Abstract
Bacillus subtilis is a useful bacterium in the food industry with applications as a starter strain for fermented food and as a probiotic. However, it is difficult to discriminate B. subtilis from other Bacillus species because of high phenotypic and genetic similarity. In this study, we employed five previously constructed multilocus sequence typing (MLST) methods for the discrimination of B. subtilis from other Bacillus species and all five MLST assays clearly distinguished B. subtilis. Additionally, the 17 housekeeping genes used in the five MLST assays also clearly distinguished B. subtilis. The pyruvate carboxylase (pyrA) and shikimate dehydrogenase (aroE) genes were selected for the discrimination of B. subtilis because of their high number of polymorphic sites and the fact that they displayed the lowest homology among the 17 housekeeping genes. Specific primer sets for the pyrA and aroE genes were designed and PCR products were specifically amplified from B. subtilis, demonstrating the high specificity of the two housekeeping genes for B. subtilis. This species-specific PCR method provides a quick, simple, powerful, and reliable alternative to conventional methods in the detection and identification of B. subtilis.
Keywords: Bacillus subtilis, 16S rRNA gene, multilocus sequence typing, pyrA, aroE
Introduction
Bacillus subtilis is a spore-forming bacterium that can withstand a range of extreme environmental conditions [1]. B. subtilis has been detected in diverse habitats such as soil, air, and within plants [1]. Its spore-forming properties also permit entrance into the gastrointestinal tract of animals, where it can form vegetative cells from spores, thereby sporulating again [2, 3]. Thus, research into the application of B. subtilis in vaccine delivery into the gastrointestinal tract or as a probiotic has been conducted [4-6] .
B. subtilis has been detected in several types of fermented soybeans in East Asia, such as meju and doenjang in Korea, douchi in China, and natto and miso in Japan [7-11]. B. subtilis exhibits extracellular amylase and protease activities [12, 13]. These activities influence the production of amino acids and flavor compounds during soybean fermentation [14-16]. It is well known that these enzymatic activities contribute toward the quality and sensory properties of fermented soybeans [14, 17]. B. subtilis also produces several bacteriocins [18] and has therefore been used as a starter culture for soybean fermentation [19], as well as a commercial fungicide (Taegro; B. subtilis var. amyloliquefaciens strain FZB24; Novozymes, Denmark).
B. subtilis is generally regarded as a safe bacterium because of its long history of use in the food industry. It also produces several industrially-important enzymes such as xylanase, lichenase, cellulose, and pectinase. These enzymes, produced from non-genetically-modified B. subtilis, can be applied in the food industry [20]. Although much research into the commercial value of B. subtilis has been conducted, including applications in the food industry and in vaccine development, [5, 20], studies on methods to distinguish B. subtilis from other Bacillus species are lacking and most of them are identified methods after DNA purification such as restriction fragment length polymorphism or randomly amplified polymorphic DNA analysis [21-24]. For the commercial use of B. subtilis, it is necessary to ensure the specific isolation of this species from other related species. In this study, we developed a method to specifically distinguish B. subtilis and thereby ensure its purity as a resource.
Materials and Methods
Culture Conditions of Bacillus Species
Bacillus species were cultured in Tryptic Soy Agar (TSA; Difco, USA) and Tryptic Soy Broth (TSB; Difco) at 37°C for 18 h to ensure that the traits of this organism were maintained.
Biochemical Characterization of Bacillus Species
Bacillus species were characterized biochemically using a commercially available API 50 CHB/E system according to the manufacturer's instructions (BioMérieux, France). For the biochemical analysis, strains were incubated in TSB at 37°C for 18 h according to the manufacturer's instructions and adjusted to an optical density (OD600) of 0.6. The bacterial suspension was added to API 50 CHB/E medium 1% (w/v), inoculated onto a API 50CH strip, and then incubated under aerobic conditions at 37°C for 24 and 48 h. The phenol red indicator ensures that the strip turns yellow when acid is produced by fermentation using the carbohydrates added to the strip during incubation. Finally, the results were analyzed using the online software apiweb (https://apiweb.biomerieux.com) by submitting negative and positive responses according to the reference color reading table.
Comparative Genomics of Bacillus Species
For comparative genomic analysis of closely related Bacillus species, the genome sequence data of six B. subtilis, three Bacillus siamensis, five Bacillus velezensis, four Bacillus amyloliquefaciens, and three Bacillus atrophaeus strains were obtained from the NCBI database (http://ncbi.nlm.nih.gov/genomes) (Table 1). Phylogenetic analyses of the 16S rRNA gene, housekeeping genes, and multilocus sequence typing (MLST) sequences were performed using the maximum likelihood algorithm of the MEGA 7.0 software. The number of alleles and polymorphic sites, the discriminatory power (DP), and the typing efficiency (TE) of these housekeeping genes were analyzed using MLSTest software (http://www.ipe.unsa.edu.ar/software). TE is defined as the number of genotypes per polymorphic site for each housekeeping gene [25]. DP is the likelihood that two strains differentiate when randomly selected from a population of unrelated strains [25]. The number of non-synonymous (dN) and synonymous (dS) nucleotide substitutions per site was estimated using MEGA 7.0 software [26].
Table 1.
Bacillus strains for comparative genomic analysis and 16S rRNA homology.
| Species | Strain | Accession No. | 16S rRNA homology (%) | No. of polymorphic sites |
|---|---|---|---|---|
| B. subtilis | KCCM 32835T* | NZ_CP020102 | 100.0 | (Reference strain) |
| B. subtilis | PS832 | NZ_CP010053 | 100.0 | 0 |
| B. subtilis | HRBS-10TDI13 | NZ_CP015222 | 99.9 | 2 |
| B. subtilis | GFR-12 | NZ_CP032852 | 99.9 | 2 |
| B. subtilis | 2RL2-3 | NZ_CP032857 | 99.8 | 3 |
| B. subtilis | SRCM102748* | NZ_CP028212 | 99.8 | 3 |
| B. velezensis | KMU01* | NZ_CP063768 | 99.8 | 3 |
| B. velezensis | B268 | NZ_CP053764 | 99.7 | 4 |
| B. velezensis | S4 | NZ_CP050424 | 99.7 | 4 |
| B. velezensis | KKLW | NZ_CP054714 | 99.7 | 5 |
| B. velezensis | DMB06* | NZ_CP083763 | 99.5 | 7 |
| B. velezensis | KCTC 13012T* | - | - | - |
| B. siamensis | SCSIO 05746 | NZ_CP025001 | 99.7 | 5 |
| B. siamensis | SDLI1 | NZ_CP013950.1 | 99.7 | 5 |
| B. siamensis | B28* | NZ_CP066219 | 99.5 | 7 |
| B. siamensis | KCTC 13613T* | - | - | - |
| B. amyloliquefaciens | MT45 | NZ_CP011252 | 99.6 | 6 |
| B. amyloliquefaciens | RD7-7 | NZ_CP016913 | 99.6 | 6 |
| B. amyloliquefaciens | YP6 | NZ_CP032146 | 99.5 | 7 |
| B. amyloliquefaciens | KCCM 40764T* | NC_014551 | 99.5 | 8 |
| B. amyloliquefaciens | KCCM 12090* | - | - | - |
| B. atrophaeus | SRCM101359 | NZ_CP021500 | 99.2 | 11 |
| B. atrophaeus | GQJK17 | NZ_CP022653 | 99.2 | 12 |
| B. atrophaeus | BA59 | NZ_CP024051 | 99.1 | 13 |
*Used for API and/or PCR analysis.
Application of Species Particular Oligonucleotide Primer
To differentiate B. subtilis from other Bacillus species, two genes, aroE and pycA, were selected based on MLST. B. subtilis-specific primer sets were designed (Table 2). Genomic DNA of Bacillus species was extracted using a DNeasy tissue kit (Qiagen, Germany). Amplification of the aroE and pycA genes was performed using the primer sets aroE-F/-R and pycA-F/-R, respectively. The PCR conditions were as follows: an initial denaturation step at 95°C for 5 min, followed by 30 cycles consisting of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec, then a post-extension step at 72°C for 5 min, and finally holding at 16°C in a T3000 Thermocycler (Biometra, Germany). Amplified PCR products were migrated on a 1.5% agarose gel.
Table 2.
Oligonucleotide primer sequences for the identification of B. subtilis.
| Primer | Sequence (5'→3') | Expected size (bp) |
|---|---|---|
| pycA-F | GTC TTC CGT TCA GGA AAG GC | 233 |
| pycA-R | GAT CTC CCG TTT GGA TCG GCT C | |
| aroE-F | GGG GAA GGC TTC GTG AAG TC | 278 |
| aroE-R | CCC ACA GAC GTT GTA TGG ATG |
Results and Discussion
Comparison of the 16S rRNA Sequence of B. subtilis with those of other Bacillus Species
The entire 16S rRNA gene sequence of B. subtilis KCCM 32835T showed >99.1% similarity with the corresponding sequences from B. amyloliquefaciens, B. siamensis, B. velezensis, and B. atrophaeus (Table 1). There were 0–3 polymorphic sites in this gene sequence among B. subtilis strains, 3–7 polymorphic sites among B. velezensis strains (Tables 1 and S1), and 11–13 polymorphic sites among B. atrophaeus strains showing 99.1%–99.2% similarity (Tables 1 and S1). High similarity and the low number of polymorphic sites within the 16S rRNA gene among Bacillus species have led to misidentification when classifying B. subtilis [27, 28]. For this reason, these five Bacillus species cannot be clearly distinguished based on the 16S rRNA gene alone.
Biochemical Characterization of B. subtilis and other Bacillus Species
To biochemically identify B. subtilis, the API 50 CHB/E system is recommended. However, using the API identification table, B. subtilis and B. amyloliquefaciens presented together and could not be distinguished, and the other three species analyzed (i.e., B. siamensis, B. velezensis, and B. atrophaeus) were not presented. This may be a result of insufficient API data on these species or difficulties with classifying these particular species into API 50 CHB system.
In this experiments, none of the species used erythritol, D-arabinose, L-xylose, D-adonitol, methyl-BD-xylopyranoside, D-galactose, L-sorbose, rhamnose, dulcitol, α-methyl-D-mannoside, melezitose, xylitol, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, gluconate, 2-keto-gluconate, or 5-keto-gluconate, but all species used ribose, D-glucose, D-fructose, mannitol, sorbitol, α-methyl-D-glucoside, amygdalin, esculin, salicin, cellobiose, maltose, sucrose, raffinose, and starch. Overall, B. subtilis showed high substrate usability, while B. amyloliquefaciens showed low substrate usability. However, despite slight differences between strains, there were no clear differences between species (Table 3). These results suggested that the API 50 CHB/E biochemical assay is unable to accurately discriminate B. subtilis from other Bacillus species.
Table 3.
Phenotypic characteristics of Bacillus species as analyzed by the API 50 CHB/E system.
| Substrate | B. subtilis | B. siamensis | B. velezensis | B. amyloliquefaciens | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| KCCM 32835T | SRCM 102748 | KCTC 13613T | B28 | KMU01 | KCTC 13012T | DMB06 | KCCM 40764T | KCCM 12090 | |
| GLYcerol | + | + | + | + | + | + | - | w | + |
| L ARAbinose | + | + | + | + | + | + | + | - | - |
| D XYLose D-XYLose | + | w | + | + | + | + | + | - | - |
| D-MaNnosE | + | + | - | - | + | + | + | + | + |
| INOsitol | + | + | + | + | + | + | + | - | - |
| N-Acethyl-Glucosamine | w | - | + | - | + | + | - | + | + |
| ARButin | + | + | + | + | + | + | - | + | + |
| LACtose | w | w | w | + | + | + | + | + | - |
| MELibiose | + | + | - | - | - | - | + | - | - |
| TREhalose | + | + | - | - | + | + | + | - | - |
| INUlin | + | + | - | - | - | - | w | - | - |
| GLYcogen | + | + | + | + | + | + | + | - | - |
| GENtiobiose | w | w | - | - | - | + | - | w | w |
Abbreviations: +: positive reaction; −: negative reaction; w: weak reaction (slight change).
Comparison of MLST Schemes for B. subtilis
MLST is a useful approach for distinguishing bacterial species based on nucleotide sequences [29] and a public MLST scheme (pubMLST) for B. subtilis was developed using seven housekeeping genes [30] (Table 4). In addition, three further MLST schemes (S1–S3) for B. subtilis and one MLST scheme (L1) for B. licheniformis have been developed [30-34]. In the S1 scheme, the housekeeping gene is the same as that in the pubMLST, but the concatenated order is different [31]. The S2 scheme uses nine housekeeping genes, two more than in the pubMLST [32]. In all five MLST schemes, seven to nine housekeeping genes are used and all were able to distinguish B. subtilis from other Bacillus species on phylogenetic trees (Fig. 1). Indeed, the five MLST schemes showed >80.00% similarity between B. subtilis and other closely related Bacillus species. These results confirmed that MLST can more accurately distinguish between Bacillus species than the 16S rRNA gene sequences (Fig. 1 and Table S2).
Table 4.
Five MLST methods for the analysis of Bacillus species.
| Method | Concatenated order of genes for MLST | Target species | Reference |
|---|---|---|---|
| pubMLST | glpF, ilvD, pta, purH, pycA, rpoD, tpiA | B. subtilis | [30] |
| S1 | rpoD, glpF, ilvD, ptA, tpiA, pycA, purH | B. subtilis | [31] |
| S2 | gyrA, gyrB, purH, glpF, pycA, ilvD, rpoD, tpiA, pta | B. subtilis | [32] |
| S3 | gyrB, adk, pycA, pyrE, sucC, mutL, aroE | B. subtilis | [33] |
| L1 | adk, ccpA, glpF, gmk, ilvD, pur, spo0A, tpi | B. paralicheniformis, B. licheniformis | [34] |
Fig. 1. Phylogenetic analysis using five multilocus sequencing typing schemes.
Data were compared using simple matching coefficients and were clustered by the maximum likelihood method. Branches with bootstrap values of 50% have been collapsed. The scale represents the pairwise distances expressed as the percentage of dissimilarity.
Although the five MLST schemes were more discriminatory in terms of identifying B. subtilis from closely related Bacillus species, the analysis of seven or nine housekeeping genes is labor-intensive. Therefore, the contribution of each housekeeping gene in identifying B. subtilis from closely related Bacillus species was analyzed. The phylogenetic trees generated for each housekeeping gene were all able to clearly distinguish B. subtilis from other Bacillus species (Fig. S1).
The allelic variation was analyzed for each gene sequence and the number of polymorphic sites within each gene ranged from 78 (adk) to 1075 (pycA), and the number of allelic genes ranged from 11 (adk) to 19 (pycA) (Table 5). Although the number of polymorphic sites varied, the dN/dS ratio for each housekeeping gene showed no significant difference. The average dN/dS ratio across all MLST genes was 0.4044, and it was thereby assumed that these genes were not under positive selective pressure (i.e., selection is against amino acid changes). For the pycA gene, the diversity in the amino acid sequence was lower compared with highly polymorphic sites. These findings were also evident in the TE (Table 5). In the five related Bacillus species analyzed, the TE of the 17 housekeeping genes ranged from 0.018 (pycA) to 0.141 (adk) (Table 5), whereas the DP did not differ significantly among these housekeeping genes, remaining at >0.935. These results suggested that the 17 housekeeping genes may be powerful markers for the discrimination of B. subtilis from other Bacillus species.
Table 5.
Characteristics of housekeeping genes in 21 B. subtilis strains.
| Housekeeping gene | Length (bp) | No. of alleles | No. of polymorphic sites | dN/dS | Typing efficiency (TE) | Discriminatory power (DP) |
|---|---|---|---|---|---|---|
| adk | 654 | 11 | 78 | 0.4057 | 0.141 | 0.935 |
| aroE | 843 | 18 | 322 | 0.3909 | 0.056 | 0.983 |
| ccpA | 1005 | 16 | 269 | 0.4046 | 0.059 | 0.974 |
| glpF | 828 | 17 | 247 | 0.4122 | 0.069 | 0.978 |
| gmk | 615 | 15 | 136 | 0.4143 | 0.110 | 0.965 |
| gyrA | 2466 | 17 | 718 | 0.4098 | 0.024 | 0.978 |
| gyrB | 1917 | 17 | 532 | 0.4116 | 0.032 | 0.978 |
| ilvD | 1677 | 18 | 458 | 0.4057 | 0.039 | 0.983 |
| mutL | 1892 | 16 | 648 | 0.3972 | 0.025 | 0.970 |
| pta | 972 | 15 | 232 | 0.3973 | 0.065 | 0.965 |
| purH | 1539 | 17 | 428 | 0.4011 | 0.040 | 0.978 |
| pycA | 3450 | 19 | 1075 | 0.3990 | 0.018 | 0.991 |
| pyrE | 651 | 14 | 228 | 0.4015 | 0.061 | 0.961 |
| rpoD | 1122 | 16 | 218 | 0.4300 | 0.073 | 0.974 |
| spo0A | 804 | 16 | 187 | 0.4068 | 0.086 | 0.970 |
| sucC | 1158 | 15 | 219 | 0.4048 | 0.068 | 0.965 |
| tpiA | 762 | 16 | 114 | 0.3823 | 0.140 | 0.952 |
Specific Oligonucleotide Primers for the Detection of B. subtilis by PCR
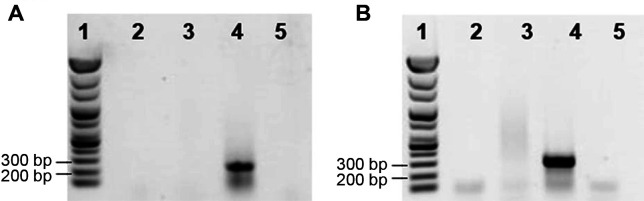
From the above results, it was confirmed that MLST and each of the housekeeping genes could distinguish B. subtilis from other Bacillus species. However, this method can only be applied after analyzing the nucleotide sequence of B. subtilis for isolation. Therefore, to more easily distinguish B. subtilis, a primer capable of identifying this species specifically was designed and its integrity was confirmed by PCR. Among the 17 housekeeping genes, pycA had the most alleles with 1075 polymorphic sites. The pycA nucleotide identity among B. subtilis strains was 98.9%–100%, compared with 79.7%–82.1% among other Bacillus species (Table S3). Therefore, we proposed that pycA was an appropriate gene to distinguish B. subtilis from other Bacillus species. Nucleotide sequences that could be distinguished were detected through comparative analysis, and a primer was designed to this sequence. PCR analysis confirmed amplification of B. subtilis DNA but not the DNA of other Bacillus species (Fig. 2).
Fig. 2. Bacillus subtilis species-specific PCR fragments of the pycA and aroE genes.
A pycA gene, B aroE gene. Lane 1: 100 bp ladder; Lane 2: Bacillus amyloliquefaciens KCCM 40764T; Lane 3: Bacillus siamensis KCTC 13613T; Lane 4: Bacillus subtilis KCCM 32835T; Lane 5: Bacillus velezensis KCTC 13012T.
The aroE gene sequence showed the lowest homology across strains among the 17 housekeeping genes. In B. subtilis, the aroE gene showed 98.8%–99.9% similarity among strains (Table S3). By contrast, other Bacillus species showed 67.0%–74.6% similarity in the aroE full sequence (Table S3). Hence, primers were designed against a partial sequence of the aroE gene, and it was confirmed that only B. subtilis DNA was amplified by PCR. In the above experiment, only two housekeeping genes, pycA and aroE, among 17 genes were applied to discriminate of B. subtilis. However as shown in table 5, we assumed that other 15 genes might also be possessed the potential for discrimination.
To assess the range of specificity of the PCR assay, the primer sets for the pycA and aroE genes were used in PCR analysis of 32 Bacillus strains, including eight B. subtilis strains. Amplicons for the pycA and aroE genes were only detected with B. subtilis strains (Fig. S2), and this assay may therefore have important implications for the accurate discrimination of B. subtilis from fermented food-derived Bacillus species.
As a result of the limitations of conventional approaches to B. subtilis identification, which include 16S rRNA gene sequence analysis and biochemical analysis, an auxiliary method was needed. MLST, and the housekeeping genes analyzed using this method, can clearly distinguish B. subtilis from other Bacillus species. In the current study, we showed that the pycA and aroE genes can be effectively used to screen for B. subtilis and clearly discriminate this species from other Bacillus species. These results confirmed that PCR amplification using our B. subtilis-specific primer set offers a quick, simple, powerful, and reliable method for accurately identifying B. subtilis from other Bacillus species.
Supplemental Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) [NRF-2019R1A2C1003639]. We thank Edanz (https://www.edanz.com/ac) for editing a draft of this manuscript.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Earl AM, Losick R, Kolter R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008;16:269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batista MT, Souza RD, Paccez JD, Luiz WB, Ferreira EL, Cavalcante RC, et al. Gut adhesive Bacillus subtilis spores as a platform for mucosal delivery of antigens. Infect. Immun. 2014;82:1414–1423. doi: 10.1128/IAI.01255-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leser TD, Knarreborg A, Worm J. Germination and outgrowth of Bacillus subtilis and Bacillus licheniformis spores in the gastrointestinal tract of pigs. J. Appl. Microbiol. 2008;104:1025–1033. doi: 10.1111/j.1365-2672.2007.03633.x. [DOI] [PubMed] [Google Scholar]
- 4.Hong HA, Duc LH, Cutting SM. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 2005;29:813–835. doi: 10.1016/j.femsre.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Ullah J, Jia J. Progress in Bacillus subtilis spore surface display technology towards environment, vaccine development, and biocatalysis. J. Mol. Microbiol. Biotechnol. 2017;27:159–167. doi: 10.1159/000475177. [DOI] [PubMed] [Google Scholar]
- 6.Lee NK, Kim WS, Paik HD. Bacillus strains as human probiotics: characterization, safety, microbiome, and probiotic carrier. Food Sci. Biotechnol. 2019;28:1297–1305. doi: 10.1007/s10068-019-00691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JH, Kim TW, Lee H, Chang HC, Kim HY. Determination of microbial diversity in meju, fermented cooked soya beans, using nested PCR-denaturing gradient gel electrophoresis. Lett. Appl. Microbiol. 2010;51:388–394. doi: 10.1111/j.1472-765X.2010.02906.x. [DOI] [PubMed] [Google Scholar]
- 8.Kim TW, Lee JH, Kim SE, Park MH, Chang HC, Kim HY. Analysis of microbial communities in doenjang, a Korean fermented soybean paste, using nested PCR-denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 2009;131:265–271. doi: 10.1016/j.ijfoodmicro.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen T, Wang M, Li S, Wu Q, Wei H. Molecular identification of microbial community in surface and undersurface douchi during postfermentation. J. Food Sci. 2014;79:M653–M658. doi: 10.1111/1750-3841.12417. [DOI] [PubMed] [Google Scholar]
- 10.Homma K, Wakana N, Suzuki Y, Nukui M, Daimatsu T, Tanaka E, et al. Treatment of natto, a fermented soybean preparation, to prevent excessive plasma vitamin K concentrations in patients taking warfarin. J. Nutr. Sci. Vitaminol. 2006;52:297–301. doi: 10.3177/jnsv.52.297. [DOI] [PubMed] [Google Scholar]
- 11.Onda T, Yanagida F, Tsuji M, Shinohara T, Yokotsuka K. Time series analysis of aerobic bacterial flora during miso fermentation. Lett. Appl. Microbiol. 2003;37:162–168. doi: 10.1046/j.1472-765X.2003.01371.x. [DOI] [PubMed] [Google Scholar]
- 12.Dash BK, Rahman MM, Sarker PK. Molecular identification of a newly isolated Bacillus subtilis BI19 and optimization of production conditions for enhanced production of extracellular amylase. Biomed Res. Int. 2015;2015:859805. doi: 10.1155/2015/859805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbieri G, Albertini AM, Ferrari E, Sonenshein AL, Belitsky BR. Interplay of CodY and ScoC in the regulation of major extracellular protease genes of Bacillus subtilis. J. Bacteriol. 2016;198:907–920. doi: 10.1128/JB.00894-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi SH, Hong SP. Characteristics of bacterial strains with desirable flavor compounds from Korean traditional fermented soybean paste (Doenjang) Molecules. 2021;26:5067. doi: 10.3390/molecules26165067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen T, Nguyen CH. Determination of factors affecting the protease content generated in fermented soybean by Bacillus subtilis 1423. Energy Rep. 2020;6:831–836. doi: 10.1016/j.egyr.2019.11.011. [DOI] [Google Scholar]
- 16.Shukla S, Lee JS, Park HK, Yoo JA, Hong SY, Kim JK, et al. Effect of novel starter culture on reduction of biogenic amines, quality improvement, and sensory properties of doenjang, a traditional Korean soybean fermented sauce variety. J. Food Sci. 2015;80:M1794–M1803. doi: 10.1111/1750-3841.12942. [DOI] [PubMed] [Google Scholar]
- 17.Dajanta K, Chukeatirote E, Apichartsrangkoon A. Improvement of thua nao production using protein-rich soybean and Bacillus subtilis TN51 starter culture. Ann. Microbiol. 2012;62:785–795. doi: 10.1007/s13213-011-0319-1. [DOI] [Google Scholar]
- 18.Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 2019;10:302. doi: 10.3389/fmicb.2019.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubo Y, Rooney AP, Tsukakoshi Y, Nakagawa R, Hasegawa H, Kimura K. Phylogenetic analysis of Bacillus subtilis strains applicable to natto (fermented soybean) production. Appl. Environ. Microbiol. 2011;77:6463–6469. doi: 10.1128/AEM.00448-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y, Liu C, Fang H, Zhang D. Bacillus subtilis: a universal cell factory for industry, agriculture, biomaterials and medicine. Microb. Cell Fact. 2020;19:173. doi: 10.1186/s12934-020-01436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cihan AC, Tekin N, Ozcan B, Cokmus C. The genetic diversity of genus Bacillus and the related genera revealed by 16S rRNA gene sequences and ardra analyses isolated from geothermal regions of turkey. Braz. J. Microbiol. 2012;43:309–324. doi: 10.1590/S1517-83822012000100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di J, Chu Z, Zhang S, Huang J, Du H, Wei Q. Evaluation of the potential probiotic Bacillus subtilis isolated from two ancient sturgeons on growth performance, serum immunity and disease resistance of Acipenser dabryanus. Fish Shellfish Immunol. 2019;93:711–719. doi: 10.1016/j.fsi.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez MJ, Gorgoroso F, Reginensi SM, Olivera JA, Bermudez J. Polyphasic identification of closely related Bacillus subtilis and Bacillus amyloliquefaciens isolated from dairy farms and milk powder. J. Microbiol. Biotechnol. Food Sci. 2013;2:2326–2331. [Google Scholar]
- 24.Xie S, Yu H, Wang Q, Cheng Y, Ding T. Two rapid and sensitive methods based on TaqMan qPCR and droplet digital PCR assay for quantitative detection of Bacillus subtilis in rhizosphere. J. Appl. Microbiol. 2020;128:518–527. doi: 10.1111/jam.14481. [DOI] [PubMed] [Google Scholar]
- 25.Roman F, Iniguez AM, Yeo M, Jansen AM. Multilocus sequence typing: genetic diversity in Trypanosoma cruzi I (TcI) isolates from Brazilian didelphids. Parasit. Vectors. 2018;11:107. doi: 10.1186/s13071-018-2696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal A, Gupta S, Yadav AK, Nema RK, Ansari K, Biswas D. Molecular and phylogenetic analysis of Chikungunya virus in central India during 2016 and 2017 outbreaks reveal high similarity with recent New Delhi and Bangladesh strains. Infect. Genet. Evol. 2019;75:103940. doi: 10.1016/j.meegid.2019.103940. [DOI] [PubMed] [Google Scholar]
- 27.Ashe S, Maji UJ, Sen R, Mohanty S, Maiti NK. Specific oligonucleotide primers for detection of endoglucanase positive Bacillus subtilis by PCR. 3 Biotech. 2014;4:461–465. doi: 10.1007/s13205-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang CH, Chang MT, Huang L, Chu WS. Development of a novel PCR assay based on the gyrase B gene for species identification of Bacillus licheniformis. Mol. Cell. Probes. 2012;26:215–217. doi: 10.1016/j.mcp.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Boers SA, van der Reijden WA, Jansen R. High-throughput multilocus sequence typing: bringing molecular typing to the next level. PLoS One. 2012;7:e39630. doi: 10.1371/journal.pone.0039630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST. org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le XT, Pham DT, Pham TA, Tran TT, Khuat TH, Le HQ, et al. Exploration of genetic diversity of Bacillus spp. from industrial shrimp ponds in Vietnam by multi-locus sequence typing. Fish Aquatic Sci. 2019;22:17. doi: 10.1186/s41240-019-0132-5. [DOI] [Google Scholar]
- 32.Boka B, Manczinger L, Kocsube S, Shine K, Alharbi NS, Khaled JM, et al. Genome analysis of a Bacillus subtilis strain reveals genetic mutations determining biocontrol properties. World J. Microbiol. Biotechnol. 2019;35:52. doi: 10.1007/s11274-019-2625-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabore D, Gagnon M, Roy D, Sawadogo-Lingani H, Diawara B, LaPointe G. Rapid screening of starter cultures for maari based on antifungal properties. Microbiol. Res. 2018;207:66–74. doi: 10.1016/j.micres.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Jeong DW, Lee B, Lee H, Jeong K, Jang M, Lee JH. Urease characteristics and phylogenetic status of Bacillus paralicheniformis. J. Microbiol. Biotechnol. 2018;28:1992–1998. doi: 10.4014/jmb.1809.09030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.