Abstract
Two major infectious forms of vaccinia virus (VACV) have been described: the intracellular mature virion (IMV), and the extracellular enveloped virion (EEV). Due to their stability in the environment, IMVs play a predominant role in host-to-host transmission, whereas EEVs play an important role in dissemination within the host. In a previous report, we demonstrated that mice vaccinated with VACV L1R (IMV immunogen) and A33R (EEV immunogen) were protected from a lethal poxvirus challenge. Vaccination with a combination of both genes conferred greater protection than either gene alone, suggesting that an immune response against both IMV and EEV is advantageous. Here, we report that in mice individually administered DNA vaccines with two different VACV immunogens, A27L (IMV immunogen) or B5R (EEV immunogen), failed to significantly protect; however, vaccination with a combination of both genes conferred a high level of protection. Mice were completely protected when vaccinated with a combination of four VACV genes (A27L + A33R + L1R + B5R). Rhesus macaques vaccinated with this four-gene-combination developed appropriate antibody responses to each protein. Antibody responses elicited by this vaccine cross-reacted with monkeypox virus orthologous proteins. These data indicate that a gene-based vaccine comprised of the VACV A27L + A33R + L1R + B5R genes may be a useful candidate to protect against other orthopoxviruses, including those that cause monkeypox and smallpox.
Keywords: Vaccinia, Monkeypox, Smallpox, Orthopoxvirus, Vaccination, Protection, Mice, Macaques, DNA vaccine
Introduction
The vaccine used by the World Health Organization to eradicate naturally occurring smallpox disease consists of live vaccinia virus administered by scarification. This vaccine is still used today to protect laboratory workers from accidental VACV infection, and has also been stockpiled as a defense against new poxvirus outbreaks, especially an accidental or malicious re-introduction of smallpox. Despite their great importance, the VACV immunogens that protect against variola virus, the causative agent of smallpox, remain largely unknown. Our goal has been to identify protective VACV immunogens and then to use this information to augment poxvirus countermeasures.
Vaccinia, genus orthopoxvirus, family Poxviridae, is a large (>200 nm) virus containing a linear double-stranded DNA genome of ∼200 kbp encoding ∼250 genes (reviewed in Moss, 2001). Other orthopoxviruses share significant homology with vaccinia. For example, variola virus and vaccinia share ∼150 open reading frames with >90% amino acid identity Goebel et al 1990, Massung et al 1994. There are two infectious forms of poxviruses, the intracellular mature virion (IMV) and the extracellular enveloped virion (EEV). The poxviral IMV assembles in the cytoplasm and consists of a core particle, which contains the genome and numerous enzymes, wrapped in a membrane. At least 11 proteins are associated with the proteinaceous IMV surface membrane: A14.5L (p ∼ 10 kDa) (Betakova et al., 2000), E10R (p12 kDa) (Senkevich et al., 2000), I5L (p13 kDa) (Takahshi et al., 1994), A13L (p14 kDa) Takahashi et al 1994, Salmons et al 1997, A27L (p14 kDa) (Rodriguez and Esteban, 1987), A9L (p ∼ 18 kDa) (Yeh et al., 2000), A14L (p17–25 kDa) (Takahshi et al., 1994), A17L (p23–29 kDa) (Takahshi et al., 1994), and L1R (p25–29 kDa) (Franke et al., 1990), D8L (p34 kDa) (Niles and Seto, 1988), and H3L (p35 kDa) (Chertov et al., 1991). As IMV accumulate, they can be released from dying cells or alternatively can be wrapped in additional trans-Golgi-derived membranes and released from the infected cells as EEV. At least five proteins are associated with the EEV lipid bilayer membrane: A33R (gp23–28) (Roper et al., 1996), A34R (gp22–34 kDa) (Duncan and Smith, 1992), F13L (p37 kDa) (Hirt et al., 1986), B5R (gp42 kDa) Engelstad et al 1992, Isaacs et al 1992, and A56R (gp85 kDa hemagluttinin) (Shida, 1986). A36R (p43–50) was previously thought to be associated with the EEV membrane (Parkinson and Smith, 1994); however, a recent report provides evidence that A36R is a component of the membrane that wraps intracellular enveloped particles within the cell but is not associated with EEV released from the infected cell (van Eijl et al., 2000). Similarly, F12L (p65 kDa) was found to co-purify with EEV; however, it remains to be determined if this protein is a component of EEV particles (Zhang et al., 2000). Some EEV remain attached to the cell surface and play a role in short-range cell-to-cell spread. Some are released into the medium and play a role in long-range dissemination within the host Payne 1980, Blasco and Moss 1992.
Individual vaccinia genes have been cloned, expressed in Escherichia coli, or in baculovirus systems, and the exogenously expressed proteins have been tested for immunogenicity and protective efficacy in mice. Lai et al. (1991) demonstrated that vaccination with the E. coli-expressed A27L elicited neutralizing antibodies (NAbs) and protected mice against a lethal challenge with vaccinia. Demkowicz et al. (1992) confirmed that vaccination with E. coli-expressed A27L completely protected mice, and also demonstrated that vaccination with the core proteins A10L or A4L was partially protective. Galmiche et al. (1999) demonstrated that vaccination with baculovirus-expressed EEV proteins, B5R or A33R, protected mice against a lethal vaccinia challenge, whereas vaccination with E. coli-expressed A34R or A36R did not.
DNA vaccination technology has also been used to evaluate the immunogenicity and protective efficacy of individual vaccinia endogenously expressed proteins. DNA vaccination with A33R or B5R, but not A34R or A36R, protected mice against a lethal intranasal (i.n.) challenge with the IHD-J strain of vaccinia (Galmiche et al., 1999). We previously demonstrated that DNA vaccination of mice with a combination of IMV and EEV genes (L1R and A33R, respectively) protected mice from challenge (strain WR, intraperitoneal [i.p.] route) more effectively than vaccination with either gene alone (Hooper et al., 2000).
The mechanisms by which the aforementioned VACV immunogens (L1R, A27L, A33R, B5R, A10L, A4L) confer protective immunity against vaccinia in mice are not completely known. Targets of IMV-neutralizing antibodies include proteins encoded by five of the 11 known IMV genes: A27L (Rodriguez and Esteban, 1987), L1R (Ichihashi et al., 1994 [misidentified as A17L]; Wolffe et al., 1995), D8L (Hsiao et al., 1999), H3L (Gordon et al., 1991 [misidentified as H6R]), and A17L (Wallengren et al., 2001). Passive transfer data for L1R-, D8L-, H3L-, and A17L-specific antibodies have not been reported; however, we have unpublished data demonstrating that passive transfer of a L1R-specific MAb, MAb-7D11, 1 day before challenge, protected BALB/c mice from a lethal challenge (VACV-WR, i.p.). There have been at least three reports that A27L-specific monoclonal antibodies (MAbs) can protect mice in passive transfer experiments Czerny and Mahnel 1990, Czerny et al 1994, Ramirez et al 2002. One EEV gene, B5R, has been reported to be the target of antibodies that neutralize EEV Galmiche et al 1999, Law and Smith 2001. Passive transfer of anti-B5R hyperimmune mouse or rabbit sera protected 40–60% of mice challenged i.n. with vaccinia, strain IHD-J (Galmiche et al., 1999) Thus, it is possible that IMV or EEV neutralization plays a role in protection observed after vaccination with L1R, A27L, or B5R. Passive transfer of anti-A33R hyperimmune mouse sera, but not rabbit sera, protected 80% of mice challenged i.n. with vaccinia, strain IHD-J (Galmiche et al., 1999). The non-neutralizng anti-A33R response might elicit protection by directing lysis of infected cells. In support of this, an A33R-specific MAb (MAb-1G10) directed lysis of vaccinia-infected cells in the presence of guinea pig complement, as measured by chromium release assay (A. Schmaljohn, unpublished data). The mechanism underlying the partial protection observed in mice vaccinated with the core proteins A10L and A4L is unknown, but likely involves cell-mediated-immunity (Demkowicz et al., 1992).
In our previous study, DNA vaccination with a combination of L1R and A33R not only conferred complete protection, but also appeared to protect nearly as well as scarification with live VACV as measured by prevention of weight loss (Hooper et al., 2000). We hypothesized that the high level of protection was due to the fact that immune responses generated by this combination vaccine targeted the infection at several levels. IMV introduced in the initial challenge, as well as IMV released from infected cells, was neutralized by L1R-specific NAbs; and A33R-specific antibodies targeted the EEV and possibly infected cells expressing A33R on their surfaces.
The objective of our work was to further test DNA vaccines comprised of different combinations of IMV and EEV immunogens for immunogenicity and protective efficacy in mice, and expand this work into nonhuman primates.
Results
Cloning the VACV A27L and B5R genes into a naked-DNA vector and transient expression in cell culture
The A27L and B5R genes from vaccinia virus, NYBH strain Connaught (VACV-CONN), were cloned into the DNA vaccine plasmid pWRG7077 to create constructs pWRG/A27L and pWRG/B5R, respectively. Radio-immunoprecipitation experiments were performed to evaluate the expression products. The predicted 14 kDa and 42 kDa proteins were expressed from pWRG/A27L and pWRG/B5R, respectively (data not shown).
DNA vaccination of mice with pWRG/B5R elicits a non-NAb response, and vaccination with pWRG/A27L elicits a NAb response
To determine the immunogenicity of the expression products, groups of nine mice each were vaccinated with pWRG/A27L or pWRG/B5R with a gene gun (experiment 1). The mice were vaccinated at weeks 0, 3, and 5. As positive controls, nine mice were vaccinated by tail scarification with VACV-CONN. Serum was collected 2 weeks after the third DNA vaccination, or 8 weeks after scarification, and tested for the presence of antibodies against B5R or A27L (Fig. 1). All mice vaccinated with pWRG/B5R developed vaccinia-specific antibodies as measured by a vaccinia-infected-cell-lysate ELISA (geometric mean end-point titer [GMT] = 1728; range, 200–12800) (Fig. 1A), and eight of nine were positive by a B5R-specific ELISA (GMT = 588; range < 200–1600) (Fig. 1B). Only eight of nine scarified mice developed a B5R-specific antibody response and the response was twofold lower than the responses in the DNA-vaccinated mice, as measured by B5R-specific ELISA (GMT = 246; range < 200–400) (Fig. 1B); however, all samples were positive on the vaccinia infected-cell-lysate ELISA (GMT = 14703; range 6400–25600) (Fig. 1A). All of the A27L-vaccinated mice had anti-vaccinia antibodies as measured by a vaccinia infected-cell-lysate ELISA (GMT = 2540; range 200–6400); moreover, all of the mice produced IMV-neutralizing antibodies (PRNT50 GMT = 470, range 40–1280) as measured by PRNT (Fig. 1C). All of the scarified mice developed NAbs (PRNT50 GMT = 1174, range 640–2560) (Fig. 1C). The B5R-vaccinated mice did not develop NAb as measured by our PRNT (data not shown); however, it should be noted that the PRNT used in this study measures antibodies that neutralize IMV but not EEV. Thus, our B5R and A27L DNA vaccine plasmids were immunogenic in BALB/c mice.
Fig. 1.

Antibody responses elicited by gene gun vaccination with pWRG/A27L or pWRG/B5R. Sera from vaccinated mice were tested for vaccinia-specific antibodies using (A) vaccinia-infected-cell-lysate ELISA, and (B) B5R-specific ELISA. The mean O.D. values and standard deviations for the nine mice in each group are shown. (C) PRNT were performed on sera from mice vaccinated with pWRG/A27L (solid lines) or scarified (dashed lines) to evaluate the vaccinia-neutralizing antibody response. Representative serum samples diluted 1:40 from mice vaccinated with a negative control plasmid are shown as symbols.
DNA vaccination of mice with a combination of both A27L and B5R elicits better protection than vaccination with either immunogen alone
To determine if the response elicited by pWRG/A27L or pWRG/B5R was protective, we performed a second experiment (experiment 2) in which mice were vaccinated with plasmids alone, or in combination, and then challenged. The mice were vaccinated at weeks 0, 3, and 5. A group of 10 mice vaccinated with an irrelevant plasmid served as the negative control, and a group of VACV-scarified mice was the positive control. We also vaccinated a group of mice with a combination of four plasmids: two IMV genes, A27L and L1R; and two EEV genes, B5R and A33R. Three weeks after the final vaccination, serum was collected and the mice were challenged with vaccinia. Serum collected on the day of challenge was evaluated for immunogen-specific antibody responses (Fig. 2A). All of the mice vaccinated with pWRG/A27L alone developed NAbs (PRNT50 GMT = 486), but only one of 10 survived challenge. Similarly, all of the mice vaccinated with pWRG/B5R developed anti-B5R antibodies (GMT = 2111), but only four of 10 survived challenge. In contrast, all of the mice vaccinated with a combination of pWRG/A27L and pWRG/B5R developed antibodies to both immunogens, and all survived challenge. Similarly, all of the mice vaccinated with the four-gene combination survived. Nine of 10 negative control mice died and all of the positive control mice survived Fig. 2A.
Fig. 2.

Protection experiments in mice. (A–C) Mice were vaccinated with a DNA vaccine containing the indicated immunogen(s) or scarified with live VACV-CONN. Sera were collected after the final vaccination and the animals were challenged i.p. with 5 × 108 PFU of VACV-WR. The prechallenge sera were evaluated for NAbs by PRNT, and for anti-B5R or anti-A33R antibodies by protein-specific ELISA. In panel C an infected-cell-lysate ELISA was used to measure anti-A27L and anti-B5R responses. PRNT and ELISA end-point titers for individual mice in each group are shown as bars. Filled bars represent animals that did not survive challenge. Numbers to the right of filled bars indicate the day of death. In groups where positive antibody responses were detected, geometric mean titers (GMT) are shown. NT, not tested. (D–E) For groups where all mice were protected, weight loss was used as a measure of relative animal health. The average % of starting weight at times after challenge is shown. Panels A, B, and D contain data from challenge Experiment 2; panels C and E contain data from challenge Experiment 3.
As part of the same experiment, we also tested four other immunogen combinations (groups 7–10, Fig. 2B). For the IMV/EEV combinations, 10 of 10 mice vaccinated with B5R + L1R, and seven of 10 mice vaccinated with A27L + A33R, survived challenge. Only four of 10 mice vaccinated with the two IMV immunogens, A27L + L1R, survived. Eight of 10 mice vaccinated with the two EEV immunogens, B5R + A33R survived.
To confirm and expand our findings in mice, we performed a third experiment (experiment 3) where we repeated groups 1–6 (see Figure 2B), vaccinated with L1R alone, and vaccinated a group of mice with cartridges that contained all four genes precipitated on different gold beads (see Methods). Mice were vaccinated at weeks 0, 3, and 6, and then challenged 3 weeks after the third vaccination. In experiment 3, the scarified controls were vaccinated 3 weeks before challenge.
Antibody responses were measured by PRNT or by infected-cell-lysate ELISA (Fig. 2C). All of the mice vaccinated with A27L developed NAb as measured by PRNT, GMT = 470. All of the mice vaccinated with either A27L or B5R developed antibodies as measured by infected-cell-lysate ELISA. Fewer than half of the mice vaccinated with either B5R or A27L alone were protected, whereas all but one of the mice vaccinated with a combination of B5R and A27L were protected (Fig. 2C). The mouse that died in this group died later than controls (day 7) and appeared completely healthy on day 6. Whether each immunogen was loaded in a separate cartridge or as a mixture in the same cartridge, all 17 of the mice vaccinated with the four-gene-combination (B5R + A27L + L1R + A33R) were protected. This experiment confirmed that vaccination with a combination of A27L and B5R was protective, and vaccination with A27L, B5R, L1R, and A33R, on the same or in different cartridges, completely protected mice from a lethal challenge.
DNA vaccination of mice with a combination of four genes results in less weight loss than scarification, or vaccination with combinations of two genes
For groups with 100% survival in experiment 2 (Fig. 2A and 2B) and experiment 3 (Fig. 2C), weight loss was measured to determine the relative protective efficacy of the different vaccines (Fig. 2D and 2E). Protected mice lost as much as 12% of their initial weight during the first 1-3 days after challenge. Mice vaccinated with the four-gene-combination lost the least amount of weight (<8%) and regained weight faster than mice in the other groups. Mice vaccinated with live VACV-CONN by scarification lost the greatest percentage of initial weight and regained the weight more slowly. Although a few mice had slightly ruffed fur on days 1 and 2 after challenge, all of the mice vaccinated with the four-gene-combinations shown in Fig. 2D and 2E regained smooth fur within 3 days after challenge.
DNA vaccine is immunogenic in nonhuman primates
Two independent experiments were performed to determine if vaccination with the plasmids (pWRG/L1R + pWRG/A27L + pWRG/A33R + pWRG/B5R) could elicit antibody responses in nonhuman primates. In the first experiment, three rhesus macaques were vaccinated with the four-gene-combination. As negative controls, three monkeys were vaccinated with the vector plasmid containing irrelevant DNA; and as positive controls, three monkeys were vaccinated subcutaneously (s.c.) twice at 42-day intervals with live vaccinia. The second experiment was identical to the first; however, an additional group of three monkeys was vaccinated with pWRG/L1R alone, and the positive control group was vaccinated with the human smallpox vaccine (Dryvax) once by scarification by the method used to vaccinate humans. Three weeks after the fourth gene gun vaccination, serum was collected and tested for the presence of specific antibodies. Six different serological assays were performed: PRNT, vaccinia-infected cell lysate ELISA, vaccinia-virion ELISA, A33R-specific immunostaining, B5R-specific immunostaining, and RIPA. The results are shown in Fig. 3.
Fig. 3.

Candidate DNA vaccine elicits antibody responses against IMV and EEV in nonhuman primates. (A) Monkeys were vaccinated with a four-gene-combination, L1R alone, live VACV by the s. c. route, or by scarification. PRNT were performed to evaluate the NAb response. The last dilution reducing plaque number by either 50% or 80% are shown. Vaccinia-infected-cell lysate ELISA and vaccinia-purified virion ELISA were performed and the end-point titers were determined. Anti-B5R and anti-A33R immunostaining assays were performed to determine the highest dilution of sera resulting in positive protein-specific staining. Open bars were not expected to be positive and served as negative controls. For samples that were negative by immunostaining but suspected to be positive, we performed RIPA. A + symbol above a bar indicates this sample was positive by RIPA, a +/− symbol indicates a weak band was detected by RIPA, and a ? indicates the sample is negative by RIPA. (B) Representative RIPA data using sera from two monkeys vaccinated s.c. with the four-gene-combination (RC114 and CH93), with live VACV (CH21 and AA016), or with live VACV by Dryvax scarification (CH86 and CH39) are shown. Each lane represents a RIPA involving the indicated monkey serum and antigen in the form of cell lysate from cells transfected with a plasmid expressing either L1R, A33R, B5R, A27L, or a control plasmid with no insert (−); or cell lysate from COS cells infected with VACV-WR. A serum from a naive monkey (normal sera) served as a negative control. L1R was not detected using these sera. (C) Sera from monkeys vaccinated with L1R alone (CH63, CH74, and CH79) contained antibodies that immunoprecipitated L1R. L1R-specific MAb-7D11 served as a positive control. A + indicates immunoprecipitation from cells transfected with pWRG/L1R, and − indicates immunoprecipitation from cells transfected with a control plasmid with no insert, pWRG7077. (D) Serum from a monkey vaccinated with L1R (CH63), or a negative control (CH85), or L1R-specific MAb-7D11, or a negative control MAb-3d7 were tested by RIPA using VACV-WR infected-cell lysate as the source of antigen. A + indicates immunoprecipitation from cells infected with VACV-WR, and − indicates immunoprecipitation from mock infected cells. Molecular mass markers (M) are shown in kDa on the left and the position of specific vaccinia immunogens are shown on right.
Of the six monkeys vaccinated with the four-gene combination, three had NAbs, five were positive in the vaccinia-infected cell lysate ELISA, and the same five were positive by the vaccinia-virion ELISA. Five of the six monkeys had B5R-specific antibodies and only three of six had A33R-specific antibodies, as measured by immunostaining. The RIPA was a more sensitive assay for detecting B5R- and A33R-specific antibodies than ELISA or immunostaining. Representative gels for monkeys vaccinated with DNA by gene gun, live VACV by s.c., or scarification, are shown in Fig. 3B. The RIPA results indicated that all six monkeysvaccinated with the four-gene combination developed antibodies to B5R and A33R. Monkey CH80 exhibited a very weak response to all four genes.
All of the positive control monkeys had NAbs and were positive by both vaccinia-infected cell lysate ELISA and vaccinia-virion ELISA. Two of the monkeys vaccinated s.c. with live VACV failed to develop levels of A33R-specific antibody detectable by immunostaining, but RIPA revealed there was a weak A33R response. Similarly, one monkey vaccinated by scarification with Dryvax developed a B5R-specific antibody response detectable only by RIPA, but not by immunostaining.
We were unable to detect a L1R-specific antibody response in any of the monkeys vaccinated with the four-gene-combination, and the L1R response in monkeys vaccinated with live VACV was almost undetectable by immunoprecipitation. To determine if the pWRG/L1R plasmid was immunogenic in monkeys, we vaccinated three animals with L1R alone. All three monkeys developed NAbs responses (Fig. 3A) and their sera immunoprecipitated detectable levels of L1R expressed in cells transfected with pWRG/L1R (Fig. 3C).
To prove that DNA vaccination of monkeys with pWRG/L1R elicited an antibody response that could precipitate authentic L1R, we performed RIPA in which serum from a representative monkey vaccinated with pWRG/L1R was used to immunoprecipitate the L1R protein from VACV-WR infected-cell lyate. Serum from monkey CH63 precipitated a protein with the same apparent molecular weight as that precipitated by a L1R-specific monoclonal antibody (Fig. 4D). This band was not precipitated when negative control antibodies were used.
Fig. 4.

Comparison of VACV, MPOV, and VARV A27L; A33R, L1R, and B5R orthologs. The amino acid sequences of the proteins encoded by our DNA vaccine plasmids (VACV-CONN) were compared with VACV-WR, MPOV-Z79, MPOV-Z96, VARV-IND, and VARV-GAR orthologs. Amino acids that were identical in all viruses are boxed in black. Positions where variation occurs are either shown boxed in gray (conserved substitutions), white (nonconserved substitutions), or as a dash (deletion). Numbers at left are amino acid position. MPOV-Z79, MPOV- Z96, VACV-CONN, VACV-WR, VARV-IND, and VARV-GAR A27L ortholog accession numbers are submitted AY160186, NP_536566, AY160184, P11258, NP_042178, and D72167, respectively; A33R ortholog accession numbers are AY160188, NP_536572, AAF63733, BAA01805, CAA47507, and B72168, respectively; L1R ortholog accession numbers are submitted AY160187, NP_536507, AAF63732, P07612, NP_042117, and 672159, respectively; B5R ortholog accession numbers are submitted AY160189, NP_536594, submitted AY160185, Q01227, NP_042219, and E72150, respectively.
Together, these data demonstrate that a candidate DNA vaccine containing four vaccinia genes was immunogenic in nonhuman primates. The NAb responses to the IMV immunogens and the antibody responses to two EEV immunogens in some, but not all, of the DNA vaccinated monkeys were similar to those attained by vaccination with live VACV.
VACV DNA vaccine elicits antibodies in nonhuman primates that cross-react with monkeypox virus orthologs
In the future we plan to test the capacity of DNA vaccination with this four-gene-combination (A27L + A33R + L1R + B5R) to protect nonhuman primates against an appropriate challenge. It is likely the challenge will involve high dose i.v. injections of monkeypox virus, which has been shown to cause severe, and sometimes lethal, monkeypox disease in rhesus monkeys (McConnel et al., 1964). We were interested in determining if the antibody responses elicited by vaccination with the VACV A27L, A33R, L1R, or B5R genes would cross-react with the monkeypox virus orthologs. To test this, we cloned the A27L, A33R, L1R and B5R orthologous genes from monkeypox virus, strain Zaire 79 (MPOV-Z79), into pWRG7077 to yield plasmids: pMPOX/A27Lo, pMPOX/A33Ro, pMPOX/L1Rho and pMPOX/B5Ro, where ‘o’ indicates ortholog. The monkeypox virus genes were sequenced and the amino acid sequences were compared with the A27L, A33R, L1R, and B5R contained in our VACV-CONN DNA vaccine plasmids (Fig. 4). Included in the comparison are sequences from: VACV-WR, the virus used in our mouse challenge studies; a recently published monkeypox virus, strain Zaire 96 (Shchelkunov et al., 2001); a variola major, strain India (VARV-IND); and a variola minor, strain Garcia (VARV-GAR) (Fig. 4). All of these viruses possess orthologs to the VACV-CONN A27L, A33R, L1R and B5R, and all of the genes are highly conserved. All of the proteins are at least 90% identical. Our VACV-CONN immunogens of interest were all at least 93% identical to the MPOV and VARV strains evaluated.
RIPA were performed to answer two questions: 1) Were proteins of the predicted size produced from the plasmids containing the four different monkeypox virus genes? and 2) Did antibody responses elicited by DNA vaccination with plasmids containing the VACV-CONN A27L, A33R, L1R, and B5R genes cross-react with the monkeypox virus orthologs? We transfected COS cells with the plasmids and performed RIPA using serum from a monkey previously vaccinated with the four vaccinia genes (monkey CH93). Serum from CH93 immunoprecipitated the MPOV-Z79 A33R, B5R, and A27L orthologs, but as in the case of the vaccinia proteins, there were insufficient levels of L1R-specific antibodies to immunoprecipitate the L1R ortholog. To determine if pMPOX/L1Ro produced the correct protein, and to determine if the antibody response elicited by vaccination with the VACV L1R was capable of cross-reacting with the MPOV L1R ortholog, serum from a monkey vaccinated with the pWRG/L1R alone (monkey CH63) was tested by RIPA. Serum from CH63 immunoprecipitated the MPOV-Z79 L1R ortholog (Fig. 5). Thus, antibodies elicited by vaccination with the VACV A27L, A33R, L1R, and B5R bind to the orthologous proteins of monkeypox virus.
Fig. 5.
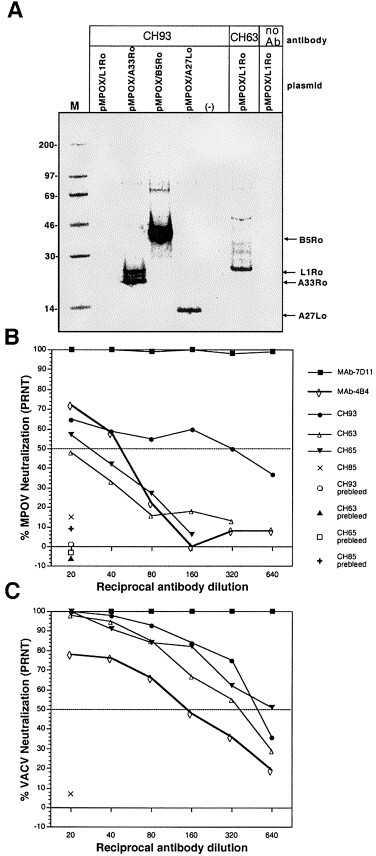
Antibodies elicited by DNA vaccination with vaccinia-based plasmids react with monkeypox virus orthologous proteins. (A) COS cells were transfected with DNA vaccine plasmids containing the monkeypox virus L1R, A33R, B5R, A27L ortholog, or empty vector; pMPOX/L1Ro, pMPOX/A33Ro, pMPOX/B5Ro, pMPOX/A27Lo, (−), respectively. RIPA were performed using serum from a monkey, CH93, vaccinated with the four-gene-combination (plasmids, pWRG/L1R, pWRG/A33R, pWRG/B5R, and pWRG/A27L). Serum from a monkey vaccinated with pWRG/L1R alone, CH63, was used to immunoprecipitate monkeypox virus L1R ortholog. A no antibody (no Ab) control was included in the monkeypox virus L1R ortholog RIPA. Molecular mass markers (M) are shown in kDa on the left and the position of specific vaccinia immunogens are shown on right. The o (e.g., L1Ro) indicates ortholog. PRNT were performed to detect (B) MPOV-neutralizing antibodies or (C) vaccinia-neutralizing antibodies in the sera from DNA vaccinated monkeys or relevant MAbs. Serum samples collected before vaccination (prebleeds) were also tested.
We tested the capacity of the serum from DNA vaccinated monkeys to cross-neutralize monkeypox virus. Serum from a representative monkey vaccinated with the four-gene-combination or pWRG/L1R alone, CH93 and CH63, respectively, was evaluated in a monkeypox virus PRNT. We also tested serum from a monkey vaccinated by scarification with VACV (Dryvax) (monkey CH65), and serum from a negative control monkey (CH85). CH93, CH63, and CH65 all neutralized monkeypox virus; however, the PRNT50 for CH63 was less then 20, and the PRNT for CH93 and CH65 were 40 and 320, respectively (Fig. 5B). None of the monkey sera (lowest dilution tested was 1:20) neutralized more than ∼70% of the plaques. The same monkey sera were tested in a vaccinia PRNT. These sera neutralized vaccinia more efficiently than monkeypox virus (Fig. 5C). The vaccinia PRNT80/50 titers of CH93, CH63, and CH65 were 160/320, 80/320, 160/640, respectively (Fig. 3, Fig. 5). Two vaccinia-specific MAbs were also tested for capacity to neutralize monkeypox virus. The vaccinia MAbs included the L1R-specific MAb-7D11 and the A27L-specific MAb-4B4. MAb-4B4 and MAb-7D11 neutralized both monkeypox virus and vaccinia virus (Fig. 5B and 5C). The PRNT50 titer of MAb-4B4 was 40 for monkeypox virus and 80 for vaccinia virus. The PRNT80 titer of MAb-7D11 was 40960 for monkeypox virus and 81920 for vaccinia virus (data not shown).
Discussion
Previously we reported that DNA vaccination with a combination of IMV- and EEV-specific immunogens conferred an enhanced level of protection compared to vaccination with either immunogen alone (Hooper et al., 2000). Here, we demonstrated that two other IMV- and EEV-specific immunogens, A27L and B5R, similarly conferred better protection when administered in combination. In two independent experiments, vaccination with A27L or B5R individually protected fewer than half the mice, whereas vaccination with a combination of the two immunogens protected 10 of 10 mice in the first experiment and 9 of 10 mice in the second.
DNA vaccination with A27L alone failed to protect despite the production of NAb titers as high as or higher than those elicited by scarification with live VACV. The NAb PRNT titers elicited in the A27L DNA-vaccinated mice were remarkably consistent: in experiments 1, 2, and 3, the GMTs of the A27L-vaccinated groups were 470, 485, and 470, respectively. The NAb responses in mice scarified with VACV-CONN in the same three experiments were less consistent: in experiments 1, 2, and 3, the GMTs were 1174, 243, and 49, respectively. The low PRNT in the experiment 3 scarified group might be attributed to a 3 week, rather than 8 week, duration between scarification and serum collection.
Despite the relatively robust NAb response elicited by DNA vaccination with A27L, 90% of the mice did not survive a VACV-WR, i.p. challenge. This was not expected because in two published studies, vaccination with E. coli-expressed A27L protein plus adjuvant protected BALB/c mice from a VACV-WR, i.p. challenge Lai et al 1991, Demkowicz et al 1992. In the Lai study, the titer that neutralized 80% of the plaques (PRNT80) in serum pools from protected groups ranged from 256–1024; however, in our study the PRNT80 ranged from 40–640 (data not shown). Demkowitz reported the presence of NAb after vaccination with E. coli-expressed protein, but did not provide titers. It is possible that higher NAb levels can account for the protection observed in the E. coli-expressed A27L vaccine studies. Our data indicate that anti-A27L NAb titers below 1280 are insufficient to protect against a lethal intraperitoneal challenge of VACV-WR. Others have reported that some, but not all, A27L-specific MAbs protected mice in passive transfer experiments Czerny and Mahnel 1990, Ramirez et al 2002. The protection observed in passive transfer studies could be the result of higher antibody concentrations achieved by direct transfer of purified antibody, or in subtle differences in the challenge model. Alternatively, the properties (e.g., epitope specificity, binding affinity, isotype) of the NAb response elicited by DNA vaccination could be different from those of the protective MAb. The A27L gene in our vaccine (from VACV-CONN) differs from the challenge strain, VACV-WR, at three amino acid positions; however, this strain difference cannot account for the absence of protection because the sera from A27L DNA vaccinated mice neutralized VACV-WR (data not shown). Alternatively, the differences in the nature of the immune responses elicited by vaccination with exogenously expressed protein (plus adjuvant) versus endogenously expressed protein (i.e., DNA vaccination) could account for the different levels of protection.
Our B5R DNA vaccine protection results differ somewhat from those reported previously. Whereas we observed an overall 37% protection rate after DNA vaccination with B5R, Galmiche et al. (1999) observed 80% protection rate. Because different challenge models were used in these studies (i.e., VACV-WR by i.p. versus VACV-IHD-J by i.n.), it is not possible to directly compare the results. One might expect a greater involvement of the EEV form of vaccinia in the i.n. model because the course of disease is extended: peak weight loss and time-to-death was ∼2 days later in the i.n. model than in the i.p. model, suggesting the first rounds of replication are insufficient to cause death. In our study, there was no correlation between B5R-specific antibody levels in the B5R-vaccinated mice and survival.
Our previous L1R + A33R data (Hooper et al., 2000) and the A27L + B5R data reported here are currently the only studies in which combinations of recombinant vaccinia immunogens have been tested for protective efficacy in animal models. Experiments testing the capacity of inactivated vaccinia (IMV) to elicit protective immunity demonstrated that an immune response to inactivated IMV, including neutralizing antibody, was sufficient to protect against lethal rabbitpox, but was insufficient to protect against intradermal challenge and lesion formation (Boulter et al., 1971). From these data, the authors concluded that viral replication and production of EEV was necessary to elicit protective immunity. In general, our data indicate that vaccination with a combination of IMV and EEV-specific immunogens confers a greater degree of protection than vaccination with only IMV or EEV immunogens. In experiment 2, Fig. 2A–B, 27 of 30 mice vaccinated with an IMV/EEV combination survived challenge, whereas only 17 of 40 mice vaccinated exclusively with IMV or EEV immunogens survived challenge. However, an IMV/EEV combination was not always necessary for protection because eight of 10 mice vaccinated with the B5R and A33R (an EEV/EEV combination) survived; and A33R alone or L1R alone can provide protection from a lethal challenge (Hooper et al., 2000).
We have focused on immunogens believed to elicit humoral responses that contribute to protection; however, it is likely that T-cell responses to as yet unknown vaccinia immunogens contribute significantly to immunity after smallpox vaccination. Patients with T-cell deficiency and apparently normal antibody responses have been diagnosed with progressive vaccinia virus infections after smallpox vaccination, suggesting a role of T-cells in controlling vaccinia virus infection (O’Connell et al., 1964). We have evaluated only antibody responses to our four-gene-combination vaccine; nevertheless, it is possible that one or all of the immunogens elicit protective T-cell-mediated responses. Demkowitz et al. (1992) reported that inoculation of BALB/c with purified E. coli-expressed A27L protein plus adjuvant not only elicited NAbs, but also elicited cellular proliferative immune responses. To our knowledge, information on cytotoxic T-cell responses to A27L, L1R, A33R, or B5R has not been reported.
Having found that vaccination with a combination of L1R and A33R, or A27L and B5R, protected mice from VACV-WR, we reasoned that a candidate vaccine containing all four genes might result in an even more potent vaccine. In two independent experiments, a total of 27 of 27 mice vaccinated with the four-gene-combination survived challenge, with minimal weight loss. Complete protection was achieved when the four genes were on different gold beads delivered to adjacent sites on the abdomen, or on different gold beads mixed together in the same cartridge and delivered to the same sites on the abdomen. The latter method indicates that it will be possible to make a single gene gun cartridge that contains all four genes. These data are important because we previously found that combining A33R on the same gold beads as L1R eliminated the immunogenicity of L1R (i.e., no NAb response) (Hooper et al., 2000). We have subsequently found that the interference is at the level of translation because L1R is not expressed in cells co-transfected with pWRG/L1R and pWRG/A33R as measured by RIPA (data not shown). For practical purposes, this indicates that a gene-based vaccine incorporating A33R must be designed to minimize delivery of A33R to the same cells as L1R. We achieved this by loading our plasmids of interest on different gold beads, then mixing the beads, as opposed to mixing the DNA then coating gold with the mixture. We surmise that delivery of the mixed beads results in fewer cells containing both the L1R and A33R plasmids.
The promising efficacy of this four-gene-combination vaccine in mice prompted us to test the vaccine for immunogenicity in a nonhuman primate model. We determined that vaccination of rhesus monkeys with the four-gene-combination DNA vaccine elicited antibodies against at least three of the four immunogens, as measured by gene-specific assays (i.e., RIPA, B5R-specific ELISA, A33R-specific ELISA). An anti-L1R response was not specifically detected in the four-gene-combination vaccinated monkeys; however, we know the pWRG/L1R plasmid is immunogenic in rhesus because monkeys vaccinated with pWRG/L1R alone developed NAb responses (PRNT50 ranged from 160–640) and all had sufficient antibody levels to immunoprecipitate the L1R protein, as well as the monkeypox virus L1R ortholog.
By redundant targeting of both the IMV and EEV, it may be possible to develop a gene-based vaccine that protects not only against vaccinia but also against other orthopox viruses. Comparison of VACV-CONN A27L, A33R, L1R, and B5R genes with the orthologous genes in monkeypox virus and variola virus indicate that all four genes encode highly conserved amino acids. VACV and MPOV-279 A27L, A33R, L1R, and B5R orthologous proteins are 95%, 94%, 98%, and 97% identical, respectively; VACV and VARV-IND are 97%, 94%, 99%, and 93% identical, respectively. More importantly, we found that DNA vaccination of rhesus monkeys with the VACV A27L, A33R, L1R, and B5R genes elicited antibodies that cross-reacted with all four monkeypox virus orthologs as measured by RIPA and PRNT. We did not test the capacity of these antibodies to bind to the variola virus orthologs; however, sequence analysis indicates that vaccinia virus is actually as or more similar to variola virus than monkeypox virus for A27L, A33R, and L1R.
Rhesus monkeys are susceptible to monkeypox virus and it is possible to protect these animals by scarifying with the human smallpox vaccine (McConnell et al., 1964). Future investigations will determine if DNA vaccination with the four-gene-combination can protect rhesus monkeys against monkeypox. Depending on the outcome of challenge studies, a gene-based vaccine could be improved by adding additional protective immunogens (e.g., protective T-cell epitopes) and/or by changing the vector, route of administration, vaccination schedule, etc. Also, it will be important to build assays of cell-mediated immunity into future studies. Work along these lines may ultimately provide an alternative poxvirus vaccine bereft of the known side effects and serious adverse reactions (e.g., inadvertent inoculation including ocular implantation, erythema multiform, eczema vaccinatum, vaccinial keratitis, generalized vaccinia, progressive vaccinia, and postvaccinial encephalitis) associated with the currently licensed live vaccinia-based smallpox vaccine.
Materials and methods
Viruses and cells
Vaccinia virus, strain Connaught (VACV-CONN) (a vaccine strain derived from the New York City Board of Health strain) (McClain et al., 1997) and strain Western Reserve (VACV-WR) (ATCC VR-119), and monkeypox virus strain Zaire 79 (MPOV-Z79) (provided by John Huggins, USAMRIID) were maintained in VERO cell (ATCC CRL-1587) monolayers grown in Eagle minimal essential medium, containing 5% heat-inactivated fetal bovine serum, 1% antibiotics (100 U/ml penicillin, 100 mg/ml streptomycin, and 50 mg/ml gentamicin), 10 mM HEPEs (cEMEM). COS cells (ATCC CRL 1651), used for transient expression experiments, were maintained in cEMEM.
Antibodies
One L1R-specific monoclonal antibody (MAb-7D11), and one A27L-specific monoclonal antibody (MAb-4B4), as ascitic fluids, were used. During the course of the studies reported here, we identified the target of MAb-4B4 using our plasmid expressing A27L, pWRG/A27L (data not shown). VACV-CONN hyperimmune mouse ascitic fluid (VACV HMAF) and normal human serum was also used. A hantavirus-specific monoclonal antibody (MAb-3d7) was used as a negative control in some experiments.
Cloning orthopox virus genes into naked-DNA expression plasmids
PWRG/A27L and pWRG/B5R
VACV-CONN DNA was purified by standard methods and used as template for PCR. PCR primer design was based on the published VACV-Copenhagen sequence (accession number M35027). The A27L forward primer was 5′-GCCGGCGGCCGCGCCACCATGGACGGAACTCTTTTCCCCGGA and the reverse primer was 5′-GCGCAGATCTTTACTCATATGGACGCCGTCCAG; the B5R forward primer was 5′-GCCGGCGGCCGCGCCACCATGAAAACGATTTCCGTTGTTACG and the reverse primer was 5′-GCGCAGATCTTTACGGTAGCAATTTATGGAACT. A NotI site (italicized) was incorporated in the forward primers and a BglII site (italicized) was incorporated into the reverse primers. Start codons are shown in bold. The A27L and B5R genes were PCR-amplified using VENT polymerase (NEB), cut with NotI and BglII, and cloned into the NotI-BglII site of pWRG7077 (Schmaljohn et al., 1997).
PWRG/L1R (x) and pWRG/A33R (x)
Previously we had cloned the VACV-CONN L1R and A33R genes into the NotI site of pWRG7077 (Hooper et al., 2000). During the course of subsequent studies, we discovered that there were ∼100 cloning artifact nucleotides (derived from SIV nef gene) between the BamHI and BglII sites of the pWRG7077 vector. To remove those nucleotides from our L1R and A33R constructs for future vaccine studies, we recloned the VACV-CONN L1R and A33R genes using a strategy that removed the unwanted sequence. The L1R forward primer was 5′-GCCGGCGGCCGCGCCACCATGGGTGCCGCAGCAAGCATACAG and the reverse primer was 5′-GCGCAGATCTTCAGTTTTGCATATCCGTGGTAG; the A33R forward primer was 5′-GCCGGCGGCCGCGCCACCATGATGACACCAGAAAACGACGAA and the reverse primer was 5′-GCGCAGATCTTTAGTTCATTGTTTTAACACAAA. The L1R and A33R genes were PCR-amplified from VACV-CONN DNA, cut with NotI and BglII, and cloned into the NotI-BglII site of pWRG7077 to yield pWRG/L1R (x) and pWRG/A33R (x), respectively. In this paper, these plasmids will be referred to as pWRG/L1R and pWRG/A33R, to avoid confusion.
Pmpox/a27lo, pmpox/a33ro, pmpox/l1ro, pmpox/b5ro
Monkeypox virus (MPOV-Z79) DNA was purified by standard methods. The aforementioned primers used to PCR-amplify and clone the VACV-CONN A27L, and L1R were used to clone the monkeypox virus orthologous genes. The monkeypox virus A33R gene ortholog (A35R) could not be amplified using the VACV-based primers. We designed new primers based on the DNA sequence from the recently published monkeypox virus, strain Zaire 96 genome (Shchelkunov et al., 2001). The primers were designed to anneal to the external sequence flanking the A33R ortholog. The forward primer was 5′-GCCGGCGGCCGCGCCACCATATAAATAACATTTATTATCATG, and the reverse primer was 5′-GCGCAGATCTTATTAAGCGATTTCATTTATTTA. The gene was PCR-amplified using VENT, cut with NotI and BglII, and cloned into the NotI-BglII site of pWRG7077. To clone the monkeypox virus B5R ortholog, we used a modified pWRG7077 vector in which the NotI-BglII sequence had been excised and replaced with a 100 nucleotide multiple cloning site (5′-GGCCGCTAGTTATTGCTCAGCGGTGGCAGCAGCCAACTCACCTTCCTTTCCCCCTTTGTTAGCAGCCGGATCAAGCTTCGAAGCTTCGAATTCCATGGTACCAGCTGCA). We used the VACV-based B5R forward primer (see above) and a new reverse primer, 5′-GCGCTTCGAATTACGGTAGCAATTTATGGAACT, containing a BstBI site (italicized). The gene was PCR-amplified using PLATINUM Tag High Fidelity polymerase (Invitrogen), cut with NotI and BstBI, and cloned into the NotI-BstBI site of pWRG7077 (NotI-BglII/MCS).
The VACV-CONN A27L and B5R genes, and the MPOV-Z79 A27L, A33R, L1R, and B5R orthologous genes were PCR amplified and sequenced directly using an ABI 377 or 3100 sequencer. In addition, the cloned genes and cloning junctions were sequenced for each DNA vaccine plasmid construct. Sequences were submitted to GeneBank.
Transient expression
Plasmid DNA was transfected into COS cell monolayers (60–80% confluent) using Fugene6 reagent as described by the manufacturer. After 24 hr, monolayers were radiolabeled with Promix (200 mCi per T-25 flask, 35S-methionine and [35S]-cysteine; Amersham) for ∼4 hr and immunoprecipitated as described previously (Hooper et al., 2000). Transfected cells were lysed on ice for 5 min with a modified RIPA buffer: 4% Zwittergent (Calbiochem) 0.5 M NaCl, 1 mM EDTA, 10 mM Tris, pH 8, and protease inhibitors (Complete; Boehringer Mannheim). Lysates were combined with the indicated antibody (previously incubated 1 h with unlabeled COS cell lysate) and incubated overnight at 4°C. Lysate-antibody mixtures were combined with protein A sepharose (CL-4B; Sigma), incubated at 4°C for 30 min, and then washed three times with lysis buffer and once with 10 mM Tris, pH 8.0. Sample buffer (125 mM Tris [pH 8.0], 1% sodium dodecyl sulphate, 10% glycerol, 0.01% bromophenol blue containing 2% 2-mercaptoethanol) was added and the samples were boiled for 2 min. Samples were run on 4 to 12% bis-Tris sodium dodecyl sulphate polyacrylamide gel electrophoresis gradient gels with 2-N (morpholino) ethane sulfonic acid running buffer (NuPAGE), at a 200-V constant voltage.
Vaccination with the gene gun
Cartridges for the gene gun were prepared as described previously Eisenbraun et al 1993, Schmaljohn et al 1997, Hooper et al 2000. Briefly, plasmid DNA was precipitated onto ∼2 μm diameter gold beads (Degussa), 1–5 μg of DNA per 1 mg of gold, which were then coated on the inner surface of Tefzel tubing (McMaster-Carr). Cartridges containing the four-gene-combination were made by loading the four different plasmid preparations on gold in separate tubes. After precipitating the DNA and washing the gold, we pooled the four DNA gold slurries together, mixed, and used them to coat Tefzel tubing. The tubing was cut into 0.5 inch cartridges. When completed, each cartridge contained 0.5–1 μg of DNA coated on 0.5 mg of gold.
To vaccinate animals, abdominal fur was removed with clippers and DNA-coated gold was administered to non-overlapping sites on the abdominal epidermis by using the gene gun (Powderject Delivery Device, Powderject, Inc.) at 400 p.s.i. as described previously (Pertmer et al., 1995). When the animal was vaccinated with a single immunogen (i.e., one plasmid construct), mice received two and monkeys received eight gene gun administrations. Mice vaccinated with two immunogens in different cartridges received one gene gun administration per plasmid. When the animal was vaccinated with four immunogens in different cartridges, mice received one and monkeys received two administrations per plasmid. Mice vaccinated with four immunogens in the same cartridge received four gene gun administrations.
Vaccination with vaccinia virus vaccines
Mice scarification
A 10 μl drop of PBS containing 8 × 106 PFU of VACV-CONN was placed ∼1 cm from the base of the tail and then scratched into the tail (∼15 3 mm scratches) using a needle on a tuberculin syringe.
Monkeys vaccinated subcutaneously with live vaccinia virus
Rhesus macaques were vaccinated with a recombinant vaccinia virus (expressing the hantavirus S and M genes) by the method used to vaccinate humans in a phase II clinical trial (McClain et al., 2000). The vaccine consisted of 3.4 × 107 PFU in 0.5 ml of PBS injected subcutaneously into the right lateral upper arm with a 26 G 3/8-in. needle. After 42 days, the monkeys received an identical vaccination on the left arm. These monkeys were part of a hantavirus DNA vaccine study which was reported previously (Hooper et al., 2001).
Monkey vaccination with vaccinia virus (Dryvax) by scarification
Rhesus macaques were vaccinated with the human smallpox vaccine (Dryvax, Wyeth Laboratories, Marietta, PA) by the protocol outlined in the vaccine insert. The vaccine (control no. 4008257) was reconstituted in the diluent provided (lot no. 4008258). The vaccination site (right lateral upper arm) was prepared by removing the hair using clippers, and wiped with an alcohol swab. A bifurcated needle was dipped in the vaccine and used to administer the vaccine by 15 pricks. Successful vaccination was confirmed 1 week after administration by the presence of a characteristic lesion.
Plaque reduction neutralization test (PRNT)
The vaccinia PRNT using VACV-CONN infected-cell lysate as the virus was performed exactly as described previously (Hooper et al., 2000). Monkeypox virus PRNT were performed in an identical manner to the vaccinia PRNT except MPOV-Z79 infected-cell lysate was the virus source, and the infections were stained with crystal violet on day 5 instead of day 3.
Vaccinia ELISA
Vaccinia infected-cell-lysate ELISA
Vaccinia-infected-cell lysate was prepared as follows. Vero cell monolayers grown in roller bottles were infected with VACV-CONN at a multiplicity of infection of 1, cell monlayers were harvested on day 4, cells were pelleted by spinning 90 min at 10,000 g, the pellet was resuspended in 10 mM Tris pH 8 (2 ml per roller bottle), Dounce homogenized, and the nuclei were removed by spinning 5 min at 1,000 g. The lysate was subjected to three rapid freeze-thaws, sonicated for 1 min on ice three times, and pelleted by spinning 40 min at 20,000 g. The pellet was resuspended in 10 mM Tris pH 8.0 (0.5 ml per roller bottle) and titration experiments were performed to determine the concentration of vaccinia infected-cell-lysate antigen to be used in ELISA.
Vaccinia purified virion ELISA antigen
The vaccinia infected-cell-lyate (above) was purified on 20–60% (weight/weight sucrose/TE) sucrose gradients spun at 82.7K g for 2.5 hr.
ELISA
96-well ELISA plates (Costar) were coated with antigen and dried overnight. Mock antigen preparations were prepared in an identical manner as the actual antigens; however, uninfected monolayers were used. Plates were washed with wash buffer (PBS + 0.05% tween-20) and then blocked 1 h at 37°C with blocking buffer (wash buffer containing 5% fetal bovine serum and 3% goat serum). Serum samples diluted in blocking buffer were added 100 μl per well to antigen or mock antigen containing wells and incubated 1 h at 37°C. Plates were washed three times and 100 μl per well of the appropriate peroxidase-labeled secondary antibody was added (peroxidase-labeled goat anti-mouse, IgG [Sigma], peroxidase-labeled goat anti-monkey IgG [KPL]). After a 1 h incubation at 37°C, the plates were washed and incubated 30 min at room temperature with 100 μl of 2,2′-azino bis-3-ethylbenzthiazoline-6-sulfonic acid (ABTS) substrate. The reactions were stopped by adding 100 μl per well of 0.2 N phosphoric acid and the O.D. at 405 nm was determined by an ELISA plate reader. For each serum dilution series, O.D. values from mock antigen wells were subtracted from the experimental values to give specific O.D. values. Each specific O.D. value represented the average value of two independent dilution series. End-point titers were determined as the highest dilution with an absorbance value greater than the mean absorbance value from negative control plasmid (pWRG7077)-vaccinated animals plus three standard deviations.
Anti-A33R and anti-B5R ELISA
The anti-A33R ELISA was described previously (Hooper et al., 2000). The anti-B5R ELISA was performed in an identical manner except the pWRG/A33R DNA was replaced with the pWRG/B5R DNA. Briefly, COS cell monolayers, grown in 96 well cell culture plates, were transfected with pWRG/A33R or pWRG/B5R (0.2 mg/well) using Fugene 6 or were mock transfected. After ∼24 h the monolayers were fixed with 1:1 acetone:methanol for 2 min, rinsed with PBS, incubated 1 h with primary antibody diluted in PBS + 3% FBS, rinsed with PBS, incubated 30 min with peroxidase labeled goat anti-mouse antibody (SIGMA, cat. no. A4416) or peroxidase-labeled goat anti-monkey antibody (KPL, cat. no. 074-11-021) diluted in PBS + 3% FBS, rinsed, and finally, incubated with ABTS substrate as described above. After ∼30 min, reactions were stopped and O.D. values were determined as described above. Optical density values from mock-transfected wells were subtracted from those of transfected wells to determine the specific O.D. for each sample. End-point titers were determined as the highest dilution with an absorbance value greater than the mean absorbance value from negative control plasmid (pWRG7077) -vaccinated animals plus three standard deviations.
A33R and B5R immunostaining
Anti-A33R and anti-B5R antibodies in the serum from vaccinated monkeys were detected by the following immunostaining procedure. Six-well plates containing COS cell monolayers at ∼80% confluency were transfected with 2 μg of either pWRG/A33R, or pWRG/B5R, or mock transfected with Fugene 6. After ∼24 hr, the monolayers were fixed with 1:1 acetone:methanol for 2 min, rinsed with PBS, incubated 1 h with serial twofold dilutions of primary antibody diluted in PBS + 3%FBS, rinsed with PBS, incubated 30 min with peroxidase-labeled goat anti-monkey antibody (KPL) diluted in PBS plus 3% fetal bovine serum, rinsed, and incubated with dianisidine (SIGMA) substrate as described by Roper et al., 1996. Endpoint titers were determined by identifying the highest antibody dilution that resulted in positively stained (brown) cells.
Challenge experiment
Mice were injected i.p. with 0.2 ml of PBS containing 5 × 108 PFU of VACV-WR (12 LD50) (clarified infected-cell lysate) with a 0.5 mm × 16 mm needle. Mice were weighed before challenge and on each day thereafter for 2 weeks. Terminal bleeds were obtained from survivors 3 to 4 weeks after challenge. This research was conducted in accordance with procedures described in the Guide for the Care and Use of Laboratory Animals (National Institute of Health, 1996). The facilities are fully accredited by the American Association for Accreditation of Laboratory Animal Care.3
Acknowledgements
The gene gun (Powderject delivery device) and pWRG7077 were provided by Powderject Vaccine, Inc. The modified pWRG7077, pWRG7077 (NotI-BglIIMCS), was constructed by Alexander Dekonenko. The smallpox vaccine (Dryvax) was provided by the Centers for Disease Control and Prevention in Atlanta, GA. We thank A. Schmaljohn for helpful discussions and C. Schmaljohn for critical comments on the manuscript.
References
- Betakova T., Wolffe E.J., Moss B. The vaccinia virus A14.5L gene encodes a hydrophobic 53-amino-acid virion membrane protein that enhances virulence in mice and is conserved among vertebrate poxviruses. J. Virol. 2000;7:4085–4092. doi: 10.1128/jvi.74.9.4085-4092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R., Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R., Sisler J.R., Moss B. Egress of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin orthology domain of the A34R gene. J. Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter E.A., Zwartouw H.T., Titmuss H.J., Maber H.B. The nature of the immune state produced by inactivated vaccinia virus in rabbits. Am. J. Epidemiol. 1971;94:612–620. doi: 10.1093/oxfordjournals.aje.a121360. [DOI] [PubMed] [Google Scholar]
- Chertov O.Y.u, Telezhinskaya I.N., Zaitseva E.V., Golubeva T.B., Zinov’ev V.V., Ovechkina L.G., Mazkova L.B., Malygin E.G. Amino acid sequence determination of vaccinia virus immunodominant protein p35 and identification of the gene. Biomed. Sci. 1991;2:151–154. [PubMed] [Google Scholar]
- Czerny C.P., Johann S., Holzle L., Meyer H. Epitope detection in the envelope of intracellular naked orthopox viruses and identification of encoding genes. Virology. 1994;200:764–777. doi: 10.1006/viro.1994.1240. [DOI] [PubMed] [Google Scholar]
- Czerny C.P., Mahnel H. Structural and functional analysis of orthopoxvirus epitopes with neutralizing monoclonal antibodies. J. Gen. Virol. 1990;71:2341–2352. doi: 10.1099/0022-1317-71-10-2341. [DOI] [PubMed] [Google Scholar]
- Demkowicz W.E., Maa J.S., Esteban M. Identification and characterization of vaccinia virus genes encoding proteins that are highly antigenic in animals and are immunodominant in vaccinated humans. J. Virol. 1992;66:386–398. doi: 10.1128/jvi.66.1.386-398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.A., Smith G.L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbraun M.D., Fuller D.H., Haynes J.R. Examination of parameters affecting the elicitation of humoral immune responses by particle bombardment-mediated genetic immunization. DNA Cell Biol. 1993;12:791–797. doi: 10.1089/dna.1993.12.791. [DOI] [PubMed] [Google Scholar]
- Engelstad M., Howard S.T., Smith G.L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- Franke C.A., Wilson E.M., Hruby D.E. Use of a cell-free system to identify the vaccinia virus L1R gene product as the major late myristylated virion protein M25. J. Virol. 1990;64:5988–5996. doi: 10.1128/jvi.64.12.5988-5996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmiche M.C., Goenaga J., Wittek R., Rindisbacher L. Neutralizing and protective antibodies directed against vaccinia virus envelope antigens. Virology. 1999;254:71–80. doi: 10.1006/viro.1998.9516. [DOI] [PubMed] [Google Scholar]
- Goebel S.J., Johnson G.P., Perkus M.E., Davis S.W., Winslow J.P., Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. , 517–563. [DOI] [PubMed] [Google Scholar]
- Gordon J., Mohandas A., Wilton S., Dales S. A prominent antigenic surface polypeptide involved in the biogenesis and function of the vaccinia virus envelope. Virology. 1991;181:671–686. doi: 10.1016/0042-6822(91)90901-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt P., Hiller G., Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J.W., Custer D.M., Thompson E., Schmaljohn C.S. DNA vaccination with the Hantaan virus M gene protects hamsters against three of four HFRS hantaviruses and elicits high-titer neutralizing antibody response in rhesus monkeys. J. Virol. 2001;75:8469–8477. doi: 10.1128/JVI.75.18.8469-8477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J.W., Custer D.M., Schmaljohn C.S., Schmaljohn A.L. DNA vaccination with vaccinia L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology. 2000;266:329–339. doi: 10.1006/viro.1999.0096. [DOI] [PubMed] [Google Scholar]
- Hsiao J.C., Chung C.S., Chang W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 1999;73:8750–8761. doi: 10.1128/jvi.73.10.8750-8761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Takahashi T., Oie M. Identification of a vaccinia virus penetration protein. Virology. 1994;202:834–843. doi: 10.1006/viro.1994.1405. [DOI] [PubMed] [Google Scholar]
- Isaacs S.N., Wolffe E.J., Payne L.G., Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.F., Gong S.C., Esteban M. The purified 14 kilodalton envelope protein of vaccinia virus produced in Escherichia coli induces virus immunity in animals. J. Virol. 1991;65:5631–5635. doi: 10.1128/jvi.65.10.5631-5635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M., Smith G.L. Antibody neutralization of the extracellular enveloped form of vaccinia virus. Virology. 2001;280:132–142. doi: 10.1006/viro.2000.0750. [DOI] [PubMed] [Google Scholar]
- Massung R.F., Liu L.I., Qi J., Knight J.C., Yuran T.E., Kerlavage A.R., Parsons J.M., Venter J.C., Esposito J.J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- McClain D.J., Harrison S., Yeager C.L., Cruz J., Ennis F.A., Gibbs P., Wright M.S., Summers P.L., Arthur J.D., Graham J.A. Immunological responses to vaccinia vaccines administered by different parenteral routes. J. Infect. Dis. 1997;175:756–763. doi: 10.1086/513968. [DOI] [PubMed] [Google Scholar]
- McClain D.J., Summers P.L., Harrison S.A., Schmaljohn A.L., Schmaljohn C.S. Clinical evaluation of a vaccinia vectored Hantaan virus vaccine. J. Med. Virol. 2000;60:77–85. doi: 10.1002/(sici)1096-9071(200001)60:1<77::aid-jmv13>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- McConnell S., Herman Y.F., Mattson D.E., Huxsoll D.L., Lang M., Yager R.H. Protection of rhesus monkeys against monkeypox by vaccinia virus immunization. Am. J. Vet. Res. 1964;25:192–195. [PubMed] [Google Scholar]
- Moss B. In: Knipe D.M., Howley P.M., editors. Vol. 4. Lippincott-Raven; Philadelphia: 2001. Poxviridae: the viruses and their replication; pp. 2849–2883. (Fields Virology). [Google Scholar]
- Niles E.G., Seto J. Vaccinia virus gene D8 encodes a virion transmembrane protein. J. Virol. 1988;62:3772–3778. doi: 10.1128/jvi.62.10.3772-3778.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell C.J., Karzon D.T., Barron A.L., Plaut M.E., Ali V.M. Progressive vaccinia with normal antibodies: a case possibly due to deficient cellular immunity. Ann. of Intern. Med. 1964;60:282–289. doi: 10.7326/0003-4819-60-2-282. [DOI] [PubMed] [Google Scholar]
- Parkinson J.E., Smith G.L. Vaccinia virus gene A36R encodes a M(r) 43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- Payne L.G. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 1980;50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- Pertmer T.M., Eisenbraun M.D., McCabe D., Prayaga S.K., Fuller D.H., Haynes J.R. Gene gun-based nucleic acid immunization: elicitation of humoral and cytotoxic T lymphocyte responses following epidermal delivery of nanogram quantities of DNA. Vaccine. 1995;13:1427–1430. doi: 10.1016/0264-410x(95)00069-d. [DOI] [PubMed] [Google Scholar]
- Ramirez J.C., Tapia E., Esteban M. Administration to mice of a monoclonal antibody that neutralizes the intracellular mature virus form of vaccinia virus limits virus replication efficiently under prophylactic and therapeutic conditions. J Gen Virol. 2002;83:1059–1067. doi: 10.1099/0022-1317-83-5-1059. [DOI] [PubMed] [Google Scholar]
- Rodriguez J.F., Esteban M. Mapping and nucleotide sequence of the vaccinia virus gene that encodes a 14-kilodalton fusion protein. J. Virol. 1987;61:3550–3554. doi: 10.1128/jvi.61.11.3550-3554.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper R.L., Payne L.G., Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmons T., Kuhn A., Wylie F., Schleich S., Rodriguez J.R., Rodriguez D., Esteban M., Griffiths G., Locker J.K. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 1997;71:7404–7420. doi: 10.1128/jvi.71.10.7404-7420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn C., Vanderzanden L., Bray M., Custer D., Meyer B., Li D., Rossi C., Fuller D., Fuller J., Haynes J., Huggins J. Naked DNA vaccines expressing the prM and E genes of Russian spring summer encephalitis virus and Central European encephalitis virus protect mice from orthologous and heterologous challenge. J. Virol. 1997;71:9563–9569. doi: 10.1128/jvi.71.12.9563-9569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich T.G., Weisberg A.S., Moss B. Vaccinia virus E10R protein is associated with the membranes of intracellular mature virions and has a role in morphogenesis. Virology. 2000;278:244–252. doi: 10.1006/viro.2000.0656. [DOI] [PubMed] [Google Scholar]
- Shchelkunov S.N., Totmenin A.V., Babkin I.V., Safronov P.F., Ryazankina O.I., Petrov N.A., Gutorov V.V., Uvarova E.A., Mikheev M.V., Sisler J.R., Esposito J.J., Jahrling P.B., Moss B., Sandakhchiev L.S. Human monkeypox and smallpox viruses: genomic comparison. FEBS Lett. 2001;509:66–70. doi: 10.1016/S0014-5793(01)03144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida H. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology. 1986;150:451–462. doi: 10.1016/0042-6822(86)90309-0. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Oie M., Ichihashi Y. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- van Eijl H., Hollinshead M., Smith G.L. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology. 2000;271:26–36. doi: 10.1006/viro.2000.0260. [DOI] [PubMed] [Google Scholar]
- Wallengren K., Risco C., Krijnse-Locker J., Esteban M., Rodriguez D. The A17L gene product of vaccinia virus is exposed on the surface of IMV. Virology. 2001;290:143–152. doi: 10.1006/viro.2001.1131. [DOI] [PubMed] [Google Scholar]
- Wolffe E.J., Vijaya S., Moss B. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology. 1995;211:53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]
- Yeh W.W., Moss B., Wolffe E.J. The vaccinia virus A9L gene encodes a membrane protein required for an early step in virion morphogenesis. J. Virol. 2000;74:9701–9711. doi: 10.1128/jvi.74.20.9701-9711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.H., Wilcock D., Smith G.L. Vaccinia virus F12L protein is required for actin tail formation, normal plaque size, and virulence. J. Virol. 2000;74:11654–11662. doi: 10.1128/jvi.74.24.11654-11662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


