Abstract
Background
Next-generation cancer immunotherapies are designed to broaden the therapeutic repertoire by targeting new immune checkpoints including lymphocyte-activation gene 3 (LAG-3) and T cell immunoglobulin and mucin-domain containing-3 (TIM-3). Yet, the molecular and cellular mechanisms by which either receptor functions to mediate its inhibitory effects are still poorly understood. Similarly, little is known on the differential effects of dual, compared with single, checkpoint inhibition.
Methods
We here performed in-depth characterization, including multicolor flow cytometry, single cell RNA sequencing and multiplex supernatant analysis, using tumor single cell suspensions from patients with cancer treated ex vivo with novel bispecific antibodies targeting programmed cell death protein 1 (PD-1) and TIM-3 (PD1-TIM3), PD-1 and LAG-3 (PD1-LAG3), or with anti-PD-1.
Results
We identified patient samples which were responsive to PD1-TIM3, PD1-LAG3 or anti-PD-1 using an in vitro approach, validated by the analysis of 659 soluble proteins and enrichment for an anti-PD-1 responder signature. We found increased abundance of an activated (HLA-DR+CD25+GranzymeB+) CD8+ T cell subset and of proliferating CD8+ T cells, in response to bispecific antibody or anti-PD-1 treatment. Bispecific antibodies, but not anti-PD-1, significantly increased the abundance of a proliferating natural killer cell subset, which exhibited enrichment for a tissue-residency signature. Key phenotypic and transcriptional changes occurred in a PD-1+CXCL13+CD4+ T cell subset, in response to all treatments, including increased interleukin-17 secretion and signaling toward plasma cells. Interestingly, LAG-3 protein upregulation was detected as a unique pharmacodynamic effect mediated by PD1-LAG3, but not by PD1-TIM3 or anti-PD-1.
Conclusions
Our in vitro system reliably assessed responses to bispecific antibodies co-targeting PD-1 together with LAG-3 or TIM-3 using patients’ tumor infiltrating immune cells and revealed transcriptional and phenotypic imprinting by bispecific antibody formats currently tested in early clinical trials.
Keywords: biomarkers, tumor; immunotherapy; clinical trials as topic
WHAT IS ALREADY KNOWN ON THIS TOPIC.
Several recent preclinical studies show improved efficacy when combining blockade of programmed cell death protein 1 (PD-1) and either T cell immunoglobulin and mucin-domain containing-3 (TIM-3) or lymphocyte-activation gene 3 (LAG-3), compared with single PD-1 blockade, but much still remains unknown about the mechanistic consequences of dual checkpoint blockade, especially in comparison to single checkpoint blockade.
Further insights into such mechanisms are required to better design future therapeutic strategies.
WHAT THIS STUDY ADDS
In this study, we assessed the single-cell level phenotypical and transcriptional consequences of treatment with anti-PD-1 and with PD1-TIM3 and PD1-LAG3 bispecific antibodies—which are currently being tested in different phase I/II clinical trials for patients with advanced cancer and are designed to engage exhausted T cells—using patient-derived tumor infiltrating immune cells ex vivo.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
These data show that intratumoral T cells exposed to single or dual immune checkpoint blockade are capable to alter their transcriptome, phenotype and functionality and likely adopt an active role in shaping the tumor microenvironment.
Prospectively, these changes may serve as future biomarkers to predict responses to immune checkpoint blockade.
Moreover, LAG-3 surface rearrangement and overexpression was uncovered as a key consequence of LAG-3 blockade, which may be monitored in the clinic to confirm therapeutic antibody binding or efficacy.
Background
Immune checkpoint inhibitors targeting cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) or programmed cell death protein 1 (PD-1) and its ligand programmed death-ligand 1 (PD-L1) are revolutionizing cancer therapy. In the last years, based on prolongation of overall survival and durable responses observed even in pretreated patients, regulatory approval has been obtained for various indications, treatment combinations and therapy lines.1–3 One key mechanism known to drive successful immune checkpoint inhibition is reinvigoration of exhausted T cells.4 T cell exhaustion is a dysfunctional state characterized by defined transcriptional and chromatin landscapes resulting in impaired effector functions and high expression of inhibitory receptors including PD-1, T cell immunoglobulin and mucin-domain containing-3 (TIM-3) and lymphocyte-activation gene 3 (LAG-3), among others.5 6 Furthermore, overexpression of TIM-3 and LAG-3 may be a compensatory mechanism associated with secondary or delayed resistance to anti-PD-1/PD-L1 therapies.7 8
Although TIM-3 and LAG-3 appear to be important for T cell exhaustion and successful checkpoint inhibition, their signaling pathways and the consequence of their blockade in patients, and especially of their combined blockade with PD-1, are still not fully understood.9 In particular, TIM-3 is found on activated T cells, regulatory T cells (Tregs) and innate immune cells including dendritic cells (DCs), natural killer (NK) cells and monocytes, and is described to bind to galectin-9, carcinoembryonic antigen-related cell adhesion molecule 1, phosphatidylserine and high mobility group box 1.9 LAG-3 is a transmembrane protein receptor structurally similar to CD4, expressed on activated T cells, Tregs, B cells and NK cells. It is known to bind to major histocompatibility complex class II molecules as well as to galectin-3, liver sinusoidal endothelial cell lectin and fibrinogen-like protein 1.9–13
While the exact signaling pathways of these receptors are still under investigation, both LAG-3 and TIM-3, similarly to PD-1, have been shown to be physiologically required to prevent overactivation and autoimmune reactions by regulating T cell function.9 10 Additionally, LAG-3 has been shown to promote Treg cell responses,9 10 while TIM-3 is known to restrain antitumor immunity by regulating inflammasome activation and the yclic GMP-AMP synthase; stimulator of interferon genes (cGAS-STING) pathway in intratumoral DCs.14 15
In the context of cancer immunotherapy, strategies aimed at single or combined blockade of different immune checkpoints including PD-1, TIM-3 and/or LAG-3 are therefore object of active preclinical and clinical investigation. Indeed, dual blockade of PD-1 and either TIM-3 or LAG-3 results in improved efficacy, compared with single PD-1 blockade in several preclinical models.16–20 Furthermore, recent evidence shows a greater clinical benefit when combining LAG-3 and PD-1 antibodies (Abs), compared to anti-PD-1 alone, in patients with previously untreated advanced melanoma.21
One particular therapeutic approach to simultaneously block two inhibitory receptors is the use of bispecific Abs (bsAbs), which are molecules designed to recognize and engage distinct cell surface receptor epitopes.22–24 In particular, bsAbs targeting PD-1 and either TIM-3 or LAG-3 (later PD1-TIM3 and PD1-LAG3, respectively25 26), currently being tested in different phase I/II clinical trials for advanced cancer (NCT04140500, NCT04785820, NCT05116202, NCT04524871, NCT03708328), are designed to re-invigorate exhausted T cells by co-targeting potentially non-redundant checkpoints.
In this work, for the first time, we examined the phenotypical and transcriptional landscape of human tumor infiltrating immune cells treated with PD1-TIM3, PD1-LAG3 and anti-PD-1. Using an ex vivo approach, we aimed o identify pharmacodynamic biomarkers associated with dual checkpoint blockade, to directly compare it to single checkpoint blockade, and to inform current and future clinical investigations.
Results
Identification of human tumor suspensions responsive ex vivo to PD1-TIM3, PD1-LAG3 and anti-PD-1
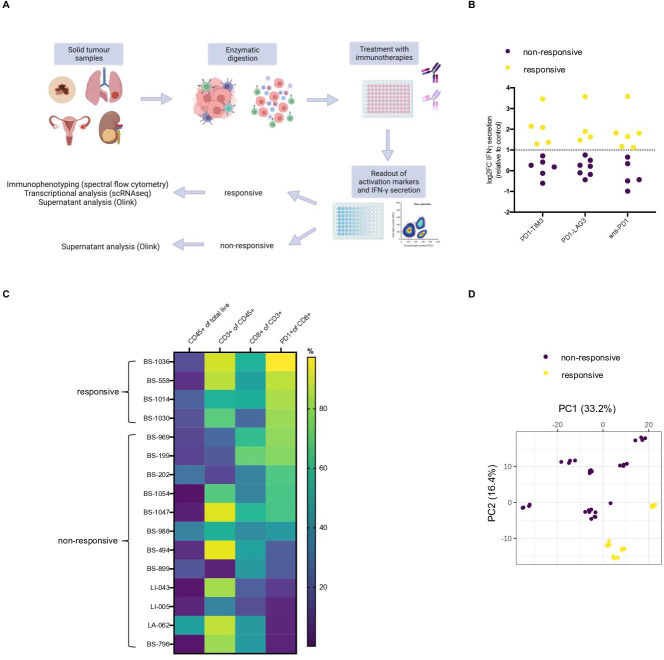
Using the experimental conditions outlined in figure 1A, we first identified functional responses in resected tumor lesions from patients with cancer exposed ex vivo to PD1-TIM3, PD1-LAG3 and anti-PD-1. A non-targeting monoclonal antibody (mAb; DP47) and single PD-1-blocking mAb (modified pembrolizumab) served as controls; all mAbs used in this assay contained the P329G-LALA mutated Fc backbone, rendering them Fc-gamma receptor-binding inert.27 28 Specifically, 21 tumor single cell suspensions (later: tumor infiltrating immune cells), derived from 19 tumor resections and 2 pleural effusions obtained from patients with cancer, were treated with bsAbs, anti-PD-1 or control isotype for 96 hours (online supplemental tables S1, S2). Activation and proliferation markers CD25 and Ki67 were then measured on CD8+ T cells by flow cytometry to assess treatment-induced responses (online supplementals figure S1A and table S1). Additionally, we investigated the secretion of gamma interferon (IFN-γ) on exposure to PD1-TIM3, PD1-LAG3 and anti-PD-1 (figure 1B), the induction of which correlated with changes in activation of CD8+ T cells, measured by either CD25 or Ki67 expression (online supplemental figure S1B). Tumor infiltrating immune cells were considered responsive to PD1-TIM3, PD1-LAG3 or anti-PD-1 if a ≥twofold increase (ie, log2 fold change (log2FC) ≥1) in IFN-γ secretion was observed, compared o the control (figure 1B). Treatment with anti-PD-1 resulted in six responses, while treatments with PD1-TIM3 and PD1-LAG3 resulted in five and four responses, respectively (figure 1B). Immunophenotyping at baseline showed high PD-1 expression on CD8+ T cells on the responsive samples (figure 1C), whereas non-responsive samples expressed lower levels of PD-1 (figure 1C). Of note, we observed positive correlations between the FC in IFN-γ induced by each compound and the baseline PD-1 expression in each sample assessed (online supplemental figure S1C). Using 11 samples, we conducted an unbiased multiplex supernatant analysis of 659 soluble markers from inflammation and oncology panels using Olink technology.29 Principal component analysis (PCA) showed separation of the samples considered responsive from the samples classified as non-responsive, allowing us to further confirm a differential response and validate our classification (figure 1D).
Figure 1.
Tumor infiltrating immune cells from selected patients show increased cytokine secretion and T cell activation in response to anti-PD-1, PD1-TIM3 and PD1-LAG3. (A) Schematic depicting treatment of tumor infiltrating immune cells from solid tumor (n=19) and pleural effusion (n=2) samples with bsAbs, anti-PD-1 or control isotype. Treatment (96 hours) was followed by assessment of activation by flow cytometry or IFN-γ, classification into responsive or non-responsive samples and further characterization by immunophenotyping, single-cell transcriptional analysis or multiplex supernatant analysis. (B) Log2 fold change (log2FC) in IFN-γ secretion comparing each treated sample (with levels of secretion above limit of detection, n of patients=11) to the isotype control. A threshold of 1 log2FC (dotted line) was applied to identify patient samples to be considered responsive to in vitro treatment. One responsive patient-derived pleural effusion sample (BS199) was excluded from further analyses due to differences in sample preparation. (C) Heatmap showing percentage of expression of selected surface markers in each patient tumor suspension samples (n=16) used in the in vitro experiments containing measurable live CD45+ cells. (D) PCA plot showing separation of tumor suspension samples (n=11, samples with sufficient supernatant that passed quality check) based on multiplex Olink supernatant analysis of 659 protein markers. Classification into responsive or non-responsive is indicated by the color legend. bsAbs, bispecific antibodies; IFN, interferon; LAG-3, lymphocyte-activation gene 3; PCA, principal component analysis; PD-1, programmed cell death protein 1; TIM-3, T cell immunoglobulin and mucin-domain containing-3.
jitc-2022-005548supp004.pdf (12.2MB, pdf)
We next focused on the four patients defined as functionally responsive to all three treatments (BS558, BS1014, BS1030, BS1036, online supplemental tables S1 and S2), in order to directly compare single with dual checkpoint inhibition within samples derived from the same patients and investigate any differential responses to treatment. Our in-depth characterization included multicolor flow cytometry, single cell RNA sequencing (scRNAseq) and multiplex supernatant analysis, to mechanistically understand the consequences of PD1-TIM3 and PD1-LAG3 treatment.
Multidimensional profiling of tumor infiltrating immune cells responsive to PD1-TIM3, PD1-LAG3 and anti-PD-1 treatment
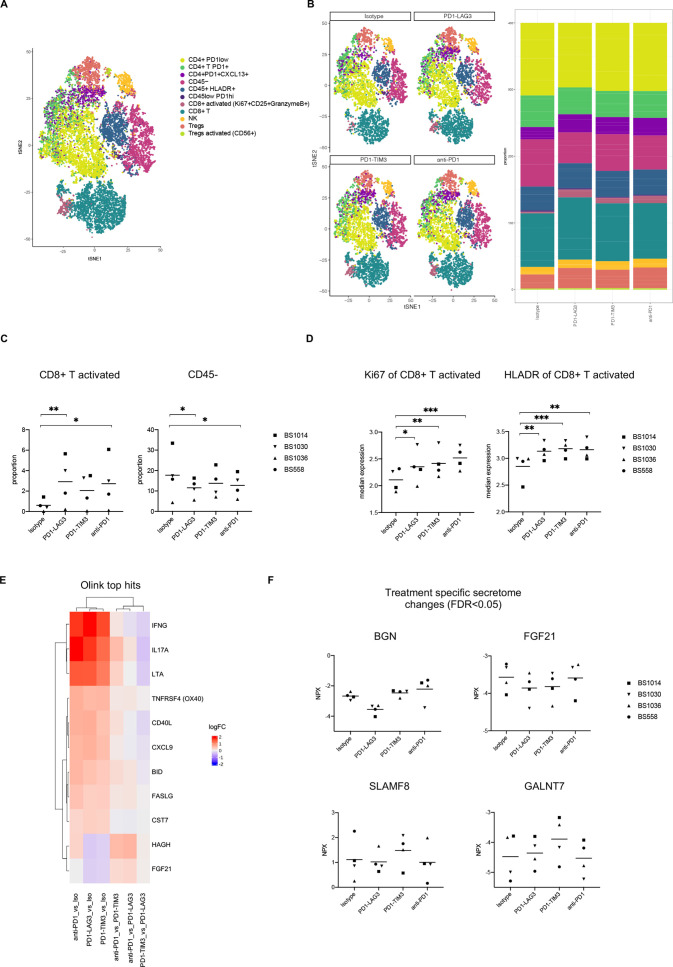
We used a 24-color spectral flow cytometry panel (online supplemental table S3) to investigate phenotypic changes induced by PD1-TIM3, PD1-LAG3 or single PD-1 blockade on the four responsive samples. Using FlowSOM clustering on logicle transformed markers expression (as previously described30) (online supplemental figure S2A), we defined the cell populations indicated in figure 2A, namely CD4+ PD-1low, CD4+ PD-1+, CD4+ PD-1+CXCL13+, CD45–, CD45+ HLA-DR+, CD45lowPD-1hi, CD8+ activated (HLA-DR+CD25+GranzymeB+), CD8+ T, NK, Tregs and Tregs activated (CD56+). We observed an increase in the proportion of activated CD8+ T and a decrease in CD45– cells on bsAb or single anti-PD-1 treatment, in line with our previous results (figure 2B, C). The decrease in CD45– cells may partly suggest killing or decreased survival of tumor cells within the treated cultures. Additionally, the activated CD8+ population presented significant upregulation of Ki67 and HLA-DR comparing each treatment with control (figure 2D). Overall, the effects were consistent across patients with some variations in the relative frequencies of different immune cell populations (online supplemental figure S2B). We assessed the expression of the inhibitory receptors PD-1, TIM-3 and LAG-3 on CD8+ and CD4+ T cells from the four patient samples, before treatment (online supplemental figure S2C). We observed high levels of expression of PD-1 in CD8+ (86.6%±7.4) and CD4+ T cells (66.1%±6.4) from all samples as well as expression of TIM-3 (36.4%±26.3 on CD8+; 7.1%±6 on CD4+) and LAG-3 (10.8%±10.9 on CD8+; 12.7%±5 on CD4+) and their co-expression with PD-1 (33.5%±26.4 PD-1+TIM-3+ on CD8+; 7.5%±6 PD-1+TIM-3+ on CD4+; 9%±10 PD-1+LAG-3+ on CD8+; 8.7%±5.2 PD-1+LAG-3+ on CD4+) (online supplemental figure S2C). Of note, BS1036—obtained from a patient with metastatic melanoma progressing on combined treatment with ipilimumab and nivolumab—showed the highest levels of TIM-3 expression as well as co-expression of TIM-3 and PD-1 on CD8+ T cells (online supplemental figure S2C, online supplemental table S2). In order to further dissect the immunological responses to PD1-TIM3, PD1-LAG3 and anti-PD-1, we next visualized the top differentially regulated proteins from the multiplex supernatant analysis of responsive samples (figure 2E). As expected, IFN-γ appeared to be the top differentially upregulated protein, comparing each treatment with the control isotype, independently validating our previous (ELISA) results (figure 2E, online supplemental figure S2D). Figure 2E shows significantly upregulated soluble proteins shared by PD1-TIM3, PD1-LAG3 and anti-PD-1, including interleukin-17A (IL-17A), lymphotoxin, TNF Receptor Superfamily Member 4 (ie, OX40), CD40-ligand (CD40L) and chemokine (C-X-C motif) ligand 9 (figure 2E). In contrast, figure 2F exhibits treatment-specific effects, including downregulation of Biglycan after treatment with PD1-LAG3, upregulation of GalNAc transferase 7 and SLAM Family Member 8 after treatment with PD1-TIM3. Fibroblast growth factor 21 was downregulated after PD1-TIM3 and PD1-LAG3 treatments, but not by anti-PD-1 (figure 2F). Taken together, these data show an increase in activated and proliferating CD8+ T cells in response to treatment with PD1-TIM3, PD1-LAG3 or anti-PD-1; in addition, secretome analysis uncovered treatment-specific effects which may include direct and indirect downstream consequences of single and dual checkpoint blockade.
Figure 2.
Multidimensional immune-profiling of tumor suspensions that show ex vivo responsiveness to bsAbs or anti-PD-1 treatment. (A) tSNE plots of pooled treatment conditions from the four responsive patients showing cell-type annotation (treatment of 96 hours; n patients=4, n treatments=4, total n analyzed samples=16). (B) tSNE plots showing identified cell types for pooled patient samples, split by treatment (left). Bar plots showing the proportion of each identified cell type out of all the cells in each treatment (patients are pooled, right). (C) Proportion of CD8+ T and CD45– cells from the four responsive tumor suspension in different treatment conditions. Clustering was conducted using FlowSOM (*p<0.05, **p<0.01, one-way ANOVA with multiple comparisons). (D) Median marker intensity of Ki67 and HLA-DR on the activated CD8+ population in the four responsive tumor suspensions in different treatment conditions. Clustering was conducted using FlowSOM (*p<0.05, **p<0.01, ***p<0.001, one-way ANOVA with multiple comparisons). (E) Heatmap of top significantly differentially regulated proteins, obtained by running a linear model using limma, from the multiplex supernatant analysis, in the four responsive tumor suspensions across different treatment versus control comparisons. (F) Normalized protein expression (NPX) of indicated soluble markers in the four responsive tumor suspensions which show significant differences (FDR <0.05) in different treatment versus control comparisons (BGN: comparing PD1-LAG3 with all other treatments; FGF21: comparing PD1-TIM3 or PD1-LAG3 with isotype or anti-PD1; GALNT7: comparing PD1-TIM3 with all other treatments; SLAMF8: comparing PD1-TIM3 with PD1-LAG3) measured by Olink analysis. ANOVA, analysis of variance; bsAbs, bispecific antibodies; BGN, Biglycan; CD40L, CD40-ligand; CXCL9, chemokine (C-X-C motif) ligand 9; FDR, false discovery rate; FGF21, fibroblast growth factor 21; GALNT7, GalNAc transferase 7; IFN, interferon; IL-17A, interleukin-17A; LAG-3, lymphocyte-activation gene 3; LOD, limit of detection; LTA, lymphotoxin; PD-1, programmed cell death protein 1; SLAMF8, SLAM Family Member 8; TIM-3, T cell immunoglobulin and mucin-domain containing-3; TNFRSF4, TNF Receptor Superfamily Member 4; tSNE, t-distributed stochastic neighbor embedding.
Single-cell transcriptional analysis of tumor infiltrating immune cells responsive to PD1-TIM3, PD1-LAG3 and anti-PD-1 in vitro
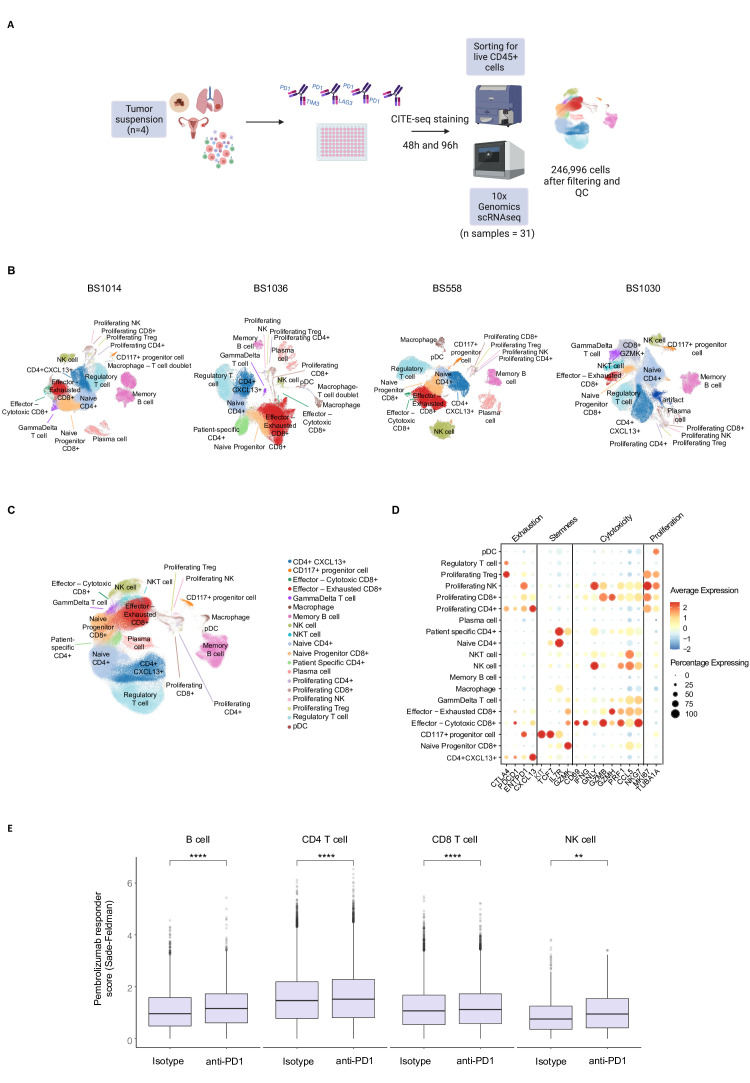
Next, in order to investigate the transcriptional landscape of immune cells exposed to single or dual checkpoint blockade, we conducted scRNAseq of tumor infiltrating immune cells (CD45+) sorted from responsive samples after 48 and 96 hours of exposure to PD1-TIM3, PD1-LAG3, anti-PD-1 and the isotype control (figure 3A). We additionally performed Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) for nine immune cell lineage markers. A total of 31 individual samples were ultimately used to perform scRNA-seq using the 10x Genomics Chromium 3’ platform (figure 3A). The sequencing statistics and average reads per cell are detailed in online supplemental file S1. After applying patient-specific filtering thresholds, we retained a total of 246 996 single live cells from the four responsive tumor suspensions. We initially performed dimensionality reduction and clustering separately for each patient (figure 3B, online supplemental figure S3A-D) to retain patient-specific clusters and avoid forceful integration of cell types. To perform a signature-based automated cell type annotation of each patient’s cluster, we used a single cell analysis toolkit platform (Besca31). A custom-made overlapping cluster ID was subsequently applied to allow merging of the cells from each patient (online supplemental file S2). We then merged the annotated clusters and batch-corrected the cells for the patient effect using batch balanced k nearest neighbors (BBKNN)32 (figure 3C, online supplemental figure S4A-B).
Figure 3.
Single-cell transcriptional analysis of the activity of PD1-TIM3, PD1-LAG3 and anti-PD-1 in vitro. (A) Schematic depicting treatment of tumor suspensions from the four responsive solid tumor samples with bsAbs, anti-PD-1 or control isotype for 48 and 96 hours, followed by CITE-seq staining and sorting for CD45+ cells before running 10x Genomics 3’ scRNAseq protocol. After filtering and quality check, a total of 246 996 CD45+ cells were ultimately obtained from 31 individual samples, while one sample failed after 10x chip loading. (B) uniform manifold approximation and projection (UMAP) of CD45+ cells, split by each patient (indicated above). Cell types were annotated using a custom ‘overlapping cluster ID’ to then merge the samples deriving from different patients. (C) UMAP of final filtered and annotated cells, with all patients merged and batch adjusted as described in the methods. (D) Dot plot showing the average gene expression and the percentage of cells expressing the genes indicated at the bottom, per each annotated cell type. The genes were categorized as exhaustion, stemness, cytotoxicity and proliferation markers. (E) Box plots showing a significant difference (Wilcoxon paired signed rank test) in the PD-1 response score, calculated as indicated in the methods using previously published data,33 between the indicated cell types in the isotype control (left) or anti-PD-1-treated (right) conditions (**p<0.01, ****p<0.0001). bsAbs, bispecific antibodies; LAG-3, lymphocyte-activation gene 3; NK, natural killer; PD-1, programmed cell death protein 1; scRNAseq, single cell RNA sequencing; IM-3, T cell immunoglobulin and mucin-domain containing-3.
jitc-2022-005548supp001.xlsx (17.2KB, xlsx)
jitc-2022-005548supp002.xlsx (17.1KB, xlsx)
The resulting 19 clusters included several distinct T cell, NK cell, B cell, CD117+ progenitor-like and myeloid cell populations as well as specific proliferating T cell and NK cell clusters (figure 3C). Matched CITE-seq allowed confirmation of the protein expression of nine main lineage markers (online supplemental figure S4C). Figure 3D shows the average expression of known exhaustion, stemness, cytotoxicity and proliferation markers on each cluster, in accordance with our cluster annotation, indicating that expected gene expression patterns are observed in different proliferating, naïve or progenitor (TCF7-expressing) and exhausted tumor-infiltrating cell types. In order to investigate whether the exposure to anti-PD-1 in the four responsive samples was inducing similar transcriptional effects observed in patients treated with anti-PD-1, we derived a response score using a published scRNAseq dataset of tumors from patients with melanoma treated with pembrolizumab33 (online supplemental figure S4D). By testing enrichment of the PD-1 responder score in different B cells, CD4+, CD8+ T cells and NK cells in our dataset, we found that this was significantly higher in the anti-PD-1-treated condition (figure 3E), compared with isotype control. These data indicate that the single-cell level transcriptional response observed in the responsive samples upon exposure to anti-PD-1 is similar to that of patients which respond to pembrolizumab, further validating our in vitro approach.
Therefore, we next set out to investigate treatment-specific effects on the abundance and transcriptional profile of each identified cell type.
Differential changes in cell-type abundance driven by single or dual checkpoint inhibition
Having defined the cell-types within our scRNAseq dataset of responsive tumor infiltrating immune cells, we next investigated whether we could see changes in cell-type abundance by running a linear model, comparing each treatment with the control isotype.
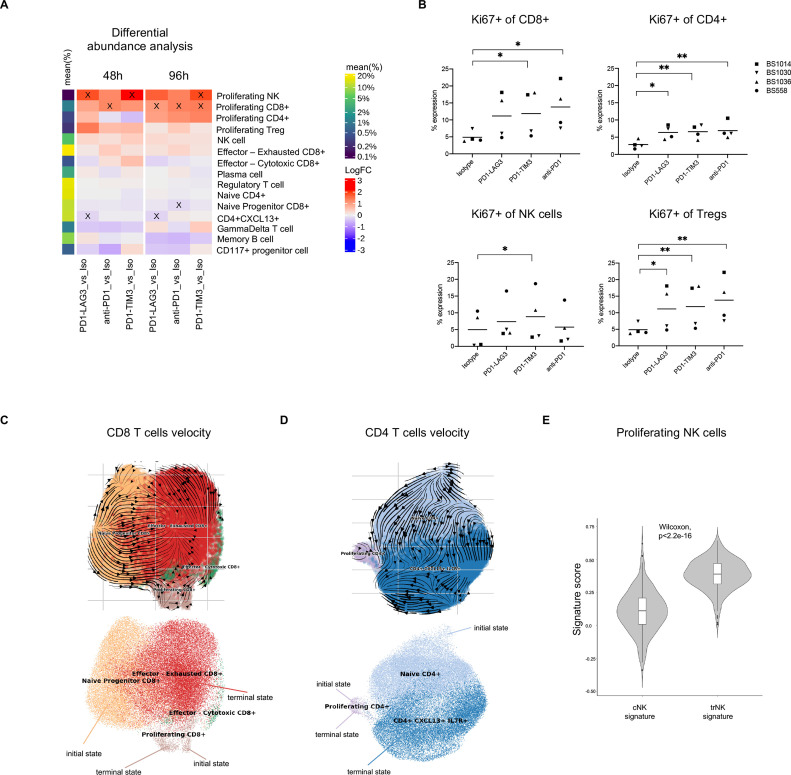
We observed significant increases in the proportion of proliferating cell subsets, consistent across patients, in response to PD1-TIM3, PD1-LAG3 or anti-PD-1 treatment (figure 4A, X indicates an adjusted p value <0.05), a finding that is in accordance with the significantly higher Ki67 expression on different T cells and NK cells clusters which we measured by flow cytometry (figure 4B).
Figure 4.
RNA velocity toward a proliferating state is enhanced in human tumor infiltrating immune cells treated with single or dual inhibitory receptor blockade. (A) Differential abundance analysis of cell types in different conditions and at different timepoints across patients. The heatmap shows the log fold change (logFC) between each treatment-control comparison indicated at the bottom (per timepoint). Significant (FDR <0.1) changes are indicated with an X. Mean frequency (%) of each cell type out of the total number of cells is also indicated in the color legend. (B) Percentage expression of Ki67 on different T cell or NK cell clusters, manually gated (*p<0.05, **p<0.01, one-way ANOVA with multiple comparisons). (C) UMAP of CD8+ T cell subsets from pooled conditions (all treated cells from the 96-hour timepoint) with projected RNA velocity vectors (top) and putative initial and terminal (bottom) states. (D) UMAP of conventional CD4+ T cell subsets (excluding Treg cells) from pooled conditions (all treated cells from the 96-hour timepoint) with projected RNA velocity vectors (top) and putative initial and terminal (bottom) states. (E) Violin plot showing cNK or trNK signature score distribution in cells from the proliferating NK cluster, calculated using AddModuleScore function within Seurat in R; p value was determined with a Wilcoxon paired test. ANOVA, analysis of variance; cNK, conventional NK; FDR, false discovery rate; NK, natural killer; trNK, tissue-resident NK; Treg, regulatory T cell.
Surprisingly, most significant changes in abundance, namely in the proportion of CD8+ proliferating cells (figure 4A), and Ki67 expression (figure 4B) were shared between bsAbs and anti-PD-1, indicating that the observed changes in proliferation were mainly driven by PD-1 blockade alone. However, PD1-TIM3, at both timepoints, and PD1-LAG3, at 96 hours, significantly increased the proportion of proliferating NK cells (figure 4A), in which we observed HAVCR3 (TIM-3) and LAG3 expression (online supplemental figure S4E). Additionally, PD1-LAG3 increased the proportion of proliferating CD4+ T cells more rapidly than other treatments (ie, already at the 48-hour timepoint), although this effect was not significant (figure 4A). Importantly, PD1-LAG3 treatment significantly decreased the proportion of CD4+CXCL13+ cells at both timepoints, while anti-PD-1 significantly decreased the proportion of the naïve progenitor (TCF7-expressing) CD8+ cluster at 96 hours. The observed decreases may suggest changes in the fate of each cluster toward different states, such as the proliferating state (ie, a decrease of a cell type may result in the increase of another cell type). In order to better understand these findings, we next set out to investigate the cell trajectories of CD8+ and conventional CD4+ T cells within our dataset.
Differentiation toward a proliferating state is enhanced in human tumor infiltrating immune cells treated with single or dual inhibitory receptor blockade
To investigate where each proliferating cluster may be positioned in an underlying biological trajectory, we performed scVelo34 on all CD8+ T cell clusters and separately on conventional CD4+ T cell clusters (from the 96-hour timepoint). Our analysis revealed two putative initial states and two terminal states in each lineage (figure 4C, D). The inferred RNA velocity dynamics of the CD8+ T cell clusters are shown in figure 4C and suggest that the ‘initial state’ naïve progenitor CD8+ T cell (which expresses TCF7) can differentiate toward ‘terminal state’ effector—exhausted cluster and partly toward the ‘terminal state’ proliferating cluster (figure 4C, online supplemental figure S5A). Furthermore, the proliferating CD8+ cluster was classified as both putative initial and terminal state and the RNA velocity dynamics point toward a self-amplifying, cycling state (figure 4C, online supplemental figure S5A). Similarly, in the CD4+ lineage, we identified naïve and proliferating as initial states and CD4+CXCL13+ and proliferating clusters as terminal states (figure 4D, online supplemental figure S5B), with the proliferating cluster also appearing to be self-amplifying (figure 4D, online supplemental figure S5B).
As we did not observe multiple NK cell clusters in our dataset which would have allowed us to calculate putative biological trajectories, we characterized the proliferating NK cell cluster by investigating the enrichment for previously described NK core signature genes associated with either a conventional phenotype (cNK) or with tissue-residency (trNK). Interestingly, we observed a significant enrichment of the trNK signature, which includes genes such as ITGA1 and ITGAE, CD69 and ENTPD1, in the proliferating NK cell cluster, indicating that these cells may be differentiating from or into tissue resident-like NK cells (figure 4E). Taken together, our cell abundance and trajectory analyses suggest that treatment with either PD1-TIM3, PD1-LAG3 or anti-PD-1 shifts the balance of either CD8+ and CD4+ T cells toward a proliferating, self-amplifying state, rather than toward the exhausted (for CD8+) or PD-1+CXCL13+ (for CD4+) cell-types, and we additionally observed that PD1-TIM3 and PD1-LAG3-expanded proliferating NK cells present tissue-residency features.
Cluster-specific pathway and gene level changes in response to PD1-TIM3, PD1-LAG3 and anti-PD-1 treatment
In order to further characterize the transcriptional response to bsAbs or anti-PD-1 treatment, we investigated pathway and gene level changes by conducting pseudobulk-based differential gene expression analysis using the scRNAseq dataset.
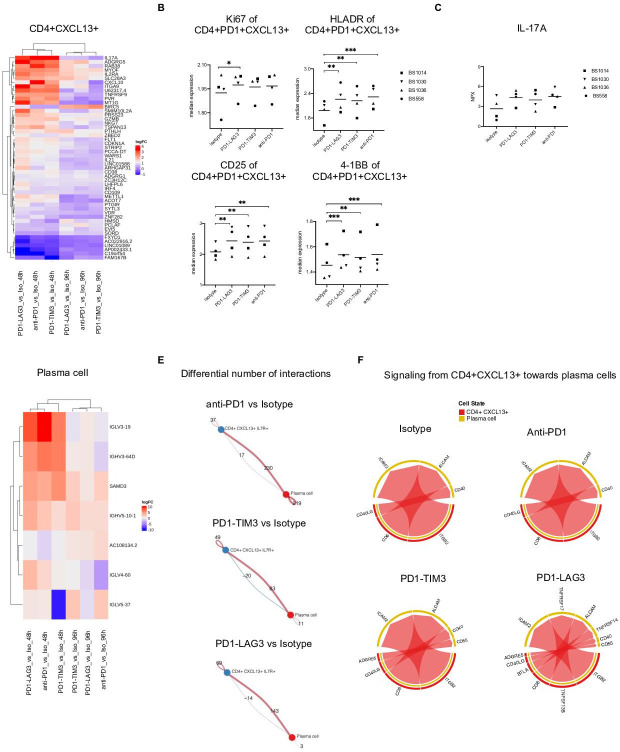
The full list of differentially expressed genes that were changing in response to treatment in each cluster is reported in online supplemental file S3. Importantly, we observed the most significantly different gene expression changes in the CD4+CXCL13+ cluster, in response to all treatments. Figure 5A shows the top differentially expressed genes which were significant in any of the comparison (each treatment compared with control isotype) within this cluster. Differentially expressed genes in response to all treatments included IL17A, ADGRG5, RAB28, MYOF, IL2RA and CXCL10, while IL-21 was found upregulated in response to PD1-LAG3 (figure 5A). Accordingly, we observed protein expression changes in the CD4+CXCL13+ population by flow cytometry; namely, increases in the expression of Ki67 between PD1-LAG3 and isotype and increase in the expression of HLA-DR, CD25 and 4-1BB between each treatment and isotype (figure 5B, online supplemental figure S6A). Additionally, the analysis of differentially expressed genes indicates that the increased levels of IL-17A in response to bsAbs or anti-PD-1 (figure 5C, figure 2E) are likely to be due to secretion by CD4+CXCL13+ cells.
Figure 5.
Transcriptional changes are induced by single or dual checkpoint blockade within CD4+CXCL13+ cells which influence interaction with plasma cells. (A) Top differentially expressed genes showing strongest change among all contrasts (average logCPM >0, adjusted p value <0.05) in the CD4+CXCL13+ cluster. (B) Median marker intensity of Ki67, HLA-DR, 4-1BB and CD25 on the CD4+CXCL13+ population in the four responsive tumor suspension in different treatment conditions. Clustering was conducted using FlowSOM (*p<0.05, **p<0.01, ***p<0.001, one-way ANOVA with multiple comparisons). (C) of IL-17A measured by Olink technology in the supernatant of the four responsive tumor suspension across different treatment versus control comparisons (FDR <0.05, comparing each treatment with control isotype). (D) Top differentially expressed genes showing strongest change among all contrasts (average logCPM >0, adjusted p value <0.05) in the plasma cell cluster. (E) Circle plots showing the differential number of interactions between the CD4+CXCL13+ and the plasma cell clusters, in each treatment compared with control, at 96 hours. Calculated using Cellchat.39 (F) Chord diagram of ligand-receptor pairs were mediating the communications from CD4+CXCL13+ toward plasma cells, in each treatment or control condition, at 96 hours, calculated using Cellchat.39 ANOVA, analysis of variance; CPM, counts per million; FDR, false discovery rate; IL-17A, interleukin-17A; LAG-3, lymphocyte-activation gene 3; PD-1, programmed cell death protein 1; TIM-3, T cell immunoglobulin and mucin-domain containing-3.
jitc-2022-005548supp003.xlsx (439.8KB, xlsx)
Moreover, we observed upregulation of immunoglobulin genes, such as IGLV3-19, IGHV3-64 and IGLV4-60 in the plasma cell cluster, in response to all treatments, which may suggest an increase in immunoglobulin production (figure 5D). Of note, the majority of gene level changes was stronger after 48 hours of treatment in all comparisons (figure 5A, D).
Using the pseudobulk analysis output, we additionally calculated the normalized enrichment score for each gene set from the MSigDB Hallmark collection35 within the identified clusters, comparing each treatment with the control isotype. Our analysis revealed positive enrichment for several different gene sets, including IL2-STAT5, IL6-JAK-STAT3, INFLAMMATORY RESPONSE and INTERFERON-ALPHA RESPONSE in distinct clusters (online supplemental figure S6B). Importantly, the changes, compared with control isotype, were overall shared by either bsAb or anti-PD-1 treatment and we observed the most changes in the CD4+CXCL13+, Treg cell and plasma cell clusters, in accordance with our pseudobulk gene expression analysis (online supplemental figure S6B). Of note, among the CD8+ clusters, we mainly observed differences, shared by all treatments, in the naïve progenitor population (TCF7-expressing) and not in the effector—exhausted or cytotoxic populations (online supplemental figure S6B), potentially suggesting differences in plasticity or susceptibility to perturbation between these CD8+ clusters.
Cell-cell interactions between CD4+CXCL13+ cells and plasma cells are enhanced on PD1-TIM3, PD1-LAG3 or anti-PD-1 treatment of tumor infiltrating immune cells
Our differential gene expression analysis revealed, within the CD4+CXCL13+ cluster, upregulation of IL-17 by all treatments and IL-21 by PD1-LAG3 (figure 5A, C), which are genes encoding for cytokines known to enhance B cell/plasma cell responses.36–38 We therefore hypothesized potential communication, including via these cytokines, between CD4+CXCL13+ and plasma cells, in which we observed upregulation of immunoglobulin genes by all treatments (figure 5D). This hypothesis led us to investigate whether treatment with bsAbs or anti-PD-1 determined changes in cell-cell interactions between these cell types within our in vitro system. Using CellChat,39 we found an increase in the number of signaling interactions, from CD4+CXCL13+ cells to plasma cells, in each treatment, compared with the isotype (figure 5E). Anti-PD-1 treatment determined the highest increase in interactions, followed by PD1-LAG3 and PD1-TIM3 (figure 5E).
We next visualized which ligand-receptor pairs were mediating the communications from CD4+CXCL13+ toward plasma cells, in each treatment condition (figure 5F). While most receptor-ligand pairs were shared across treatments and control, including CD40- CD40L, our analysis identified putative PD1-TIM3 and PD1-LAG3 treatment-specific pairs namely ADREG5 (CD97)-CD55 in both treatments, and TNFRSF17-TNFRSF13B as well as TNFRSF14-BTLA in PD1-LAG3 (figure 5F). Of note, CD40L was found among the top differentially upregulated proteins in all treatments within the multiplex supernatant analysis (figure 2E, online supplemental figure S6C). We additionally assessed whether CD4+CXCL13+ cells may be directly affected by single or dual inhibitory receptor blockade, by visualizing the expression of PDCD1 (PD-1), HAVCR2 (TIM-3) and LAG3 in our dataset. We observed high gene co-expression of all three receptors in this cluster, as well as in the effector—exhausted, effector—cytotoxic CD8+ clusters and the proliferating clusters (online supplemental figure S6D). Taken together, these data suggest that single or dual checkpoint blockade may directly trigger PD-1, TIM-3 and LAG-3-expressing CD4+CXCL13+ to secrete B cell-activating cytokines, including IL-17, IL-21 and CD40 ligand.
LAG-3 surface upregulation is an anti-LAG-3-specific effect which does not require anti-PD-1
Our scRNAseq analysis identified transcriptional and cell-abundance changes, many of which were shared in response to PD1-TIM3, PD1-LAG3 and anti-PD-1. This indicates that most of the molecular effects observed may be first driven by blockade of PD-1. Given that our tools, namely our specific Ab formats and our in vitro system, offered a unique opportunity to directly compare single with dual checkpoint inhibition and tease out potential differential responses, we set out to identify further phenotypic effects which may distinguish anti-PD-1 treatment from dual PD-1-TIM-3/LAG-3 blockade, by conducting additional mining of our flow cytometry dataset.
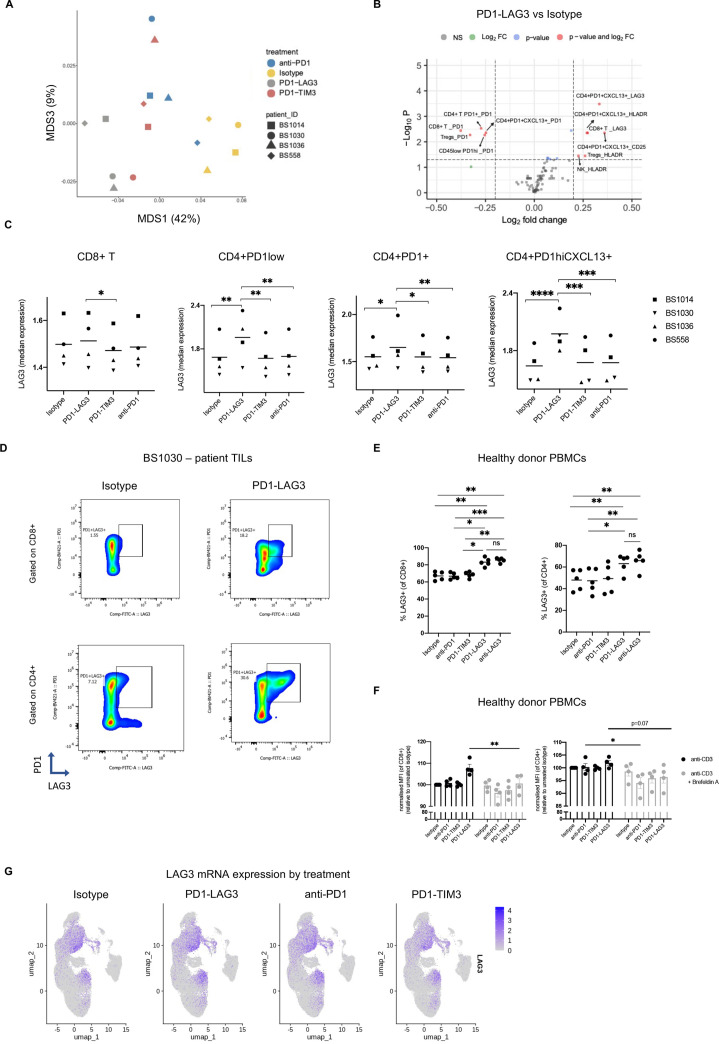
We applied the scaled and transformed30 fluorescence intensities of each marker on all cell populations (defined as described above) and batch-adjusted by patient (using ComBat in R40). We then visualized the similarity between all control and treated samples with a multidimensional scaling (MDS) plot (figure 6A, online supplemental figure S7A). As expected, we observed a clear separation of control isotype from the treated samples (online supplemental figure S6A, up). Of note, we observed a separation of the PD1-LAG3-treated samples from samples treated with either PD1-TIM3 or anti-PD-1 (figure 6A). This separation was even more striking when using the median fluorescence intensities of manually gated populations (online supplemental figure S7A, down, gating as per online supplemental figure S7B). As these observations may highlight a novel mechanism worthy of future exploration, we elucidated this effect by running a linear mixed model to identify PD1-TIM3/LAG3-specific differentially expressed markers on each FlowSOM clustered population (online supplemental figure S7C, figure 6B). The full list of differentially expressed markers on each population is indicated in online supplemental tables S4 and S5. Using this method, we identified LAG-3 upregulation on CD8+ and CD4+PD-1+CXCL13+ clusters among the top differentially expressed markers (figure 6B). This upregulation was even more evident when running a linear mixed model on manually gated populations (online supplemental figure S7D, gated as per online supplemental figure S7B). We next visualized LAG-3 marker expression and observed striking PD1-LAG3-driven upregulation on different subsets of CD8+ T and CD4+ T cell clusters (figure 6C). Of note, such upregulation was most pronounced in the CD4+CXCL13+ cluster (figure 6C). Figure 6D shows a representative dot plot of LAG-3 and PD-1 expression on CD8+ and CD4+ T cells from patient BS1030, treated with isotype control or PD1-LAG3.
Figure 6.
LAG-3 surface upregulation is an anti-LAG-3-specific effect which does not require anti-PD-1. (A) Multidimensional scaling plots of each indicated sample, based on the scaled and transformed fluorescence intensities of each marker on all cell populations and batch-adjusting by patient (using ComBat in R40), indicating the distance in similarity between samples. (B) Volcano plot depicting differentially expressed markers (logFC >|1.5|, adjusted p value <0.05) within each cell subset across patients in the PD1-LAG3-treated condition compared with the isotype control. The color legend indicates significant values according to the indicated thresholds. The values were obtained by running a linear mixed model using the median marker intensities of each population based on FlowSOM clustering. (C) Median marker intensity of LAG-3 on different T cell populations in the four responsive tumor suspensions in different treatment conditions. Clustering was conducted using FlowSOM (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, one-way ANOVA with multiple comparisons). (D) Representative dot plots showing expression of PD-1 and LAG-3 on CD8+ or CD4+ T cells from patient BS1030 tumor suspension in indicated treatment conditions. (E) Frequency of LAG-3+CD8+ (left) or CD4+ (right) T cells from healthy donor PBMCs, treated with isotype control, anti-PD-1, PD1-TIM3, PD1-LAG3 or an anti-LAG-3 antibody. P value was obtained by running two-way ANOVA with multiple comparisons (*p<0.05). Each dot indicates and independent PBMC donor (n=4). (F) Geometric MFI of LAG-3 surface expression on CD8+ (left) or CD4+ (right) T cells from anti-CD3 (clone OKT3) prestimulated healthy donor PBMCs, treated with isotype control, anti-PD-, PD1-TIM3 or PD1-LAG3 in the presence or absence of Brefeldin A. P value was obtained by running two-way ANOVA with multiple comparisons (*p<0.05). MFI is normalized to the isotype control, each dot indicates and independent PBMC donor (n=3). (G) UMAP projections showing average LAG-3 expression in all cells deriving from different treatment conditions. ANOVA, analysis of variance; FC, fold change; LAG-3, lymphocyte-activation gene 3; MFI, mean fluorescence intensity; PBMC, peripheral blood mononuclear cell; PD-1, programmed cell death protein 1; TIM-3, T cell immunoglobulin and mucin-domain containing-3.
To dissect the contribution of anti-PD-1 in the observed LAG-3 upregulation following PD1-LAG3 treatment, we treated anti-CD3 pre-activated healthy donor peripheral blood mononuclear cells (PBMCs) with isotype control, anti-PD-1, PD1-TIM3, PD1-LAG3 and a single LAG-3-blocking Ab. Interestingly, we observed LAG-3 upregulation in the PD1-LAG3 or single anti-LAG-3 conditions, but not with the isotype, anti-PD-1 or with PD1-TIM3 (figure 6E).
Furthermore, in order to better understand the mechanism of LAG-3 upregulation in response to PD1-LAG3, we treated healthy donor PBMCs with isotype, anti-PD-1, PD1-TIM3 or PD1-LAG3 in the presence or absence of protein transport inhibitor Brefeldin A (figure 6F). Abrogation of protein transport to the cell surface inhibited treatment-induced LAG-3 upregulation. Of note, we did not observe upregulation of LAG3 mRNA in the PD1-LAG3-treated condition (figure 6G).
Our results reveal LAG-3 upregulation as an anti-LAG-3-specific effect which does not require anti-PD-1 and is likely due to transport and reorganization of pre-existing intracellular stores of LAG-3 molecules to the cell surface, rather than de novo transcription, in response to LAG-3 blockade.
Discussion
Inhibitory receptors (immune checkpoints) are found upregulated and co-expressed on CD8+ T cells and correlate with increasing states of dysfunction and stages of cancer.4 41 Increased immune checkpoint co-expression may also constitute a compensatory resistance mechanism in the context of anti-PD-1 treatment.42 43 Indeed, a metastatic melanoma sample from a patient pretreated with nivolumab and ipilimumab (BS1036) included in this study, presented very high level of co-expression of TIM-3 and PD-1 on CD8+ T cells. Targeting multiple inhibitory receptors is an attractive new strategy in cancer immunotherapy with preclinical and clinical evidence showing a significant benefit in (co)-targeting PD-1, TIM-3, LAG-3 and TIGIT (T cell immunoreceptor with immunoglobulin and ITIM domains), among other immune checkpoints, using different modalities.9 21 44 While such studies ultimately show an advantage to targeting multiple inhibitory receptors in terms of efficacy,9 18 45 clear differential responses which distinguish single from dual checkpoint blockade are still unknown.
As both the bsAbs and the anti-PD-1 mAb used in this study present with the same PD-1-targeting moiety,25–27 our study offered a unique opportunity to directly compare single with dual checkpoint inhibition and tease out such potential differential responses.
Our study uncovered several protein-level and transcriptional patterns of response which were shared by single and dual checkpoint blockade and may therefore be primarily PD-1-driven. Additionally, we described dual checkpoint blockade-driven effects, including expansion of proliferating NK cells, downstream secretome changes and upregulation of surface LAG-3 protein, among other differential responses which we here discuss in depth.
As functional T cell exhaustion and its reversal are highly dependent on T cells localized within the tumor,4 we examined whether PD1-TIM3 and PD1-LAG3 were able to stimulate tumor infiltrating immune cells freshly obtained from patients with cancer. We used a combination of IFN-γ as a measure of T cell activity46 together with a flow cytometric assessment of T cell activation markers Ki67 and CD25. Using this approach, we screened 21 patient-derived tumor single cell suspensions for signals of T cell activity under single or dual inhibitory receptor blockade, with 4 out of 21 patient samples showing such signals of activation in the absence of additional stimulation. We focused on samples responsive ex vivo to PD1-TIM3, PD1-LAG3 and anti-PD-1, (ie, samples which showed increased IFN-γ secretion as well as activated and/or proliferating T cells) to directly compare single with dual checkpoint inhibition within samples derived from the same patients. We observed specific treatment-induced changes in the immune microenvironment at the cellular, secretome and transcriptional level. These were specific to the responsive samples in contrast to the non-responsive ones. Similarly, we previously observed that tumor-resident immune cells can be reactivated ex vivo and that the immunological responses measured ex vivo can be predictive of clinical response.47
Our analysis was confirmed using a published dataset of anti-PD-1 responder patients,33 which together with our PCA analysis of 659 soluble markers, further corroborates the idea that our responsive samples present specific cellular and transcriptional features predictive of clinical responses. This analysis therefore served as a confirmation for our experimental approach, validating it as an appropriate system to model responses to checkpoint inhibition. The changes measured in our system could be informative for pharmacodynamic analyses in current and future clinical trials which recruit patients with different indications in escalating dose levels. Mirroring the latter, we decided to include patients with different tumor types in our cohort. Yet, while this potentially increases heterogeneity, it may however introduce variations in our analysis.
Although we focused our mechanistic comparisons on the four patients that responded to all treatments (PD1-TIM3, PD1-LAG3 and anti-PD-1), our in vitro approach identified six samples responsive to treatment with anti-PD-1, while treatments with PD1-TIM3 and PD1-LAG3 resulted in five and four responses, respectively.
We believe differences in number of in vitro responses across treatments could be at least partly explained by differential PD-1, TIM-3 and LAG-3 expression. Indeed, we observed significant correlations between the FC in IFN-γ induced by each compound to the baseline PD-1 expression in each sample assessed, suggesting that the higher the PD-1 expression the higher is the response to PD-1 blocking. Furthermore, our assessment of TIM-3, LAG-3 and PD-1 expression on the responsive samples shows that PD-1 expression is consistently higher on CD8+ and CD4+ T cells in all patients, compared with TIM-3 and LAG-3 expression levels, which were more variable across patients. These data may at least partly explain why anti-PD-1 treatment resulted in the highest number of in vitro responses.
Importantly, our immunophenotyping and scRNAseq analyses identify the presence of a CD4+CXCL13+ cluster which is also found activated and undergoes a number of pathway and gene-level changes on bsAbs or anti-PD-1 treatment. Several lines of evidence from the literature point to a role for CD4+CXCL13+ Tfh or intratumoral Tfh-like cells, besides CD8+PD1hiCXCL13+ T cells, to influence other immune cells within the tumor microenvironment and responses to checkpoint inhibition.38 48 49 49 50 50 51
The chemokine CXCL13 and Tfh cells within the tumor microenvironment play key roles as B cell attractants and modulators of tertiary lymphoid structures, respectively,52 53 with the latter being implicated as a prognostic factor for improved patient survival and clinical outcomes under immunotherapy in recent studies.52 53 Indeed, our scRNAseq analysis revealed that the responsive tumor samples contained large B cell and plasma cell clusters. The presence of these and the CD4+CXCL13+ (Tfh) cluster may be an important determinant of these samples’ potential to respond to treatment in vitro.
As mentioned above, the CD4+CXCL13+ Tfh cluster undergoes several gene, pathway-level and phenotypic changes under treatment, measured in our scRNAseq and flow cytometry analyses. Specifically, we observed display of an activated phenotype with increased HLA-DR, 4-1BB and CD25 surface expression and upregulation of genes for cytokines IL-21, with PD1-LAG3, and IL-17, with all treatments. In parallel, we observed upregulation of immunoglobulin genes in the plasma cell cluster, in response to each treatment, at the 48 hours timepoint. We additionally observed significantly increased secretion of IL-17 and CD40L, which can activate professional antigen presenting cells including B cells,54 55 in response to either bsAbs or anti-PD-1.
It has been shown that B cells can undergo differentiation toward Ab-secreting plasma cells and memory B cells in response to signals from Tfh cells, including IL-21 and IL-17,36–38 48 56 while the existence of B cell effector responses and tumor-specific Abs have been shown to correlate with responses to immune checkpoint inhibitors in renal cell cancer.57
Our analyses using CellChat showed an increase in the numbers of cell-cell interactions, in response to each treatment, between CD4+CXCL13+ and plasma cells and subtle treatment-specific differences. Taken together, our data support the hypothesis that single or dual checkpoint blockade triggers activation of Tfh cells, likely by directly binding to them, and their release of cytokines, such as IL-17 or CD40L (with all treatments) and IL-21 (with PD1-LAG3), which in turn may stimulate B cells to secrete tumor-specific Abs.
Our deeper immunophenotyping by flow cytometry revealed increased activated or cytotoxic states of CD8+ T cells as well as NK cells in response to bsAbs or an anti-PD-1 Ab, together with a decrease in CD45– cells which may indicate reinvigoration of cytotoxic T cells and consequential tumor cell killing. This is in accordance with previous studies in mouse models, demonstrating efficacy of PD-1-targeting, TIM-3-targeting and LAG-3-targeting approaches mediated by effector CD8+ TILs.18 45 58 Although we observed phenotypic changes at the protein level, our single-cell analyses revealed a lack of evident gene-level and pathway-level transcriptional changes in different CD8+ T cell subsets in response to treatments. This phenomenon may be partly dependent on our experimental set-up, which lacks influx and efflux of cells, unlike in a systemic setting, where key events occurring in lymph nodes are known to be important in anti-PD-1/PD-L1 efficacy.59 60 Additionally, it is possible that the bsAbs may primarily target peripheral rather than tumor-resident CD8+ T cells, as inferred from the clonal replacement of tumor-specific T cells and peripheral T cell expansion observed in patients treated with immune checkpoint inhibitors.61 62
Interestingly, PD1-LAG3 treatment significantly decreased the proportion of CD4+CXCL13+ cells and anti-PD-1 significantly decreased the proportion of the naïve progenitor CD8+. We hypothesized that these decreases may reflect differentiation of different cell types toward distinct fates. Our trajectory analyses identified initial naïve and proliferating cell states for either CD4+ or CD8+ lineages and terminal CXCL13+ or exhausted states, respectively. Furthermore, we identify the proliferating subsets as an additional putative terminal state, which indicates that these cells may exist in a self-amplifying, static state. A ‘burned-out’ highly proliferative CD8+ T cell subset was previously found in patients and associated with resistance to anti-PD-1 therapy in another study.63 Importantly, the increase of proliferating CD8+ T cells, either in the tumor or the periphery, has been previously shown in patients treated with anti-PD-1 Abs.64–66 Whether the proliferating CD8+ T cells in our study are a dysfunctional subset or mediators of the response to treatment remains to be explored.
Additionally, we show that PD1-TIM3 and PD1-LAG3 significantly increase the overall proportion of a proliferating NK cell cluster. In particular, we found that the proliferating NK cell subset is enriched for a tissue-residency signature, a finding which is in accordance with previous data from our group, in which we show an association between the presence of tissue-resident NK cells and response to immunotherapies in patients.67
Finally, with our flow cytometric analysis, we identified one distinctive effect in LAG-3 surface upregulation in different CD4+ and CD8+ T cell subsets as a biomarker of response to anti-LAG-3 treatment, which does not require anti-PD-1. Similarly, in a mouse model, a bsAb targeting LAG-3 and PD-L1 was shown to enhance T cell activation and increase soluble LAG-3 in plasma.68 Such increase in soluble LAG-3 may be driven by increased surface expression of LAG-3, as we observe in our study, and its shedding. Our data further indicates that the mechanism by which anti-LAG-3 determines upregulation of LAG-3 surface expression is not by de novo transcription, but rather by conformational changes and reorganization of intracellular stores to the surface. It remains to be investigated whether LAG-3 upregulation occurs in treated patients, it may be a relevant biomarker to monitor responses to treatment and should be considered in the design of treatment schedules.
In conclusion, our study provides a detailed resource of the phenotypical and transcriptional landscapes of human tumor-infiltrating immune cells following treatment with novel clinical bsAbs PD1-TIM3 and PD1-LAG3, as well as with anti-PD-1. Using multi-omic analyses of human tumor samples we developed and validated an experimental system suitable to investigate functional responses to checkpoint inhibition on a single cell level. We identified key cellular and molecular drivers of response to bsAbs treatment, including major transcriptional and cell-networking changes on CD4+CXCL13+ Tfh cells and expansion of proliferating NK cells, and identified LAG-3 surface upregulation as a key consequence of anti-LAG-3 treatment. Prospectively, our findings may aid in the development of therapeutic and biomarker strategies and in the prediction of responses to immune checkpoint blockade.
Methods
Primary human PBMCs and tumor samples
Human PBMCs were isolated from healthy donor (Blood Bank, University Hospital Basel, Switzerland) whole blood by density gradient centrifugation, using Histopaque (Sigma-Aldrich, 10771), and then frozen in liquid nitrogen, using fetal bovine serum (FBS) (Pan Biotech, P30-5500) and 10% dimethyl sulfoxide (Sigma D2650). Tumor samples or pleural effusions were obtained from patients with cancer undergoing tumor resections at University Hospital Basel or Kantonsspital Baselland Liestal, Switzerland. Patient characteristics are summarized in online supplemental tables S1 and S2. Tumor samples were mechanically dissociated and digested into single cell suspensions using accutase (Innovative Cell Technologies, AT-104), collagenase IV (Worthington, LS004188), hyaluronidase (Sigma-Aldrich, H6254) and DNAse type IV (Sigma-Aldrich, D5025). Single-cell suspensions were then stored in liquid nitrogen until further use. In the in vitro assays, single-cell suspensions derived from cancer samples were maintained in RPMI medium containing L-glutamine (Sigma-Aldrich, R8758) supplemented with 1x penicillin/streptomycin (Sigma-Aldrich, P4333), 1 mM pyruvate (Sigma-Aldrich), 1% non-essential amino acids (NEAA); Sigma-Aldrich) and 10% FBS (Pan Biotech, P30-5500).
Treatment of patient samples with therapeutic antibodies
To set up the in vitro treatment of patient samples, 300,000 single cells from the tumor suspensions, prepared as described above, were seeded in 90 µL of RPMI medium containing L-glutamine (Sigma-Aldrich, R8758) supplemented with 1x penicillin/streptomycin (Sigma-Aldrich, P4333), 1 mM pyruvate (Sigma-Aldrich), 1% NEAA (Sigma-Aldrich) and 10% FBS (Pan Biotech, P30-5500) in a 96-well flat bottom plate; 10 µL of 10× concentrated Abs (optimal dose calculated based on unpublished observations), PD1-TIM3 (RO7121661, RG7768, Roche), PD1-LAG3 (RO7247669, RG6139, Roche), anti-PD-1 (pembrolizumab, human IgG1 containing the P329G-LALA mutation, Roche) or DP47 isotype control (Roche) were then added to the cultures to obtain a final concentration of 10 µg/mL. The treated cells were incubated at 37°C, 5% CO2 for 48 or 96 hours before flow cytometry, cytokine analysis or scRNAseq preparation.
Flow cytometry
Treated PBMCs or tumor suspensions were centrifuged at 400 g for 5 min in 96-well plates. They were then stained with a fixable live/dead Zombie dye (BioLegend, 1:200) and blocked with a human Fc receptor binding inhibitor (Invitrogen, 1:100) for 20 min at 4°C. After further centrifugation, cells were stained for cell surface antigens using the fluorophore-conjugated antihuman Abs listed in online supplemental table S6 diluted in FACS buffer (phosphate-buffered saline (PBS), 0.5 mM EDTA, 2% fetal calf serum, 10% sodium azide (NaN3)), for 20 min at 4°C. After two washes with FACS buffer, cells were fixed with intracellular (IC) fix buffer (eBioscience, 00-8222-49) for 20 min at room temperature. Alternatively, for intranuclear staining, cells were further permeabilized and fixed using Invitrogen Fixation/Perm diluent (00-5223-56) and then stained intranuclearly with Perm buffer using the Abs listed in online supplemental table S6. After two washes in Perm buffer, the samples were resuspended in FACS buffer and acquired using Aurora (Cytek). FlowJo (V.10.7.1) was used for standard analysis and R (V.4.1.0) within RStudio (V.1.4.1717) was used for differentially discovery as previously published, using logicle transformation of the acquired data.30
Analysis of LAG-3 protein expression on PBMCs
To measure LAG-3 expression on PBMCs, 96-well plates were coated with PBS containing 5 μg/mL anti-CD3 (clone OKT3, Biolegend 317347) and incubated at 37°C, 5% CO2 for 2 hours. Healthy donor PBMCs were then thawed and seeded at a density of 500,000 cells per well in 90 µL, using RPMI medium containing L-glutamine (Sigma-Aldrich, R8758) supplemented with 1x penicillin/streptomycin (Sigma-Aldrich, P4333) 1 mM pyruvate (Sigma-Aldrich, S8636), 1% NEAA (Sigma-Aldrich, M7145) and 10% FBS (Pan Biotech, P30-5500). Next, 10 µL of either PD1-LAG3, PD1-TIM3, anti-PD-1 (mutated pembrolizumab containing the P329G-LALA mutation, Roche), a parental bivalent anti-LAG-3 AB (RO7223584, Roche) or DP47 isotype were added to reach a final concentration of 10 µg/mL. Cells were incubated for 2 days at 37°C, 5% CO2 after which LAG-3 was measured on CD8+ or CD4+ T cells by flow cytometry, as described above.
To investigate the role of intracellular trafficking of LAG-3, healthy donor PBMCs were thawed and seeded in 96-well U-bottom plates at a density of 300,000 cells per well, in the presence of 5 µg/mL anti-CD3 (clone OKT3, Biolegend 317347), with treatment Abs or isotype and with or without protein trafficking inhibitor Brefeldin A (final 1x concentration, BioLegend, 420601). After 4 hours of incubation at 37°C, 5% CO2, CD8+ and CD4+ T cells were stained for LAG-3 expression, using a non-competitive LAG-3 Ab (LS Bio, LS-B2237-50), as described above.
Measurement of cytokine production
For measurement of cytokine release from either PBMCs or tumor suspensions, 96-well plates containing treated cells were centrifuged at 1500 rpm for 5 min and the cell culture supernatant was collected and stored at −20°C until further use. IFN-γ concentration was measured using a BD OptEIA Human IFN-γ ELISA (BD Biosciences, 555142) following the manufacturer’s instructions.
For multiplex supernatant analysis, samples were analyzed using the proximity extension assays, 96-plex immunoassay developed by Olink Proteomics (Uppsala, Sweden),29 using the Oncology and Inflammation panels. Internal standard and external (blank medium) controls were used for quality check and data were normalized and rescaled to log2 scale using the Normalized Protein eXpression (NPX) software, Olink NPX Manager (V.2.1.0.224). Additional data preprocessing and statistical quality check were performed, including the normality of the NPX values, outlier detection, data variability, the missing data and protein with low detectability (<limit of detection (LOD)). The data were filtered and 659 proteins were kept which had at least 30% (n≥27) observed samples with NPX values above LOD. Further analyses were conducted using limma,69 mixed-effect linear model with the individual-level variability accounted, to compare each Ab treatment with the control, where the p values from moderated t-tests and adjusted false discovery rate for multiple testing were obtained.
Cell sorting and scRNAseq library preparation
For FACS and scRNAseq library preparation, single cells from four tumor samples, treated with either PD1-TIM3, PD1-LAG3, anti-PD-1 (mutated pembrolizumab containing the P329G-LALA mutation, Roche) or DP47 isotype were collected after 48 and 96 hours as described above. Cells were stained using a Zombie Aqua fixable dye (BioLegend, 423101, 1:200) and an antihuman CD45 Ab (clone QA17A19, BioLegend, 393409, 1:100) in PBS 10% bovine serum albumin. Together with the surface antihuman CD45 staining, cells were stained with a TotalSeq-B Human TBNK Cocktail (BioLegend, 399902), using 1 µg of Ab cocktail per 1×106 cells in 100 µL staining volume, following the manufacturer’s instructions. Cells were then filtered using 35 μm nylon mesh-screen filter caps and CD45+ live cells were sorted in PBS 10% FBS using a FACSMelody machine (BD Bioscience). Sorted single cells were then counted and up to 20,000 cells were loaded onto a 10x Genomics Chromium NEXT GEM chip G. Libraires were prepared following the 10x Genomics protocol for 3′ gene expression profiling with feature barcoding (CG000206, Rev A) and complementary DNA or library quality was assessed using a 4200 TapeStation System (Agilent). A total of 31 individual libraries with matched feature (CITE-seq) libraries were prepared (from four patients treated with four treatments each acquired at two timepoints), with one sample (BS558_PD1-TIM3_48h) failed at preparation due to chip wetting failure. Samples were sequenced using an Illumina NovaSeq 6000 instrument aiming to obtain 50,000 reads per cell.
scRNAseq analysis
For scRNAseq data analysis, Cell Ranger V.5.0 was used for demultiplexing, alignment, filtering, barcode counting and unique molecular identifier (UMI) counting. Besca’s standard workflow,31 based on Scanpy,70 was then used on the obtained count matrices to perform quality checks and downstream analysis. A minimum of 800 genes per barcode and a minimum 1500 counts per barcode were used as filtering parameters to remove barcodes that likely did not correspond to viable cells. Thresholds for the proportion of mitochondrial genes (4%–8%) were also applied separately on each group of samples deriving from each patient to remove cells that were likely dead. Doublets were then removed using DoubletDetection.71 Per-cell normalization (counts per 10,000 (cp10k)), count depth and mitochondrial gene content regression, scaling, highly variable gene selection, PCA-based neighborhood analysis and clustering were then performed separately by patient. Besca’s signature-based automated cell type annotation allowed hierarchical classification and annotation of the identified clusters. A further overlapping custom annotation (online supplemental file 2) was user defined to allow comparison of the different patient samples. All samples were then merged and batch corrected for the patient effect using BBKNN.32
CellChat39 was used to investigate cell-cell receptor-ligand interactions. For pseudubulk and cell type abundance analyses, normalized (cp10k) counts were aggregated per cell type (pseudobulk) and the proportion of each cell type out of all cells was calculated per sample (abundance); differential gene expression and differential abundance analyses were then run comparing each treatment with the isotype control at each timepoint. We used limma69 72 for differential gene expression analysis and diffcyt/voom for differential abundance analysis.73 74
Gene Set Enrichment Analysis was run on the pseudobulk outputs for each cell type, using fgsea and msigdbr35 in R V.4.1.0. RNA velocity analysis was conducted using scVelo34 and CellRank.75 Enrichment of previously published NK signatures76 was tested using Seurat72 AddModuleScore function. To generate the anti-PD-1 responder signature, we obtained published data of patients with melanoma treated with anti-PD-133 and followed a previously published approach.77 Briefly, we identified genes which were upregulated and downregulated on anti-PD-1 treatment for patients classified as responders and separately for the non-responders. We then filtered the sets of upregulated and downregulated genes, specific to responders, by removing any genes which showed altered expression in the non-responders, to create response-specific upregulated and downregulated genes.
We then compared the normalized-expression values of the upregulated genes against the downregulated genes in each cell, using a t-test. The absolute value of the t-statistic from this test, showing the magnitude of difference between the upregulated and downregulated genes in our dataset, was used as the anti-PD-1 responder score.
Data visualization was conducted using either Scanpy, Seurat, CellChat, scVelo, CellRank or GraphPad Prism (V.9.0.2).
Statistical analysis
Data were plotted and statistical analysis was conducted using GraphPad Prism (V.9.0.2) or R (V.4.1.0) in RStudio (V.1.4.1717).
Acknowledgments
We would like to acknowledge the advice from the Genomics Facility of the University of Basel and the Department of Biosystems Science and Engineering, ETH Zurich. We thank the Visceral Surgery Laboratory at University Hospital Basel for providing resources used to address questions during the peer-review process.
Footnotes
Twitter: @natoli_marina
PG and FJ contributed equally.
HK and AZ contributed equally.
Contributors: Conceptualization: MN, CK, LC-D, AZ, HK. Methodology: MN, KH, PG, FJ, IID, ZJ, MG, MT, DM, AZw, PW, SS, PU, CK, LC-D. Resources: MW, DL, VH-S, KM, RR, LT. Investigation: MN, KH, PG, FJ, IID, ZJ, PH, DM. Visualization: MN, KH, IID, ZJ, DM. Supervision: AZ, HK. Writing—original draft: MN. Writing—review and editing: MN, KH, PG, FJ, PH, ZJ, IID, MG, MT, DM, CK, LC-D, HK, AZ. Guarantors: MN, HK, AZ. PG and FJ contributed equally to this work. HK and AZ contributed equally to this work as joint senior authors. MN, HK and AZ are co-corresponding authors.
Funding: University of Basel Research grant 3BM1082 (MN). Swiss National Science Foundation 320030_162575 (AZ).
Competing interests: KH, PG, IID, FJ, DM, AZw, PW, SS, PU, CK, LC-D and HK are current or former employees of F. Hoffmann-La Roche and declare patent applications and stock ownership with Roche. AZ received consulting/advisor fees from Bristol-Myers Squibb, Merck Sharp & Dohme, Hoffmann-La Roche, NBE Therapeutics, Secarna, ACM Pharma and Hookipa and maintains further non-commercial research agreements with Secarna, Hookipa, Roche and Beyondsprings. All other authors declare they have no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Raw and aligned sequencing data generated for this work are available from the Gene Expression Omnibus database (GEO, accession number GSE208113). All other data are available in the main text or the supplementary materials.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
This study was approved by Ethikkommission Nordwestschweiz and University Hospital Basel (reference EK321/10). Participants gave informed consent to participate in the study before taking part.
References
- 1.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8:1069–86. 10.1158/2159-8290.CD-18-0367 [DOI] [PubMed] [Google Scholar]
- 2.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350–5. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang AC, Zappasodi R. A decade of checkpoint blockade immunotherapy in melanoma: understanding the molecular basis for immune sensitivity and resistance. Nat Immunol 2022;23:660–70. 10.1038/s41590-022-01141-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thommen DS, Schumacher TN. T cell dysfunction in cancer. Cancer Cell 2018;33:547–62. 10.1016/j.ccell.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baitsch L, Baumgaertner P, Devêvre E, et al. Exhaustion of tumor-specific CD8⁺ T cells in metastases from melanoma patients. J Clin Invest 2011;121:2350–60. 10.1172/JCI46102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zippelius A, Batard P, Rubio-Godoy V, et al. Effector function of human tumor-specific CD8 T cells in melanoma lesions: a state of local functional tolerance. Cancer Res 2004;64:2865–73. 10.1158/0008-5472.CAN-03-3066 [DOI] [PubMed] [Google Scholar]
- 7.Gettinger S, Choi J, Hastings K, et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov 2017;7:1420–35. 10.1158/2159-8290.CD-17-0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyama S, Akbay EA, Li YY, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun 2016;7:10501. 10.1038/ncomms10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson AC, Joller N, Kuchroo VK. Lag-3, tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity 2016;44:989–1004. 10.1016/j.immuni.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenfeld AJ, Hellmann MD. Acquired resistance to immune checkpoint inhibitors. Cancer Cell 2020;37:443–55. 10.1016/j.ccell.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Sanmamed MF, Datar I, et al. Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG-3. Cell 2019;176:334–47. 10.1016/j.cell.2018.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guy C, Mitrea DM, Chou P-C, et al. LAG3 associates with TCR-CD3 complexes and suppresses signaling by driving co-receptor-Lck dissociation. Nat Immunol 2022;23:757–67. 10.1038/s41590-022-01176-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maruhashi T, Sugiura D, Okazaki I-M, et al. Binding of LAG-3 to stable peptide-MHC class II limits T cell function and suppresses autoimmunity and anti-cancer immunity. Immunity 2022;55:912–24. 10.1016/j.immuni.2022.03.013 [DOI] [PubMed] [Google Scholar]
- 14.Dixon KO, Tabaka M, Schramm MA, et al. Tim-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature 2021;595:101–6. 10.1038/s41586-021-03626-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Mingo Pulido Álvaro, Hänggi K, Celias DP, et al. The inhibitory receptor tim-3 limits activation of the cGAS-STING pathway in intra-tumoral dendritic cells by suppressing extracellular DNA uptake. Immunity 2021;54:e1157–67. 10.1016/j.immuni.2021.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Munger ME, Veenstra RG, et al. Coexpression of tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood 2011;117:4501–10. 10.1182/blood-2010-10-310425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngiow SF, von Scheidt B, Akiba H, et al. Anti-tIM3 antibody promotes T cell IFN-γ-mediated antitumor immunity and suppresses established tumors. Cancer Res 2011;71:3540–51. 10.1158/0008-5472.CAN-11-0096 [DOI] [PubMed] [Google Scholar]
- 18.Sakuishi K, Apetoh L, Sullivan JM, et al. Targeting tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med 2010;207:2187–94. 10.1084/jem.20100643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wierz M, Pierson S, Guyonnet L, et al. Dual PD1/LAG3 immune checkpoint blockade limits tumor development in a murine model of chronic lymphocytic leukemia. Blood 2018;131:1617–21. 10.1182/blood-2017-06-792267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gestermann N, Saugy D, Martignier C, et al. LAG-3 and PD-1+LAG-3 inhibition promote anti-tumor immune responses in human autologous melanoma/T cell co-cultures. Oncoimmunology 2020;9:1736792. 10.1080/2162402X.2020.1736792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tawbi HA, Schadendorf D, Lipson EJ, et al. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N Engl J Med 2022;386:24–34. 10.1056/NEJMoa2109970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surowka M, Schaefer W, Klein C. Ten years in the making: application of crossmab technology for the development of therapeutic bispecific antibodies and antibody fusion proteins. MAbs 2021;13:1967714. 10.1080/19420862.2021.1967714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrigues Mantuano N, Natoli M, Zippelius A, et al. Tumor-associated carbohydrates and immunomodulatory lectins as targets for cancer immunotherapy. J Immunother Cancer 2020;8:e001222. 10.1136/jitc-2020-001222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin S, Sun Y, Liang X, et al. Emerging new therapeutic antibody derivatives for cancer treatment. Signal Transduct Target Ther 2022;7:39. 10.1038/s41392-021-00868-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Codarri-Deak L, Seeber S, Perro M. AACR, 2020. [Google Scholar]
- 26.Codarri-Deak L, Weber PA, Seeber S. SITC. Maryland, US, 2019. [Google Scholar]
- 27.Junker F, Gulati P, Wessels U, et al. A human receptor occupancy assay to measure anti-PD-1 binding in patients with prior anti-PD-1. Cytometry A 2021;99:832–43. 10.1002/cyto.a.24334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlothauer T, Herter S, Koller CF, et al. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng Des Sel 2016;29:457–66. 10.1093/protein/gzw040 [DOI] [PubMed] [Google Scholar]
- 29.Assarsson E, Lundberg M, Holmquist G, et al. Homogenous 96-plex pea immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014;9:e95192. 10.1371/journal.pone.0095192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowicka M, Krieg C, Crowell HL, et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res 2017;6:748. 10.12688/f1000research.11622.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mädler SC, Julien-Laferriere A, Wyss L, et al. Besca, a single-cell transcriptomics analysis toolkit to accelerate translational research. NAR Genom Bioinform 2021;3:lqab102. 10.1093/nargab/lqab102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polański K, Young MD, Miao Z, et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 2020;36:964–5. 10.1093/bioinformatics/btz625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sade-Feldman M, Yizhak K, Bjorgaard SL, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018;175:998–1013. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergen V, Lange M, Peidli S, et al. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 2020;38:1408–14. 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- 35.Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu H-C, Yang P, Wang J, et al. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol 2008;9:166–75. 10.1038/ni1552 [DOI] [PubMed] [Google Scholar]
- 37.Mitsdoerffer M, Lee Y, Jäger A, et al. Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc Natl Acad Sci U S A 2010;107:14292–7. 10.1073/pnas.1009234107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui C, Wang J, Fagerberg E, et al. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell 2021;184:6101–18. 10.1016/j.cell.2021.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using cellchat. Nat Commun 2021;12:1088. 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics 2007;8:118–27. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 41.Thommen DS, Schreiner J, Müller P, et al. Progression of lung cancer is associated with increased dysfunction of T cells defined by coexpression of multiple inhibitory receptors. Cancer Immunol Res 2015;3:1344–55. 10.1158/2326-6066.CIR-15-0097 [DOI] [PubMed] [Google Scholar]
- 42.Huang R-Y, Francois A, McGray AR, et al. Compensatory upregulation of PD-1, LAG-3, and CTLA-4 limits the efficacy of single-agent checkpoint blockade in metastatic ovarian cancer. Oncoimmunology 2017;6:e1249561. 10.1080/2162402X.2016.1249561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shayan G, Srivastava R, Li J, et al. Adaptive resistance to anti-PD1 therapy by tim-3 upregulation is mediated by the PI3K-Akt pathway in head and neck cancer. Oncoimmunology 2017;6:e1261779. 10.1080/2162402X.2016.1261779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciraolo E, Althoff S, Ruß J, et al. Simultaneous genetic ablation of PD-1, LAG-3, and Tim-3 in CD8 T cells delays tumor growth and improves survival outcome. Int J Mol Sci 2022;23. 10.3390/ijms23063207. [Epub ahead of print: 16 Mar 2022]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurtulus S, Madi A, Escobar G, et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1-CD8+ tumor-Infiltrating T cells. Immunity 2019;50:181–94. 10.1016/j.immuni.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castro F, Cardoso AP, Gonçalves RM, et al. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol 2018;9:847. 10.3389/fimmu.2018.00847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voabil P, de Bruijn M, Roelofsen LM, et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat Med 2021;27:1250–61. 10.1038/s41591-021-01398-3 [DOI] [PubMed] [Google Scholar]
- 48.Niogret J, Berger H, Rebe C, et al. Follicular helper-T cells restore CD8+-dependent antitumor immunity and anti-PD-L1/PD-1 efficacy. J Immunother Cancer 2021;9:e002157. 10.1136/jitc-2020-002157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Chen H, Mo H, et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 2021;39:1578–93. 10.1016/j.ccell.2021.09.010 [DOI] [PubMed] [Google Scholar]
- 50.Thommen DS, Koelzer VH, Herzig P, et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 2018;24:994–1004. 10.1038/s41591-018-0057-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Workel HH, Lubbers JM, Arnold R, et al. A Transcriptionally distinct CXCL13+CD103+CD8+ T-cell population Is associated with B-cell recruitment and neoantigen load in human cancer. Cancer Immunol Res 2019;7:784–96. 10.1158/2326-6066.CIR-18-0517 [DOI] [PubMed] [Google Scholar]
- 52.Dieu-Nosjean M-C, Giraldo NA, Kaplon H, et al. Tertiary lymphoid structures, drivers of the anti-tumor responses in human cancers. Immunol Rev 2016;271:260–75. 10.1111/imr.12405 [DOI] [PubMed] [Google Scholar]
- 53.Trüb M, Zippelius A. Tertiary lymphoid structures as a predictive biomarker of response to cancer immunotherapies. Front Immunol 2021;12:674565. 10.3389/fimmu.2021.674565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennedy MK, Mohler KM, Shanebeck KD, et al. Induction of B cell costimulatory function by recombinant murine CD40 ligand. Eur J Immunol 1994;24:116–23. 10.1002/eji.1830240118 [DOI] [PubMed] [Google Scholar]
- 55.Schoenberger SP, Toes RE, van der Voort EI, et al. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature 1998;393:480–3. 10.1038/31002 [DOI] [PubMed] [Google Scholar]
- 56.Olatunde AC, Hale JS, Lamb TJ. Cytokine-skewed Tfh cells: functional consequences for B cell help. Trends Immunol 2021;42:536–50. 10.1016/j.it.2021.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meylan M, Petitprez F, Becht E, et al. Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity 2022;55:527–41. 10.1016/j.immuni.2022.02.001 [DOI] [PubMed] [Google Scholar]
- 58.Woo S-R, Turnis ME, Goldberg MV, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res 2012;72:917–27. 10.1158/0008-5472.CAN-11-1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dammeijer F, van Gulijk M, Mulder EE, et al. The PD-1/PD-L1-checkpoint restrains T cell immunity in tumor-draining lymph nodes. Cancer Cell 2020;38:685–700. 10.1016/j.ccell.2020.09.001 [DOI] [PubMed] [Google Scholar]
- 60.Fransen MF, Schoonderwoerd M, Knopf P, et al. Tumor-draining lymph nodes are pivotal in PD-1/PD-L1 checkpoint therapy. JCI Insight 2018;3. doi: 10.1172/jci.insight.124507. [Epub ahead of print: 06 12 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu TD, Madireddi S, de Almeida PE, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 2020;579:274–8. 10.1038/s41586-020-2056-8 [DOI] [PubMed] [Google Scholar]
- 62.Yost KE, Satpathy AT, Wells DK, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 2019;25:1251–9. 10.1038/s41591-019-0522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanmamed MF, Nie X, Desai SS, et al. A burned-out CD8+ T-cell subset expands in the tumor microenvironment and curbs cancer immunotherapy. Cancer Discov 2021;11:1700–15. 10.1158/2159-8290.CD-20-0962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang AC, Orlowski RJ, Xu X, et al. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med 2019;25:454–61. 10.1038/s41591-019-0357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang AC, Postow MA, Orlowski RJ, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60–5. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kamphorst AO, Pillai RN, Yang S, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 2017;114:4993–8. 10.1073/pnas.1705327114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirchhammer N, Trefny MP, Natoli M, et al. NK cells with tissue-resident traits shape response to immunotherapy by inducing adaptive antitumor immunity. Sci Transl Med 2022;14:eabm9043. 10.1126/scitranslmed.abm9043 [DOI] [PubMed] [Google Scholar]
- 68.Kraman M, Faroudi M, Allen NL, et al. FS118, a bispecific antibody targeting LAG-3 and PD-L1, enhances T-cell activation resulting in potent antitumor activity. Clin Cancer Res 2020;26:3333–44. 10.1158/1078-0432.CCR-19-3548 [DOI] [PubMed] [Google Scholar]
- 69.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gayoso A, Shor J, Carr AJ. Dana DoubletDetection (version v3.0) Zenodo; 2020. [Google Scholar]
- 72.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Law CW, Chen Y, Shi W, et al. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weber LM, Nowicka M, Soneson C, et al. Diffcyt: differential discovery in high-dimensional cytometry via high-resolution clustering. Commun Biol 2019;2:183. 10.1038/s42003-019-0415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lange M, Bergen V, Klein M, et al. CellRank for directed single-cell fate mapping. Nat Methods 2022;19:159–70. 10.1038/s41592-021-01346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crinier A, Milpied P, Escalière B, et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity 2018;49:971–86. 10.1016/j.immuni.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Creighton CJ, Fu X, Hennessy BT, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res 2010;12:R40. 10.1186/bcr2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005548supp004.pdf (12.2MB, pdf)
jitc-2022-005548supp001.xlsx (17.2KB, xlsx)
jitc-2022-005548supp002.xlsx (17.1KB, xlsx)
jitc-2022-005548supp003.xlsx (439.8KB, xlsx)
Data Availability Statement
Data are available in a public, open access repository. Raw and aligned sequencing data generated for this work are available from the Gene Expression Omnibus database (GEO, accession number GSE208113). All other data are available in the main text or the supplementary materials.