Abstract
Background
Bilateral breast cancer (BBC), as well as ovarian cancer, are significantly associated with germline deleterious variants in BRCA1/2, while BRCA1/2 germline deleterious variants carriers can exquisitely benefit from poly (ADP-ribose) polymerase (PARP) inhibitors. However, formal genetic testing could not be carried out for all patients due to extensive use of healthcare resources, which in turn results in high medical costs. To date, existing BRCA1/2 deleterious variants prediction models have been developed in women of European or other descent who are quite genetically different from Asian population. Therefore, there is an urgent clinical need for tools to predict the frequency of BRCA1/2 deleterious variants in Asian BBC patients balancing the increased demand for and cost of cancer genetics services.
Methods
The entire coding region of BRCA1/2 was screened for the presence of germline deleterious variants by the next generation sequencing in 123 Chinese BBC patients. Chi-square test, univariate and multivariate logistic regression were used to assess the relationship between BRCA1/2 germline deleterious variants and clinicopathological characteristics. The R software was utilized to develop artificial neural network (ANN) and nomogram modeling for BRCA1/2 germline deleterious variants prediction.
Results
Among 123 BBC patients, we identified a total of 20 deleterious variants in BRCA1 (8; 6.5%) and BRCA2 (12; 9.8%). c.5485del in BRCA1 is novel frameshift deleterious variant. Deleterious variants carriers were younger at first diagnosis (P = 0.0003), with longer interval between two tumors (P = 0.015), at least one medullary carcinoma (P = 0.001), and more likely to be hormone receptor negative (P = 0.006) and HER2 negative (P = 0.001). Area under the receiver operating characteristic curve was 0.903 in ANN and 0.828 in nomogram modeling individually (P = 0.02).
Conclusion
This study shows the spectrum of the BRCA1/2 germline deleterious variants in Chinese BBC patients and indicates that the ANN can accurately predict BRCA deleterious variants than conventional statistical linear approach, which confirms the BRCA1/2 deleterious variants carriers at the lowest costs without adding any additional examinations.
Supplementary information
The online version contains supplementary material available at 10.1186/s12885-022-10160-y.
Keywords: Bilateral breast cancer, BRCA1, BRCA2, Germline deleterious variant, Artificial neural network
Background
Bilateral breast cancer (BBC), categorized as synchronous and metachronous disease, is observed in 2-11% of breast cancer cases [1, 2]. Patients with breast cancer have a 2-20% chance of developing a contralateral breast cancer (CBC), either synchronously detected, or as a metachronous cancer [3]. The increased number of breast cancer cases and improved survival after the first BC diagnosis contribute to the current higher incidence of BBC. Several factors are thought to be associated with the occurrence and development of bilateral breast cancer, such as early age at diagnosis, histology, family history, and especially the presence of germline deleterious variants that include BRCA1/2, PALB2, CDH1, and CHEK2. Studies have reported that the prognosis of BBC patients is similar or worse than unilateral breast cancer (UBC) patients [4–7].
BRCA1 and BRCA2 genes are involved in homologous recombination repair. Germline BRCA1 and BRCA2 loss-of-function variants predispose to development of breast cancer. Previous research has demonstrated that the BRCA1/2 deleterious variant frequency in BBC (29.6%) is significantly higher than the rate in unselected breast cancer (5.4%) [8, 9]. Of note, patients with bilateral breast cancer are suggested by National Comprehensive Cancer Network (NCCN) Guidelines to undergo further genetic risk evaluation [10]. Identifying BRCA1/2 deleterious variants carriers could not only shed light on adjusting chemotherapy schemes, but also contribute to the prevention of ovarian cancer and offspring onset. Secondary analyses of the GeparOcto and GeparSixto Randomized Clinical Trial have revealed higher pathological complete response (pCR) rates in BRCA1 and BRCA2 deleterious variants carriers [11, 12].
However, in low- and middle- income countries, only a small number of studies that included Chinese bilateral breast cancer patients have been reported and related clinical characteristics have not always been well clarified. Thus, taking uncovered health insurance together, Chinese physicians are always cautious over recommending expensive genetic risk evaluation. Many models have been developed for predicting the likelihood of carrying germline BRCA deleterious variants using data of patients with breast cancer. However, these models underestimate BRCA1/2 deleterious variants carriers and cannot distinguish well between carriers and non-carriers in Asian breast cancer patients [13]. What’s more, although BBC status has been considered as an indicator of BRCA deleterious variants in Asian populations, models are built on data from breast cancer patients, instead of the BBC population [14–16].
In this study, we performed the next generation sequencing for all exons of BRCA1 and BRCA2 in 123 Chinese BBC patients to analyze the relationship between BRCA1/2 germline deleterious variants and characteristics of BBC. We aimed to construct a user-friendly model to predict the risk of BRCA1/2 deleterious variants in Chinese BBC patients.
Methods
Study patients
We conducted a retrospective study of patients diagnosed with BBC who were treated in the Fujian Medical University Union Hospital from 2005 to 2021. In this study BBC was classified as metachronous bilateral breast cancer (MBBC) and synchronous bilateral breast cancer (SBBC), with an interval between the first and contralateral breast cancer of ≥ 2 years and < 2 years, respectively. All patients were histopathologically confirmed by at least two pathologists. Patients with metastasis that occurred before or at the same time as bilateral breast cancer were excluded. Clinicopathological characteristics were obtained after informed consent that included menstrual history, reproductive history, lactation history, family history, ER (estrogen receptor), PR (progesterone receptor), HER2 (human epidermal growth factor receptor 2), and chemotherapy or radiation status. About 5ml of peripheral venous blood was collected individually.
DNA extraction and sequencing
The genomic DNA was isolated from peripheral blood lymphocytes using Large amount of whole blood genomic DNA extraction kits (DP2202, Bioteke, China) according to the manufacturer’s instructions. DNA purity and concentration were assessed by the NanoDrop2000 spectrophotometer (Thermo Fisher Scientific) and DNA quality was assessed by agarose gel electrophoresis.
Library preparation and sequencing of all coding regions and exon-intron boundaries of the BRCA1 and BRCA2 genes were performed through next generation sequencing (Illumina Novaseq) by shanghai aita gene technology Co.Ltd with Human BRCA1/BRCA2 Gene Mutation Detection Kit. The libraries were quantified by Qubit™ dsDNA HS Assay Kit (Invitrogen) and the size and quantity of the captured library were assessed by Bioanalyzer 2100 instrument (Agilent). The sequencing results were then aligned to the BRCA1 (NM_007294.3) and BRCA2 (NM_000059.3) reference sequences for mutation detection using the Burrows-Wheeler Alignment tool, which further recalibrated by the Genome Analysis Toolkit (GATK) and annotated by ANNOVAR (http://www.openbioinformatics.org/annovar/). Classification of variants was performed according to ACMG criteria [17]. Benign variant and variants of uncertain significance were excluded in our study. All the pathogenic mutations detected by next generation sequencing assay were validated via Sanger sequencing on the ABI 3730XL platform (Life Technologies), which was described in our previous study [18].
Statistical analysis
The chi-square tests or Fisher’s exact tests for categorical variables and the Mann-Whitney U tests for continuous variables were used to analyze the differences in clinicopathological characteristics between deleterious variants carriers and non-carriers. Univariate and multivariate logistic regression analysis were utilized to assess the association between clinicopathological characteristics and carrying a deleterious variants. The nomogram and artificial neural network algorithms were conducted using the R library, rms (6.3-0) and RSNNS (0.4–14) package, individually [19]. To evaluate the feasibility and performance of the artificial neural network, a conventional multivariate logistic regression model was also constructed for comparison. The logistic regression model was developed using factors selected by univariate analysis as well. The predictive accuracy of the two models were estimated by receiver operating characteristic (ROC). Comparison of ROC curves was carried out using the method described by DeLong et al. [20]. SAS software, version 9.4 (SAS Institute), IBM SPSS Statistics 22.0 software (IBM Corporation) and the R version 4.1 software (The R Foundation for Statistical Computing) were used in this study [21–23]. A P value of < 0.05 was adopted as statistical significance.
Results
Clinicopathological characteristics of bilateral breast cancer patients
Among the 123 BBC patients, the clinicopathological characteristics of deleterious variants carriers and non-carriers are summarized in Table 1. When diagnosed as primary breast cancer for the first time, BRCA deleterious variants carriers were younger than non-carriers, median age 41.5 years vs. 48.0 years (P = 0.0003) respectively. We also performed statistical analysis and found that the interval when the proportion of metachronous bilateral breast cancer patients among the BRCA carriers was significantly higher than in the non-carriers from 2 years to 5 years. Furthermore, 45.0% (9/20) of the BRCA deleterious variants carriers were diagnosed with at least one triple negative breast cancer (TNBC) tumor, 15.0% (3/20) with at least one medullary carcinoma, 25.0% (5/20) with both tumors’ hormone receptor negative, 70.0% (14/20) both HER2 negative tumors and 20.0% (4/20) both TNBC, which was higher than that in non-carriers respectively. However, menophania, menopausal status and family history did not differ significantly between patients with and without BRCA1/2 deleterious variants. In brief, deleterious variant carriers were more likely to be hormone receptor negative, HER2 negative breast cancer, or medullary carcinoma positive.
Table 1.
Clinical-pathological characteristics of deleterious variants carriers and non-carriers
| Characteristics | BRCA carriers | Non-carriers | P |
|---|---|---|---|
| Median age at first diagnosis (year) | 41.5 (28–57) | 48.0 (31–85) | 0.0003 |
| Median menophania (year) | 15.0 (12–19) | 15.0 (11–20) | 0.833 |
| Menopausal status | 4/20 (20.0%) | 32/96 (33.3%) | 0.241 |
| Median lactation (month) | 8.5 (0–32) | 10 (0–48) | 0.358 |
| Family history | 4/20 (20.0%) | 12/103 (11.7%) | 0.310 |
| Median interval time (year) | 5.2 (0-17.4) | 2.1 (0-27.8) | 0.393 |
| Interval time ≥ 1y | 16/20 (80.0%) | 57/103 (55.3%) | 0.040 |
| Interval time ≥ 2y | 16/20 (80.0%) | 52/103 (50.5%) | 0.015 |
| Interval time ≥ 3y | 15/20 (75.0%) | 49/103 (47.6%) | 0.025 |
| Interval time ≥ 4y | 13/20 (65.0%) | 36/103 (35.0%) | 0.012 |
| Interval time ≥ 5y | 12/20 (60.0%) | 30/103 (29.1%) | 0.008 |
| Interval time ≥ 6y | 8/20 (40.0%) | 25/103 (24.3%) | 0.146 |
| Interval time ≥ 7y | 6/20 (30.0%) | 21/103 (20.4%) | 0.342 |
| Interval time ≥ 8y | 3/20 (15.0%) | 19/103 (18.4%) | 0.713 |
| Interval time ≥ 9y | 2/20 (10.0%) | 16/103 (15.5%) | 0.522 |
| First tumor size ≥ T2 | 10/18 (55.6%) | 55/94 (58.5%) | 0.816 |
| First tumor nodal status negative | 11/18 (61.1%) | 43/93 (46.2%) | 0.248 |
| Second tumor size ≥ T2 | 6/18 (33.3%) | 38/103 (36.9%) | 0.772 |
| Second tumor nodal status negative | 12/20 (60.0%) | 68/102 (66.7%) | 0.566 |
| Same histologic type in both tumors | 11/18 (61.1%) | 64/89 (71.9%) | 0.361 |
| Same HR status in both tumors | 14/19 (73.7%) | 55/84 (65.5%) | 0.492 |
| Same HER2 status in both tumors | 14/14 (100%) | 41/62 (66.1%) | 0.011 |
| At least one HR (-) tumor | 10/20 (50.0%) | 39/101 (38.6%) | 0.343 |
| At least one HER2 (-) tumor | 19/20 (95.0%) | 82/100 (82.0%) | 0.146 |
| At least one TNBC | 9/20 (45.0%) | 16/100 (16.0%) | 0.004 |
| At least one medullary carcinoma | 3/20 (15.0%) | 1/103 (1.0%) | 0.001 |
| Both HR (-) tumors | 5/20 (25.0%) | 6/103 (5.8%) | 0.006 |
| Both HER2 (-) tumors | 14/20 (70.0%) | 33/103 (32.0%) | 0.001 |
| Both TNBCs | 4/20 (20.0%) | 0/101 (0.0%) | <0.0001 |
The prevalence and spectrum of deleterious variants in BRCA1 and BRCA2 genes
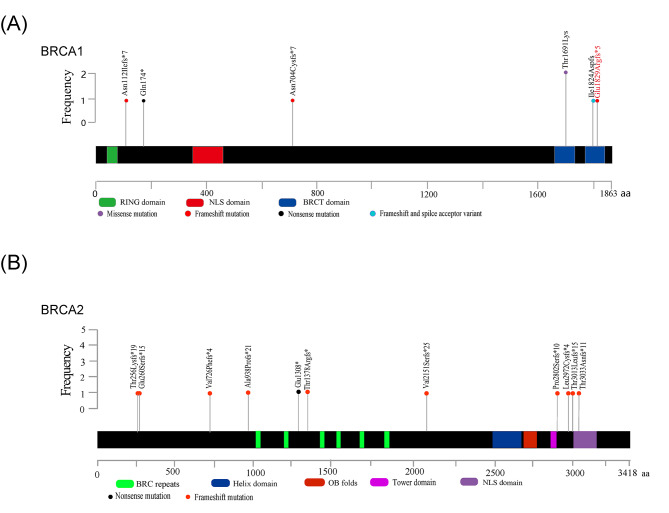
Of all the 123 participants, 20 patients who carried a deleterious variant were identified, eight (6.5%) in the BRCA1 gene and 12 (9.8%) in the BRCA2 gene (Fig. 1; Table 2). All deleterious variants were detected once except for the missense variant (c.5072 C > A) and the frameshift variant (c.9097dupA). As shown in Table 2, a total of 18 deleterious variants are listed (seven in BRCA1 and 11 in BRCA2), including 13 frameshift variants (c.335delA, c.2110_2111delAA, c.5485delG, c.767_768delCA, c.774_775delAA, c.2175delA, c.2808-2811del, c.4133_4136del, c.6448dupA, c.8399_8400insA, c.8915delT, c.9037delA and c.9097dupA), two nonsense variants (c.520 C > T and c.3922G > T), one frameshift variant & splice acceptor variant (c.5470_5477delATTGGGCA), one intron variant (c.213-12 A > G), and one missense variant (c.5072 C > A). Deleterious variants, c.5485delG in BRCA1 and c.8399_8400insA in BRCA2 were not listed in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) or dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP) [24, 25]. However, c.8399_8400insA in BRCA2 was already reported [26]. All sequence variants were shown in supplementary table.
Fig. 1.
Schematic diagram of the spectrum of BRCA1(A) and BRCA2(B) germline mutation detected in our study. Intronic mutation rs80358163 in BRCA1 was not shown in the schematic diagram
Table 2.
Deleterious variants identified in 123 bilateral breast cancer patients of this study
| Gene | Location | dbSNP Id | HGVS.c | HGVS.p | Consequence Type | Variation ID | Significance | No. of observations |
|---|---|---|---|---|---|---|---|---|
| BRCA1 | Intron 4 | rs80358163 | c.213-12 A > G | / | Intron Variant | 37,450 | Pathogenic | 1 |
| BRCA1 | Exon 6 | rs886040119 | c.335del | p.Asn112Ilefs*7 | Frameshift | 266,359 | Pathogenic | 1 |
| BRCA1 | Exon 7 | rs1567806048 | c.520 C > T | p.Gln174* | Nonsense | 575,899 | Pathogenic | 1 |
| BRCA1 | Exon 10 | rs80357814 | c.2110_2111del | p.Asn704Cysfs*7 | Frameshift | 54,462 | Pathogenic | 1 |
| BRCA1 | Exon 16 | rs80357034 | c.5072 C > A | p.Thr1691Lys | Missense | 37,627 | Likely Pathogenic | 2 |
| BRCA1 | Exon 23 | rs80357973 | c.5470_5477del | p.Ile1824Aspfs | Frameshift & splice acceptor variant | 55,591 | Pathogenic | 1 |
| BRCA1 | Exon 23 | Novel | c.5485del | p.Glu1829Argfs*5 | Frameshift | / | Pathogenic | 1 |
| BRCA2 | Exon 9 | rs80359670 | c.767_768del | p.Thr256Lysfs*19 | Frameshift | 52,382 | Pathogenic | 1 |
| BRCA2 | Exon 9 | rs75096777 | c.774_775del | p.Glu260Serfs*15 | Frameshift | 188,425 | Pathogenic | 1 |
| BRCA2 | Exon 11 | rs276174819 | c.2175del | p.Val726Phefs*4 | Frameshift | 926,187 | Pathogenic | 1 |
| BRCA2 | Exon 11 | rs80359351 | c.2808_2811del | p.Ala938Profs*21 | Frameshift | 9,322 | Pathogenic | 1 |
| BRCA2 | Exon 11 | rs80358638 | c.3922G > T | p.Glu1308* | Nonsense | 37,867 | Pathogenic | 1 |
| BRCA2 | Exon 11 | rs80359430 | c.4133_4136del | p.Thr1378Argfs* | Frameshift | 51,602 | Likely Pathogenic | 1 |
| BRCA2 | Exon 11 | rs80359594 | c.6448dup | p.Val2151Serfs*25 | Frameshift | 52,107 | Pathogenic | 1 |
| BRCA2 | Exon 19 | / | c.8399_8400insA | p.Pro2802Serfs*10 | Frameshift | / | Pathogenic | 1 |
| BRCA2 | Exon 22 | rs397508019 | c.8915del | p.Leu2972Cysfs*4 | Frameshift | 52,701 | Pathogenic | 1 |
| BRCA2 | Exon 23 | / | c.9037del | p.Thr3013Leufs*15 | Frameshift | 1,454,156 | Pathogenic | 1 |
| BRCA2 | Exon 23 | rs397507419 | c.9097dup | p.Thr3033Asnfs*11 | Frameshift | 38,208 | Pathogenic | 2 |
Univariate logistic regression analysis for bilateral breast cancer patients
Fifteen clinicopathological characteristics factors were analyzed in a univariate analysis. We show in Table 3 that the deleterious variant status was significantly correlated with six factors including age at the first diagnosis (P = 0.0012), interval between diagnosis of the two tumors (P = 0.0211), at least one TNBC (P = 0.0056), at least one medullary carcinoma (P = 0.0146), both tumors HR negative (P = 0.0114) and both tumors HER2 negative (P = 0.0026). TNBC is one specific type of breast cancer by lacking of the expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2), which exists multicollinearity in HR (-) and HER2 (-). Therefore, the variable “at least one TNBC” was not used in further analysis.
Table 3.
Univariable logistic regression analysis between clinical-pathological characteristics and BRCA1/2 mutations
| Characteristics | OR | 95% CI | P |
|---|---|---|---|
| Age (year) | |||
| > 45 | Reference | ||
| ≤ 45 | 6.088 | 2.042–18.149 | 0.0012 |
| Menophania (year) | |||
| > 13 | Reference | ||
| ≤ 13 | 1.750 | 0.627–4.884 | 0.2853 |
| Menopausal status | |||
| Postmenopausal | Reference | ||
| Pre-menopausal | 0.500 | 0.154–1.619 | 0.2476 |
| Lactation time (month) | |||
| > 10 | Reference | ||
| ≤ 10 | 1.922 | 0.669–5.521 | 0.225 |
| Family history | |||
| negative | Reference | ||
| positive | 1.896 | 0.543–6.618 | 0.3159 |
| Interval time (year) | |||
| < 2 | Reference | ||
| ≥ 2 | 3.922 | 1.228–12.532 | 0.0211 |
| First tumor size | |||
| ≤T2 | Reference | ||
| ≥T2 | 0.886 | 0.321–2.449 | 0.8160 |
| First tumor nodal status | |||
| positive | Reference | ||
| negative | 1.827 | 0.651–5.126 | 0.2521 |
| Second tumor size | |||
| ≤T2 | Reference | ||
| ≥T2 | 0.855 | 0.297–2.465 | 0.772 |
| Second tumor nodal status | |||
| positive | Reference | ||
| negative | 0.750 | 0.280–2.008 | 0.567 |
| Same histologic type in both tumors | |||
| no | Reference | ||
| yes | 0.614 | 0.214–1.762 | 0.3643 |
| Same HR status in both tumor | |||
| no | Reference | ||
| yes | 1.476 | 0.484–4.505 | 0.4939 |
| Same HER2 status in both tumor | |||
| no | Reference | ||
| yes | > 999.999 | < 0.001->999.999 | 0.9534 |
| At least one HR (-) tumor | |||
| no | Reference | ||
| yes | 1.590 | 0.607–4.168 | 0.3457 |
| At least one HER2 (-) tumor | |||
| no | Reference | ||
| yes | 4.171 | 0.524–33.204 | 0.1773 |
| At least one TNBC | |||
| no | Reference | ||
| yes | 4.296 | 1.533–12.038 | 0.0056 |
| At least one medullary carcinoma | |||
| no | Reference | ||
| yes | 18.000 | 1.768-183.287 | 0.0146 |
| Both HR (-) tumor | |||
| no | Reference | ||
| yes | 5.389 | 1.461–19.883 | 0.0114 |
| Both HER2 (-) tumor | |||
| no | Reference | ||
| yes | 4.949 | 1.746–14.030 | 0.0026 |
| Both TNBC | |||
| no | Reference | ||
| yes | > 999.999 | < 0.001->999.999 | 0.9707 |
Model development and comparison for predicting the risk of BRCA deleterious variants in BBC
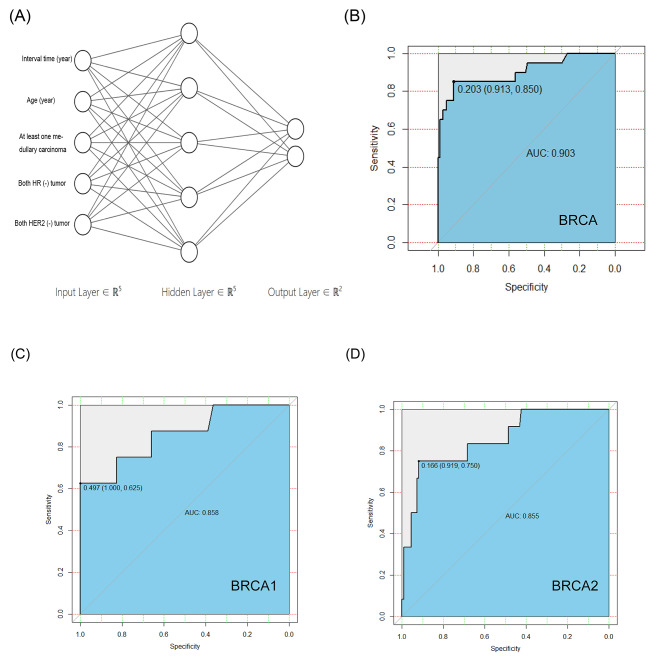
To keep raw data as much as possible, variables identified by univariate analysis were regarded as continuous variables as possible and incorporated to build the artificial neural network model. An input layer composed by five neurons mentioned above and the hidden layer was with 5 neurons (Fig. 2 A). A multivariate analysis was subsequently performed in Table 4 and these predictors were together included to build the logistic regression model.
Fig. 2.
ANN for predicting the risk of BRCA1/2 germline mutation in Chinese bilateral breast cancer patients. (A) Three layers of the ANN model. The accuracy of ANN in BRCA (B), BRCA1(C) and BRCA2(D) germline mutation prediction are assessed by ROC curve
Table 4.
Multivariable logistic regression analysis between clinical histopathological characteristics and BRCA1/2 mutations
| Characteristics | Effect | S.E. | OR | 95% CI | P |
|---|---|---|---|---|---|
| Interval time (year) | -0.0209 | 0.0543 | 0.979 | 0.88–1.089 | 0.6998 |
| Age (year) | -0.1415 | 0.0497 | 0.868 | 0.787–0.957 | 0.0045 |
| At least one medullary carcinoma | 2.6974 | 1.6657 | 14.841 | 0.56704–388.41 | 0.1054 |
| Both HR (-) tumor | 1.2346 | 1.0064 | 3.4369 | 0.47807–24.708 | 0.2199 |
| Both HER2 (-) tumor | 1.7107 | 0.62778 | 5.533 | 1.6166–18.938 | 0.0064 |
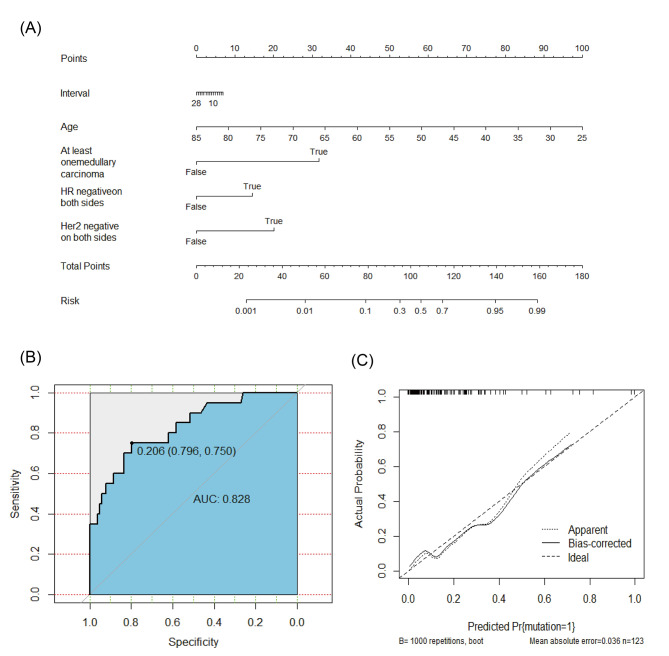
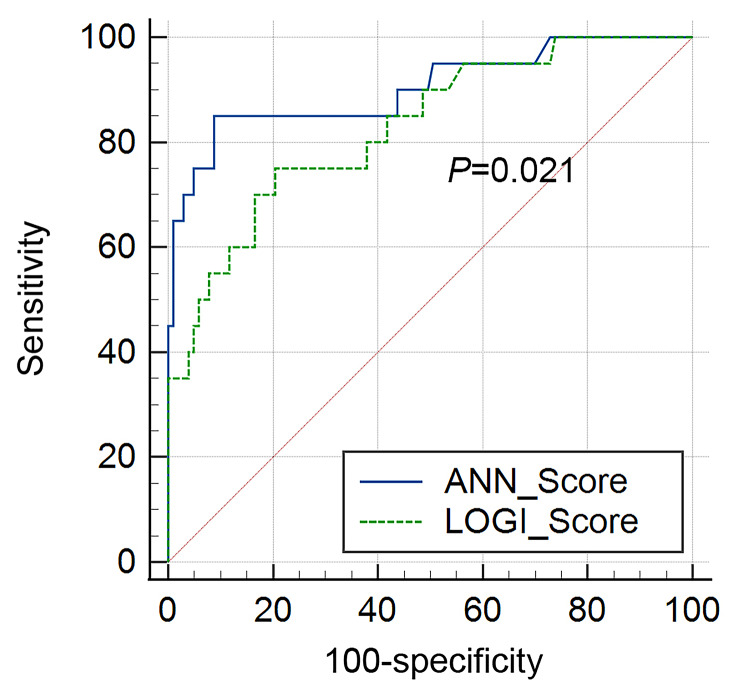
The AUC for the artificial neural network model (Fig. 2B-D) was 0.903 (95% C.I. = 0.836–0.949, 0.858 for BRCA1 and 0.855 for BRCA2), while the AUC for nomogram predicting the risk of BRCA1/2 germline deleterious mutation in Chinese bilateral breast cancer patients was 0.828 (95% C.I. = 0.750–0.890, Fig. 3). Predictive performance of the artificial neural network was superior to that of the logistic regression model (P = 0.021) (Fig. 4). We applied a cutoff of 0.203 for artificial neural network and achieved a sensitivity of 91.3% and a specificity of 85.0%, while logistic regression nomogram model on BRCA1 and BRCA2 separately showed excellent predictive performance in BRCA1 (AUC 0.929) in supplementary Fig. 1 and Fig. 2.
Fig. 3.
Nomogram for predicting the risk of BRCA1/2 germline mutation in Chinese bilateral breast cancer patients. (A) Nomogram model. (B) The accuracy of nomogram in BRCA1/2 germline mutation prediction is assessed by ROC curve, which shows good performance (Area under curve, 0.828). (C) The calibration curve of the nomogram in predicting BRCA1/2 germline mutation
Fig. 4.
Comparison of ROC curve between the artificial neural network and logistic regression models
Discussion
In our study, variables identified in univariate analysis were incorporated within the modeling design for predicting BRCA deleterious variants in BBC patients in China, including age at the first diagnosis, interval between the two tumors, at least one TNBC, at least one medullary carcinoma, both tumors HR negative and both tumors HER2 negative. Our ANN model showed an effective capability for outcome prediction with an AUC of 0.903. Besides, the artificial neural network model is more accurate than the logistic regression model (AUC = 0.828, P = 0.021). These findings show the feasibility and validity for predicting BRCA deleterious variant in BBC patients in China. To our knowledge, this is the first study of the BRCA1/2 deleterious variant spectrum and characteristics in Chinese BBC patients and/or reporting model to predict the risk of deleterious variant status, which showed great potential application value in health economics.
BRCA1/2 deleterious variants were detected in 20 of 123 BBC patients (16.3%) in our study. The frequency was higher than in unselected breast cancer patients (5.4%) [9], while not significantly differed from other BBC reports from China, 16.3% vs. 23.3%, 12.2% and 12.5%, respectively [9, 27, 28]. Likewise, the prevalence of deleterious variants in our study is similar to Korean and Bulgarian BBC patients, but lower than Polish BBC patients [8, 29, 30]. Our study also found that 12 (9.8%) BBC patients carried BRCA2 deleterious variants, higher than eight (6.5%) identified with BRCA1 deleterious variants, which was similar to other Chinese studies that evaluated unselected breast cancer patients [27, 28, 31]. The BRCA1 deleterious variants are more likely to occur in Caucasians, particularly in Latin American and Polish patients [8, 32]. In addition, exon 11 of the BRCA2 gene was the most frequently mutated. This is consistent with the results of previous studies [27, 33]. No damaging variants were located in exon 11 of BRCA1 gene. The most prevalent deleterious variant in our study was BRCA1 c. 5470_5477del in was detected only once and BRCA2 c. 9097dupA was detected twice while they have been frequently reported in Chinese populations [9, 28]. The BRCA1 c.5470_5477del is founder for the Chinese Han population and is associated with poor prognosis [34]. It is very important that our study also identified novel frameshift variant, c.5485delG in BRCA1, which was found in first diagnosed at 28 years old as metachronous bilateral triple negative breast cancer patient with unilateral medullary carcinoma.
Patients with two breast cancer lesions, including bilateral breast cancer, are recommended for further genetic risk evaluation by NCCN Guidelines [10]. There is a multitude of well known BRCA mutation-carrier prediction models like BRCAPRO, Myriad and BOADICEA [35–37]. BRCAPRO is a Bayesian statistical mode based on data from white individuals and Myriad II is an empirical model based on the testing experience of Myriad Genetics Laboratorie, while both of them underpredicted Asian carriers by two-fold and showed less accurate discrimination between Asian carriers and noncarriers. BOADICEA incorporating genetic and nongenetic risk factors is also with the AUC of 0.73 [13–16]. Chinese clinicians face serious difficulties in estimating the probability of patients carrying a deleterious variant for the propose of conducting genetic risk evaluation for those high-risk patients, especially BBC patients. What’s more, in the whole world, few models can perform well in confirming the BRCA1/2 deleterious variant carriers in BBC patients at the lowest cost [38].
ANN have been successfully applied to address a variety of clinical problems, which provides a powerful and accurate predictive method superior to traditional statistical methods [39–41]. Previously, several studies have confirmed the potential of artificial neural network models in predicting gene deleterious variants, including EGFR and BRAF [31, 42, 43]. We collected a variety of easily accessible clinical characteristics and developed easy-to-use tool for decision-making by physicians and patients, which not add any costly and time-consuming upfront testing procedures. Therefore, an ANN model may be widely applied and popularized, especially in developing countries.
Our study also has some limitations. First, the number of our BBC patients is small, and they derived from a single center in China. Therefore, this model is not appropriate for all situations. A larger scale study and further external validation are required to fully determine its universal utility. Second, the model simply provides an assessment of individual risk for BRCA1/2 deleterious variants while specific treatment recommendations are not offered. Future targeted research projects should address these limitations.
Conclusion
In our retrospective study of 123 BBC cases, the spectrum of the BRCA1/2 germline deleterious variant in Chinese BBC patients was well elaborated. This study shows and provides effective ANN modeling for predicting the risk of a BRCA1/2 deleterious variant in BBC patients without adding any additional examinations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the comments of the editor and reviewers that helped to improve the study.
Author contributions
Chuan Wang and Fangmeng Fu conceived and designed the study. Wenzhe Zhang, Shengmei Li, Xuan Jin, Weifeng Cai, Kun Zhang and Yeyuan Huang collected the data. Liuwen Yu, Jialin Hou and Peng Zhou analyzed the data. Minyan Chen, Bangwei Zeng, Wenhui Guo and Yuxiang Lin offer technical assistance. Yan Li, Lili Chen, Jinxing Lv and Xiaobin Chen wrote the manuscript. All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Funding
This work was supported by Joint Funds for the Innovation of Science and Technology, Fujian Province [grant numbers 2018Y9205, 2019Y9103]; Joint Key Funds for the Health and Education of Fujian Province [grant number 2019-WJ-23].
Data Availability
The datasets used and/or analysed during the current study are available in the BioProject repository [PRJNA859288].
Declarations
Ethics approval
The study protocol was approved by ethics committee of Fujian Medical University Union Hospital, written informed consent was obtained from all subjects before their participation. All the procedures performed in studies involving human participants adhere to The Code of Ethics of the World Medical Association (Declaration of Helsinki).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yan Li, Lili Chen, Jinxing Lv and Xiaobin Chen contributed equally to this work.
Contributor Information
Chuan Wang, Email: chuanwang1968@outlook.com.
Fangmeng Fu, Email: ffm@fjmu.edu.cn.
References
- 1.Hartman M, Czene K, Reilly M, et al. Incidence and prognosis of synchronous and metachronous bilateral breast cancer. J Clin Oncol. 2007;25:4210–6. doi: 10.1200/JCO.2006.10.5056. [DOI] [PubMed] [Google Scholar]
- 2.Jobsen JJ, van der Palen J, Ong F, Riemersma S, Struikmans H. Bilateral breast cancer, synchronous and metachronous; differences and outcome. Breast Cancer Res Treat. 2015;153:277–83. doi: 10.1007/s10549-015-3538-5. [DOI] [PubMed] [Google Scholar]
- 3.Alkner S, Bendahl PO, Ferno M, Manjer J, Ryden L. Prediction of outcome after diagnosis of metachronous contralateral breast cancer. BMC Cancer. 2011;11:114. doi: 10.1186/1471-2407-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwentner L, Wolters R, Wischnewsky M, Kreienberg R, Wockel A. Survival of patients with bilateral versus unilateral breast cancer and impact of guideline adherent adjuvant treatment: a multi-centre cohort study of 5292 patients. Breast. 2012;21:171–7. doi: 10.1016/j.breast.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Kheirelseid EA, Jumustafa H, Miller N, et al. Bilateral breast cancer: analysis of incidence, outcome, survival and disease characteristics. Breast Cancer Res Treat. 2011;126:131–40. doi: 10.1007/s10549-010-1057-y. [DOI] [PubMed] [Google Scholar]
- 6.Kuo WH, Yen AM, Lee PH, et al. Cumulative survival in early-onset unilateral and bilateral breast cancer: an analysis of 1907 Taiwanese women. Br J Cancer. 2009;100:563–70. doi: 10.1038/sj.bjc.6604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichol AM, Yerushalmi R, Tyldesley S, et al. A case-match study comparing unilateral with synchronous bilateral breast cancer outcomes. J Clin Oncol. 2011;29:4763–8. doi: 10.1200/JCO.2011.35.0165. [DOI] [PubMed] [Google Scholar]
- 8.Rogozinska-Szczepka J, Utracka-Hutka B, Grzybowska E, et al. BRCA1 and BRCA2 mutations as prognostic factors in bilateral breast cancer patients. Ann Oncol. 2004;15:1373–6. doi: 10.1093/annonc/mdh352. [DOI] [PubMed] [Google Scholar]
- 9.Deng M, Chen HH, Zhu X, et al. Prevalence and clinical outcomes of germline mutations in BRCA1/2 and PALB2 genes in 2769 unselected breast cancer patients in China. Int J Cancer. 2019;145:1517–28. doi: 10.1002/ijc.32184. [DOI] [PubMed] [Google Scholar]
- 10.www.nccn.org/NCCN guidelines insights: Genetic/Familial high-risk assessment: breast, ovarian and pancreatic, version 2.2022.
- 11.Pohl-Rescigno E, Hauke J, Loibl S, et al. Association of Germline Variant Status With Therapy Response in High-risk Early-Stage Breast Cancer: A Secondary Analysis of the GeparOcto Randomized Clinical Trial. JAMA Oncol. 2020. [DOI] [PMC free article] [PubMed]
- 12.Hahnen E, Lederer B, Hauke J, et al. Germline Mutation Status, Pathological Complete Response, and Disease-Free Survival in Triple-Negative Breast Cancer: Secondary Analysis of the GeparSixto Randomized Clinical Trial. JAMA Oncol. 2017;3:1378–85. doi: 10.1001/jamaoncol.2017.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurian AW, Gong GD, Chun NM, et al. Performance of BRCA1/2 mutation prediction models in Asian Americans. J Clin Oncol. 2008;26:4752–8. doi: 10.1200/JCO.2008.16.8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ang BH, Ho WK, Wijaya E, et al. Predicting the Likelihood of Carrying a BRCA1 or BRCA2 Mutation in Asian Patients With Breast Cancer. J Clin Oncol. 2022;40:1542–51. doi: 10.1200/JCO.21.01647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung FH, Wang YA, Jian JW, et al. Evaluating BRCA mutation risk predictive models in a Chinese cohort in Taiwan. Sci Rep. 2019;9:10229. doi: 10.1038/s41598-019-46707-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Zhao H, Zheng Y, et al. DrABC: deep learning accurately predicts germline pathogenic mutation status in breast cancer patients based on phenotype data. Genome Med. 2022;14:21. doi: 10.1186/s13073-022-01027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Fu F, Huang M, Lv J, Zhang W, Wang C. The spectrum of BRCA1 and BRCA2 mutations and clinicopathological characteristics in Chinese women with early-onset breast cancer. Breast Cancer Res Treat. 2020;180:759–66. doi: 10.1007/s10549-020-05573-x. [DOI] [PubMed] [Google Scholar]
- 19.Bergmeir C, Benítez JM. Neural Networks in R Using the Stuttgart Neural Network Simulator: RSNNS. ournal of Statistical Software. 2012 46(7):1–26.
- 20.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 21.SAS. http://www.sas.com.
- 22.IBM SPSS. http://www-01.ibm.com/software/uk/analytics/spss.
- 23.The R Project for Statistical Computing. https://www.r-project.org.
- 24.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherry ST1, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan Y, Hu H, Peng Y, et al. Detection of inherited mutations for hereditary cancer using target enrichment and next generation sequencing. Fam Cancer. 2015;14:9–18. doi: 10.1007/s10689-014-9749-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Sun J, Chen J, et al. Comprehensive analysis of BRCA1 and BRCA2 germline mutations in a large cohort of 5931 Chinese women with breast cancer. Breast Cancer Res Treat. 2016;158:455–62. doi: 10.1007/s10549-016-3902-0. [DOI] [PubMed] [Google Scholar]
- 28.Lang GT, Shi JX, Hu X, et al. The spectrum of BRCA mutations and characteristics of BRCA-associated breast cancers in China: Screening of 2,991 patients and 1,043 controls by next-generation sequencing. Int J Cancer. 2017;141:129–42. doi: 10.1002/ijc.30692. [DOI] [PubMed] [Google Scholar]
- 29.Kang E, Seong MW, Park SK, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast Cancer Res Treat. 2015;151:157–68. doi: 10.1007/s10549-015-3377-4. [DOI] [PubMed] [Google Scholar]
- 30.Dodova RI, Mitkova AV, Dacheva DR, et al. Spectrum and frequencies of BRCA1/2 mutations in Bulgarian high risk breast cancer patients. BMC Cancer. 2015;15:523. doi: 10.1186/s12885-015-1516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Meng H, Yao L, et al. Germline Mutations in Cancer Susceptibility Genes in a Large Series of Unselected Breast Cancer Patients. Clin Cancer Res. 2017;23:6113–9. doi: 10.1158/1078-0432.CCR-16-3227. [DOI] [PubMed] [Google Scholar]
- 32.Hall MJ, Reid JE, Burbidge LA, et al. BRCA1andBRCA2mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009;115:2222–33. doi: 10.1002/cncr.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayraktar S, Jackson M, Gutierrez-Barrera AM, et al. Genotype-Phenotype Correlations by Ethnicity and Mutation Location in BRCA Mutation Carriers. Breast J. 2015;21:260–7. doi: 10.1111/tbj.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng H, Yao L, Yuan H, et al. BRCA1 c.5470_5477del, a founder mutation in Chinese Han breast cancer patients. Int J Cancer. 2020;146:3044–52. doi: 10.1002/ijc.32877. [DOI] [PubMed] [Google Scholar]
- 35.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancersusceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–58. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–90. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 37.Antoniou AC, Pharoah PP, Smith P, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91:1580–90. doi: 10.1038/sj.bjc.6602175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ready KJ, Vogel KJ, Atchley DP, et al. Accuracy of the BRCAPRO model among women with bilateral breast cancer. Cancer. 2009;115:725–30. doi: 10.1002/cncr.24102. [DOI] [PubMed] [Google Scholar]
- 39.Burke HB, Goodman PH, Rosen DB, et al. Artificial neural networks improve the accuracy of cancer survival prediction. Cancer. 1997;79:857–62. doi: 10.1002/(SICI)1097-0142(19970215)79:4<857::AID-CNCR24>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 40.Cucchetti A, Piscaglia F, Grigioni AD, et al. Preoperative prediction of hepatocellular carcinoma tumour grade and micro-vascular invasion by means of artificial neural network: a pilot study. J Hepatol. 2010;52:880–8. doi: 10.1016/j.jhep.2009.12.037. [DOI] [PubMed] [Google Scholar]
- 41.Ziada AM, Lisle TC, Snow PB, Levine RF, Miller G, Crawford ED. Impact of different variables on the outcome of patients with clinically confined prostate carcinoma: prediction of pathologic stage and biochemical failure using an artificial neural network. Cancer. 2001;91:1653–60. doi: 10.1002/1097-0142(20010415)91:8+<1653::AID-CNCR1179>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 42.Qin X, Wang H, Hu X, Gu X, Zhou W. Predictive models for patients with lung carcinomas to identify EGFR mutation status via an artificial neural network based on multiple clinical information. J Cancer Res Clin Oncol. 2020;146:767–75. doi: 10.1007/s00432-019-03103-x. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Yang Y, Wang Y, Fan Q. Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction with Counter Propagation Artificial Neural Network. Molecules. 2019;24. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available in the BioProject repository [PRJNA859288].