Abstract
Brain extraction is an indispensable step in neuro-imaging with a direct impact on downstream analyses. Most such methods have been developed for non-pathologically affected brains, and hence tend to suffer in performance when applied on brains with pathologies, e.g., gliomas, multiple sclerosis, traumatic brain injuries. Deep Learning (DL) methodologies for healthcare have shown promising results, but their clinical translation has been limited, primarily due to these methods suffering from i) high computational cost, and ii) specific hardware requirements, e.g., DL acceleration cards. In this study, we explore the potential of mathematical optimizations, towards making DL methods amenable to application in low resource environments. We focus on both the qualitative and quantitative evaluation of such optimizations on an existing DL brain extraction method, designed for pathologically-affected brains and agnostic to the input modality. We conduct direct optimizations and quantization of the trained model (i.e., prior to inference on new data). Our results yield substantial gains, in terms of speedup, latency, through-put, and reduction in memory usage, while the segmentation performance of the initial and the optimized models remains stable, i.e., as quantified by both the Dice Similarity Coefficient and the Hausdorff Distance. These findings support post-training optimizations as a promising approach for enabling the execution of advanced DL methodologies on plain commercial-grade CPUs, and hence contributing to their translation in limited- and low- resource clinical environments.
Keywords: Low resource environment, Deep learning, Segmentation, CNN, Convolutional neural network, Brain extraction, Brain tumor, Glioma, Glioblastoma, BraTS, OpenVINO, BrainMaGe, GaNDLF
1. Introduction
One of the most important first steps in any neuro-imaging analysis pipeline is brain extraction, also known as skull-stripping [1,2]. This process removes all non-brain portions in a brain scan and leaves the user with the portion of the image that is of maximal interest, i.e., the brain tissue and all associated pathologies. This step is an indispensable pre-processing operation that has a direct effect on subsequent analyses, and also used for de-identification purposes [3]. Enabling this to run on clinical workstations could have a tremendously positive impact on automated clinical workflows. The effects of the quality of brain extraction in downstream analyses have been previously reported, for studies on tumor segmentation [4-6] and neuro-degeneration [7].
This study specifically focuses on glioblastoma (GBM), which is the most aggressive type of adult brain tumors. GBM has poor prognosis despite current treatment protocols [8,9], and its treatment and management is often problematic with a necessity of requiring personalized treatment plans. To improve the treatment customization process, computational imaging and machine learning based assistance could prove to be highly beneficial. One of the key steps for this would be to enable a robust approach to obtain the complete region of immediate interest irrespective of the included pathologies that would result in an improved computational workflow.
While deep learning (DL) has been showing promising results in the field of semantic segmentation in medical imaging [4,10-17], the deployability of such models poses a substantial challenge, mainly due to their computational footprint. While prior work on brain extraction has focused on stochastic modeling approaches [1,2,18], modern solutions leveraging DL have shown great promise [12,15]. Unfortunately, models trained for this application also suffer from such deployment issues, which in turn reduces their clinical translation.
In recent years, well-known DL frameworks, such as PyTorch [19] and TensorFlow [20] have enabled the democratization of DL development by making the underlying building blocks accessible to the wider community. They usually require the help of moderately expensive computing with DL acceleration cards, such as Graphical Processing Units (GPUs) [21] or Tensor Processing Units (TPUs) [22]. While these frameworks will work on sites with such computational capacity (i.e., GPUs and TPUs), deploying them to locations with low resources is a challenge. Most DL-enabled studies are extremely compute intensive, and the complexity of the pipeline makes them very difficult to deploy, especially in tightly controlled clinical environments. While cloud-based solutions could be made available, patient privacy is a major health system concern, which requires multiple legal quandaries to be addressed prior to uploading data to the cloud. However, the availability of such approaches for local inexpensive compute solutions would be the sole feasible way for their clinical translation.
Quantizing neural networks can reduce the computational time required for the forward pass, but more importantly can reduce the memory burden during the time of inference. Post-quantization, a high precision model is reduced to a lower bit resolution model, thus reducing the size of the model. The final goal is to leverage the advantages of quantization and optimization, while maintaining the segmentation performance of the full precision floating point models as much as possible. Such methods can facilitate the reduction of the required memory to save and infer the generated model [23].
In this paper, we take an already published DL method, namely Brain Mask Generator (BrainMaGe)1 [15], and make it usable for low resource environments, such as commercial-grade CPUs with low memory, and older generation CPUs by leveraging the advantages of quantization and optimization for performance improvements. We provide a comprehensive evaluation of the observed performance improvements across multiple CPU configurations and quantization methods for the publicly available TCGA-GBM dataset [6,24,25], as well as a private testing dataset.
2. Methods
2.1. Data
We identified and collected n = 864 multi-parametric magnetic resonance images (mpMRI) brain tumor scans from n = 216 GBM patients from both private and public collections. The private collections included n = 364 scans, from n = 91 patients, acquired at the Hospital of the University of Pennsylvania (UPenn). The public data is available through The Cancer Imaging Archive (TCIA) [24] and comprises of the pre-operative mpMRI scans of The Cancer Genome Atlas Glioblastoma (TCGA-GBM, n = 125) [6,25] collection. The final dataset (Table 1) included n = 864 mpMRI scans from n = 216 subjects with 4 structural modalities for each subject available, namely T1-weighted pre- & post-contrast (T1, & T1Gd), T2-weighted (T2) and T2 fluid attenuated inversion recovery (FLAIR). Notably, the multi-institutional data of the TCGA-GBM collection is highly heterogeneous, including scan quality, slice thickness between different modalities, scanner parameters. For the private collection data, the T1 scans were taken with high axial resolutions. The brain masks for the private collection data were generated internally and went through rigorous manual quality control, while the brain masks for the TCGA-GBM data were provided through the International Brain Tumor Segmentation (BraTS) challenge [4-6,26-28].
Table 1.
The distribution of all the datasets used in the study.
| Dataset | No. of subjects | No. of mpMRI scans |
|---|---|---|
| TCGA-GBM | 125 | 500 |
| UPenn | 91 | 364 |
| Total | 216 | 864 |
2.2. Data Pre-processing
All DICOM scans were converted to the Neuroimaging Informatics Technology Initiative (NIfTI) [29] file format to facilitate computational analysis, following the well-accepted pre-processing protocol of the BraTS challenge [4-6,26-28]. Specifically, all the mpMRI volumes were reoriented to the left-posterior-superior (LPS) coordinate system, and the T1Gd scan of each patient was rigidly (6 degrees of freedom) registered and resampled to an isotropic resolution of 1mm3 based on a common anatomical atlas, namely SRI24 [30]. We chose this atlas [30] as the common anatomical space, following the convention suggested by the BraTS challenge. The remaining scans (i.e., T1, T2, FLAIR) of each patient were then rigidly co-registered to this resampled T1Gd scan by first obtaining the rigid transformation matrix to T1Gd, then combining with the transformation matrix from T1Gd to the SRI24 atlas, and resampling. For all the image registrations we used the “Greedy”2 tool [31], which is a central processing unit (CPU)-based C++ implementation of the greedy diffeomorphic registration algorithm [32]. Greedy is integrated into the ITK-SNAP3 segmentation software [33,34], as well as into the Cancer Imaging Phenomics Toolkit (CaPTk)4 [35-39]. We further note that use of any non-parametric, non-uniform intensity normalization algorithm [40-42] to correct for intensity non-uniformities caused by the inhomogeneity of the scanner’s magnetic field during image acquisition, obliterates the T2-FLAIR signal, as it has been previously reported [5]. Thus, taking this into consideration, we intentionally apply the N4 bias field correction approach [41] in all scans temporarily’ to facilitate an improved registration of all scans to the common anatomical atlas. Once we obtain the transformation matrices for all the scans, then we apply these transformations to the non-bias corrected images. This complete pre-processing is available through CaPTk, as officially used for the BraTS challenge (Fig. 1).
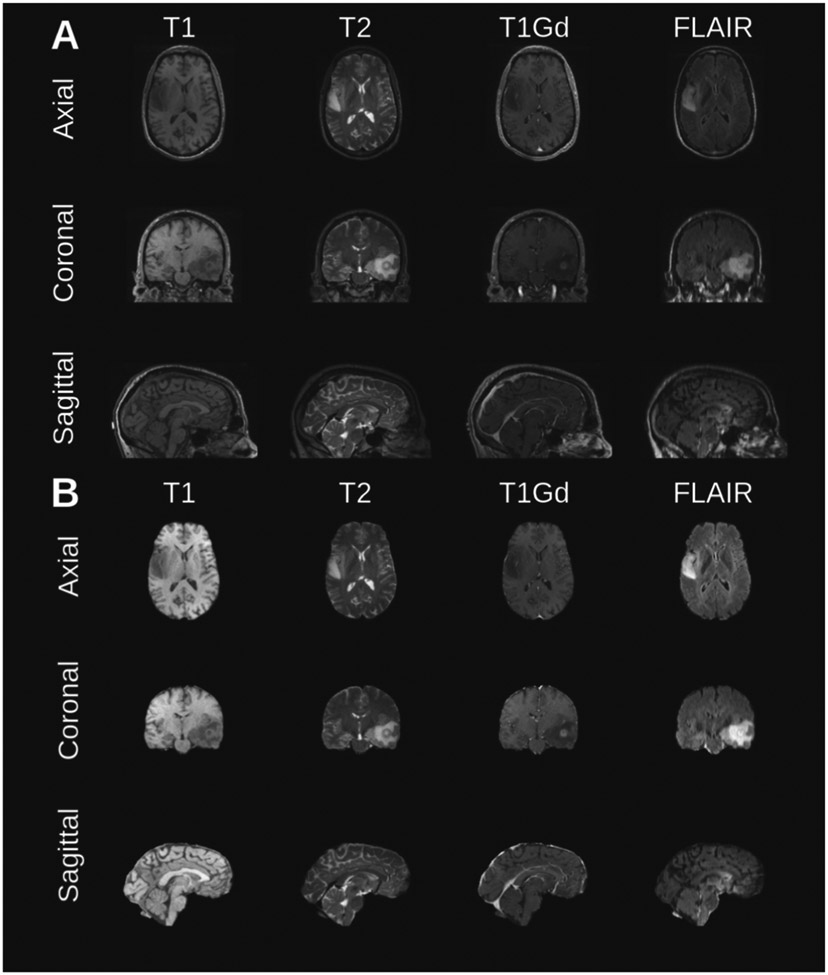
Fig. 1.
Example of MRI brain tumor scan from a randomly selected subject from the test set. The Original scans (A) include the skull and other non-brain tissues, and (B) the corresponding scan slices depicting only the brain.
2.3. Network Topology
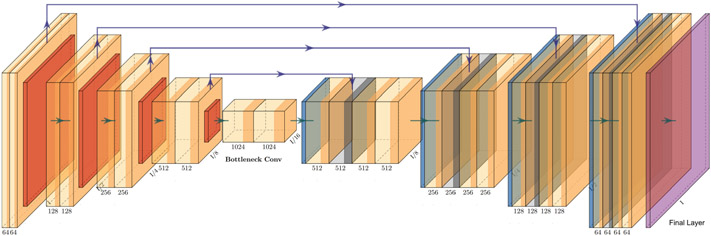
We have used the 3D implementation [10], of the widely-used network topology of U-Net [44], with added residual connections between the encoder and the decoder, to improve the backpropagation process [10,13,15,44-46]. The actual topology used here is highlighted in Fig. 2. The U-Net topology has been extensively used in semantic segmentation of both 2D and 3D medical imaging data. The U-Net consists of an encoder, which contains convolutional layers and downsampling layers, a decoder offering upsampling layers (applying transpose convolution layers), and convolutional layers. The encoder-decoder structure contributes towards automatically capturing information at varying resolutions and scales. There is an addition of skip connections, which includes concatenated feature maps paired across the encoder and the decoder layer, to improve context and feature re-usability. The residual connections utilize additional information from previous layers (across the encoder and decoder) that enable a segmentation performance boost.
Fig. 2.
The U-Net topology with residual connections from GaNDLF was used for this study. Figure was plotted using PlotNeuralNet [43].
2.4. Inference Optimizations
In this work, we used the OpenVINO toolkit (OV) for the optimizations of the BrainMaGe model. First, in order to provide estimates of scalability of the model performance in low resource environments, we conduct a comparison between the inference performance of the optimized OV model with that of the PyTorch framework. We further show a comparison of the optimized model performance across various hardware configurations typically found in such environments. We then showcase further performance improvements obtained through post-training quantization of the model and perform similar comparisons across different hardware configurations. In summary, for the BrainMaGe model, we explored both (i) conversion from PyTorch to the optimized model with an additional intermediate conversion to ONNX, which lead to an intolerable accuracy drop during the PyTorch to ONNX conversion step, and (ii) direct conversion from PyTorch to the model’s optimized intermediate representation format.
2.4.1. OpenVINO Toolkit
OV is a neural network inference optimization toolkit [47], which provides inference performance optimizations for applications using computer vision, natural language processing, and recommendation systems, among others. Its main components are two: 1) A model optimizer and 2) an inference engine. The OV model optimizer, provides conversion from a pre-trained network model trained in frameworks (such as PyTorch and TensorFlow) into an intermediate representation (IR) format that can be consumed by its second main component, i.e., its inference engine. Other types of formats that are supported include the ONNX format. Hence, for frameworks like TensorFlow and PyTorch, there is an intermediate conversion step that can be performed offline. While support for direct conversion from the PyTorch framework is limited, there are specific extensions [48] that enable this. The OV inference engine, provides optimized implementations for common operations found in neural networks, such as convolutions, and pooling operations. OV also provides graph level optimizations, such as operator fusion and optimizations for common neural network patterns through the Ngraph library [49]. These optimizations can provide direct improvements in the execution time of the model, enabling the latter for low- (or limited-) resource environments with tight compute constraints.
2.5. Network Quantization
Quantization is an optimization technique that has been adopted in recent times, to improve inference performance of neural network models [50,51]. It involves a conversion from a high precision datatype to a lower-precision datatype. In this study, we specifically discuss the quantization of a 32-bit floating point (FP32) model to an 8-bit integer (INT8) model as provided by Eq. 1:
| (1) |
where the scale factor provides a mapping of the FP32 values to the low-precision range. The zerooffset provides a representation of the FP32 zero value to an integer value [52,53].
We have explored leveraging quantization for further improvements in inference, while maintaining the model’s segmentation performance. Quantization has many benefits, including (i) speedup improvements, and (ii) reduction of memory utilization. There are two popular approaches to model quantization, namely:
Quantization-aware training [54], which involves training the neural network with fake quantization operations inserted in the network graph. The fake quantization nodes are able to learn the range of the input tensors and hence this serves as a simulation of the quantization.
Post-training quantization [55], which is the idea where the quantization process is performed post-training, but prior to the actual inference. A subset of the training dataset is selected for calibration, and this dataset is used to learn the minimum and maximum ranges of the input weights and activations for tensor quantization.
In this study, we have focused on exploring post-training quantization using the OV AccuracyAware technique [56], which provides model optimizations while explicitly limiting the segmentation performance drop. The intuition of the method is that the quantization is targeted towards all eligible layers in the topology. However, if a segmentation performance drop is observed, greater than the user-specified threshold, the layers that contribute the most to the segmentation performance drop are iteratively reverted back to the original datatype, until the desired segmentation performance level is achieved.
2.5.1. Quantitative Evaluation
The segmentation performance of the model is quantitatively evaluated according to (i) the Dice Similarity Coefficient [57] (a widely used and accepted metric for quantifying segmentation results [58]), (ii) the 95th percentile of the (symmetric) Hausdorff Distance (commonly used in Biomedical Segmentation challenges) (iii) memory utilization, and (iv) inference performance (latency). We further report the model performance for each stage of optimization, i.e., for the 1) baseline PyTorch implementation, 2) OV optimized FP32 model, and 3) OV optimized model converted to INT8 format through the post-training quantization step (Table 4). It is important to note that quantization to lower precision formats, such as INT8, typically results in a small drop in segmentation performance but this is highly dependent on the dataset. In our case, we do not notice any loss in segmentation performance after converting the model to the OV optimized model format.
Table 4.
Summary of accuracy, memory utilization and performance (latency) on the hardware configuration 4: Core(TM) i7-1185G7 @ 3.00 GHz.
| DL framework | Version | Precision | Average dice score |
Average Hausdorff distance |
Memory utilization (normalized) |
Avg. latency speedup (normalized) |
|---|---|---|---|---|---|---|
| PyTorch | 1.5.1 | FP32 | 0.97198 | 2.6577 ± 3.0 | 1 | 1 |
| 1.9.0 | FP32 | 0.97198 | 2.6577 ± 3.0 | 0.769 | 3.8 | |
| OpenVINO | 2021.4 | FP32 | 0.97198 | 2.6577 ± 3.0 | 1.285 | 7.1 |
| INT8 | 0.97118 | 2.7426 ± 3.1 | 0.907 | 23.3 |
2.6. Experimental Design
In favor of completeness, we chose five hardware platforms from various CPU generations, to benchmark our various model configurations. We ran inference benchmarks on all five hardware platforms with n = 132 images from the TCGA-GBM dataset. The results are reported based on average of running inferences on these images with a batch size of n = 1. See Tables 2 and 3 for the detailed hardware and software configurations.
Table 2.
The detailed hardware configurations used in for our experiments. Hyperthreading and turbo was enabled for all.
| Config 1 | Config 2 | Config 3 | Config 4 | Config 5 | |
|---|---|---|---|---|---|
| Platform | Kaby Lake | Coffee Lake | Ice Lake -U | Tiger Lake | Cascade Lake |
| CPU | Core(TM) i5-7400 CPU @ 3.00 GHz |
Core(TM) X-GOLD 626 CPU @ 2.60 GHz | Core(TM) i7-1065G7 CPU @ 1.30GHz | Core(TM) i7-1185G7 CPU @ 3.00GHz | Xeon(R) Gold 6252N CPU @ 2.30 GHz |
| # Nodes, # Sockets | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 2 |
| Cores/socket, Threads/socket | 4, 4 | 8, 16 | 4, 8 | 4, 8 | 24, 48 |
| Mem config: type, slots, cap, speed | DDR4, 2, 4 GB, 2133MT/s | DDR4, 2, 8 GB, 2667MT/s | LPDDR4, 2, 4 GB, 3733MT/s | DDR4, 2, 8 GB, 3200MT/s | DDR4, 12, 16 GB, 2933MT/s |
| Total memory | 8 GB | 16 GB | 8 GB | 16 GB | 192 GB |
| Advanced technologies | AVX2 | AVX2 | AVX2, AVX512, DL Boost (VNNI) | AVX2, AVX512, DL Boost (VNNI) | AVX2, AVX512, DL Boost (VNNI) |
| TDP | 90 W | 95 W | 15 W | 28 W | 150 W |
Table 3.
Details of the topology implementation. We used the 3D-ResU-Net architecture with 1 input channel, 2 output classes, and number of initial filters as 16.
| Framework | OpenVINO 2021.4 | PyTorch 1.5.1, 1.9.0 |
|---|---|---|
| Libraries | nGraph/MKLDNN | MKLDNN |
| Model | Resunet_ma.xml, Resunet_ma.bin | Resunet_ma.pt |
| Input shape | (1, 1, 128, 128, 128) | (1, 1, 128, 128, 128) |
| Precision | FP32, INT8 | FP32, INT8 |
3. Results
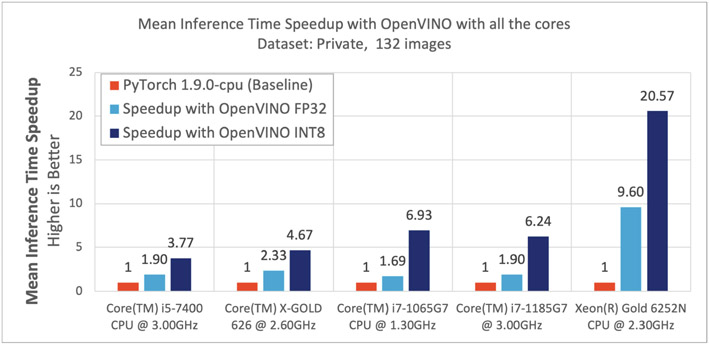
Of particular interest are the results obtained using the Hardware Configuration 4 (Core(TM) i7-1185G7 @ 3.00 GHz machine), which describes the current generation of hardware available in the consumer market. We further summarize the results obtained from all hardware configurations, in Fig. 3. Table 4 shows the summary of these metrics running on the hardware configuration 4, using the n = 132 images from the public dataset. We also compare the results obtained using PyTorch v.1.5.1 and PyTorch v.1.9.0. Notably the dynamic quantization methodology on PyTorch v.1.9.0 did not yield any performance improvement. With FP32 precision, the performance between the PyTorch and the OV models is identical. Although memory utilization is slightly better with PyTorch v.1.9.0, the inference performance (latency) is 1.89x better with OV. When assessing the INT8 quantized/OV model, the performance drop is negligible, with comparable memory utilization, but with a 6.2x boost in ‘latency’, when compared to PyTorch v.1.9.0. The memory utilization and the model performance are similar across the hardware configurations, with some variations in ‘latency’. On the client hardware platforms (Configurations 1, 2, 3, and 4), with OV FP32 precision, we observed up to 2.3x improvements in latency. The OV INT8 precision yielded further speedups up to 6.9x. On server hardware platforms (Configuration 5), with OV FP32 precision, we observed upto 9.6x speedup and with the INT8 precision we observed a speedup up to 20.5x. Figure 3 illustrates the speedup per configuration, and Fig. 4 highlights some example qualitative results. The additional boost in performance with INT8 quantized model in Configurations 3, 4, and 5, is due to the hardware platform’s advanced features, i.e., AVX512 & Intel DL Boost technology [59,60].
Fig. 3.
Speedup across different platforms using all the cores available on a processor.
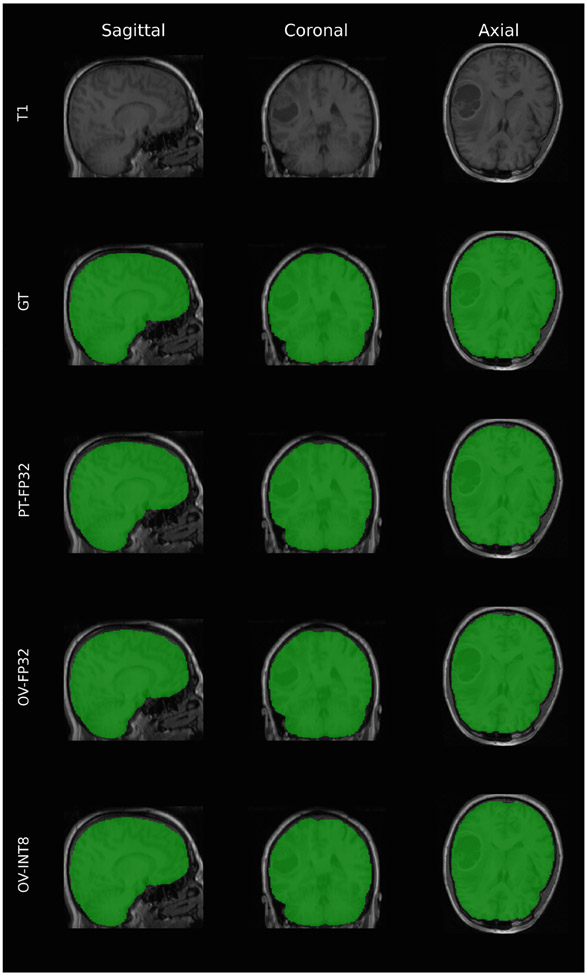
Fig. 4.
Qualitative comparison of results for one of the subjects with high resolution T1 scans across the 3 visualization slices. “GT” is the ground truth mask, “PT-FP32” is the mask generated by the original PyTorch FP32 model, “OV-FP32” is the output of the optimized model in FP32, and “OV-INT8” is the output of the optimized model after quantizing to INT8.
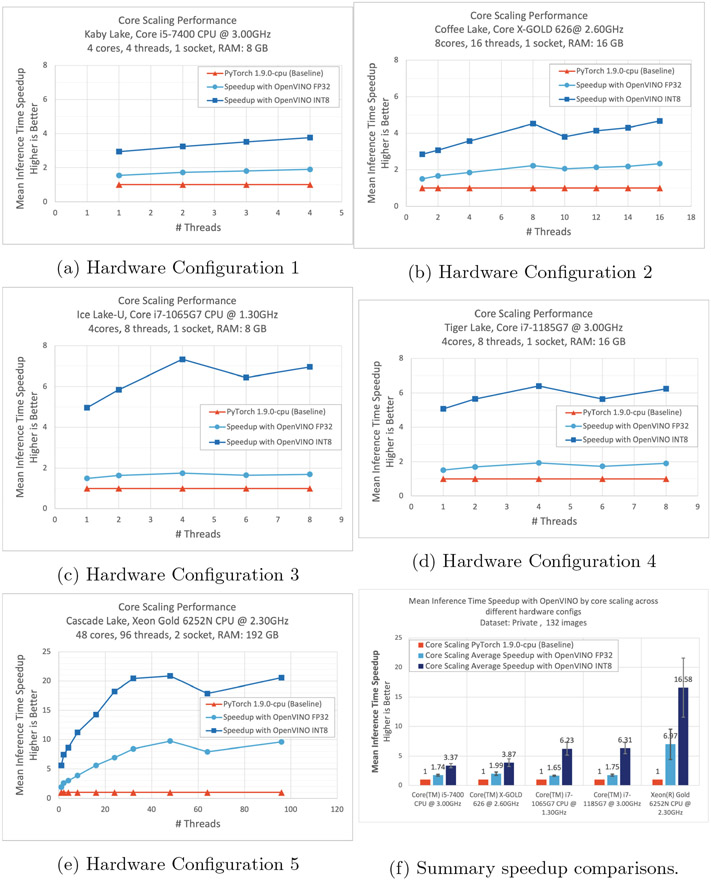
3.1. Core Scaling Improvements Across Various CPUs
Additionally, we performed a core scaling performance benchmarking to determine the scalability aspects of the model and the hardware. By limiting the number of threads to run the inference, we performed benchmarking on all the hardware configurations. Figure 5 shows a trend of increased performance with the increase in the number of threads. A slight drop in speedup can be observed if the number of threads assigned is greater than the number of physical cores. This is due to the imbalance and over-subscription of the threads. When varying the number of threads for inference, the memory utilization and accuracy are similar to running on all the threads available. The performance of both the PyTorch and the OV models improved with the increase in the number of threads allocated to the inference. However, the speedup achieved with the OV optimized FP32 and INT8 models, over PyTorch, is substantial and can be observed on all hardware configurations. Figure 5f shows the average inference time speedup achieved by limiting the number of threads on different hardware configurations.
Fig. 5.
Core scaling performance improvements, across various hardware configurations, shown in (a–e). The average speedup across all hardware configurations, and comparison with the PyTorch baseline performance (f).
4. Discussion
In this study, we investigated the potential contributions of mathematical optimizations of an already trained Deep Learning (DL) segmentation model, to enable its application in limited-/low-resource environments. We specifically focused on a MRI modality agnostic DL method, explicitly designed and developed for the problem of brain extraction in the presence of diffuse gliomas [14,15]. We explored these mathematical optimizations, in terms of their potential model improvements on 1) execution time, for different hardware configurations (i.e., speedup, Fig. 3), 2) speedup, as a function of increasing number of CPU cores for all the hardware configuration we considered (Fig. 5), 3) memory requirements (Table 4), and 4) segmentation performance. Our results yield a distinct speedup, and a reduction in computational requirements, while the segmentation performance remains stable, thereby supporting the potential of the proposed solution for application in limited-/low-resource environments.
For these intended inference time optimizations (i.e., applied in the already trained model), we have particularly focused on using the post-training quantization technique. We observe that the largest improvement in terms of speedup was obtained from the post-training quantized INT8 model, which ended up being > 23x faster than the native single-precision implementations, while producing a negligible segmentation performance drop as measured by both the Dice Similarity Coefficient and the Hausdorff distance (Table 4). Post training quantization is the quickest method of obtaining the quantized INT8 model and is desirable in situations where the “accuracy” (i.e., segmentation performance) drop is minimal, as well as within an acceptable threshold. In scenarios where the “accuracy” drop is greater than the acceptable threshold, quantization aware training could be an alternative approach to help in obtaining such potential improvements. However, such optimization (quantization aware training) would require model re-training.
The total number of parameters of the BrainMaGe 3D-ResU-Net model are 8.288 × 106, for which the number of Floating point operations per second (Flops) required for the OV FP32 model are 350.72665 × 109, whereas for the OV INT8 model the number of Flops required are 2.09099 × 109 and number of Integer operations per second (Iops) required are 348.63566 × 109. We observed that approximately 99.4% of Flops have been converted to Iops in the optimized INT8 model, resulting in two major computational benefits: (i) With lower precision (INT8), there is an improved data transfer speed through the memory hierarchy due to better cache utilization and reduction of bandwidth bottle-necks, thus enabling to maximize the compute resources; (ii) With hardware advanced features [59,60], the number of compute operations per second (OPS) are higher, thus reducing the total compute time. These two benefits of reduced memory bandwidth and higher frequency of OPS with the lower precision model resulted in substantial improvements (Table 4).
In favor of transparency and reproducibility, we make publicly available the optimized BrainMaGe brain extraction model, through its original repository5. Furthermore, a more generalized solution will also be made publicly available through the Generally Nuanced Deep Learning Framework (GaNDLF)6 [13], towards enabling scalable end-to-end clinically-deployable workflows.
We consider the immediate future work as a three-fold: 1) performance evaluation of quantization aware training compared against post-training quantization; 2) extended evaluation on a larger multi-institutional dataset [61,62], as well as evaluation of additional network topologies; 3) a comprehensive analysis covering additional hardware configurations; 4) assessment of the potential contributions of these mathematical optimizations for varying DL workloads, beyond segmentation and towards regression and classification tasks in the healthcare domain.
Acknowledgments.
Research reported in this publication was partly supported by the National Cancer Institute (NCI) and the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health (NIH), under award numbers NCI:U01CA242871 and NINDS:R01NS042645. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH.
Footnotes
github.com/pyushkevich/greedy, hash: 1a871c1, Last accessed: 27/May/2020.
itksnap.org, version: 3.8.0, last accessed: 27/May/2020.
www.cbica.upenn.edu/captk, version: 1.8.1, last accessed: 11/February/2021.
References
- 1.Smith SM: “Bet: Brain extraction tool,” FMRIB TR00SMS2b, Oxford Centre for Functional Magnetic Resonance Imaging of the Brain). Department of Clinical Neurology, Oxford University, John Radcliffe Hospital, Headington, UK: (2000) [Google Scholar]
- 2.Smith SM: Fast robust automated brain extraction. Hum. Brain Mapp 17(3), 143–155 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz CG, et al. : Identification of anonymous MRI research participants with face-recognition software. N. Engl. J. Med 381(17), 1684–1686 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakas S, et al. : Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprint arXiv:1811.02629 (2018) [Google Scholar]
- 5.Bakas S, et al. : Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4, 170117 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakas S, et la.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. Cancer Imaging Arch. (2017). 10.7937/K9/TCIA.2017.KLXWJJ1Q [DOI] [Google Scholar]
- 7.Gitler AD, Dhillon P, Shorter J: Neurodegenerative disease: models, mechanisms, and a new hope. Disease Models Mech. 10, 499–502 (2017). 28468935[pmid] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostrom QT, Rubin JB, Lathia JD, Berens ME, Barnholtz-Sloan JS: Females have the survival advantage in glioblastoma. Neuro-oncol. 20, 576–577 (2018). 29474647[pmid] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrlinger U, et al. : Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): a randomised, open-label, phase 3 trial. Lancet 393, 678–688 (2019) [DOI] [PubMed] [Google Scholar]
- 10.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O: 3D U-net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham: (2016). 10.1007/978-3-319-46723-8.49 [DOI] [Google Scholar]
- 11.Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH: NNU-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021) [DOI] [PubMed] [Google Scholar]
- 12.Isensee F, et al. : Automated brain extraction of multisequence MRI using artificial neural networks. Hum. Brain Mapp 40(17), 4952–4964 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pati S, et al. : Gandlf: a generally nuanced deep learning framework for scalable end-to-end clinical workflows in medical imaging. arXiv preprint arXiv:2103.01006 (2021) [Google Scholar]
- 14.Thakur SP: Skull-stripping of glioblastoma MRI scans using 3D deep learning. In: Crimi A, Bakas S (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 57–68. Springer, Cham: (2020). 10.1007/978-3-030-46640-4_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakur S, et al. : Brain extraction on MRI scans in presence of diffuse glioma: multi-institutional performance evaluation of deep learning methods and robust modality-agnostic training. Neuroimage 220, 117081 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhalerao M, Thakur S: Brain tumor segmentation based on 3D residual U-net. In: Crimi A, Bakas S (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 218–225. Springer, Cham: (2020). 10.1007/978-3-030-46643-5_21 [DOI] [Google Scholar]
- 17.Kamnitsas K, et al. : Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal 36, 61–78 (2017) [DOI] [PubMed] [Google Scholar]
- 18.Leung KK, et al. : Brain maps: an automated, accurate and robust brain extraction technique using a template library. Neuroimage 55(3), 1091–1108 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paszke A, et al. : Pytorch: an imperative style, high-performance deep learning library. In: Advances in Neural Information Processing Systems, pp. 8026–8037 (2019) [Google Scholar]
- 20.Abadi M, et al. : TensorFlow: a system for large-scale machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI), vol. 16, pp. 265–283 (2016) [Google Scholar]
- 21.Lin HW, Tegmark M, Rolnick D: Why does deep and cheap learning work so well? J. Stat. Phys 168, 1223–1247 (2017) [Google Scholar]
- 22.Jouppi NP, et al. : In-datacenter performance analysis of a tensor processing unit (2017) [Google Scholar]
- 23.Hubara I, Courbariaux M, Soudry D, El-Yaniv R, Bengio Y: Quantized neural networks: training neural networks with low precision weights and activations (2016) [Google Scholar]
- 24.Clark K, et al. : The cancer imaging archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarpace L, et al. : Radiology data from the cancer genome atlas glioblastoma multiforme [TCGA-GBM] collection. Cancer Imaging Arch. 11(4), 1 (2016) [Google Scholar]
- 26.Menze BH, et al. : The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakas S, et al. : Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. Cancer Imaging Arch. (2017). 10.7937/K9/TCIA.2017.GJQ7R0EF [DOI] [Google Scholar]
- 28.Baid U, et al. : The RSNA-ASNR-MICCAI brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint arXiv:2107.02314 (2021) [Google Scholar]
- 29.Cox R, et al. : A (sort of) new image data format standard: Nifti-1: We 150. Neuroimage 22 (2004) [Google Scholar]
- 30.Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A: The sri24 multichannel atlas of normal adult human brain structure. Hum. Brain Mapp 31, 798–819 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yushkevich PA, Pluta J, Wang H, Wisse LE, Das S, Wolk D: Fast automatic segmentation of hippocampal subfields and medial temporal lobe subregions in 3 tesla and 7 tesla t2-weighted MRI. Alzheimer’s & Dementia: J. Alzheimer’s Assoc 12(7), P126–P127 (2016) [Google Scholar]
- 32.Joshi S, Davis B, Jomier M, Gerig G: Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage 23, S151–S160 (2004) [DOI] [PubMed] [Google Scholar]
- 33.Yushkevich PA, et al. : User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3), 1116–1128 (2006) [DOI] [PubMed] [Google Scholar]
- 34.Yushkevich PA, et al. : User-guided segmentation of multi-modality medical imaging datasets with ITK-snap. Neuroinformatics 17(1), 83–102 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davatzikos C, et al. : Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging 5(1), 011018 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathore S, et al. : Brain cancer imaging phenomics toolkit (brain-CaPTk): an interactive platform for quantitative analysis of glioblastoma. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 133–145. Springer, Cham: (2018). 10.1007/978-3-319-75238-9_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pati S, et al. : The cancer imaging phenomics toolkit (CaPTk): technical overview. In: Crimi A, Bakas S (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 380–394. Springer, Cham: (2020). 10.1007/978-3-030-46643-5_38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fathi Kazerooni A, Akbari H, Shukla G, Badve C, Rudie JD, Sako C, Rathore S, Bakas S, Pati S, Singh A, et al. , ”Cancer imaging phenomics via captk: multi-institutional prediction of progression-free survival and pattern of recurrence in glioblastoma,” JCO clinical cancer informatics, vol. 4, pp. 234–244, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rathore S, et al. : Multi-institutional noninvasive in vivo characterization of IDH, 1p/19q, and egfrviii in glioma using neuro-cancer imaging phenomics toolkit (neuro-captk). Neuro-oncol. Adv 2(Supplement_4), iv22–iv34 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sled JG, Zijdenbos AP, Evans AC: A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging 17(1), 87–97 (1998) [DOI] [PubMed] [Google Scholar]
- 41.Tustison NJ, et al. : N4itk: improved N3 bias correction. IEEE Trans. Med. Imaging 29(6), 1310–1320 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen CT, Iglesias JE, Van Leemput K: N3 bias field correction explained as a Bayesian modeling method. In: Cardoso MJ, Simpson I, Arbel T, Precup D, Ribbens A (eds.) BAMBI 2014. LNCS, vol. 8677, pp. 1–12. Springer, Cham: (2014). 10.1007/978-3-319-12289-2_1 [DOI] [Google Scholar]
- 43.Iqbal H: Harisiqbal88/plotneuralnet v1.0.0, December 2018 [Google Scholar]
- 44.Ronneberger O, Fischer P, Brox T: U-net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham: (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 45.Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C: The importance of skip connections in biomedical image segmentation. In: Carneiro G, et al. (eds.) LABELS/DLMIA -2016. LNCS, vol. 10008, pp. 179–187. Springer, Cham: (2016). 10.1007/978-3-319-46976-8_19 [DOI] [Google Scholar]
- 46.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016) [Google Scholar]
- 47.Openvino™ toolkit overview (2021). https://docs.openvinotoolkit.org/latest/index.html
- 48.Pytorch extension for openvino™ model optimizer (2021). https://github.com/openvinotoolkit/openvino_contrib/tree/master/modules/mo_pytorch [Google Scholar]
- 49.Cyphers S, et al. : Intel nGraph: an intermediate representation, compiler, and executor for deep learning. arXiv preprint arXiv:1801.08058 (2018) [Google Scholar]
- 50.Wu H, Judd P, Zhang X, Isaev M, Micikevicius P: Integer quantization for deep learning inference: principles and empirical evaluation. CoRR, vol. abs/2004.09602 (2020) [Google Scholar]
- 51.Choukroun Y, Kravchik E, Yang F, Kisilev P: Low-bit quantization of neural networks for efficient inference. In: ICCV Workshops, pp. 3009–3018 (2019) [Google Scholar]
- 52.Quantization algorithms. https://intellabs.github.io/distiller/algo_quantization.html
- 53.Int8 inference (2021). https://oneapi-src.github.io/oneDNN/dev_guide_inference_int8.html
- 54.Tailor SA, Fernandez-Marques J, Lane ND: Degree-quant: quantization-aware training for graph neural networks. arXiv preprint arXiv:2008.05000 (2020) [Google Scholar]
- 55.Fang J, Shafiee A, Abdel-Aziz H, Thorsley D, Georgiadis G, Hassoun JH: Post-training piecewise linear quantization for deep neural networks. In: Vedaldi A, Bischof H, Brox T, Frahm J-M (eds.) ECCV 2020. LNCS, vol. 12347, pp. 69–86. Springer, Cham: (2020). 10.1007/978-3-030-58536-5_5 [DOI] [Google Scholar]
- 56.Openvino™ toolkit accuracyaware method (2021). https://docs.openvinotoolkit.org/latest/workbench_docs_Workbench_DG_Int_8_Quantization.html
- 57.Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC: Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans. Med. Imaging 13(4), 716–724 (1994) [DOI] [PubMed] [Google Scholar]
- 58.Reinke A, et al. : Common limitations of performance metrics in biomedical image analysis. Med. Imaging Deep Learn (2021) [Google Scholar]
- 59.Arafa M, et al. : Cascade lake: next generation intel Xeon scalable processor. IEEE Micro 39(2), 29–36 (2019) [Google Scholar]
- 60.Lower numerical precision deep learning inference and training (2018). https://software.intel.com/content/www/us/en/develop/articles/lower-numerical-precision-deep-learning-inference-and-training.html
- 61.Davatzikos C, et al. : Ai-based prognostic imaging biomarkers for precision neuro-oncology: the respond consortium. Neuro-oncol. 22(6), 886–888 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakas S, et al. : iGlass: imaging integration into the glioma longitudinal analysis consortium. Neuro Oncol. 22(10), 1545–1546 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]