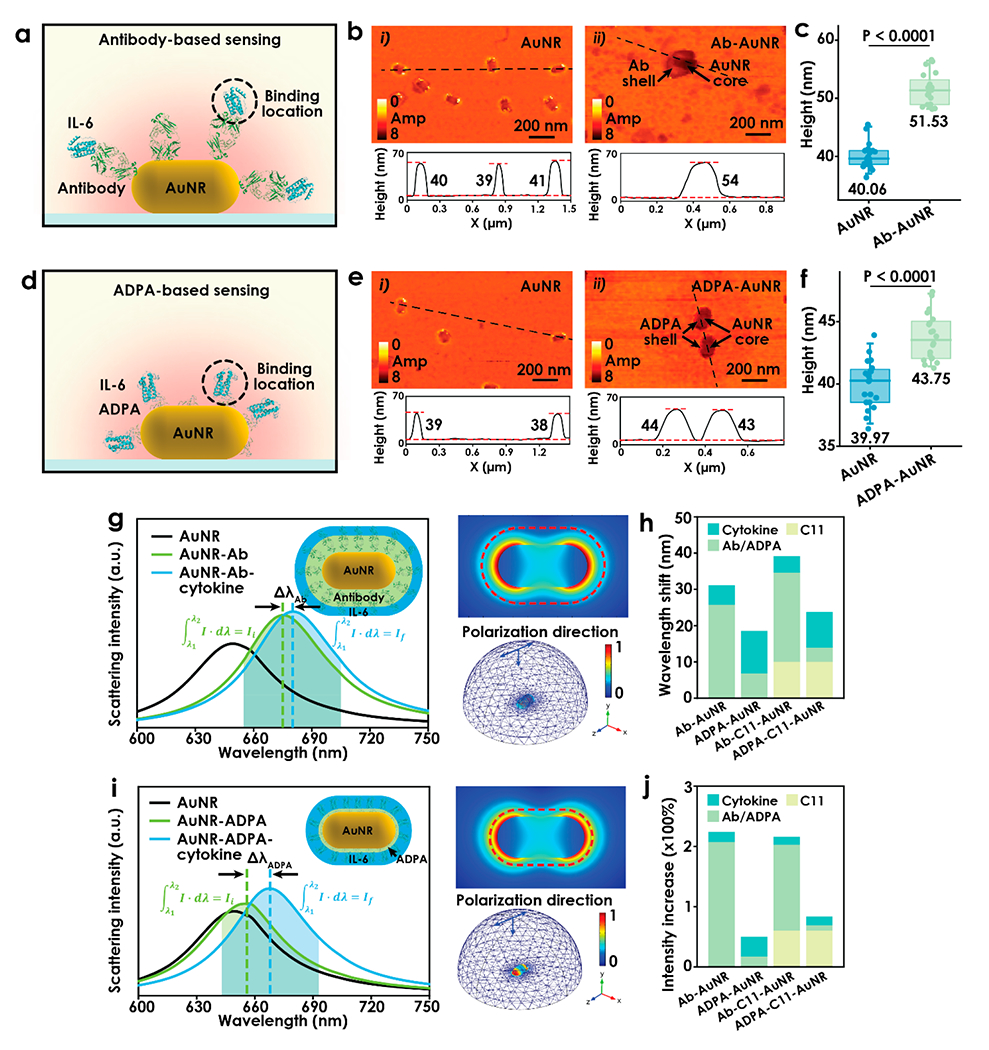
(a, d) Respective illustrations of typical antibody- and APDA-antigen binding scenarios in LSPR sensing scheme, where the binding occurs at different locations from AuNR-dielectric interface. (b, e) Respective representative s-SNOM images of isolated AuNRs patterned on the substrate and antibody/ADPA directly functionalized on AuNR. Bottom shows cross-sectional profiles of particles taken along the dashed line. (c, f) Statistics of particle heights of AuNR/Ab-AuNR and AuNR/APDA. Each dot represents an individual measurement of the particles (n = 20). Box plots depict the 25th percentile, median, and 75th percentile; whiskers show the 95% confidence intervals. (g, i) Predicted scattering spectra of AuNR, AuNR coated with Ab/ADPA, and after binding with cytokine. A 2D illustration of the model is given in the top-right corner in the plot, and 2D and 3D electromagnetic field distributions are given in the top-right and bottom-right corners, respectively. (h) Comparison of scattering resonance wavelength shift induced in each step. (j) Comparison of relative scattering intensity increase in each step.
We first performed a series of experimental characterizations to confirm the direct immobilization of ADPA on AuNRs yields a thinner recognition layer for enhanced plasmonic sensing. The bare Cit-AuNRs before surface functionalization were imaged by scanning electron microscopy (SEM), showing high monodispersity with a dimension of ~40 nm in width and ~80 nm in length (Figure S3 in the Supporting Information). After functionalization with ADPA Scheme 1, Scheme 2, or anti-IL-6 antibody, the size and surface properties of the AuNRs were characterized by dynamic light scattering (DLS), zeta potential (ξ) analysis, and scattering-type scanning near-field optical microscopy (s-SNOM).46 The DLS measurements of bare AuNRs showed an average hydrodynamic size of 87.3 nm in length (Table 2), which is consistent with the result from SEM (Figure S3). The immobilization of Scheme 1 and Scheme 2 resulted in small size increases of 4.2 and 2.3 nm, respectively. Meanwhile, antibody-coated AuNRs displayed a larger hydrodynamic size increase of 12.3 nm, indicating the formation of a thinner ADPA layer on the AuNRs than that of antibody. Zeta-potential measurements revealed the as-expected positive surface charges of +19.5 and +11.0 mV for Scheme 1 and Scheme 2 ADPAs, and −10.6 mV for anti-IL-6-Antibodies at pH 7.4, respectively (Table 2). The functionalization of ADPAs neutralized the surface charge of the bare AuNR (−29.2 mV) and resulted in a less-charged complex under physiological conditions (−10.3 mV for Scheme 1-AuNR, −13.5 mV for Scheme 2-AuNR). Successful complex formation of antibody or ADPAs on AuNRs was further verified with s-SNOM by scanning the images at the characteristic infrared spectrum (1660 cm−1) of amide I band from peptides and proteins (Figures 2b and 2e).53 Specifically, the well-dispersed bare AuNRs were imaged with bright hot spots at the apexes, showing enhanced localized electrical fields due to the antenna effect. The dense dark coating observed for the antibody-AuNR (Ab-AuNR, Figure 2b) and ADPA-AuNR (Figure 2e) originated from the stretching vibration of the amide I band at 1660 cm−1, confirming the presence of antibody and ADPAs on the AuNRs. The height profiles displayed an average value of 40.1 nm for isolated AuNRs. This height increased to 51.5 nm (Figure 2c) upon antibody assembly, while only 43.8 nm (Figure 2f) upon ADPA assembly, indicating a significantly thinner recognition layer of ADPA-AuNRs. Note that the dimensions of NPs imaged by s-SNOM appeared to be slightly larger than the SEM results, which is mainly due to the relatively large radius (~35 nm) of the tip apex used in our experiment.53
Table 2.
Hydrodynamic Size and Zeta Potential of Antibody, Peptide Aptamers, AuNR, and the Corresponding Conjugates
| antibody | Scheme 1 | Scheme 2 | AuNR | Antibody-AuNR | Scheme 1-AuNR | Scheme 2-AuNR | |
|---|---|---|---|---|---|---|---|
| size (nm) | 23.6 ± 0.3 | 2.4 ± 0.8 | 2.8 ± 1.0 | 87.3 ± 3.8 | 99.6 ± 2.3 | 91.5 ± 1.9 | 89.6 ± 8.2 |
| zeta potential (mV) | −10.6 ± 3.5 | 19.5 ± 1.0 | 11.7 ± 2.1 | −29.2 ± 2.2 | −16.4 ± 1.4 | −10.3 ± 0.7 | −13.5 ± 1.7 |
We then conducted finite-element analysis (FEA) simulations on a AuNR complexed with varied probes to validate the effect of the recognition layer thickness on plasmonic sensing. Based on our SEM and s-SNOM results, a 40 nm × 80 nm AuNR model was constructed, and its scattering resonance peak was calculated to be at ~649 nm (Figure 2g). The local refractive index change induced by the presence of a recognition layer generated a shift in resonance wavelength (Δλ) and a relative scattering intensity increase (ΔI/I0) of the AuNRs. As shown in Figures 2g–j, the formation of a dense packed antibody layer on the AuNR resulted in a large resonance wavelength shift of 25.8 nm, while the ADPA-AuNR displayed an ~9.0 nm red-shift in resonance peak. Subsequently, the binding of a single layer of IL-6 on Ab-AuNR induced a small change in Δλ (5.3 nm) and a corresponding ΔI/I0 of 16.4%. In contrast, the IL-6 layer coated on the ADPA-AuNR yielded a red-shift of the resonance peak of 11.6 nm and a much stronger scattering intensity increase of 32.1%, highlighting the significance of a thinner capture layer for enhanced LSPR response to analyte binding.
In practical sensing applications, conjugation of antibodies on AuNPs often requires a thiolated linker (i.e., thiolated alkane HS-(CH2)11-COOH, (C11)) to form a monolayer35 on NPs to allow subsequent surface function (Figure S2 in the Supporting Information). To understand how our ADPA-based direct immobilization strategy affects the sensor performance, we compared the plasmonic signal (Δλ and ΔI/I0) upon IL-6 binding in four conjugation models (Figures 2g, 2i, and S4 in the Supporting Information). As indicated in Figures 2h and 2j, the ADPA-AuNR elicits enhanced plasmonic response in two ways: (i) small-sized ADPA-based sensor induces stronger plasmonic signals than conventional antibody-functioned LSPR sensor; (ii) direct functionalization of probes brings the binding close to NP surface by eliminating additional surface layers, leading to notable higher signals in all models. The coupling of these two effects have thus drastically altered the calculated plasmonic enhancement for the bound IL-6 layer, achieving a 3.3-fold intensity increase in comparison to conventional Ab-AuNR sensor conjugates. It is worth noting that all the simulation results are based on the assumption that all the antibodies and ADPAs are oriented in a manner to sterically favor the IL-6 binding on the sensor surface.
ADPA-LSPRi Immunoassay for Serum IL-6 Detection.
In comparison to conventional spectrum-based approaches, intensity-based LSPRi platforms have recently been reported as one of the most sensitive label-free cytokine immunoassays with limits of detection (LODs) down to 11.3–45.6 pg/mL.23 As these LSPRi immunoassays involve exclusive use of covalently linked antibodies, the substitution with ADPAs has the potential to further push down the detection limits and allow ultrasensitive real-time measurement. Hence, we devised the computationally designed anti-IL-6 ADPAs to construct an ADPA-LSPRi immunoassay to demonstrate enhanced label-free IL-6 cytokine detection in serum. As shown in Figure S5 in the Supporting Information, the workflow of the immunoassay involves AuNR immobilization, direct ADPA functionalization, and on-chip sensing. Notably, with the scalable prefabrication of the AuNR patterned chips that can be stored for at least a few weeks, the subsequent one-step ADPA functionalization requires only 1 h of incubation to prepare the immunoassay ready for on-chip protein quantification. Figure 3a presents the real-time sensing curves of IL-6 in serum using the ADPA-LSPRi immunoassays. The introduction of IL-6 (100 pg/mL) resulted in relative intensity changes (ΔI/I0) of 2.5% for ADPA-Scheme 1, 2.1% for ADPA-Scheme 2, and 0.7% for antibody after 30 min IL-6 incubation and 5 min buffer washing. Both ADPA schemes showed excellent selectivity toward IL-6 (see Figures 3b and 3c). The LSPRi heat maps and corresponding calibration curves of the three probes were established by measuring a set of serum samples with varied IL-6 concentrations in three replicates (Figures 3d–f). The ADPA-LSPRi immunoassay displayed significantly higher IL-6 sensitivity (signal magnitude that corresponds to unit concentration change of IL-6) at the low concentration region with 0.25%/pg mL−1 for ADPA-Scheme 1 and 0.17%/pg mL−1 for ADPA-Scheme 2, compared to antibody-LSPRi immunoassay with 0.06%/pg mL−1. The LODs (defined as three times the standard deviation of a blank sample over the slope of the calibration curve, 3σ/S) were calculated as 4.6 pg/mL for ADPA-Scheme 1 and 6.7 pg/mL for ADPA-Scheme 2, which were 8-fold lower than the antibody-LSPRi immunoassay (37.4 pg/mL). Such enhanced sensing performance on the low level of IL-6 can be ascribed to the thinner recognition layer formed by the smaller peptides, as predicted in our optical simulations. Moreover, the ADPA-LSPRi immunoassay exhibited a wider dynamic range that spans across ~6 orders (5 to 1 × 106 pg/mL), compared to their antibody counterpart (37 to 5 × 104 pg/mL). This is likely due to the combined contribution of larger binding site density on gold surface and uniform desired orientation of ADPAs via cysteine-targeted immobilization, offering higher accessibility of IL-6 molecules to the sensing surface. The ADPA-LSPRi immunoassays were further validated in a blind test, together with the “gold standard” ELISA by measuring the IL-6 levels in 10 unknown samples in cell culture medium (see Figure S6 in the Supporting Information). The results from our immunoassay and ELISA were in good agreement (a correlation coefficient of R2= 0.99 was observed), demonstrating the computationally derived ADPAs as valid probes for LSPRi immunoassay to provide enhanced sensing capability for label-free cytokine detection to achieve real-time immune monitoring.8
Figure 3.
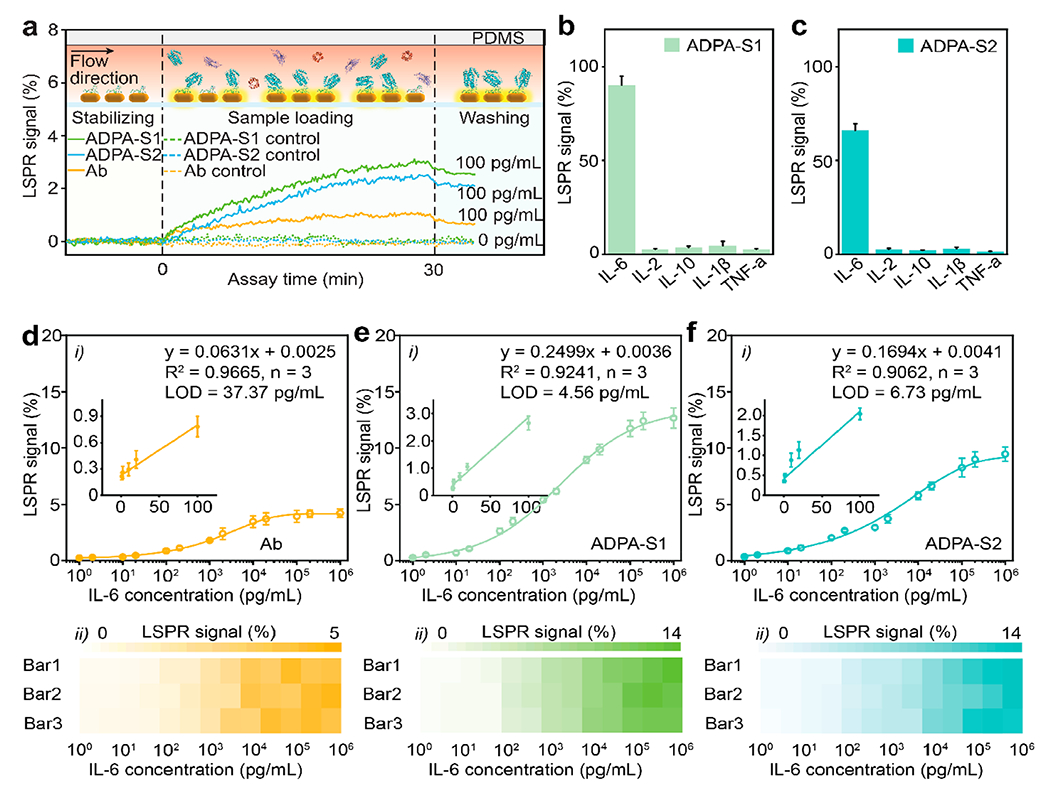
(a) Real-time monitoring of scattering light intensity change during a LSPRi immunoassay. (Top) Cross-sectional views of the channel during baseline establishment, sample loading, and washing process. (b, c) Selectivity test for ADPA-Scheme 1 and ADPA-Scheme 2. (d–f) Calibration curves for Ab (panel (d)), ADPA-Scheme 1 (panel (e)), and ADPA-Scheme 2 (panel (f)) LSPRi immunoassay. Insets show the corresponding linear regions of the calibration curves. The bottom panels present the corresponding heatmaps of the LSPR signals.
On-Chip Analysis of SARS-CoV-2 S-Protein-Induced Inflammation.
To unveil the potential of the ADPA-LSPRi as a translation approach for in situ cytokine monitoring under clinically relevant conditions, we performed on-chip cytokine detection for quantitative analysis SARS-CoV-2 S-protein-induced inflammation using in vitro and patient samples. At the onset of COVID-19, the pathophysiology of SARS-CoV-2 infected lung is worsened with a hyperinflammatory response, eliciting excessive release of inflammatory cytokines such as IL-6.54,55 In an attempt to create a biomimetic immune status of COVID-19 patients upon viral infection, we incubated epithelial cells and/or macrophages with SARS-CoV-2 and conducted time-course measurements (confirmed by ELISA, as shown in Figure S7 in the Supporting Information) of IL-6 production, in response to different doses of S-protein utilizing our ADPA-LSPRi immunoassay (Figure 4a.i). The isolated macrophages, comparing to epithelial cells and control samples, showed elevated immune response with increased SARS-CoV-2 concentration (Figures 4b and 4c). When cocultured with epithelial cells, the macrophages exerted an acute inflammatory response by the viral S-proteins with a significantly heightened IL-6 expression overtime. This is primarily due to a positive feedback loop between epithelial cells and macrophages initiated by the binding of viral S-proteins on host cells via angiotensin-converting enzyme 2 (ACE2),56 which activates a series of intercellular pathways and results in the upregulation of pro-inflammatory cytokines (i.e., IL-6). The pro-inflammatory cytokines attract macrophages to the site of infection, exacerbating the inflammation response by further production of IFN-γ and establishing the positive feedback loop. We further examined the immune status of COVID-19 patients by measuring the IL-6 concentration in 16 blood samples with our ADPA-LSPRi immunoassay. These human serum specimens were collected from two groups of COVID-19 severe patients with and without immunomodulatory treatment of tocilizumab (Figure 4a.ii) from the University of Michigan Hospital (patients or their authorized representatives provided informed consent for the use of biospecimens, as approved by the Institutional Review Board of the University of Michigan (HUM000179668)). The serum specimens were separated from the whole blood samples, following the standard procedure, and directly used for ADPA-LSPRi immunoassay without dilution. The results from the 9 samples of COVID-19 patients that had not received any treatment showed a range of 10.4–1452.4 pg/mL with an average IL-6 level of 374.3 pg/mL (Figure 4d). For 7 patients after treatments, IL-6 level was elevated to the range of 91.5–5277.4 pg/mL with an average of 1250.1 pg/mL. The elevated serum IL-6 might be explained by the inhibited receptor-mediated clearance of serum IL-6, induced by the tocilizumab.57–59 All the results above from ADPA-LSPRi immunoassay were validated by the ELISA method (Figure S8 in the Supporting Information), which again prove the feasibility of ADPA-LSPRi immunoassay for accurate quantification of cytokines across a wide concentration range in clinical diagnosis.
Figure 4.
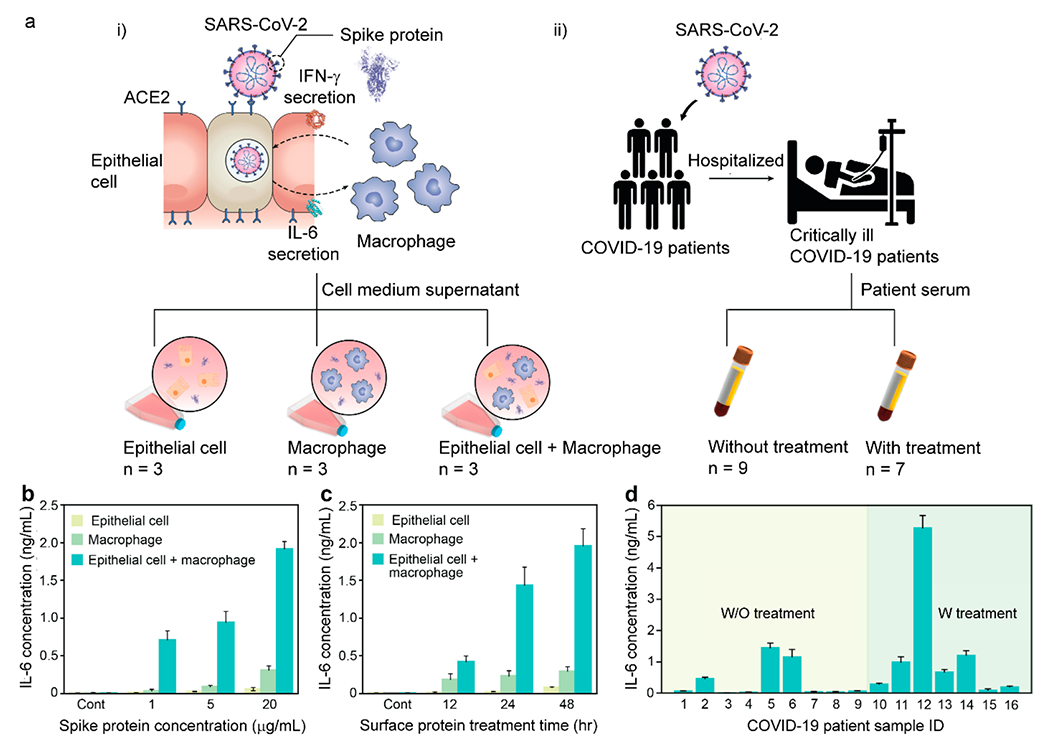
(a) (i) Illustration of SARS-CoV-2 infected host cells with a positive cytokine feedback loop and (ii) blood test workflow for COVID-19 patient. Macrophages cocultured with epithelial cells were stimulated by SARS-CoV-2 surface protein and the resulting cell medium was collected for ADPA-LSPRi immunoassay. IL-6 secretion levels under (b) different concentrations of surface protein and (c) incubation times. (d) Blood test results using ADPA-LSPRi immunoassay for patient stratification.
CONCLUSIONS
In conclusion, we have demonstrated a computation-assisted rationale design of ADPAs as a translational sensing interface for sensitive label-free detection of IL-6 in pathogenic inflammation. In place of traditional approach for anti-IL-6 PA generation, the anti-IL-6 ADPAs were readily constructed by assembling three CDR loop structures with designed spacers to form the much-simplified antibody alternatives. The tiny-sized ADPAs allowed proximity of target molecule to AuNR surface down to 1.5 nm upon recognition with high selectivity, adequate affinity, and ample binding sites, which greatly advanced the nanoplasmonic sensing of IL-6 in a label-free setting. Such enhancement was verified by both FEA simulation and experimental results, yielding a LOD of 4.6 pg/mL and wide dynamic range for IL-6 that surpassed most of the antibody-LSPRi immunoassays. The ADPA-LSPRi immunoassay showed high consistency with the “gold standard” ELISA by testing under physiological conditions and in COVID-19 patient stratification. Most significantly, this computational approach can be applied as a general method to produce the corresponding ADPAs from antibodies for any emerging biomarkers. Coupling the label-free nature of the LSPRi with the excellent sensing performance enabled by APDAs, the ADPA-LSPRi immunoassay could establish a new paradigm for rapid, high-throughput diagnosis, and can be readily integrated with microfluidic systems to provide in situ, multiparametric, point-of-care analysis. Further investigations in the robustness of the approach in constructing ADPAs for target analytes, the scalable production of designed ADPAs, the system variation in immunoassay fabrication, and the reproducibility of assay performance in complex biological medium are essential to overcome the limitations to provide practical clinical tools to better understand the human physiology and improve disease treatment for better health care.
EXPERIMENTAL SECTION
Computational Design of Antibody-Derived Peptide Aptamers.
Based on the X-ray crystallography structure of the antibody–antigen complex (Figure S1a in the Supporting Information), coordinates of three heavy-chain CDR loops were obtained as the starting template for designing ADPAs. Since CDR2 happens to be positioned between CDR1 and CDR3 in space and the C-terminus of CDR2 is in close proximity to the N-terminus of the other two loops (Figure S1b), two sequence design schemes were selected to connect three loops–Scheme 1: Cys-CDR3-Cys-Linker-Cys-CDR1-Cys-Linker-CDR2-Cys, and Scheme 2: Cys-CDR2-Cys-Linker-Cys-CDR1-Cys-Linker-CDR3-Cys. Here, cystines (Cys) were introduced to flank the CDR loops and serve as anchors to bind to AuNPs via Au–S bonding. Loop structures were generated using Modeler.60 All-atom DMD simulations were performed to assess whether the designed peptides bind IL-6 as in the original antibody.61 DMD simulations of proteins interacting with gold and silver nanoparticles (AuNPs and AgNPs, respectively) have been described in detail elsewhere.62,63 Briefly, a (1 1 1) gold surface with five atom layers (e.g., Figure 1a) was used to model the local environment of AuNP for APDA binding. The major physical interactions considered for Au and Au interaction with proteins include van der Waals solvation energy, and the electrostatic interaction of “image” charges, where the corresponding interaction parameters were adopted from the first-principles based GolP force field.64
Surface Plasmon Resonance Analysis.
A bare gold sensor was thoroughly cleaned in piranha solution (30 mL of 98% sulfuric acid, 10 mL of 30% hydrogen peroxide) for 10 min, followed by 15 min ultrasonication in 50:50 (v/v) ethanol/H2O. A quantity of 30 μL of Scheme 1 or Scheme 2 stock solution (1 mg/mL) was added into 60 μL of 10 mM TCEP solution for disulfide bond reduction. The mixture was incubated in darkness for 1 h and diluted to 1 mg/L with pH 7.4 PBS. The obtained solution was injected at a rate of 30 μL/min through the bare gold sensor until a surface coverage of 83.3 pg/mm2 was achieved. The sensor surface was passivated by 6-mercaptohexanol (3 mM) in pH 7.4 PBS for 30 min, and thoroughly washed with PBST buffer (pH 7.4 PBS, 0.05% Tween-20). The affinity analysis was performed by challenging the ADPA functionalized sensor with a series of standard concentrations of IL-6 in PBST buffer (250 μL) at a flow speed of 30 μL/min under room temperature. After each run, the sensor was regenerated by flowing NaOH solution (pH 12.3) for 30 s, which showed repeatable baseline recovery and minimal effect on probe activity within six replicates. The sensograms were fitted using a one-to-one interaction model in the TraceDrawer software version 1.5 (Ridgeview Instruments, Uppsala, Sweden). The kinetic parameters, including the association constant (ka), dissociation constant (kd), and equilibrium dissociation constant (KD = kd/ka) were determined by the model fitting.
Dynamic Light Scattering and Zeta Potential Measurements.
A quantity of 200 μL of freshly prepared sample solution (bare AuNRs, ADPAs, antibodies, ADPA-AuNRs, or antibody-AuNRs) was diluted with 600 μL of deionized (DI) water and transferred into a capillary zeta cell (Malvern). The measurement of zeta potential and hydrodynamic size was performed by Zetaiszer Nano ZS90, according to standard procedures.
Scattering-Type Scanning Near-Field Optical Microscopy.
The morphology of AuNR complex was characterized by nano-IR images from scattering-type scanning near-field optical microscopy (s-SNOM, Neaspec). The samples were prepared by drop-casting 1 mL AuNR, ADPA-AuNR, or antibody-AuNR solution onto a silicon wafer and dried at room temperature. The tapping-mode atomic force microscopy (AFM) was performed with a tapping amplitude of 40 nm and tapping frequency at 250 kHz. The AFM tip with a radius of 10 nm was coated with PtIr5. A monochromatic IR quantum cascade laser (QCL) was used to illuminate the AFM tip during the imaging. The backscattered optical signal off the tip was collected and demodulated at the third harmonics of the tapping frequency to obtain genuine near-field optical responses.
Fabrication of LSPRi Sensors.
Polydimethylsiloxane (PDMS) prepolymer was prepared by mixing 1:10 (m/m) silicone elastomer base and curing agent (SYLGARD 184). The resulting mixture was poured over silicon molds and degassed for 30 min, followed by 4 h of curing at 65 °C. Glass slides were washed repeatedly by DI water, dried thoroughly at 65 °C, and cleaned in the piranha solution (the mixture of 98% H2SO4 and 30% H2O2) to remove organic residues. Following 15 min of ultrasonication, cleaned glass slides were placed in a closed desiccator with 0.5 mL APTES placed nearby for 48 h. Further APTES fixation was accomplished by heating the obtained slides at 100 °C for 30 min. The prepared PDMS layers with microfluidic channels were detached from silicon molds and attached to the APTES-modified glass slides. Cit-AuNR solution was loaded into the microfluidic channels at 1 μL/mL for 8 min and incubated for 12 h, allowing the negatively charged Cit-AuNRs to be immobilized onto the positively charged APTES-modified glass slides via electrostatic interactions. Thorough washing was performed to remove unbounded AuNRs. 50 μg/mL probe solution (antibody or ADPA) was subsequently injected at 1 μL/mL for 8 min. Following 1 h of incubation, excessive probes were washed away by pH 7.4 PBS. The sensor surface was passivated by 1% BSA in pH 7.4 PBS for 1 h. The PDMS layers used above were discarded and new layers of PDMS was applied perpendicularly onto the as-prepared nanoplasmonic chips for the following LSPRi immunoassay.
LSPRi On-Chip Sensing.
The prepared nanoplasmonic chips were placed on a motorized stage (ProScan III, Prior Scientific). An oil dark-field condenser lens was used and gently attached to the bottom side of the nanoplasmonic chip with immersion oil. Under dark-field mode, only scattering light was allowed to pass through an objective lens (10×, Nikon Instruments, Inc.), filtered by a band-pass filter (645–695 nm, Chroma), and recorded by an electron multiplying charged coupled device (EMCCD) (Photometrics Evolve 512, Teledyne Photometrics) in a 10-s temporal resolution. The collected images were analyzed by a customized MATLAB code for real-time LSPR signals. Prior to a single measurement, pH 7.4 PBS solution (cell culture medium used in cell analysis experiment) was flowed through the microfluidic channels to stabilize the baselines. A series of standard IL-6 samples spiked in human serum were injected into loading channels for 30 min (Figure 3a bottom) with LSPRi signals (ΔI/I0, the scattering light intensity change (ΔI) from target barcode region divided by the initial scattering light intensity of the same area (I0)) recorded in real time. pH 7.4 PBS solution was subsequently injected for another 5 min to remove nonspecifically bound serum components. The calibration curve was obtained by fitting the data into the dose–response model in Origin.
Cell Culture and SARS-CoV-2 Surface Protein Stimulation.
A human leukemia monocytic cell line (THP-1) was cultured in Roswell Park Memorial Institute (RPMI) 1640 medium with 10% fetal bovine serum and 1% penicillin-streptomycin in a 5% CO2 humidified incubator at 37 °C. Macrophages were differentiated from THP-1 by 48 h of incubation in the aforementioned cell culture medium with 100 ng/mL phorbol 12-myristate-13-acetate (PMA). Lung epithelial cells (ATCC) were cultured in Keratinocyte serum-free medium (Gibco) with 0.05 mg/mL bovine pituitary extract (Gibco), 5 ng/mL epidermal growth factor (Gibco), and 10 ng/mL cholera toxin (Invitrogen). The above macrophages (106 cells/mL) and epithelial cells (106 cells/mL) were seeded in a 12-well plate overnight prior to SARS-CoV-2 surface protein stimulation. SARS-CoV-2 surface proteins were added at varying concentrations (1 μg/mL to 20 μg/mL) for varying incubation time (0–48 h). The cell culture supernatants were subsequently collected for IL-6 analysis.
Finite Element Analysis (FEA) Simulation.
The electromagnetic simulations of AuNR, ADPA-AuNR, and antibody-AuNR were performed by multiphysics simulation software (COMSOL). The dimension parameters of AuNR, antibody, and ADPA were obtained from the SEM and DLS results. The simulation model constructed a 40 nm × 80 nm AuNR, with a shell of recognition layer (12 nm thickness for antibody; 1.5 nm for ADPA), where a far-field domain was defined within a sphere with a diameter equivalent to the incident light wavelength. A perfectly matched layer was set as the boundary condition of scattering. A polarized electric field was defined parallel to the longitudinal direction of the AuNR. The frequency-dependent dielectric constant was calculated from the Lorentz–Drude model, where the relative permittivity of the antibody and ADPA was set as 2.25.2
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH (through Nos. MIRA R35GM133795 (to P.C.) and R01CA243001, to W.C.) and NSF (through Nos. CAREER CBET-1943302 (to P.C.), CBET-2030828 (to F.D.), and OIA-2033454 (to S. Dai)). The authors appreciate the support and guidance in patient serum sample measurements provided by Dr. Benjamin H. Singer (Department of Internal Medicine, University of Michigan, Ann Arbor, MI).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.2c14748.
Supplementary figures regarding DMD simulation, SEM characterization, zeta potential analysis, FEA simulation and ELISA results47 (PDF)
Animation of the possible binding between the designed ADPA and IL-6 on the AuNP surface (MPG)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsami.2c14748
The authors declare no competing financial interest.
Contributor Information
Jiacheng He, Materials Research and Education Center, Auburn University, Auburn, Alabama 36849, United States.
Lang Zhou, Materials Research and Education Center, Auburn University, Auburn, Alabama 36849, United States.
Gangtong Huang, Department of Physics and Astronomy, Clemson University, Clemson, South Carolina 29634, United States.
Jialiang Shen, Materials Research and Education Center, Auburn University, Auburn, Alabama 36849, United States.
Wu Chen, Department of Drug Discovery and Development, Harrison School of Pharmacy, Auburn University, Auburn, Alabama 36849, United States.
Chuanyu Wang, Materials Research and Education Center, Auburn University, Auburn, Alabama 36849, United States.
Albert Kim, Center for Medicine, Health, and Society, Vanderbilt University, Nashville, Tennessee 37235, United States.
Zhuoyu Zhang, Department of Mechanical and Aerospace Engineering and Department of Biomedical Engineering, New York University, Brooklyn, New York 11201, United States.
Weiqiang Cheng, Department of Mechanical and Aerospace Engineering and Department of Biomedical Engineering, New York University, Brooklyn, New York 11201, United States.
Siyuan Dai, Materials Research and Education Center, Auburn University, Auburn, Alabama 36849, United States.
Feng Ding, Department of Physics and Astronomy, Clemson University, Clemson, South Carolina 29634, United States.
Pengyu Chen, Materials Research and Education Center, Auburn University, Auburn, Alabama 36849, United States.
REFERENCES
- (1).Mangalmurti N; Hunter CA Cytokine Storms: Understanding Covid-19. Immunity 2020, 53 (1), 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chen P; Huang N-T; Chung M-T; Cornell TT; Kurabayashi K Label-Free Cytokine Micro-and Nano-Biosensing Towards Personalized Medicine of Systemic Inflammatory Disorders. Adv. Drug Delivery Rev 2015, 95, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Del Valle DM; Kim-Schulze S; Huang H-H; Beckmann ND; Nirenberg S; Wang B; Lavin Y; Swartz TH; Madduri D; Stock A; et al. An Inflammatory Cytokine Signature Predicts Covid-19 Severity and Survival. Nat. Med 2020, 26 (10), 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Qin C; Zhou L; Hu Z; Zhang S; Yang S; Tao Y; Xie C; Ma K; Shang K; Wang W; Tian D-S Dysregulation of Immune Response in Patients with Coronavirus 2019 (Covid-19) in Wuhan, China. Clin. Infect. Dis 2020, 71 (15), 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tan M; Liu Y; Zhou R; Deng X; Li F; Liang K; Shi Y Immunopathological Characteristics of Coronavirus Disease 2019 Cases in Guangzhou, China. Immunology 2020, 160 (3), 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Whetton AD; Preston GW; Abubeker S; Geifman N Proteomics and Informatics for Understanding Phases and Identifying Biomarkers in Covid-19 Disease. J. Proteome Res 2020, 19 (11), 4219–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Shang L; Zhao J; Hu Y; Du R; Cao B On the Use of Corticosteroids for 2019-Ncov Pneumonia. Lancet 2020, 395 (10225), 683–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Russell SM; Alba-Patiño A; Barón E; Borges M; Gonzalez-Freire M; De La Rica R Biosensors for Managing the Covid-19 Cytokine Storm: Challenges Ahead. ACS sensors 2020, 5 (6), 1506–1513. [DOI] [PubMed] [Google Scholar]
- (9).Zhou L; Chen P; Simonian A Advanced Biosensing Towards Real-Time Imaging of Protein Secretion from Single Cells. In Biosensors-Current and Novel Strategies for Biosensing; IntechOpen, 2020. [Google Scholar]
- (10).Liu C; Chu D; Kalantar-Zadeh K; George J; Young HA; Liu G Cytokines: From Clinical Significance to Quantification. Advanced Science 2021, 8 (15), 2004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Qavi AJ; Washburn AL; Byeon J-Y; Bailey RC Label-Free Technologies for Quantitative Multiparameter Biological Analysis. Anal. Bioanal. Chem 2009, 394 (1), 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mayer KM; Hafner JH Localized Surface Plasmon Resonance Sensors. Chem. Rev 2011, 111 (6), 3828–3857. [DOI] [PubMed] [Google Scholar]
- (13).He J; Jiang Y; Peng J; Li C; Yan B; Wang X Fast Synthesis of Hierarchical Cuprous Oxide for Nonenzymatic Glucose Biosensors with Enhanced Sensitivity. J. Mater. Sci 2016, 51 (21), 9696–9704. [Google Scholar]
- (14).Kaur B; Kumar S; Kaushik BK Recent Advancements in Optical Biosensors for Cancer Detection. Biosens. Bioelectron 2022, 197, 113805. [DOI] [PubMed] [Google Scholar]
- (15).Waterhouse DJ; Fitzpatrick CR; Pogue BW; O’Connor JP; Bohndiek SE A Roadmap for the Clinical Implementation of Optical-Imaging Biomarkers. Nature biomedical engineering 2019, 3 (5), 339–353. [DOI] [PubMed] [Google Scholar]
- (16).Khanmohammadi A; Aghaie A; Vahedi E; Qazvini A; Ghanei M; Afkhami A; Hajian A; Bagheri H Electrochemical Biosensors for the Detection of Lung Cancer Biomarkers: A Review. Talanta 2020, 206, 120251. [DOI] [PubMed] [Google Scholar]
- (17).Cui F; Zhou Z; Zhou HS Measurement and Analysis of Cancer Biomarkers Based on Electrochemical Biosensors. J. Electrochem. Soc 2020, 167 (3), 037525. [Google Scholar]
- (18).Chalklen T; Jing Q; Kar-Narayan S Biosensors Based on Mechanical and Electrical Detection Techniques. Sensors 2020, 20 (19), 5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang T; Yang H; Qi D; Liu Z; Cai P; Zhang H; Chen X Mechano-Based Transductive Sensing for Wearable Healthcare. Small 2018, 14 (11), 1702933. [DOI] [PubMed] [Google Scholar]
- (20).Zhu J; He J; Verano M; Brimmo AT; Glia A; Qasaimeh MA; Chen P; Aleman JO; Chen W An Integrated Adipose-Tissue-on-Chip Nanoplasmonic Biosensing Platform for Investigating Obesity-Associated Inflammation. Lab Chip 2018, 18 (23), 3550–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Cai Y; Zhu J; He J; Yang W; Ma C; Xiong F; Li F; Chen W; Chen P Magnet Patterned Superparamagnetic Fe3o4/Au Core–Shell Nanoplasmonic Sensing Array for Label-Free High Throughput Cytokine Immunoassay. Adv. Healthcare Mater 2019, 8 (4), 1801478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Javed I; He J; Kakinen A; Faridi A; Yang W; Davis TP; Ke PC; Chen P Probing the Aggregation and Immune Response of Human Islet Amyloid Polypeptides with Ligand-Stabilized Gold Nanoparticles. ACS Appl. Mater. Interfaces 2019, 11 (11), 10462–10471. [DOI] [PubMed] [Google Scholar]
- (23).Chen P; Chung MT; McHugh W; Nidetz R; Li Y; Fu J; Cornell TT; Shanley TP; Kurabayashi K Multiplex Serum Cytokine Immunoassay Using Nanoplasmonic Biosensor Microarrays. ACS Nano 2015, 9 (4), 4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Oh B-R; Huang N-T; Chen W; Seo JH; Chen P; Cornell TT; Shanley TP; Fu J; Kurabayashi K Integrated Nanoplasmonic Sensing for Cellular Functional Immunoanalysis Using Human Blood. ACS Nano 2014, 8 (3), 2667–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bensebaa F Optoelectronics. In Interface Science and Technology, Vol. 19; Elsevier, 2013; pp 429–479. [Google Scholar]
- (26).Willets KA; Van Duyne RP Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu. Rev. Phys. Chem 2007, 58 (1), 267–297. [DOI] [PubMed] [Google Scholar]
- (27).Zimmermann I; Egloff P; Hutter CA; Kuhn BT; Bräuer P; Newstead S; Dawson RJ; Geertsma ER; Seeger MA Generation of Synthetic Nanobodies against Delicate Proteins. Nat. Protoc 2020, 15 (5), 1707–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Mascini M; Palchetti I; Tombelli S Nucleic Acid and Peptide Aptamers: Fundamentals and Bioanalytical Aspects. Angew. Chem, Int. Ed 2012, 51 (6), 1316–1332. [DOI] [PubMed] [Google Scholar]
- (29).Reverdatto S; Burz DS; Shekhtman A Peptide Aptamers: Development and Applications. Curr. Top. Med. Chem 2015, 15 (12), 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pardon E; Laeremans T; Triest S; Rasmussen SG; Wohlkönig A; Ruf A; Muyldermans S; Hol WG; Kobilka BK; Steyaert J A General Protocol for the Generation of Nanobodies for Structural Biology. Nat. Protoc 2014, 9 (3), 674–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Klarenbeek A; Blanchetot C; Schragel G; Sadi AS; Ongenae N; Hemrika W; Wijdenes J; Spinelli S; Desmyter A; Cambillau C; et al. Combining Somatic Mutations Present in Different in Vivo Affinity-Matured Antibodies Isolated from Immunized Lama Glama Yields Ultra-Potent Antibody Therapeutics. Protein Engineering, Design and Selection 2016, 29 (4), 123–133. [DOI] [PubMed] [Google Scholar]
- (32).Blanchetot C; De Jonge N; Desmyter A; Ongenae N; Hofman E; Klarenbeek A; Sadi A; Hultberg A; Kretz-Rommel A; Spinelli S; et al. Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (Il-6) Antibodies. J. Biol. Chem 2016, 291 (26), 13846–13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Nusz GJ; Curry AC; Marinakos SM; Wax A; Chilkoti A Rational Selection of Gold Nanorod Geometry for Label-Free Plasmonic Biosensors. ACS Nano 2009, 3 (4), 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Anker JN; Hall WP; Lyandres O; Shah NC; Zhao J; Van Duyne RP Biosensing with Plasmonic Nanosensors. Nanoscience and Technology: A Collection of Reviews from Nature Journals 2009, 308–319. [Google Scholar]
- (35).Haes AJ; Zou S; Schatz GC; Van Duyne RP Nanoscale Optical Biosensor: Short Range Distance Dependence of the Localized Surface Plasmon Resonance of Noble Metal Nanoparticles. J. Phys. Chem. B 2004, 108 (22), 6961–6968. [Google Scholar]
- (36).Krieger F; Möglich A; Kiefhaber T Effect of Proline and Glycine Residues on Dynamics and Barriers of Loop Formation in Polypeptide Chains. J. Am. Chem. Soc 2005, 127 (10), 3346–3352. [DOI] [PubMed] [Google Scholar]
- (37).Van Rosmalen M; Krom M; Merkx M Tuning the Flexibility of Glycine-Serine Linkers to Allow Rational Design of Multidomain Proteins. Biochemistry 2017, 56 (50), 6565–6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Basak S; Saikia N; Dougherty L; Guo Z; Wu F; Mindlin F; Lary JW; Cole JL; Ding F; Bowen ME Probing Interdomain Linkers and Protein Supertertiary Structure in Vitro and in Live Cells with Fluorescent Protein Resonance Energy Transfer. J. Mol. Biol 2021, 433 (5), 166793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yang J; Wang B; You Y; Chang W-J; Tang K; Wang YC; Zhang W; Ding F; Gunasekaran S Probing the Modulated Formation of Gold Nanoparticles–Beta-Lactoglobulin Corona Complexes and Their Applications. Nanoscale 2017, 9 (45), 17758–17769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Javed I; Sun Y; Adamcik J; Wang B; Kakinen A; Pilkington EH; Ding F; Mezzenga R; Davis TP; Ke PC Cofibrillization of Pathogenic and Functional Amyloid Proteins with Gold Nanoparticles against Amyloidogenesis. Biomacromolecules 2017, 18 (12), 4316–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Javed I; Peng G; Xing Y; Yu T; Zhao M; Kakinen A; Faridi A; Parish CL; Ding F; Davis TP; Ke PC; Lin S Inhibition of Amyloid Beta Toxicity in Zebrafish with a Chaperone-Gold Nanoparticle Dual Strategy. Nat. Commun 2019, 10 (1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhou JO; Zaidi HA; Ton T; Fera D The Effects of Framework Mutations at the Variable Domain Interface on Antibody Affinity Maturation in an Hiv-1 Broadly Neutralizing Antibody Lineage. Front. Immunol 2020, 11, 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ramezani F; Amanlou M; Rafii-Tabar H Comparison of Amino Acids Interaction with Gold Nanoparticle. Amino Acids 2014, 46 (4), 911–920. [DOI] [PubMed] [Google Scholar]
- (44).Liu X; Li L; Wang Q; Jiang F; Zhang P; Guo F; Liu H; Huang J A Novel Humanized Anti-Interleukin-6 Antibody Hz0408b with Anti-Rheumatoid Arthritis Therapeutic Potential. Front. Immunol 2022, 12, 816646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kedem O; Vaskevich A; Rubinstein I Critical Issues in Localized Plasmon Sensing. J. Phys. Chem. C 2014, 118 (16), 8227–8244. [Google Scholar]
- (46).Busch RT; Karim F; Weis J; Sun Y; Zhao C; Vasquez ES Optimization and Structural Stability of Gold Nanoparticle–Antibody Bioconjugates. ACS omega 2019, 4 (12), 15269–15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Oliveira JP; Prado AR; Keijok WJ; Antunes PWP; Yapuchura ER; Guimarães MCC Impact of Conjugation Strategies for Targeting of Antibodies in Gold Nanoparticles for Ultrasensitive Detection of 17β-Estradiol. Sci. Rep 2019, 9 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ruiz G; Tripathi K; Okyem S; Driskell JD Ph Impacts the Orientation of Antibody Adsorbed onto Gold Nanoparticles. Bioconjugate Chem 2019, 30 (4), 1182–1191. [DOI] [PubMed] [Google Scholar]
- (49).Wang A; Perera YR; Davidson MB; Fitzkee NC. Electrostatic Interactions and Protein Competition Reveal a Dynamic Surface in Gold Nanoparticle–Protein Adsorption. J. Phys. Chem. C 2016, 120 (42), 24231–24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Treuel L; Brandholt S; Maffre P; Wiegele S; Shang L; Nienhaus GU. Impact of Protein Modification on the Protein Corona on Nanoparticles and Nanoparticle–Cell Interactions. ACS Nano 2014, 8 (1), 503–513. [DOI] [PubMed] [Google Scholar]
- (51).Del Caño R; Gisbert-González JM; González-Rodríguez J; Sánchez-Obrero G; Madueño R; Blázquez M; Pineda T. Effective Replacement of Cetyltrimethylammonium Bromide (Ctab) by Mercaptoalkanoic Acids on Gold Nanorod (Aunr) Surfaces in Aqueous Solutions. Nanoscale 2020, 12 (2), 658–668. [DOI] [PubMed] [Google Scholar]
- (52).Zhou S; Huo D; Goines S; Yang T-H; Lyu Z; Zhao M; Gilroy KD; Wu Y; Hood ZD; Xie M; Xia Y Enabling Complete Ligand Exchange on the Surface of Gold Nanocrystals through the Deposition and Then Etching of Silver. J. Am. Chem. Soc 2018, 140 (38), 11898–11901. [DOI] [PubMed] [Google Scholar]
- (53).Amenabar I; Poly S; Nuansing W; Hubrich EH; Govyadinov AA; Huth F; Krutokhvostov R; Zhang L; Knez M; Heberle J; Bittner AM; Hillenbrand R Structural Analysis and Mapping of Individual Protein Complexes by Infrared Nanospectroscopy. Nat. Commun 2013, 4 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Dosch SF; Mahajan SD; Collins AR. Sars Coronavirus Spike Protein-Induced Innate Immune Response Occurs Via Activation of the Nf-Kb Pathway in Human Monocyte Macrophages in Vitro. Virus Res 2009, 142 (1–2), 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Wang W; Ye L; Ye L; Li B; Gao B; Zeng Y; Kong L; Fang X; Zheng H; Wu Z Up-Regulation of 11-6 and Tnf-A Induced by Sars-Coronavirus Spike Protein in Murine Macrophages Via Nf-Kb Pathway. Virus Res 2007, 128 (1–2), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Tay MZ; Poh CM; Rénia L; MacAry PA; Ng LF The Trinity of Covid-19: Immunity, Inflammation and Intervention. Nature Reviews Immunology 2020, 20 (6), 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Nishimoto N; Terao K; Mima T; Nakahara H; Takagi N; Kakehi T. Mechanisms and Pathologic Significances in Increase in Serum Interleukin-6 (Il-6) and Soluble Il-6 Receptor after Administration of an Anti–Il-6 Receptor Antibody, Tocilizumab, in Patients with Rheumatoid Arthritis and Castleman Disease. Blood 2008, 112 (10), 3959–3964. [DOI] [PubMed] [Google Scholar]
- (58).Song Y; Ye Y, Su S-H; Stephens A; Cai T; Chung M-T; Han MK; Newstead MW; Yessayan L; Frame D; et al. A Digital Protein Microarray for Covid-19 Cytokine Storm Monitoring. Lab Chip 2021, 21 (2), 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Hoiland RL; Stukas S; Cooper J; Thiara S; Chen LYC; Biggs CM; Hay K; Lee AYY; Shojania K; Abdulla A; Wellington CL; Sekhon MS. Amelioration of Covid-19 Related Cytokine Storm Syndrome: Parallels to Chimeric Antigen Receptor-T Cell Cytokine Release Syndrome. Br. J. Hamaetol 2020, 190, e150–el54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Fiser A; Sali A Modloop: Automated Modeling of Loops in Protein Structures. Bioinformatics 2003, 19 (18), 2500–2501. [DOI] [PubMed] [Google Scholar]
- (61).Ding F; Tsao D; Nie H; Dokholyan NV Ab Initio Folding of Proteins with All-Atom Discrete Molecular Dynamics. Structure 2008, 16 (7), 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Huma Z.-e.; Javed I; Zhang Z; Bilal H; Sun Y; Hussain SZ; Davis TP; Otzen DE; Landersdorfer CB; Ding F; Hussain I; Ke PC. Nanosilver Mitigates Biofilm Formation Via Fapc Amyloidosis Inhibition. Small 2020, 16 (21), 1906674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Wang B; Seabrook SA; Nedumpully-Govindan P; Chen P; Yin H; Waddington L; Epa VC; Winkler DA; Kirby JK; Ding F; Ke PC Thermostability and Reversibility of Silver Nanoparticle–Protein Binding. Phys. Chem. Chem. Phys 2015, 17 (3), 1728–1739. [DOI] [PubMed] [Google Scholar]
- (64).Wright LB; Rodger PM; Corni S; Walsh TR. Golp-Charmm: First-Principles Based Force Fields for the Interaction of Proteins with Au (111) and Au (100). J. Chem. Theory Comput 2013, 9 (3), 1616–1630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


