Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) causes severe infections and poses a global healthcare challenge. The utilization of novel molecules which confer synergistical effects to existing MRSA-directed antibiotics is one of the well-accepted strategies in lieu of de novo development of new antibiotics. Thymol is a key component of the essential oil of plants in the Thymus and Origanum genera. Despite the absence of antimicrobial potency, thymol is known to inhibit MRSA biofilm formation. However, the anti-MRSA activity of thymol analogs is not well characterized. Here, we assessed the antimicrobial activity of several thymol derivatives and found that 4-chloro-2-isopropyl-5-methylphenol (chlorothymol) has antimicrobial activity against MRSA and in addition it also prevents biofilm formation. Chlorothymol inhibited staphyloxanthin production, slowed MRSA motility, and altered bacterial cell density and size. This compound also showed a synergistic antimicrobial activity with oxacillin against highly resistant S. aureus clinical isolates and biofilms associated with these isolates. Our results demonstrate that chlorinated thymol derivatives should be considered as a new lead compound in anti-MRSA therapeutics.
Keywords: MRSA, thymol derivatives, chlorothymol, antimicrobial, synergistic effect, biofilm
Introduction
Since the discovery of penicillin, many types of antibiotics have been developed and used [1]. Antibiotics are therapeutic agents that inhibit bacterial growth or kill bacteria. Due to the indiscriminate use of antibiotics, significant selection pressure has been applied to many bacteria [2]. Staphylococcus aureus is a pathogen that is widespread in humans and has various virulence factors; it can infiltrate body tissues and cause infection [3]. Although many antibiotics have been used to treat S. aureus infections, most of them are ineffective against methicillin-resistant S. aureus (MRSA), which first emerged in 1962 and has spread worldwide over time [4]. mecA, which is part of the staphylococcal chromosome cassette mec (SCCmec), exists only in MRSA and expresses penicillin-binding protein 2a (PBP2a), which weakens β-lactam sensitivity [5]. Patients infected with MRSA suffer from severe illness and high mortality rates, and their treatment is difficult [6]. Therefore, MRSA has emerged as a global healthcare issue [7]. MRSA can be broadly classified as healthcare-associated MRSA (HA-MRSA), community-associated MRSA (CA-MRSA), and livestock-associated MRSA (LA-MRSA) [8, 9]. Although HA-MRSA was initially noted for its high pathogenicity and mortality rate, CA-MRSA, which is highly infectious and resistant to different antibiotics, has recently emerged as a major problem [10]. The most well-known CA-MRSA strains are LAC (USA300) and MW2 (USA400) [11].
MRSA adapts to the surrounding environment and protects itself by expressing various virulence factors [12]. In particular, unlike other S. aureus strains, MRSA protects itself from antibiotics by expressing PBP2a and β-lactamases and forms biofilms that are impermeable to antibiotics [13, 14]. The protection of persister cells by biofilms is a problem in the complete treatment of MRSA [15]. Staphyloxanthin, a golden carotenoid pigment produced by S. aureus, is a virulence factor [16]. This pigment not only protects cells from host reactive oxygen species (ROS) and neutrophils but also reduces cell membrane fluidity and prevents the penetration of antimicrobial substances [17, 18]. S. aureus does not have movable appendages such as pili or flagella, but it can move through secretion of an amphiphilic substance called phenol-soluble modulin (PSM) [19]. These characteristics can enhance the transmission of MRSA [20]. In addition, PSM itself functions as a virulence factor, causing human cell lysis, inflammatory responses, and biofilm development [21]. Expression of these virulence factors is a great obstacle to patient treatment and helps the bacteria infect hosts [6, 20]. Therefore, inhibition of these virulence factors is important in treating MRSA infections. Compounds such as essential oils have been studied to assess their ability to inhibit these virulence factors [22]. Here, we attempted to identify a novel inhibitory compound that can be used in combination with antibiotics.
Thymol (2-isopropyl-5-methylphenol), a monoterpenoid compound, is a key component of the essential oil of many plants belonging to the Thymus and Origanum genera [23]. Thymol is widely used for fragrance, food flavoring, and dental treatment [24]. It also has antioxidant, antimicrobial, antitussive, expectorant, antispasmodic, and antibacterial effects [25]. It inhibits a wide range of bacteria, such as Escherichia coli, Salmonella enterica, and S. aureus [26]. In addition, according to a recent study, thymol may have sarA-dependent antibiofilm efficacy against MRSA [27].
Carvacrol, a structural isomer of thymol, has antibacterial properties [24]. Carvacrol and thymol exert antibacterial effects through cell membrane disruption, biofilm reduction, inhibition of motility, membrane-bound ATPase, and efflux pumps [24]. Although the effective and diverse antibacterial effects of thymol are well known, little is known about the antibacterial effects of thymol derivatives on MRSA. Therefore, we not only studied the effects of thymol derivatives on CA-MRSA strains LAC and MW2 but also extended the scope to clinical strains to evaluate the antibacterial effect of thymol derivatives against more diverse MRSA strains. Lastly, we analyzed the various phenotypes of the cells that appeared when thymol derivatives were added.
Materials and Methods
Strains, Media, Materials, and Culture Conditions
Wild-type (WT) strains of S. aureus USA300-0114 (LAC), USA400 (MW2), and their Δagr mutant strains were obtained from Dr. Michael Otto at the Pathogen Molecular Genetics Section, Laboratory of Bacteriology, National Institute of Allergy and Infectious Diseases, U.S. National Institutes of Health. The ΔsarA transposon mutant JE2 and the clinical strains were obtained from Jae-Seok Kim, M.D., PhD, Department of Laboratory Medicine, Kangdong Sacred Heart Hospital, Hallym University College of Medicine, Seoul, Korea. Carvacrol (5-isopropyl-2-methylphenol), thymol iodide ([4-(4-iodooxy-2-methyl-5-propan-2-ylphenyl)-5-methyl-2-propan-2-ylphenyl] hypoioditel), 4-isopropyl-3-methylphenol, and chlorothymol (4-chloro-2-isopropyl-5-methylphenol) (Sigma Aldrich, USA) were used as thymol derivatives. The cells were cultured in liquid tryptic soybean broth (TSB). For preculture, a single colony of the strain from a TSB agar plate was inoculated with 5 ml of TSB medium using a sterilized inoculation loop. After that, 1% (v/v) of the cell culture suspension was inoculated into TSB for subsequent cell cultivation at 37°C in a shaking incubator (200 rpm) or a 96-well plate without shaking [28].
Antimicrobial Susceptibility and Biofilm Formation
To investigate the antimicrobial susceptibility and biofilm formation, 200 μl of culture broth containing serially diluted oxacillin was prepared in a 96-well plate. Precultured cells were inoculated (1% v/v), and the plate was incubated at 37°C for 24 h without shaking. Cell optical density was measured using a 96-well plate reader (Thermo Fisher Scientific, USA). Biofilm formation was analyzed using crystal violet staining [29]. After the supernatant was carefully removed, biofilm fixation was performed with methanol and subsequently air-dried for 24 h. Thereafter, a 0.2% crystal violet solution was added to each well to stain the biofilm for 5 min. The remaining dye was removed and washed twice with distilled water. Finally, absorbance was measured at 595 nm using a 96-well microplate reader (Thermo Fisher Scientific) [28]. As chlorothymol is insoluble in common solvents, including water, ethanol was used as the solvent. Due to the potential toxic effect of ethanol on the cells, the same amount of ethanol without the compound was added to the control, and an inhibitory effect test was performed [30]. Ethanol did not show any inhibitory effect on cell growth even at the volume used with a compound concentration of 512 μg/ml (Fig. S1).
Motility Assay in a Soft Agar Plate
To determine the change in motility caused by the addition of chlorothymol, we conducted a previously reported soft agar assay [31]. A 20 μl aliquot of precultured cells was centrifuged and resuspended in the same volume of phosphate-buffered saline (PBS). Aliquots (2 μl) of the mixture were dropped onto the center of a 0.24%TSB agar plate and incubated for 10 h at 37°C. All experiments were performed in triplicate [28].
Staphyloxanthin Extraction and Quantification
Cells were grown in 5 ml of TSB with shaking (200 rpm) for 6, 12, 18, and 24 h at 37°C and harvested by centrifugation (3,000 ×g, 20 min). Each sample was then subjected to methanol extraction. The cell pellet was washed once with PBS and centrifuged. After the supernatant was completely removed, the pellet was resuspended in 500 μl methanol and incubated at 55°C for 20 min for staphyloxanthin extraction. Following centrifugation, 200 μl of staphyloxanthin in methanol was obtained. To prevent contamination with the cell pellet, the pigment-containing extracts were filtered through a 0.2 μm syringe filter (Chromdisc, Korea). The amount of pigment in each sample was determined immediately by measuring the optical density at 470 nm using a plate reader spectrometer (Thermo Fisher Scientific) [28].
Semiquantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Preculture was conducted using 5 ml of TSB, initiated using a single colony from a TSB agar plate, in a shaking incubator at 37°C overnight. Cells were cultured in 5 ml TSB with 1% (v/v) inoculum in a shaking incubator at 37°C for 24 h to extract total RNA. The cells were centrifuged at 3,521 ×g for 20 min. Total RNA was extracted using TRIzol reagent, and reverse transcription was performed using Superscript IV Reverse Transcriptase (Invitrogen Co., USA) to generate cDNA, according to the manufacturer’s instructions. Primer Express software v3.0.1 was used to design primers from Thermo Fisher Scientific. Then, 150 bp of amplicons generated from these primers were used to compare gene expression levels. PCR cycle number optimization was performed before semiquantitative PCR to determine the saturated gene expression levels of DNA gyrase subunit B (gyrB, the endogenous control) for each template. The optimal cycle number was set to 25 to enable further comparative gene expression analysis. Semiquantitative PCR was conducted using LA Taq with GC buffer I (Takara Bio, Japan) using the methods described in the manual [32].
Fractional Inhibitory Concentration (FIC) Index Analysis
The FIC index analysis was used to mathematically express the effect of the combination of two other antibacterial agents. The FIC of each antibacterial agent (A and B) was calculated as follows:
The FIC index (ΣFIC) is the sum of FICA and FICB and is a numerical value of the degree of interaction between the two substances. FIC index of less than 0.5 indicates synergism, 0.5–1 signifies an additive effect, 1–2 means indifference, and higher than 2 indicates antagonism [33,34]. We compared the FIC index values of each well of a 96-well plate and used them to determine the optimal concentration combination of the two antibacterial agents.
Scanning Electron Microscopy (SEM) Analysis of Cell Morphology
For SEM analysis, after 24 h of cultivation, 1 ml of each sample was collected by centrifugation (3,000 ×g, 10 min), washed three times with phosphate buffer (pH 6.8–7.0), and fixed with 2% buffered glutaraldehyde overnight. Glutaraldehyde was added after centrifugation, and the residual glutaraldehyde was removed by washing with phosphate buffer three times. The samples were dehydrated using a gradient of ethanol (v/v) (50%, 70%, 95%, and 100%). Different ratios of ethanol and hexamethyldisilazane (HMDS; 2:1, 1:1, 1:2 v/v) were used for chemical drying; 100% HMDS was used in the final step, and the mixture of HMDS and the samples was mounted on specimen stubs. The HMDS was evaporated overnight in a fume hood. The samples were then coated with gold dust at 5 mA for 300 s, and backscattered electron (BSE) images were acquired using SEM (TM4000Plus; Hitachi, Tokyo, Japan) with an accelerating voltage of 5–15 kV [35].
MRSA Biofilm Killing
The MRSA biofilm-killing activity of the thymol derivatives was assessed as previously described [36]. S. aureus MW2 was grown overnight in TSB broth at 37°C. The overnight culture of cells was diluted 1:200 in TSB supplemented with 0.2% glucose and 3% NaCl. A 13 mm mixed cellulose ester membrane (GSWP01300; EMD Millipore, USA) was placed in each well of a 24-well plate. A 100 μl aliquot of the diluted overnight culture was dispensed into each well and cultured statically in a 37°C incubator. After 24 h, the biofilm-forming membranes were washed three times with 1 ml PBS. Then, 1 ml of PBS containing the desired concentration of antibiotics was added to each well and incubated at 37°C for 24 h. The membranes were then washed three times with 1 ml PBS, placed in 1.5-ml microcentrifuge tubes containing 1 ml PBS, and sonicated in a Bronson ultrasonic bath for 10 min. The samples were serially diluted with PBS and spot-plated on Mueller-Hinton II agar plates. The plate was incubated at 37°C and the colonies were counted to estimate the number of surviving cells.
Membrane Permeability Assay
The membrane permeability of MRSA was assessed by using the membrane-impermeable DNA-binding dye SYTOX Green (cat. No. S7020; Invitrogen) as previously described [37]. Exponential-phase MRSA MW2 cells were washed three times with PBS and adjusted to an OD600 of 0.5. SYTOX Green was added to the washed cells at a final concentration of 5 μM. The samples were incubated for 30 min at room temperature in the dark. A 50 μl aliquot of the sample was added to each well of a black 96-well plate containing the indicated concentrations of the compounds. Fluorescence was measured at room temperature using a multimode plate reader (Cytation 5; BioTek, USA or Infinite M200 Pro microplate reader; Tecan Group Ltd., Mannedorf, Switzerland) at excitation and emission wavelengths of 485 nm and 525 nm, respectively. The Infinite M200 plate reader was equipped at Ewha Drug Development Research Core Center. All experiments were conducted in triplicate.
Results
Antibacterial Activity of Thymol Derivatives
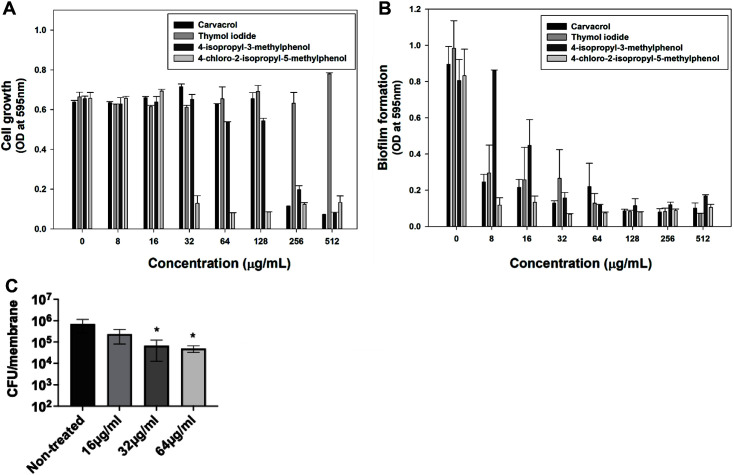
Although thymol itself has no inhibitory effect on cell growth, we attempted to measure the antibacterial activity of four derivatives. The results suggest that these derivatives have different properties from thymol [27]. To determine their antibacterial activity, their effects on cell growth and biofilm formation were evaluated in the LAC strain. After culturing the cells for 24 h, absorbance was measured at 595 nm. Carvacrol and 4-isopropyl-3-methylphenol showed a minimum inhibitory concentration (MIC) of 512 μg/ml (Fig. 1A). However, thymol iodide showed no growth inhibition effect, even at 512 μg/ml. Chlorothymol had a MIC of 32 μg/ml, showing the most outstanding effect of the four derivatives.
Fig. 1. Antibacterial activity and anti-biofilm potency of thymol derivatives against MRSA at various concentrations.
(A) Growth inhibitory activity against S. aureus LAC by various concentrations of thymol derivatives was quantified as a measure of OD595. (B) Inhibition of S. aureus LAC biofilm formation by thymol derivatives was assessed by staining with 0.2% crystal violet. Statistical analysis involved 240 ANOVA (with the level of significance at 5%). (C) S. aureus MW2 biofilms formed on a 13-mm membrane were treated with the indicated concentrations of chlorothymol for 24 h. The remaining survival cells were enumerated by serial dilution and plating on CaMH II agar plates. The level of detection was 2 × 102 CFU/membrane. Statistical differences between control and treated groups were analyzed by one-way ANOVA and post hoc Tukey test (*p < 0.05).
All four derivatives inhibited biofilm formation in a dose-dependent manner. In particular, chlorothymol almost completely inhibited MRSA biofilm formation even at a sub-MIC of 8 μg/ml (Fig. 1B). Next, we assessed the bactericidal activity of chlorothymol on MRSA cells in mature biofilms. As shown in Fig. 1C, chlorothymol at the MIC level of 32 μg/ml led to an approximately 1-log decrease in viability of MRSA biofilm cells. These results indicate that in contrast to thymol, chlorothymol not only inhibits MRSA biofilm formation but also has potency against mature MRSA biofilms. Since chlorothymol exhibited promising antimicrobial activity, we further investigated its effects on MRSA.
Inhibitory Effect of Chlorothymol on Motility and Staphyloxanthin Production
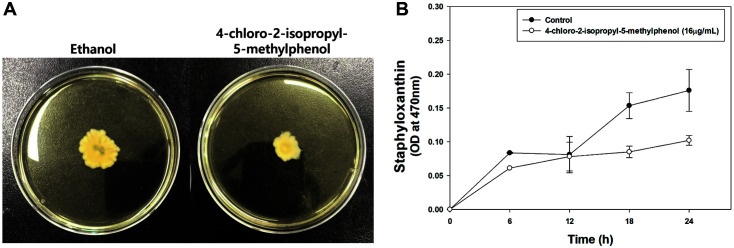
Motility characteristics of S. aureus play an important role in biofilm formation and host colonization, which are essential for cell survival and adaptation [38]. S. aureus was originally considered non-motile; however, it has recently been shown to move through spreading and comet formation under certain conditions, such as on a soft agar plate [19, 31]. It can move through the production of virulence factors called PSMs, amphiphilic α-helical peptides with surfactant properties [38, 39]. We assessed the effect of chlorothymol on the motility of the LAC strains on soft agar plates. When the compound was added, motility significantly decreased compared to the control. The same amount of ethanol was added to the control as to the compound solvent, which did not affect cell growth. (Fig. 2A).
Fig. 2. Inhibitory effect of chlorothymol on motility and staphyloxanthin production.
(A) The experiment was performed in triplicate with similar results. (B) Statistical analysis was performed by applying 240 ANOVA with the level of significance at 5%.
Staphyloxanthin functions as a barrier from the host immune system [16, 17, 41]. It is also known that thymol has staphyloxanthin inhibitory potential against MRSA [42]. Therefore, we evaluated the change in staphyloxanthin production when 16 μg/ml chlorothymol was added. Each sample was harvested at 6-hour intervals to extract staphyloxanthin (the OD595 value of each sample was set to 5, and the cell amount was adjusted equally). The decrease in staphyloxanthin production in the sample with chlorothymol showed a tendency to increase over time (Fig. 2B).
Effect of Chlorothymol on Major Virulence-Related Genes
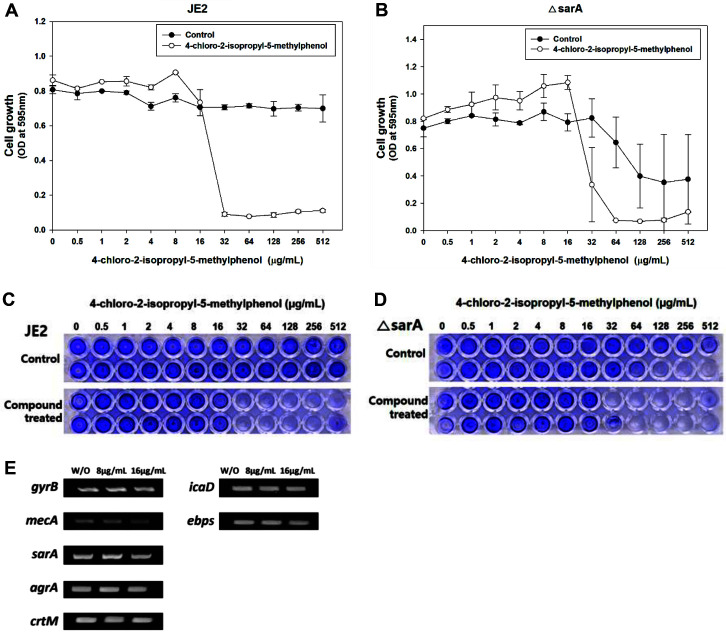
Thymol and its analog carvacrol alter the expression of several S. aureus virulence factors including the global regulator sarA and the accessory gene regulator agr [27, 43, 44]. Thus, we sought to determine whether chlorothymol also modulates S. aureus virulence factors. Since thymol inhibits S. aureus biofilm formation in a sarA-dependent way [27] and sarA regulates agr [45], we assessed the role of sarA and agr on the antimicrobial activity of chlorothymol using sarA and agr deletion mutants, respectively. The MIC of chlorothymol was 32 μg/ml against both the parental control strain JE2 and its sarA deletion mutant (Figs. 3A and 3B). This compound completely inhibited the biofilm formation of both MRSA JE2 and the sarA mutant at 32 μg/ml (Figs. 3C and 3D). Consistently, the MIC of chlorothymol was 32 μg/ml against both the parental control strain LAC and its agr deletion mutant; its biofilm inhibition concentration was approximately 16 μg/ml (data not shown). These results indicate that unlike thymol, chlorothymol inhibits MRSA growth and biofilm formation in either sarA- or agr-independent manners.
Fig. 3. Effect of chlorothymol on ΔsarA mutant and semiquantitative RT-PCR of different virulence factorrelated genes.
(A, B) Statistical analysis was performed by applying 240 ANOVA with the level of significance at 5%. (C, D) Images below show post crystal violet staining in 96-well plates cultivated at 37°C for 24 h. (E) Semi-quantitative PCR of mecA, sarA, agrA, crtM, icaD, and ebps genes. gyrB is used as an endogenous control.
Next, we evaluated the expression levels of virulence factor genes, such as sarA, agrA, the antibiotic resistance gene mecA [46], the diapophytoene synthase gene crtM, the intercellular adhesion gene icaD, and elastin-binding protein gene ebpS [47, 48] after treating S. aureus with chlorothymol at 8 and 16 μg/ml. There was no significant difference in the expression levels of these genes (Fig. 3E), indicating that unlike thymol and carvacrol, chlorothymol does not target these virulence-related genes.
Synergetic Effect of Chlorothymol with Oxacillin
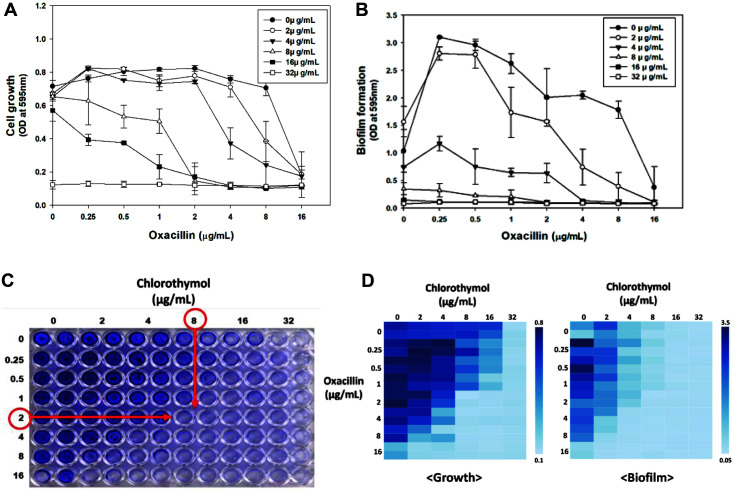
As the antibacterial effect of thymol derivatives was confirmed, the synergetic effect of thymol derivatives with other antibiotics was evaluated. Cell growth inhibition was observed by adding 8 μg/ml oxacillin and various concentrations of thymol derivatives to the LAC strain. Carvacrol showed a MIC of 64 μg/ml (Fig. S3A). We found a synergetic effect compared to 256 μg/ml when carvacrol was used alone. In the case of thymol iodide, when used alone, even at a concentration of 512 μg/ml, there was no significant inhibitory effect on LAC, and the combination with oxacillin showed no synergistic effects (Fig. S3B). In the case of 4-isopropyl-3-methylphenol, the MIC was reduced to 128 μg/ml when combined with oxacillin, compared to the MIC of 512 μg/ml alone (Fig. S3C). In the case of chlorothymol, the inhibitory effect was strong enough to show MIC, even at a concentration of 8 μg/ml (Fig. S3D). We found a significant synergetic effect with oxacillin compared to 32 μg/ml when chlorothymol was used alone.
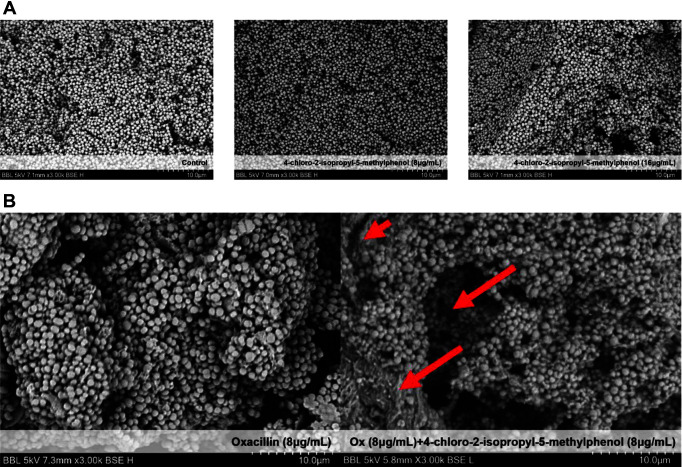
Therefore, we conducted a checkerboard assay to determine the optimal concentration for the synergetic effects of oxacillin and chlorothymol. Using the checkerboard assay, we calculated the FIC index values. The lowest FIC index value of cell growth was approximately 0.3125 (each concentration was oxacillin 2 μg/ml, chlorothymol 8 μg/ml), which was less than 0.5. This confirmed a synergetic effect between oxacillin and chlorothymol (Figs. 4A and 4D). When 2 μg/ml oxacillin and 8 μg/ml chlorothymol were used together, we found that inhibition of growth and biofilm was greatly increased. (Figs. 4B-4D). We found that a significant MIC reduction effect could be obtained with the minimal use of antibiotics. To check how the surface of the cell changes with chlorothymol, the cell size and density were observed using a SEM. When chlorothymol was added at concentrations of 8 and 16 μg/ml, there was a decrease in cell density and growth (Fig. 5A). However, there was no significant change in cell size up to 16 μg/ml chlorothymol.
Fig. 4. Synergetic effects between oxacillin and chlorothymol.
(A, B) Statistical analysis was performed by applying 240 ANOVA with the level of significance at 5%. (C) Images below show post crystal violet staining in 96-well plates cultivated at 37°C for 24 h. (D) The results of the checkerboard assay for confirming the synergetic effect of oxacillin and chlorothymol were presented as heatmaps. Each legend represents the absorbance value at OD595.
Fig. 5. Effects of cell size, density, and synergetic effect with oxacillin when chlorothymol was added, as evaluated using SEM.
(A) Control and LAC with 8 μg/ml and 16 μg/ml of chlorothymol were incubated in a shaking incubator at 37°C for 24 h. (B) Control and LAC, to which 8 μg/ml of oxacillin and 8 μg/ml of chlorothymol were added, were incubated in a shaking incubator at 37°C for 24 h.
The synergistic effects of oxacillin and chlorothymol were confirmed by SEM. When only oxacillin was used, the cell size was mostly uniformly distributed, but when treated with 8 μg/ml chlorothymol, not only the density of cells but also the size of many cells decreased (Fig. 5B). As a result, we found through SEM that chlorothymol had its own antibacterial effect as well as a synergetic effect with oxacillin.
Effect of Chlorothymol on Clinical Strains-Synergetic Effect with Oxacillin
The antimicrobial activity of chlorothymol on several clinical strains isolated from patients was also evaluated. Most clinical strains showed strong resistance even at high concentrations of oxacillin (Table 1). Ethanol alone was used on control cells and did not significantly affect cell growth or biofilm formation in the clinical strains, indicating there was no impact of ethanol use on the chlorothymol results (Figs. S2A and S2B). When chlorothymol was added, cell growth was significantly inhibited in most clinical strains at 32–64 μg/ml (Table 2); only MRSA 12779 was resistant. A biofilm inhibition test was also performed on clinical strains. When chlorothymol was added, biofilm formation by all clinical strains, including MRSA 12779, was inhibited to below 128 μg/ml (Table 3).
Table 1.
Clinically isolated strain characteristics and oxacillin MIC.
| Name | Type | SCCmec Type | Oxacillin MIC (μg/ml) | Spa Type | MLST (ST) |
|---|---|---|---|---|---|
| 2065 | MRSA | III | 1024 | t037 | 239 |
| 6230 | MRSA | IV | 128 | t324 | 72 |
| 6288 | MRSA | III | 1024 | t037 | 239 |
| 7557 | MRSA | II | 1024 | t9353 | 5 |
| 7875 | MRSA | IV | 128 | t664 | 72 |
| 8471 | MRSA | II | 1024 | t9353 | 5 |
| 9291 | MRSA | II | 1024 | t601 | 5 |
| 12779 | MRSA | II | 1024 | t2460 | 5 |
| 14278 | MRSA | II | 1024 | t9353 | 5 |
| 14459 | MRSA | IV | 1024 | t324 | 72 |
| 28984 | MSSA | - | 1 | - | 30 |
| 28985 | MRSA | IV | 64 | - | 30 |
Table 2.
MIC of chlorothymol and 16 μg/ml of chlorothymol and oxacillin for clinically isolated strains.
| Name | Type Ch | lorothymol MIC (μg/ml) | Oxacillin MIC (μg/ml) | 16 μg/ml Chlorothymol with Oxacillin MIC (μg/ml) |
|---|---|---|---|---|
| 2065 | MRSA | 64 | 1024 | 64 |
| 6230 | MRSA | 32 | 128 | 16 |
| 6288 | MRSA | 64 | 1024 | 32 |
| 7557 | MRSA | 64 | 1024 | 512 |
| 7875 | MRSA | 64 | 128 | 0.5 |
| 8471 | MRSA | 64 | 1024 | 512 |
| 9291 | MRSA | 64 | 1024 | 512 |
| 12779 | MRSA | - | 1024 | 256 |
| 14278 | MRSA | 64 | 1024 | 512 |
| 14459 | MRSA | 64 | 1024 | 0.5 |
| 28984 | MSSA | 64 | 1 | 0.5 |
| 28985 | MRSA | 64 | 64 | 0.5 |
Table 3.
BIC of chlorothymol and 16 μg/ml of chlorothymol and oxacillin for clinically isolated strains.
| Name | Type Ch | lorothymol BIC (μg/ml) | Oxacillin BIC (μg/ml) | 16 μg/ml Chlorothymol with Oxacillin BIC (μg/ml) |
|---|---|---|---|---|
| 2065 | MRSA | 128 | 1024 | 0.5 |
| 6230 | MRSA | 64 | 128 | 16 |
| 6288 | MRSA | 64 | 1024 | 8 |
| 7557 | MRSA | 16 | 1024 | 0.5 |
| 7875 | MRSA | 64 | 512 | 512 |
| 8471 | MRSA | 64 | 1024 | 256 |
| 9291 | MRSA | 32 | 1024 | 128 |
| 12779 | MRSA | 32 | 512 | 64 |
| 14278 | MRSA | 32 | 1024 | 256 |
| 14459 | MRSA | 32 | 512 | 512 |
| 28984 | MSSA | 32 | 0.5 | 0.5 |
| 28985 | MRSA | 16 | 512 | 128 |
The biofilm inhibition concentration (BIC) was set to an OD595 value less than 0.3. Statistical analysis was performed by applying 240 ANOVA with a level of significance of 5%.
Additionally, we evaluated the synergetic effect of chlorothymol and oxacillin in clinical strains. In previous studies, 16 μg/ml chlorothymol showed little cell growth inhibition in clinical strains. However, by combining the same concentration of chlorothymol and various concentrations of oxacillin, we succeeded in inhibiting the growth of all strains (Table 2). In addition, although MRSA strain 12779 was resistant to chlorothymol, we were able to inhibit its growth by the combination of 16 μg/ml chlorothymol with 256 μg/ml oxacillin (Table 2). Consistently, chlorothymol acted synergistically with oxacillin against the biofilm formation of these clinical MRSA strains (Table 3).
Chlorothymol Disrupts Membrane Integrity of MRSA
Membrane-active antimicrobials often exhibit bactericidal activity against MRSA biofilms and synergism with other antibiotics [37, 49]. Thus, we hypothesized that chlorothymol targets the bacterial membrane. To test this, we treated MRSA MW2 cells with a range of concentrations of chlorothymol and measured the cellular uptake of the membrane-impermeable DNA-binding fluorescent dye SYTOX Green. As shown Fig. 6, chlorothymol induced rapid membrane permeabilization of MRSA MW2 cells. This result indicates that the antimicrobial activity of chlorothymol results from disruption of membrane integrity.
Fig. 6. Chlorothymol induces rapid membrane permeabilization of S. aureus MW2.
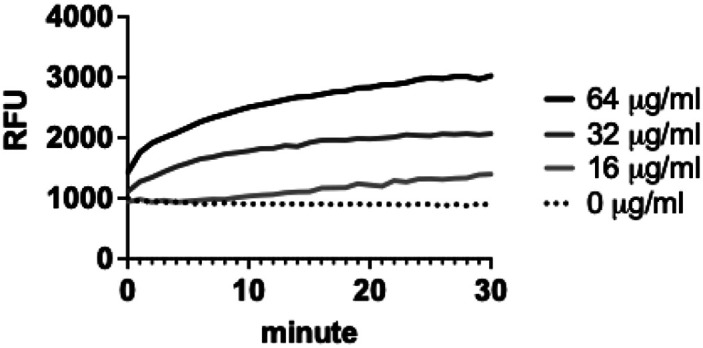
After treating MRSA MW2 cells with the indicated concentrations of chlorothymol, the uptake of SYTOX Green was measured by detecting fluorescence intensity (Ex 485 nm, Em 525 nm). The graph represents three independent experiments. Error bars are not shown for clarity.
Discussion
In this study, we assessed the antibacterial effects of four thymol derivatives against MRSA and identified that chlorothymol had the most potent effects. We found that chlorothymol not only inhibited MRSA growth, it also prevented MRSA biofilm formation, killed MRSA biofilm cells, decreased MRSA motility and the production of staphyloxanthin, and functioned synergistically with oxacillin. Additionally, chlorothymol alone or in combination with oxacillin was effective in most clinical strains.
Thymol inhibited the expression of sarA in MRSA and inhibited the expression of other sarA-regulated genes [27]. As a result, the expression of ica genes was inhibited and biofilm formation was suppressed [27]. However, our semiquantitative RT-PCR results revealed that chlorothymol did not affect the expression of icaD or sarA. Further unlike thymol, chlorothymol did not change the expression levels of virulence genes. Therefore, we concluded that the anti-biofilm potency of chlorothymol contributes to its antimicrobial activity.
The antimicrobial synergism between chlorothymol and oxacillin most likely results from the chlorothymol-induced membrane disruption and subsequent promotion of cellular uptake of oxacillin. Indeed, the disruption of the bacterial cytoplasmic membrane is known to increase the permeability of oxacillin[49]. Chlorothymol with hydrophobic properties has a strong affinity with the membrane lipid bilayer, which may facilitate its embedment into the lipid bilayers and eventually cause the reduction of permeability barrier function [50]. Thus, it is highly possible that the concentration of oxacillin increases in chlorothymol-treated MRSA cells.
This study determined the thymol derivative chlorothymol to have the best antibacterial effect among the four thymol derivatives. Chlorothymol significantly inhibited the growth and virulence factors of MRSA. In addition, it showed a synergistic effect with oxacillin and inhibited the growth of all clinical strains. Considering the antimicrobial and adjuvant properties of chlorothymol, chlorothymol and other thymol derivatives have potential to be developed as new therapeutics against deadly MRSA infections.
Supplemental Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
This research was supported by Research Program to solve social issues of the National Research Foundation of Korea (NRF)s funded by the Ministry of Science and ICT, South Korea [grant number 2017M3A9E4077234] and [NRF- 2022R1A2C2003138, NRF-2019M3E6A1103979]. W.K. was supported by the National Research Foundation of Korea (NRF) Grant (2020R1C1C1008842, 2018R1A5A2025286).
Footnotes
Conflicts of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Alanis AJ. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 2005;36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Imperial ICVJ, Ibana JA. Addressing the antibiotic resistance problem with probiotics: reducing the risk of its double-edged sword effect. Front. Microbiol. 2016;7:1983. doi: 10.3389/fmicb.2016.01983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyake Y, Iwai1 T, Sugai M, Miura1 K, Suginaka H, Nagasaka1 N. Incidence and characterization of Staphylococcus aureus from the tongues of children. J. Dent. Res. 1991;70:1045–1047. doi: 10.1177/00220345910700070501. [DOI] [PubMed] [Google Scholar]
- 4.Verhoef J, Beaujean D, Blok H, Baars A, Meyler A, van der Werken C, et al. A Dutch approach to methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 1999;18:461–466. doi: 10.1007/s100960050324. [DOI] [PubMed] [Google Scholar]
- 5.Peacock SJ, Paterson GK. Mechanisms of methicillin resistance in Staphylococcus aureus. Ann. Rev. Biochem. 2015;84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 6.Blot SI, Vandewoude KH, Hoste EA, Colardyn FA. Outcome and attributable mortality in critically Ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Arch. Intern. Med. 2002;162:2229–35. doi: 10.1001/archinte.162.19.2229. [DOI] [PubMed] [Google Scholar]
- 7.Pantosti A, Venditti M. What is MRSA? Eur. Resp. J. 2009;34:1190–1196. doi: 10.1183/09031936.00007709. [DOI] [PubMed] [Google Scholar]
- 8.Boswihi SS, Udo EE. Methicillin-resistant Staphylococcus aureus? an update on the epidemiology, treatment options and infection control. Curr. Med. Res. Pract. 2018;8:18–24. doi: 10.1016/j.cmrp.2018.01.001. [DOI] [Google Scholar]
- 9.Cuny C, Wieler LH, Witte W. Livestock-associated MRSA: the impact on humans. Antibiotics. 2015;4:521–543. doi: 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diep BA, Otto M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008;16:361–369. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto T, Nishiyama A, Takano T, Yabe S, Higuchi W, Razvina O, et al. Community-acquired methicillin-resistant Staphylococcus aureus: community transmission, pathogenesis, and drug resistance. J. Infect. Chemother. 2010;16:225–254. doi: 10.1007/s10156-010-0045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto M. MRSA virulence and spread. Cell. Microbiol. 2012;14:1513–1521. doi: 10.1111/j.1462-5822.2012.01832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilke MS, Lovering AL, Strynadka NCJ. β-Lactam antibiotic resistance: a current structural perspective. Curr. Opin. Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Cascioferro S, Carbone D, Parrino B, Pecoraro C, Giovannetti E, Cirrincione G, et al. Therapeutic strategies to counteract antibiotic resistance in MRSA biofilm-associated infections. ChemMedChem. 2021;16:65–80. doi: 10.1002/cmdc.202000677. [DOI] [PubMed] [Google Scholar]
- 15.Khatoon Z, McTiernan CD, Suuronen EJ, Mah T-F, Alarcon EI, Alarcon Bacterial EI. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon. 2018;4:e01067. doi: 10.1016/j.heliyon.2018.e01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt DC, Holden MTG, Tong SYC, Castillo-Ramirez S, Clarke L, Quail MA, et al. A very early-branching staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol. Evol. 2011;3:881–895. doi: 10.1093/gbe/evr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dharmaraja AT. Role of Reactive Oxygen Species (ROS) in therapeutics and drug resistance in cancer and bacteria. J. Med. Chem. 2017;60:3221–3240. doi: 10.1021/acs.jmedchem.6b01243. [DOI] [PubMed] [Google Scholar]
- 18.Xue L, Chen YY, Yan Z, Lu W, Wan D, Zhu H. Staphyloxanthin: a potential target for antivirulence therapy. Infect. Drug Res. 2019;12:2151–2160. doi: 10.2147/IDR.S193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollitt EJG, Crusz SA, Diggle SP. Staphylococcus aureus forms spreading dendrites that have characteristics of active motility. Sci. Rep. 2015;5:17698. doi: 10.1038/srep17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui H, Zhang C, Li C, Lin L. Inhibition mechanism of cardamom essential oil on methicillin-resistant Staphylococcus aureus biofilm. LWT. 2020;122:109057. doi: 10.1016/j.lwt.2020.109057. [DOI] [Google Scholar]
- 21.Cheung GYC, Joo HS, Chatterjee SS, Otto M. Phenol-soluble modulins - critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 2014;38:698–719. doi: 10.1111/1574-6976.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao S, Young G, Oberg C, Nakaoka K. Inhibition of methicillin-resistant Staphylococcus aureus (MRSA) by essential oils. Flavour Fragr. J. 2008;23:444–449. doi: 10.1002/ffj.1904. [DOI] [Google Scholar]
- 23.Marchese A, Orhan IE, Daglia M, Barbieri R, di Lorenzo A, Nabavi SF, et al. Antibacterial and antifungal activities of thymol: a brief review of the literature. Food Chem. 2016;210:402–414. doi: 10.1016/j.foodchem.2016.04.111. [DOI] [PubMed] [Google Scholar]
- 24.Kachur K, Suntres Z. The antibacterial properties of phenolic isomers, carvacrol and thymol. Crit. Rev. Food Sci. Nutr. 2020;60:3042–3053. doi: 10.1080/10408398.2019.1675585. [DOI] [PubMed] [Google Scholar]
- 25.Salehi B, Mishra AP, Shukla I, Sharifi-Rad M, Contreras M del M, Segura-Carretero A, et al. Thymol, thyme, and other plant sources: health and potential uses. Phytother. Res. 2018;32:1688–1706. doi: 10.1002/ptr.6109. [DOI] [PubMed] [Google Scholar]
- 26.Oussalah M, Caillet S, Saucier L, Lacroix M. Inhibitory effects of selected plant essential oils on the growth of four pathogenic bacteria: E. coli O157:H7, Salmonella Typhimurium, Staphylococcus aureus and Listeria monocytogenes. Food Control. 2007;18:414–420. doi: 10.1016/j.foodcont.2005.11.009. [DOI] [Google Scholar]
- 27.Valliammai A, Selvaraj A, Yuvashree U, Aravindraja C, Karutha Pandian S. sarA-dependent antibiofilm activity of thymol enhances the antibacterial efficacy of rifampicin against Staphylococcus aureus. Front. Microbiol. 2020;11:1744. doi: 10.3389/fmicb.2020.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HS, Song HS, Lee HJ, Kim SH, Suh MJ, Cho JY, et al. Comparative study of the difference in behavior of the accessory gene regulator (agr) in USA300 and USA400 community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) J. Microbiol. Biotechnol. 2021;31:1060–1068. doi: 10.4014/jmb.2104.04032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi TR, Song HS, Han YH, Park YL, Park JY, Yang SY, et al. Enhanced tolerance to inhibitors of Escherichia coli by heterologous expression of cyclopropane-fatty acid-acyl-phospholipid synthase (cfa) from Halomonas socia. Bioprocess Biosys. Eng. 2020;43:909–918. doi: 10.1007/s00449-020-02287-8. [DOI] [PubMed] [Google Scholar]
- 30.Peters BM, Ward RM, Rane HS, Lee SA, Noverr MC. Efficacy of ethanol against Candida albicans and Staphylococcus aureus polymicrobial biofilms. Antimicrob. Agents Chemother. 2013;57:74–82. doi: 10.1128/AAC.01599-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollitt EJG, Diggle SP. Defining motility in the Staphylococci. Cell. Mol. Life Sci. 2017;74:2943–2958. doi: 10.1007/s00018-017-2507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song HS, Bhatia SK, Choi TR, Gurav R, Kim HJ, Lee SM, et al. Increased antibiotic resistance of methicillin-resistant Staphylococcus aureus USA300 Δpsm mutants and a complementation study of Δpsm mutants using synthetic phenol-soluble modulins. J. Microbiol. Biotechnol. 2021;31:115–122. doi: 10.4014/jmb.2007.07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botelho MG. Fractional inhibitory concentration index of combinations of antibacterial agents against cariogenic organisms. J. Dent. 2020;28:565–570. doi: 10.1016/S0300-5712(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 34.Meletiadis J, Pournaras S, Roilides E, Walsh TJ. Defining fractional inhibitory concentration index cutoffs for additive interactions based on self-drug additive combinations, Monte Carlo simulation analysis, and in vitro-in vivo correlation data for antifungal drug combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2010;54:602–609. doi: 10.1128/AAC.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park YL, Bhatia SK, Gurav R, Choi TR, Kim HJ, Song HS, et al. Fructose based hyper production of poly-3-hydroxybutyrate from Halomonas sp. YLGW01 and impact of carbon sources on bacteria morphologies. Int. J. Biol. Macromol. 2020;154:929–936. doi: 10.1016/j.ijbiomac.2020.03.129. [DOI] [PubMed] [Google Scholar]
- 36.Kim W, Zou G, Hari TPA, Wilt IK, Zhu W, Galle N, et al. A selective membrane-targeting repurposed antibiotic with activity against persistent methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 2019;116:16529–16534. doi: 10.1073/pnas.1904700116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SM, Zou G, Kim H, Kang M, Ahn S, Heo HY, et al. Antimicrobial activity of the membrane-active compound nTZDpa is enhanced at low pH. Biomed. Pharmacother. 2022;150:112977. doi: 10.1016/j.biopha.2022.112977. [DOI] [PubMed] [Google Scholar]
- 38.Muthukrishnan G, Masters EA, Daiss JL, Schwarz EM. Mechanisms of immune evasion and bone tissue colonization that make Staphylococcus aureus the primary pathogen in osteomyelitis. Curr. Osteoporo. Rep. 2019;17:395–404. doi: 10.1007/s11914-019-00548-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsompanidou E, Denham EL, Becher D, de Jong A, Buist G, van Oosten M, et al. Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl. Environ. Microbiol. 2013;79:886–895. doi: 10.1128/AEM.03157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Y, Joo HS, Nair V, Le KY, Otto M. Do amyloid structures formed by Staphylococcus aureus phenol-soluble modulins have a biological function? Int. J. Med. Microbiol. 2018;308:675–682. doi: 10.1016/j.ijmm.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winterbourn CC, Kettle AJ, Hampton MB. Reactive oxygen species and neutrophil function. Ann. Rev. Biochem. 2016;85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 42.Valliammai A, Selvaraj A, Muthuramalingam P, Priya A, Ramesh M, Pandian SK. Staphyloxanthin inhibitory potential of thymol impairs antioxidant fitness, enhances neutrophil mediated killing and alters membrane fluidity of methicillin resistant Staphylococcus aureus. Biomed. Pharmacother. 2021;141:111933. doi: 10.1016/j.biopha.2021.111933. [DOI] [PubMed] [Google Scholar]
- 43.Selvaraj A, Valliammai A, Muthuramalingam P, Priya A, Suba M, Ramesh M, et al. Carvacrol targets SarA and CrtM of methicillin-resistant Staphylococcus aureus to mitigate biofilm formation and staphyloxanthin synthesis: an in vitro and in vivo approach. ACS Omega. 2020;5:31100–31114. doi: 10.1021/acsomega.0c04252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan PF, Foster SJ. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. Am. Soc. Microbiol. 1998;23:6232–6241. doi: 10.1128/JB.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Queck SY, Jameson-Lee M, Villaruz AE, Bach THL, Khan BA, Sturdevant DE, et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shang Wei Wu, de Lencastre H, Tomasz A. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 2001;183:2417–2424. doi: 10.1128/JB.183.8.2417-2424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue L, Chen YY, Yan Z, Lu W, Wan D, Zhu H. Staphyloxanthin: a potential target for antivirulence therapy. Infect. Drug Res. 2019;12:2151–2160. doi: 10.2147/IDR.S193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemati M, Hermans K, Devriese LA, Maes D, Haesebrouck F. Screening of genes encoding adhesion factors and biofilm formation in Staphylococcus aureus isolates from poultry. Avian Pathol. 2009;38:513–517. doi: 10.1080/03079450903349212. [DOI] [PubMed] [Google Scholar]
- 49.Phitaktim S, Chomnawang M, Sirichaiwetchakoon K, Dunkhunthod B, Hobbs G, Eumkeb G. Synergism and the mechanism of action of the combination of α-mangostin isolated from Garcinia mangostana L. and oxacillin against an oxacillin-resistant Staphylococcus saprophyticus. BMC Microbiol. 2016;16:195. doi: 10.1186/s12866-016-0814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kowalczyk A, Przychodna M, Sopata S, Bodalska A, Fecka I. Thymol and thyme essential oil-new insights into selected therapeutic applications. Molecules. 2020;25:4125. doi: 10.3390/molecules25184125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.