Abstract
The eradication of smallpox and the cessation of global vaccination led to the increased prevalence of human infections in Central Africa. Serologic and protein-based diagnostic assay for MPXV detection is difficult due to cross-reactive antibodies that do not differentiate between diverse orthopoxvirus (OPXV) species. A previously characterized monoclonal antibody (mAb 69-126-3-7) against MPXV [1] was retested for cross-reactivity with various OPXVs. The 14.5 kDa band protein that reacted with mAb 69-126-3 was identified to be MPXV A29 protein (homolog of vaccinia virus Copenhagen A27). Amino acid sequence analysis of the MPXV A29 with other OPXV homologs identified four amino acid changes. Peptides corresponding to these regions were designed and evaluated for binding to mAb 69-126-3 by ELISA and BioLayer Interferometry (BLI). Further refinement and truncations mapped the specificity of this antibody to a single amino acid difference in a 30-mer peptide compared to other OPXV homologs. This particular residue is proposed to be essential for heparin binding by VACV A27 protein. Despite this substitution, MPXV A29 bound to heparin with similar affinity to that of VACV A27 protein, suggesting flexibility of this motif for heparin binding. Although binding of mAb 69-126-3-7 to MPXV A29 prevented interaction with heparin, it did not have any effect on the infectivity of MPXV. Characterization of 69-126-3-7 mAb antibody allows for the possibility of the generation of a serological based species-specific detection of OPXVs despite high proteomic homology.
Keywords: Monkeypox virus, Vaccinia virus, Orthopoxvirus, Antibody, Heparin binding
Introduction
Orthopoxviruses are a genus of DNA viruses of which four species are known to cause disease in humans: vaccinia virus (VACV), cowpox virus (CPXV), variola virus (VARV), and monkeypox virus (MPXV). Monkeypox virus is a zoonotic virus, endemic to central and Western Africa that can cause smallpox-like symptoms in humans. The virus was initially identified in non-human primate rash lesions in 1958 and first identified in humans in 1970 during the smallpox eradication efforts. Incidence of MPXV infections in the Democratic Republic of the Congo (DRC), where the majority of cases occur, has increased as much as 20-fold since the end of smallpox vaccination in 1980 (Rimoin et al., 2010). The first case of human MPXV in the USA occurred in 2003 and was linked to the handling of prairie dogs that had been housed with imported African rodents (Reed et al., 2004). Similar to VARV or smallpox, MPXV disease presents with fever 10–12 days after exposure, followed by rash 2–3 days later. This rash progresses from macular to papular, vesicular, and finally pustular phases, similar to smallpox. This makes differentiation between these diseases based on clinical presentation difficult. While smallpox was declared eradicated by the World Health Organization in 1979, it is still a concern as a potential biological weapon or potential accidental release. The methods for laboratory confirmed diagnostics include electron microscopy, isolation of virus on chorioallantonic membranes (CAM), antibody-based assays, and viral DNA assays (Jezek and Fenner, 1988). Of these, only CAM and DNA assays are capable of providing a species-specific diagnosis (Li et al., 2006, Li et al., 2010, Olson et al., 2004, Shchelkunov et al., 2011), which require sensitive reagents and equipment. An antibody based serological assay is ideal for diagnostics and surveillance, but due to antigen conservation within OPXV genomes, most serum is cross-reactive and cannot differentiate between individual species of OPXVs. Recent work utilizing animal and human sera samples have identified the primary immune targets within the poxvirus proteome and demonstrated that the antibody response is redundant, cross-reactive, and cross-protective, which is perfect for vaccine design, but far from optimal for species-specific antibody assays (Benhnia et al., 2008, Davies et al., 2007, Jezek and Fenner, 1988, Keasey et al., 2010, Keckler et al., 2011, Townsend et al., 2013).
Previous work identified a MPXV-specific monoclonal antibody (Roumillat et al., 1984). This antibody, 69-126-3-7, was shown to be non-neutralizing and to bind to a 14.5 kD band under reducing conditions; however, the protein target was not identified. To explore this antibody׳s potential use as a diagnostic tool, it was imperative to identify its target protein and epitope. In this study, we have identified the protein target to be MPXV A29 protein (VACV A27 ortholog) and mapped its binding epitope.
Extensive work has been accomplished towards understanding the multiple functions and interactions of VACV A27. It has been reported that VACV A27 exists in complex with A17, A26, and A25, though A27 does not interact directly with A25 ( Fig. 1) (Howard et al., 2008, Rodriguez et al., 1993, Wang et al., 2014). It binds heparin on the cell surface (Chung et al., 1998). It, in complex with A17, can mediate cell fusion (Gong et al., 1990, Kochan et al., 2008). This A27-mediated fusion requires binding to glycosaminoglycans (GAGs), such as heparin or heparan sulfate on the cell surface and fusion is inhibited by the presence of A26 (Ching et al., 2009, Hsiao et al., 1998). MPXV and some strains of VACV, like many other OPXVs, are resistant to heparin inhibition, indicating that binding to heparin is not required for entry and VACV A27 mediated fusion that is heparin/heparan sulfate dependent more likely to be involved in cell-to-cell spread via cell fusion (Bengali et al., 2012, Hsiao et al., 1998). A27 is also required for intracellular mature virus (IMV) trafficking on microtubules within the cell and the formation of intracellular enveloped virus (IEV) in the Golgi (Rodriguez and Smith, 1990, Sanderson et al., 2000). Additionally, the A27/A17 complex likely serves as an anchor to the viral membrane for A26 and A25 in the formation of A-type inclusion bodies in OPXV viruses that form inclusion bodies such as some strains of cowpox virus (Howard et al., 2010).
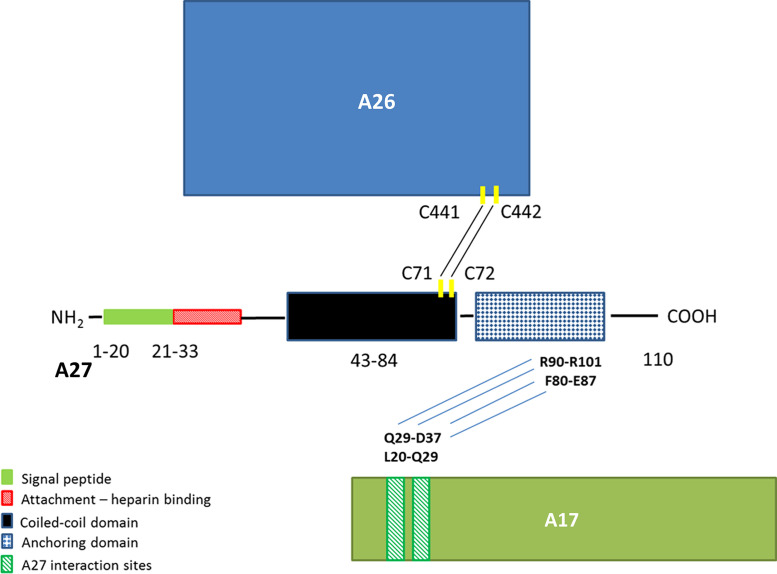
Fig. 1.
Diagram of VACV A27 functional domains. Functional regions: signal peptide (29), HBS (4, 15), coiled-coil (34), and anchoring domains (37) are modeled highlighting the heparin binding domain in red. Interaction with VACV A26 and A17 are both shown. Interaction with VACV A26 is shown by the formation of two disulfide bonds between VACV A26 C441 and C442 with VACV A27 C71 and C72 respectively (3). Binding between VACV A27 and VACV A17 occurs at two binding sites on A17. VACV A27 amino acids R90 and R101 bind to one site on A17 through VACV A17 amino acids Q29 and D37. The other binding site is between VACV A27 F80 and E87 and VACV A17 L20 and Q29 (37).
VACV A27 is a 110 amino acid protein that consists of a signal peptide, an attachment domain or heparin binding site, a fusion domain, a coiled-coil domain, and an anchoring domain as depicted in Fig. 1 (Rodriguez et al., 1991, Vázquez and Esteban, 1999). Structural studies indicate that in the monomer the N-terminal amino acids form a random and fairly flexible coil that is followed by a rigid α-helical region (Lin et al., 2002). The native protein exists as a hexamer and the α-helical region forms a coiled-coil in the self-assembling oligomerization region. Even though the heparin binding site (HBS) is found at the N-terminus, the oligomeric structure is required for heparin binding (Ho et al., 2005). The “KKPE” sequence within the HBS has been shown to be required for binding to heparin and this binding is sequence specific rather than a charge requirement (Shih et al., 2009). Extensive work has been done with VACV A27, but many questions still remain and similar studies have not been done with other OPXV orthologs. Identification of MPXV A29 specific monoclonal antibody and its epitope within the HBS demonstrate flexibility in amino acid requirement for heparin interaction.
Results
Characterization of the protein target of mAb 69-126-3-7
When originally isolated and identified, mAb 69-126-3-7 was observed to bind to an approximately 14.5 kD protein by Western blot analysis (Roumillat et al., 1984). Select MPXV proteins <50 kD were obtained and screened against 69-126-3-7 ( Fig. 2A). Reactivity was found specific only for MPXV protein A29, and not to MPXV proteins A35, B6, or M1, or to vaccinia ortholog A27. As further confirmation, binding was measured using BLI technology on the forteBio Octet system. Binding to MPXV A29 was specific and little dissociation was observed while binding to the VACV ortholog was comparable to background (Fig. 2B). To ensure the specificity was, in fact, due to differences in the orthologous proteins and not an artifact caused by choice in expression system/conditions or alterations in protein structure triggered by the addition of a 6X-his tag, COS7 cells were transfected with clones of either, MPXV A29, VACV A27, or the empty plasmid vector. Using mAb 69-126-3-7, specific binding was found to cells transfected with a plasmid expressing MPXV A29 and negative when transfected with a plasmid expressing VACV A27 or with an empty vector as measured by IFAT (Fig. 2C). Ten additional unrelated monoclonal antibodies including VARV-specific antibodies, H3L-specific antibodies, and antibodies raised against MPXV were used as controls and did not exhibit any reactivity with either MPXV A29 or VACV A27 (data not shown).
Fig. 2.
Binding is specific to MPXV A29. (A) Western blot: MPXV proteins A29, A35, B6, M1 and VACV A27 were screened for binding by mAb 69-126-3-7. Due to limited reagent, B6 was loaded at ¼ of the concentration as the other proteins in the coomassie gel shown below. The antibody reacted with MPXV A29 only. (B) BLI analysis: MPXV A29 and VACV A27 were tested for binding on the octet system and binding was both strong and specific to MPXV A29. (C) IFAT: Expression plasmids containing either VACV A27 or monkeypox A29 were transfected into COS cells and the cells were screened for expression using 69-126-3-7. Empty vector and unrelated antibodies were used as controls. Binding was specific to cells transfected with the MPXV A29 expression plasmid.
Significant work has been accomplished in identifying new OPXVs and characterizing the genealogical relationship amongst existing and newly identified orthopoxviruses in the time since mAb antibody 69-126-3-7 was initially screened for specificity (Roumillat et al., 1984). To ensure that mAb 69-126-3-7 was truly specific to MPXV, an alignment encompassing VACV A27 orthologs from all currently identified OPXVs was compiled ( Fig. 3A). All possible sequence variations were chosen to ensure that there was no possible cross-reactivity of mAb 69-126-3-7 with any non-MPXV orthopoxvirus. The A27 orthologs of MPXV, VACV, VARV, Camelpox, Ectromelia, Taterapox, and 3 strains of CPXV were tested for reactivity with mAb 69-126-3-7 by immunoblot. Results confirm that mAb 69-126-3-7 reacts only with MPXV not with any other orthopoxviruses tested (Fig. 3B).
Fig. 3.
Sequence variability in VACV A27. (A) Alignment: As new orthopoxvirus sequences have been identified since the initial studies were done, an alignment incorporating all currently identified orthopoxvirus sequences of MPXV A29 orthologs was done and all viruses that demonstrated sequence variation were analyzed by Western blot to confirm specificity. (B) Western blot: Viruses representing all possible sequence variations within the orthologs of VACV A27 were screened for binding by mAb 69-126-3-7. Lanes were loaded as follows: MPXV-USA2003-044, VACV-Wyeth, VARV-BGD75-Banu, CPXV-FIN2000-MAN, CPXV-NOR1994-MAN, CPXV-UK2000-K2984, CMLV-M96, ECTV-Mos, and TATV-DAH68. Binding was specific to MPXV. The bottom portion of the figure represents virus loading. Each semi-pure virus was loaded at equal loading concentrations and screened with VIG at a 1:500 dilution. Differences in band intensity are likely due to differential reactivity by VIG and slight variation in cellular protein contaminants during the virus purification process.
Identification of the MPXV A29 epitope
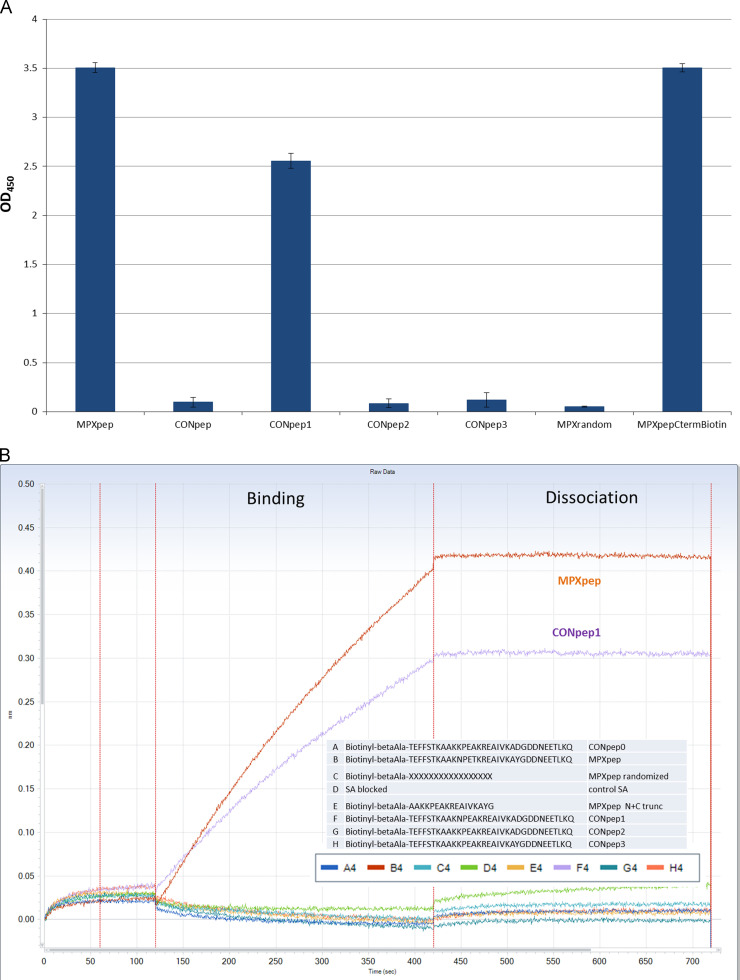
Alignment of amino acid sequence of vaccinia A27, MPXV A29, and other OPXV orthologs indicate only 4 amino acids differ in MPXV compared to all other OPXVs considered (Fig. 3A). The sequence for MPXV A29 was then analyzed for predicted immunogenicity using the Hopps–Wood and Kyte–Doolittle hydropathy prediction models and 3 of the 4 amino acids were predicted to be within immunogenic regions (Hopp and Woods, 1981, Kyte and Doolittle, 1982). As the epitope was likely to be linear because the antibody recognized protein on a nitrocullose membrane blotted from a denaturing gel, this information was used to design a series of peptides ( Table 1). Peptides representing the MPXV sequence and the OPXV consensus sequence were generated synthetically along with 3 additional peptides matching the consensus with the exception of one of the four different amino acid substitutions found only in the MPXV sequence. These peptides were initially screened for reactivity to mAb 69-126-3-7 by ELISA ( Fig. 4A). Monoclonal antibody 69-126-3-7 bound specifically to the MPXpep and CONpep1, which contained the single K→N substitution, indicating that the specificity of binding is conferred by a single amino acid difference between MPXV A29 and all other orthologs. A C-terminal biotinylated MPXpep and a randomized sequence peptide (negative) were used as controls. To confirm the ELISA results, binding was measured on the forteBio Octet system (Fig. 4B). Similarly to the ELISA results, the antibody bound to MPXpep and CONpep1 with no significant binding to the consensus peptide or the other two substitution peptides.
Table 1.
Peptides.
| MPXpep | Biotin-beta alanine-TEFFSTKAAKNPETKREAIVKAYGDDNEETLKQ |
| CONpep | Biotin-beta alanine-TEFFSTKAAKKPEAKREAIVKADGDDNEETLKQ |
| CONpep1 | Biotin-beta alanine-TEFFSTKAAKNPEAKREAIVKADGDDNEETLKQ |
| CONpep2 | Biotin-beta alanine-TEFFSTKAAKKPETKREAIVKADGDDNEETLKQ |
| CONpep3 | Biotin-beta alanine-TEFFSTKAAKKPEAKREAIVKAYGDDNEETLKQ |
| MPXpepNtrunc | Biotin-beta alanine-TKAAKNPETKREAIVKAYGDDNEETLKQ |
| MPXpepCtrunc | Biotin-beta alanine-TEFFSTKAAKNPETKREAIVKAYGDDN |
| MPXpepN+C | Biotin-beta alanine-TKAAKNPETKREAIVKAYGDDN |
| MPXpepNtrunc1 | Biotin-beta alanine-STKAAKNPETKREAIVKAYGDDNEETLKQ |
| MPXpepNtrunc2 | Biotin-beta alanine-FSTKAAKNPETKREAIVKAYGDDNEETLKQ |
| MPXpepNtrunc3 | Biotin-beta alanine-FFSTKAAKNPETKREAIVKAYGDDNEETLKQ |
| MPXpepNtrunc4 | Biotin-beta alanine-EFFSTKAAKNPETKREAIVKAYGDDNEETLKQ |
| MPXpep-Random | Biotin-beta alanine-VTIKEYTATQRKLNFNEKDKESPEAADKTAEGF |
| MPXpepCtermBiotin | TEFFSTKAAKNPETKREAIVKAYGDDNEETLKQ-beta alanine-biotin |
Fig. 4.
ELISA and BLI of peptides. (A) ELISA: 5 biotinylated peptides were plated onto a streptavidin coated ELISA plate in replicates of 4 or coated to octet sensor and assayed for binding by the antibody 69-126-3-7. 2 additional peptides were used as controls. The antibody bound to the MPX peptide (MPXpep) and did not bind to the consensus peptide (CONpep) confirming the use of peptides in these assays. When testing the single amino acid substitution peptides (CONpep1, CONpep2, and CONpep3), the antibody bound only to CONpep1. (B) BLI: 5 biotinylated peptides and the 2 control peptides were bound to the streptavidin coated sensors and tested for binding by mAb 69-126-3-7. The mAb bound strongly and specifically to MPXpep and CONpep1.
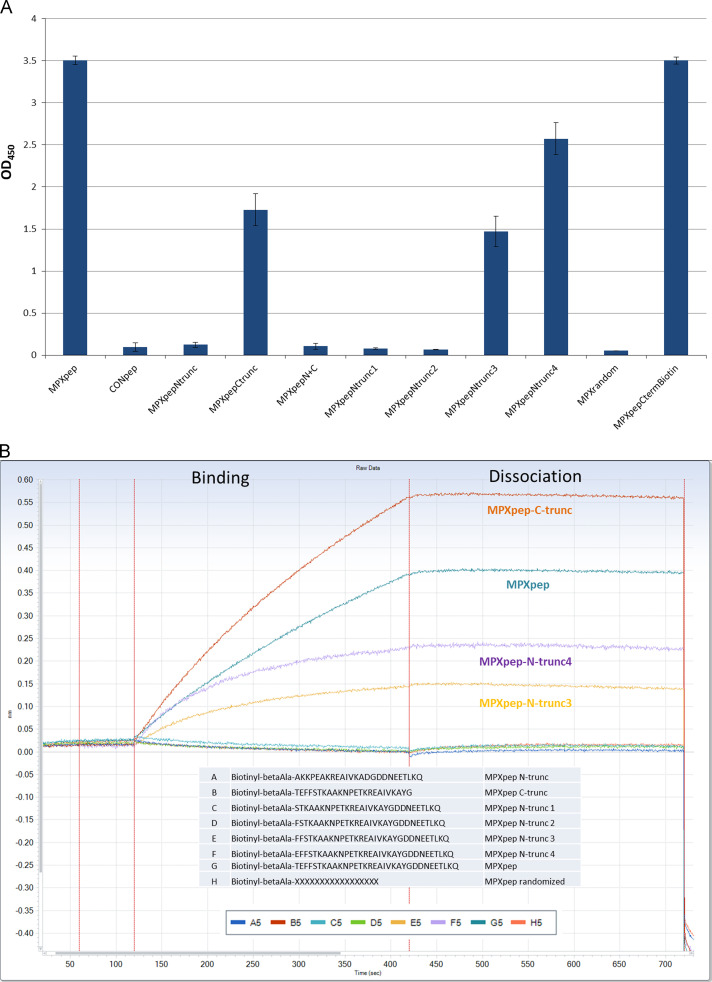
To further refine the epitope and address minimal length required for binding, peptides were designed that incorporated truncations at the N-terminus, the C-terminus, or both ends of the peptide of recognition (Table 1). Analysis by ELISA indicates that the C-terminal truncation was tolerated while the N-terminal truncation was not ( Fig. 5A and B). To investigate this further, each of the 5 amino acids removed from the N-truncated peptide was added back individually creating peptides that are each one amino acid longer than the previous until the full length MPXpep sequence was reached. As seen in Fig. 5A by ELISA and in Fig. 5B by BLI, binding was restored by approximately 40% with the addition of three amino acids and by 70% with the addition of four amino acids to the N-truncated peptide. The highest level of binding required the full length N-terminal region of the MPXpep.
Fig. 5.
ELISA and BLI of truncated peptides. To try and identify the minimal region required for antibody binding, 2 truncations were made (5 amino acids from the N-terminus and separately 6 amino acids from the C-terminus of the peptide) and screened for binding. In both ELISA (A) and BLI analysis (B), the C-terminal truncation was tolerated while the N-terminal truncation was not. To further refine the role of the N-terminal amino acids in antibody binding, a single amino acid was added back to the N-truncated peptide one at time. Interestingly binding was minimally restored with addition of 3 amino acids with an increase in binding signal with the restoration of 4 amino acids, but all 5 n-terminal amino acids were required for binding signals equivalent to MPXpep.
Heparin binding
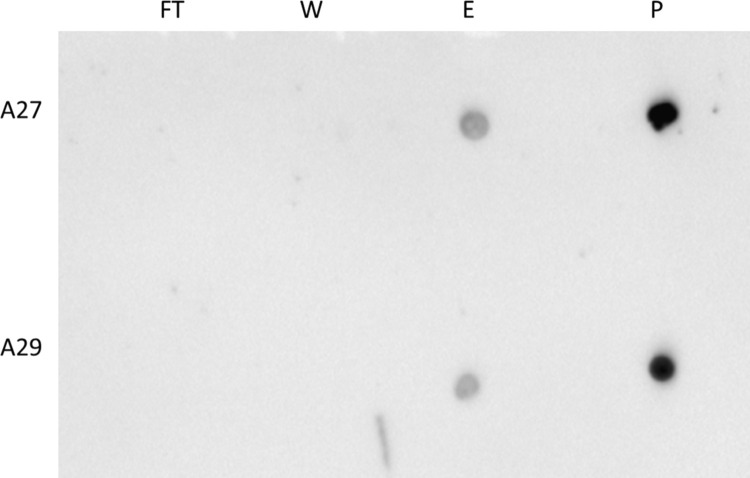
The HBS of VACV A27 has been well characterized. Specifically, it has been noted that the conserved “KKPE” sequence was required for heparin binding and this requirement is sequence specific. As MPXV is the only OPXV to differ in this sequence, we wanted to investigate whether MPXV A29 could bind heparin. To determine if MPXV A29 bound heparin, A29 and VACV A27 were run across a heparin column and the flow-through, wash, and 2 M NaCL eluate were blotted onto a membrane and probed with a polyclonal anti-A27 antibody known to bind both A29 and A27. Both proteins were found only in the eluate ( Fig. 6). In order to examine the heparin binding affinities associated with these two proteins (MPXV A29 and VACV A27), disassociation kinetics were determined. Dissociation constants (K D) were calculated for both proteins on the Octet system. Results indicate that MPXV A29 (11.0 nM) exhibited similar affinity for heparin as VACV A27(6.3 nM) as shown in Table 2.
Fig. 6.
Protein binding to heparin and heparin inhibition dot blots, MPXV A29 and VACV A27, were run over a heparin column. (A) The flow through (FT), wash (W) and eluate or elution (E) were along with purified protein (P) were dotted onto nitrocellulose membrane. This was probed with polyclonal rabbit α-A27 antibody at a 1:10,000 dilution. Protein was visible in E and the loading control, but none in the FT or the W for both proteins, indicating that A29 binds heparin regardless of the amino acid difference in the heparin binding site. (B) Antibody 69-126-3-7 was incubated with 10 µg of MPXV at a ratio of antibody:protein of 1:2, 1:1, 2:1, and 5:1 and run over a heparin column. Flow through (FT), representing what did not bind to the column, and Eluate or Elution, representing what did bind, were concentrated and screened with a polyclonal α-A27 antibody. Inhibition of binding to heparin was seen as early as 1:2, but almost complete inhibition of binding was not observed until 5:1.
Table 2.
BLI binding kinetics of MPXV A29 and VACV A27.
| A29L hexamer 84 kDa | A27L hexamer 84 kDa | |
|---|---|---|
| Cycle 1 | 9.8 nM (r2=0.94) | 5 nM (r2=0.97) |
| Cycle 2 | 11.3 nM (r2=0.94) | 7 nM (r2=0.96) |
| Cycle 3 | 12 nM (r2=0.95) | 7 nM (r2=0.96) |
To corroborate that this region was necessary for heparin binding, MPXV A29 was incubated with mAb 69-126-3-7 at protein:antibody rations of 2:1, 1:1, 1:2, and 1:5 and then run across a heparin column. The flow-through and 2 M NaCl eluate were blotted and visualized by probing with polyclonal anti-A27 antibody ( Fig. 7). Inhibition of heparin binding was observable at 2:1, but complete inhibition was not seen until 1:5 protein to antibody ratios. This combined data indicated that the antibody binds to the HBS and is capable of blocking MPXV A29 binding to heparin.
Fig. 7.
Heparin inhibition dot blot. Antibody 69-126-3-7 was incubated with 10 µg of MPXV at a ratio of antibody:protein of 1:2, 1:1, 2:1, and 5:1 and run over a heparin column. Flow through (FT), representing what did not bind to the column, and Eluate or Elution, representing what did bind, were concentrated and screened with a polyclonal α-A27 antibody. Inhibition of binding to heparin was seen as early as 1:2, but almost complete inhibition of binding was not observed until 5:1.
Previous work had indicated that the HBS alone was not sufficient for binding to heparin (Ho et al., 2005). To investigate whether increased peptide length could affect binding to heparin or if the oligomeric structure was indeed required, MPXpep, CONpep, and a randomized sequence peptide, each of which was 30 amino acids in length, were run across a heparin column and screened for binding to heparin using BLI. No significant binding above background was seen either to the column or to heparin demonstrating that increasing peptide length did not affect heparin binding (data not shown).
Discussion
Here we demonstrate that mAb 69-126-3-7 binds to MPXV A29. This was established using three different techniques including immunoblot, IFA, and biolayer interferometry. These experiments also show specificity for MPXV by the absence of immunoblot reactivity to other OPXV׳s tested including VACV, VARV, Camelpox virus, Ectromelia virus, Taterapox virus, and 3 strains of CPXV confirming the ELISA results previously reported (Bengali et al., 2012). Amino acid sequence alignments indicated that there were only four amino acid differences specific to MPXV when compared to OPXVs protein orthologs. Of these, only three were found within a hydrophilic, and therefore likely exposed and immunogenic, region. Peptides were designed that encompassed this region and that represented the MPXV sequence, the conserved sequence, and a series of point mutations within the conserved sequence to confirm the use of peptides in binding assays and to investigate the role of each amino acid in mAb specificity. Using both ELISA and BLI, binding to this region of the protein was found to be specific to MPXV and binding dependent on a single amino acid difference between the MPXV sequence and the conserved sequence. Interestingly, this change from lysine to asparagine at amino acid position 27 is found, not only within the HBS, but specifically at the second position with the conserved “KKPE” sequence previously determined to be stringently required for heparin binding. This led to the obvious question of whether MPXV bound heparin in the same way as VACV and other OPXVs. As binding to heparin and heparan sulfate is redundant in multiple poxvirus proteins, it is unlikely that such a small change within a single protein would have an overwhelming effect on the entire virus. For that reason, we addressed the question of whether MPXV A29 was capable of binding heparin similarly to VACV A27 despite the amino acid difference within the key “KKPE” sequence at the protein level rather than the virus level. Studies of binding on a heparin column and BLI show that both VACV A27 and MPXV A29 bind to heparin with similar affinity regardless of the sequence changes in the HBS and the “KKPE” sequence. The original reports that define the sequence requirement used alanine substitutions to explore the role of the sequence in binding. This is standard protocol when investigating the contributions of single amino acids to structure or function within a protein as alanine is a small amino acid that is well tolerated in most proteins and its effects on structure are usually minimal if a particular amino acid is not required (Morrison and Weiss, 2001). While lysine and asparagine do not have the same charge since the lysine side chain is positively charged at pH 7.4, their side chains are of similar length and both are capable of forming hydrogen bonds, one of the mechanisms proposed for binding to heparin, making it entirely feasible that asparagine would be tolerated and have minimal effect on protein function (Shih et al., 2009).
Increasing peptide length from the 10 amino acids of the heparin binding site to 30 amino acids did not increase peptide binding to heparin and supports work by Ho et al., in which the authors used either a peptide of the HBS or an almost full-length recombinant protein to test heparin binding. The authors concluded that the oligomeric region of A27 was required for heparin binding. This was supported by additional experiments in which they perturbed the oligomeric region and observed loss in heparin binding (Ho et al., 2005). Finally, to establish that the epitope for mAb 69-126-3-7 was within the HBS, inhibition assays were performed and as the ratio of antibody to protein increased, heparin binding decreased, indicating that the specific mAb epitope is within the HBS. In short, while the actual HBS is required for heparin binding, some amino acid substitutions are tolerated more than others and the oligomeric region is also required for MPXV A29 or VACV A27 binding to heparin.
The characterization of a MPXV-specific antibody and the identification of an epitope in which specificity is conferred by a single amino acid difference in such a highly conserved protein are exciting in that it lends itself towards the design of OPXV species-specific serological assays. To date though other MPXV-specific antibodies have been identified, there have been reports of only one species-specific serological based assay (Dubois et al., 2012, Golden and Hooper, 2008, Hammarlund et al., 2005). This assay shows promise, but it has only been tested in its ability to differentiate between VACV and MPXV and has not been screened against other OPXV potential human pathogens such as various strains of cowpox virus or variola virus. The use of species-specific peptides to design serological assays that can differentiate between multiple orthopoxvirus infections in humans would be ideal for both diagnostic and survey purposes. Species-specific serological survey assays would provide support for the current estimations of MPXV prevalence. The ability to diagnose OPXV infections in an outbreak situation with a technique that requires little in the way of special equipment and reagents and that is also rapid and not cost prohibitive would make early identification in a MPXV outbreak easier and allow for a targeted, rapid and effective response thus saving lives as the fatality rate for MPXV ranges from 10% in the western strain to 30% in the Congo strain. The use of specifically designed peptides and proteins that are both unique and immunogenic will aid significantly in achieving the goal of sensitive, yet specific serological assays for OPXVs.
Materials and methods
Protein expression and purification and peptide synthesis
A27 and A29 were expressed in a baculovirus system with a C-terminal 6X His-tag and purified on a nickel column by Chesapeake-Perl (Savage, MD). A35, B6, and M1 were kindly provided by Genoveffa Franchini at NIH (Bethesda, MD). All peptides were synthesized by the Biotechnology Core facility at the Centers for Disease Control and Prevention and purified by HPLC using Fmoc/tBu solid-phase peptide synthesis and were characterized by analytical reversed-phase U-HPLC and mass spectrometry. The peptides were used in the form of their trifluoroacetate salts. Unless otherwise stated, all peptides contain an N-terminal biotin moiety followed by β-alanine.
Viruses
MPXV-USA2003-044 (#DQ011153), VACV-Wyeth, VARV-BGD75-Banu (#DQ437581), CPXV-FIN2000-MAN (#HQ420893), CPXV-NOR1994-MAN (#HQ420899), CPXV-UK2000-K2984 (#HQ420900), CMLV-M96 (#NC_003391), ECTV-Mos (#NC_004105), and TATV-DAH68 (#NC_008291) were grown up in BSC-40 cells and purified using a sucrose cushion.
Immunoblot
All proteins and viruses were diluted in SDS, β-mercaptoethanol, and sample buffer. After heat inactivation at 100 °C for 10 min, samples were separated on a 4–20% SDS-PAGE and transferred to PVDF. Membranes were probed with antibody at a 1:10,000 dilution (100 ng/mL) in Superblock T20 (Thermo Scientific). After 3×10 min wash in PBST, membrane is probed with goat α-mouse (KPL) at 1:1000 in blocking buffer and detected with Immun-Star Western C (Bio-Rad).
Indirect fluorescent antibody tests (IFAT)
This assay was performed as described previously (Custer et al., 2003). Briefly, COS cells grown on 15 mm diameter glass coverslips in 12-well culture plates and were transfected with 0.5 μg/well of DNA vaccine plasmid pWRG/A27, pWRG/A29 and empty vector pWRG/7077. After 48 h, cells were washed with PBS (pH 7.4) and fixed with acetone for 10 min. Cells were rinsed 3× with PBS and incubated in blocking buffer (PBS+5% goat serum). Primary antibodies were diluted in blocking buffer to 5 μg/mL and cells incubated in 500 μL for 1 h at 37 °C. Cells were washed 3× PBS and then incubated with secondary fluorescent antibody (KPL 02-18-07) at 1:400 and Hoechst as a counter stain at 1:1000. Cells were incubated at 37 °C for 30 min and then washed 3× PBS and rinsed 1× deionized water and then placed on a drop of fluorescent mounting medium (DAKO) on glass slides. The cells were observed with a Nikon E600 fluorescence microscope. Ten additional antibodies were screened as controls including VARV-specific antibodies (E2:B7;B5, E2:D4:E6, and E2:G3:C6), H3L-specific antibodies (3B6:F4:G7, 3B6:F4:D7, and 3B6:F4:E7), and unrelated MPXV antibodies (69-178-3-10, 1B6E12H9, 6F6-2-G8H8, and 6F6-2-G8E12).
Binding assays/biolayer interferometry (BLI)
Octet analysis was conducted at temperature control at 30 °C in PBS buffer. Streptavidin (SA) sensors were pre-wet for 10 min in buffer prior to use and microplates used in the Octet were filled with 200 µl of sample or buffer and agitated at 700 rpm. SA-coated tips were saturated with 20 µg/mL biotinylated synthetic peptides. Typical capture levels were 1.10±0.15 nm within a row of eight tips with the standard deviation within the instrument noise. Peptide coated sensors were incubated with mAb 69-126-3-7 at a concentration of (200 nM) and binding was allowed to occur for approximately 300 s and allowed to dissociate for 500 s. Dissociation buffer was used only once to prevent non-specific binding. Other mAbs were used as a control. For binding kinetics to heparin, SA-coated tips were saturated with 50 µg/mL biotinylated heparin. A 300 nM titration of VACV A27 or MPXV A29 was bound for 300 s and allowed to dissociate for 500 s in PBS buffer. Dissociation buffer was used only once to prevent non-specific binding. Blank binding cycles containing only peptide were used to correct for baseline drift. Peptides were compared in the same experiment by coupling each onto its own tips in triplicate. Shift data from the Octet were exported for processing and analysis in Data Analysis 6.4. To deduce a direct binding affinity via the kinetic rate constants (K D=k off/k on, where K D=equilibrium dissociation rate constant, k on=association rate constant, and k off=dissociation rate constant) the buffer subtracted Octet data were fit globally to a simple 1:1 Langmuir model.
ELISA
Peptides were diluted in PBS and incubated overnight at 4 °C in streptavidin high-binding 96-well plates per manufacturer׳s directions (Thermo Scientific, MA) at a concentration of 25 ng/µL. Plates are incubated for 1 h at room temperature with Superblock T20 (Fisher Scientific). Blocking buffer is removed and 69-126-3-7 is diluted 1:10,000 (10 ng/mL) in blocking buffer and added to wells and incubated at 37 °C for 1 h. Plates are washed 3× with PBST and incubated with HRP conjugated goat α-mouse (KPL) at a 1:1000 dilution in blocking buffer at 37 °C for 1 h. After washing 3× with PBST, plates are incubated with TMB substrate solution for 3 min and reaction is stopped with Stop solution. Signal is read at 650 nm after shaking. All peptides are done in replicates of 4 and values are averaged and standard error is calculated.
Heparin binding assay
GE Healthcare HiTrap Heparin column (Thermo Scientific) was equilibrated with 10 column volumes (10 mL) of 10 mM sodium phosphate buffer, pH 7.0 (binding buffer) per manufacturer׳s directions. 10 μg of sample was diluted in 1 mL of binding buffer and run across column 3×. Flow through (FT) was collected after 3rd time off the column. Column was washed with 10 column volumes of binding buffer. Wash (W) was collected. Samples were eluted with 10 column volumes of binding buffer plus 2 M NaCl (elution buffer). Elution was collected (E). FT, W, and E were concentrated to less than 50 μL using Millipore and then Pierce 10 kD concentrators. The Millipore spins were conducted for 20-30 min at 2500 rpm, 25 °C. The final 2 spins of E were done with concentrated sample and 7 mL PBS to promote buffer exchange. The Pierce spins were done with 200 μL of concentrated sample placed in Pierce concentrator and topped off with PBS. The samples were further concentrated to 20–50 μL by spinning at room temperature for 3.5 min. 1 µL of each was blotted on a nitrocellulose membrane and allowed to dry. Membrane is then incubated as described in immunoblot protocol.
Inhibition assay
10 μg of A29 was incubated with 69-126-3-7 antibody at 2:1, 1:1, 1:2, and 1:5 ratios in 1 mL 10 mM sodium phosphate buffer, pH 7 (binding buffer) while rocking at 4 °C for 1 h. Protein:antibody solutions were then run across the column and blotted as described in the heparin binding assay protocol.
Acknowledgments and funding
We would like to thank Melissa McDonald and Chuck Boozer for peptide synthesis and George Buchman from Chesapeake PERL, Inc. for protein purification. We would like to thank Ginny Emerson and PS Satheshkumar for helpful scientific discussions. This research was supported in part by an appointment to the Research Participation Program at the Centers for Disease Control and Prevention administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and CDC.
References
- Bengali Z., Satheshkumar P.S., Moss B. Orthopoxvirus species and strain differences in cell entry. Virology. 2012;433:506–512. doi: 10.1016/j.virol.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhnia M.R.-E.-I., McCausland M.M., Su H.-P., Singh K., Hoffmann J., Davies D.H., Crotty S. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J. Virol. 2008;82:3751–3768. doi: 10.1128/JVI.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching Y.-C., Chung C.-S., Huang C.-Y., Hsia Y., Tang Y.-L., Chang W. Disulfide bond formation at the c termini of vaccinia virus a26 and a27 proteins does not require viral redox enzymes and suppresses glycosaminoglycan-mediated cell fusion. J. Virol. 2009;83:6464–6476. doi: 10.1128/JVI.02295-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.-S., Hsiao J.-C., Chang Y.-S., Chang W. A27l protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 1998;72:1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custer D.M., Thompson E., Schmaljohn C.S., Ksiazek T.G., Hooper J.W. Active and passive vaccination against hantavirus pulmonary syndrome with andes virus m genome segment-based DNA vaccine. J. Virol. 2003;77:9894–9905. doi: 10.1128/JVI.77.18.9894-9905.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D., Molina D., Wrammert J., Miller J., Hirst S., Mu Y., Felgner P. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- Dubois M.E., Hammarlund E., Slifka M.K. Optimization of peptide-based elisa for serological diagnostics: a retrospective study of human monkeypox infection. Vector-Borne Zoonotic Dis. 2012;12:400–409. doi: 10.1089/vbz.2011.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.W., Hooper J.W. Heterogeneity in the a33 protein impacts the cross-protective efficacy of a candidate smallpox DNA vaccine. Virology. 2008;377:19–29. doi: 10.1016/j.virol.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S., Lai C., Esteban M. Vaccinia virus induces cell fusion at acid ph and this activity is mediated by the n-terminus of the 14-kDa virus envelope protein. Virology. 1990;178:81–91. doi: 10.1016/0042-6822(90)90381-z. [DOI] [PubMed] [Google Scholar]
- Hammarlund E., Lewis M.W., Carter S.V., Amanna I., Hansen S.G., Strelow L.I., Slifka M.K. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat. Med. 2005;11:1005–1011. doi: 10.1038/nm1273. [DOI] [PubMed] [Google Scholar]
- Ho Y., Hsiao J.-C., Yang M.-H., Chung C.-S., Peng Y.-C., Lin T.-H., Tzou D.-L.M. The oligomeric structure of vaccinia viral envelope protein a27l is essential for binding to heparin and heparan sulfates on cell surfaces: a structural and functional approach using site-specific mutagenesis. J. Mol. Biol. 2005;349:1060–1071. doi: 10.1016/j.jmb.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Hopp T., Woods K. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA. 1981;73:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A.R., Senkevich T.G., Moss B. Vaccinia virus a26 and a27 proteins form a stable complex tethered to mature virions by association with the a17 transmembrane protein. J. Virol. 2008;82:12384–12391. doi: 10.1128/JVI.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A.R., Weisberg A.S., Moss B. Congregation of orthopoxvirus virions in cytoplasmic a-type inclusions is mediated by interactions of a bridging protein (a26p) with a matrix protein (atip) and a virion membrane-associated protein (a27p) J. Virol. 2010;84:7592–7602. doi: 10.1128/JVI.00704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao J.-C., Chung C.-S., Chang W. Cell surface proteoglycans are necessary for a27l protein-mediated cell fusion: identification of the n-terminal region of a27l protein as the glycosaminoglycan-binding domain. J. Virol. 1998;72:8374–8379. doi: 10.1128/jvi.72.10.8374-8379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezek Z., Fenner F. Vol. 17. Karger; Basel, Switzerland: 1988. Human monkeypox; pp. 1–140. (Monographs in Virology). [Google Scholar]
- Keasey S., Pugh C., Tikhonov A., Chen G., Schweitzer B., Nalca A., Ulrich R.G. Proteomic basis of the antibody response to monkeypox virus infection examined in cynomolgus macaques and a comparison to human smallpox vaccination. PLoS One. 2010;5:e15547. doi: 10.1371/journal.pone.0015547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckler M.S., Carroll D.S., Gallardo-Romero N.F., Lash R.R., Salzer J.S., Weiss S.L., Damon I.K. Establishment of the black-tailed prairie dog (Cynomys ludovicianus) as a novel animal model for comparing smallpox vaccines administered preexposure in both high- and low-dose monkeypox virus challenges. J. Virol. 2011;85:7683–7698. doi: 10.1128/JVI.02174-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan G., Escors D., González J.M., Casasnovas J.M., Esteban M. Membrane cell fusion activity of the vaccinia virus a17–a27 protein complex. Cell. Microbiol. 2008;10:149–164. doi: 10.1111/j.1462-5822.2007.01026.x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Li Y., Olson V.A., Laue T., Laker M.T., Damon I.K. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. 2006;36:194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhao H., Wilkins K., Hughes C., Damon I.K. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo basin strain DNA. J. Virol. Methods. 2010;169:223–227. doi: 10.1016/j.jviromet.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.-H., Chia C.-M., Hsiao J.-C., Chang W., Ku C.-C., Hung S.-C., Tzou D.-L.M. Structural analysis of the extracellular domain of vaccinia virus envelope protein, a27l, by NMR and CD spectroscopy. J. Biol. Chem. 2002;277:20949–20959. doi: 10.1074/jbc.M110403200. [DOI] [PubMed] [Google Scholar]
- Morrison K.L., Weiss G.A. Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 2001;5:302–307. doi: 10.1016/s1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- Olson V.A., Laue T., Laker M.T., Babkin I.V., Drosten C., Shchelkunov S.N., Meyer H. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J. Clin. Microbiol. 2004;42:1940–1946. doi: 10.1128/JCM.42.5.1940-1946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K.D., Melski J.W., Graham M.B., Regnery R.L., Sotir M.J., Wegner M.V., Damon I.K. The detection of monkeypox in humans in the western hemisphere. N. Engl. J. Med. 2004;350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- Rimoin A.W., Mulembakani P.M., Johnston S.C., Lloyd Smith J.O., Kisalu N.K., Kinkela T.L., Muyembe J.-J. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the democratic republic of congo. Proc. Natl. Acad. Sci. USA. 2010;107:16262–16267. doi: 10.1073/pnas.1005769107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez D., Rodriguez J.R., Esteban M. The vaccinia virus 14-kilodalton fusion protein forms a stable complex with the processed protein encoded by the vaccinia virus a17l gene. J. Virol. 1993;67:3435–3440. doi: 10.1128/jvi.67.6.3435-3440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J.-R., Rodriguez D., Esteban M. Structural properties of HIV-1 Env fused with the 14-kDa vaccinia virus envelope protein. Virology. 1991;181:742–748. doi: 10.1016/0042-6822(91)90910-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez J.F., Smith G.L. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by golgi membrane and egress. Nucleic Acids Res. 1990;18:5347–5351. doi: 10.1093/nar/18.18.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumillat L.F., Patton J.L., Davis M.L. Monoclonal antibodies to a monkeypox virus polypeptide determinant. J. Virol. 1984;52:290–292. doi: 10.1128/jvi.52.1.290-292.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson C.M., Hollinshead M., Smith G.L. The vaccinia virus a27l protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol. 2000;81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- Shchelkunov S.N., Shcherbakov D.N., Maksyutov R.A., Gavrilova E.V. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. J. Virol. Methods. 2011;175:163–169. doi: 10.1016/j.jviromet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih P.-C., Yang M.-S., Lin S.-C., Ho Y., Hsiao J.-C., Wang D.-R., Tzou D.-L.M. A turn-like structure “KKPE” segment mediates the specific binding of viral protein a27 to heparin and heparan sulfate on cell surfaces. J. Biol. Chem. 2009;284:36535–36546. doi: 10.1074/jbc.M109.037267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend M.B., Keckler M.S., Patel N., Davies D.H., Felgner P., Damon I.K., Karem K.L. Humoral immunity to smallpox vaccines and monkeypox virus challenge: proteomic assessment and clinical correlations. J. Virol. 2013;87:900–911. doi: 10.1128/JVI.02089-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez M.-I., Esteban M. Identification of functional domains in the 14-kilodalton envelope protein (a27l) of vaccinia virus. J. Virol. 1999;73:9098–9109. doi: 10.1128/jvi.73.11.9098-9109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.-R., Hsiao J.-C., Wong C.-H., Li G.-C., Lin S.-C., Yu S.S.-F., Tzou D.-L.M. Vaccinia viral protein a27 is anchored to the viral membrane via a cooperative interaction with viral membrane protein a17. J. Biol. Chem. 2014;289:6639–6655. doi: 10.1074/jbc.M114.547372. [DOI] [PMC free article] [PubMed] [Google Scholar]