Abstract
The Brassicaceae family includes many economically important crop species, as well as cosmopolitan agricultural weed species. In addition, Arabidopsis thaliana, a member of this family, is used as a molecular model plant species. The genus Brassica is mesopolyploid, and the genus comprises comparatively recently originated tetrapolyploid species. With these characteristics, Brassicas have achieved the commonly accepted status of model organisms for genomic studies. This paper reviews the rapid research progress in the Brassicaceae family from diverse omics studies, including genomics, transcriptomics, epigenomics, and three-dimensional (3D) genomics, with a focus on cultivated crops. The morphological plasticity of Brassicaceae crops is largely due to their highly variable genomes. The origin of several important Brassicaceae crops has been established. Genes or loci domesticated or contributing to important traits are summarized. Epigenetic alterations and 3D structures have been found to play roles in subgenome dominance, either in tetraploid Brassica species or their diploid ancestors. Based on this progress, we propose future directions and prospects for the genomic investigation of Brassicaceae crops.
Introduction
Brassicaceae, often called Cruciferae or the mustard family, comprises 4636 known species in 340 genera [1]. Many of them have been domesticated as important crops for agriculture, ornaments, or condiments, some of which are also of medicinal significance. This family includes species with both ancient and recent polyploidies, as well as species with relatively small genomes, such as the model plant species Arabidopsis thaliana. A key agricultural genus of the Brassicaceae family is the Brassica genus, in which the six most commonly known members are three diploid species, Brassica rapa (A genome, n = 10), Brassica nigra (B genome, n = 8), and Brassica oleracea (C genome, n = 9), and three allotetraploid species, Brassica juncea (AB genome, n = 18), Brassica napus (AC genome, n = 19), and Brassica carinata (BC genome, n = 17). The genomic relationships of these six representative members have been defined as the ‘Triangle of U’.
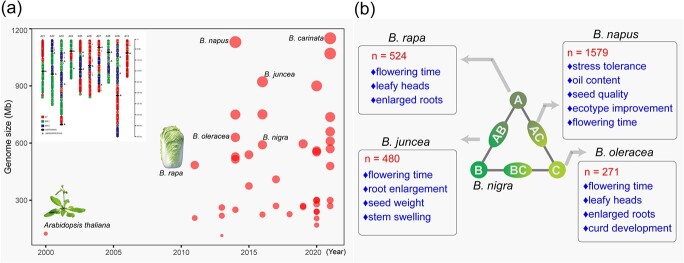
With the rapid advances in sequencing technology, more and more Brassicaceae species have been sequenced and assembled into high-quality reference genomes. Using the latest statistics from 2022, 43 species in Brassicaceae have been sequenced (https://plabipd.de/timeline_view.ep) (Fig. 1a, Table S1, see online supplementary material). After the genome assembly of the first Brassicaceae species, A. thaliana, it took more than 10 years for the second one, B. rapa [2], to be sequenced in 2011. The release of the B. rapa genome sequence not only warrants further analysis of gene functions within B. rapa, but also provides an important reference for the study of the polyploidization of Brassicaceae species and the evolution of members of the ‘Triangle of U’. After this, it took another 10 years for all the other members of the ‘Triangle of U’ to be sequenced [3–8]. In the last five years, a quickly increasing number of genomes of species from Brassicaceae have been decoded. Some representative species, e.g. B. napus, B. oleracea, and B. rapa, have obtained high-quality reference genomes following multiple rounds of genome upgrades [9–12], while some less popular species have also recently been decoded [13–24] (Table S1, see online supplementary material).
Figure 1.
Overview of genomic studies in Brassicaceae. Studies on the whole-genome sequencing of Brassicaceae species. a Non-redundant statistics of sequenced species in Brassicaceae. The red dot represents each sequenced species, varying in size according to the genome size. The details of these sequenced species are provided in Table S1 (see online supplementary material). The image in the upper-left corner shows the composition of three subgenomes in mesopolyploid Brassica rapa [10]. b Studies on important agronomic traits in Brassica crops using population-scale resequencing strategies. n, total number of resequenced accessions within each species.
The release of these reference genomes has bolstered the exploration of genomic variation, modification, and regulation in Brassicaceae species. During the past five years, the major Brassica crops have been resequenced, scaling up to populations of several hundred accessions. Moreover, investigations using whole-genome bisulfite sequencing (WGBS), high-throughput chromatin conformation capture (Hi-C), chromatin immunoprecipitation assays with sequencing (ChIP-seq), Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), and DNA Affinity Purification and sequencing (DAP-seq) have been reported for Brassicaceae plants. These have significantly improved our cognizing of the genome modification and regulation of gene expression. Recently, de novo sequencing of multiple accessions of several Brassica crops has brought investigations of genomic variation into the pan-genomics era, and RNA sequencing (RNA-seq) has evolved to explore single-cell and spatial transcriptomes. Here, we summarize the recent progress of genomics investigations of Brassicaceae species, with a focus on their application in cultivated crops.
Variation in Brassicaceae genomes
A. thaliana has been taken as a model organism for basic plant research. Intraspecific genome variation study on a large scale started at the beginning of 2008 by launching the 1001 Genomes Project as a pioneer for plant species-wide diversity research [25]. The aim of the project was to describe detailed whole-genome sequence variation in at least 1001 accessions of A thaliana. A variation map derived from resequencing of 1135 A. thaliana accessions was published in 2016 [26]. As important cultivated crops, Brassica and Raphanus species have received significant attention in the characterization of genomic variations. With the exception of B. carinata, all Brassica crops have been subjected to large-scale genome resequencing in different laboratories [27–31], while pan-genomes have been constructed for B. napus, B. oleracea, B. rapa, and Raphanus [11, 27, 32, 33]. The in-depth analysis of whole-genome variation data has provided not only the genomic features explaining the domestication of extreme morphotypes but has also identified specific genes and variants contributing to important agronomic traits.
Genomic variation revealed by population-scale resequencing in Brassica
Due to multiple rounds of genome duplication, Brassica species show unique genomic variations. In the past five years, hundreds of accessions within each of the Brassica crops have been resequenced (Fig. 1b). The genomes of the three diploid members of the ‘Triangle of U’ were all derived from an extra whole-genome triplication (WGT) of the tPCK ancestors, which led to three subgenomes, namely, Least Fractionated (LF), More Fractionated (MF1), and Most Fractionated (MF2). While LF is the least variable subgenome, the WGT increased the genomic variation contributing to the diversified morphotypes in Brassica species [34]. During the intraspecific diversification of the A and C genomes, it was found that the gene repertoire, transposable element (TE) content, and the number of variations varied greatly among individuals [27, 33]. This phenomenon has also been observed in the allotetraploid species B. napus (AACC) and B. juncea (AABB) [29, 31]. Investigations in both B. napus and B. juncea established the same conclusion: A subgenomes in both allotetraploid species have a higher frequency of genetic recombination and maintain higher nucleotide diversity than either the C or B subgenomes. However, the mechanism is yet to be clarified.
Brassica crops have been domesticated to a number of extreme morphotypes, such as the leafy heads in Chinese cabbage (B. rapa ssp. pekinensis) and cabbage (B. oleracea var. capitata); enlarged roots in turnip (B. rapa ssp. rapa), rutabaga (B. napus var. rapifera), and root mustard (B. juncea ssp. napiformis); enlarged stems in kohlrabi (B. oleracea var. gongyloides) and tuber mustard (B. juncea var. tumida); and thickened inflorescences in cauliflower (B. oleracea var. botrytis) and broccoli (B. oleracea var. italica). In 2016, the domestication of leafy-head and tuber-forming Brassicas was investigated by resequencing 199 B. rapa accessions and 119 B. oleracea accessions [28]. In that study, not only selection sweeps of the domestication of leafy heads and root/stem enlargement were identified but also candidate genes for these traits were pinpointed. Moreover, homoeologous genes were selected in parallel in both species during trait domestication for leafy head and root/stem enlargement. Cai et al. genotyped structure variations (SVs) across 524 diverse B. rapa accessions and found that four SV-containing genes (BrPIN3.3, BrMYB95.3, BrFL5.1, and BrSAL4.2) might be involved in the formation of leafy heads [27]. Very recently, Sun et al. identified two genes with chloroplast-related functions that are responsible for the yellow leaves traits using phenotype screening and resequencing of a large-scale ethyl methane sulfonate (EMS) mutant population [35]. This study demonstrated that the strategy of combination phenotypic and genotypic screening is powerful in the detection of candidate genes for target traits.
By analysing selection signatures for root mustard in B. juncea, 14 candidate genes were identified as being involved in the storage roots formation. These genes include CDC48A4 and EXP1 genes and genes involved in cell division, cell expansion, and the regulation of auxin signaling. This research also highlighted the subgenomic prevalence of selective sweeps in the Aj subgenome (A subgenome in B. juncea) over the Bj subgenome (B subgenome in B. juncea) [29]. Cauliflower curd composes of thousands of inflorescence meristems with floral arrested. By analysing resequencing data of 104 accessions of cauliflower and 167 accessions of the other morphotypes of B. oleracea, Guo et al. detected dozens of selected SVs and associated genes that are potentially involved in the curd formation and enlargement [36]. Additionally, variants associated with seed yield, oil content, erucic acid, and glucosinolates in seeds, as well as seed weight, were also revealed in oil Brassica crops (Fig. 1b).
Genome-wide association analysis (GWAS) is a widely used approach for exploring the sequence variants associated with complex traits in crops. The Brassica 60 k SNP array has been widely used to genotype B. napus natural populations in GWAS research [37–40]. Very recently, based on large-scale genome resequencing of 403 diverse rapeseed accessions, Hu et al. traced the genomic basis of agronomic traits during modern rapeseed breeding. A total of 628 causative candidate genes were identified for 56 agronomic traits in B. napus by GWAS [41]. Although GWAS is powerful in genome-widely detecting of genetic variations associated with target traits, the resolution of GWAS is generally not sufficient to directly determine the causal genes. Therefore, the combination of GWAS and transcriptome-wide association studies (TWAS) provides more powerful approaches for genetic dissection of target traits. Similarly, Tang et al. developed a gene prioritization framework based on multi-omics data and information from A. thaliana to prioritize the causal gene BnPMT6 for seed oil content in B. napus [42].
Flowering time, as one of the most important agronomic traits, has been a specific focus in investigations of trait domestication or GWAS analyses in Brassica crops. Su et al. (2018) resequenced 194 Chinese cabbage accessions representing spring, summer, and autumn ecotypes [43]. They identified that the sequence variations in the cis elements of the BrVIN3.1 promoter contribute to varied vernalization responses in different ecotypes of Chinese cabbage [43]. Wu et al. resequenced 991 B. napus accessions and found that single nucleotide polymorphisms (SNPs) in the promoter regions of FT and FLC orthologs specifically in line with the three rapeseed ecotype groups [31]. Similarly, in B. rapa, a nonsynonymous mutation at the 58th nucleotide of exon1 and a splicing site mutation in intron 6 of BrFLC1 contributed to flowering time variations [44, 45]. In B. juncea, two SNPs in SRR1 and five SNPs in VIN3 were identified as being closely associated with flowering time [29] using population-scale resequencing strategies.
Pan-genomes are the new references for mining genomic variations
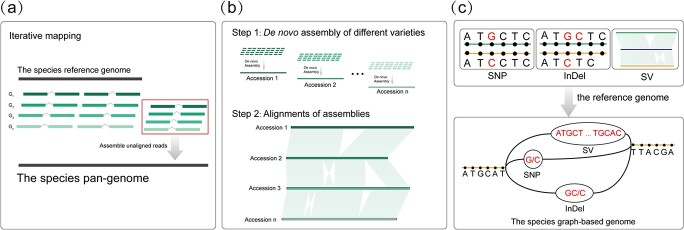
The pan-genomes of important Brassicaceae crops have been constructed using three popular approaches. The first involves aligning reads from a sequenced accession onto the reference genome and then assembling the unaligned reads into novel contigs (Fig. 2a). This ‘map-to-pan’ strategy was employed in constructing the B. oleracea pan-genome [33]. The second is de novo assembly of the genomes of diverse varieties, while the third is to construct a species graph-based genome (Fig. 2b). The pan-genome of A. thaliana [46] was constructed using the second strategy, while the pan-genomes of Raphanus [32] and B. rapa [27] were de-novo assembled using diverse varieties and a graph-based genome strategy (Fig. 2c). The iterative assembly for constructing the pan-genome is cost-effective, as an iterative assembly fills up gene sequences that are absent in the reference genome, and the accessions are sequenced on a low-cost short-read sequencing platform. However, a pan-genome constructed entirely using short reads largely limits the exploration of complex structural variations. In recent years, the development of long-read sequencing and graph-based genome strategies has resolved this limitation, and species are moving toward population-scale long-read sequencing [47, 48].
Figure 2.
Strategies commonly used for mining genomic variations in the pan-genome era. a Construction of a pan-genome using an iterative mapping approach. b Construction of a pan-genome by assembling the genomes of diverse varieties. c Construction of a pan-genome by integrating genomic variations and the reference genome.
Pan-genome analysis can reveal hidden genomic variations. As early as 2016, the B. oleracea pan-genome was published by assembling short reads [33]. It was found that 18.7% of genes in the pan-genome were composed of dispensable genes, including genes related to major agronomic traits, such as disease resistance, flowering time, etc. [33]. Long-read sequencing technologies were employed for constructing the pan-genomes of A. thaliana [46], B. rapa [27], and Raphanus [32], revealing more hidden genomic variations in these three species. For example, the A. thaliana pan-genome constructed by assembling seven A. thaliana accessions revealed that ~1900 genes were absent from the reference genome [46]. Zhang et al. (2021) constructed a pan-genome of Raphanus that included 11 accessions from domesticated, wild, and weedy radishes [32]. In the Raphanus pan-genome, the number of SVs is ~26 × 103 per sample, which is similar to the number in soybean pan-genome [32]. However, the size of the radish genome is only half of that of the soybean genome, and the SV density of Raphanus is twice that of the soybean genome. Cai et al. (2021) constructed a pan-genome consisting of 18 B. rapa accessions from six morphotypes. It revealed that each genome contains 15.14%–37.39% of sequences that were not syntenic with the Chiifu reference genome. A total of 33.24–56.7 Mb insertions and 35.75–58.84 Mb deletions (size ≥50 bp) were detected in the B. rapa pan-genome. Further analysis indicated that SV highly associated to the morphotype domestication in B. rapa [27]. Transposable elements (TEs) are major components of eukaryotic genomes. Recently, Cai et al. (2022) developed a novel pipeline to detect TE insertion polymorphisms (TIPs) on a population scale via combination of the B. rapa pan-genome and resequencing data from 524 B. rapa accessions and revealed that the TIPs in TIP-containing genes had been selected more strongly than non-synonymous SNPs [49]. In summary, the graph-based pan-genomes of Brassicaceae species will serve as a useful reference for GWAS or domestication analysis at the SV level, providing significant advantages over SNP-based analysis using a single genome as the reference.
In addition, Brassica genus-wide pan-genomes were established. The first and foremost advantage of the genus-wide pan-genomes is facilitating gene content description under a framework using systematic nomenclature proposed by the Multinational Brassica Genome Project [50]. One of the unresolved questions in the evolution of Brassica genomes is the mechanism by which LF subgenome exhibits less gene loss (fractionation) than the MF1 and MF2 subgenomes. It has been proposed that Brassica mesohexaploidy had occurred via a two-step process, according to the gene density in the three subgenomes of B. rapa [2]. The different methylation levels between the LF and MF subgenomes further supported this hypothesis [51]. The genus-wide pan-genomes for Brassica enabled updating Ancestral Crucifer karyotype (ACK) block organization, and provided further evidence of two-step pathways in the Brassica genome evolution.
Homoeologous exchanges as important resources for generating novelties in brassica allopolyploids
Homoeologous exchanges (HEs) specifically describe the exchanges of chromosome segments in allopolyploids, which occur from crossover formation between homoeologous genomes [52, 53]. They have been suggested as important resources for increasing genomic variations and generating new phenotypes to be selected for domestication in allopolyploid species. Homoeologous exchanges can result in one of the homoeologous DNA fragments becoming fixed over the other, consequently leading to copy number variation (CNV) or presence-absence variation (PAV) of the genes [54, 55]. HEs have been reported as prevalent events in many allopolyploid species, including rapeseed [3], peanut [56], bread wheat [57], polyploid rice [58], and other wild species [59]. Among these species, many in-depth studies regarding HE mechanisms and impacts on phenotypes have been conducted on B. napus. The HE phenomenon was first reported in synthetic lines, which extended back to at least 1995 [60], and later, the association of HE with flowering time diversification after several generations was also established [52, 61–64]. Recently, owing to the rapid development of the genomic era, many studies have been conducted at a large population scale and have discovered that HEs have a much higher frequency in different domesticated varieties than expected [64–66]. Additionally, HEs have been identified as the major cause of gene PAV in different populations [54], and their impacts on gene expressional changes were demonstrated to be proportional to the gene copy number changes [53, 54]. The genes affected by HEs have also been shown to be responsible for many important trait diversities in Brassica polyploids, including flowering time [55, 67], leaf morphology [63], seed glucosinolates content [64], and disease resistance [54], suggesting the potential role of HEs in generating phenotypic novelties for domestication.
Some important genes responsible for HE have been reported in B. napus. Gonzalo et al. showed that reducing MSH4 copy number prevents meiotic crossovers between non-homologous chromosomes in B. napus [68]. Recently, by applying both quantitative trait locus (QTL) mapping and cytogenetic analysis in a resynthesized segregating B. napus population, Higgins et al. (2021) successfully identified several important quantitative loci for controlling HEs. The major locus BnaPh1 on chromosome A9 contributed 32–58% observed variation of homoeologous recombination, and the genes found in the locus would facilitate the identification of the causal and new genes for controlling successful meiotic adaptation in polyploids [69]. Compared to the artificially synthesized B. napus, natural B. napus plants have much lower frequency of HEs. Reports on genes that reduce HE in B. napus suggest that natural allopolyploid plants have undergone selection to reduce the frequency of HE. Further studies on HEs and their mechanisms could ultimately offer new breakthroughs for improving diversity in polyploid crops [70, 71].
Domestication of Brassica crops revealed by genomics analysis
Brassica comprises about 35 species of mainly annual herbs, with some perennial herbs and small shrubs. Cultivated Brassicas are not only used for different purposes, such as fresh and preserved vegetables, vegetable oils, and condiments, but are also cultivated worldwide in different climatic conditions. Moreover, Brassica species possess high genome plasticity, allowing for the domestication of crops with extremely high morphological variability. Similar traits, such as the traits of enlarged root/stem and leafy head, have been domesticated in different Brassica species in parallel. All of these features render Brassica an ideal system for investigating crop domestication and artificial selection.
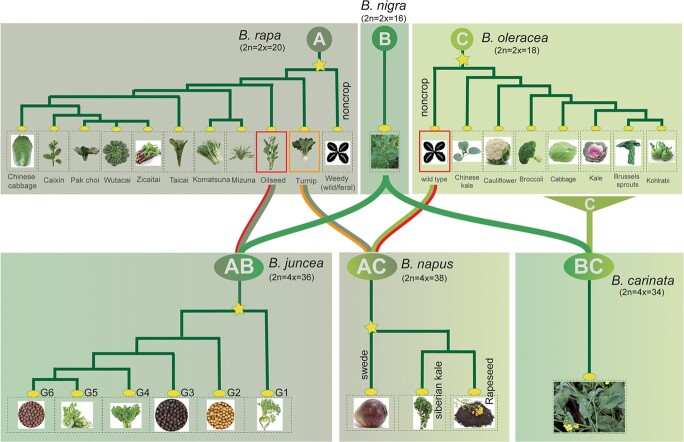
It is widely accepted that turnip was the first domesticated B. rapa crop type [28, 72]. Previously, Song et al. [73] proposed that Europe was the primary center of domestication for turnip and turnip rape, while China was the second center where various Asian vegetable crops, including Chinese cabbage, pak choi, wutacai, mizuna, and komatsuna, were domesticated (Fig. 3). Recently, by analysing the evolution of BrFLC1 in diverse B. rapa crops, it was found that the ancient crops (e.g. turnip, turnip rape) were skewed toward carrying the haplotype of late flowering; the crops that differentiated in China were biased toward carrying the early flowering haplotype, supporting two independent centers of origin of B. rapa [44]. B. oleracea has been domesticated into even more extreme morphotypes (Fig. 3). Previously, various origins of cultivated B. oleracea have been proposed, including a single origin from wild B. oleracea in western Europe, as well as triple and even multiple origins involving related wild species [74]. Recently, Mabry et al. proposed that the Aegean endemic B. cretica is the closest living relative of cultivated B. oleracea, supporting a single origin of cultivation in the Eastern Mediterranean region [75]. Leaf heading and enlarged root/stem traits were domesticated respectively in B. rapa and B. oleracea in parallel. One study proved that parallel selection at the subgenome level played a critical role in the domestication of parallel traits in these two different species [28].
Figure 3.
Crop domestication in Brassica. ‘A’, ‘B’, ‘C’, ‘AB’, ‘AC’, and ‘BC’ represent the genomes of Brassica rapa, B. nigra, B. oleracea, B. juncea, B. napus, and B. carinata, respectively. The phylogenetic relationships of Brassica crops were based on information published previously by McAlvay et al. [72], Cai et al. [27], Cheng et al. [28], Lu et al. [30], Yang et al. [5], and Kang et al. [29]. The stars represent the boom of morphotypes in target species. Background colors of A, B, and C genomes are olive green, green, and grass green, respectively; the background colors of AB, AC, and BC genomes are the gradient background colors representing their progenitors.
Based on the ‘Triangle of U’ model, the pairwise hybridizations of B. rapa and B. nigra formed B. juncea, of B. rapa and B. oleracea formed B. napus, and of B. nigra and B. oleracea formed B. carinata. Establishing the comparative genomics platform for the species of the ‘Triangle of U’ allowed the examination of the dynamics of polyploid evolution and the impact of subgenome dominance in domestication and agronomical improvement. However, as each of the ancestor species possesses diverged morphotypes, the most direct ancestor morphotype has yet to be defined for each of the allotetraploid species. A recent genomics investigation supported that the Aj subgenome (A subgenome in B. juncea) was derived from B. rapa ssp. tricolaris [29], and the An subgenome (A subgenome in B. napus) was derived from European turnip (Fig. 3) [5]. This implicates that B. juncea and B. napus evolved from independent geographical origins, as their progenitor B. rapa ssp. tricolaris was distributed in Asia, while European turnip was distributed in Europe. As for the Cn subgenome (C subgenome in B. napus), Lu et al. proposed that it is derived from the common ancestor of kohlrabi, cauliflower, broccoli, and Chinese kale [30]. By analysing 480 B. juncea accessions collected from 38 countries, Kang et al. proposed that West Asia is most likely to be the single origin of B. juncea. Subsequently, various B. juncea vegetable and oil crops formed through spontaneous gene mutations and introgressions along three independent routes of eastward expansion [29]. It was proposed that B. napus was formed ~7500 years ago [3] via natural interspecific hybridization of European turnip and wild B. oleracea (Fig. 3). Subsequently, the three ecotypes of winter, spring, and semi-winter formed. It was reported that winter B. napus was first domesticated in Europe [76]. Spring B. napus was developed in Europe and spread to England [77], and the semi-winter ecotypes were mainly cultivated in China via introduction from Europe [30, 78]. In the case of B. carinata, the relationship between the Bc and Cc subgenomes is greater than that between other two tetraploid subgenomes (Bj and Cn) and their respective diploid parents [6].
Epigenetics and 3D structure of Brassicaceae genomes
Over the past decades, plant linear genomes and epigenetic modifications have been studied extensively. More recently, three-dimensional (3D) genome structures in plants began to be rapidly unveiled. Accumulating evidence has revealed that not only epigenetic modification in the linear DNA sequence but also 3D genome architecture play an important role in determining genome organization, genome functionality, and gene expression regulation [79]. With abundant genetic resources and updated reference genome sequences in Brassicaceae plants, advances in sequencing technology and multidisciplinary methods facilitated the study epigenetic regulation ad 3D genome architecture and their relationships with genome complexity and subgenome dominance in Brassicaceae.
Epigenetics of Brassicaceae genomes
Epigenetics is the study of any potentially stable and heritable change in gene expression or cellular phenotype that does not involve the changes in DNA sequence [80]. The underlying mechanisms of epigenetic regulation are mediated by DNA methylation, histone modification, non-coding RNA, etc. In the 1001 Epigenomes Project of A. thaliana, 1107 high-quality single-base resolution methylomes and 1203 transcriptomes of A. thaliana have been established [81]. It revealed that geographic origin is highly related to genome-wide DNA methylation levels and altered gene expression caused by epialleles, although the genetic basis of methylation variation is highly complex [81].
The Brassica genus represents a fascinating model for epigenetic studies because of its unique genomic variations [2, 51]. To date, many epigenetic studies on Brassica have been conducted not only at the subgenome level but also across duplicated gene copies. These studies suggest that epigenetic modifications may be the determinants of the subgenome dominance and functional diversification of duplicated genes. In diploid Brassica, global analysis of the DNA methylation profile showed that the DNA methylation level was similar or higher in B. rapa compared to that in B. oleracea but was much higher than that of A. thaliana, probably resulting from the difference in genome structure, such as the difference in the amount and distribution of TEs and repeat sequences [82, 83]. The three subgenomes of B. oleracea show imbalanced DNA methylation, with the dominant LF subgenome exhibiting the lowest levels of DNA methylation. Moreover, the triplicated gene copies appear to have independent DNA methylation patterns, and the non-syntenic genes have significantly enhanced DNA methylation [84]. In B. rapa, the single-copy retained genes were found to have significantly higher DNA methylation compared to those of genes retained in pairs or triplets [83]. Generally, the gene expression level is negatively associated with DNA methylation. These results suggest that DNA methylation variations in Brassica play a role in subgenome dominance and biased gene retention and the expressional diversification of duplicated genes [84].
Genome-wide profiling of histone H3 lysine methylation in B. rapa found that H3K4me3 and H3K36me3 are enriched in the transcription start sites [85]. Genes with H3K4me3 and H3K36me3 marks have higher expression levels but a low degree of tissue specificity [85]. In contrast, H3K27me3 marks are correlated with decreased or low gene expression or high tissue-specific gene expression [86, 87]. Furthermore, the distribution of H3K36me3 and H3K27me3 vary between homoeologous paired genes, which result in their variations in gene expression levels or tissue specificity, and eventually result in their sub-functionalization [85–87].
Comparative analysis of TE and 24-nt small RNA in B. rapa showed that the distribution of TEs is imbalanced among subgenomes, which is also reflected between the flanking regions of homoeologous gene pairs. These findings suggest that the biased distribution of TEs and the targeting of 24-nt small RNAs both involved in the dominant expression phenomenon at a subgenome scale or among the homoeologous gene copies [88, 89].
In contrast to diploid Brassica, epigenetic alterations are thought to be involved in polyploidization events of allotetraploid Brassica. For example, in the B. napus genome, epigenetic modifications are imbalanced not only between the An and Cn subgenomes but also between homoeologous gene pairs. The Cn subgenome has a higher methylation level than that of the An subgenome, possibly resulting from higher TEs density in the Cn subgenome [3]. Comparative analysis among B. rapa, B. oleracea, and B. napus showed that histone H3 methylation and DNA methylation differ among these three Brassica species, which might be attributed mainly to differences in genome structure rather than ploidy level [90]. A recent study in B. napus found that the An subgenome has a higher level of active epigenetic marks and a lower level of inhibitory epigenetic marks compared with the Cn subgenome. Meanwhile, the distributions of histone modifications between homoeologous gene pairs reflect their biased expression patterns [91].
In addition, some studies on Brassica found that epigenetic modification such as DNA methylation played roles in genomic stability. It has been revealed that synthetic allotetraploid AACC undergoes higher DNA methylation changes than its diploid parents. This status might be highly correlated with the genomic instability of newly synthesized allotetraploid [92]. Comparative analysis of natural and synthetic B. napus detected that the most obvious difference in DNA methylation patterns was CHG methylation levels, which were significantly lower in synthetic rapeseed than those in natural B. napus [93]. Moreover, the genes related to DNA repair and nucleotide metabolism display differential expression patterns and CHG methylation levels between natural and synthetic B. napus, thereby suggesting that the genomic instability of newly synthesized allotetraploid plants is associated with DNA methylation changes and the disruption of the DNA repair system [93].
3D structure of Brassicaceae genomes
Over the past decade, the development of chromatin conformation capture (3C)-based technologies, especially Hi-C, has enabled the exploration of the hierarchical 3D structure of Brassicaceae genomes.
As reported, several types of 3D chromosomal organization, such as compartments A/B, topologically associating domains (TADs), KNOT structure, and chromatin loops, shape Brassicaceae genomes and play roles in gene expression regulation and genome function. Global analysis of high-order chromatin organization revealed that the chromatin regions of Brassicaceae could be partitioned into two compartments, namely A and B. In Arabidopsis and Brassica, the A compartment mainly overlaps with active euchromatic regions, while the B compartment mainly constitutes heterochromatin regions [94, 95]. Moreover, epigenetic modifications have been correlated with the compartmentalization of Brassicaceae genomes. For example, H3K4me2 is a typical marker euchromatin, while the distribution of H3K9me2,3 in euchromatin or heterochromatin seems to be species-specific [90]. TADs have not been detected in A. thaliana [94, 96, 97]. However, TADs are the most prominent feature and conserved between B. rapa and B. oleracea [95]. TAD boundaries were found to be significantly enriched in active epigenetic marks and highly transcribed genes in B. rapa and B. oleracea [95]. KNOT is the most intriguing 3D structure in the A. thaliana genome and has been reported to be enriched with TEs and involved in strong long-range interactions [94, 96, 97]. It was observed that Knot Engaged Elements (KEEs) or Interactive Heterochromatic Islands (IHIs) involved in KNOT structure greatly expanded in B. rapa but contracted in B. oleracea [94–96, 98]. Chromatin loops have been widely detected in plant genomes, which represent long- and short-range interactions. In Arabidopsis, a strong chromatin loop formation was observed between the 5′ promoter and downstream of 3′ end of the FLC locus, and this chromatin loop of FLC is disrupted after cold exposure, suggesting that the chromatin loops may be involved in the transcription of the FLC gene [99].
Emerging evidence suggests that 3D structure also plays significant roles in gene expression regulation, biased gene retention, and subgenome dominance in Brassicaceae genomes. Characterizing the nuclear organization of B. rapa and B. oleracea, Xie et al. observed that homoeologs retained on the dominant subgenome (LF) exhibited significantly stronger interaction strength than that of submissive subgenomes (MF1 and MF2) [95], which was consistent with the subgenome dominance phenomenon [51]. In addition, they also found that homoeologs retained in doublets or triplets are more likely to physically interact [95]. Furthermore, the interacting homoeolog pairs exhibited significantly higher similarity in epigenetic modifications and Gene Ontology patterns than those non-interacting homoeolog pairs in both B. rapa and B. oleracea [95]. These results suggested that the chromatin interactions of retained homoeologs are correlated with their biased retention and subgenome dominance and tend to be co-regulated with highly similar epigenetic modifications in Brassica [95].
It is worthwhile noting that most of the epigenetic datasets or the 3D structure in Brassicaceae were acquired from pooled tissues and only represent the average patterns of various cell types, which may ignore their dynamic changes in different cell types, leading to inaccurate results. Therefore, high-resolution single-cell strategies for epigenomic profiling or 3D structure capturing are needed to provide more specific and accurate data in Brassicaceae genomes. In addition, future studies should address the epigenomic or 3D genome structure and its functional roles in plant growth and development.
Exploring gene expression at different levels
To explore gene expression (especially for analysing differential gene expression) at the genome scale, RNA-seq is a popular tool that is shaping our understanding of genomic function [100]. RNA-seq can capture transcriptomic dynamics during developmental stages and physiological changes under different conditions [101]. Meanwhile, RNA-seq has been driven by the development of technologies in specific niches [100]. The analysis of gene expression from traditional RNA-seq (referred to as bulk RNA-seq here) reveals the average expression of genes from bulk tissues and/or cells. Single-cell transcriptome enables the determination of gene expression at single-cell level. The spatial transcriptome may record spatial information, enabling the investigation of tissue architecture. However, some limitations in these methods should be considered. Bulk RNA-seq cannot resolve the expression of genes from special cell types, and both bulk RNA-seq and single-cell transcriptome lose the spatial contents of expressed genes. Spatial transcriptome analysis may become the routine toolkit in the future.
In the past few years, bulk RNA-seq has been widely used for studies in Brassicaceae plants. Typically, bulk RNA-seq is used to explore expression characteristics of genes at different developmental stages or under different treatments [102–109]. To explore the genetics and evolution of polyploid crops, different methods have been developed for bulk RNA-seq analysis. The subgenome dominance in B. oleracea was revealed by transcriptome and methylome profiling [109]. A high-density SNP linkage map and associated transcriptomics have been developed to investigate the genetic complexity in B. napus [110–111]. The GWAS and RNA-seq analyses of 505 inbred lines identifies hundreds of genes associated with seed oil content of B. napus and experiments of the homologous gene pair of BnPMT6s demonstrates that they negatively regulate seed oil content [42]. Meanwhile, homoeologous exchanges in allopolyploid genomes (AACC and AABB) were investigated by mRNAseq-based visualization [64]. Moreover, to reveal the rule of evolution in plant, a study has been conducted to explore the origin and diversification of B. napus using the comprehensive RNA-seq and organellar data [112].
To increase the resolution of gene expression profiling of specific tissue, laser capture microdissection (LCM) technology has been applied to isolate cell populations for RT-PCR or bulk sequencing [113, 114]. The study of papilla cell-expressed genes from A. thaliana, Arabidopsis halleri, and B. rapa by LCM coupled with RNA-sequencing (LCM-seq) identified specific genes involved in plant reproduction and development [115]. In B. napus, the distinct transcriptional feature expression of genes among the epidermis, cortex, and vasculature cells of the funiculus organ were analysed using LCM-seq, which revealed the coordination of these tissue systems to support seed development [116]. The comparative analysis of radish root-tissue- and stage-specific transcriptomes that were generated by LCM-seq and the previously reported transcriptomic data of Arabidopsis roots identified the evolutionary conserved stress-response gene-regulatory network; the network analysis identified that ERF-1 may be the novel key regulator of cambial activities [117]. LCM-seq enabled gene expression analysis with a higher spatial resolution than bulk RNA-seq. The drawback of LCM-seq is that a more limited number of cells can be analysed [113], and the difference between individual cells may remain unresolved after obtaining the average of gene expression profiles from cells [118].
The successful application of high-throughput single-cell transcriptome technologies in plant science in recent years has facilitated the discovery of the cell atlas at the single-cell level, providing new insights into cell heterogeneity and cellular function and helping us to understand the fundamental aspects of plant life [118–122]. These technologies are mainly classified into two types, namely single-cell RNA sequencing (scRNA-seq) and single-nucleus RNA sequencing (snRNA-seq). The droplet-based scRNA-seq approach includes three systems, namely inDrop, Drop-seq, and 10X Genomic Chromium [123]. The construction of scRNA-seq and snRNA-seq libraries relies on the isolation of protoplasts and nuclei, respectively. The major steps of data analysis include quality control, normalization, dimensionality reduction, cell type identification, and visualization [120–126]. The analytical results from both scRNA-seq and snRNA-seq offer opportunities to investigate plant cell identity and the function of tissues and organs [120].
For different tissues/organs of A. thaliana, including the roots [125–137], seedlings [138], the vegetative shoot apex [139], leaves [140–142], flowers [138], and seeds [143], the expression of genes at the single-cell resolution has been reported. These complex tissues/organs consist of diverse cell types. In A. thaliana, expressed genes from nine, eight, seven, ten, and five major cell types were identified in the roots, vegetative shoot apex, leaves, flowers, and seeds, respectively (Table 1). Some cell types can be further divided into several sub-cell types, such as four sub-cell types (pericycle, procambium, phloem, and xylem) for stele cells in the roots (Table 1). For each cell type, single-cell transcriptomic analysis provides cell type-specific expression genes, known as marker genes, for cell type discovery. Typically, marker genes are expressed differently among cell types but do not demonstrate an absolutely cell type-specific expression [119]. For each cell type, trajectory methods provide the possibility of refining cell-type identification and cell developmental transitions, such as the detection of protoxylem and metaxylem for xylem cell lineages [133]. Meanwhile, single-cell analysis provides an opportunity for inferring transcriptional factor regulatory networks, thereby elucidating the genetic coordination among cells [127–140].
Table 1.
Discovery of cell types in Arabidopsis thaliana by high-throughput single-cell transcriptome analysis.
| Tissues/Organs | Major cell types | Sub-cell types |
|---|---|---|
| Roots | (1) root cap cell, (2) trichoblasts (i.e. root hair cell), (3) atrichoblast (i.e. non-hair cell), (4) columella, (5) cortex, (6) endodermis, (7) stele cell, (8) quiescent center, (9) meristematic cell | (7) stele cell: (7.1) pericycle, (7.2) procambium, (7.3) phloem: (7.3.1) phloem procambium, (7.3.2) sieve element, (7.3.3) companion cell. (7.4) xylem: (7.4.1) protoxylem, (7.4.2) metaxylem |
| Vegetative shoot apex | (1) mesophyll cell, (2) shoot meristematic cell, (3) epidermal cell, (4) proliferating cell, (5) vascular cell, (6) guard cell, (7) companion cell, (8) shoot endodermis | — |
| Leaves | (1) mesophyll cell, (2) epidermis, (3) guard cell, (4) hydathode, (5) vascular cell, (6) meristemoid cell, (7) pavement cell | (5) vascular cell: (5.1) bundle sheath, (5.2) xylem, (5.3) phloem, (5.4) procambium, (5.5) companion cell |
| Flowers | (1) meristem, (2) anther, (3) perianth, (4) internode, (5) vasculature, (6) epidermis, (7) carpel, (8) mesophyll, (9) inflorescence axis, (10) stigma | — |
| Seeds | (1) peripheral endosperm (2) micropylar endosperm, (3) chalazal endosperm, (4) embryo proper, (5l) seed coat | (5) seed coat: (5.1) chalazal seed coat, (5.2) general seed coat |
Studies on A. thaliana at the single-cell level provide opportunities and challenges for future studies on other species in Brassicaceae. The reported pipelines can be adopted for performing single-cell transcriptomic analysis. The reported marker genes in A. thaliana could be used to annotate cell types in other species by inferring the expression of their orthologs. However, several challenges remain, which are as follows: (i) orthologs of A. thaliana marker genes may not be conserved in other species [144], which may require experiments such as RNA in situ hybridization to confirm the discovery of cell types; (ii) the novel or rare cell types that exist in other species may be difficult to uncover, as no suggestion can be inferred from the previously reported cells; and (iii) the prior knowledge of non-model plants may be limited.
For complex tissues/organs in plants, their architecture is linked to biological function. Although a single-cell transcriptome provides the expression of genes at the single-cell level, the precise locations (i.e. the spatial contents) of the cells or tissues are lost. The development of spatial transcriptome technology enables the exploration of tissue architecture [145]. The single-cell spatial transcriptome of A. thaliana leaves can identify upper and lower epidermal cells, as well as the spatial developmental trajectories of vascular cells and guard cells [146], which provides opportunities to identify cells with limited knowledge of marker genes and to explore cell interactions and communications.
Integration of Brassicaceae genomic information
With the enormous amount of sequencing data becoming publicly available in the post-genomics era, the development of tools for the efficient use of these sequencing data has become a pertinent research direction, with databases remaining one of the best tools. In recent years, in addition to the well-known Arabidopsis Information Resource, TAIR [147], many excellent databases have been developed for non-model Brassicaceae species, such as the Brassica Database (BRAD) [148], B. napus Pan-genome Information Resources (BnPIR) [149], gene expression database for Brassica crops (BrassicaEDB) [150], and Genomic Variation Database of B. napus (BnaGVD) [151].
The Brassicaceae database BRAD contains 36 reference genomes from 26 species [148]. It generates a table of syntenic genes of all genomes based on A. thaliana and B. rapa that is made available to the user. New features have been added such as the phylogenetic tree and sequence alignment of syntenic genes, homology comparison between two genomic fragments, primer design, retrieval of variant loci, and genomic sequence retrieval.
The developing knowledge system for Brassicaceae BrassiBase [152] includes cross-referenced information on the accurate enumeration of all species, genera, and tribes, chromosome numbers, genome sizes, morphological characteristics, and biological traits. The B. napus pan-genome information resource BnPIR [149] is a comprehensive database constructed based on the B. napus pan-genome (eight reference genomes) and 1688 rapeseed resequencing data. It was also the first pan-genome database for Brassicaceae crops, with GBrowse synteny and the pan-genome browser as key tools for using pan-genome data. The gene expression database for Brassica crops BrassicaEDB [150], a database focusing on gene expression in Brassica, specifically provides transcriptomic data and expression information for genes of B. napus. The genomic variation database of B. napus BnaGVD [151] specifically includes 34 591 899 high-quality SNPs and 12 281 923 high-quality InDels. It also provides tools for extracting annotations across 1007 accessions of worldwide rapeseed germplasm. These genomic databases that integrate omics data have facilitated studies on Brassicaceae species in the post-genomic era. The increasing amount of publicly available omics data will provide the opportunity for developing databases such as PLAZA [153] and Plant-ImputeDB [154] for comparative genomics and genotyping studies on Brassicaceae species.
Although these genomic databases have greatly increased the efficiency of the usage of genomic and other omics data, there are few tools for integrating genomic data with phenotypic data, which would greatly facilitate our breeding efforts. The GWAS Atlas database contains genotype–phenotype associations for understanding the genetic architecture of traits in B. napus [155], though very limited phenotypic data are available. Databases providing comprehensive phenotype information with powerful analysis tools for other Brassicaceae species are necessary.
Future directions and prospects
Advances in genomics technologies have greatly promoted investigations into the genomes of Brassicaceae species. Population genomics will benefit from the greatly reduced cost for sequencing data production. It is now possible to resequence at a population scale of thousands or even over ten thousand accessions. Such studies will allow more accurate and reliable identification of genes associated with traits for Brassicaceae crops with much higher sensitivity. Pan-genomes of Brassicaceae crop species will be constructed to near completion with large numbers of representative accessions. Evidently, greater effort is still needed to integrate the rich genomic information with phenomic and metabolomic data to dissect interesting traits. Therefore, deep learning will play an important role in genomic interpretation of Brassicaceae crops in the future.
The combination of sequencing technologies with small-scale sample indexing, investigation of gene expression, genome modification, and 3D genome interaction is facilitating single-cell analysis. The newly developed spatial transcriptome analysis has a resolution at the scale of a few micrometers. Such a sub-cell scale analysis of gene expression provides us with a powerful tool for resolving the gene functions and networks of gene regulation. Emerging applications of these new technologies in A. thaliana are fascinating, and we are expecting digital 3D gene expression maps of different developmental stages at the cell level in Brassicaceae species.
Protein structure information greatly facilitates the resolution of gene functions. Recently, with the dramatic development of deep learning, the accurate prediction of protein structures became possible. DeepMind released the AlphaFold2, which has a protein structure prediction accuracy that competes with experimental structures in most cases and greatly outperforms other methods [156]. DeepMind and EMBL-EBI developed the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk), which provides 992 316 protein structures, including Arabidopsis and B. napus proteins. The Baker lab at the University of Washington developed RoseTTaFold, which is slightly less accurate than AlphaFold2, but 100 times faster than AlphaFold2 and has lower hardware requirements [157]. RoseTTaFold has made it practical to build comprehensive protein structure databases for important Brassica species.
Based on the original motivation to study crop genomes, the achieved knowledge will push the genetic improvement of crops. Some important mechanisms unraveled by the investigation of trait domestication in Brassica species that are featured with duplicated genomes are of important value for directing breeding programs. During the trait domestication of B. rapa and B. oleracea, some of the homoeologous genes were selected in parallel [28]. This mechanism allowed us to propose a molecular design strategy on ‘combined selection of homoeologous genes’ for the genetic improvement of complex traits in polyploid or paleopolyploid species. Combining specific homoeologous genes significantly improved bolting tolerance and anti-cancer sulforaphane content in B. rapa [158–159]. We expect extensive application of this strategy to explore homoeologous genes.
Moreover, utilizing the wild and distant species is an important practice to introduce valuable alien genetic variation or genes into cultivated crops. Cheng et al. proposed a multi-vertex model to describe the possibility of crossing different Brassiceae species, which experienced the same genome triplication as Brassica [160]. This model provided a framework for utilizing the wild and distant species within the Brassiceae tribe. However, to facilitate successful utilization of the alien genomes, it is important to obtain high-quality genome sequences for as many Brassiceae species as possible.
Overall, with the dramatically fast development of sequencing technologies and bioinformatics, the investigation of Brassicaceae species is becoming increasingly intensive. The achieved knowledge will ultimately promote the breeding of Brassicaceae crops.
Acknowledgements
This work was funded by the National Key Research and Development Program of China (2021YFF1000101), the Central Public-Interest Scientific Institution Basal Research Fund (Y2020PT21), and the Agricultural Science and Technology Innovation Program (ASTIP). This research was conducted in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China and the Sino-Dutch Joint Lab of Horticultural Genomics Technology, Beijing.
Author contributions
J.W. organized the manuscript and drafted and revised the manuscript with J.L. and R.L.; X.C., L.Z., X.G., T.W., and H.C. contributed to the collection of the published multi-genomics data of Brassicaceae genomes; X.W. organized and revised the manuscript. All authors read and approved the final manuscript.
Conflict of interests
The authors declare that they have no conflict of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Jian Wu, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Jianli Liang, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Runmao Lin, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Xu Cai, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Lei Zhang, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Xinlei Guo, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Tianpeng Wang, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Haixu Chen, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Xiaowu Wang, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Reference
- 1. Francis A, Lujan-Toro BE, Warwick SIet al. Update on the Brassicaceae species checklist. Biodivers Data J. 2021;9:e58773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X, Wang H, Wang Jet al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43:1035–9. [DOI] [PubMed] [Google Scholar]
- 3. Chalhoub B, Denoeud F, Liu Set al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–3. [DOI] [PubMed] [Google Scholar]
- 4. Liu S, Liu Y, Yang Xet al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang J, Liu D, Wang Xet al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet. 2016;48:1225–32. [DOI] [PubMed] [Google Scholar]
- 6. Song X, Wei Y, Xiao Det al. Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in brassica. Plant Physiol. 2021;186:388–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yim WC, Swain ML, Ma Det al. The last missing piece of the Triangle of U: the evolution of the tetraploid Brassica carinata genome. Plant Cell. 2022;koac249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perumal S, Koh CS, Jin Let al. A high-contiguity Brassica nigra genome localizes active centromeres and defines the ancestral brassica genome. Nature Plants. 2020;6:929–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai X, Wu J, Liang Jet al. Improved Brassica oleracea JZS assembly reveals significant changing of LTR-RT dynamics in different morphotypes. Theor Appl Genet. 2020;133:3187–99. [DOI] [PubMed] [Google Scholar]
- 10. Zhang L, Cai X, Wu Jet al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic Res. 2018;5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song JM, Guan Z, Hu Jet al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants. 2020;6:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen X, Tong C, Zhang Xet al. A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol J. 2021;19:615–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin MY, Koppers N, Denton Aet al. Whole genome sequencing and assembly data of Moricandia moricandioides and M. arvensis. Data Brief. 2021;35:106922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alabi N, Wu Y, Bossdorf Oet al. Genome report: a draft genome of Alliaria petiolata (garlic mustard) as a model system for invasion genetics. G3 (Bethesda). 2021;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang C, Ying H, Yang Xet al. The Cardamine enshiensis genome reveals whole genome duplication and insight into selenium hyperaccumulation and tolerance. Cell Discov. 2021;7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rellstab C, Zoller S, Sailer Cet al. Genomic signatures of convergent adaptation to Alpine environments in three Brassicaceae species. Mol Ecol. 2020;29:4350–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang L, Ma Y, Jiang Jet al. A chromosome-scale reference genome of Lobularia maritima, an ornamental plant with high stress tolerance. Hortic Res. 2020;7:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu Q, Ma Y, Mandáková Tet al. Genome evolution of the psammophyte Pugionium for desert adaptation and further speciation. Proc Natl Acad Sci USA. 2021;118:e2025711118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nowak MD, Birkeland S, Mandáková Tet al. The genome of Draba nivalis shows signatures of adaptation to the extreme environmental stresses of the Arctic. Mol Ecol Resour. 2021;21:661–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mishra B, Ploch S, Runge Fet al. The genome of Microthlaspi erraticum (Brassicaceae) provides insights into the adaptation to highly calcareous soils. Front Plant Sci. 2020;11:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang M, Wu H, Yang Qet al. A chromosome-scale genome assembly of Isatis indigotica, an important medicinal plant used in traditional Chinese medicine: An Isatis genome. Hortic Res. 2020;7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang W, Zhang L, Mandáková Tet al. The chromosome-level genome sequence and karyotypic evolution of Megadenia pygmaea (Brassicaceae). Mol Ecol Resour. 2021;21:871–9. [DOI] [PubMed] [Google Scholar]
- 23. Yang Q, Bi H, Yang Wet al. The genome sequence of Alpine Megacarpaea delavayi identifies species-specific whole-genome duplication. Front Genet. 2020;11:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bell L, Chadwick M, Puranik Met al. The Eruca sativa genome and Transcriptome: a targeted analysis of Sulfur metabolism and Glucosinolate biosynthesis pre and postharvest. Front Plant Sci. 2020;11:525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;10:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alonso-Blanco C, Andrade J, Becker Cet al. 1, 135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai X, Chang L, Zhang Tet al. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021;22:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng F, Sun R, Hou Xet al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet. 2016;48:1218–24. [DOI] [PubMed] [Google Scholar]
- 29. Kang L, Qian L, Zheng Met al. Genomic insights into the origin, domestication and diversification of Brassica juncea. Nat Genet. 2021;53:1392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu K, Wei L, Li Xet al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat Commun. 2019;10:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu D, Liang Z, Yan Tet al. Whole-genome Resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Mol Plant. 2019;12:30–43. [DOI] [PubMed] [Google Scholar]
- 32. Zhang X, Liu T, Wang Jet al. Pan-genome of Raphanus highlights genetic variation and introgression among domesticated, wild, and weedy radishes. Mol Plant. 2021;14:2032–55. [DOI] [PubMed] [Google Scholar]
- 33. Golicz AA, Bayer PE, Barker GCet al. The pangenome of an agronomically important crop plant Brassica oleracea. Nat Commun. 2016;7:13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng F, Wu J, Wang X. Genome triplication drove the diversification of Brassica plants. Hortic Res. 2014;1:14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun X, Li X, Lu Yet al. Construction of a high-density mutant population of Chinese cabbage facilitates the genetic dissection of agronomic traits. Mol Plant. 2022;15:913–24. [DOI] [PubMed] [Google Scholar]
- 36. Guo N, Wang S, Gao Let al. Genome sequencing sheds light on the contribution of structural variants to Brassica oleracea diversification. BMC Biol. 2021;19:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Helal M, Gill RA, Tang Met al. SNP- and haplotype-based GWAS of flowering-related traits in Brassica napus. Plants (Basel). 2021;10:2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Körber N, Bus A, Li Jet al. Agronomic and seed quality traits dissected by genome-wide association mapping in Brassica napus. Front Plant Sci. 2016;7:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu K, Xiao Z, Jian Het al. A combination of genome-wide association and transcriptome analysis reveals candidate genes controlling harvest index-related traits in Brassica napus. Sci Rep. 2016;6:36452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun F, Liu J, Hua Wet al. Identification of stable QTLs for seed oil content by combined linkage and association mapping in Brassica napus. Plant Sci. 2016;252:388–99. [DOI] [PubMed] [Google Scholar]
- 41. Hu J, Chen B, Zhao Jet al. Genomic selection and genetic architecture of agronomic traits during modern rapeseed breeding. Nat Genet. 2022;54:694–704. [DOI] [PubMed] [Google Scholar]
- 42. Tang S, Zhao H, Lu Set al. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol Plant. 2021;14:470–87. [DOI] [PubMed] [Google Scholar]
- 43. Su T, Wang W, Li Pet al. A genomic variation map provides insights into the genetic basis of spring Chinese cabbage (Brassica rapa ssp. pekinensis) selection. Mol Plant. 2018;11:1360–76. [DOI] [PubMed] [Google Scholar]
- 44. Liu J, Cai X, Li Yet al. Selection on BrFLC1 is related to intraspecific diversity of Brassica rapa vegetables. Horticulturae. 2021;7:247. [Google Scholar]
- 45. Yuan YX, Wu J, Sun RFet al. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J Exp Bot. 2009;60:1299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiao WB, Schneeberger K. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat Commun. 2020;11:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. De Coster W, Weissensteiner MH, Sedlazeck FJ. Towards population-scale long-read sequencing. Nat Rev Genet. 2021;22:572–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Della Coletta R, Qiu Y, Ou Set al. How the pan-genome is changing crop genomics and improvement. Genome Biol. 2021;22:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cai X, Lin R, Liang Jet al. Transposable element insertion: a hidden major source of domesticated phenotypic variation in Brassica rapa. Plant Biotechnol J. 2022;20:1298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He Z, Ji R, Havlickova Let al. Genome structural evolution in brassica crops. Nat Plants. 2021;7:757–65. [DOI] [PubMed] [Google Scholar]
- 51. Cheng F, Wu J, Fang Let al. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One. 2012;7:e36442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gaeta RT, Chris Pires J. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol. 2010;186:18–28. [DOI] [PubMed] [Google Scholar]
- 53. Lloyd A, Blary A, Charif Det al. Homoeologous exchanges cause extensive dosage-dependent gene expression changes in an allopolyploid crop. New Phytol. 2018;217:367–77. [DOI] [PubMed] [Google Scholar]
- 54. Hurgobin B, Golicz AA, Bayer PEet al. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol J. 2018;16:1265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schiessl S-V, Katche E, Ihien Eet al. The role of genomic structural variation in the genetic improvement of polyploid crops. Crop J. 2019;7:127–40. [Google Scholar]
- 56. Bertioli DJ, Jenkins J, Clevenger Jet al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat Genet. 2019;51:877–84. [DOI] [PubMed] [Google Scholar]
- 57. Zhang Z, Gou X, Xun Het al. Homoeologous exchanges occur through intragenic recombination generating novel transcripts and proteins in wheat and other polyploids. Proc Natl Acad Sci U S A. 2020;117:14561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu Y, Lin F, Zhou Yet al. Genomic mosaicism due to homoeologous exchange generates extensive phenotypic diversity in nascent allopolyploids. Natl Sci Rev. 2021;8:nwaa277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Henry IM, Dilkes BP, Tyagi Aet al. The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell. 2014;26:181–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Song K, Lu P, Tang Ket al. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci U S A. 1995;92:7719–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schranz ME, Osborn TC. De novo variation in life-history traits and responses to growth conditions of resynthesized polyploid Brassica napus (Brassicaceae). Am J Bot. 2004;91:174–83. [DOI] [PubMed] [Google Scholar]
- 62. PIRES JC, ZHAO J, SCHRANZ MEet al. Flowering time divergence and genomic rearrangements in resynthesized brassica polyploids (Brassicaceae). Biol J Linn Soc. 2004;82:675–88. [Google Scholar]
- 63. Gaeta RT, Pires JC, Iniguez-Luy Fet al. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. He Z, Wang L, Harper ALet al. Extensive homoeologous genome exchanges in allopolyploid crops revealed by mRNAseq-based visualization. Plant Biotechnol J. 2017;15:594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Higgins EE, Clarke WE, Howell ECet al. Detecting de novo Homoeologous recombination events in cultivated Brassica napus using a genome-wide SNP Array. G3 (Bethesda). 2018;8:2673–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stein A, Coriton O, Rousseau-Gueutin Met al. Mapping of homoeologous chromosome exchanges influencing quantitative trait variation in Brassica napus. Plant Biotechnol J. 2017;15:1478–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schiessl S, Huettel B, Kuehn Det al. Post-polyploidisation morphotype diversification associates with gene copy number variation. Sci Rep. 2017;7:41845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gonzalo A, Lucas MO, Charpentier Cet al. Reducing MSH4 copy number prevents meiotic crossovers between non-homologous chromosomes in Brassica napus. Nat Commun. 2019;10:2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Higgins EE, Howell EC, Armstrong SJet al. A major quantitative trait locus on chromosome A9, Bna Ph1, controls homoeologous recombination in Brassica napus. New Phytol. 2021;229:3281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sourdille P, Jenczewski E. Homoeologous exchanges in allopolyploids: how Brassica napus established self-control. New Phytol. 2021;229:3041–3. [DOI] [PubMed] [Google Scholar]
- 71. Ferreira de Carvalho J, Stoeckel S, Eber Fet al. Untangling structural factors driving genome stabilization in nascent Brassica napus allopolyploids. New Phytol. 2021;230:2072–84. [DOI] [PubMed] [Google Scholar]
- 72. McAlvay AC, Ragsdale AP, Mabry MEet al. Brassica rapa domestication: untangling wild and feral forms and convergence of crop Morphotypes. Mol Biol Evol. 2021;38:3358–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Song K, Osborn TC, Williams PH. Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs): 3. Genome relationships in brassica and related genera and the origin of B. oleracea and B. rapa (syn. Campestns). Theor Appl Genet. 1990;79:497–506. [DOI] [PubMed] [Google Scholar]
- 74. Warwick SI. Brassicaceae in Agriculture. In: Schmidt R, Bancroft I, eds. Genetics and Genomics of the Brassicaceae. New York: Springer, 2011,33–65. [Google Scholar]
- 75. Mabry ME, Turner-Hissong SD, Gallagher EYet al. The evolutionary history of wild, domesticated, and feral Brassica oleracea (Brassicaceae). Mol Biol Evol. 2021;38:4419–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. S, G.O.-C.C.P . Origin and domestication. Dev Plant Genet Breed. 1999;4:25. [Google Scholar]
- 77. Heslop-Harrison P. Genetics, genomics and breeding of oilseed brassicas. Ann Bot. 2013;112:vi–i. [Google Scholar]
- 78. Qian W, Meng J, Li Met al. Introgression of genomic components from Chinese Brassica rapa contributes to widening the genetic diversity in rapeseed (B. napus L.), with emphasis on the evolution of Chinese rapeseed. Theor Appl Genet. 2006;113:49–54. [DOI] [PubMed] [Google Scholar]
- 79. Dogan ES, Liu C. Three-dimensional chromatin packing and positioning of plant genomes. Nat Plants. 2018;4:521–9. [DOI] [PubMed] [Google Scholar]
- 80. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–8. [DOI] [PubMed] [Google Scholar]
- 81. Kawakatsu T, Huang SSC, Jupe Fet al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166:492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cokus SJ, Feng S, Zhang Xet al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen X, Ge X, Wang Jet al. Genome-wide DNA methylation profiling by modified reduced representation bisulfite sequencing in Brassica rapa suggests that epigenetic modifications play a key role in polyploid genome evolution. Front Plant Sci. 2015;6:836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Parkin IAP, Koh C, Tang Het al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014;15:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mehraj H, Takahashi S, Miyaji Net al. Characterization of histone H3 lysine 4 and 36 tri-methylation in Brassica rapa L. Front Plant Sci. 2021;12:659634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Akter A, Takahashi S, Deng Wet al. The histone modification H3 lysine 27 tri-methylation has conserved gene regulatory roles in the triplicated genome of Brassica rapa L. DNA Res. 2019;26:433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Paya-Milans M, Poza-Viejo L, Martin-Uriz PSet al. Genome-wide analysis of the H3K27me3 epigenome and transcriptome in Brassica rapa. Gigascience. 2019;8:giz147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cheng F, Sun C, Wu Jet al. Epigenetic regulation of subgenome dominance following whole genome triplication in Brassica rapa. New Phytol. 2016;211:288–99. [DOI] [PubMed] [Google Scholar]
- 89. Woodhouse MR, Cheng F, Pires JCet al. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci USA. 2014;111:5283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Braszewska-Zalewska A, Bernas T, Maluszynska J. Epigenetic chromatin modifications in brassica genomes. Genome. 2010;53:203–10. [DOI] [PubMed] [Google Scholar]
- 91. Zhang Q, Guan P, Zhao Let al. Asymmetric epigenome maps of subgenomes reveal imbalanced transcription and distinct evolutionary trends in Brassica napus. Mol Plant. 2021;14:604–19. [DOI] [PubMed] [Google Scholar]
- 92. Lukens LN, Pires JC, Leon Eet al. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 2006;140:336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yin L, Zhu Z, Huang Let al. DNA repair- and nucleotide metabolism-related genes exhibit differential CHG methylation patterns in natural and synthetic polyploids (Brassica napus L.). Hortic Res. 2021;8:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Grob S, Schmid MW, Grossniklaus U. Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of drosophila. Mol Cell. 2014;55:678–93. [DOI] [PubMed] [Google Scholar]
- 95. Xie T, Zhang FG, Zhang HYet al. Biased gene retention during diploidization in brassica linked to three-dimensional genome organization. Nat Plants. 2019;5:822–32. [DOI] [PubMed] [Google Scholar]
- 96. Feng S, Cokus SJ, Schubert Vet al. Genome-wide hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol Cell. 2014;55:694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu C, Wang C, Wang Get al. Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res. 2016;26:1057–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Grob S, Grossniklaus U. Invasive DNA elements modify the nuclear architecture of their insertion site by KNOT-linked silencing in Arabidopsis thaliana. Genome Biol. 2019;20:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Crevillen P, Sonmez C, Wu Zet al. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32:140–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20:631–56. [DOI] [PubMed] [Google Scholar]
- 101. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ziegler DJ, Khan D, Kalichuk JLet al. Transcriptome landscape of the early Brassica napus seed. J Integr Plant Biol. 2019;61:639–50. [DOI] [PubMed] [Google Scholar]
- 103. Sun X, Basnet RK, Yan Zet al. Genome-wide transcriptome analysis reveals molecular pathways involved in leafy head formation of Chinese cabbage (Brassica rapa). Hortic Res. 2019;6:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Greenham K, Guadagno CR, Gehan MAet al. Temporal network analysis identifies early physiological and transcriptomic indicators of mild drought in Brassica rapa. elife. 2017;6:e29655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bhardwaj AR, Joshi G, Kukreja Bet al. Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 2015;15:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang X, Liu Y, Fang Zet al. Comparative transcriptome analysis between broccoli (Brassica oleracea var. italica) and wild cabbage (Brassica macrocarpa Guss.) in response to Plasmodiophora brassicae during different infection stages. Front Plant Sci. 2016;7:1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Shah S, Weinholdt C, Jedrusik Net al. Whole-transcriptome analysis reveals genetic factors underlying flowering time regulation in rapeseed (Brassica napus L.). Plant Cell Environ. 2018;41:1935–47. [DOI] [PubMed] [Google Scholar]
- 108. Du L, Li C, Su Ret al. Transcriptome profiling reveals candidate genes involved in stem swelling of tumorous stem mustard. Hortic Plant J. 2020;6:158–66. [Google Scholar]
- 109. Parkin IA, Koh C, Tang Het al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014;15:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bancroft I, Morgan C, Fraser Fet al. Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat Biotechnol. 2011;29:762–6. [DOI] [PubMed] [Google Scholar]
- 111. Harper AL, Trick M, Higgins Jet al. Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotechnol. 2012;30:798–802. [DOI] [PubMed] [Google Scholar]
- 112. An H, Qi X, Gaynor MLet al. Transcriptome and organellar sequencing highlights the complex origin and diversification of allotetraploid Brassica napus. Nat Commun. 2019;10:2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chen X, Teichmann SA, Meyer KB. From tissues to cell types and Back: single-cell gene expression analysis of tissue architecture. Annu. Rev. Biomed. Data Sci. 2018;1:29–51. [Google Scholar]
- 114. Kerk NM, Ceserani T, Tausta SLet al. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Osaka M, Matsuda T, Sakazono Set al. Cell type-specific Transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing. Plant Cell Physiol. 2013;54:1894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Chan AC, Khan D, Girard IJet al. Tissue-specific laser microdissection of the Brassica napus funiculus improves gene discovery and spatial identification of biological processes. J Exp Bot. 2016;67:3561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hoang NV, Choe G, Zheng Yet al. Identification of conserved gene-regulatory networks that integrate environmental sensing and growth in the root cambium. Curr Biol. 2020;30:2887–2900.e7. [DOI] [PubMed] [Google Scholar]
- 118. Shaw R, Tian X, Xu J. Single-cell Transcriptome analysis in plants: advances and challenges. Mol Plant. 2021;14:115–26. [DOI] [PubMed] [Google Scholar]
- 119. Seyfferth C, Renema J, Wendrich JRet al. Advances and opportunities in single-cell transcriptomics for plant research. Annu Rev Plant Biol. 2021;72:847–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ryu KH, Zhu Y, Schiefelbein J. Plant cell identity in the era of single-cell transcriptomics. Annu Rev Genet. 2021;55:479–96. [DOI] [PubMed] [Google Scholar]
- 121. Rich-Griffin C, Stechemesser A, Finch Jet al. Single-cell transcriptomics: a high-resolution avenue for plant functional genomics. Trends Plant Sci. 2020;25:186–97. [DOI] [PubMed] [Google Scholar]
- 122. Rhee SY, Birnbaum KD, Ehrhardt DW. Towards building a plant cell atlas. Trends Plant Sci. 2019;24:303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhang X, Li T, Liu Fet al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol Cell. 2019;73:130–142.e5. [DOI] [PubMed] [Google Scholar]
- 124. Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–45. [DOI] [PubMed] [Google Scholar]
- 125. Denyer T, Ma X, Klesen Set al. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev Cell. 2019;48:840–852.e5. [DOI] [PubMed] [Google Scholar]
- 126. Farmer A, Thibivilliers S, Ryu KHet al. Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol Plant. 2021;14:372–83. [DOI] [PubMed] [Google Scholar]
- 127. Gala HP, Lanctot A, Jean-Baptiste Ket al. A single-cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana. Plant Cell. 2021;33:2197–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CMet al. Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell. 2019;31:993–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Long Y, Liu Z, Jia Jet al. Flsn RNA-seq: protoplasting-free full-length single-nucleus RNA profiling in plants. Genome Biol. 2021;22:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ryu KH, Huang L, Kang HMet al. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 2019;179:1444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Shahan R, Hsu CW, Nolan TMet al. A single cell Arabidopsis root atlas reveals developmental trajectories in wild type and cell identity mutants. Dev Cell. 2022;57:543–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Shulse CN, Cole BJ, Ciobanu Det al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 2019;27:2241–2247.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wendrich JR, Yang BJ, Vandamme Net al. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science. 2020;370:eaay4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhang TQ, Xu ZG, Shang GDet al. A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol Plant. 2019;12:648–60. [DOI] [PubMed] [Google Scholar]
- 135. Coate JE, Farmer AD, Schiefelbein JWet al. Expression partitioning of duplicate genes at single cell resolution in Arabidopsis roots. Front Genet. 2020;11:596150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Turco GM, Rodriguez-Medina J, Siebert Set al. Molecular mechanisms driving switch behavior in xylem cell differentiation. Cell Rep. 2019;28:342–351.e4. [DOI] [PubMed] [Google Scholar]
- 137. Roszak P, Heo JO, Blob Bet al. Cell-by-cell dissection of phloem development links a maturation gradient to cell specialization. Science. 2021;374:eaba5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Sunaga-Franze DY, Muino JM, Braeuning Cet al. Single-nucleus RNA sequencing of plant tissues using a nanowell-based system. Plant J. 2021;108:859–69. [DOI] [PubMed] [Google Scholar]
- 139. Zhang TQ, Chen Y, Wang JW. A single-cell analysis of the Arabidopsis vegetative shoot apex. Dev Cell. 2021;56:1056–1074.e8. [DOI] [PubMed] [Google Scholar]
- 140. Kim JY, Symeonidi E, Pang TYet al. Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell. 2021;33:511–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Liu Z, Zhou Y, Guo Jet al. Global dynamic molecular profiling of stomatal lineage cell development by single-cell RNA sequencing. Mol Plant. 2020;13:1178–93. [DOI] [PubMed] [Google Scholar]
- 142. Lopez-Anido CB, Vatén A, Smoot NKet al. Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Dev Cell. 2021;56:1043–1055.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Picard CL, Povilus RA, Williams BPet al. Single nucleus analysis of Arabidopsis seeds reveals new cell types and imprinting dynamics. bioRxiv. 2020. [Google Scholar]
- 144. Liu Q, Liang Z, Feng Det al. Transcriptional landscape of rice roots at the single-cell resolution. Mol Plant. 2021;14:384–94. [DOI] [PubMed] [Google Scholar]
- 145. Rao A, Barkley D, Franca GSet al. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596:211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Xia K, Sun HX, Li Jet al. Single-cell stereo-seq enables cell type-specific spatial transcriptome characterization in Arabidopsis leaves. Dev Cell. 2022;57:1299–310. [DOI] [PubMed] [Google Scholar]
- 147. Huala E, Dickerman AW, Garcia-Hernandez Met al. The Arabidopsis information resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Chen H, Wang T, He Xet al. BRAD V3.0: an upgraded Brassicaceae database. Nucleic Acids Res. 2022;50:D1432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Song JM, Liu DX, Xie WZet al. BnPIR: Brassica napus pan-genome information resource for 1689 accessions. Plant Biotechnol J. 2021;19:412–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Chao H, Li T, Luo Cet al. BrassicaEDB: a gene expression database for Brassica crops. Int J Mol Sci. 2020;21:5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yan T, Yao Y, Wu Det al. BnaGVD: a genomic variation database of rapeseed (Brassica napus). Plant Cell Physiol. 2021;62:378–83. [DOI] [PubMed] [Google Scholar]
- 152. Kiefer M, Schmickl R, German DAet al. Brassi Base: introduction to a novel knowledge database on Brassicaceae evolution. Plant Cell Physiol. 2014;55:e3. [DOI] [PubMed] [Google Scholar]
- 153. Van Bel M, Diels T, Vancaester Eet al. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018;46:D1190–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Gao Y, Yang Z, Yang Wet al. Plant-imputeDB: an integrated multiple plant reference panel database for genotype imputation. Nucleic Acids Res. 2021;49:D1480–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Tian D, Wang P, Tang Bet al. GWAS Atlas: a curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020;48:D927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Jumper J, Evans R, Pritzel Aet al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Baek M, DiMaio F, Anishchenko Iet al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wu J, Wei K, Cheng Fet al. A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol. 2012;12:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Liu Z, Liang J, Zheng Set al. Enriching Glucoraphanin in Brassica rapa through replacement of BrAOP2.2/BrAOP2.3 with non-functional genes. Front Plant Sci. 2017;8:1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Cheng F, Liang J, Cai Cet al. Genome sequencing supports a multi-vertex model for Brassiceae species. Curr Opin Plant Biol. 2017;36:79–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.