Abstract
Orthopoxviruses include many important pathogens such as variola major virus, camelpox, buffalopox, monkeypox, cowpox, and variola minor viruses. This group of viruses also includes vaccinia virus, which is extensively used in human vaccine development. Genomes of orthopoxviruses encode proteins with sequences similar to human regulators of complement activation (RCA) that contain tandem short consensus repeats (SCRs). We employed phylogenetic tree analysis to evaluate the structural relationships among SCRs of orthopoxvirus RCA-like proteins and those of human complement regulators. The human complement RCA proteins analyzed were factor H (FH), C4 binding protein alpha chain, membrane cofactor protein (MCP), decay accelerating factor (DAF), and complement receptors type 1 (CR1) and 2 (CR2). Sequences of key poxvirus regulators of complement activation, vaccinia virus complement control protein (VCP), smallpox inhibitor of complement enzymes (SPICE), and cowpox inflammation modulatory protein (IMP) were similar to SCRs 1 through 5 of C4 binding protein, alpha chain, and they were also clustered with other homologous repeats of MCP, DAF, CR1, CR2, and FH. Phylogenetic clustering of RCA sequences suggested that poxvirus complement regulators VCP, SPICE, and IMP arose from a single ancestral sequence that shared similarity with all human regulators of complement activation. Any changes in poxvirus complement regulators leading to the enhancement of their ability to regulate complement activation likely resulted from new mutations in the viral lineages.
Abbreviations: B5R, vaccinia virus B5R protein; C3, complement component C3; C3a, C3a cleavage fragment; C3b, C3b cleavage fragment; C3d, C3d cleavage fragment; C3dg, C3dg cleavage fragment; C3L, vaccinia virus complement control protein (product of the C3L gene of vaccinia virus, strain WR); C4, complement component C4; C4b, C4b cleavage fragment; C4bp, C4 binding protein; C4bpα, C4 binding protein, α chain; C4bpβ, C4 binding protein, β chain; C5, complement component C5; CR1, complement receptor type 1; CR2, complement receptor type 2; DAF, decay accelerating factor; D12L, smallpox inhibitor of complement enzymes (product of the D12L gene of the variola major virus, strain India-1967); D17L, inflammation modulatory protein (product of the D17L gene of the cowpox virus, strain GRI-90); D15L, smallpox inhibitor of complement enzymes (product of D15L gene of the variola major virus, strain Bangladesh-1975); FH, factor H; iC3b, iC3b cleavage fragment; IgG1, immunoglobulin G subclass 1; IMP, inflammation modulatory protein; MCP, membrane cofactor protein; ps-hr, plaque size-host range gene; RCA, regulators of complement activation; SCR, short consensus repeat; SPICE, smallpox inhibitor of complement enzymes; TBR, tree bisection-reconnection; VCP, vaccinia virus complement control protein
Keywords: Regulator of complement activation, Poxvirus, Vaccinia complement control protein, Smallpox inhibitor of complement enzymes, Inflammation modulatory protein
1. Introduction
The human complement system plays a highly important role in host defense against pathogens, in regulation of humoral immune response, and in inflammation Müller-Eberhard (1985). The early stages of complement activation may be initiated via three different pathways: the classical pathway, the alternative pathway, and the mannan-binding lectin pathway. These pathways lead to the assembly of distinct multi-component protein enzymatic complexes, C3 convertases. The C3 convertases are serine proteases that proteolytically cleave complement component C3 into two fragments, C3a and C3b. After the C3 cleavage, the three initiating pathways converge on a common series of steps that lead to the formation of the C5 convertase, assembly of the terminal complement components in the membranes of pathogens, cell lysis, and recruitment and activation of phagocytes (Müller-Eberhard, 1985, Pier et al., 2004).
To avoid injury to host tissue and to prevent uncontrolled activation, the activation of complement is tightly regulated by several mechanisms (Hourcade et al., 2000). Some of these mechanisms regulate the formation and stability of C3 convertases (Hourcade et al., 2002), and they involve a group of soluble and cell membrane-anchored regulatory proteins, referred to as regulators of complement activation (RCA). The RCA proteins consist of partially homologous tandem short consensus repeats (SCRs) of 60–70 amino acids (Stehle and Larvie, 2003). The two important human soluble plasma complement regulators are C4 binding protein (C4bp) and factor H (FH) that modulate the C3 activation in the classical and the alternative pathways, respectively (Gigli et al., 1979, Pangburn and Müller-Eberhard, 1985). Structurally, C4bp contains 8 disulfide linked chains. Seven of these chains are identical (C4 binding protein α chain, or C4bpα), and they regulate complement activation. Each C4bpα chain consists of 8 short consensus repeats. The non-identical β chain of C4 binding protein (C4bpβ) participates in coagulation (Blom, 2002). The single polypeptide chain of human FH consists of 20 SCRs (Zipfel et al., 2002).
Human membrane-anchored RCA proteins that regulate complement activation to prevent uncontrolled C3 activation include decay accelerating factor (DAF), membrane cofactor protein (MCP) and complement receptor type 1 (CR1). Both DAF and MCP contain 4 SCRs, while CR1 protein has different allelic variants, ranging from 23 to 44 SCRs (Hourcade et al., 2000). Recent analysis of genomic DNA of the human CR1 gene suggested evidence for additional SCRs that are not expressed (McLure et al., 2004a, McLure et al., 2004b).
In addition to regulation of complement activation, some RCA proteins perform other important biological functions in immune regulation. For example, MCP participates in modulation of T-lymphocyte-mediated immune responses, while CR1 serves as a cofactor for factor I mediated proteolysis of iC3b to generate C3dg, which may affect IgG1 responses and elicit memory cells (Hourcade et al., 2000, Kemper et al., 2001, Marie et al., 2002). Another RCA-related protein, complement receptor type 2 (CR2), participates in B-lymphocyte activation, and it is also expressed on other cells of the immune system. It interacts with cleavage fragments iC3b, C3dg, and C3d (Ross et al., 1973, Stehle and Larvie, 2003). CR2 protein has allelic variants that consist of 15 and 16 SCRs (Fujisaku et al., 1989), and additional non-expressed SCR duplications have been identified in the CR2 gene (McLure et al., 2004a, McLure et al., 2004b). Additional SCR-containing proteins with less established function or function different from complement regulation are also known (Krushkal et al., 2000, Zipfel et al., 2002).
Viral group Orthopoxviridae includes variola major virus, a highly virulent agent that causes smallpox. Many other orthopoxviruses also pose a serious health threat, e.g. camelpox, buffalopox, monkeypox, cowpox, and variola minor viruses (Georges and Georges-Courbot, 1999, McFadden, 2005). Another orthopoxvirus, vaccinia virus, is extensively used in human vaccine development and in biomedical research. The genomes of orthopoxviruses encode virulence-related proteins that can regulate complement activation. These proteins consist of four SCRs and are similar in sequence to human regulators of complement activation (Kotwal et al., 1998a, Kotwal et al., 1998b, Kotwal, 2000, Lee et al., 2003). It has been suggested that viral complement regulators originated from a host genome as a result of horizontal gene transfer (Kirkitadze et al., 1999, Uvarova and Shchelkunov, 2001). In vaccinia virus, an important virulence factor that regulates human complement activation is vaccinia virus complement control protein (VCP) (Kotwal and Moss, 1988, Kotwal, 2000). VCP is a major secretory protein of cells infected with vaccinia virus, and it also exists in a membrane-bound form (Rosengard et al., 2002). VCP inhibits both the alternative and the classical pathways, it can bind both C3b and C4b, and it blocks complement-mediated antibody-induced virus neutralization. It also binds to heparin and heparan sulfate proteoglycans, blocking chemotactic signals (Kotwal et al., 1990, Sahu et al., 1998, Smith et al., 2000, Smith et al., 2003).
Other orthopoxviruses express VCP homologs that are important for immune modulation and virulence. The VCP homolog in variola major virus, the most virulent orthopoxvirus, is termed the smallpox inhibitor of complement enzymes (SPICE) (Kotwal, 2000, Lee et al., 2003). SPICE differs from VCP at 11 amino acid sites (Rosengard et al., 2002). It is highly efficient in inactivating both the classical and the alternative pathways of the complement system, and it shows strong preference to human complement inactivation as opposed to other mammalian species (Rosengard et al., 2002). The VCP homolog in cowpox virus is the inflammation modulatory protein (IMP), which downregulates complement and allows the infected tissue to evade inflammation (Kotwal et al., 1998a, Kotwal et al., 1998b).
The relationship of VCP, SPICE, and IMP to human complement regulators has not been fully elucidated. A comparable level of sequence similarity has been observed for SCRs 1–4 of VCP to the four repeats of MCP, the first four SCRs of C4bpα, and SCRs 1–4, 8–11, and 15–18 of CR1; SCRs 1–3 of VCP are also similar to SCRs 2–4 of DAF (Kotwal and Moss, 1988, Kotwal et al., 1990, Kirkitadze et al., 1999, Kotwal, 2000, Uvarova and Shchelkunov, 2001). These N-terminal domains in the human RCA proteins share sequence similarity to one another (Krushkal et al., 2000) and many of them are functionally important (Dahlback et al., 1983, Gordon et al., 1995, Hourcade et al., 2000, Zipfel et al., 2002). Three-dimensional structural similarity of VCP repeats to individual modules of CR1 and FH has been observed, while the structure of the VCP domain arrangement is similar to that of MCP (Wiles et al., 1997, Kirkitadze et al., 1999, Kotwal, 2000, Murthy et al., 2001, Stehle and Larvie, 2003). Functionally, modules in FH, C4bpα, and VCP contain conserved heparin binding sites (Smith et al., 2000, Blom, 2002, Zipfel et al., 2002, Ganesh et al., 2004). Like CR1, VCP is capable of regulating both the alternative and the classical pathways of the complement system. However, VCP has a number of functional differences from both FH and CR1 (Sahu et al., 1998). Thus, the structural and functional similarity between VCP and several human complement regulators makes it difficult to identify the human RCA protein to which VCP may be most closely related.
In the present report, we describe the use of phylogenetic tree inference to investigate the relationships among individual short consensus repeats of vaccinia proteins VCP and B5R, their homologs in other orthopoxviruses, and human complement proteins FH, C4bpα, MCP, DAF, CR1, and CR2. Phylogenetic analysis of individual repeats, which uses information from multiple sequences, is better suited for interpreting the relationships among RCA-like proteins than is the direct sequence comparison of viral proteins (Krushkal et al., 1998, Krushkal et al., 2000).
2. Materials and methods
2.1. Sequence retrieval and alignment
Due to the high degree of sequence divergence among human and poxvirus regulators of complement activation (Kotwal et al., 1990, Krushkal et al., 1998), our analysis was restricted to protein sequences. We investigated complement proteins with established functional roles. Sequences of human complement regulators were collected from GenBank as described previously (Krushkal et al., 2000). These were sequences of factor H (FH; Genbank accession no. CAA68704); C4 binding protein, α chain (C4bpα; AAA36507); membrane cofactor protein (MCP; P15529); decay accelerating factor (DAF; P08174); complement receptor type 1 (CR1; P17927); and complement receptor type 2 (CR2; P20023). Poxvirus protein sequences were collected from GenBank (http://www.ncbi.nlm.nih.gov/) using the protein–protein BLAST (blastp) similarity searches to full length C4bpα and FH. Included in analysis were vaccinia virus complement control protein (VCP, or C3L gene product) of vaccinia virus, strain WR (Genbank accession no. P68638); smallpox inhibitor of complement enzymes (SPICE) of two strains of variola major virus: strain India-1967 (D12L gene product; GenBank accession no. NP_042056) and Bangladesh-1975 (D15L gene product; T28450); and inflammation modulatory protein (IMP) of the cowpox virus, strain GRI-90 (IMP, or D17L gene product; CAA64102). The list of sequences analyzed and the E-values of blastp similarity searches are provided on the supplemental World Wide Web site for this report (http://www.utmem.edu/prevmed/pm/scr.html).
Individual SCRs of human and viral proteins were aligned together. The non-SCR regions and short inter-SCR stretches of several amino acids were excluded from analysis. Each poxvirus SCR was manually aligned against the previously completed alignment of human SCRs (Krushkal et al., 2000). Alignment of the entire set of human and poxvirus SCRs is available online at the supplemental web site for this report (http://www.utmem.edu/prevmed/pm/scr.html).
Sequence logos of short consensus repeats of aligned human and viral SCRs were inferred using the online software Weblogo at http://weblogo.berkeley.edu (Crooks et al., 2004).
2.2. Phylogenetic analysis
Individual short consensus repeats were grouped by two hierarchical clustering algorithms, the neighbor-joining and parsimony methods of phylogenetic tree inference. Due to the short length of SCRs (60–70 amino acids), bootstrap was not applied to avoid large sampling errors. Instead, results of tree inference were verified by comparing the clustering of repeats in the neighbor-joining tree and the consensus of parsimony trees. A phylogenetic tree was inferred from individual SCRs by the neighbor-joining method using phylogenetic analysis software MEGA v. 2.1 (Kumar et al., 2001). Distances between pairs of sequences were corrected for multiple amino acid substitutions and variation of substitution rate among sites using the gamma model (Krushkal et al., 1998, Krushkal et al., 2000). The value of the shape parameter for the gamma distribution of substitution rate among sites used was 0.93, as estimated previously from human C4bpα and FH (Krushkal et al., 1998). A phylogenetic tree was also inferred by the parsimony method using the TBR branch-swapping heuristic search method with 100 replicates of a random order of taxon addition using PAUP* software version 4.8 (Swofford, 2000). Both neighbor-joining and parsimony trees were midpoint rooted. Insertions and deletions (gaps) were excluded from all analyses in pairs of sequences under comparison.
3. Results
3.1. Phylogenetic clustering of short consensus repeats
Parsimony tree analysis of human complement regulators and orthopoxvirus proteins VCP, SPICE and IMP resulted in 157 most parsimonious trees with a score of 1986 each. A strict consensus tree (i.e., a summary tree with 100% support) inferred from these trees is shown in Fig. 1 . The neighbor-joining tree inferred from the short consensus repeats of VCP, SPICE, IMP, and human complement regulators is presented on the supplementary web site for this report.
Fig. 1.
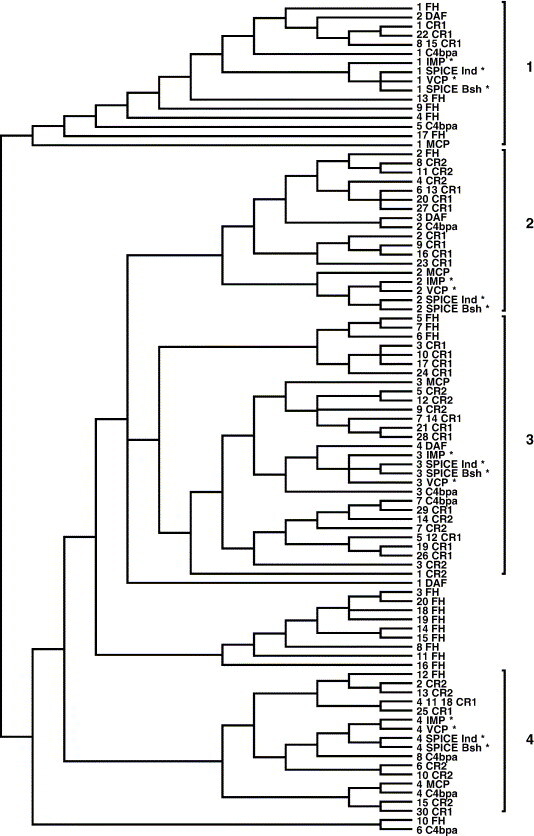
A strict (100%) consensus of 157 equally most parsimonious phylogenetic trees inferred by PAUP* from 91 short consensus repeats of VCP, SPICE, IMP, and human complement regulators. Clusters of repeats similar to viral SCRs 1–4 are shown by brackets. C4bpa indicates C4 binding protein, α chain (C4bpα). SPICE_Ind, smallpox inhibitor of complement enzymes (SPICE) from variola major, strain India. SPICE_Bsh, SPICE from variola major, strain Bangladesh. All other abbreviations of human and orthopoxvirus complement regulators are as described in Materials and methods. Asterisks (*) indicate viral SCRs.
Both consensus of the parsimony trees and the neighbor-joining tree showed that viral proteins VCP, SPICE, and IMP were more similar to one another than to human complement regulators, suggesting that the poxvirus complement regulators had a single precursor. The results of phylogenetic clustering are summarized in Table 1 listing those SCRs of human complement regulators that were clustered with each repeat of poxvirus proteins in both the neighbor-joining tree and the consensus of parsimony trees. While an overall pattern of similarity was observed between SCRs 1–4 of VCP and functionally important N-terminal SCRs of human complement regulators, the phylogenetic trees did not show any single human complement regulator to be the closest to viral proteins. It was clear from the trees inferred that VCP, SPICE, and IMP were similar to SCRs 1–4 of MCP and 1–5 of C4bpα. For example, both neighbor-joining and parsimony trees showed that SCR 1 of VCP, SPICE, and IMP were similar to SCRs 1 and 5 of C4bpα. SCR 2 of VCP and its poxvirus homologs was similar to SCR 2 of C4bpα. SCR 3 of VCP, SPICE, and IMP was similar to SCR 3 of C4bpα, and SCR 4 of VCP and its homologs in other poxviruses was similar to SCR 4 of C4bpα. Viral SCRs 1–3 were similar to SCRs 2–4 of DAF. No similarity to DAF repeats was found for viral SCR 4, consistent with an earlier observation that the homolog of SCR 4 of C4bpα and MCP is not present in DAF (Krushkal et al., 2000, McLure et al., 2004a, McLure et al., 2004b). Similarity to CR1 and CR2 was also consistent with previously observed relationships of SCRs in those proteins to repeats in C4bpα and MCP (Fujisaku et al., 1989, Krushkal et al., 2000, McLure et al., 2004a, McLure et al., 2004b). Phylogenetic trees also showed similarity of VCP, SPICE, and IMP to SCRs 1, 2, 4–6, 9, 12, 13, and 17 in factor H, which are important for either cofactor activity, decay accelerating activity, C3b binding, or a combination of these activities (Gordon et al., 1995, Pangburn et al., 2000, Zipfel et al., 2002).
Table 1.
Summary of clustering of short consensus repeats of human complement regulators with each repeat of viral proteins
| Viral proteins | Viral SCR | Most similar repeats in human complement regulatorsa |
|||||
|---|---|---|---|---|---|---|---|
| C4bpα | MCP | DAF | CR1 | CR2 | FH | ||
| VCP, SPICE, IMP | 1 | 1, 5 | 1 | 2 | 1, 8, 15, 22 | NM | 1, 4, 9, 13, 17 |
| 2 | 2 | 2 | 3 | 2, 6, 9, 13, 16, 20, 23, 27 | 4, 8, 11 | 2 | |
| 3 | 3 | 3 | 4 | 3, 5, 7, 10, 12, 14, 17, 19, 21, 24, 26, 28, 29 | 3, 5, 7, 9, 12, 14, 29 | 5, 6 | |
| 4 | 4 | 4 | Noneb | 30 | 2, 6, 10, 13, 15 | 12 | |
For each human complement regulator, short consensus repeats that were clustered with each poxvirus SCR 1, 2, 3, and 4 are listed. Only those clusterings between pairs of human and viral SCRs are listed that were observed in both tree inference methods used, the neighbor-joining tree and the strict consensus of 157 parsimony trees.
None indicates that neither tree inference method clustered this viral SCR with any repeats from this human complement regulator. The detailed trees showing all clusterings are provided in Fig. 1 and online (see supplementary online material for this report).
3.2. Sequence logos of viral and human repeats
Sequence logos of viral and human SCRs presented on the supplementary web page for this manuscript identified amino acids conserved within each group of SCRs, in agreement with previous reviews by other authors (Hourcade et al., 2000, Kotwal, 2000). These conserved sites were in most cases shared between the human and viral sequences, despite the overall sequence divergence between the orthopoxvirus and human SCRs. The positions in which the most common conserved amino acid exceeded 1 bit in both poxvirus and human sequence logos were C2, P8, N13, G14, Y/F32, G37, V/I40, Y/F42, C44, G47, Y/F54, L56, G60, C68, G76, W81, P87, and C91. Here, the first letter indicates the most common amino acid or amino acids and the second number indicates the position number in the SCR alignment provided in the online supplement. Further studies of shared and divergent sequence sites within similar groups of SCRs presented in Table 1 may identify the combinations of sequence variants that would explain the functional differences among SCRs.
4. Discussion
4.1. Phylogenetic similarity between viral and human short consensus repeats
Phylogenetic analysis indicated that VCP, SPICE, and IMP are similar to functional N-terminal SCRs in virtually all functional human complement regulators, including both C4bpα and factor H, which have likely diverged a very long time ago (Krushkal et al., 1998, Krushkal et al., 2000). Similarity of VCP, SPICE, and IMP was also observed to additional functionally important SCRs of FH. For example, SCRs of poxvirus complement regulators were similar to repeats 12 and 13 of FH that participate in interaction with C3b and affect decay acceleration activity (Pangburn et al., 2000, Zipfel et al., 2002). This conserved sequence similarity of viral complement regulators to functional SCRs in human complement regulators suggests that the ancestor of the present-day RCA proteins in orthopoxviruses was capable of some complement regulation.
Interestingly, SCRs 1 and 2 of VCP, SPICE, and IMP were consistently clustered with SCRs 1 and 2, respectively, of C4bpα. SCR 1 of poxvirus complement regulators was also clustered with SCR 13 of FH in both parsimony and neighbor-joining trees, and it was also clustered with SCR 7 of FH in the neighbor-joining tree. All these SCRs of C4bpα and FH contain probable heparin binding sites (Blom, 2002, Zipfel et al., 2002). While SCRs 1 and 2 of VCP have some heparin-binding activity (Smith et al., 2000), recent studies suggested that SCR 4 of VCP is likely the most important repeat for heparin binding by VCP (Smith et al., 2003, Ganesh et al., 2004). However, phylogenetic trees presented here show that SCR 4 of VCP is divergent from SCRs 7, 13, and 20 of FH and from SCRs 1–3 of C4bpα that have been implicated in heparin binding (Blom, 2002, Zipfel et al., 2002). This may suggest that the functional similarity of SCRs of VCP to those of FH and C4bpα in heparin binding activity may be due to convergent evolution, independent of their sequence evolution. Another possibility is that a small number of residues rather than the sequence of entire SCRs may affect the heparin binding properties. In the VCP protein homolog of monkeypox virus, a large portion of SCR 4 is absent (Uvarova and Shchelkunov, 2001), suggesting that SCRs 1–3 rather than 4 may be most functionally important, in agreement with the sequence similarity observed in the present study. However, Isaacs et al. (2003) used mouse monoclonal antibodies to show that SCR 4 is functionally important for interaction of VCP with C3b/C4b, contrary to its absence in monkeypox virus. Further sequence analysis comparisons and experimental mutational analysis are needed to elucidate the exact functional role of individual amino acids and their combinations in different SCRs of poxvirus complement regulators.
4.2. Possible origin of poxvirus complement regulators
While VCP, SPICE, and IMP showed resemblance to the functional N-terminal repeats of human complement regulators, these viral proteins were much more similar in their protein sequence to one another than to any human complement regulators analyzed. These results are in agreement with those from a previous study (Kotwal et al., 1998a, Kotwal et al., 1998b) that showed a separation between sequences of orthopoxvirus complement regulators and mammalian C4bpα proteins on phylogenetic trees from the entire sequences that were not divided into individual SCRs. The phylogenetic trees inferred in the present study show in detail that sequences of each short consensus repeat of poxvirus complement regulators are phylogenetically distinct and form a separate clade on the tree. This suggests that the viral complement regulators VCP, SPICE, and IMP likely had a single ancestor, in agreement with the hypothesis proposed by Kotwal et al., 1998a, Kotwal et al., 1998b. Therefore, any changes in the poxvirus complement regulators since their divergence from that ancestor represent new mutations. A striking example of such mutations can be found in SPICE. SPICE is a variola major virus protein that is nearly 100-fold more potent than VCP in inactivating human C3b and 6-fold more potent in inactivating C4b (Rosengard et al., 2002). Phylogenetic tree analysis showed that not only were SPICE and VCP much more similar to one another than to human complement regulators, but also that none of the SCRs 1–4 of SPICE were closer to the ancestral node of viral proteins VCP, SPICE, and IMP than any of the repeats of VCP. One can conclude that SPICE was able to greatly enhance its ability to regulate human complement by acquiring new mutations after the precursor gene of viral complement regulators was introduced into poxviruses.
The evolutionary origin of poxvirus RCA proteins VCP, SPICE, and IMP can be explained under different scenarios. One possibility is that the ancestor of the present orthopoxvirus complement regulators was acquired a very long time ago in evolutionary history, possibly even prior to the duplication and split between C4bpα and factor H (Krushkal et al., 2000). This explanation would be consistent with an observation that VCP, SPICE, and IMP each have four SCRs, similar to an N-terminal four-SCR stretch conserved in C4bpα, MCP, and CR1. However, a second, more plausible explanation consistent with the results of phylogenetic inference is that orthopoxviruses originally acquired the ancestral viral complement regulator from a species highly divergent from humans. This scenario is in agreement with the close relationship among sequences of different orthopoxviruses. In the present study, we observed a low degree of divergence among complement regulator sequences from vaccinia, variola, and cowpox viruses. In addition, a recent detailed study of many poxvirus genomes (Gubser et al., 2004) confirmed a low degree of divergence among genome sequences of orthopoxviruses, which may be explained by the recent split among their lineages. Therefore, it is more likely that the common precursor of the VCP, SPICE, and IMP genes was acquired by orthopoxviruses from a species distant from human. This hypothesis is consistent with the acquisition by poxviruses of many other immune modulator genes (McFadden and Murphy, 2000). The answer to which host cellular gene gave rise to poxvirus complement regulator genes will likely come from comparisons of complement regulator sequences of multiple poxvirus species, which are now becoming available as a result of sequencing projects (McFadden, 2005), and of multiple genes from a large range of their hosts. In summary, poxviruses seem to have adapted remarkably to protect themselves against the host complement system by incorporating genes for complement regulation in their genomes and by further modifying them through the mutation process, resulting in protein products highly efficient in the regulation of the host complement.
Acknowledgements
E.C., A.E., and D.K. performed this work part of the requirement for the B.Sc. degree at Worcester Polytechnic Institute. At the time of her participation in this project, E.C. was not affiliated with Acambis, Inc, a smallpox vaccine producer, where she was later employed following the completion of this project. This work was performed without association to any commercial venture or any outside source of funding.
We express our gratitude to Dr. I. Gigli (University of Texas-Houston) for her help, comments, and suggestions in the preparation of this manuscript. We also thank Dr. R. Adkins (University of Tennessee Health Science Center) for technical assistance with this project and for help with manuscript preparation. We are also grateful to anonymous reviewers for their comments.
Received by W. Makalowski
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.gene.2005.05.008.
Appendix A. Supplementary data
References
- Blom A.M. Structural and functional studies of complement inhibitor C4-binding protein. Biochem. Soc. Trans. 2002;30:978–982. doi: 10.1042/bst0300978. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlback B., Smith C.A., Müller-Eberhard H.J. Visualization of human C4b-binding protein and its complexes with vitamin K-dependent protein S and complement protein C4b. Proc. Natl. Acad. Sci. U. S. A. 1983;80:3461–3465. doi: 10.1073/pnas.80.11.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaku A., Harley J.B., Frank M.B., Gruner B.A., Frazier B., Holers V.M. Genomic organization and polymorphisms of the human C3d/Epstein–Barr virus receptor. J. Biol. Chem. 1989;264:2118–2125. [PubMed] [Google Scholar]
- Ganesh V.K., Smith C.A., Kotwal G.J., Murthy K.H. Structure of vaccinia complement protein in complex with heparin and potential implications for complement regulation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8924–8929. doi: 10.1073/pnas.0400744101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Georges A.J., Georges-Courbot M.C. Biohazards due to orthopoxvirus: should we re-vaccinate against smallpox? Med. Trop. (Mars) 1999;59:483–487. [PubMed] [Google Scholar]
- Gigli I., Fujita T., Nussenzweig V. Modulation of the classical pathway C3 convertase by plasma proteins C4 binding protein and C3b inactivator. Proc. Natl. Acad. Sci. U. S. A. 1979;76:6596–6600. doi: 10.1073/pnas.76.12.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.L., Kaufman R.M., Blackmore T.K., Kwong J., Lublin D.M. Identification of complement regulatory domains in human factor H. J. Immunol. 1995;155:348–356. [PubMed] [Google Scholar]
- Gubser C., Hue S., Kellam P., Smith G.L. Poxvirus genomes: a phylogenetic analysis. J. Gen. Virol. 2004;85:105–117. doi: 10.1099/vir.0.19565-0. [DOI] [PubMed] [Google Scholar]
- Hourcade D., Liszewski M.K., Krych-Goldberg M., Atkinson J.P. Functional domains, structural variations and pathogen interactions of MCP, DAF and CR1. Immunopharmacology. 2000;49:103–116. doi: 10.1016/s0162-3109(00)80296-9. [DOI] [PubMed] [Google Scholar]
- Hourcade D.E., Mitchell L., Kuttner-Kondo L.A., Atkinson J.P., Medof M.E. Decay-accelerating factor (DAF), complement receptor 1 (CR1), and factor H dissociate the complement AP C3 convertase (C3bBb) via sites on the type A domain of Bb. J. Biol. Chem. 2002;277:1107–1112. doi: 10.1074/jbc.M109322200. [DOI] [PubMed] [Google Scholar]
- Isaacs S.N., Argyropoulos E., Sfyroera G., Mohammad S., Lambris J.D. Restoration of complement-enhanced neutralization of vaccinia virus virions by novel monoclonal antibodies raised against the vaccinia virus complement control protein. J. Virol. 2003;77:8256–8262. doi: 10.1128/JVI.77.15.8256-8262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper C., et al. Membrane cofactor protein (MCP; CD46) expression in transgenic mice. Clin. Exp. Immunol. 2001;124:180–189. doi: 10.1046/j.1365-2249.2001.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkitadze M.D., et al. Central modules of the vaccinia virus complement control protein are not in extensive contact. Biochem. J. 1999;344(Pt 1):167–175. [PMC free article] [PubMed] [Google Scholar]
- Kotwal G.J. Poxviral mimicry of complement and chemokine system components: what's the end game? Immunol. Today. 2000;21:242–248. doi: 10.1016/s0167-5699(00)01606-6. [DOI] [PubMed] [Google Scholar]
- Kotwal G.J., Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- Kotwal G.J., Isaacs S.N., McKenzie R., Frank M.M., Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- Kotwal G.J., Blasco R., Miller C.G., Kuntz S., Jayaraman S., Shchelkunov S.N. In: Symposium in Immunology VII: Vaccination. Eibl M.M., Huber C., Peter H.H., Wahn W., editors. Springer Verlag; Berlin: 1998. Strategies for Immunomodulation and Evasion by Microbes: Important Consideration in the Development of Live Vaccines. [Google Scholar]
- Kotwal G.J., Miller C.G., Justus D.E. The inflammation modulatory protein (IMP) of cowpox virus drastically diminishes the tissue damage by down-regulating cellular infiltration resulting from complement activation. Mol. Cell. Biochem. 1998;185:39–46. doi: 10.1023/a:1006844624825. [DOI] [PubMed] [Google Scholar]
- Krushkal J., Kemper C., Gigli I. Ancient origin of human complement factor H. J. Mol. Evol. 1998;47:625–630. doi: 10.1007/pl00013152. [DOI] [PubMed] [Google Scholar]
- Krushkal J., Bat O., Gigli I. Evolutionary relationships among proteins encoded by the regulator of complement activation gene cluster. Mol. Biol. Evol. 2000;17:1718–1730. doi: 10.1093/oxfordjournals.molbev.a026270. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lee S.-H., Jung J.U., Means R.E. ‘Complementing’ viral infection: mechanisms for evading innate immunity. Trends Microbiol. 2003;11:449–452. doi: 10.1016/j.tim.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Marie J.C., Astier A.L., Rivailler P., Rabourdin_Combe C., Wild T.F., Horvat B. Vol. 3. 2002. pp. 659–666. (Linking Innate and Acquired Immunity: Divergent Role of CD46 Cytoplasmic Domains in T Cell Induced Inflammation). [DOI] [PubMed] [Google Scholar]
- McFadden G. Poxvirus tropism. Nat. Rev. Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G., Murphy P.M. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr. Opin. Microbiol. 2000;3:371–378. doi: 10.1016/s1369-5274(00)00107-7. [DOI] [PubMed] [Google Scholar]
- McLure C.A., et al. Amino acid patterns within short consensus repeats define conserved duplicons shared by genes of the RCA complex. J. Mol. Evol. 2004;59:143–157. doi: 10.1007/s00239-004-2609-8. [DOI] [PubMed] [Google Scholar]
- McLure C.A., Williamson J.F., Stewart B.J., Keating P.J., Dawkins R.L. Genomic analysis reveals a duplication of eight rather than seven short consensus repeats in primate CR1 and CR1L: evidence for an additional set shared between CR1 and CR2. Immunogenetics. 2004;56:631–638. doi: 10.1007/s00251-004-0731-9. [DOI] [PubMed] [Google Scholar]
- Müller-Eberhard H.J. In: Complement. Müller-Eberhard H.J., Miescher P.A., editors. Springer; Berlin: 1985. Introduction and Overview; pp. 1–5. [Google Scholar]
- Murthy K.H., et al. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell. 2001;104:301–311. doi: 10.1016/s0092-8674(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Pangburn M.K., Müller-Eberhard H.J. In: Complement. Müller-Eberhard H.J., Miescher P.A., editors. Springer; Berlin: 1985. The Alternative Pathway of the Complement. [Google Scholar]
- Pangburn M.K., Pangburn K.L., Koistinen V., Meri S., Sharma A.K. Molecular mechanisms of target recognition in an innate immune system: interactions among factor H, C3b, and target in the alternative pathway of human complement. J. Immunol. 2000;164:4742–4751. doi: 10.4049/jimmunol.164.9.4742. [DOI] [PubMed] [Google Scholar]
- Pier G.B., Lyczak J.B., Wetzler L.M. Immunology, Infection, and Immunity. ASM Press; Washington, DC: 2004. pp. 85–109. [Google Scholar]
- Rosengard A.M., Liu Y., Nie Z., Jimenez R. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8808–8813. doi: 10.1073/pnas.112220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross G.D., Polley M.J., Rabellino E.M., Grey H.M. Two different complement receptors on human lymphocytes. One specific for C3b and one specific for C3b inactivator-cleaved C3b. J. Exp. Med. 1973;138:798–811. doi: 10.1084/jem.138.4.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu A., Isaacs S.N., Soulika A.M., Lambris J.D. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J. Immunol. 1998;160:5596–5604. [PubMed] [Google Scholar]
- Smith S.A., et al. Conserved surface-exposed K/R-X-K/R motifs and net positive charge on poxvirus complement control proteins serve as putative heparin binding sites and contribute to inhibition of molecular interactions with human endothelial cells: a novel mechanism for evasion of host defense. J. Virol. 2000;74:5659–5666. doi: 10.1128/jvi.74.12.5659-5666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.A., et al. Mapping of regions within the vaccinia virus complement control protein involved in dose-dependent binding to key complement components and heparin using surface plasmon resonance. Biochim. Biophys. Acta. 2003;1650:30–39. doi: 10.1016/s1570-9639(03)00189-4. [DOI] [PubMed] [Google Scholar]
- Stehle T., Larvie M. In: Innate Immunity. Ezekowitz R.A.B., Hoffman J.A., editors. Humana; Totowa, NJ: 2003. Structures of Complement Control Proteins; pp. 231–235. [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, MA: 2000. PAUP*: Phylogenetic Analysis Using Parsimomy (*and Other Methods) Version 4. [Google Scholar]
- Uvarova E.A., Shchelkunov S.N. Species-specific differences in the structure of orthopoxvirus complement-binding protein. Virus Res. 2001;81:39–45. doi: 10.1016/S0168-1702(01)00332-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles A.P., Shaw G., Bright J., Perczel A., Campbell I.D., Barlow P.N. NMR studies of a viral protein that mimics the regulators of complement activation. J. Mol. Biol. 1997;272:253–265. doi: 10.1006/jmbi.1997.1241. [DOI] [PubMed] [Google Scholar]
- Zipfel P.F., et al. Factor H family proteins: on complement, microbes and human diseases. Biochem. Soc. Trans. 2002;30:971–978. doi: 10.1042/bst0300971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


