Abstract
We have shown that Pasteurella multocida toxin (PMT) directly causes transient activation of Gqα protein that is coupled to phosphatidylinositol-specific phospholipase Cβ1 in Xenopus oocytes (B. A. Wilson, X. Zhu, M. Ho, and L. Lu, J. Biol. Chem. 272:1268–1275, 1997). We found that antibodies directed against an N-terminal peptide of PMT inhibited the toxin-induced response in Xenopus oocytes, but antibodies against a C-terminal peptide did not. To test whether the intracellular activity domain of PMT is localized to the N terminus, we conducted a deletion mutational analysis of the PMT protein, using the Xenopus oocyte system as a means of screening for toxin activity. Using PCR and conventional cloning techniques, we cloned from a toxinogenic strain of P. multocida the entire toxA gene, encoding the 1,285-amino-acid PMT protein, and expressed the recombinant toxin as a His-tagged fusion protein in Escherichia coli. We subsequently generated a series of N-terminal and C-terminal deletion mutants and expressed the His-tagged PMT fragments in E. coli. These proteins were screened for cytotoxic activity on cultured Vero cells and for intracellular activity in the Xenopus oocyte system. Only the full-length protein without the His tag exhibited activity on Vero cells. The full-length PMT and N-terminal fragments containing the first 500 residues elicited responses in oocytes, but the C-terminal 780 amino acid fragment did not. Our results confirm that the intracellular activity domain of PMT is localized to the N-terminal 500 amino acids of the protein and that the C terminus is required for entry into cells.
Pasteurella multocida toxin (PMT) is a major virulence factor associated with progressive atrophic rhinitis in domestic and wild animals (1, 12), respiratory disease in cattle and laboratory rabbits (3–6, 8, 14), and dermonecrosis and bacteremia resulting from bite wounds or animal exposure in humans (15, 16, 20, 23, 28, 35, 41). PMT is a 1,285-amino-acid protein (7, 22, 26, 32) that appears to bind to and enter mammalian cells via receptor-mediated endocytosis (34, 36) and acts intracellularly to initiate DNA synthesis and cytoskeletal rearrangements (9, 17, 18, 21, 24, 29, 36). Some of the intracellular events that have been observed upon exposure to PMT in cultured fibroblasts and osteoblasts include enhanced hydrolysis of inositolphospholipids to increase intracellular inositol phosphates and diacylglycerol (29, 30, 38); mobilization of intracellular Ca2+ pools (29, 30, 38, 40); decreased ADP-ribosylation of GRP78/BiP (39); increased protein phosphorylation (40); and tyrosine phosphorylation of p125Fak and paxillin, as well as actin stress fiber formation and focal adhesion assembly (9, 24). Recently, we used voltage-clamped Xenopus oocytes to demonstrate direct PMT-mediated stimulation of the β1 isoform of phospholipase C (PLCβ1) and the inositol 1,4,5-trisphosphate (IP3) signaling pathway and to identify the immediate intracellular target of PMT as the free, monomeric α subunit of the Gq protein (43).
During our earlier studies, we observed that specific antibodies against an N-terminal peptide of PMT, anti-toxA28–42, were able to block the PMT-mediated response in Xenopus oocytes (43). On the other hand, specific antibodies to a C-terminal peptide of PMT, anti-toxA1239–1253, did not block the activity, strongly implying that the N terminus of PMT is critical for its intracellular activity. The N-terminal half of the related cytotoxic necrotizing factor type 1 (CNF1) and CNF2 from pathogenic Escherichia coli show homology to the N terminus of PMT (10, 31). Comparisons of the amino acid sequence of PMT with those of other bacterial dermonecrotic toxins has been reported elsewhere (10, 27, 31, 42). While there is homology in the C termini of the CNFs and the dermonecrotic toxin from Bordetella bronchiseptica (DNT), the C-terminal region of PMT does not have this similarity with those of the CNFs and DNT (10, 27, 31, 42). As has been shown for PMT, both DNT and the CNFs induce DNA synthesis, as well as actin stress fiber formation and focal adhesion assembly (19, 25, 31). For the CNFs and DNT, this activity has been shown to occur through constitutive activation of the small G protein RhoA by deamidation of Gln-63 (11, 19, 31, 37). The RhoA deamidase activity was reported to be localized to the C terminus of CNF1 (27), and the receptor-binding activity was postulated to be in the N terminus. While all of these toxins have been shown to induce stress fiber formation and focal adhesion assembly, the molecular and functional organization of this group of toxins remains unclear. It is also not clear how the modification of RhoA by the CNFs and DNT and the modification of Gqα by PMT lead to each of the observed intracellular changes.
To define the functional domain of PMT responsible for intracellular activity, we cloned the entire toxA gene from P. multocida and expressed the corresponding recombinant PMT (rPMT) as a hexahistidine (His)-tagged fusion protein in E. coli. We confirmed that the full-length rPMT protein is as active as native PMT in both oocyte and cell culture assays. We then constructed a series of N- and C-terminal deletion mutants of the toxA gene and expressed the corresponding recombinant His-tagged PMT fragments (ToxAN and ToxAC, respectively) in E. coli. To screen for the functional domain of PMT responsible for activation of the intracellular Gq-PLCβ1-IP3 signaling pathway, the recombinant toxin proteins were microinjected into voltage-clamped Xenopus oocytes to observe the toxin-induced Ca2+-dependent Cl− current. Our results localize the functional domain of PMT responsible for intracellular activity to the N terminus. To determine if this functional domain alone possessed cytotoxic activity on intact cells, we also tested each of the recombinant proteins for effects on cultured Vero cells and found that only the full-length rPMT protein is able to cause the morphological effects induced by native PMT.
MATERIALS AND METHODS
Materials.
Rabbit polyclonal antisera against two synthetic peptides, comprising residues 28 to 42 of PMT (NSDFTVKGKSADEIF) and residues 1239 to 1253 of PMT (PVDDWALEIAQRNRA), were obtained from Bio-Synthesis, Inc., using multiple antigen peptide conjugation methodology. The anti-peptide immunoglobulin G (IgG) antibodies (anti-toxA28–42 and anti-toxA1239–1253) were purified by using a protein A-agarose column (PURE-1; Sigma). Goat anti-rabbit IgG antibodies conjugated to alkaline phosphatase and tetramethylrhodamine B isothiocyanate (TRITC)-conjugated phalloidin were obtained from Sigma. E. coli BL21(DE3) and the plasmid vectors pET-15b, pET-21b, and pET-33b were obtained from Novagen. E. coli XL-1 Blue was obtained from Stratagene. TA-cloning kits, E. coli TOP10 and competent TOP10F′ One-Shot cells, and plasmid vectors pTrcHisC and pCR3-Uni were obtained from Invitrogen. LipofectAMINE, the plasmid vector pROEX-1, and tissue culture media, sera, and reagents were obtained from Gibco/BRL. The plasmid vector pEGFP-N1 was obtained from Clontech. Ni2+-nitrilotriacetic acid (NTA)-agarose and Qiaex II DNA purification kits were obtained from Qiagen. PCR primers were obtained from Bio-Synthesis and Gibco/BRL. Restriction enzymes were obtained from Gibco/BRL or New England Biolabs. Centricon, Ultra-Free, and Centricon Plus-20 concentrators were obtained from Amicon/Millipore. Xenopus laevis frogs were obtained from Xenopus I, Dexter, Mich. African green monkey kidney (Vero) cells (CCL-81) and murine Swiss 3T3 (CCL-92) and NIH 3T3 (CRL-1658) fibroblast cells were obtained from the American Type Culture Collection. P. multocida subsp. multocida 45/78 (NCTC 12178) was obtained from the National Collection of Type Cultures and grown at 37°C on Trypticase soy agar supplemented with 5% defibrinated sheep blood (BBL 11043). Native PMT was purchased from Sigma as a lyophilized powder with bovine serum albumin and resuspended in 50 mM Tris-HCl (pH 7.5) containing 5% glycerol prior to use. Native PMT, purified to homogeneity, quantified, and titered by Vero cell cytotoxicity assays as described elsewhere (8), was a generous gift from Clarence Chrisp. All other reagents were of the highest quality commercially available.
Construction of plasmids containing deletion mutants of PMT.
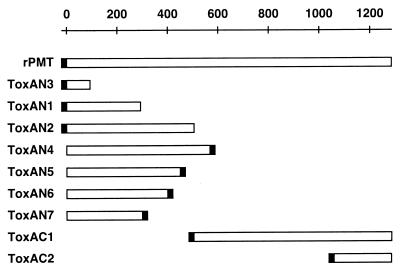
All recombinant proteins were engineered to have either an N- or a C-terminal fusion peptide containing a His tag, with all N-terminal fusion peptides also containing a thrombin proteolytic recognition site (Fig. 1). All cloning was carried out with either E. coli XL-1 Blue or E. coli TOP10F′ One-Shot cells.
FIG. 1.
Localization of deletion mutant proteins used in this study relative to full-length PMT. Open bar, sequence derived from the toxA gene; closed bar, location of the His tag in each recombinant protein.
Genomic DNA from P. multocida (NCTC 12178) was isolated as described elsewhere (2) and used as a template for PCR. Primers 1 (coding, 5′-CGCGGCAGCCATATGAAAACAAAACATTTTTTTAAC-3′) and 2 (noncoding, 5′-TATGCGAATTCATCATGCAATCCATTGTTTCAGATC-3′) were used to clone by PCR the region of the toxA gene, encoding the N-terminal 293 amino acids. The forward primer 1 incorporated an NdeI site (underscored) at the start of the gene (bold), and the reverse primer 2 incorporated an EcoRI site (underscored) and a stop codon (bold) after the codon for amino acid 293. The resulting PCR product was digested with NdeI and EcoRI, and the NdeI/EcoRI fragment was inserted into the NdeI/EcoRI sites of the expression vector pET-15b, to yield pET15b-toxAN1, which was sequence confirmed and used to express the ToxAN1 protein. Plasmid pET15b-toxAN1 was used to generate the beginning region of the toxA gene in all subsequent constructs encoding the N-terminal region of PMT due to the lack of unique and practical restriction sites in the toxA gene.
To clone the remaining portion of the toxA gene, genomic DNA from P. multocida (NCTC 12178) was exhaustively digested with HpaII. A 15-kb HpaII fragment, shown by Southern blotting (2) to hybridize to a DNA probe generated from the NdeI/EcoRI insert from pET15b-toxAN1 (data not shown), was used as a template to obtain the entire toxA gene by a three-step process using PCR. In the first step, two plasmids containing two overlapping fragments of the toxA gene were generated by TA cloning. Primers 1 and 3 (noncoding, 5′-AAGACGTACCAGACGAATTTTATGT-3′) were used to clone the region of the toxA gene encoding the N-terminal 521 amino acids into the TA-cloning vector pCR3-Uni to yield pCR3-Uni(1&3). Primers 4 (coding, 5′-GTAGGAGAAGAATTACATTCTATCTCA-3′) and 5 (noncoding, 5′-GAATTGGGTACCTTATAGTGCTCTTGTTAAGCGAGG-3′), with a KpnI site (underscored) and a stop codon (bold) incorporated into primer 5, were used to clone the region of toxA encoding the C-terminal 793 amino acids into the TA-cloning vector pCR3-Uni to yield pCR3-Uni(4&5). In the second step, pCR3-Uni(1&3) was digested with StuI and XhoI, and the resulting StuI/XhoI fragment was inserted into the StuI/XhoI sites of pET15b-toxAN1. Then an NcoI/XhoI toxA gene fragment from the resulting plasmid was subcloned into the expression vector pTrcHisC to yield pTHC-toxAN2, which was used to express the ToxAN2 protein. In the third step, pCR3-Uni(4&5) was digested with XhoI and KpnI, and the resulting XhoI/KpnI toxA gene fragment was inserted into the XhoI/KpnI sites of pTHC-toxAN2 to yield pTHC-toxA, which was used to express the full-length rPMT protein.
A BamHI linker was inserted into the StuI site of pET15b-toxAN1, and a BamHI/EcoRI vector fragment from pET-15b was inserted into the BamHI/EcoRI sites of pET15b-toxAN1, to yield pET15b-toxAN3, which was used to express the ToxAN3 protein. The NdeI/EcoRI insert from pET15b-toxAN1 was fragment exchanged into pET-21b to yield pET21b-toxAN1. A NotI linker was inserted into the SnaBI site of pTHC-toxA, and a StuI/NotI insert from pTHC-toxA was subcloned into the StuI/NotI sites of pET21b-toxAN1 to generate pET21b-toxAN4, which was used to express the ToxAN4 protein. The XhoI/KpnI insert from pCR3-Uni(4&5) was subcloned into the XhoI/KpnI sites of the expression vector pTrcHisC to yield pTHC-toxAC1, which was used to express the ToxAC1 protein. The SpeI/KpnI insert from pCR3-Uni(4&5) was subcloned into the SpeI/KpnI sites of the expression vector pROEX-1 to generate pROEX1-toxAC2, which was used to express the ToxAC2 protein. With the pTHC-toxA plasmid serving as a DNA template for PCR, primers 6 (coding, 5′-CGCCGCGCCATGGCAACAAAACATTTTTTTAAC-3′) and 7 (noncoding, 5′-TGCTGCGGCCGCGCTGAATAATGCAGATTG-3′), primers 6 and 8 (noncoding, 5′-GCGGCCGCTATTGCAGATAACTGTG-3′), and primers 6 and 9 (noncoding, 5′-GCGGCCGCTTTGTTATCTTTTAAATGCCAT-3′), with an NcoI site (underscored) incorporated at the start (bold) of the gene in primer 6 and an NotI site (underscored) incorporated at the end of the gene fragment in primers 7, 8, and 9, were used to generate by PCR three toxA-containing fragments that were ligated into the TA-cloning vector pCR3-Uni to yield pCR3-Uni(6&7), pCR3-Uni(6&8), and pCR3-Uni(6&9), respectively. NcoI/NotI inserts from each of the three plasmids were then subcloned into the expression vector pET33b to yield pET33b-toxAN5, pET33b-toxAN6, and pET33b-toxAN7, respectively, which in turn were used to express the corresponding ToxAN5, ToxAN6, and ToxAN7 proteins.
Expression of recombinant toxin proteins in E. coli.
Recombinant proteins were expressed in E. coli BL21(DE3) for constructs generated in expression vector pET15b, pET21b, pET33b, or pROEX-1 or in E. coli TOP10 for constructs generated in expression vector pTrcHisC. Seed flasks containing 50 ml of LB with the appropriate antibiotic, ampicillin (100 μg/ml) or kanamycin (25 μg/ml), were inoculated with the desired bacterial colonies and incubated with vigorous shaking overnight at 37°C. Fermentation flasks containing 500 ml of LB with ampicillin (100 μg/ml) or kanamycin (25 μg/ml) were inoculated with 8 to 10 ml of the overnight seed culture and incubated with vigorous shaking at 37°C until an optical density at 600 nm of 0.6 to 0.8 was reached. Expression of the recombinant proteins was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and growth was continued for 3 h. Cells were harvested by centrifugation at 5,000 × g for 10 min in a Sorvall RC-5C Plus centrifuge. Cell pellets were suspended in lysis buffer (100 ml per 2.5-liter starting culture volume; 50 mM potassium phosphate [pH 7.5] containing 100 mM KCl, 0.1% Tween 20, 5% glycerol, 5 μg each of pepstatin, leupeptin, and aprotinin per ml, 2 mM benzamidine, 2 mM phenylmethylsulfonyl fluoride, 0.2 mg of lysozyme per ml, 200 U of RNase A, and 1,500 U of DNase I). The cell suspension was sonicated for four rounds of 1-min pulses at a duty cycle of 0.7 in a Brau-Sonic-U cell disruptor, with 1-min cooling intervals on ice. The cellular debris was removed by centrifugation at 20,000 × g for 90 min, and the supernatants were immediately used as cell extracts for further purification.
Purification of recombinant toxin proteins.
The His tag affinity handle allowed for essentially one-step purification of the full-length and deletion mutant toxins by nickel chelate affinity chromatography. Cell extracts were passed twice through a 5-ml/Ni2+-NTA-agarose column (Qiagen). The column was washed twice with 50 ml of Ni column buffer (50 mM potassium phosphate [pH 7.5], 100 mM KCl, 0.1% Tween 20, 5% glycerol, 10 mM imidazole), followed by stepwise elution with 50 ml each of Ni column buffer containing 20, 30, 60, 110, 160, and 500 mM imidazole. Column fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting, and fractions containing the toxin proteins were pooled and concentrated in Centricon/Millipore concentrators to remove the imidazole and to exchange the buffer with 50 mM Tris-HCl (pH 7.5) containing 5% glycerol. To further desalt the samples, the concentrated solutions were passed through a PD-10 desalting column (Pharmacia).
SDS-PAGE and Western analysis.
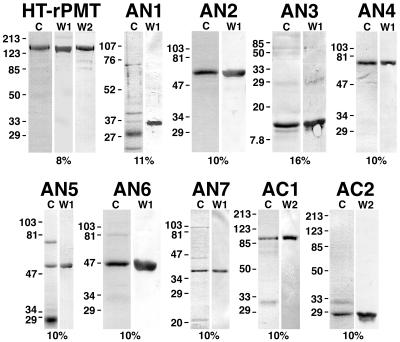
To analyze dilute protein fractions or fractions containing high salt concentrations, samples were precipitated with an equal volume of cold 20% trichloroacetic acid and microcentrifuged at 15,000 × g at 4°C for 20 min, washed twice with cold acetone, and then air dried. Protein pellets were redissolved in a mixture of 100 mM Tris-HCl (pH 9.0), SDS-PAGE sample buffer, and tracking dye. Alternatively, SDS-PAGE sample buffer and tracking dye were added directly to protein samples. The protein samples were then separated by SDS-PAGE on 8, 10, 11, or 16% gels. Protein bands were visualized by Coomassie or silver staining and by Western blotting. For Western blot analysis, proteins were first transferred onto nitrocellulose membrane and immunoblotted with rabbit polyclonal antibodies to PMT, followed by secondary goat anti-rabbit IgG antibodies conjugated to alkaline phosphatase. The images of gels and Western blots shown in Fig. 2 and 3 were obtained by using a ScanJet 3C (Hewlett-Packard) with DeskScan II (Hewlett-Packard) image acquisition software, and the graphics were produced by using Adobe Photoshop on an Apple Macintosh G3 computer.
FIG. 2.
Coomassie-stained SDS-polyacrylamide gel and Western blot analysis of Ni2+ chelate affinity-purified samples of rPMT and recombinant PMT fragments used in this study. The toxin protein sample analyzed is indicated above each set of gels. Lanes: C Coomassie-stained SDS-polyacrylamide gels; W1 and W2, the corresponding Western blots with anti-toxA28–42 and anti-toxA1239–1253, respectively, as primary antibodies. The percentage of acrylamide used for the gel is indicated below each set of gels. In each lane, 3 to 5 μg of protein was loaded. The positions of molecular weight markers are indicated in kilodaltons at the left. HT-rPMT denotes rPMT before removal of the His tag. The ToxAN2 and ToxAN3 samples shown were after removal of the His tag, and the ToxAN1 sample shown was before removal of the His tag.
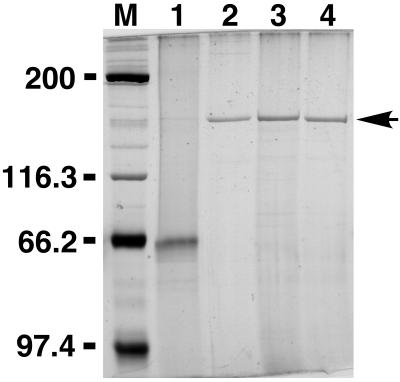
FIG. 3.
SDS-PAGE comparison of native and recombinant PMT protein samples. Shown is a silver-stained SDS–8% polyacrylamide gel. Lanes: M, molecular weight markers (positions indicated in kilodaltons at the left); 1, 1.0 μg of native PMT (concentration based on that provided by supplier [Sigma]); 2, 0.1 μg of highly purified, native PMT; 3, 0.1 μg of rPMT before removal of the His tag; 4, 0.1 μg of rPMT after removal of the His tag.
Removal of the His tag by thrombin treatment.
The His tag peptide was removed from recombinant toxin proteins possessing N-terminal His tags (rPMT, ToxAN2, and ToxAN3) by thrombin treatment. The His-tagged protein (4 to 5 mg) and biotinylated thrombin (5 U; Novagen) in thrombin cleavage buffer (500 μl; 20 mM Tris-HCl [pH 8.4], 150 mM NaCl, 2.5 mM CaCl2) were incubated at room temperature for 16 to 20 h. Completion of reaction was assessed by SDS-PAGE analysis. To terminate the reaction, streptavidin-agarose beads (20 μl; Novagen) were added and the suspension was incubated for an additional hour with gentle shaking. The beads were pelleted by centrifugation at 400 × g for 30 s, and the supernatant was diluted with Ni column buffer (1 to 2 ml) and passed twice through a 2- to 3-ml Ni2+-NTA-agarose column (Qiagen) to remove the His tag peptide. Samples were desalted on PD-10 columns (Pharmacia) against 50 mM Tris-HCl (pH 7.5) containing 5% glycerol and concentrated in Centricon concentrators.
Quantitation of recombinant toxin proteins.
The concentration of His-tagged rPMT was determined by Coomassie Plus protein assay (Pierce), with bovine serum albumin as a standard. The concentration of the commercial native PMT sample was determined by image quantitation analysis of silver-stained SDS-polyacrylamide gels, using His-tagged rPMT as a standard. The concentrations of rPMT without the His tag, ToxAN2 without the His tag, ToxAN4, ToxAN5, and ToxAC1 were determined by image quantitation analysis of the appropriate bands on Western blots with either anti-toxA28–42 or anti-toxA1239–1253 as the primary antibody, using His-tagged rPMT as a standard. The concentrations of ToxAN3 without the His tag, ToxAN6, ToxAN7, and ToxAC2 were determined by image quantitation analysis of the appropriate bands on Western blots with either anti-toxA28–42 or anti-toxA1239–1253 as the primary antibody, using ToxAN4 as a standard. Western blots were imaged as described above, and images were quantitated by using NIH Image (National Institutes of Health) image analysis software and Microsoft Office Excel (Microsoft) data analysis software.
Cell culture assays.
Cultured cells were maintained at 37°C and 10% carbon dioxide in Dulbecco modified Eagle medium (DMEM; Gibco/BRL), pH 7.4, supplemented with 10% heat-inactivated fetal bovine serum (Gibco/BRL) for Vero cells and NIH 3T3 cells) or 15% heat-inactivated calf serum (Gibco/BRL) for Swiss 3T3 cells, containing penicillin G (100 U/ml) and streptomycin (100 μg/ml) (Gibco/BRL). After trypsinization, cells were plated onto 12-well plates (Falcon) at a density of 4 × 104 (Vero cells) or 2 × 104 (Swiss 3T3 cells) per well, and incubation was continued with a change of medium after 24 h. Vero cells and Swiss 3T3 cells were allowed to reach confluency 5 days after plating, and then the medium was changed to DMEM–1% fetal bovine or calf serum (pH 7.4) containing antibiotics. Two days after quiescence with low-serum media, the medium was replaced with filter-sterilized, low-serum medium containing 200 ng of the native PMT, rPMT, and ToxAN4 proteins per ml, as well as controls containing the corresponding heat-inactivated proteins and no protein. The cells were visualized after 1, 4, and 6 days of toxin exposure for morphological changes by phase-contrast microscopy using an Olympus IX-70 inverted microscope. Heat-inactivated controls of the protein samples were prepared by heating the samples at 70°C for 45 min.
Transfection of NIH 3T3 cells with mammalian expression plasmids containing an N-terminal fragment of PMT or green fluorescent protein (GEP).
Cells were cotransfected with a 1:1 ratio of pCR3-Uni(1&3) and pEGFP-N1 or with pEGFP-N1 alone, using LipofectAMINE reagent (Gibco/BRL) according to the manufacturer’s protocol. Briefly, 1 day prior to transfection, NIH 3T3 cells were plated at a density of 2 × 105 cells/well on six-well plates in antibiotic-free medium. DNA plasmids (1 μg of each) in 100 μl of Opti-MEM (Gibco/BRL) and 4 μl of LipofectAMINE reagent in 100 μl of Opti-MEM were combined, incubated at room temperature for 15 min, and then added to each well containing 800 μl of Opti-MEM. After 3 h of incubation at 37°C, 1 ml of antibiotic-free, low-serum medium was added, and cells were further incubated at 37°C for 24 h prior to staining and visualization.
Fluorescence staining of transfected NIH 3T3 cells expressing an N-terminal fragment of PMT.
The cells were washed twice with phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde in PBS for 10 min, washed with PBS, permeabilized with 0.5% Triton X-100 in PBS for 10 min, washed twice with PBS, incubated for 1 h at room temperature with TRITC-conjugated phalloidin (2 μM; Sigma) in PBS, washed twice with PBS, and visualized by fluorescence microscopy using an Olympus IX-70 inverted microscope, equipped with U-MWB and U-MWIG fluorescence cubes for observing GFP and TRITC, respectively. Images were captured by using NIH Image 1.61, and micrographs were generated by using Adobe Photoshop 4.0.
Oocyte preparation.
Adult female X. laevis frogs (Xenopus I) were anesthetized by immersion in a 0.1% tricaine methanesulfonate (Sigma), adjusted to pH 7.4 with NaHCO3. A small incision was made on one side of the abdomen to remove several ovarian lobes. The lobes were gently torn apart and immersed in a Ca2+-free OR-2 solution (100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES-Tris [pH 7.5]). Oocytes were defolliculated by incubation with collagenase (2 mg/ml; Sigma type 1A) at room temperature (22 to 24°C) for 1 to 2 h. The oocytes were then washed five times with OR-2 solution and five times with a modified Barth’s solution [88 mM NaCl–1 mM KCl–2.4 mM NaHCO3–0.3 mM Ca(NO3)2–0.4 mM CaCl2–0.8 mM MgSO4–15 mM Tris-HCl (pH 7.6) containing penicillin G (100 μg/ml) and streptomycin (100 μg/ml)]. Stage 5–6 oocytes were selected and stored at 18°C in modified Barth’s solution.
Two-microelectrode voltage clamp.
Two microelectrodes, made by a vertical puller (model PP-83, Narishige) and filled with 3 M KCl to give a resistance of 1.5 to 2.0 megaohms, were used for voltage clamping. Voltage clamp experiments were performed in a continuously perfused bath (10 ml/min) at room temperature (22 to 24°C). The bath was connected through an Ag-AgCl-agar-3 M KCl bridge to the two-electrode voltage clamp system (TEV-200 voltage clamp; Dagan). Membrane currents were measured in normal Ringer’s solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 11.8 mM CaCl2, 5 mM HEPES-NaOH [pH 7.4]). Data acquisition was performed by using MacLab/8s (ADInstruments) linked to a Macintosh Quadra 800 computer with Chart and Scope application software (ADInstruments). Graphics were obtained by using Cricket Graph III (Computer Associates).
Oocyte microinjections.
Microinjections of toxins, toxin fragments, and antibodies were performed under voltage-clamped conditions at −80-mV holding potential, using a pulse-controlled microinjector (Dagan model PMI-200). Toxins and other reagents were prepared in 50 mM Tris-HCl (pH 7.5) with 5% glycerol prior to injection. For experiments involving anti-toxA28–42 inhibition of ToxAN4, protein A-purified antibodies were passed through a PD10 column in 50 mM Tris-HCl (pH 7.5) with 5% glycerol and concentrated such that an equal volume could neutralize the rPMT response in oocytes. This antibody preparation was then incubated in a 1:1 ratio with ToxAN4 on ice for 1 h prior to injection.
Data analysis and determination of EC50s.
In all experiments reported here, each group of oocytes was tested for positive responses elicited by rPMT. All results are expressed as the mean ± standard error of the maximal inward current assayed in oocytes from the same donor group. For the titration experiments, the same group of oocytes from a given donor was used for all doses tested. The number of oocytes used for data analysis at each dose is given in parentheses in Fig. 5. Fifty percent effective concentrations (EC50s) were calculated with SigmaPlot (Jandell Scientific) by fitting data points to a logistic function f(x) = a/{1 + exp[b*(x − c)]}, where a is the maximum response observed, b is the slope, and c is the EC50. The EC50 curves shown in Fig. 5 were plotted by using Cricket Graph III (Computer Associates).
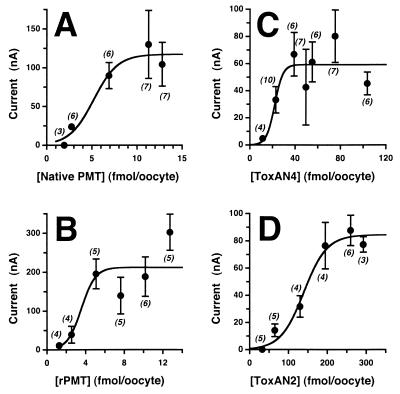
FIG. 5.
EC50 curves for toxin proteins. Shown are curves of the dose dependence of the toxin-induced response in voltage-clamped oocytes for native PMT (A), rPMT (B), ToxAN4 (C), and ToxAN2 (D). The mean ± standard error of the relative response induced by injection of toxin is shown for each data point, with the total number of oocytes tested for each point given in parentheses. Since the average maximal response observed varied from donor group to donor group, all data points for each EC50 curve were obtained with the same batch of oocytes from a single donor. EC50s were determined from the data by using SigmaPlot (see Materials and Methods) and are summarized in Table 1.
Protein sequence and structure comparison between PMT and the CNFs and DNT.
Primary sequence comparison between PMT, CNF1, CNF2, and DNT was performed by using the BestFit sequence analysis program from the Genetics Computer Group Wisconsin Package with BLOSUM30 and BLOSUM62 amino acid alignment scoring matrices. The secondary structure prediction for PMT and CNF1 was performed with the GOR-IV program available at http://absalpha.dcrt.nih.gov:8008/gor.html. The area plot shown in Fig. 7 was generated from the GOR-IV analysis results, with an additional smoothing window of 11 amino acid residues for the helical probability, 5 amino acid residues for the extended strand probability, and the remainder for the coil probability.
FIG. 7.
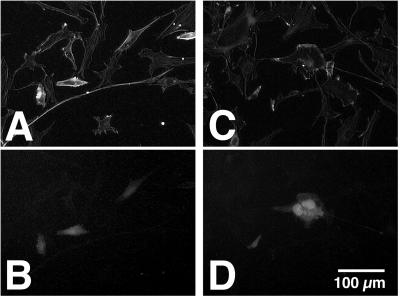
Effects of expressing an N-terminal fragment of PMT in NIH 3T3 cells. Shown are representative fluorescence micrographs of transfected NIH 3T3 cells expressing the N-terminal 521 amino acids of PMT and GFP (A and B) or GFP alone (C and D). Cells were fixed and stained with TRITC-phalloidin and visualized for actin (A and C) or GFP (B and D).
RESULTS
Construction of N-terminal and C-terminal deletion mutants of PMT.
To define the specific region of PMT responsible for activation of the intracellular Gqα-PLCβ1-IP3 signaling pathway, we cloned the toxA gene, encoding the full-length PMT protein, and designed a series of N- and C-terminal deletion mutants (Fig. 1). Expression plasmids containing the entire toxA gene or various deletion fragments thereof were constructed by using PCR and conventional cloning techniques. A plasmid containing the full-length toxA gene, pTHC-toxA, was generated by splicing three overlapping toxA fragments from cloning vectors containing the corresponding PCR products, pET15b-toxAN1, pCR3-Uni(1&3), and pCR3-Uni(4&5). The N- and C-terminal deletion mutants were obtained from regions of the toxA gene encoding the N- and C-terminal portions of the PMT protein. The deletion mutants were designed such that all regions of the entire protein would be included in at least one of the deletion fragments. The ToxAN4 fragment containing the N-terminal 568 amino acids of PMT included the region showing homology with the CNFs. Sequentially shorter N-terminal fragments, which could be used to further define the activity domain, were also designed; these included the N-terminal 505, 450, 400, 300, 293, and 93 amino acids of PMT (ToxAN2, ToxAN5, ToxAN6, ToxAN7, ToxAN1, and ToxAN3, respectively). The two C-terminal fragments included the C-terminal 780 amino acids of PMT (ToxAC1) and the C-terminal 226 amino acids (ToxAC2). The various toxA deletion fragments were subcloned into E. coli expression vectors which incorporated a His tag affinity handle at either the N- or C terminus of the protein, as indicated in Fig. 1.
Expression, purification, and quantitation of rPMT and PMT deletion mutant proteins.
We expressed the recombinant toxin proteins under IPTG-inducible promoters in E. coli as His-tagged fusion peptides for ease in purification. The His tag allowed for essentially one-step purification of the proteins, thereby reducing the amount of sample handling and decreasing the likelihood of protein denaturation or inactivation. During the course of our initial studies, we observed that the His tag placed at the N terminus of rPMT interfered with the activity of the protein in the Xenopus oocyte assay system, as well as in tissue culture assays (data not shown). Hence, following Ni2+ chelate chromatography, we opted to remove the His tag by thrombin cleavage from all of the N-terminal fragments having His tag at their N-termini, including rPMT, ToxAN1, ToxAN2, and ToxAN3. The ToxAN1 protein with a His tag at the N terminus was difficult to purify (Fig. 2) and was not stable to subsequent thrombin treatment, and we could not obtain quantities adequate to assay for activity. ToxAN7 was similar in length to ToxAN1. The His tag was placed at the C termini of ToxAN4, ToxAN5, ToxAN6, and ToxAN7. The C-terminal His tag was not removed from these recombinant proteins. For ToxAC1 and ToxAC2, the N-terminal His tag was placed at the N terminus but was not removed.
The degree of purification achieved by use of the His tag affinity handle varied somewhat among the proteins. Representative Coomassie-stained SDS-polyacrylamide gels and Western blots of each of the toxin samples used in this study are shown in Fig. 2. Because of the differences in sample purity, we chose to quantitate the protein concentration of each toxin sample by Western blot analysis, comparing the sample against a standard curve generated from purified His-tagged rPMT or ToxAN4 of known concentration.
Comparison of rPMT with native PMT.
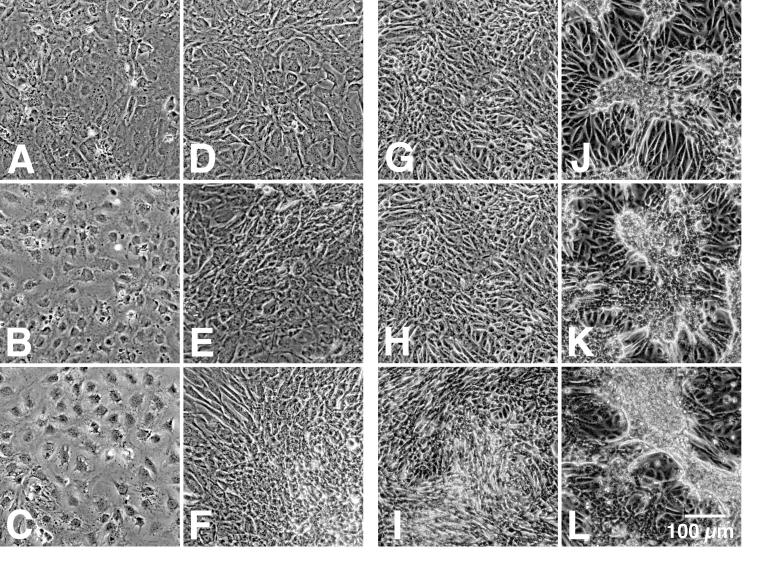
Figure 3 shows a silver-stained SDS-polyacrylamide gel of rPMT before and after thrombin cleavage in comparison to native PMT from two different sources. Anti-toxA28–42 and anti-toxA1239–1253 were reactive with recombinant toxin samples in Western blots (Fig. 2). Native and recombinant toxins gave comparable responses in oocyte experiments (Fig. 4A and B), although the highly purified PMT and rPMT samples were at least 20-fold more active than the commercial native PMT sample. However, after determining the concentrations of the impure native PMT samples by analyzing silver-stained SDS-polyacrylamide gels, using purified rPMT as a standard, we found that the commercial native PMT was as active as rPMT. When the EC50s were adjusted accordingly, the native and recombinant toxins exhibited similar responses in oocytes, with EC50s of 5.2 and 3.6 fmol/oocyte, respectively (Fig. 5A and B). Anti-toxA28–42, but not anti-toxA1239–1253, specifically inhibited the activities of both native and recombinant toxin samples in oocyte experiments (reference 43 and data not shown). As depicted in Fig. 6, rPMT caused dramatic morphological effects on confluent, quiescent Vero cells, showing focus formation surrounded by enlarged cells, as well as proliferative effects on confluent, quiescent Swiss 3T3 cells. Native PMT caused similar effects (data not shown), although the rPMT sample consistently appeared to be more potent than the commercial native PMT sample. The results from these studies are summarized in Table 1.
FIG. 4.
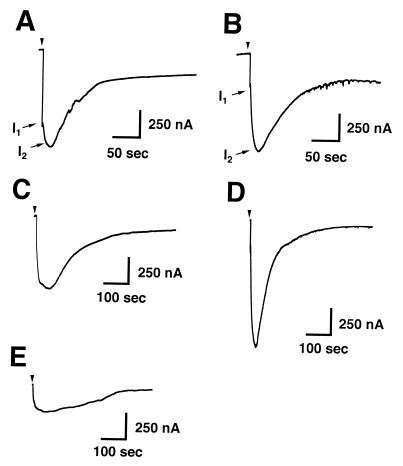
Toxin-induced Ca2+-dependent Cl− currents in Xenopus oocytes. Shown are representative traces of strong responses elicited in voltage-clamped oocytes injected with native PMT (2 fmol; A), rPMT (21 fmol; B), ToxAN4 (330 fmol; C), ToxAN2 (520 fmol; D), and ToxAN5 (1,320 fmol; E). I1 denotes the first peak current from mobilization of intracellular Ca2+; I2 denotes the second peak current from Ca2+ influx through capacitative Ca2+ channels on the plasma membrane.
FIG. 6.
Effects of recombinant PMT on confluent, quiescent Swiss 3T3 and Vero cells. Shown are representative phase-contrast micrographs of confluent, quiescent Swiss 3T3 (left-hand pairs of panels) and Vero cells (right-hand pairs of panels) treated as described in Materials and Methods without (A to C and G to I) or with (D to F and J to L) rPMT (200 ng/well) for 18 h (A, D, G, and J), 96 h (B, E, H, and K), or 144 h (C, F, I, and L).
TABLE 1.
Deletion mutational analysis of PMT activity
| Toxin protein | PMT residues | Calculated mol wt | Activity
|
EC50 (fmol/oocyte) | |
|---|---|---|---|---|---|
| Vero cell assay | Oocyte assay | ||||
| Native PMT | 1–1285 | 146,391 | Yes | Yes | 5.2 |
| rPMT | 1–1285 | 146,673 | Yesa | Yesa | 3.6 |
| ToxAN3 | 1–93 | 11,028 | Noa | Noa | |
| ToxAN1 | 1–293 | 34,576 | NTb | NTb | |
| ToxAN2 | 1–505 | 60,464 | Noa | Yesa | 140 |
| ToxAN4 | 1–568 | 67,077 | No | Yes | 22 |
| ToxAN5 | 1–450 | 53,329 | No | Yes | NDc |
| ToxAN6 | 1–400 | 48,176 | No | No | |
| ToxAN7 | 1–300 | 36,438 | No | No | |
| ToxAC1 | 505–1285 | 92,370 | No | No | |
| ToxAC2 | 1059–1285 | 29,355 | No | No | |
Tested after removal of the His tag.
NT, not tested (not stable to thrombin treatment).
ND, not determined (only weak responses observed with injection of up to 3 pmol/oocyte).
Activities of N- and C-terminal deletion fragments of PMT.
The C-terminal PMT fragments, ToxAC1 and ToxAC2, and the smaller N-terminal fragments, ToxAN3, ToxAN6, and ToxAN7, exhibited no detectable responses in the oocyte assay at concentrations as high as 1 pmol/oocyte. In contrast, ToxAN4, consisting of the N-terminal 568 amino acids, gave reproducible responses in oocytes (Fig. 4C) with an EC50 of 22 fmol/oocyte (Fig. 5C), compared to 3.6 fmol/oocyte for the full-length rPMT. ToxAN2, consisting of the N-terminal 505 amino acids, also showed a reproducible activity (Fig. 4D), with an EC50 of 140 fmol/oocyte (Fig. 5D), about 40-fold lower than that for full-length rPMT. ToxAN5, consisting of the N-terminal 450 amino acids, gave detectable responses in oocytes (Fig. 4E), but the responses were so weak that it was not possible to accurately determine an EC50 for this mutant protein with injection of as much as 3 pmol. Unlike the full-length rPMT, none of the N- or C-terminal deletion mutants exhibited an effect on cultured Vero cells (data not shown). The results from these studies are summarized in Table 1.
To determine whether the N terminus of PMT could also trigger downstream signaling events in mammalian fibroblasts, leading to changes in cellular morphology, a mammalian expression plasmid containing the N-terminal 521 amino acids of PMT under the cytomegalovirus promoter was cotransfected into NIH 3T3 cells along with a reporter plasmid containing GFP under the same promoter. In vitro translation assays using pCR3-Uni(1&3) were also performed to ensure that the construct was indeed capable of expressing truncated toxin protein (data not shown). We found that cells cotransfected with both plasmids underwent significant morphological responses compared to control cells transfected with reporter plasmid alone. As shown in Fig. 7, control cells expressing only the GFP protein have a characteristic spread-out fibroblast morphology, whereas cells expressing the N-terminal fragment of PMT and GFP have a more retracted and spindly appearance, with evidence of more intense TRITC-phalloidin staining.
DISCUSSION
Evidence from previous studies suggests that the mode of action of PMT involves binding to cell surface receptors and internalization via receptor-mediated endocytosis (34, 36), followed by activation of various signaling pathways (29, 30, 38–40, 43), leading eventually to morphological and proliferative events (9, 17, 18, 21, 24, 29, 36). In our earlier studies, we demonstrated that intact PMT protein could directly activate the Gqα-PLCβ1-IP3 signaling pathway in Xenopus oocytes (43). Using this system, we bypassed the cell-binding and internalization process by direct microinjection of the toxin, apparently without the requirement for further intracellular processing or activation. In the present work, we report the cloning, expression, and purification of fully active recombinant PMT from E. coli. This full-length recombinant protein has an activity in oocytes similar to that of the native toxin isolated from P. multocida. We have also shown that rPMT has an effect on cultured Vero cells similar to that of the native toxin.
We have demonstrated that a recombinant deletion mutant consisting of the N-terminal 568 amino acids of PMT could elicit a PMT-like response in oocytes, while two mutants consisting of the C-terminal 780 and 226 amino acids gave no detectable response at higher doses. Weaker responses were also observed with mutants consisting of the N-terminal 505 and 450 amino acids but not with shorter N-terminal fragments. We thus conclude that the functional domain responsible for activating the intracellular Gqα-PLCβ1-IP3 signaling pathway in Xenopus oocytes is localized to the N terminus of PMT.
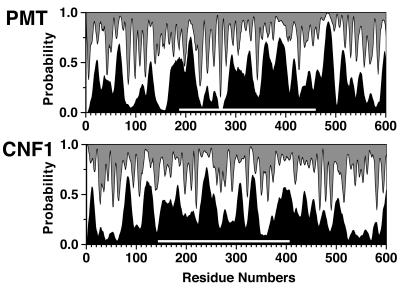
The N-terminal 500 residues of PMT have 24 to 27% homology to the N-terminal 500 residues of CNF1 and CNF2 (10, 31). As can be seen from the area plot shown in Fig. 8, the predicted secondary structures of PMT and CNF1 also have limited similarity in this region, particularly in the region with 30% homology between residues 200 and 450 of PMT. We have shown that deletion mutations made within this conserved region resulted in loss of intracellular activity. With localization of the intracellular activity domain to the N-terminal 500 amino acid residues of PMT, the sequence and structural similarity in the N-terminal region between the two toxins suggests that CNF1 and CNF2 may have an intracellular activity similar to that of PMT. However, there are sufficient differences between the toxins within this region, as indicated in the secondary structure prediction profiles (Fig. 8), for it to be feasible that they have different intracellular activities and target preferences. CNF1 and CNF2 possess deamidase activity on RhoA (11, 19, 31, 37). The deamidase activity domain of CNF1 has been reported to be located in the C-terminal 300 residues (27), which shares about 20% homology with the C-terminal 300 residues of DNT but has no discernible sequence similarity with PMT. It is possible that the N-terminal region of CNF1 with homology to the N-terminal region of PMT possesses an intracellular activity different from the activity associated with the C-terminal 300 residues of CNF1.
FIG. 8.
Area plot of secondary structure prediction for PMT and CNF1. Shadings represent the probabilities for helix (black), for extended or beta strand (gray), and for the remainder (white). White bars indicates the region with 30% sequence identity between the two proteins.
In our previous study (43), we showed that antibodies specific for the N terminus of PMT blocked the PMT-induced response in oocytes, but antibodies against the C terminus did not. The N-terminal toxin fragments ToxAN4 and ToxAN2 are able to elicit responses in oocytes similar to those elicited by full-length rPMT. Antibodies to the N terminus of PMT also blocked the ToxAN4-induced response in oocytes, yet ToxAN4 had no effect on cultured Vero cells. Moreover, none of the deletion mutants exhibited an effect on Vero cells. This is consistent with earlier findings that deletion mutants lacking N-terminal (Δ28-149 and Δ175-247) or C-terminal (Δ1130-1285) portions of the PMT protein had no toxic activity in assays for cytopathicity on embryonic bovine lung (EBL) cells, dermonecrosis in guinea pigs, or mouse lethality (33). Previous studies also showed that monoclonal antibodies recognizing an epitope in the C-terminal 155 amino acids of PMT blocked the cytopathic effect of PMT on EBL cells (13, 33). The C-terminal fragments ToxAC1 and ToxAC2 were inactive in the oocyte assay and had no effect on cultured Vero cells, thus supporting the notion that only the N terminus of PMT has intracellular activity and the C terminus of PMT is required for toxin entry into cells. Further study is required to determine the precise biochemical activity of PMT that induces the Gqα-PLCβ1-IP3 signaling pathway and to understand how this PMT-associated activity and the CNF-associated deamidase activity could result in the observed changes in cells caused by these toxins.
ACKNOWLEDGMENTS
This work was supported by grant AI38396 from the National Institutes of Health/NIAID (to B.A.W.).
We thank Xinjun Zhu, Brooke Murphy, Tina Caserta, April Buehler, James Burgett, and Jennifer Williams for technical assistance. We especially thank Adrian Corbett for helpful discussions and generous assistance in setting up the electrophysiological equipment, Hangjun Zhan for helpful discussions and assistance in the initial cloning stages of this project, and Steven Berberich for assistance with in vitro translation assays. We are grateful to Clarence Chrisp for generously providing purified samples of native PMT.
REFERENCES
- 1.Ackermann M R, DeBey M C, Register K B, Larson D J, Kinyon J M. Tonsil and turbinate colonization by toxigenic and nontoxigenic strains of Pasteurella multocida in conventionally raised swine. J Vet Diagn Invest. 1994;6:375–377. doi: 10.1177/104063879400600318. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Barbour E K, Nabbut N H, Hamadeh S K, Al-Nakhli H M. Bacterial identity and characteristics in healthy and unhealthy respiratory tracts of sheep and calves. Vet Res Commun. 1997;21:421–430. doi: 10.1023/a:1005855318665. [DOI] [PubMed] [Google Scholar]
- 4.Biberstein, E. L. 1990. Our understanding of the Pasteurellaceae. Can. J. Vet. Res. 54(Suppl.):S78–S82. [PubMed]
- 5.Biberstein E L, Jang S S, Kass P H, Hirsh D C. Distribution of indole-producing urease-negative pasteurellas in animals. J Vet Diagn Invest. 1991;3:319–323. doi: 10.1177/104063879100300408. [DOI] [PubMed] [Google Scholar]
- 6.Bisgaard M. Ecology and significance of Pasteurellaceae in animals. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;279:7–26. doi: 10.1016/s0934-8840(11)80487-1. [DOI] [PubMed] [Google Scholar]
- 7.Buys W E, Smith H E, Kamps A M, Kamp E M, Smits M A. Sequence of the dermonecrotic toxin of Pasteurella multocida ssp. multocida. Nucleic Acids Res. 1990;18:2815–2816. doi: 10.1093/nar/18.9.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chrisp C E, Foged N T. Induction of pneumonia in rabbits by use of a purified protein toxin from Pasteurella multocida. Am J Vet Res. 1991;52:56–61. [PubMed] [Google Scholar]
- 9.Dudet L I, Chailler P, Dubreuil J D, Martineau-Doize B. Pasteurella multocida toxin stimulates mitogenesis and cytoskeleton reorganization in Swiss 3T3 fibroblasts. J Cell Physiol. 1996;168:173–182. doi: 10.1002/(SICI)1097-4652(199607)168:1<173::AID-JCP21>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun. 1993;61:4909–4914. doi: 10.1128/iai.61.11.4909-4914.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 12.Foged, N. T. 1992. Pasteurella multocida toxin. The characterization of the toxin and its significance in the diagnosis and prevention of progressive atrophic rhinitis in pigs. Acta Pathol. Microbiol. Immunol. Scand. 100(Suppl. 25):1–56. [PubMed]
- 13.Foged N T. Quantitation and purification of the Pasteurella multocida toxin by using monoclonal antibodies. Infect Immun. 1988;56:1901–1906. doi: 10.1128/iai.56.8.1901-1906.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frymus T, Bielecki W, Jakubowski T. Toxigenic Pasteurella multocida in rabbits with naturally occurring atrophic rhinitis. Zentrbl Veterinaermed Reihe B. 1991;38:265–268. doi: 10.1111/j.1439-0450.1991.tb00870.x. [DOI] [PubMed] [Google Scholar]
- 15.Garcia V F. Animal bites and Pasteurella infections. Pediatr Rev. 1997;18:127–130. doi: 10.1542/pir.18-4-127. [DOI] [PubMed] [Google Scholar]
- 16.Griego R D, Rosen T, Orengo I F, Wolf J E. Dog, cat, and human bites: a review. J Am Acad Dermatol. 1995;33:1019–1029. doi: 10.1016/0190-9622(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 17.Gwaltney S M, Galvin R J, Register K B, Rimler R B, Ackermann M R. Effects of Pasteurella multocida toxin on porcine bone marrow cell differentiation into osteoclasts and osteoblasts. Vet Pathol. 1997;34:421–430. doi: 10.1177/030098589703400506. [DOI] [PubMed] [Google Scholar]
- 18.Higgins T E, Murphy A C, Staddon J M, Lax A J, Rozengurt E. Pasteurella multocida toxin is a potent inducer of anchorage-independent cell growth. Proc Natl Acad Sci USA. 1992;89:4240–4244. doi: 10.1073/pnas.89.10.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horiguchi Y, Inoue N, Masuda M, Kashimoto T, Katahira J, Sugimoto N, Matsuda M. Bordetella bronchiseptica dermonecrotizing toxin induces reorganization of actin stress fibers through deamidation of Gln-63 of the GTP-binding protein Rho. Proc Natl Acad Sci USA. 1997;94:11623–11626. doi: 10.1073/pnas.94.21.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbert W T, Rosen M N. Pasteurella multocida infection due to animal bites. Am J Public Health. 1970;60:1103–1108. doi: 10.2105/ajph.60.6.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jutras I, Martineau-Doize B. Stimulation of osteoclast-like cell formation by Pasteurella multocida toxin from hemopoietic progenitor cells in mouse bone marrow cultures. Can J Vet Res. 1996;60:34–39. [PMC free article] [PubMed] [Google Scholar]
- 22.Kamps A M, Kamp E M, Smits M A. Cloning and expression of the dermonecrotic toxin gene of Pasteurella multocida ssp. multocida in Escherichia coli. FEMS Microbiol Lett. 1990;55:187–190. doi: 10.1016/0378-1097(90)90192-s. [DOI] [PubMed] [Google Scholar]
- 23.Klein N C, Cunha B A. Pasteurella multocida pneumonia. Semin Respir Infect. 1997;12:54–56. [PubMed] [Google Scholar]
- 24.Lacerda H M, Lax A J, Rozengurt E. Pasteurella multocida toxin, a potent intracellularly acting mitogen, induces p125FAK and paxillin tyrosine phosphorylation, actin stress fiber formation, and focal contact assembly in Swiss 3T3 cells. J Biol Chem. 1996;271:439–445. doi: 10.1074/jbc.271.1.439. [DOI] [PubMed] [Google Scholar]
- 25.Lacerda H M, Pullinger G D, Lax A J, Rozengurt E. Cytotoxic necrotizing factor 1 from Escherichia coli and dermonecrotic toxin from Bordetella bronchiseptica induce p21(rho)-dependent tyrosine phosphorylation of focal adhesion kinase and paxillin in Swiss 3T3 cells. J Biol Chem. 1997;272:9587–9596. doi: 10.1074/jbc.272.14.9587. [DOI] [PubMed] [Google Scholar]
- 26.Lax A J, Chanter N. Cloning of the toxin gene from Pasteurella multocida and its role in atrophic rhinitis. J Gen Microbiol. 1990;136:81–87. doi: 10.1099/00221287-136-1-81. [DOI] [PubMed] [Google Scholar]
- 27.Lemichez E, Flatau G, Bruzzone M, Boquet P, Gauthier M. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol Microbiol. 1997;24:1061–1070. doi: 10.1046/j.1365-2958.1997.4151781.x. [DOI] [PubMed] [Google Scholar]
- 28.Morris J T, McAllister C K. Bacteremia due to Pasteurella multocida. South Med J. 1992;85:442–443. doi: 10.1097/00007611-199204000-00027. [DOI] [PubMed] [Google Scholar]
- 29.Mullan P B, Lax A J. Pasteurella multocida toxin is a mitogen for bone cells in primary culture. Infect Immun. 1996;64:959–965. doi: 10.1128/iai.64.3.959-965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy A C, Rozengurt E. Pasteurella multocida toxin selectively facilitates phosphatidylinositol 4,5-bisphosphate hydrolysis by bombesin, vasopressin, and endothelin. Requirement for a functional G protein. J Biol Chem. 1992;267:25296–25303. [PubMed] [Google Scholar]
- 31.Oswald E, Sugai M, Labigne A, Wu H C, Fiorentini C, Boquet P, O’Brien A D. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia coli modifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc Natl Acad Sci USA. 1994;91:3814–3818. doi: 10.1073/pnas.91.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen S K. The complete nucleotide sequence of the Pasteurella multocida toxin gene and evidence for a transcriptional repressor, TxaR. Mol Microbiol. 1990;4:821–830. doi: 10.1111/j.1365-2958.1990.tb00652.x. [DOI] [PubMed] [Google Scholar]
- 33.Petersen S K, Foged N T, Bording A, Nielsen J P, Riemann H K, Frandsen P L. Recombinant derivatives of Pasteurella multocida toxin: candidates for a vaccine against progressive atrophic rhinitis. Infect Immun. 1991;59:1387–1393. doi: 10.1128/iai.59.4.1387-1393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettit R K, Ackermann M R, Rimler R B. Receptor-mediated binding of Pasteurella multocida dermonecrotic toxin to canine osteosarcoma and monkey kidney (Vero) cells. Lab Investig. 1993;69:94–100. [PubMed] [Google Scholar]
- 35.Raffi F, Barrier J, Baron D, Drugeon H B, Nicolas F, Courtieu A L. Pasteurella multocida bacteremia: report of thirteen cases over twelve years and review of the literature. Scand J Infect Dis. 1987;19:385–393. doi: 10.3109/00365548709021670. [DOI] [PubMed] [Google Scholar]
- 36.Rozengurt E, Higgins T, Chanter N, Lax A J, Staddon J M. Pasteurella multocida toxin: potent mitogen for cultured fibroblasts. Proc Natl Acad Sci USA. 1990;87:123–127. doi: 10.1073/pnas.87.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 38.Staddon J M, Barker C J, Murphy A C, Chanter N, Lax A J, Michell R H, Rozengurt E. Pasteurella multocida toxin, a potent mitogen, increases inositol 1,4,5-trisphosphate and mobilizes Ca2+ in Swiss 3T3 cells. J Biol Chem. 1991;266:4840–4847. [PubMed] [Google Scholar]
- 39.Staddon J M, Bouzyk M M, Rozengurt E. Interconversion of GRP78/BiP. A novel event in the action of Pasteurella multocida toxin, bombesin, and platelet-derived growth factor. J Biol Chem. 1992;267:25239–25245. [PubMed] [Google Scholar]
- 40.Staddon J M, Chanter N, Lax A J, Higgins T E, Rozengurt E. Pasteurella multocida toxin, a potent mitogen, stimulates protein kinase C-dependent and -independent protein phosphorylation in Swiss 3T3 cells. J Biol Chem. 1990;265:11841–11848. [PubMed] [Google Scholar]
- 41.Waldor M, Roberts D, Kazanjian P. In utero infection due to Pasteurella multocida in the first trimester of pregnancy: case report and review. Clin Infect Dis. 1992;14:497–500. doi: 10.1093/clinids/14.2.497. [DOI] [PubMed] [Google Scholar]
- 42.Walker K E, Weiss A A. Characterization of the dermonecrotic toxin in members of the genus Bordetella. Infect Immun. 1994;62:3817–3828. doi: 10.1128/iai.62.9.3817-3828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson B A, Zhu X, Ho M, Lu L. Pasteurella multocida toxin activates the inositol triphosphate signaling pathway in Xenopus oocytes via Gqα-coupled phospholipase C-β1. J Biol Chem. 1997;272:1268–1275. doi: 10.1074/jbc.272.2.1268. [DOI] [PubMed] [Google Scholar]