Abstract
Monkeypox is a zoonotic disease with clinical manifestations similar to smallpox in humans. Since May 13, 2022, an increasing number of suspected and confirmed cases have been reported, affecting non-endemic regions across the globe. More strikingly, reports from the current outbreak reveal unique aspects regarding transmission dynamics and an unprecedented, rapidly expanding and sustained community transmission. As demonstrated through the still-ongoing COVID-19 pandemic, genomic surveillance has been an essential resource for monitoring and tracking the evolution of pathogens of public health relevance. Herein, we performed a phylogenomic analysis of available Monkeypox virus (MPXV) genomes to determine their evolution and diversity. Our analysis revealed that all MPXV genomes grouped into three monophyletic clades: two previously characterized clades and a newly emerging clade harboring genomes from the ongoing 2022 multi-country outbreak with 286 genomes comprising the hMPXV-1A clade and the newly classified lineages: A.1 (n = 6), A.1.1 (n = 1), A.2 (n = 3) and B.1 (n = 262), where lineage B.1 includes all MPXV genomes from the 2022 outbreak. Finally, it was estimated that B.1 lineage of this clade emerged in Europe on 03/02/2022 [95%CI = 11/13/2021 to 05/10/2022]. The exceptional surge of cases and the broader geographical expansion suggest multifactorial factors as drivers of the current outbreak dynamics. Such factors may include the cessation of smallpox vaccination and its potential spread across particular networks. Integrating pertinent epidemiological information with genomic surveillance information will help generate real-time data to help implement adequate preventive and control measures by optimizing public health decisions to mitigate this outbreak.
Keywords: Monkeypox virus, Outbreak, Phylogenomic analysis, B.1 lineage
1. Introduction
Monkeypox (MPX) is a zoonotic disease caused by the Monkeypox virus (MPXV) (Poxviridae: Orthopoxvirus) [1] which is endemic to Central and Western-African countries [2]. It is a double-stranded DNA genome consisting of ∼197 kb with ∼190 non-overlapping coding genes [3], which are responsible for its various biological characteristics (immunomodulation, host preference/diversity and viral pathogenicity) [4]. Clinical features of MPXV are similar to those caused by smallpox but with lower mortality, case fatality and complication rates [5]. This virus was first identified in monkeys but it can infect a wide range of host species, including rodents such as rats, mice, and squirrels [6,7] and a large variety of non-human primate species, porcupines and domestic pigs [8]. Many aspects of the ecology of MPXV remain yet to be elucidated, most notably those concerning the mechanisms and possible routes of transmission [9]. Classically, the virus spreads mainly through prolonged exposure to respiratory droplets or close physical contact and exposure to infected body fluids [1]. Indirect transmission through direct contact with faeces and faeces carried by flies has also been suggested as possible sources of transmission among wild chimpanzees [10]. Furthermore, recently, a new pattern of spread among sexual networks, specifically among men who have sex with men (MSM), has underscored sexual contact as a main spreading route for the virus [11] amongst other potential sources [12].
Five major MPXV outbreaks have been reported, occurring in 1970, 1996–97, 2003, and 2018. The most recent of them, is the still-ongoing multi-country outbreak 2022, causing more than 6000 cases in over 50 non-endemic countries on multiple continents [1,13]. The origin of most of these outbreaks can be traced and associated with infections originating from African countries. During the first two outbreaks, many African countries reported high numbers of MPX-associated cases, prompting governmental organizations to rapidly implement mass vaccination campaigns to halt transmission and reduce infection rates [5]. As a direct consequence, many countries instituted broad MPX surveillance systems to monitor and follow epidemiological dynamics, which since 1986, became restricted to geographical regions linked to specific outbreaks [5,14]. Since the implementation of such surveillance programs, an ever-increasing trend of MPXV cases has been reported, including imported cases outside Africa to other continents [15]. The increasing number of cases and broadening geographical expansion have prompted revisiting the many aspects of MPXV ecology, its epidemiology and the genetic determinants for virulence and transmissibility through genomics and phylodynamic approaches to formulate better disease control and prevention measures.
From an evolutionary standpoint, MPXV originated approximately 3500 years ago within the Old World orthopoxviruses clade, undergoing independent evolution and further separation to other genetic variants such as Clade 2 (former West African clade) approximately 600 hundred years ago [16]. In terms of its genome architecture, historically, MPXV has been classified in two main clades (West African and Central African clades). However, a new proposal for classifying MPXV has been established to group isolates into three clades [11], each with distinct geographical, clinical, genomic and epidemiological differences [17,18]. These clades correlate with the different epidemiological MPXV outbreaks. For instance, the first two clades include the majority of isolates linked to outbreaks from the Democratic Republic of Congo (DRC) (MPXV Clade 1), formerly known as the Congo Basin clade, and from West Africa (MPXV Clade 2). Isolates from these clades, which are responsible for most of the natural transmission cycles and endemism in Africa, reveal different transmission, pathobiologic patterns and distinct clinical outcomes that further demonstrate differentiation amongst them [8]. Clade 1 (Congo Basin Clade) is more clinically severe, with higher mortality rates and exhibited increased transmissibility.
Conversely, Clade 2 MPXV isolates are associated with milder infection, lower mortality rates and reduced transmissibility [19]. Interestingly, clade 3 (MPXV Clade 3) includes isolates originating from the 2017-to-2019 outbreaks [20] and genomes from the most recent 2022 outbreak diverging emerging lineages that are currently under investigation [20]. The main differences among these clades are associated with coding regions that relate mainly to immunomodulatory and host recognition antigenic determinants such as H3L and B21R [17,21,22]. This sharp divergence and early signs of sub-clustering may be associated with the diverse epidemiological landscapes among outbreaks (geography and demography), which could have driven different microevolutionary events redrawing the contemporary genomic architecture of MPXV [23].
Based on the geographic distribution and epidemiological features of MPXV, cases from outside Africa are considered rare and mainly linked to animal trade or international travel [15,24]. However, a recent increase in the number of MPX cases has been reported from non-endemic countries setting off global alarms and warnings alongside the COVID-19 pandemic [25]. Furthermore, this recent multi-country outbreak, unlike previous ones, appears to be following unusual trends, given that most MPXV-associated cases appear not to be linked to travel history to endemic territories or known exposure to infected subjects [26]. This current and unprecedented pattern of global dispersal has led international and national authorities to rapidly launch regional and global surveillance efforts to understand better the gaps and intricacies defining this multi-country outbreak dynamics. However, to date, the genomic features and phylogenetic relationships of the viruses responsible for this new outbreak remain to be deciphered. Therefore, this study aims to characterize the phylogenetic diversity of MPXV genomes from this recent outbreak, performing a comparative genomic analysis through publicly available genomes to provide further insights necessary to inform global surveillance efforts.
2. Materials and methods
We analyzed 337 high-quality and complete MPXV genomes available in GISAID and Viral NCBI databases by June 29, 2022 (Table S1). The analyses of these genomes were carried out according to the following scheme: An alignment using NextClade v2.1.0 tool [25], a descriptive analysis of the genomes studied and the number of confirmed MPXV cases [26] and an analysis of the phylogenomic relationships of genome sequences publicly available. For phylogenomic analysis, the aligned dataset obtained from NextClade was used to build a maximum likelihood (ML) in IQ-TREE multicore v1.6.12 [27], using the best substitution model and other parameters by default.
Additionally, we estimated the potential introduction date and dispersion dynamics of MPXV using TreeTime software [28], which considers a fixed clock rate of 9x10−6 (SD = 10.2x10−6) [20,29], a strict clock (SC) under a coalescent tree skyline prior and a root step to minimize residuals in a root-to-tip. The obtained tree was graphically represented in Interactive Tree Of Life (iTOL) v5 [30] where the clades were assigned using the intrataxa classification system that most accurately describes the viral diversity and includes viruses from the animal reservoir and previous human outbreaks, encompassing MPXV Clades 1, 2 and 3 as described in Ref. [15].
3. Results
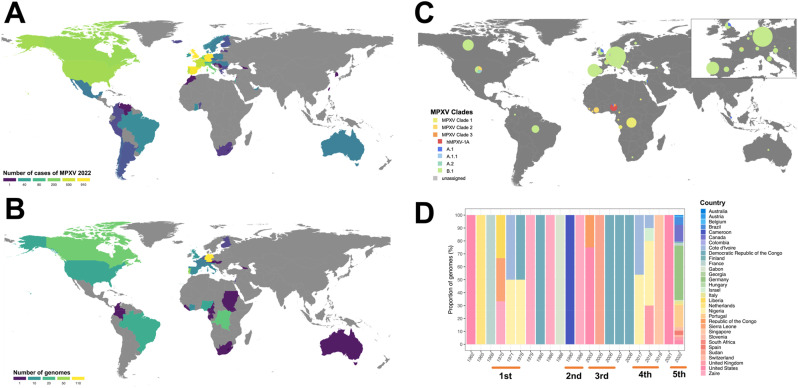
We analyzed 337 MPXV genomes from GISAID and NCBI corresponding to the various MPXV outbreaks (Fig. 1 and Table S1). To date, the ongoing outbreak of MPXV seems to be affecting non-endemic countries, where England, Spain and Germany have the highest reported cases of MPXV (Fig. 1A). Moreover, most of genome sequences publicly available come from non-endemic countries such as Germany (n = 110, 32.6%) and Portugal (n = 45, 13.4%) (Fig. 1B). Additionally, most genomes from these non-endemic areas belong to the B.1 lineage of the MPXV Clade 3 (n = 261, 77.4%; Fig. 1C), whereas in the endemic areas the genomes were from MPXV Clades 1 (n = 35, 10.4%) and 2 (n = 10, 2.5%; Fig. 1C). At a temporal scale, we found that from 1970 to 1996 (1st and 2nd outbreaks), MPXV was circulating in endemic countries such as DRC, Nigeria and Cameroon (Fig. 1D). However, after the 2003 outbreak (3rd outbreak), genomes started to be reported in non-endemic countries, which increased during the ongoing outbreak of MPXV (Fig. 1D).
Fig. 1.
Descriptive analyses of monkeypox virus. (A) Geographical distribution of the current MPXV outbreak 2022. (B) Geographical distribution of available MPXV complete genomes for each country. (C) Distribution of clades and lineages associated with the complete and available MPXV genomes. (D) Temporal variations in the proportions of MPXV genomes reported for each country taking into account the five major MPXV outbreaks.
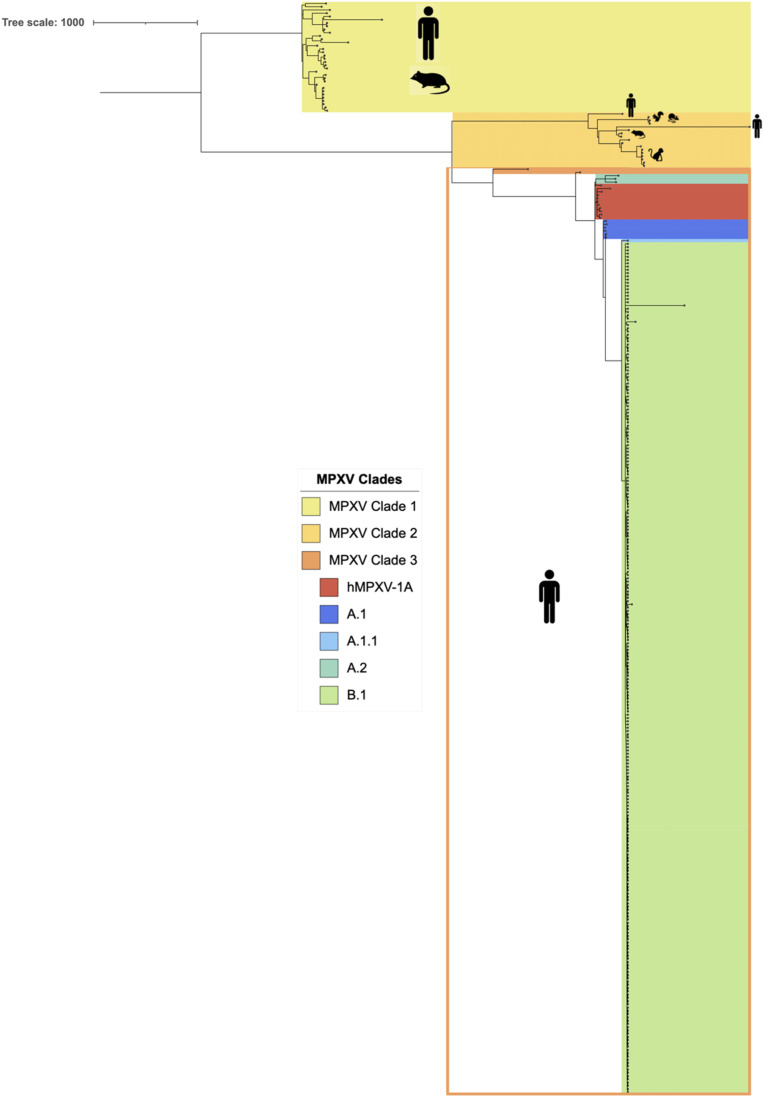
The maximum-likelihood time-scaled phylogeny (ML-TSP) obtained from Tree-Time (Fig. 2 ) showed clustering in the main three clades reported for MPXV that includes viral sequences from the animal reservoirs and previous human outbreaks. These clades were monophyletic and included the following number of genomes (Fig. 2): with 35 human MPXV genomes associated with the 1970 to 2017 outbreaks in Central Africa, MPXV Clade 2 with 16 genomes from the 1970 to 2017 outbreaks in different countries and from other sources and MPXV Clade 3 with 286 genomes from MPXV Clade 3 belonged to the last two epidemiological outbreaks of MPXV (2017–2022). The latter clade comprises the hMPXV-1A clade and the newly classified lineages: A.1 (n = 6), A.1.1 (n = 1), A.2 (n = 3) and B.1 (n = 262), where lineage B.1 includes all MPXV genomes from the 2022 outbreak.
Fig. 2.
Phylogenomic analysis of monkeypox virus. The figure shows the ML tree with the phylogenomic relationships of MPXV genomes. These genomes were grouped according to the current MPXV classification proposed by Happi et al.
The time-scaled phylogeny obtained from Tree-Time showed the introduction date based on a node-date assignment from the Most Recent Common Ancestor (MRCA) (Fig. 2). This analysis showed that MPXV Clade 1 was the most ancestral cluster, in contrast, the hMPXV-1A clades are the most recent (Fig. 2). Finally, it is estimated that B.1 lineage of this clade emerged in Europe on 03/02/2022 [95%CI = 11/13/2021 to 05/10/2022].
4. Discussion
Amid the COVID-19 pandemic, the recent emergence and ever-increasing number of MPXV cases across non-endemic countries have raised international alarms [31]. Despite that African researchers have been warning for years about the growing number of human MPXV cases reported across Central and West African countries [32]. Now, the sudden emergence of MPXV cases in several regions, including the Americas, Europe, Eastern Mediterranean, and Western Pacific (Fig. 1A), has raised many questions about the natural evolution of this current multi-country outbreak, particularly in regards to its unusual enhanced human-to-human transmission among patients with no travel history to endemic areas, suggesting the occurrence of undetected transmission chains with multiple sources of introduction [33]. In addition, the number of confirmed cases has increased over time such that, as of June 15, 2103 cases have been reported in 42 countries, with the majority of reports from Europe (84%) and the Americas (12%) [31].
Several hypotheses have been put forward to explain the upsurge of recent cases in Africa and other non-endemic areas. One hypothesis suggests that the cessation of mass vaccination against smallpox during the 1980s, which conferred up to 85% cross-immunity against MPXV, increased human susceptibility to the virus [34,35]. In turn, this has forced selection pressure over MPXV, favoring the development of immune evasion mechanisms and leading to an enhanced virus transmissibility [[34], [35], [36]]. Another hypothesis is that the acquisition of non-synonymous mutations associated with coding regions for predicted host recognition elements could represent a source of fitness adaptation for the virus [30,37]. For example, H3L, a viral envelope protein involved in the attachment to human target cells, poxvirus internalization and a critical epitope for host immune system recognition, has shown variability of 21 amino acids out of 324 amino acids (6.5% of the complete protein sequence), when compared to the H3L protein of the variola virus. This variability seems to unveil differences between the natural history of variola virus, which has been restricted to humans, compared to MPXV, which exhibits a broad host range, and continued transiting through species-to-species interfaces leading potentially to host-switch affinity and possible adaptation and enhanced transmission efficiency of MPXV to humans [38].
Regardless of the hypothesis, MPXV seems to have undergone a stochastic evolutionary event leading to the further diversification of MPXV Clade 3 due to its dispersal to other geographical locations and possible adaptation to new hosts across new regions. On the other hand, consistent with Gigante's et al. study, we found that the most common ancestor for these divergent clades traces back to a 1971 Nigerian genome (Nigeria 1971, KJ642617.1) [39]. Therefore, this would support the possibility of a diversification event due to MPXV spreading to non-endemic regions and expanding its range of traditional hosts to ignite the latest outbreak. Furthermore, the recent diversification of the clade perceives evidence of such a scenario with all its lineages (Fig. 2), which reveal distinctive monophyletic groupings conferred by their genomic characteristics [20].
One of the lineages that have received the most attention is B.1, given its close relationship to the most recent human outbreak of MPX by forming a divergent branch descendant from viruses within the A.1 lineage [40]. The occurrence of specific mutations in B.1 genomes compared to those from related viruses in 2018–2019 cases (Fig. 2) has drawn the attention of researchers due to its segregation into this divergent phylogenetic branch, signalling accelerated micro-evolutionary events leading to possible enhanced human-to-human transmission [40]. Indeed, 46 B.1-specific SNPs have been observed, including 24 non-synonymous mutations that separate the clade from the closest reference sequence. For example, the immunogenic surface glycoprotein B21R is considered relevant as it was previously described as an essential antibody target with several critical immunodominant epitopes featuring amino changes D209 N, P722S, and M1741I that alter host interaction and favor transmission [41]. Therefore, considering the continued and fast-paced evolutionary characteristics of MPXV lineage B.1 and its accelerated spread, genomic surveillance efforts should be maintained to help visualize genomic modifications among present circulating lineages, track and predict aspects of transmission dynamics, and monitor other mutational events leading to the phenotypic expression of immune evasion determinants or enhanced transmission elements contributing further to the outbreak spread.
Although the present outbreak's origin has not yet been determined, MPX is still mainly considered a zoonotic disease with limited human-to-human transmission [8]. However, the appearance of a series of mutations in such a short period [30], a feature not frequently observed in orthopoxviruses related to MPXV [22], is suggestive of rapid adaptive evolution to its host and fitness advantage for procurement of sustained (human-to-human) transmission. Indeed, recent studies suggest that the 2017 human MPX epidemic favoured the emergence of a single human clade. This hypothesis arose from the phylogenetic reconstruction of genomes involved in the last human outbreak in 2022, which exhibited a predominance of GA→AA and TC→TT changes in their viral genome [42]. Such dinucleotide changes are distinctive of APOBEC3 enzymatic activity, a family of deaminases which act on single-stranded DNA to mutate the complementary strand and inactivate the virus. That raises the question of whether this family of enzymes may be the primary genetic driver behind MPXV's current diversification and hastened adaptation to the human host, thus favouring the emergence of a human-specific clade and underscoring the importance of closely monitoring the potential genome-editing activity of APOBEC and its modulating effects in MPXV infections.
As for the emergence of MPXV (B.1 lineage) responsible for the ongoing outbreak, its estimated time of emergence in the European continent has been predicted to have occurred in as early as March 2022. This finding could indicate that B.1 emerged and spread across the European continent generating the first cases of MPXV, which subsequently spread to other continents such as Oceania and the Americas. Moreover, results from our analyses suggest a more recent emergence for the 2022 MPXV than that reported by Nextstrain (May 03) (https://nextstrain.org/monkeypox/hmpxv1?dmin=2021-07-03&label=clade:B.1 (accessed on 01 July 2022)), which performed the dating analysis from the genomes of the MPXV Clade 3 lineages. Regardless of the analysis, B.1 is an emerging lineage of MPXV that to date has been dispersed in different countries causing an increase in the number of MPX cases but with a low mortality rate. Given the epidemiological impact and genomic features of this new outbreak, it is necessary to perform genomic surveillance on this virus in order to design disease control and prevention mechanisms in non-endemic regions.
5. Conclusion
In summary, all genomes belonging to the current human MPXV outbreak of 2022 reveal a distinct monophyletic lineage compared to those from previous MPXV outbreaks. Our analyses suggest that the virus may have emerged in Europe as early as March 2022. An intriguing aspect is that the current 2022 MPXV exhibits a striking divergence from its related viral predecessors from 2018 to 2019. This heightened mutational signature contrasts with the ordinarily occurring rate observed in Orthopoxviruses, signalling an accelerated evolutionary path. Likewise, evidence suggests that some of these genomic changes could favor more efficient transmission and dispersion mechanisms, which is reflected in the increased number of cases reported across different regions suggesting that this may be associated with sexual transmission, however this is speculative and further studies are needed. Likewise, it is possible that mechanisms intrinsic to the host, such as the APOBEC enzymatic editing, could be driving this swift viral evolution favouring the appearance of a host-specific clade with enhanced human-to-human transmission. With an ever-increasing number of cases reported globally and sustained spread across different countries, genomic variability is expected to continue to rise in its transmission efficiency.
The paucity of data as to the origin and routes of transmission of this current outbreak demands further investigations to provide appropriate epidemiological insights and tools to predict better disease dynamics and prevent further spread. It is also time to anticipate several potential scenarios, such as spillover to new animal reservoirs in these new niches of expansion and the establishment of emerging epizootic events that may contribute to disease persistence in non-endemic regions. Genomic epidemiology initiatives should be strengthened with particular focus on the broader interface between humans and susceptible animal species, human-to-human interactions in anticipation of human or zoonotic amplification events and the establishment of cryptic transmission routes leading to the establishment and endemism of MPXV in previously unaffected regions. The COVID-19 pandemic has highlighted the value of translating genomic findings into meaningful tools to track transmission dynamics and predict the emergence of rapidly-evolving pathogens like the 2022-MPXV. Given the particular characteristics of this outbreak, it is essential to continue genomic surveillance efforts to identify and inform genomic changes of the virus to help developing timely prevention and control measures.
Funding source
None.
CRediT authorship contribution statement
Nicolas Luna: Methodology, Validation, Formal analysis, Writing – original draft, Writing – review & editing. Angie L. Ramírez: Methodology, Validation, Formal analysis, Writing – original draft, Writing – review & editing. Marina Muñoz: Methodology, Formal analysis, Writing – review & editing. Nathalia Ballesteros: Methodology, Formal analysis, Writing – review & editing. Luz H. Patiño: Methodology, Formal analysis, Writing – review & editing. Sergio Andres Castañeda: Methodology, Formal analysis, Writing – review & editing. D. Katterine Bonilla-Aldana: Methodology, Formal analysis, Writing – review & editing. Alberto Paniz-Mondolfi: Conceptualization. Juan David Ramírez: Conceptualization, Validation, Writing – original draft.
Declaration of competing interest
Nothing to declare.
Acknowledgements
We thank all the authors around the world that have deposited their MPXV genomes in GISAID. Following GISAID policy, the authors have been acknowledged in Table S1.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tmaid.2022.102402.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Alakunle E, Moens U, Nchinda G, Okeke MI. Monkeypox Virus in Nigeria: Infection Biology, Epidemiology, and Evolution. Viruses [Internet] 2020;12:1257. Available from:: https://www.mdpi.com/1999-4915/12/11/1257. [DOI] [PMC free article] [PubMed]
- 2.Bonilla-Aldana D.K., Rodriguez-Morales A.J. Is monkeypox another reemerging viral zoonosis with many animal hosts yet to be defined? Vet Q. 2022;42:148–150. doi: 10.1080/01652176.2022.2088881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shchelkunov S.N., Totmenin A.V., Safronov P.F., Mikheev M.V., Gutorov V.V., Ryazankina O.I., et al. Analysis of the monkeypox virus genome. Virol [Internet] 2002;297:172–194. doi: 10.1006/viro.2002.1446. https://linkinghub.elsevier.com/retrieve/pii/S0042682202914467 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kugelman J.R., Johnston S.C., Mulembakani P.M., Kisalu N., Lee M.S., Koroleva G., et al. Genomic variability of monkeypox virus among humans, democratic republic of the Congo. Emerg Infect Dis. 2014;20 doi: 10.3201/eid2002.130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen E., Kantele A., Koopmans M., Asogun D., Yinka-Ogunleye A., Ihekweazu C., et al. Human monkeypox. Infect Dis Clin. 2019;33:1027–1043. doi: 10.1016/j.idc.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sklenovská N., van Ranst M. Emergence of monkeypox as the most important Orthopoxvirus infection in humans. Front Public Health. 2018;6 doi: 10.3389/fpubh.2018.00241. https://www.frontiersin.org/article/10.3389/fpubh.2018.00241/full [Internet]: Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damon I.K. Status of human monkeypox: clinical disease, epidemiology and research. Vaccine. 2011;29:D54–D59. doi: 10.1016/j.vaccine.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Parker S., Nuara A., Buller R.M.L., Schultz D.A. Human monkeypox: an emerging zoonotic disease. Future Microbiol. 2007;2:17–34. doi: 10.2217/17460913.2.1.17. [DOI] [PubMed] [Google Scholar]
- 9.Abdelmoez Farahat R, Sah R, El-Sakka AA, Yasmine Benmelouka A, Kundu M, Labieb F, et al. Human monkeypox disease (MPX). [DOI] [PMC free article] [PubMed]
- 10.Patrono L.v., Pléh K., Samuni L., Ulrich M., Röthemeier C., Sachse A., et al. Monkeypox virus emergence in wild chimpanzees reveals distinct clinical outcomes and viral diversity. Nat Microbiol. 2020;5:955–965. doi: 10.1038/s41564-020-0706-0. [DOI] [PubMed] [Google Scholar]
- 11.Endo A., Murayama H., Abbott S., Ratnayake R., Pearson C.A.B., Edmunds W.J., et al. May 2022. Heavy-tailed sexual contact networks and the epidemiology of monkeypox outbreak in non-endemic regions.http://medrxiv.org/content/early/2022/06/13/2022.06.13.22276353.abstract medRxiv [Internet] 2022;2022.06.13.22276353. Available from: [DOI] [PubMed] [Google Scholar]
- 12.Ogoina D., Izibewule J.H., Ogunleye A., Ederiane E., Anebonam U., Neni A., et al. The 2017 human monkeypox outbreak in Nigeria—report of outbreak experience and response in the Niger delta university teaching hospital, Bayelsa state, Nigeria. PLoS One. 2019;14 doi: 10.1371/journal.pone.0214229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunge E.M., Hoet B., Chen L., Lienert F., Weidenthaler H., Baer L.R., et al. The changing epidemiology of human monkeypox—a potential threat? A systematic review. PLoS Neglected Trop Dis. 2022;16 doi: 10.1371/journal.pntd.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durski K.N., McCollum A.M., Nakazawa Y., Petersen B.W., Reynolds M.G., Briand S., et al. Emergence of monkeypox — west and central Africa, 1970–2017. MMWR. Morbidity Mortality Week Rep. 2018;67:306–310. doi: 10.15585/mmwr.mm6710a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauldin M.R., McCollum A.M., Nakazawa Y.J., Mandra A., Whitehouse E.R., Davidson W., et al. Exportation of monkeypox virus from the African continent. J Infect Dis. 2022;225:1367–1376. doi: 10.1093/infdis/jiaa559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babkin I.v. Babkina IN, tikunova N v. An update of Orthopoxvirus molecular evolution. Viruses. 2022;14:388. doi: 10.3390/v14020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berthet N, Descorps-Declère S, Besombes C, Curaudeau M, Nkili Meyong AA, Selekon B, et al. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. Scientific Reports [Internet] 2021;11:13085. Available from:: http://www.nature.com/articles/s41598-021-92315-8. [DOI] [PMC free article] [PubMed]
- 18.Likos A.M., Sammons S.A., Olson V.A., Frace A.M., Li Y., Olsen-Rasmussen M., et al. A tale of two clades: monkeypox viruses. J Gen Virol. 2005;86:2661–2672. doi: 10.1099/vir.0.81215-0. [DOI] [PubMed] [Google Scholar]
- 19.Jezek Z., Szczeniowski M., Paluku K.M., Mutombo M. Human monkeypox: clinical features of 282 patients. JID (J Infect Dis) 1987;156:293–298. doi: 10.1093/infdis/156.2.293. [DOI] [PubMed] [Google Scholar]
- 20.Happi C., Adetifa I., Mbala P., Njouom R., Nakoune E., Happi A., et al. 2022. Urgent need for a non-discriminatory and non-stigmatizing nomenclature for monkeypox virus [Internet]https://virological.org/t/urgent-need-for-a-non-discriminatory-and-non-stigmatizing-nomenclature-for-monkeypox-virus/853 [cited 2022 Jun 14];Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shchelkunov S.N., Totmenin A.V., Safronov P.F., Mikheev M.V., Gutorov V.V., Ryazankina O.I., et al. Analysis of the monkeypox virus genome. Virology. 2002;297:172–194. doi: 10.1006/viro.2002.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole Á., Rambaut A. 2022. Initial observations about putative APOBEC3 deaminase editing driving short-term evolution of MPXV since 2017 [Internet]https://virological.org/t/initial-observations-about-putative-apobec3-deaminase-editing-driving-short-term-evolution-of-mpxv-since-2017/830 [cited 2022 Jun 19];Available from: [Google Scholar]
- 23.Gomes J.P., Isidro J., Borges V., Pinto M., Sobral D., Santos J., et al. 2022. Multi-country outbreak of Monkeypox virus: genetic divergence and first signs of microevolution; pp. 1–8. [Google Scholar]
- 24.Borsky S., Hennighausen H., Leiter A., Williges K. CITES and the zoonotic disease content in international wildlife trade. Environ Resour Econ. 2020;76:1001–1017. doi: 10.1007/s10640-020-00456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zumla A., Valdoleiros S.R., Haider N., Asogun D., Ntoumi F., Petersen E., et al. Monkeypox outbreaks outside endemic regions: scientific and social priorities. Lancet Infect Dis. 2022 doi: 10.1016/S1473-3099(22)00354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization . 2022. Disease outbreak news; multi-country monkeypox outbreak in non-endemic countries [Internet]https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON385 [cited 2022 Jun 11];Available from: [Google Scholar]
- 27.Aksamentov I., Roemer C., Hodcroft E., Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Software. 2021;6:3773. [Google Scholar]
- 28.Kraemer M.U.G., Tegally H., Pigott D.M., Dasgupta A., Sheldon J., Wilkinson E., et al. The Lancet Infectious Diseases; 2022. Tracking the 2022 monkeypox outbreak with epidemiological data in real-time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagulenko P., Puller V., Neher R.A. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4 doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Firth C., Kitchen A., Shapiro B., Suchard M.A., Holmes E.C., Rambaut A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol. 2010;27:2038–2051. doi: 10.1093/molbev/msq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization . 2022. Multi-country monkeypox outbreak: situation update [Internet]https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON392 [cited 2022 Jun 15];Available from: [Google Scholar]
- 32.Kozlov M. Monkeypox in Africa: the science the world ignored. Nature. 2022 doi: 10.1038/d41586-022-01686-z. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z. Monkeypox: a potential global threat? J Med Virol. 2022 doi: 10.1002/jmv.27884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fine P.E.M., Jezek Z., Grab B., Dixon H. The transmission potential of monkeypox virus in human populations. Int J Epidemiol. 1988;17:643–650. doi: 10.1093/ije/17.3.643. https://academic.oup.com/ije/article-lookup/doi/10.1093/ije/17.3.643 [Internet]: Available from. [DOI] [PubMed] [Google Scholar]
- 35.Fenner F. The global eradication of smallpox. Med J Aust. 1980;1:455–456. doi: 10.5694/j.1326-5377.1980.tb135034.x. https://onlinelibrary.wiley.com/doi/abs/10.5694/j.1326-5377.1980.tb135034.x [Internet]: Available from. [DOI] [PubMed] [Google Scholar]
- 36.Gomes J.P., Isidro J., Borges V., Pinto M., Sobral D., Santos J., et al. 2022. Multi-country outbreak of Monkeypox virus: genetic divergence and first signs of microevolution; pp. 1–8.https://virological.org/t/multi-country-outbreak-of-monkeypox-virus-genetic-divergence-and-first-signs-of-microevolution/806 Available from: [Google Scholar]
- 37.O'Toole Á., Rambaut A. 2022. Update to observations about putative APOBEC3 deaminase editing in the light of new genomes from USA. [Google Scholar]
- 38.Giorgi F.M., Pozzobon D., di Meglio A., Mercatelli D. 2022. Genomic analysis of the recent monkeypox outbreak.http://biorxiv.org/content/early/2022/06/14/2022.06.01.494368.abstract bioRxiv [Internet] 2022: 06.01.494368. Available from. [DOI] [PubMed] [Google Scholar]
- 39.Gigante C.M., Korber B., Seabolt M.H., Wilkins K., Davidson W., Rao A.K., et al. 2022. Multiple lineages of Monkeypox virus detected in the United States, 2021- 2022.http://biorxiv.org/content/early/2022/06/11/2022.06.10.495526.abstract bioRxiv [Internet] 2022. 06.10.495526. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isidro J., Borges V., Pinto M., Sobral D., Santos J.D., Nunes A., et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med. 2022 doi: 10.1038/s41591-022-01907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.World Health Organization . 2022. Multi-country monkeypox outbreak: situation update [Internet]https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON392 [cited 2022 Jun 14];Available from: [Google Scholar]
- 42.Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.