Abstract
Pollen development is crucial for the fruit setting process of tomatoes, but the underlying regulatory mechanism remains to be elucidated. Here, we report the isolation of one HD-Zip III family transcription factor, SlHB8, whose expression levels decreased as pollen development progressed. SlHB8 knockout using CRISPR/Cas9 increased pollen activity, subsequently inducing fruit setting, whereas overexpression displayed opposite phenotypes. Overexpression lines under control of the 35 s and p2A11 promoters revealed that SlHB8 reduced pollen activity by affecting early pollen development. Transmission electron microscopy and TUNEL analyses showed that SlHB8 accelerated tapetum degradation, leading to collapsed and infertile pollen without an intine and an abnormal exine. RNA-seq analysis of tomato anthers at the tetrad stage showed that SlHB8 positively regulates SPL/NZZ expression and the tapetum programmed cell death conserved genetic pathway DYT1–TDF1–AMS–MYB80 as well as other genes related to tapetum and pollen wall development. In addition, DNA affinity purification sequencing, electrophoretic mobility shift assay, yeast one-hybrid assay and dual-luciferase assay revealed SlHB8 directly activated the expression of genes related to pollen wall development. The study findings demonstrate that SlHB8 is involved in tapetum development and degradation and plays an important role in anther development.
Introduction
Pollen development consists of two phases: microsporogenesis and microgametogenesis [1, 2]. During microsporogenesis, archesporial cells convert into pollen mother cells [1, 2]. The mother cells transformed into a tetrad of haploid microspores via meiosis [1, 2]. The tapetum, a layer of secretory cells, undergoes programmed cell death (PCD), releasing the callose and other cell wall-degrading enzymes into the locule; this divides tetrad into individual microspores by digesting the cell walls which are composed of the polysaccharide callose [3]. Defects in the tapetum generation and degradation usually result in pollen abnormal development and decreased fertility [4, 5].
In Arabidopsis and rice, the molecular mechanisms as well as key genes regulating tapetum development have been well illuminated [3, 6–11]. During early tapetum development, SPOROCYTELESS/NOZZLE (SPL/NZZ), EXCESS MICROSPORO CYTES1/EXTRASPORO GENOUS CELLS (EMS1/EXS), and TAPETUM DETERMINANT1 (TPD1) play key roles in determining the tapetal generation [12–15]. At the late microsporogenesis stage, the tapetum undergoes a tapetal cell death process that is controlled by the conserved genetic pathway DYT1–TDF1–AMS–MYB80–MS1 [11]. The loss-of-function mutants dyt1, tdf1, ams, and ms1 show delayed tapetal degradation, whereas the myb80 mutantdisplays a precocious tapetum degeneration phenotype [8, 16–20]. The pollen wall increases microspore survival, helps pollen to resist environmental stresses and provides pollen–stigma recognition. The pollen wall consists of exine and intine layers [21]. The exine, which is made up of lipid-like materials is synthesized by the sporophytic tapetum. Defectives in tapetum development and degradation result in pollen wall malformations [21]. TDF1, AMS, MYB80, and MS1 are also involved in pollen wall development, with different roles in exine and sexine formation and transcription regulation of tapetum-specific genes related to pollen wall development such as MS188, TEK, ABDG26, exl5/6, CYP703A2, LAP5/6, and so on [10, 16, 20, 22–24].
In tomato, few reports have been published on the genes involved in pollen development, and among them, even fewer have examined the regulators of the conserved PCD genetic pathway [17, 25–32]. SlDYT1 (MS10) was the first isolated regulator in the tomato tapetum development; the ms10 mutant fails to produce fertile pollen, and all PCD pathway members as well as genes related to sporopollenin synthesis are down-regulated [17]. Another Arabidopsis homolog gene bHLH89/90 isolated was MS32, whose loss-of-function resulted in delayed tapetum degradation. In addition, the genes involved in PCD are all down-regulated in the ms32 mutant [26]. Moreover, SlPIF4 was reported to induce pollen activity under cold conditions by interacting with the conserved tapetum PCD genetic pathway [25]. Although the conserved PCD genetic pathway was predicted to be present in tomato, other regulators involved in this process remain to be clarified.
The class III homeodomain-leucine zipper (HD-Zip III) transcription factor family were reported to determine the ad/abaxial polarity of leaves, anthers, vascular organs, and developing embryos [13, 33–37]. Five HD-Zip III genes including PHABULOSA (PHB)/ATHB14, REVOLUTA (REV), PHAVOLUTA (PHV)/ATHB9, INCURVATA4/CORONA/ATHB15, and ATHB8 were identified in the Arabidopsis genome [38]. These HD-Zip III genes show overlapping expression and exhibit redundant functions. Single loss-of-function of these genes gives non-obvious phenotypes. Simultaneous mutation of REV, PHV, and PHB affected meristem formation and seedlings structure [38, 39]. HD-Zip III mRNA can be degraded by the microRNA miR165/6 [40]. Up-regulation of HD-Zip III genes via disruption of microRNA regulation sites result in strong developmental phenotypes [37, 41, 42]. Gain-of-function of PHB and PHV displayed leaves with damaged polarity and abnormal appearance [41]. MiR166-PHB-SPL/NZZ module regulates the stamen polarity through modifying the boundary thickness [13]. In cucumber, CsSPL formed a complex with CsPHB and CsWUS to orchestrate sex organ development in an unidentified regulatory pathway [12]. There are six HD-Zip III genes in tomato, including Solyc11g069470 (SlREV), Solyc08g066500 (SlHB8), Solyc12g044410, Solyc03g120910 (SlHB15A), Solyc02g024070, and Solyc02g069830 [43]. These genes are negatively regulated by miR166; overexpression of Sly-pre-miR166b down-regulates all six HD-Zip III genes, and the plant bears a fruit with another fruit developing inside or parthenocarpic fruit [44]. Overexpression of SlREV does not result in a discernable plant phenotype, but its overexpression via disruption of microRNA regulation leads to ectopic flower formation and fused fruit [44]. SlHB15A regulates parthenocarpic fruit set under cold conditions via miR166-mediated recessive dosage sensitivity [43]. Although the effect of SlREV and SlHB15A genes on fruit development has been clarified, its role in anthers need to be investigated.
Here, we isolated one HD-Zip III family transcription factor, SlHB8, whose loss-of-function increased pollen activity, inducing fruit setting. SlHB8 overexpression showed the opposite phenotype. In order to clarify the function of SlHB8 in pollen development, cytobiology and molecular biology technologies were carried out on the anthers at different development stages. The results proved SlHB8 was a negative regulator during pollen development by mediating the tapetum development through conserved genetic pathway DYT1–TDF1–AMS–MYB80.
Results
SlHB8 shows a pollination-dependent expression pattern
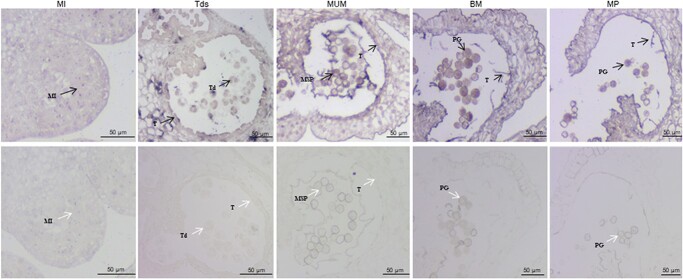
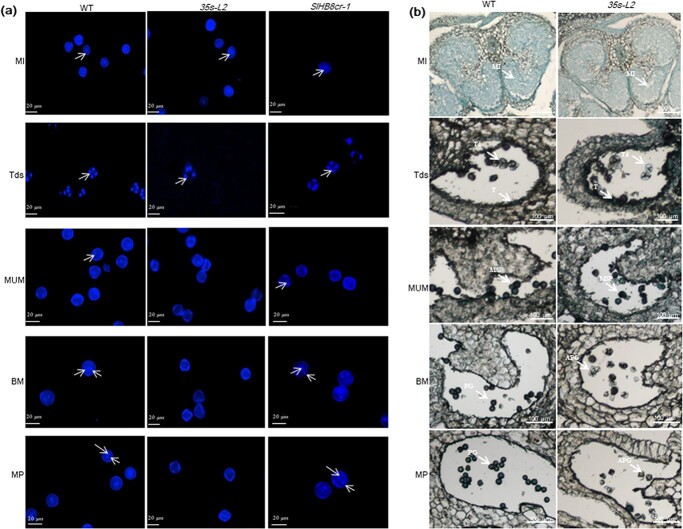
The SlHB8 transcription factor belongs to the HD-ZIP III transcription factor family, as it contains the four conserved domains: HD, bZip, START, and PAS [45]. SlHB8 is expressed in all tissues, such as the root, stem, leaves, flowers, mature green fruits, breaker fruits, and red fruits, showing highest expression level in stem and lowest expression level in the red rip fruit (Fig. S1a, see online supplementary material). The highest expression level in the stem is in line with the enlarged stem diameter of SlHB8 knocking out lines [46]. Here, SlHB8 was detected in the petal, sepal, stamen, and carpel flower organs and showed the highest expression level in sepals, as evidenced by qPCR (Fig. S1b, see online supplementary material). During stamen development, SlHB8 expression decreased (Fig. S1c, see online supplementary material). The in situ hybridization result showed that SlHB8 transcripts were found in the microspores and tapetum cells from microspore mother cell stage to the mature pollen stage (Fig. 1), indicating its role in the pollen development. SlHB8 is induced after treatment with auxin, gibberellic acid, and artificial pollination [45]. When the fruit set succeeded, SlHB8 expression levels increased when compared to ovaries before pollination (Fig. S1d, see online supplementary material). These data suggest that SlHB8 is important for stamen development and the fruit setting process.
Figure 1.
RNA in situ hybridization of SlHB8 during anther development of wild-type tomato plant. Anthers at MI, Tds, MUM, BM and MP stages were cross-sectioned for hybridization with antisense (upper) and sense (lower) probes of SlHB8. Black and white arrows indicate positive and negative in situ hybridization signals for SlHB8 transcripts respectively. BM: binucleate microspore stage; MI: microspore mother cell stage; MP: mature pollen stage; MSP: microspore pollen; MUM: middle uninucleate microspore stage; PG: pollen grain; T: tapetum; Td: tetrad; Tds: tetrad stage. Bars = 50 μm.
Knocking out SlHB8 via CRISPR/Cas9 promotes fruit set rate and pollen activity in tomato
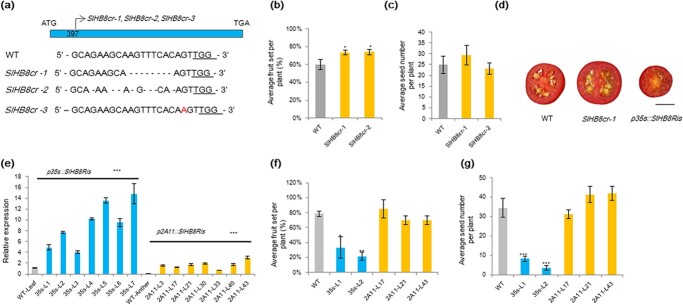
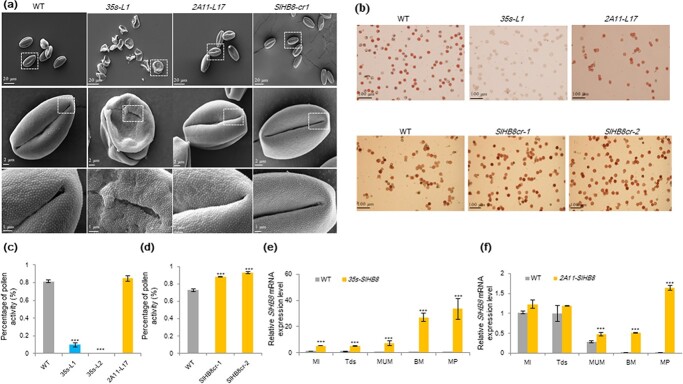
To determine its role in anther development and the fruit setting process, we knocked out SlHB8 using CRISPR/Cas9; SlHB8-specific primers and sequencing were used to verify the knockout effect. Three mutant types were obtained, including two with an 8 bp deletion in the CDS and one with a 1 bp insertion (Fig. 2a). Phenotyping was performed for the two deletion lines. Compared with the WT, the fruit set rates were higher in the SlHB8 knockout lines (Fig. 2b), but the fruit size, weight, and seed number (Fig. 2c and dFig. S2a and b, see online supplementary material) did not change. Pollen activity, pollen tube length, and the pollen tube germination rate were higher in the SlHB8 knockout lines than that in the WT (Fig. 3a,b–d, Fig. S2c–e, see online supplementary material), and the anther width was thinner (Fig. S2f, see online supplementary material).
Figure 2.
Phenotyping of SlHB8 gene overexpression and knockout plants. a Three SlHB8 gene knockout lines were established using CRISPR/Cas9. Fruit set rate (b) and the number of seeds per fruit (c) of SlHB8 knockout plants. d Photos of cross-section of red ripe fruits of wild-type, SlHB8 gene knockout and SlHB8 overexpression lines. Scale bar = 1 cm. e Expression levels of SlHB8 under control of the 35 s and 2A11 promoters in the SlHB8 overexpression lines. Ubi was used as a reference gene. Fruit set rate (f) and the number of seeds per fruit (g) of SlHB8 gene overexpression plants. The error bars denote SE; *P < 0.5, **P < 0.01, ***P < 0.001 (Student’s t-test; compared with the WT). 35 s-L1, promoter 35 s-driven SlHB8 overexpression line 1; 2A11-L17, promoter 2A11-driven SlHB8 overexpression line 17.
Figure 3.
Pollen viability and pollen morphology of SlHB8 gene knockout and overexpression lines. a Scanning electron micrographs of pollen grains from wild-type, SlHB8 gene knockout and SlHB8 overexpression lines under control of the 35 s and 2A11 promoters. BM: binucleate microspore stage; MI: microspore mother cell stage; MP: mature pollen stage; MUM: middle uninucleate microspore; Tds: tetrad stage. From the top to the bottom, the scale bar indicated 20 μm, 2 μm, 1 μm, respectively. b Pollen viability after TTC staining. Scale bar = 100 μm. c, d Percentage of pollen viability in wild-type and SlHB8 transgenic lines. e, f Expression levels of SlHB8 in the transgenic tomato lines p35s::SlHB8Ris and p2A11::SlHB8Ris during anther development. Ubi was used as reference gene. Expression level in MI stage flower bud was used as control. The error bars denote SE; ***P < 0.001 (Student’s t-test; compared with the WT).
Overexpression of SlHB8 results in pollen abortion and seedless fruits
An miRNA166 target site was found in the SlHB8 gene, and thus individual SlHB8 overexpression lines under control of the 35S and fruit-specific 2A11 promoters were generated by mutating the miRNA166 target site [47]; transgenic plants were verified by qPCR, with seven p35S::SlHB8Ris and seven p2A11::SlHB8Ris lines showing overexpressed SlHB8 levels (Fig. 2e). Most p35s::SlHB8Ris overexpression lines produced seedless fruits (Fig. 2d); therefore, only two lines (line 1 and 2) were collected from a few seeds [47]. Compared with the WT, the p35::SlHB8Ris overexpression transgenic plants bore fewer fruits (Fig. 2f) and the fruit size, weight, and seed number (Fig. 2g, Fig. S3a and b, see online supplementary material) were significantly reduced. All p2A11::SlHB8Ris overexpression lines displayed phenotypes – such as fruit set, seed number, fruit size, and fruit weight – similar to those of the WT tomato plants (Fig. 2f and g, Fig. S3a and b, see online supplementary material).
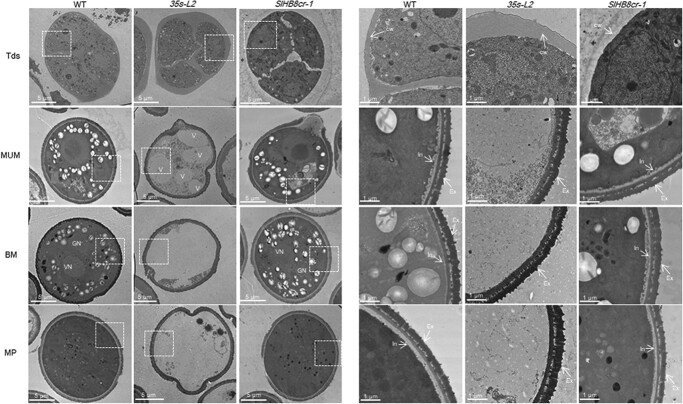
Because pollen viability and ovule development affects the fruit set rate and seed number, which were reduced in the L1 and L2 lines, reciprocal cross experiments between p35s::SlHB8Ris and the WT as well as p2A11::SlHB8Ris and the WT were performed. When the WT was used as the female parent and p35s::SlHB8Ris as the male parent, the fruit set rate reached 2.7%; however, the fruit set rate decreased to 93.5% when this was switched (Table 1). In contrast to p35s::SlHB8Ris, the fruit set rate remained similar regardless of the pollen donor when WT and p2A11::SlHB8Ris were crossed (Table 1). These results indicate that the pollen varieties were mainly responsible for the lower fruit set rate and reduced seed number. We thus assessed the viability of mature pollen grains at the anthesis stage using the TTC method. Via microscopy, pollen grains of p35s::SlHB8Ris were found defective, whereas those of p2A11::SlHB8Ris and SlHB8-cr were similar to those of the WT (Fig. 3a and b). In addition to pollen viability, pollen shape, pollen tube length, and pollen tube germination capacity showed observable differences between p35s::SlHB8Ris and the WT (Fig. 3a–c, Fig. S3c–e, see online supplementary material), and the p35s::SlHB8Ris anther width was thinner (Fig. S3f, see online supplementary material). By contrast, pollen viability, pollen tube length, pollen tube germination capacity, and morphology of the p2A11::SlHB8Ris lines were similar to those of the WT (Fig. 3a–c, Fig. S3c–e, see online supplementary material). SEM was then employed to observe the whole structure of mature pollens at the anthesis stage. Unlike the round and regularly shaped WT pollen grains, the p35s::SlHB8Ris transgenic pollen grains were irregular, shrunken, and collapsed. The pollen grain surface was also different from that of the WT (Fig. 3a). No significant differences were observed between p2A11::SlHB8Ris, SlHB8cr, and the WT (Fig. 3a). Next, transmission electron microscopy (TEM) was used to investigate the ultrastructural changes of the aborted p35s::SlHB8Ris pollen grains; the exine layer of irregular pollen grains was wider in p35s::SlHB8Ris than in the WT, and the intine layer between the plasma membrane and exine was absent in the p35s::SlHB8Ris lines (Fig. 4).
Table 1.
Cross-fertilization assay.
| SlHB8-ox♂ x WT♀ | WT♂ x SlHB8-ox♀ | 2A11-SlHB8♂ x WT♀ | WT♂ x 2A11-SlHB8♀ | |
|---|---|---|---|---|
| Fruit set | 2.70% | 93.50% | 60.60% | 50% |
| Hybrid number | 37 | 31 | 33 | 30 |
| Setted fruits number | 1 | 29 | 20 | 15 |
Emasculated wild type flowers were fertilized with SlHB8-ox pollen and the number of setting fruit [lr1] was assessed at the ripe stage. Conversely, tomato pollen from wild type flowers was used to fertilize emasculated SlHB8-ox flowers. The same assay was also carried out on the 2A11 lines.
Figure 4.
Transmission electron microscopy of pollen development in the wild-type, SlHB8 gene knockout and p35s::SlHB8Ris lines. BM: binucleate microspore stage; cw: callose wall; Ex: exine; GN: generative nuclei; In: intine; MP: mature pollen stage; MUM: middle uninucleate microspore stage; Tds: tetrad stage; V: vacuole; VN: vegetative nuclei. The scale bar indicated 5 μm (left: whole organ) and 1 μm (right: magnified location), respectively.
Because of the different effects of SlHB8 on fruit morphology in the p35s::SlHB8Ris and p2A11::SlHB8Ris overexpression lines, we compared SlHB8 expression levels during anther development. The SlHB8 gene was overexpressed throughout anther development (from MI to MP) in p35s::SlHB8Ris (Fig. 3e), but was induced starting from the MUM stage in p2A11::SlHB8Ris (Fig. 3f). This differential expression pattern may account for the development of seedless fruit in p35s::SlHB8Ris transgenic plants.
Overexpression of SlHB8 disrupts pollen development due to early tapetal PCD
To identify the key stage at which SlHB8 regulates pollen development, we defined the pollen development stages as MI, Tds, MUM, BM, and MP (anthesis) using DAPI staining (Fig. 5). At the MI and Tds, the nuclei and tetrad formed normally both in the WT, SlHB8cr and p35s::SlHB8Ris transgenic lines. Starting from the MUM stage to the MP stage, the nucleus disappeared in the p35s::SlHB8Ris lines and the pollen shape became irregular and collapsed, which is obviously different from that of WT and SlHB8cr anthers with two nuclei and round, flush pollen grains (Fig. 5a). We further performed a set of cytological experiments to characterize the spatial and temporal cytological defects in p35s::SlHB8Ris anthers. In agreement with the above observations, histological anther sections showed that at the MI stage, the cell layer differentiation appeared similar to that of WT anthers, and a tetrad formed in both the WT, SlHB8cr and p35s::SlHB8Ris anthers during the Tds; there were no observable defects in the anthers during these two stages (Fig. 5b, Fig. S4, see online supplementary material). At the MUM stage, abnormal pollen grains with an irregular shape and vacuolation appeared in the p35s::SlHB8Ris anthers, and most of the pollen grains were aborted in the p35s::SlHB8Ris lines (Fig. 5b). Via TEM, we observed a large nucleus in the WT and SlHB8cr, which was not present in the p35s::SlHB8Ris lines (Fig. 4). Instead, there was an increased number of large vacuoles, and the intine of the pollen wall was absent during the MUM stage (Fig. 4). During the WT and SlHB8cr BM stage, the microspore contained a full cytoplasm with normal vegetative and generative nuclei (Fig. 4). However, the majority of the cytoplasm and nuclei in pollen grains of the p35s::SlHB8Ris lines were completely degraded, and only trace cytoplasmic inclusions were observed (Fig. 4). At the Tds, the p35s::SlHB8Ris anther tapetum was thinner than that of the WT and SlHB8cr-1, and tapetal cells were infertile and shrunken (Fig. 6a).
Figure 5.
Histocytological observation of pollen development in the wild-type, SlHB8 gene knockout and SlHB8 overexpression lines under control of the 35 s promoter. DAPI staining (a) and semi-thin section comparison (b) of anther and pollen development between the wild-type, SlHB8 gene knockout and 35 s-L2 lines. APG: abnormal pollen grain; BM: binucleate microspore stage; MI: microspore mother cell stage; MP: mature pollen stage; MSP: microspore; MUM: middle uninucleate microspore stage; PG: pollen grain; T: tapetum; Td: tetrad; Tds: tetrad stage. The white arrows indicate the nuclei (a). The scale bar indicated 20 μm (a) and 100 μm (b).
Figure 6.
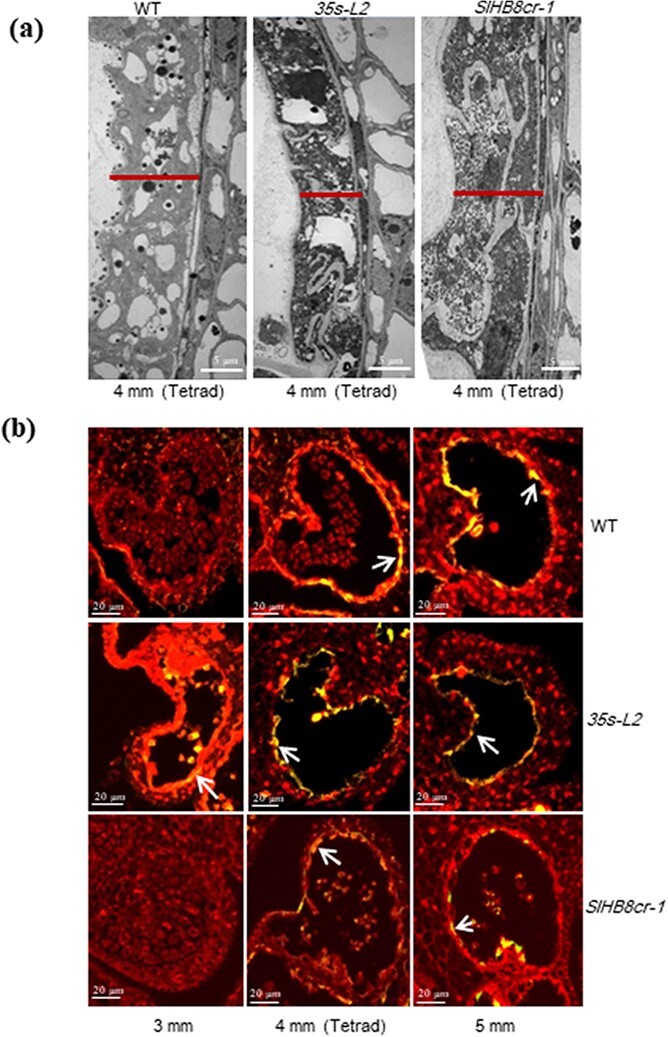
Morphology of tapetum development and programmed cell death. a Transmission electron micrographs of anthers at the tetrad stage of wild-type, SlHB8 gene knockout and SlHB8 overexpression plants under control of the 35 s promoter. Scale bars = 5 μm. b Fluorescence microscopy of DNA fragmentation detected by TUNEL assays of anthers from wild-type, SlHB8 gene knockout and 35 s-L2 line plants at different stages. Scale bars = 20 μm.
Tapetum PCD is one of the key factors affecting tapetal cell degradation and pollen development. To assess whether the infertile tapetal cells are associated with early PCD, we performed a TUNEL assay, which provides strong fluorescent signals when cells undergo massive DNA fragmentation. In the WT and SlHB8cr-1, a positive signal appeared at the 4 mm anther stage (Tds), which was observed earlier in p35s::SlHB8Ris at the 3 mm anther stage (Fig. 6b). The results indicate that SlHB8 overexpression accelerates tapetal cell degradation, leading to pollen abortion.
Primary metabolite determination in p35s::SlHB8Ris anthers at the mature pollen stage
Given that anther development is associated with metabolite levels, we determined primary metabolite content in the WT and p35s::SlHB8Ris anthers. Primary metabolites measured included amino acids and derivatives; nucleotides and derivatives; carbohydrates; indole derivatives; organic acids and derivatives; and lipids. All nucleotides and derivatives were down-regulated, and with the exception of O-rhamnoside, levels of the other three carbohydrates measured were reduced. All lipids detected were significantly up-regulated, as well as half of the organic acids and derivatives, with the other half down-regulated. All indole derivatives were down-regulated in the p35s::SlHB8Ris anthers (Fig. S5; Table S1, see online supplementary material).
Transcriptome analysis of DEGs after SlHB8 overexpression
Given that SlHB8 belongs to the HD-Zip III transcription factor family and that its up-regulation resulted in pollen abortion and anther development defects, SlHB8 was predicted to regulate pollen development by mediating the transcription of target genes. To identify such genes, comparative transcriptome analyses via RNA-seq were performed using tetrad stage flower bud. Each sample contained three biological replicates, and totally six transcriptome libraries were generated. In our study, 97.17%–97.53% of short clean reads identified from RNA-seq data were mapped to the tomato genome (Solanum lycopersicum ITAG4.0). Among the six libraries, 23 723 to 24 399 genes were annotated and 659 novel genes were identified (Table S2, see online supplementary material). The correlation among the three biological replicates was qualified by Pearson’s correlation coefficient (R2), with R2 > 0.8 as the significance cutoff. The R2 value of the three biological replicates was >0.99, indicating a high correlation (Fig. S6, see online supplementary material).
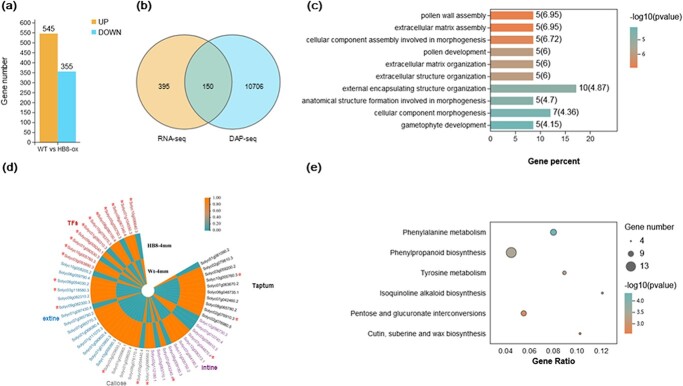
To identify DEGs in tetrad stage anthers between the WT and p35s::SlHB8Ris lines, pairwise comparisons were performed with |log2 (fold change)| > 1 and FDR < 0.05 as cutoff thresholds to filter the significant DEGs. These comparisons allowed the identification of 900 DEGs, including 355 down-regulated and 545 up-regulated genes (Fig. 7a; Table S3, see online supplementary material). Among these 900 DEGs, genes involved in the regulation of microspore protein biosynthesis, tapetum development, callose metabolism, pollen inner wall formation, pollen outer wall formation, and hormone signal transduction pathways related to pollen development were up-regulated (Fig. 7d).
Figure 7.
Identification of differentially expressed genes (DEGs) and SlHB8-targeted genes in wild-type and p35s::SlHB8Ris anthers by RNA-seq and DAP-seq. a DEGs between wild-type and p35s::SlHB8Ris anthers at the tetrad stage. b Overlap of DEGs identified by RNA-seq and SlHB8-target genes identified by DAP-seq. Gene ontology (GO) (c) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (e) analysis of up-regulated DEGs in wild-type and p35s::SlHB8Ris anthers at the tetrad stage. d Heatmaps of DEGs involved in pollen development, including DEGs related to sporopollenin biosynthesis and transport, tapetum development, pollen intine development, callose metabolites, and transcription factors. * indicates up-regulated DEGs targeted by SlHB8.
qRT-PCR was then performed using seven randomly selected genes to confirm the accuracy of RNA-seq. All seven genes exhibited similar expression pattern and high Pearson’s correlation coefficient [RNA-seq and qRT-PCR: 0.9715 (P < 0.0001)], indicating that the transcriptome data were highly reliable (Fig. S7, see online supplementary material).
As SlHB8 was predicted to be an activator, the up-regulated DEGs may represent a direct response of SlHB8 overexpression. To further understand the putative functions of these 545 up-regulated DEGs, gene ontology (GO) assignment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were carried out. Using a significant cutoff value of q ≤ 0.05, the data revealed that the 545 DEGs were only significantly enriched in the biological process term related to pollen wall assembly, pollen development, and gametophyte development (Fig. 7c; Table S4, see online supplementary material), with six KEGG pathways significantly enriched (Fig. 7e; Table S4, see online supplementary material): ‘phenylalanie metabolism’, ‘tyrosine metabolism’, ‘isoquinoline alkaloid biosynthesis’, ‘phenylpropanoid biosynthesis’, ‘pentose and glucuronate interconversions’, and ‘cutin, suberin and wax biosynthesis’. The last three pathways have been proved to be related to pollen development (Fig. 7e).
Identification of SlHB8-targeted DEGs by DAP-seq
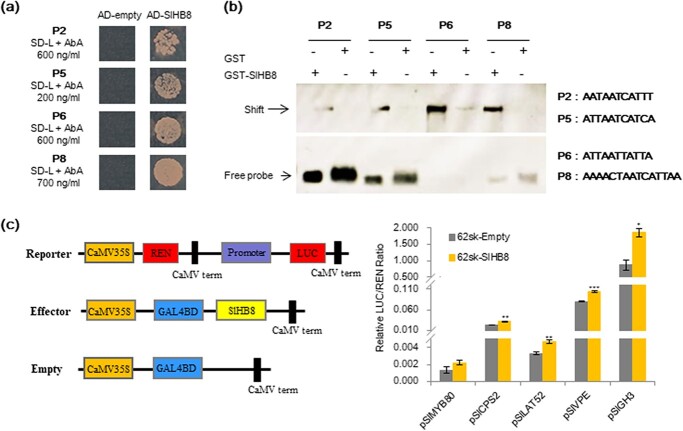
DEGs identified by RNA-seq were directly or indirectly regulated by SlHB8. To identify DEGs directly regulated by SlHB8, we used DAP-seq to identify the SlHB8-binding sites in vitro. In total, 71 504 200 bp peaks were identified in the tomato genome, including 12 627 (17.66%) peaks that were presented within 2.0 kb upstream of the annotated ORFs, 4583 (6.41%) located within 300 bp downstream of putative ORFs, and the remaining 54 293 (75.93%) distributed in introns, exons, or intergenic regions in the genome (Fig. S8; Table S5, see onlinesupplementary material). To confirm the DAP-seq results, we selected four binding elements for Y1H and EMSA assays. SlHB8 interacted with all tested elements (Fig. 8a and b). Furthermore, we performed a dual-luciferase assay using five genes (SlVPE, SlCPS2, SlLAT52, SlGH3.1, and SlMYB80) whose promoters contained the target motifs of SlHB8 and found that SlHB8 activated their expression except SlMYB80 which showed a slight up-regulation without significance (Fig. 8c). After analyzsing the genes with promoters containing the DAP-seq fragments and the SlHB8-regulated DEGs, we revealed 150 overlapping genes (Fig. 7b; Table S6, see online supplementary material). Among these genes, there were genes involved in sporopollenin biosynthesis and transport, tapetum development, hormone metabolism and signaling pathways, pollen intine development, callose metabolism and genes belong to transcription factors, indicating that SlHB8 regulates pollen development by mediating these pathways (Fig. 7d; Table S6, see online supplementary material).
Figure 8.
Validation of SlHB8 downstream target genes via yeast one-hybrid, EMSA and dual-luciferase assays. a Validation of SlHB8 binding with the four selected DAP-seq fragments using yeast one-hybrid assay. AD-empty indicates the control yeast strain transformed with the empty pGADT7 vector without SlHB8. b EMSA validation of the binding motifs in four selected DAP-seq fragments. The corresponding motif are listed beside the band. The detected bands are indicated with black arrows. c Validation of SlHB8 activation on five selected promoters using a dual-luciferase assay. The empty effector was used as control (set as 1). The ratio is presented as the mean ± SE (n = 3).
Discussion
SlHB8 regulates pollen development by disturbing tapetum PCD
The tapetum and its degradation triggered by PCD which produced enzymes, nutrients, and precursors play an important role in microspore and pollen wall development [3]. Numerous reports have shown that incorrect timing of tapetal PCD (premature or delayed degradation) and disintegration induces male sterility and pollen wall defects [3, 6–9, 16, 29]. In contrast to previous results and our findings in the WT, tapetum PCD in the SlHB8 overexpression lines occurred early in tomato anthers, before the tetrad stage, while tapetum PCD in the SlHB8 gene knockout lines appeared at the tetrad stage (Fig. 6b).
Over the past decade, the functionally conserved genetic pathway DYT1–TDF1–AMS–MYB80 in tapetum development and degradation has been identified in Arabidopsis, rice, tomato, and other crops [3, 7, 11, 17, 26]. Loss-of-function of these regulators results in aborted pollen and induces male sterility [6, 8–10, 16, 17, 26]. DYT1 is a critical transcription factor for early tapetum development and function upstream of TDF1, AMS, MYB80, TEK, and MS1 [16, 17]. TDF1 is the direct target gene of DYT1, and mutations in TDF1 severely impair tapetal development and callose dissolution. Tapetal cells in tdf1 fail to transit to the secretory type as a result of the low expression levels of MYB80 [9, 16]. AMS and MYB80 act as master regulators of pollen wall development due to its regulation on the genes related to callose degeneration (QRT3 and A6), genes related to sporopollenin biosynthesis and metabolism (CYP86C3, ACOS5, and SHT), genes related to lipid transport (LACS6 and WBC27), and pollen coat formation (EXL4-EXL6) [9, 16, 23]. Interestingly, our RNA-seq data showed that the core transcription factors that regulate tapetal PCD (SlDYT1, SlTDF1, and SlMYB80) and their multiple downstream targets (e.g. SlA6, CYP703A2, CYP704B1, SlPKSA, and SlPKSB) were up-regulated in the SlHB8 overexpression lines (Fig. 7d); SlAMS was also significantly up-regulated (fold <2) (Table S7, see online supplementary material). Moreover, DAP-seq, Y1H, and dual-luciferase assays confirmed that some genes were directly targeted by SlHB8, such as SlMYB80 and SlA6 (Fig. 8; Table S6, see online supplementary material), indicating that SlHB8 induces tapetum PCD by regulating these key tapetal pathway genes. Moreover, in the SlHB8 overexpression lines, TUNEL-positive signals were observed in the microspores before the tetrad stage (Fig. 6b), suggesting early callose generation and early secretory tapetal cell transition, which may induce early tapetum degradation. The overexpression of SlMYB80 and SlA6 also supports the hypothesis that early decretory tapetal cell transition leads to early tapetum PCD. While loss-of-function of SlHB8 has no effect on the tapetum PCD signal and expression of tapetal PCD regulators (Fig. 6b, Fig. S9c, see online supplementary material), indicating another gene may compensate the function of SlHB8 or another partner work together with SlHB8 in regulating the tapetum PCD.
SlHB8 may function upstream of SlSPL to regulate early pollen development
SPL has been reported to function in the process of sex organ development [12–14, 48, 49]. In Arabidopsis, SPL/NZZ controls early microsporocyte differentiation. Loss-of-function of SPL resulted in inhibited microsporocyte formation and deformed tapetum [13, 14]. In cucumber, CsSPL formed a complex with HD-Zip III and CsWUS to regulate anther and ovule development [12]. In tomato, the SlSPL loss-of-function mutant hydra showed sterility phenotypes both on male and female organs [49]. In developing anthers of Arabidopsis, miR165/6 acts as a regulator balancing the expression of PHB and SPL/NZZ to determine the polarity of the anthers [13]. In addition to PHB, the adaxial identity genes include the other HD-Zip III family genes (REV, PHB, PHV, CNA, and ATHB8), which are also repressed by microRNA165/6 [50]. Overexpression of SlHB8 with a mutated miR165 target site resulted in aborted pollen grains (Fig. 3a–c) and up-regulation of SlSPL (Table S3, see online supplementary material), indicating its upstream function during tomato anther development. According to the DAP-seq data, a non-SlHB8 binding site was found in the SlSPL promoter, indicating indirect regulation by SlHB8, a regulatory loop with miR166, or a loss of gene expression. DYT1, which is positively regulated by SPL and EMS1, functions downstream of SPL and partially rescues the spl phenotype [51]. DYT1 is sufficient to activate the downstream genes, such as TDF1, AMS, MYB80, TEK, and MS1, in tapetum development [16]. In the p35s::SlHB8Ris lines, the expression of DYT1, TDF1, MYB80, and TEK was induced (Fig. 7d). Furthermore, a SlHB8 binding site on the promoter of SlMYB80 was identified, but its activation by SlHB8 was not strong (Fig. 8), indicating that SlHB8 needs a partner or that the elevated expression levels of SPL, DYT1, and TDF1 contribute to the up-regulation of SlMYB80 in the p35s::SlHB8Ris lines. Overall, the results indicate that SlHB8 may function upstream of SlSPL and regulates early pollen development.
SlHB8 affects exine and intine development
The genetic pathway (DYT1–TDF1–AMS–MS188–MS1) for tapetum development is reported to be closely connected to exine formation in Arabidopsis. The key genes involved in sporopollenin formation, such as CYP703A2, CYP704B1, PKSB, and PKSA, are positively regulated by DYT1, TDF1, AMS, and MYB80 [23, 52]. Moreover, AMS regulates nexine and sexine layer formation by directly modifying the expression of TEK and MS188. The absence of TEK function results in pollen grains without the intine and exine layers [22]. In the present study, the exine and intine were affected in the p35s::SlHB8Ris lines, with an extine thicker than that of the WT and an absent intine (Fig. 4). Primary metabolite levels and the main components of sporopollenin were also altered in the SlHB8 overexpression lines (Fig. S5, see online supplementary material). SlDYT1, SlTDF1, SlMYB80, SlTEK, SlCYP703A, SlPSKB, and SlPSKA were up-regualted when SlHB8 was overexpressed (Fig. 7d), which is consistent with the abnormal pollen wall development of the p35s::SlHB8Ris lines. We did not find a SlHB8 binding site in the promoters of SlCYP704B1, SlCYP703A, SlPSKB, and SlPSKA; however, such a binding site was present in the SlMYB80 and SlTEK promoters (Table S5, see online supplementary material), indicating indirect up-regulation resulting from the elevated expression levels of SlMYB80 and SlTEK, which target CYP703A2, CYP704B1, PSKB, and PSKA in Arabidopsis.
Compared with exine development, knowledge of intine formation is lacking. Intine is secreted by the microspores and is associated with pectin, cellulose, and callose metabolism [21]. Inhibition of intine synthesis during the early stages of male gametogenesis may arrest pollen development, leading to collapsed, aborted pollen [53–55]. As the pollen tube consists of an intine layer, defects of the intine structure are accompanied by abnormal pollen tube germination [54, 56–58]. In the p35s::SlHB8Ris lines, an intine layer did not form in the shrunken, irregular, and infertile pollen grains and the germination rate was reduced (Fig. 4; Fig. 3a, Fig. S3d, see online supplementary material). UDP-sugar pyrophyllase (USP) is involved in pectin synthesis; loss-of-function of AtUSP blocks the synthesis of matrix polysaccharides, which are required for intine synthesis, resulting in pollen without an intine layer [57]. The homologous gene of USP in tomato was found to be a direct target of SlHB8 (Table S5, see online supplementary material), but its expression level was not altered in the SlHB8 overexpression line (Table S7, see online supplementary material), indicating a complex regulation of this gene. PME genes encode enzyme called pectin methylesterases which function in the de-esterification of pectin. PME genes belong to a multigene family, and some of them display a pollen-specific expression pattern. BcPME37c and BcMF23a mutations cause abnormal thickening of the pollen intine of Bactris campestris, which affects pollen germination and growth [54, 56]. Loss-of-function of PME48 leads to late pollen germination and lower germination rate [59]. In our study, four out of eight PME genes showed increased expression levels in the p35s::SlHB8Ris line (Fig. 7d), among which SlPME8 was the target gene of SlHB8 (Table S5). Polygalacturonase (PG) – whose gene family is expressed in the pollen and/or anthers – functions in the pectin degradation and cell wall disintegration. BcMF2, BcMF6, and BcMF9 are associated with intine development. Inhibition of BcMF2 or BcMF9 results in reduced PG activity and disturbed pectin metabolism in the process of intine formation [53]. In our study, two out of three PG genes showed increased expression levels in the p35s::SlHB8Ris line (Fig. 7d), among which SlPG1 was the target gene of SlHB8 (Table S5, see online supplementary material). PLLs encode Pectate lyases (or pectate transeliminases; PLs) function in the process of cell wall disintegration and are necessary for intine loosening. Down-regulation of BcPLL9 led to abnormal intine formation and delayed pollen tube growth in B. campestris ssp. chinensis [60]. The reduced expression level of BcPLL20 resulted in abnormal disproportionated of intine distribution [61]. In our study, two genes encoding the PLs (LAT56 and AT59) were down-regulated in the p35s::SlHB8Ris line (Fig. 7d), suggesting the potential regulation by SlHB8. Finally, the fasciclin-like arabinogalactan protein affects microspore development and intine formation through cellulose deposition. Down-regulation of FLA3 in plants reduces male fertility and produces collapsed pollens grains without an intine [55]. Antisense RNA transgenic lines with reduced BcMF18 levels show abnormal pollen grains lacking an intine, cytoplasm, and nuclei as well as abnormal cellulose distribution [62]. In our study, two FLA genes showed increased expression levels in the p35s::SlHB8Ris line (Fig. 7d), but none contained a SlHB8 binding site (Table S5, see online supplementary material), indicating indirect regulation. Until now, most studies on intine formation have focused on the enzymes involved in pectin, cellulose, and callose metabolism. SlHB8 is thus the first transcription factor demonstrated to exhibit direct regulation of the genes involved in intine formation.
In summary, SlHB8 exhibited space–time characteristic expression pattern from microsporocyte differentiation to the microspore generation (Fig. 1), phenotypes of the SlHB8 knockout and overexpression lines along with the RNA-seq and DAP-seq data support the notion that SlHB8 together with the conserved genetic pathway SPL–DYT1–TDF1–AMS–MS80 are instrumental in the regulation of tapetum development and degradation. Loss-of-function of SlHB8 induces pollen activity and promotes fruit setting (Figs 2b and 3b,d), but the pollen morphology and tapetum degradation were similar to that of wild-type plant (Figs 3a, 4 and 5). Moreover, the expression levels of tapetum PCD regulators were not affected in the SlHB8cr plant (Fig. S9c, see online supplementary material) indicating another partner may compensate the function of SlHB8 or work together with SlHB8 in regulating the PCD process. By contrast, SlHB8 overexpression induces the expression of these conserved pathway genes, leading to premature tapetum degradation and resulting in pollen abortion. Based on these findings, a putative regulatory mechanism was proposed (Fig. 9), where overexpression of SlHB8 resistant to miR166-induced SPL expression directly or indirectly activates DYT1, TDF1, and MYB80 expression, thereby accelerating tapetum development and degradation. Therefore, SlHB8 emerges as a factor regulating pollen development. These findings expand our understanding of the molecular factors involved in tapetum development and degradation and provide potential genes for breeding strategies aimed at controlling this important trait.
Figure 9.
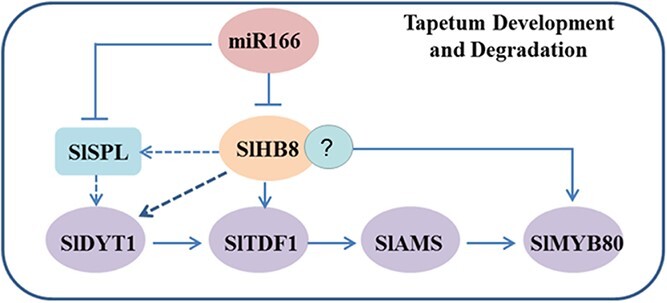
Schematic model of SlHB8-regulated genes in tapetum development and degradation.
Materials and methods
Plant material and growth conditions
SlHB8 knockout mutants were generated using CRISPR/Cas9. One single guide (sg) RNA (GCAGAAGCAAGTTTCACAGT) complementary to the coding sequence (CDS) of Solyc08g066500 was constructed into the pAGM4723 vector and transformed into Agrobacterium tumefasciens, which was used for tomato genetic transformation. Plants bearing two kinds of 8 bp deletions in the CDS and a 1 bp insertion in the CDS were obtained. The two 8 bp deletion lines were used for flower and fruit development studies. The overexpression lines p35s::SlHB8Ris and p2A11::SlHB8Ris were generated separately. The full-length CDS of SlHB8Ris was cloned into the overexpression vectors pMDC32 and 2A11, which were under control of the 35 s and 2A11 promoters, respectively, that show specific expression during the mature stages of anther and early fruit development [63]. The transgenic plants were selected on MS medium with antibiotic selection of the construction vector. Positive overexpression lines were identified by checking the expression levels of SlHB8. Experiments were conducted in an artificial climate room (25 ± 1°C) with a light:dark cycle of 16 h:8 h at the South China Agricultural University. The planting medium was a mixture of 2:1 imported peat soil and vermiculite, and plants were potted in a 10 × 10 cm planting container. Tap water with Huabao nutrient particles was used for daily watering to provide nutrition.
RNA in situ hybridization
The anthers at MI, Tds, MUM, BM, MP stages of wild-type were sampled for RNA in situ hybridization analysis. The experiment was carried out with reference to the method described [46]. All images were taken using an optical microscope (Zeiss, Oberkochen, Germany).
Quantitative reverse transcription (qRT)-PCR
Total RNA was extracted, after which the PrimeScript TM RT reagent kit (Takara Bio, Kusatsu, Japan) was used for the cDNA synthesis. qRT-PCR was carried out with SYBR PrimeScript™ RT PCR Kit II (Takara Bio) and sequence-specific primers, with ubiquitin (UBI; serial number: Solyc01g056940) as the reference gene. The relative expression levels of examined genes were computed according to the 2 −ΔΔCT method with three biological replicates.
Fruit phenotype analysis
Five plants were randomly selected, and then ten flowers from each plant were randomly selected for fruit set rate counting. The length and width of Br + 7 fruits (7 days after the breaker stage) were measured using a cursor caliper, and single fruit weight was determined using an electronic analytical balance. The number of seeds in the tomato fruits at the Br + 7 stage was also counted.
Pollen viability assay
Pollen viability was measured using the TTC method [26], and pollen germination media used was according to Yang’s method [26]. Tomato pollen from the wild-type (WT) and SlHB8 transgenic tomato plants were incubated in PGM at 25°C for 2 h. Images were taken under a Leica microscope (Leica, Wetzlar, Germany). Anthers from the WT and transgenic tomato plants were counterstained with 0.1 mg mL−1 DAPI to assess their nuclear status. The DAPI emission signals were 350 nm/460 nm. WT and transgenic lines were crossed as both paternal and maternal plants, after which fruit setting rates were statistically analysed.
Cytological characterization of anthers
Anthers from different developmental stages (MI, microspore mother cell stage; Tds, tetrad stage; MUM, middle uninucleate microspore stage; BM, binucleate microspore stage; MP, mature pollen stage) were collected during the flowering period and fixed at room temperature for 24–36 h. After paraffin embedding and sectioning, anther cell characteristics were examined using a Leica microscope. A terminal deoxynucleotidyl transferase-mediated biotin-16-dUTP nick-end labeling (TUNEL) assay was carried out using the DeadEnd™ Fluorometric TUNEL System (Promega, Madison, WI, USA), according to its handbook. The images of sections were taken under a Leica TCS SP5 fluorescence confocal scanning microscope. Emission wavelengths of 488 nm/505–545 nm and 561 nm/575–650 nm were used for the TUNEL and propidium iodide signals detection.
Electron microscopy of pollen phenotypes
For scanning electron microscopy (SEM), mature pollens from SlHB8 transgenic and wild-type plants were fixed on SEM carriers, coated with gold–palladium. Pollen images were taken under a EVO MA15 scanning electron microscope (Zeiss, Oberkochen, Germany).
Anthers at different developmental stages (MI, Tds, MUM, BM, and MP) were fixed in 4% glutaraldehyde and 2% paraformaldehyde at 4°C overnight. After washing with 0.1 M PBS (four times), the fixed pollen grains were incubated in 1.5% low melting agar, post-fixed in 1% osmium tetroxide, and dehydrated with a graded ethanol series. The samples were then transferred to acetone and embedded in Spurr’s resin (SPI, West Chester, PA, USA). Sections (70 nm thick) were cut using an ultramicrotome (UC7; Leica), collected on copper grids, and stained with uranyl acetate and lead citrate. The stained grids were then photographed with a Talos L120C electron microscope (Thermo Fisher Scientific, Waltham, MA, USA).
RNA-seq and DNA affinity purification (DAP)-seq analysis
Flower Buds at the tetrad stage were collected and frozen with liquid nitrogen for RNA extraction and transcriptome sequencing experiment which will be carried out by the company Gene Denovo Biotechnology Co., Ltd (Guangzhou, China). Fragments per kilobase of transcript per million mapped reads (FPKM) was used for calculating the expression levels of detected genes. The threshold of log2 (fold change) ≥ 1 and false discovery rate (FDR) ≤ 0.05 were used for defining the differentially expressed genes (DEGs). Transcriptome data analysis and mapping were performed using the online platform OmicShare Tools developed by Gene Denovo (www.omicshare.com/tools). Heatmaps were generated using TBtools as described in the manual [64].
The SlHB8 gene was cloned into the protein expression halo vector provided by Gene Denovo, the buds at different developmental stages (MI, Tds, MUM, BM, and MP) were graded sampled and mixed at equal ratios, frozen with liquid nitrogen for stock. Protein purification for DAP-seq and extraction and sequencing of genomic DNA were performed by Gene Denovo.
Yeast one-hybrid assay (Y1H)
The pGADT7-Rec vector with full length CDS of SlHB8 was treated as a prey vector. pAbAi vector containing multiple SlHB8 binding elements obtained from DAP-seq analysis were regarded as a bait vector. The selection of minimal inhibitory concentration of aureobasidin A was carried out after the transformation of Y1H Gold yeast strains with linearized pAbAi constructs. The binding activity of SlHB8 on the elements was examined by transforming the prey vector to the bait yeast strains which will be cultured on the SD medium lacking Leu (SD/−Leu) with or without aureobasidin A of selected concentration at 30°C for 2–3 days.
Dual-luciferase transient expression assay
To check the regulatory activity of SlHB8 on the promoters containing SlHB8 binding elements, the pGreenII 62-SK vector containing the SlHB8 CDS were treated as effector vector, and the pGreenII 0800-LUC vector with target promoters were regarded as reporter vector. These plasmids with differential ratios and composition were injected into Nicotiana benthamiana leaves via Agrobacterium tumefaciens mediation. The activities of luciferase and Renilla were measured using the Dual-Luciferase Assay Kit (Promega) according to its handbook.
Metabolite analysis
Anthers from flowers at the anthesis stage were collected, frozen in liquid nitrogen for stock. Primary metabolome analysis was performed by MetWare Biotech Ltd. ANOVA (P < 0.01) was used to identify different metabolomes between WT and SlHB8 transgenic plant.
Electrophoretic mobility shift assay (EMSA)
The pGEX-4 T-1 vector containing full-length CDS of SlHB8 was transferred into Escherichia coli strain BM Rosetta (DE3) to producing SlHB8-GST fusion protein. The SlHB8 protein purification and EMSA operational approach were according to the methods descried in the publication of Drakakaki [65].
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31902013 and 31870286); the Natural Science Foundation of Guangdong Province (2018A030310205, 2022A1515012278, 2021A1515010528, and 2017A030313114); and the General Project of Guangzhou City (201804010031). We thank Jilei Huang and Chuanhe Liu from the Instrumental Analysis & Research Center, South China Agricultural University for help with TEM sample processing and image acquisition; Juan Zhou from the Instrumental Analysis & Research Center, South China Agricultural University for help with SEM sample processing and image acquisition; Dr. Hai Zhou from the College of Biology Science of South China Agricultural University for help with TUNEL experiment.
Author contributions
C.W., Y.Y., Z.X., C.Y., Z.P., H.G., and D.S. performed the research; Y.H., R.C., and Z.L. design of the research. Y.H., G.H., D.C., and C.W. analysed the data, Y.H. and C.W. wrote the manuscript. All authors assisted with manuscript revision. All authors read and approved the final version of the manuscript.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Conflict of interest
The authors declare no conflicts of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Caiyu Wu, Key Laboratory of Horticultural Crop Biology and Germplasm Innovation in South China, Ministry of Agriculture, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
Yang Yang, Key Laboratory of Horticultural Crop Biology and Germplasm Innovation in South China, Ministry of Agriculture, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
Deding Su, Key Laboratory of Plant Hormones and Development Regulation of Chongqing, School of Life Sciences, Chongqing University, Chongqing, China; Center of Plant Functional Genomics, Institute of Advanced Interdisciplinary Studies, Chongqing University, Chongqing 400044, China.
Canye Yu, Key Laboratory of Horticultural Crop Biology and Germplasm Innovation in South China, Ministry of Agriculture, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
Zhiqiang Xian, Key Laboratory of Plant Hormones and Development Regulation of Chongqing, School of Life Sciences, Chongqing University, Chongqing, China; Center of Plant Functional Genomics, Institute of Advanced Interdisciplinary Studies, Chongqing University, Chongqing 400044, China.
Zanlin Pan, Key Laboratory of Horticultural Crop Biology and Germplasm Innovation in South China, Ministry of Agriculture, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
Hongling Guan, Key Laboratory of Horticultural Crop Biology and Germplasm Innovation in South China, Ministry of Agriculture, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
Guojian Hu, UMR990 INRA/INP-ENSAT, Université de Toulouse, Castanet-Tolosan, France.
Da Chen, Key Laboratory of Horticultural Crop Biology and Germplasm Innovation in South China, Ministry of Agriculture, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
Zhengguo Li, Key Laboratory of Plant Hormones and Development Regulation of Chongqing, School of Life Sciences, Chongqing University, Chongqing, China.
Riyuan Chen, Key Laboratory of Horticultural Crop Biology and Germplasm Innovation in South China, Ministry of Agriculture, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
Yanwei Hao, Key Laboratory of Horticultural Crop Biology and Germplasm Innovation in South China, Ministry of Agriculture, College of Horticulture, South China Agricultural University, Guangzhou 510642, China.
References
- 1. Borg M, Brownfield L, Twell D. Male gametophyte development: a molecular perspective. J Exp Bot. 2009;60:1465–78. [DOI] [PubMed] [Google Scholar]
- 2. Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parish RW, Li SF. Death of a tapetum: a programme of developmental altruism. Plant Sci. 2010;178:73–89. [Google Scholar]
- 4. Ni E, Zhou L, Li Jet al. OsCER1 plays a pivotal role in very-Long-chain alkane biosynthesis and affects plastid development and programmed cell death of Tapetum in Rice (Oryza sativa L.). Front Plant Sci. 2018;9:1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng S, Dong J, Lu Jet al. A cytosolic pentatricopeptide repeat protein is essential for tapetal plastid development by regulating OsGLK1 transcript levels in rice. New Phytol. 2022;234:1678–95. [DOI] [PubMed] [Google Scholar]
- 6. Pan XY, Yan W, Chang Zet al. OsMYB80 regulates anther development and pollen fertility by targeting multiple biological pathways. Plant Cell Physiol. 2020;61:988–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang YK, Ye H, Bai JFet al. The regulatory framework of developmentally programmed cell death in floral organs: a review. Plant Physiol Biochem. 2021;158:103–12. [DOI] [PubMed] [Google Scholar]
- 8. Phan HA, Iacuone S, Li SFet al. The MYB80 transcription factor is required for pollen development and the regulation of Tapetal programmed cell death in Arabidopsis thaliana. Plant Cell. 2011;23:2209–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu J, Chen H, Li Het al. Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008;55:266–77. [DOI] [PubMed] [Google Scholar]
- 10. Xu Y, Iacuone S, Li SFet al. MYB80 homologues in Arabidopsis, cotton and brassica: regulation and functional conservation in tapetal and pollen development. BMC Plant Biol. 2014;14:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu J, Lou Y, Xu XFet al. A genetic pathway for Tapetum development and function in Arabidopsis. J Integr Plant Biol. 2011;53:892–900. [DOI] [PubMed] [Google Scholar]
- 12. Liu XF, Ning K, Che Get al. CsSPL functions as an adaptor between HD-ZIPIII and CsWUS transcription factors regulating anther and ovule development in Cucumis sativus (cucumber). Plant J. 2018;94:535–47. [DOI] [PubMed] [Google Scholar]
- 13. Li XR, Lian H, Zhao QXet al. MicroRNA166 monitors SPOROCYTELESS/NOZZLE for building of the anther internal boundary. Plant Physiol. 2019;181:208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang W-C, Ye D, Xu Jet al. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999;13:2108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jia GX, Liu XD, Owen HAet al. Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc Natl Acad Sci U S A. 2008;105:2220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gu JN, Zhu J, Yu Yet al. DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J. 2014;80:1005–13. [DOI] [PubMed] [Google Scholar]
- 17. Jeong HJ, Kang JH, Zhao Met al. Tomato male sterile 10(35) is essential for pollen development and meiosis in anthers. J Exp Bot. 2014;65:6693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu J, Yang C, Yuan Zet al. The ABORTED MICROSPORES regulatory network is required for Postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell. 2010;22:91–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang ZB, Zhu J, Gao JFet al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 2007;52:528–38. [DOI] [PubMed] [Google Scholar]
- 20. Vizcay-Barrena G, Wilson ZA. Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J Exp Bot. 2006;57:2709–17. [DOI] [PubMed] [Google Scholar]
- 21. Ma XF, Wu Y, Zhang GF. Formation pattern and regulatory mechanisms of pollen wall in Arabidopsis. J Plant Physiol. 2021;260:153388. [DOI] [PubMed] [Google Scholar]
- 22. Lou Y, Xu XF, Zhu Jet al. The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat Commun. 2014;5:3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiong SX, Lu JY, Lou Yet al. The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana. Plant J. 2016;88:936–46. [DOI] [PubMed] [Google Scholar]
- 24. Lu JY, Xiong SX, Yin Wet al. MS1, a direct target of MS188, regulates the expression of key sporophytic pollen coat protein genes in Arabidopsis. J Exp Bot. 2020;71:4877–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pan CT, Yang D, Zhao Xet al. PIF4 negatively modulates cold tolerance in tomato anthers via temperature-dependent regulation of tapetal cell death. Plant Cell. 2021;33:2320–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu XY, Yang M, Liu Xet al. A putative bHLH transcription factor is a candidate gene for male sterile 32, a locus affecting pollen and tapetum development in tomato. Hortic Res. 2019;6:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, Zhang B, Yang Tet al. The GAMYB-like gene SlMYB33 mediates flowering and pollen development in tomato. Hortic Res. 2020;7:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perez-Martin F, Pineda B, Garcia-Sogo Bet al. Developmental role of the tomato mediator complex subunit MED18 in pollen ontogeny. Plant J. 2018;96:300–15. [DOI] [PubMed] [Google Scholar]
- 29. Yan MY, Xie DL, Cao JJet al. Brassinosteroid-mediated reactive oxygen species are essential for tapetum degradation and pollen fertility in tomato. Plant J. 2020;102:931–47. [DOI] [PubMed] [Google Scholar]
- 30. Chen LF, Yang D, Zhang Yet al. Evidence for a specific and critical role of mitogen-activated protein kinase 20 in uni-to-binucleate transition of microgametogenesis in tomato. New Phytol. 2018;219:176–94. [DOI] [PubMed] [Google Scholar]
- 31. Gan ZY, Feng Y, Wu Tet al. Downregulation of the auxin transporter gene SlPIN8 results in pollen abortion in tomato. Plant Mol Biol. 2019;99:561–73. [DOI] [PubMed] [Google Scholar]
- 32. Wang R, Shi CL, Wang Xet al. Tomato SlIDA has a critical role in tomato fertilization by modifying reactive oxygen species homeostasis. Plant J. 2020;103:2100–18. [DOI] [PubMed] [Google Scholar]
- 33. Ohashi-Ito K, Kubo M, Demura Tet al. Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol. 2005;46:1646–56. [DOI] [PubMed] [Google Scholar]
- 34. Kim J, Jung JH, Reyes JLet al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005;42:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rong FX, Chen F, Huang Let al. A mutation in class III homeodomain-leucine zipper (HD-ZIP III) transcription factor results in curly leaf (cul) in cucumber (Cucumis sativus L.). Theor Appl Genet. 2019;132:113–23. [DOI] [PubMed] [Google Scholar]
- 36. Byrne ME. Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes. PLoS Genet. 2006;2:e89–790 e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith ZR, Long JA. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature. 2010;464:423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emery JF, Floyd SK, Alvarez Jet al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–74. [DOI] [PubMed] [Google Scholar]
- 39. Prigge MJ, Otsuga D, Alonso J́Met al. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhong R, Ye Z-H. Regulation of HD-ZIP III genes by MicroRNA 165. Plant Signal Behav. 2007;2:351–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McConnell JR, Emery J, Eshed Yet al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature (London). 2001;411:709–13. [DOI] [PubMed] [Google Scholar]
- 42. Carlsbecker A, Lee JY, Roberts CJet al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clepet C, Devani RS, Boumlik Ret al. The miR166-SlHB15A regulatory module controls ovule development and parthenocarpic fruit set under adverse temperatures in tomato. Mol Plant. 2021;14:1185–98. [DOI] [PubMed] [Google Scholar]
- 44. Hu GJ, Fan J, Xian Zet al. Overexpression of SlREV alters the development of the flower pedicel abscission zone and fruit formation in tomato. Plant Sci. 2014;229:86–95. [DOI] [PubMed] [Google Scholar]
- 45. Yang Yang XZ, Riyuan C, Yanwei H. Cloning of SlHB8 gene from tomato and its response to abiotic stress. Northern Horticulture. 2019;18:10–8. [Google Scholar]
- 46. Liu X, Wu C, Su Det al. The SlHB8 acts as a negative regulator in stem development and lignin biosynthesis. Int J Mol Sci. 2021;22:13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang Yang XZ, Riyuan C, H . Construction of plant overexpression vector of SlHB8 gene in tomato and genetic transformation. Molecular Plant Breed. 2020;18:1513–9. [Google Scholar]
- 48. Ito T, Wellmer F, Yu Het al. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature. 2004;430:356–60. [DOI] [PubMed] [Google Scholar]
- 49. Rojas-Gracia P, Roque E, Medina Met al. The parthenocarpic hydra mutant reveals a new function for a SPOROCYTELESS-like gene in the control of fruit set in tomato. New Phytol. 2017;214:1198–212. [DOI] [PubMed] [Google Scholar]
- 50. Zhou GK, Kubo M, Zhong RQet al. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007;48:391–404. [DOI] [PubMed] [Google Scholar]
- 51. Zhang W, Sun Y, Timofejeva Let al. Regulation of Arabidopsis tapetum development and function by dysfunctional tapetum1 (dyt1) encoding a putative bHLH transcription factor. Development. 2006;133:3085–95. [DOI] [PubMed] [Google Scholar]
- 52. Wang K, Guo ZL, Zhou WTet al. The regulation of Sporopollenin biosynthesis genes for rapid Pollen Wall formation. Plant Physiol. 2018;178:283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang L, Ye Y, Zhang Yet al. BcMF9, a novel polygalacturonase gene, is required for both Brassica campestris intine and exine formation. Ann Bot. 2009;104:1339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiong XP, Zhou D, Xu Let al. BcPME37c is involved in pollen intine formation in Brassica campestris. Biochem Biophys Res Commun. 2019;517:63–8. [DOI] [PubMed] [Google Scholar]
- 55. Li J, Yu MA, Geng LLet al. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 2010;64:482–97. [DOI] [PubMed] [Google Scholar]
- 56. Yue XY, Lin SE, Yu YJet al. The putative pectin methylesterase gene, BcMF23a, is required for microspore development and pollen tube growth in Brassica campestris. Plant Cell Rep. 2018;37:1003–9. [DOI] [PubMed] [Google Scholar]
- 57. Schnurr JA, Storey KK, Jung HJGet al. UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta. 2006;224:520–32. [DOI] [PubMed] [Google Scholar]
- 58. Lin S, Dong H, Zhang Fet al. BcMF8, a putative arabinogalactan protein-encoding gene, contributes to pollen wall development, aperture formation and pollen tube growth in Brassica campestris. Ann Bot. 2014;113:777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Leroux C, Bouton S, Kiefer-Meyer MCet al. PECTIN METHYLESTERASE48 is involved in Arabidopsis pollen grain germination. Plant Physiol. 2015;167:367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang JJ, Yao L, Yu Yet al. PECTATE LYASE-LIKE 9 from Brassica campestris is associated with intine formation. Plant Sci. 2014;229:66–75. [DOI] [PubMed] [Google Scholar]
- 61. Jiang JJ, Yao L, Yu Yet al. PECTATE LYASE-LIKE10 is associated with pollen wall development in Brassica campestris. J Integr Plant Biol. 2014;56:1095–105. [DOI] [PubMed] [Google Scholar]
- 62. Lin S, Yue X, Miao Yet al. The distinct functions of two classical arabinogalactan proteins BcMF8 and BcMF18 during pollen wall development in Brassica campestris. Plant J. 2018;94:60–76. [DOI] [PubMed] [Google Scholar]
- 63. Van Haaren MJ, Houck CM. A functional map of the fruit-specific promoter of the tomato 2A11 gene. Plant Mol Biol. 1993;21:625–40. [DOI] [PubMed] [Google Scholar]
- 64. Chen CJ, Chen H, Zhang Yet al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–202. [DOI] [PubMed] [Google Scholar]
- 65. Drakakaki G, Zabotina O, Delgado Iet al. Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol. 2006;142:1480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.