Abstract
Background
Tumor necrosis factor receptor type 2 (TNFR2) is primarily expressed by CD4+FoxP3+ regulatory T cells (Tregs), especially those present in tumor microenvironment. There is compelling evidence that TNFR2 plays a crucial role in the activation, expansion, and phenotypic stability of Tregs and promotes tumor immune evasion. Understanding of epigenetic regulation of TNFR2 expression in Tregs may help device a novel strategy in cancer immunotherapy.
Methods
MiR-125b-5p-overexpressing or knockdown murine CD4 T cells and Tregs were constructed, and the effect of miR-125b-5p on Tregs proliferation, suppressive function and TNFR2 expression were examined. In vivo antitumor efficacy of Ago-125b-5p (miR-125b-5p agomir) was evaluated in MC38 tumor bearing mice, and tumor-infiltrating Tregs and CD8+ cytotoxic T lymphocytes (CTLs) were analyzed. RNA-seq analysis was applied to reveal the genes and signaling pathways regulated by miR-125b-5p in Tregs.
Results
In this study, we found that TNFR2 was a direct target of miR-125b-5p. Overexpression of miR-125b-5p decreased the proportion of Tregs and their expression of TNFR2 and consequently inhibited its proliferation and suppressive function by regulating the metabolism-related signaling pathways. Moreover, in colon cancer bearing mice, the administration of Ago-125b-5p markedly inhibited the tumor growth, which was associated with reduction of Tregs and increase of IFNγ+CD8+ T cells in tumor environment. Furthermore, in human colon adenocarcinoma patients, we verified that miR-125b-5p expression was downregulated, and low levels of miR-125b-5p were associated with poor prognosis. Interestingly, the expression of miR-125b-5p and TNFR2 were negatively correlated.
Conclusions
Our study for the first time found that the expression of TNFR2 by Tregs was regulated by miR-125b-5p. Our results showed that miR-125b-5p had the capacity to inhibit the expression of TNFR2 and immunosuppressive activity of Tregs and consequently enhanced the antitumor efficacy. This property of miR-125b-5p may be therapeutically harnessed in the treatment of human cancers.
Keywords: Tumor Microenvironment; CD4-Positive T-Lymphocytes; Immunotherapy; Lymphocytes, Tumor-Infiltrating
WHAT IS ALREADY KNOWN ON THIS TOPIC
Tumor necrosis factor receptor type 2 (TNFR2), one of two receptors transducing TNF biological function, preferentially expressed by the most highly suppressive CD4+FoxP3+ Tregs, including those in the tumor environment. TNFR2 signals play decisive role in the activation, expansion, function and phenotypical stability of Tregs. There is compelling evidence that targeting of TNFR2 represents a novel strategy in enhancing the efficacy of tumor immunotherapy by eliminating Treg activity.
WHAT THIS STUDY ADDS
We found TNFR2 was a direct target of miR-125b-5p in Tregs. Overexpression of miR-125b-5p markedly inhibited TNFR2 expression and suppressive function of Tregs. Furthermore, in vivo treatment with miR-125b-5p agomir showed the capacity to inhibit tumor growth in mouse models, accompanied by the decrease of Tregs and increase of CD8 cytotoxic T lymphocytes (CTLs) in tumor tissues.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
These findings for the first time provide experimental evidence that miR-125b-5p could directly regulate TNFR2 expression and suppressive function of Tregs, and this property of miR-125b-5p may be harnessed to design novel cancer immunotherapy.
Background
Overcoming the immunosuppressive tumor microenvironment (TME) is a prerequisite for an effective immunotherapy against tumor.1 The accumulation of CD4+FoxP3+ regulatory T cells (Tregs) is largely attributable to the maintenance of immunosuppressive environment in tumor tissue,2–4 and consequently, the abundance of Tregs is associated with poor prognosis of broad spectrum of human cancers, including colorectal cancer,5 lung cancer,6 and breast cancer.7 Targeting of Tregs has become a strategy to enhance the efficacy of cancer immunotherapy.
MicroRNAs (MiRNAs) are a series of highly conserved single-stranded nucleotide (20–25 nt) non-coding RNAs. The function of miRNAs is mediated by binding of RNA-induced silencing complex to complementary sequences of 3′ untranslated region (3′UTR) in the target mRNA, then inhibit the gene expression at post-transcriptional level.8–10 Through its regulation, miRNAs have been shown to act as oncogenes or tumor suppressors in cancer development and play critical roles in the regulation of immune responses.11 12 Increasing evidence demonstrated that miRNAs could regulate the development and suppressive function of Tregs. For example, miR-142-3p could impair the differentiation of Tregs and reduce Foxp3 stability in models of type 1 diabetes by directly downregulating TET2 expression.13 miR-21, a key regulator of Treg stability, is more highly expressed by Treg cells and can regulate Foxp3 expression.14 15
Our group for the first time found and reported that TNF potently promotes the activation and expansion of Tregs by interacting with TNFR2, one of TNF receptors that expressed by highly suppressive subset of Tregs.16 17 We also reported that tumor-infiltrating Tregs express markedly higher levels of TNFR2 as compared with peripheral Tregs.18 A recent single-cell RNA-sequencing study showed that in patients with metastatic melanoma, TNFR2 is one of the most upregulated genes in Tregs.19 Upregulation or downregulation of TNFR2, therefore, can modulate Treg activity for therapeutic purpose.
Recent studies showed that TNFR2 could be regulated by miRNAs. For example, protumor TNFR2 signal in gastric cancer tissue may be regulated by miR-19a and miR-103a.20 Downregulation of TNFR2 by Let-7f-5p was reported to be the molecular mechanism underlying the osteoprotective effects of plastrum testudinis extracts,21 while miR-125a-5p was purported to be a positive regulator of osteoclast formation by targeting TNFR2.22 However, the epigenetic regulation of TNFR2 expression in Tregs remains elusive.
In this study, we first predicted several potential miRNAs with pseudo binding sites in the 3′UTR of TNFR2 and found that TNFR2 was a direct target of miR-125b-5p. Through transfection of miR-125b-5p mimic or inhibitor (anti-miR-125b-5p), the effects of miR-125b-5p on TNFR2 expression by Tregs and on the proliferation and inhibitory function of Tregs were examined. Moreover, the antitumor efficacy of chemically synthesized miR-125b-5p analog (Ago-125b-5p) was determined in murine tumor models. The results of our study indicate that miR-125b-5p reduced the function of Treg cells by downregulation of TNFR2. This property of miR-125b-5p may be therapeutically harnessed in the treatment of human cancers.
Materials and methods
Clinical samples
A total of 31 pairs of colon adenocarcinoma (COAD) tissues and matched adjacent normal tissues were acquired from Tianjin Medical University Cancer Institute and Hospital (Tianjin, China). The patients were clinically and histopathologically diagnosed as COAD according to WHO criteria. None of the patients have received radiotherapy and chemotherapy before enterectomy.
Mouse, cells and reagents
Female wide type C57BL/6J mice (6–8 weeks old) were provided by the Animal Facility of University of Macau. The mouse colon cancer cell lines of CT26 and MC38 were purchased from American Type Culture Collection (ATCC). The flow cytometry fluorescent conjugated antibodies of FITC anti-mouse CD45 (30-F11), and PE-Cyanine7 anti-mouse CD4 (GK1.5) were purchased from eBioscience; PerCP-Cy5.5 anti-mouse TCRβ/CD3 (H57-597), PE anti-mouse IFN-γ (XMG1.2), and APC anti-mouse CD8a (53–6.7) were purchased from BD Pharmingen; PE anti-mouse CD120b/TNFR2 (TR75-89) was purchased from BioLegend, and APC anti-mouse Foxp3 (FJK-16s) for intracellular staining of Foxp3 was purchased from Invitrogen. Recombinant mouse IL-2 (200 µg/mL) and TNF (200 µg/mL) were obtained from BD Pharmingen. The miRNeasy Mini kit (Cat# 217004) was purchased from QIAGEN, and the PrimeScript RT reagent kit (Cat# RR047A.) was purchased from TAKARA.
Bioinformatic analysis
The online software of Targetscan (http://www.targetscan.org/), miRDB (http://mirdb.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/) and Starbase V.3.0 (http://starbase.sysu.edu.cn/) were used to predict potential miRNAs that target TNFR2 and the binding sites of miRNA in TNFR2 mRNA.
T cell purification and in vitro culture
Mouse CD4+ T cells were purified from spleen and lymph nodes using CD4 (L3T4) microbeads. CD4+CD25+ T cells were purified using the mouse CD4+CD25+ Regulatory T Cell Isolation Kit (Cat# 130091041, Miltenyi Biotec), yielding a purity of ~90% for CD4+FoxP3+ Treg cells. The purified cells were cultured in a U-bottom 96 well plate in a medium of RPMI1640 supplemented with 10% FBS, 2 mM L-glutamine, 10 mM HEPES buffer, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 1% penicillin (100 U/mL)/streptomycin (100 mg/mL), and 50 µM 2-Methylmercaptoethanol, at 37℃ in a humidified incubator with 5% CO2. For T cells culture, IL-2 (10 ng/mL, BD Pharmingen) was added into the culture medium to maintain the survival of CD4+ T cells, and TNF (10 ng/mL, BD Pharmingen) was added to activate the Tregs expansion and proliferation.
MiRNA transfection
The mimics of miR-125b-5p, let-7a-5p, let-7b-5p, let-7c-5p, let-7d-5p, let-7k, and anti-miR-125b-5p and their relative negative control (NC) were chemically synthesized by RiboBio (Guangzhou, China). MiR-125b-5p, anti-miR-125b-5p or five other miRNAs and their negative controls (miR-NC, anti-miR-NC) were transfected into CD4+ T cells or CD4+CD25+ T cells by using riboFECT transfection reagent at a working concentration of 100 nM, following the manufacturer’s instructions. The lentiviral vector GV369 expressing mouse miR-125b-5p (LV-miR-125b-5p) and its empty vector (LV-miR-NC) were generated by Genechem (Shanghai, China). The CD4+CD25+ T cells were transfected with lentivirus at a multiplicity of infection (MOI) of 30 according to the manual protocol. All cells were prepared for analysis after 48 to 72 hours of transfection.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA and miRNA were extracted and purified with miRNeasy Kit (Qiagen), and 0.5 µg of RNA was transcribed into cDNA using PrimeScript RT reagent kit with DNA erase, according to the manufacturer’s instructions. The Bulge-loop miRNA qRT-PCR primer sets for the quantification of miR-125b-5p and other miRNAs were designed and synthesized by RiboBio. The quantitative RT-PCR detection for mRNA and miRNA using TB Green Premix Ex Taq II (Takara). β-actin and U6 were used as the normalization control for mRNA and miRNA. Primers used in this study were as follows: β-actin (forward: 5′-GCTTCTTTGCAGCTCCTTCGT-3′ and reverse: 5′- CCTTCTGACCCATTCCCACC-3′); TNFR2 (forward: 5′-ACAGTGCCCGCCCAGGTTGTCTTG-3′ and reverse: 5′-GAAATGTTTCACATATTGGCCAGGAGG-3′); Foxp3 (forward: 5′- CTTATCCGATGGGCCATCCTG-3′ and reverse: 5′- GTGGAAACCTCACTTCTTGGTC-3′). Data analysis were performed using the 2-ΔΔCт methods.
Dual-luciferase reporter assay
293T cells (2×105) were seeded in a 24 well plate, then transfected with wildtype pGL3-TNFR2 3′-UTR (WT) or mutant pGL3-TNFR2 3′-UTR (MUT) and pRL-TK renilla luciferase plasmid with miR-125b-5p mimic, or miR-NC using the lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions. After 48-hour culture, the relative luciferase activity was determined by the dual-luciferase reporter assay system (Promega); the renilla luciferase value was used as the normalization for firefly luciferase value.
Cell proliferation assay
For in vitro assays of suppression of proliferation by Tregs, CD4+FoxP3+ Tregs (0.5~1×105 cells/well) were seeded in a U-bottom 96-well plate, cultured in complete RPMI-1640 medium with 10 ng/mL IL-2 and 10 ng/mL TNF, then transfected with lentivirus of miR-125b-5p (LV-miR-125b-5p) or scramble control (LV-miR-NC) for 2 days. CFSE-labeled CD4+CD25− effector T cells (Teff, 5×104 cells/well) were cocultured with 2×105 irradiated (3000 Rad) APCs (Antigen-presenting cells, T lymphocytes depleted splenocytes) and 0.5 µg/mL of soluble antimouse CD3 antibody, then Tregs were added to the wells at a desired ratio (Teff: Treg=1:1, 2:1, 4:1, 8:1). After 48~72 hours, CFSE dilution was determined by flow cytometry. The percentage of suppression by Teff/ Treg ratio was calculated by the following formula: [Y (Teffs alone) − Y (Teffs+Tregs)] / Y (Teffs alone) × 100. The Y value represents the average number of divided Teff cells.
RNA sequencing (RNA-seq)
Purified CD4+CD25+ Treg cells were cultured in RMPI1640 complete medium containing 10 ng/mL IL2 and 10 ng/mL TNF, then infected with GFP labeled lentivirus of miR-125b-5p (LV-miR-125b-5p) or scramble control (LV-miR-NC), respectively. After 2 days, FACS-sorted CD4+FoxP3+GFP+ Tregs and total RNA were also isolated. RNA sequencing libraries preparation and sequencing were conducted at Shanghai Genechem Co, Ltd. RNA libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations, and sequencing were performed on an Illumina Novaseq platform. For transcriptome sequencing, FPKM can be used as gene expression data.
Treatment of MC38 mouse model and tumor tissue processing
C57BL/6J mice (female, 6~8 weeks old) were subcutaneously implanted with 0.1 mL PBS containing 5×105 MC38 tumor cells in the right flank. When tumor size reached 50~100 mm3, the tumor-bearing mice were randomly divided into two groups (n=5 mice/group), then intratumorally injected with chemical synthetic analogs of miR-125b-5p (Ago-125b-5p) or its negative control Ago-NC (2.5 nmol/mice, respectively) twice a week for a total of five doses. The tumor size (length and width) and bodyweight of tumor-bearing mice were monitored every 3 days. The tumor size was calculated by the formula: (length×width2 /2. After indicated treatments, the tumors were excised, minced and digested in RPMI1640 medium containing collagenase IV (100 U/mL) and 0.1 mg/mL DNase, and draining lymph nodes (dLNs) or spleen were minced and pushed into a 70 µm cell strainer to create single cell suspensions. Finally, cells were washed using PBS and stained with specific diluted antibodies to determine the contribution of lymphocytes to the development of antitumor immune defense.
Inflammation cytokines assay
Mouse inflammation cytokines were detected using the mouse inflammation kit by BD Cytometric Bead Array, including mouse IL-6, IL-10, MCP-1, IFN-γ, TNF and IL-12p70. For cytokines staining, 10 µL of mouse serum from treated mice were added 10 µL of each mouse inflammation capture bead, 10 µL PE detection reagent and 20 µL PBS, then incubated at room temperature and prevented light for 2 hours. Reconstitute mouse inflammation standards by serial dilutions were used to make a standard curve. Finally, flow cytometry analysis was performed to detect the cytokines level, then convert to concentrations according to the calibration curve.
Quantitation of soluble TNFR2 by ELISA assay
A mouse TNFR2/TNFRSF1B ELISA kit was obtained from SinoBiological Inc (Cat# KIT50128, Beijing, China) for the quantitative detection of mouse soluble TNFR2 in peripheral blood serum from mice treated with Ago-125b-5p or Ago-NC. It is based on a Sandwich assay principle and carried out according to the manufacturer’s specifications. The concentration of mouse TNFR2 was calibrated according to the standard curve. All samples and standards were assayed in duplicate.
Intracellular staining and flow cytometry
After blocking non-specific FcR, cells were incubated with appropriately diluted antibodies according to the manufacturer’s instructions. For intracellular staining of Foxp3, the cells were resuspended in fixation/permeabilization buffer; for IFN-γ staining, the cells were cultured and stimulated by PMA (phorbol 12-myristate 13-acetate) and ionomycin, then resuspended in fixation/permeabilization buffer. All cells acquisition was performed using a BD Fortessa cytometer (BD Biosciences), and data were analyzed by FlowJo software (V.11.0). FACS analysis was gated on the live cells only by using a LIVE/DEAD Fixable Dead Cell Stain kit.
Statistical analysis
GraphPad Prism software (V.8.3.0) was used for data analysis. The statistical significance between two groups was analyzed by Student’s t-test, and the multiple groups’ comparison was analyzed by one-way analysis of variance. The results data were represented as mean±SD (SEM) of multiple experiments. Statistical significance was determined by *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Results
TNFR2 is a direct target of miR-125b-5p
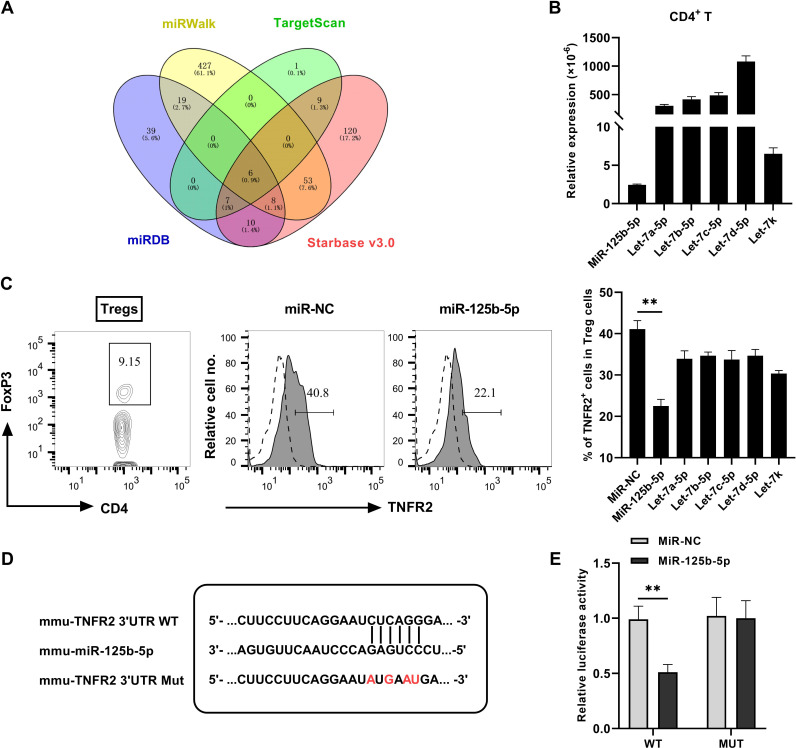
To determine the effect of miR-125b-5p on TNFR2, online resource in websites including Targetscan, miRDB, miRWalk and Starbase V.3.0 were used to predict the putative binding of miRNAs to 3′ untranslated region (3′UTR) of TNFR2. There are six common potential miRNAs obtained from the analysis of four databases, as summarized in figure 1A. The results suggested that several miRNAs might regulate TNFR2 expression. To verify the indeed miRNA that responsible for targeting TNFR2 expression, we examined the expression of miRNAs in CD4+ T cells. As shown in figure 1B, the expression of miR-125b-5p by CD4+ T cells was significantly lower as compared with five other miRNAs. Then we synthesized and transfected these six miRNAs mimic into CD4+ T cells and found that the proportion of TNFR2+ Tregs were markedly reduced. Interestingly, the overexpression of miR-125b-5p was much more potent in the inhibition of TNFR2 expression, in comparison with overexpression of five others (figure 1C, p<0.01). The 3′UTR of TNFR2 containing the complementary binding sites within miR-125b-5p was shown in figure 1D. In addition, the luciferase activities were tested in 293 T cells transfected with miR-125b-5p mimic, including wildtype (WT) or Mutant (MUT) version of predicted TNFR2 3′UTR binding sites of miR-125b-5p. We observed that the overexpression of miR-125b-5p reduced the luciferase activity in WT 3′UTR of TNFR2 but not mutated type (figure 1E). These data suggested that TNFR2 could be directly targeted by miR-125b-5p.
Figure 1.
TNFR2 is a direct target of miR-125b-5p. (A) Bioinformatics search through four public databases of miRWalk, TargetScan, miRDB and Starbase V.3.0 predicted six potential microRNAs (let-7a-5p, let-7b-5p, let-7c-5p, let-7d-5p, let-7k, miR-125b-5p) that target TNFR2, as shown by Venn diagram. (B) The relative expression of miRNAs expression in CD4+ T cells were examined by qRT-PCR. (C) Flow cytometric plot of the proportion of Tregs in CD4 T cells and TNFR2 expression by Tregs, and summary of the proportion of TNFR2+ Tregs after transfected with indicated miRNAs mimic in CD4 T cells. Representative FACS plot on the left, and the quantitative data on the right were presented as mean±SEM of three independent experiments. (D) Schematic presentation of miR-125b-5p binding sites in the 3′UTR regions of TNFR2. Sequences below indicate the mutant form of this site (red mark). (E) Dual luciferase reporter assay in 293 T cells showed miR-125b-5p inhibit the relative luciferase activity of TNFR2 vector-3′UTR (WT) compared with negative control, while miR-125b-5p had no obvious effect on mutant TNFR2 vector-3′UTR (MUT) (n=3, mean±SEM). Comparison of indicated groups, *p<0.05, **p<0.01 (Student’s t-test). TNFR2, tumor necrosis factor receptor type 2; WT, wildtype.
miR-125b-5p downregulates TNFR2 expression on Tregs
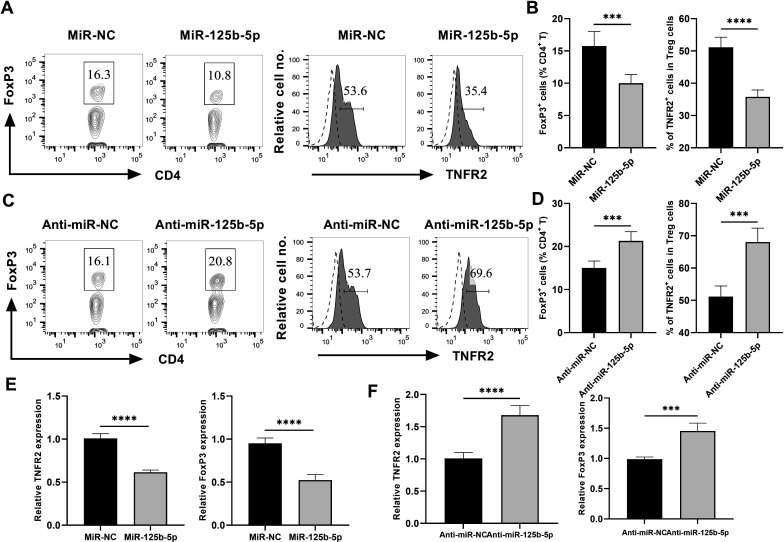
Previously, we and others showed that TNFR2 plays a crucial role in the phenotypical and functional stability of Tregs, and TNF-TNFR2 signal stimulates the activation of Tregs and enhances their suppressive activity.23 24 Thus, miR-125b-5p may reduce the stability of Tregs through downregulation of TNFR2 expression. To test this idea, CD4+ T cells were cultured in the medium containing IL-2 with or without TNF (10 ng/mL each). The result showed that TNF could markedly increase the proportion of FoxP3+ Tregs and TNFR2 expression by Tregs (online supplemental figure S1A, B p<0.001), as expected. Then, miR-125b-5p expression in CD4 T cells was overexpressed or suppressed by transfecting with miR-125b-5p mimic or anti-miR-125b-5p, respectively (the transfection efficiency was shown in online supplemental figure S1C). The result showed that the overexpression of miR-125b-5p in CD4 T cells markedly reduced the proportion of FoxP3+ Treg cells by 40%, and downregulated their TNFR2 expression by ~40% (figure 2A, B, p<0.001). In sharp contrast, silence of miR-125b-5p expression resulted in a 30% increase in the proportion of Tregs in CD4 T cells and an increase of ~25% TNFR2 expression by Tregs (figure 2C, D, p<0.01), which might be explained by the low levels of miR-125b-5b expression. Interestingly, the transcriptional levels of TNFR2 and FoxP3 expression were also markedly reduced in miR-125b-5p overexpressing Tregs (figure 2E, p<0.0001), while their expression were increased by the treatment with anti-miR-125b-5p (figure 2F, p<0.01).
Figure 2.
miR-125b-5p downregulates TNFR2 expression on Tregs. MACS-purified CD4+ T cells from pooled lymph nodes and spleen of wide type C57BL/6 J mice were cultured in the presence of IL-2 and TNF (10 ng/mL each), then cells were transfected with 100 nM miR-125b-5p mimic/inhibitor or their relative negative control (NC). Flow cytometric plot (A) and summary (B) of the proportion of FoxP3+ Tregs in CD4+ T cells and TNFR2 expression by Tregs after transfected with miR-125b-5p. Number shows the proportion of gated cells. Flow cytometric plot (C) and summary (D) of the proportion of Tregs in CD4+ T cells and TNFR2 expression by Tregs after transfected with anti-miR-125b-5p. Number shows the proportion of gated cells. The mRNA expression of TNFR2 and FoxP3 by Tregs after transfected with miR-125b-5p (E), and anti-miR-125b-5p (F) were examined through qRT-PCR. Representative FACS data were from three independent experiments with similar results. The summarized data shown in figure parts B, D, E and F were presented as mean±SEM of three independent experiments performed in triplicate (n=9). By comparison with miR-NC (or anti-miR-NC), **p<0.01, ***p<0.001, ****p<0.0001 (Student’s t-test). TNFR2, tumor necrosis factor receptor type 2.
jitc-2022-005241supp002.pdf (427.9KB, pdf)
Furthermore, overexpression of miR-125b-5p also markedly reduced the proportion of FoxP3+ Tregs in TNFR1−/− CD4+ T cells by 37%, but it only resulted in a 25% reduction of Tregs in TNFR2−/− CD4+ T cells (online supplemental figure S2A, B p<0.01), which suggesting that miR-125b-5p may also downregulate FoxP3 expression in a TNFR2-independent manners. Moreover, overexpression of miR-125b-5p markedly decreased the expression of TNFR2 by TNFR1−/− Treg cells (online supplemental figure S2C, D p<0.001). Therefore, our data clearly indicated that miR-125b-5p could significantly reduce TNFR2 and FoxP3 expression in Tregs.
jitc-2022-005241supp003.pdf (538.8KB, pdf)
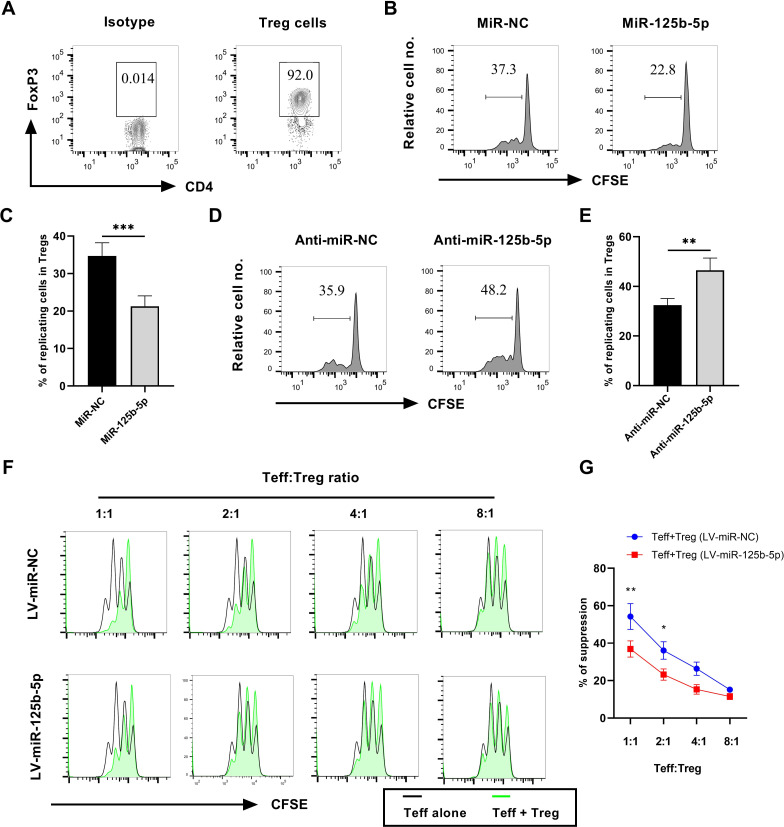
miR-125b-5p inhibits the proliferation and attenuates the suppressive activity of Tregs
There is compelling evidence that TNFR2 expression by Tregs is required for their immunosuppressive function.25 26 Thus, decreased expression of TNFR2 by miR125b-5p may attenuate Treg function. To test this possibility, Treg cells from normal C57BL/6J mice were MACS-purified based on surface expression of CD4 and CD25 and the purity of FoxP3+ Tregs >90% (figure 3A). The proliferation of Tregs was induced by stimulating with IL-2 and TNF as we previously reported.27 Results showed that the overexpression of miR-125b-5p reduced the proliferation of Tregs by 40% (figure 3B, C, p<0.01). In contrast, the proportion of proliferating Tregs was increased by 30% after the silence of miR-125b-5p (figure 3D, E, p<0.01). However, there was no difference in cellular viability between miR-125b-5p or anti-miR-125b-5p treatment and its controls (online supplemental figure S3). Therefore, we assumed that altered proliferative expansion may result in an increase or decrease in the number of Tregs, which will affect its function. Besides, transfection of miR-125b-5p or anti-miR-125b-5p did not alter the activation and proliferation of CD4+CD25− Teff cells (online supplemental figure S4A–E) and even CD8 T cells (data not shown).
Figure 3.
MiR-125b-5p inhibits the proliferation and suppressive activities of Tregs. MACS-purified CD4+CD25+ T cells from pooled lymph nodes and spleen of wide type C57BL/6J mice were cultured in the presence of IL-2 and TNF (10 ng/mL each), then transfected with 100 nM miR-125b-5p mimic/inhibitor. The proliferation of Tregs were assessed by in vitro CellTrace CFSE assay. (A) The purity of FoxP3+ Tregs in CD4+CD25+ T cells was examined by flow cytometry. Number shows the proportion of gated cells. Typical FACS analysis (B) and summary (C) of Tregs proliferation, as shown by dilution of CFSE expression (gated on FoxP3+ Tregs) by transfected with miR-125b-5p. Number in the histogram indicates the proportion of gated cells. Typical FACS analysis (D) and summary (E) of Tregs proliferation, as shown by dilution of CFSE expression (gated on FoxP3+ Tregs) by transfected with anti-miR-125b-5p. Number in the histogram indicates the proportion of gated cells. Furthermore, suppressive activity of miR-125b-5p overexpressed Tregs cocultured with CFSE-labeled CD4+CD25− Teff cells at desired ratios was measured at day 2, using an irradiated (3000 rad) APCs and soluble CD3-specific antibody assay. (F) The representative CFSE profile of Teff cells at different Teff: Treg ratios (1:1, 2:1, 4:1, 8:1) as shown in the histogram. (G) The percentage of suppression at different Teff:Treg cell ratios as shown. Representative FACS data were from three independent experiments with similar results. The summarized data shown in figure parts C, E and G were presented as mean±SEM of three independent experiments performed in triplicate (n=9). By comparison with miR-NC (or anti-miR-NC), *p<0.05, **p<0.01 (Student’s t-test).
jitc-2022-005241supp004.pdf (349.3KB, pdf)
jitc-2022-005241supp005.pdf (497.6KB, pdf)
To examine the effect of miR-125b-5p on the suppressive function of Tregs, CD4+CD25+ Treg cells were infected with LV-miR-125b-5p and cocultured with CFSE-labeled Teff cells. The result showed that the suppressive activity of miR-125b-5p-overexpressing Treg cells was markedly reduced (figure 3F, G, p<0.05), indicating that miR-125b-5p could regulate the function of Tregs which is at least partially resulted from the downregulation of TNFR2. Moreover, consistent with previous report, TNF treatment markedly reduced PD-1 expression but increased CTLA4 expression in Tregs (online supplemental figure S5A p<0.05). However, overexpression of miR-125b-5p markedly reduced CTLA4 expression, but increased PD-1 expression in Tregs (online supplemental figure S5B, p<0.01). In contrast, silence of miR-125b-5p showed an opposite effect (online supplemental figure S5C, p<0.05). Therefore, miR-125b-5p may preferentially regulate the proliferation and function of Treg cells.
jitc-2022-005241supp006.pdf (308.8KB, pdf)
miR-125b-5p inhibits metabolism-related signaling pathways in Tregs
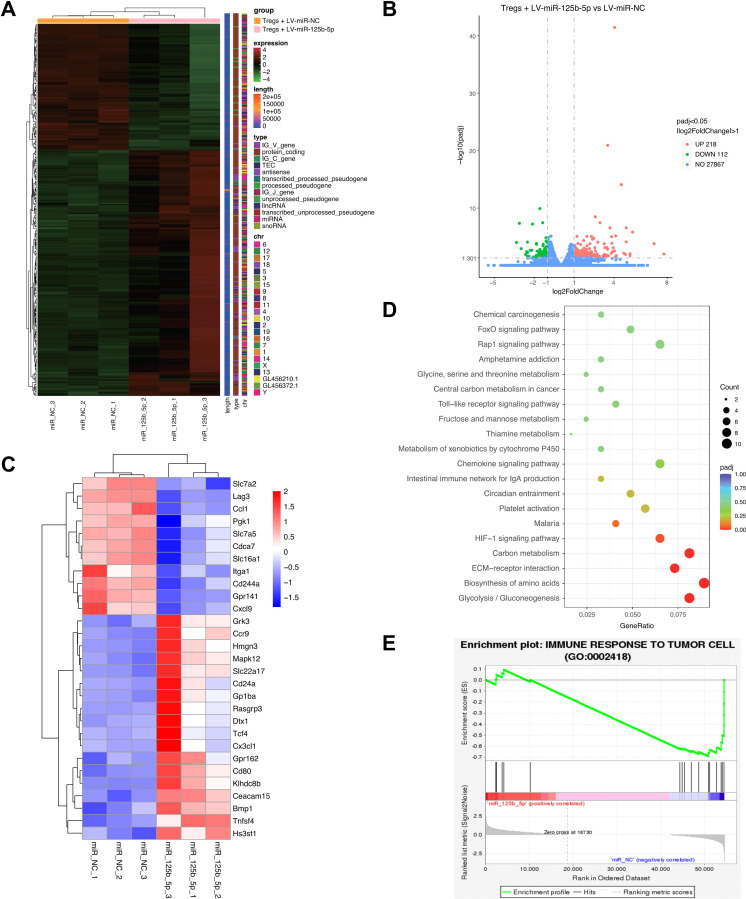
To further understand the molecular basis of regulatory effect of miR-125b-5p on Tregs, transcriptome sequence of LV-miR-125b-5p transfected Tregs showed a differentially expressed genes pattern (figure 4A), including downregulation of 112 (fold change <0.5), and upregulation of 218 genes (fold change >2) (figure 4B). As shown in figure 4C, the most upregulated genes include Tnfsf4, Bmp1, Hs3st1, Cd80, Gpr162, and the downregulated genes include Lag3, Ccl1, Pgk1, Cxcl9, Itga1, etc. The GO and KEGG pathway analysis revealed that the differentially expressed genes mostly involved in glycolysis, carbon metabolism, and HIF-1 signaling pathways, which are known regulators of cell metabolism (figure 4D). In addition, GSEA analysis showed that the expression of miR-125b-5p affect immune responses to tumor cell progression (figure 4E). Taken together, these findings further shed light on the important roles of miR-125b-5p in regulating the function of Tregs.
Figure 4.
RNA-seq analysis of differentially expressed genes in miR-125b-5p transfected Tregs. Purified CD4+CD25+ Treg cells were transfected with 100 nM LV-miR-125b-5p or its negative control (LV-miR-NC) for 2 days, then transcriptome sequencing was performed. (A) The differentially expressed genes in miR-125b-5p overexpressed Tregs compared with control cells (n=3, each group) were shown in the heatmap. (B) The differentially expressed genes were shown in volcano plot. X-axis means log2 foldchange; Y-axis means −log10pvalue. Gray dotted line represents the screening standard threshold of different expressed genes. (C) The selected most upregulated and downregulated genes were shown in heatmap. (D) KEGG pathway analysis of enriched differentially expressed genes. (E) Gene Set Enrichment Analysis (GSEA) using Tregs RNA-seq data showed that miR-125b-5p expression levels affect regulation of immune response to tumor cell progression.
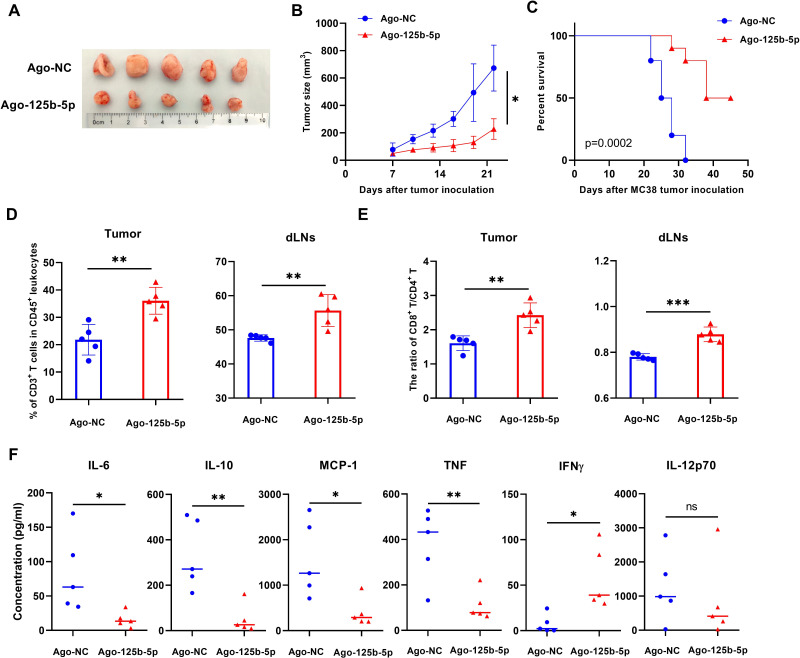
Antitumor effect of Ago-125b-5p is associated with reduction of TNFR2-expressing Tregs in murine MC38 tumor models
To further determine the effect of overexpression of miR-125b-5p on tumor growth, C57BL/6 J mice bearing established subcutaneous MC38 tumors were intratumorally injected with Ago-125b-5p, the growth of tumors was monitored. The result showed that Ago-125b-5p treatment significantly reduced the tumor growth (figure 5A, B, p<0.05) and potently elongated the tumor-bearing mice survival (figure 5C, p<0.05). The treatment with Ago-125b-5p resulted in a marked increase in the infiltration of CD3+ T lymphocytes into the tumor and draining LNs (figure 5D, p<0.01), while the ratio of CD8 versus CD4 T cells was increased in the tumor tissue as well as draining LNs (figure 5E, p<0.01). Furthermore, the treatment with Ago-125b-5p markedly reduced the serum levels of IL-6, IL-10, TNF and MCP-1, while levels of IFNγ were increased in tumor bearing mice (figure 5F, p<0.05).
Figure 5.
Therapeutic effect of Ago-125b-5p on MC38 tumor bearing mice models. C57BL/6 J mice were subcutaneously injected with 5×105 MC38 tumor cells each mouse on the right flank on day 1. From day 7, MC38 tumor bearing mice with intratumor injections of Ago-125b-5p or Ago-NC (control) every other day at 2.5 nmol/mouse as indicated for total of five doses. (A) Representative tumor size from each mouse was shown (n=5, each group). (B) The tumor growth curve was plotted (n=5, each group). (C)The tumor bearing mice survival curve (n=10 mice, each group) from two independent experiments. After the last treatment, the tumor tissue, draining lymph nodes and periphery blood serum from each mouse were collected. The tumor tissue and draining lymph nodes were digested and grinded into single cell suspensions as indicated. (D) Summary of the percentage of CD3+ T cells infiltrated in tumor tissue and draining lymph nodes. (E) The ratio of CD8:CD4 T cells in tumor tissue and draining lymph nodes. (F) The serum levels of inflammatory cytokine IL-6, IL-10, MCP-1, TNF, IFNγ and IL-12p70 from Ago-125b-5p treated mouse. The summarized data shown in figure parts D, E, and F were displayed as mean±SEM, n=5 mice of each group. All data were representatives of three separate experiments. By comparison with Ago-NC, *p<0.05, **p<0.01, ***p<0.001, (Student’s t-test).
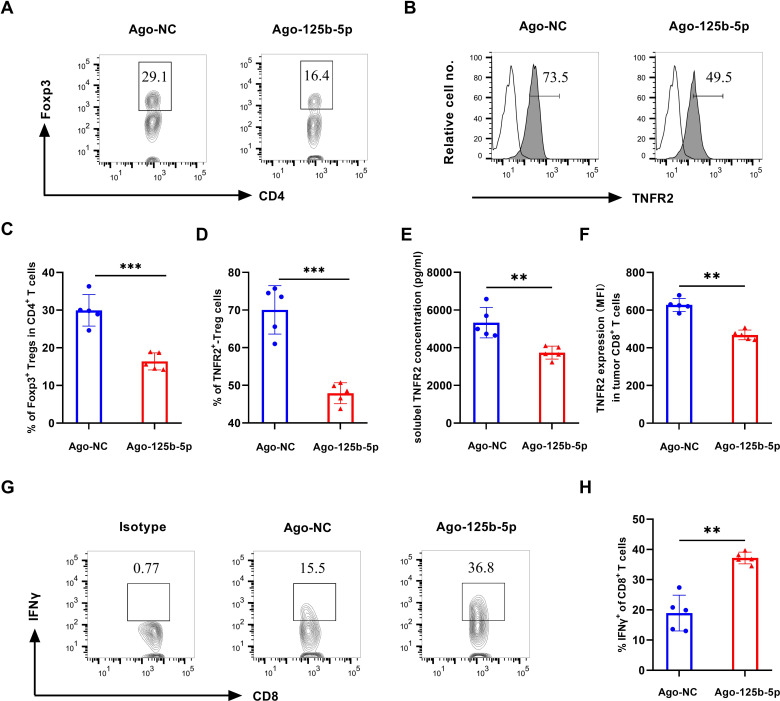
We further analyze Tregs and their TNFR2 expression in tumor environment and found that the proportion of Foxp3+ Treg cells in tumor-infiltrating CD4+ T cells was significantly decreased in mice treated with Ago-125b-5p (figure 6A, C, p<0.001), accompanied by a marked decrease of TNFR2 expression by Treg cells (figure 6B, D, p<0.001). Intriguingly, the soluble levels of TNFR2 were reduced in serum in mice treated with Ago-125b-5p (figure 6E, p<0.01). The expression of TNFR2 was also downregulated in tumor-infiltrating CD8+ T cells by Ago-125b-5p treatment (figure 6F, p<0.01), while the treatment increased the expression of IFN-γ in CD8+ T cells in tumor tissue (figure 6G, H, p<0.01) and in dLNs (data not shown). These data suggest that the reduction of TNFR2-expressing Treg cells and mobilization of CD8+ T cells were attributable at least in part to the antitumor effect of miR-125b-5p.
Figure 6.
Effect of Ago-125b-5p on the tumor infiltration Tregs and TNFR2 expression and on cytotoxic CD8 T cells. After treatment by Ago-125b-5p for five doses on MC38 tumor-bearing mice, the immune cells in tumor tissue were detected by flow cytometry. (A) Representative flow cytometric plot of the proportion of Tregs in CD4+ T cells in mouse tumor tissue after treated by Ago-125b-5p. Number shows the proportion of gated cells. (B) Representative typical histograms of TNFR2 expression in Foxp3+ Tregs. Number in the histogram indicates the proportion of gated cells. (C) Summary of the proportion of FoxP3+ Tregs in CD4 populations as shown in figure part A. (D) The expression of TNFR2-expressing cells in Tregs. (E) The concentration of soluble TNFR2 levels in serum after treated by Ago-125b-5p. (F) The mean fluorescence intensity (MFI) of TNFR2 expression by tumor-infiltrating CD8+ T cells. (G) Representative flow cytometric plot of the proportion of IFNγ+CD8+ T cells in CD8+ T cells in mouse tumor tissue after treated by Ago-125b-5p. Number shows the proportion of gated cells. (H) Summary of the proportion of IFNγ+CD8+ T cells in CD8+ T cells. The summarized data shown in figure parts C, D, E, F and H were displayed as mean±SEM, n=5 mice of each group. All data were representatives of three separate experiments. By comparison with Ago- NC, *p<0.05, **p<0.01, ***p<0.001 (Student’s t-test). TNFR2, tumor necrosis factor receptor type 2.
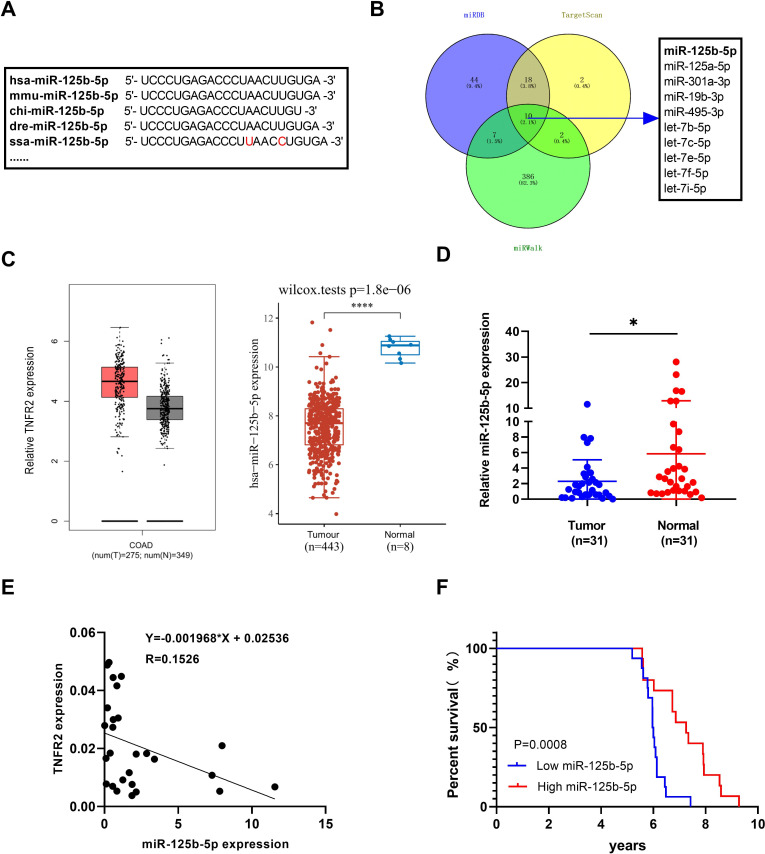
Clinical relevance of low miR-125b-5p expression may be associated with high TNFR2 expression in human colon cancer patients
Through the bioinformatics analysis, we found that the mature sequence of miR-125b-5p is highly conserved among most vertebrate species, including human and mouse (figure 7A). To predict putative miRNAs that have binding sites in 3′UTR of human TNFR2, we analyzed the online database (Targetscan, miRDB and miRWalk). The result indicated that human TNFR2 may also be directly targeted by miR-125b-5p (figure 7B). We also found that human COAD tumor tissues expressed higher levels of TNFR2 and lower levels of miR-125b-5p, as compared with adjacent normal tissues according to the data from TCGA cohort (figure 7C). Furthermore, qRT-PCR analysis on 31 pairs of colon cancer tumor tissues, we could confirm that miR-125b-5p levels were significantly decreased in tumor tissues (figure 7D, p<0.05), and the relationship between TNFR2 expression and miR-125b-5p in human cancer was shown in figure 7E. χ2 test showed that low miR-125b-5p was associated with more advanced TNM stage and lymphatic metastasis (online supplemental figure S1). Furthermore, Kaplan-Meier survival analysis indicated that poor prognosis of patients with cancer was associated with low miR-125b-5p expression (figure 7F, p=0.0008). Taken together, these results suggested that miR-125b-5p was negative correlated to TNFR2 expression in human tumor tissues, aligning well with the results of our in vitro and in vivo laboratory mouse studies.
Figure 7.
miR-125b-5p is negative associated with TNFR2 expression in human colon cancer patients. (A) The mature sequences of miR-125b-5p in human, mice and other species. (B) Bioinformatics search through public databases of miRWalk, TargetScan and miRDB predicted potential microRNAs that target human TNFR2 3’UTR, as shown by Venn diagram. (C) The relative expression of TNFR2 and miR-125b-5p in human colon adenocarcinoma (COAD) tissues and adjacent normal tissues from TCGA cohort. (D) qRT-PCR analysis of miR-125b-5p levels in human colon cancer patients (n=31; *p<0.05, Student’s t-test). (E) The linear regression analysis of correlation between miR-125b-5p and TNFR2 expression in human COAD tissues. (F) miR-125b-5p expression is associated with overall survival (OS) of colon cancer patients (p=0.0008, Kaplan-Meier analysis). TNFR2, tumor necrosis factor receptor type 2.
Discussion
Disruption of the synthesis of miRNA in Tregs results in the complete dysfunction of Tregs and induces the occurrence of inflammatory disease and autoimmunity,28–30 indicating that miRNAs are critical in the development and function of Treg cells. It was proposed that miR-155 plays a role in regulating the activity of induced Treg (iTreg) and natural Treg (nTreg) by suppression of cytokine signaling 1 (a negative regulator of IL-2/STAT5 signaling pathway).31 Acting as an immuno-metabolic regulator, miR-142–5 p was shown to be essential for controlling Treg suppressive function through the maintenance of high intracellular concentration of cAMP.32 It was also shown that miR-21 was a positive regulator, while miR-31 acting as a negative regulator of human natural Tregs by modulating Foxp3 expression.15 Moreover, miR-17 was found to inhibit the function of Tregs by downregulating the coregulators of the Foxp3 transcriptional factor,33 and NF-kB-driven miR-34a could disrupt Treg/Th17 balance via targeting Foxp3.34 In this study, we for the first time found that miR-125b-5p could negatively control Treg activity via, at least partially, downregulating TNFR2 expression.
The mature sequence of miR-125b-5p is evolutionarily conserved in diverse vertebrate species, making it an attractive target for translation research since the findings based on mouse study is likely to extrapolate to humans. Previously, the study on miR-125b-5p main focused on human cancer cells, such as hepatocellular carcinoma.35 It was found that the expression of miR-125b-5p was markedly downregulated in tumor cells and acted as a tumor suppressor.36 37 Nevertheless, miR-125b-5p was also reported to play a role in the regulation of immune responses. For example, it inhibits the activation and induces the apoptosis of human γδ T cells.38 In this study, through bioinformatics analysis, we first predicted six miRNAs that may bind with 3′UTR of TNFR2, including miR-125b-5p and five let-7 family members. Among them, miR-125b-5p most potently inhibited TNFR2 expression by CD4 T cells. In view of the potential role of miR-125b-5p in suppressing tumor growth and regulating immune cells, we further investigated the effect of miR-125b-5p on TNFR2 expression by Tregs.
The results of our study suggest that miR-125b-5p directly acts on and negatively regulates TNFR2 expression in Tregs, consequently reduces the number of Tregs and attenuates their suppressive activity. The fact that miR-125b-5p has a similar effect on Tregs in WT and TNFR1-KO T cells, while a minor but a clear effect was observed in TNFR2-KO T cells (online supplemental figure S2A, B), suggesting that miR-125b-5p can regulate Tregs through TNFR2-dependent and TNFR2-independent mechanism. Indeed, it was reported that miR-125b could directly act on Foxp3 and promote autophagy.39
TNFR2 can also be expressed by effector CD8+ T cells, which drives a cytotoxic ability to CD8+ Teff cells during the early immune response, as well as an apoptosis signal to terminate the immune response.40 Besides, the high expression of TNFR2 on cytotoxic CD8 T cells may attenuate their antitumor effector function.19 Therefore, miR-125b-5p as a direct target on TNFR2 may mitigate the negative effect of TNFR2 expression by CD8 CTLs. This notion is supported by the fact that Ago-125b-5p treatment reduced TNFR2 expression by CD8 T cells in tumor bearing mice, while promoting the effector function of CD8 CTLs as evidenced by the upregulation of IFNγ (figure 6F, G, H). Further study is needed to clarify if the activation of CD8 CTLs in Ago-125b-5p-treated mice is resulted from the down regulation of TNFR2 on CD8 T cells, or from the elimination of Tregs. In TME, myeloid-derived suppressor cells (MDSCs) with potent immunosuppressive function can also express TNFR2 and the signal of TNFR2 can stimulate the accumulation and activation of MDSCs.41 42 Whether miR-125b-5p also regulate the activity of MDSCs through decreasing TNFR2 expression should be further clarified in future study.
The soluble TNFR2 (sTNFR2) is produced by proteolytic cleavage of its membrane bound counterpart. Its level in serum is a potent immunosuppressive mediator and acts as a predictor of treatment response to inflammatory disease.43 44 Increased levels of sTNFR2 were reported to be associated with enhanced colorectal cancer risk and significantly accelerated their all-cause mortality.45 46 Therefore, serum sTNFR2 might be a powerful predictive factor for cancer treatment. This notion is also supported by the fact that, in our study, sTNFR2 were reduced remarkably by the treatment of Ago-125b-5p in tumor bearing mice, suggesting the reduction of sTNFR2 could be attributable to the mobilization of antitumor immune response.
It was reported that intracellular metabolism played crucial roles in modulating the activation and expansion of Treg cells.47 48 The activation of Tregs by TNF-TNFR2 signaling were attributable to stabilize human regulatory T cells switch to glycolysis, with increased intracellular lactate levels and glucose consumption.49 To demonstrate the potential molecular mechanism of miR-125b-5p on the activity of Tregs, we performed an RNA-seq analysis in miR-125b-5p overexpressed Tregs and found that miR-125b-5p exerted its roles on Tregs by regulating the glucose and lipid metabolism related signaling pathways, including HIF-1 signaling pathway, carbon metabolism and glycolysis (figure 4). Therefore, the overexpression of miR-125b-5p partially disrupt TNF-TNFR2 signaling on suppressing Treg cells by interfering cellular metabolism related signaling.
To date, several miRNAs-based therapeutics have reached clinical trials, including a miR-34 mimic for the treatment of cancer,50 and anti-miR-122 for the treatment of hepatitis.51 Since miRNAs are small, water soluble and can be injected subcutaneously or intravascularly, targeted delivery of miRNA with nanoparticles, plasmids, viral vectors or lipid delivery vectors into desirable site appears to be feasible and safe.52 Our future study will focus on the development of appropriate delivery methods for miR-125b-5p or its modified analogs in a preclinical and then clinical settings.
Conclusions
In summary, we for the first time found that miR-125b-5p could directly act on TNFR2. Our data favor the notion that downregulation of TNFR2 expression is at least partially attributable to the negative regulation of miR-125b-5p on Treg activity. Therefore, in addition to the known antitumor effect by directly acting on tumor cells, targeting of miR-125b-5p may represent a novel strategy in cancer immunotherapy by eliminating tumor infiltrating TNFR2-expressing Tregs.
jitc-2022-005241supp001.pdf (83.4KB, pdf)
Footnotes
Contributors: MJ and YY contribute equally to this work. All authors' contribution: conceptualization and design: MJ and XC. Data collection and analysis: MJ, YY and LN. Methodology: YY, PL, FC and PL. Investigation: YC, JZ, YW and HH. Funding acquisition: XC. Project administration: XC and HL. Writing – original draft: MJ and XC. Writing – review and editing: MJ, HL and XC. XC and HL acted as guarantors and corresponding authors for this study.
Funding: This project has been funded by The Science and Technology Development Fund, Macau SAR (FDCT, File No. 0056/2019/AFJ and 0099/2021/A2), University of Macau (File No. MYRG2019-00169-ICMS, CPG-CPG2022-00024-ICMS), Guangdong-Hong Kong-Macau Joint Lab on Chinese Medicine and Immune Disease Research (EF006/ICMS-CX/2021/GDSTC) and The 2020 Guangdong Provincial Science and Technology Innovation Strategy Special Fund (Guangdong-Hong KongMacau Joint Lab), No: 2020B1212030006.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
All clinical samples were collected from consenting individuals according to the protocols approved by the Ethics Review Board at Tianjin Medical University Cancer Institute and Hospital (No. Ek2021165). The animal study protocol was approved by Animal Research Ethics Committee of University of Macau (#UMARE-AMEND-102). Participants gave informed consent to participate in the study before taking part.
References
- 1.Jain RK. Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J Clin Oncol 2013;31:2205–18. 10.1200/JCO.2012.46.3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dees S, Ganesan R, Singh S, et al. Regulatory T cell targeting in cancer: emerging strategies in immunotherapy. Eur J Immunol 2021;51:280–91. 10.1002/eji.202048992 [DOI] [PubMed] [Google Scholar]
- 3.Li C, Jiang P, Wei S, et al. Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol Cancer 2020;19:116. 10.1186/s12943-020-01234-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paluskievicz CM, Cao X, Abdi R, et al. T regulatory cells and priming the suppressive tumor microenvironment. Front Immunol 2019;10:2453. 10.3389/fimmu.2019.02453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y-C, Mahalingam J, Chiang J-M, et al. Activated but not resting regulatory T cells accumulated in tumor microenvironment and correlated with tumor progression in patients with colorectal cancer. Int J Cancer 2013;132:1341–50. 10.1002/ijc.27784 [DOI] [PubMed] [Google Scholar]
- 6.Tao H, Mimura Y, Aoe K, et al. Prognostic potential of FOXP3 expression in non-small cell lung cancer cells combined with tumor-infiltrating regulatory T cells. Lung Cancer 2012;75:95–101. 10.1016/j.lungcan.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 7.Shou J, Zhang Z, Lai Y, et al. Worse outcome in breast cancer with higher tumor-infiltrating FOXP3+ Tregs : a systematic review and meta-analysis. BMC Cancer 2016;16:687. 10.1186/s12885-016-2732-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Yu X, Hu S, et al. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics 2009;7:147–54. 10.1016/S1672-0229(08)60044-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B, Pan X, Cobb GP, et al. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1–12. 10.1016/j.ydbio.2006.08.028 [DOI] [PubMed] [Google Scholar]
- 10.O'Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol 2018;9:402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortez MA, Anfossi S, Ramapriyan R, et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer 2019;58:244–53. 10.1002/gcc.22725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rupaimoole R, Calin GA, Lopez-Berestein G, et al. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov 2016;6:235–46. 10.1158/2159-8290.CD-15-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherm MG, Serr I, Zahm AM, et al. miRNA142-3p targets Tet2 and impairs Treg differentiation and stability in models of type 1 diabetes. Nat Commun 2019;10:5697. 10.1038/s41467-019-13587-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong L, Wang X, Tan J, et al. Decreased expression of microRNA-21 correlates with the imbalance of Th17 and Treg cells in patients with rheumatoid arthritis. J Cell Mol Med 2014;18:2213–24. 10.1111/jcmm.12353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouas R, Fayyad-Kazan H, El Zein N, et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol 2009;39:1608–18. 10.1002/eji.200838509 [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Subleski JJ, Hamano R, et al. Co-expression of TNFR2 and CD25 identifies more of the functional CD4+FOXP3+ regulatory T cells in human peripheral blood. Eur J Immunol 2010;40:1099–106. 10.1002/eji.200940022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Bäumel M, Männel DN, et al. Interaction of TNF with TNF receptor type 2 promotes expansion and function of mouse CD4+CD25+ T regulatory cells. J Immunol 2007;179:154–61. 10.4049/jimmunol.179.1.154 [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Oppenheim JJ. The phenotypic and functional consequences of tumour necrosis factor receptor type 2 expression on CD4(+) FoxP3(+) regulatory T cells. Immunology 2011;133:426–33. 10.1111/j.1365-2567.2011.03460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tirosh I, Izar B, Prakadan SM, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossi AFT, Contiero JC, Manoel-Caetano FdaS, et al. Up-regulation of tumor necrosis factor-α pathway survival genes and of the receptor TNFR2 in gastric cancer. World J Gastrointest Oncol 2019;11:281–94. 10.4251/wjgo.v11.i4.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen G-Y, Ren H, Huang J-J, et al. Plastrum testudinis extracts promote BMSC proliferation and osteogenic differentiation by regulating Let-7f-5p and the TNFR2/PI3K/AKT signaling pathway. Cell Physiol Biochem 2018;47:2307–18. 10.1159/000491541 [DOI] [PubMed] [Google Scholar]
- 22.Sun L, Lian JX, Meng S. MiR-125a-5p promotes osteoclastogenesis by targeting TNFRSF1B. Cell Mol Biol Lett 2019;24:23. 10.1186/s11658-019-0146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Wu X, Zhou Q, et al. TNFR2 is critical for the stabilization of the CD4+Foxp3+ regulatory T. cell phenotype in the inflammatory environment. J Immunol 2013;190:1076–84. 10.4049/jimmunol.1202659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomon BL, Leclerc M, Tosello J, et al. Tumor necrosis factor α and regulatory T cells in oncoimmunology. Front Immunol 2018;9:444. 10.3389/fimmu.2018.00444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Subleski JJ, Kopf H, et al. Cutting edge: expression of TNFR2 defines a maximally suppressive subset of mouse CD4+CD25+FoxP3+ T regulatory cells: applicability to tumor-infiltrating T regulatory cells. J Immunol 2008;180:6467–71. 10.4049/jimmunol.180.10.6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Oppenheim JJ. Targeting TNFR2, an immune checkpoint stimulator and oncoprotein, is a promising treatment for cancer. Sci Signal 2017;10. doi: 10.1126/scisignal.aal2328. [Epub ahead of print: 17 01 2017]. [DOI] [PubMed] [Google Scholar]
- 27.Jiang M, Liu J, Yang D, et al. A TNFR2 antibody by countering immunosuppression cooperates with HMGN1 and R848 immune stimulants to inhibit murine colon cancer. Int Immunopharmacol 2021;101:108345. 10.1016/j.intimp.2021.108345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell 2009;136:26–36. 10.1016/j.cell.2008.12.027 [DOI] [PubMed] [Google Scholar]
- 29.Gao Y, Lin F, Su J, et al. Molecular mechanisms underlying the regulation and functional plasticity of FOXP3(+) regulatory T cells. Genes Immun 2012;13:1–13. 10.1038/gene.2011.77 [DOI] [PubMed] [Google Scholar]
- 30.Vinuesa CG, Rigby RJ, Yu D. Logic and extent of miRNA-mediated control of autoimmune gene expression. Int Rev Immunol 2009;28:112–38. 10.1080/08830180902934909 [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, Zhang Q, Liu F, et al. MicroRNA-155 may affect allograft survival by regulating the expression of suppressor of cytokine signaling 1. Med Hypotheses 2011;77:682–4. 10.1016/j.mehy.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 32.Anandagoda N, Willis JC, Hertweck A, et al. microRNA-142-mediated repression of phosphodiesterase 3B critically regulates peripheral immune tolerance. J Clin Invest 2019;129:1257–71. 10.1172/JCI124725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H-Y, Barbi J, Wu C-Y, et al. MicroRNA-17 modulates regulatory T cell function by targeting co-regulators of the FOXP3 transcription factor. Immunity 2016;45:83–93. 10.1016/j.immuni.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie M, Wang J, Gong W, et al. NF-κB-driven miR-34a impairs Treg/Th17 balance via targeting FOXP3. J Autoimmun 2019;102:96–113. 10.1016/j.jaut.2019.04.018 [DOI] [PubMed] [Google Scholar]
- 35.Guo Y-N, Dong H, Ma F-C, et al. The clinicopathological significance of decreased miR-125b-5p in hepatocellular carcinoma: evidence based on RT-qPCR, microRNA-microarray, and microRNA-sequencing. Int J Clin Exp Pathol 2019;12:21–39. [PMC free article] [PubMed] [Google Scholar]
- 36.Yu T, Li G, Wang C, et al. MIR210HG regulates glycolysis, cell proliferation, and metastasis of pancreatic cancer cells through miR-125b-5p/HK2/PKM2 axis. RNA Biol 2021;18:2513–30. 10.1080/15476286.2021.1930755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Wang Y, Fan H, et al. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun 2018;504:277–82. 10.1016/j.bbrc.2018.08.172 [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Zhang S, Li Z, et al. miR-125b-5p and miR-99a-5p downregulate human γδ T-cell activation and cytotoxicity. Cell Mol Immunol 2019;16:112–25. 10.1038/cmi.2017.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Wu J, Ren J, et al. MicroRNA-125B interacts with FOXP3 to induce autophagy in thyroid cancer. Mol Ther 2018;26:2295–303. 10.1016/j.ymthe.2018.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye L-L, Wei X-S, Zhang M, et al. The Significance of Tumor Necrosis Factor Receptor Type II in CD8+ Regulatory T Cells and CD8+ Effector T Cells. Front Immunol 2018;9:583. 10.3389/fimmu.2018.00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polz J, Remke A, Weber S, et al. Myeloid suppressor cells require membrane TNFR2 expression for suppressive activity. Immun Inflamm Dis 2014;2:121–30. 10.1002/iid3.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chavez-Galan L, Vesin D, Uysal H, et al. Transmembrane Tumor Necrosis Factor Controls Myeloid-Derived Suppressor Cell Activity via TNF Receptor 2 and Protects from Excessive Inflammation during BCG-Induced Pleurisy. Front Immunol 2017;8:999. 10.3389/fimmu.2017.00999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel M, Oni L, Midgley A, et al. Increased concentration of plasma TNFR1 and TNFR2 in paediatric lupus nephritis. Lupus 2016;25:1040–4. 10.1177/0961203316631634 [DOI] [PubMed] [Google Scholar]
- 44.Parodis I, Ding H, Zickert A, et al. Serum soluble tumour necrosis factor receptor-2 (sTNFR2) as a biomarker of kidney tissue damage and long-term renal outcome in lupus nephritis. Scand J Rheumatol 2017;46:263–72. 10.1080/03009742.2016.1231339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan AT, Ogino S, Giovannucci EL, et al. Inflammatory markers are associated with risk of colorectal cancer and chemopreventive response to anti-inflammatory drugs. Gastroenterology 2011;140:799–808. 10.1053/j.gastro.2010.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Babic A, Shah SM, Song M, et al. Soluble tumour necrosis factor receptor type II and survival in colorectal cancer. Br J Cancer 2016;114:995–1002. 10.1038/bjc.2016.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beier UH, Angelin A, Akimova T, et al. Essential role of mitochondrial energy metabolism in Foxp3⁺ T-regulatory cell function and allograft survival. Faseb J 2015;29:2315–26. 10.1096/fj.14-268409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angelin A, Gil-de-Gómez L, Dahiya S, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, High-Lactate environments. Cell Metab 2017;25:1282–93. 10.1016/j.cmet.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Kivit S, Mensink M, Hoekstra AT, et al. Stable human regulatory T cells switch to glycolysis following TNF receptor 2 costimulation. Nat Metab 2020;2:1046–61. 10.1038/s42255-020-00271-w [DOI] [PubMed] [Google Scholar]
- 50.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol 2013;31:577. 10.1038/nbt0713-577 [DOI] [PubMed] [Google Scholar]
- 51.Janssen HLA, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med 2013;368:1685–94. 10.1056/NEJMoa1209026 [DOI] [PubMed] [Google Scholar]
- 52.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203–22. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005241supp002.pdf (427.9KB, pdf)
jitc-2022-005241supp003.pdf (538.8KB, pdf)
jitc-2022-005241supp004.pdf (349.3KB, pdf)
jitc-2022-005241supp005.pdf (497.6KB, pdf)
jitc-2022-005241supp006.pdf (308.8KB, pdf)
jitc-2022-005241supp001.pdf (83.4KB, pdf)
Data Availability Statement
Data are available in a public, open access repository.