ABSTRACT
Macroautophagy/autophagy is a finely-regulated process in which cytoplasm encapsulated within transient organelles termed autophagosomes is delivered to lysosomes or vacuoles for degradation. Phospholipids, particularly phosphatidic acid (PA) that functions as a second messenger, play crucial and differential roles in autophagosome formation; however, the underlying mechanism remains largely unknown. Here we demonstrated that PA inhibits autophagy through competitive inhibition of the formation of ATG3 (autophagy-related)-ATG8e and ATG6-VPS34 (vacuolar protein sorting 34) complexes. PA bound to GAPC (glyceraldehyde-3-phosphate dehydrogenase) or PGK (phosphoglycerate kinase) and promoted their interaction with ATG3 or ATG6, which further attenuated the interactions of ATG3-ATG8e or ATG6-VPS34, respectively. Structural and mutational analyses revealed the mechanism of PA binding with GAPCs and PGK3, and that GAPCs or ATG8e competitively interacted with ATG3, and PGK3 or VPS34 competitively interacted with ATG6, at the same binding interface. These results elucidate the molecular mechanism of how PA inhibits autophagy through binding GAPC or PGK3 proteins and expand the understanding of the functional mode of PA, demonstrating the importance of phospholipids in plant autophagy and providing a new perspective for autophagy regulation by phospholipids.
Abbreviation: ATG: autophagy-related; BiFC: bimolecular fluorescence complementation; co-IP: co-immunoprecipitation; Con A: concanamycin A; ER: endoplasmic reticulum; EZ: elongation zone; FRET-FLIM: fluorescence resonance energy transfer with fluorescence lifetime imaging microscopy; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GST: glutathione S-transferase; MDC: monodansylcadaverine; MZ: meristem zone; PA: phosphatidic acid; PAS: phagophore assembly site; PC: phosphatidylcholine; PE: phosphatidylethanolamine; PGK3: phosphoglycerate kinase; PtdIns3K: phosphatidylinositol 3-kinase; PLD: phospholipase D; TEM: transmission electron microscopy; TOR: target of rapamycin; VPS34: vacuolar protein sorting 34; WT: wild type; Y2H: yeast two-hybrid.
KEYWORDS: Autophagy, autophagy-related protein, competitive inhibition, glyceraldehyde-3-phosphate dehydrogenase, phosphatidic acid, phosphoglycerate kinase
Introduction
Eukaryotic cells maintain the internal homeostasis through engaging elaborate catabolic processes that recognize and recycle the superfluous or potentially dangerous cytosolic entities [1]. Macroautophagy/autophagy process encapsulates and delivers intracellular components to lysosomes or vacuoles for degradation via double-membrane vesicles termed autophagosomes. In plant cells, three distinct types of autophagic pathways have been identified: microautophagy, megaautophagy, and macroautophagy (henceforth, autophagy) [2]. Genetic screens using yeast cells identified ~36 evolutionarily conserved ATG (autophagy related) genes, most of which are required for the core autophagic machinery that generates the autophagosome [3]. Studies have shown that autophagy is associated with diverse physiological processes including nutrient starvation, senescence, hypoxia, responses to high salt, oxidative stress and drought [4–8]. Arabidopsis autophagy-defective (such as atg) mutants exhibit hypersensitivity to nutrient starvation, premature leaf senescence and shortened life span [6].
In eukaryotes, ATG proteins form four distinct functional complexes that independently or synergistically govern the autophagosome formation, including initiation, elongation, and closure [9]. During the initiation of autophagosome biogenesis, reduction of the TOR (target of rapamycin) kinase activity activates the Atg1-Atg13 protein complex, which recruits the Vps30/Atg6-phosphatidylinositol 3-kinase (PtdIns3K) complex to form the PtdIns3P-enriched phagophore assembly site (PAS) [10]. Two ubiquitin-like conjugation complexes, Atg12–Atg5-Atg16 and Atg8–phosphatidylethanolamine (PE), regulate phagophore expansion. Atg3 functions as an E2-like conjugating enzyme and the Atg12–Atg5 complex acts as an E3 ligase for conjugating Atg8 to PE [11–13]. The Atg9 complex consists of the transmembrane protein Atg9, PtdIns3P-binding protein Atg18 and Atg2, and helps the delivery of lipids to the expanding phagophore [14]. Finally, the outer membrane of the autophagosome fuses with the vacuole, which releases the inner vesicle (autophagic body) to resident hydrolases for degradation [2]. During the process, Atg8–PE is initially embedded in the inner and outer membranes of autophagosomes, and subsequently delivered to the vacuole together as part of the autophagosomes, and thus is regarded as a common marker for phagophores and mature autophagosomes [13].
PLD (phospholipase D) is a widely distributed phospholipase in prokaryotes and eukaryotes that catalyze the hydrolysis of structural phospholipids phosphatidylcholine (PC) and PE to produce phosphatidic acid (PA). There are 12 PLD genes in the Arabidopsis genome, which can be divided into 6 subtypes, α (3 members), β (2), γ (3), δ, ε (1) and ζ (2) based on sequence homology and biochemical properties [15]. As an important signaling molecule, production of PA is induced by developmental and environmental cues, and PA participates and plays vital role in various signaling pathways through binding with and affecting the activity, subcellular localization, protein interaction or complex assembly of target proteins [16–18]. Considering that PA is responsible for MTOR activation, PA has been considered as a negative regulator of autophagy in animals [19,20]. Although studies showed that PA might function in several stages of autophagy in mammalian cells, whether PA affects autophagy in plant cells and how PA exactly functions remains unclear.
Glycolysis is a metabolic pathway that provides energy in the form of ATP and reducing power, pyruvate molecules to fuel the mitochondrial tricarboxylic acid cycle, and precursors for secondary metabolism [21]. Because GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and PGK (phosphoglycerate kinase) are involved in both photosynthetic and glycolytic reactions in the same compartment, the functional characteristics of them are crucial [22]. There are eight GAPDHs (chloroplastic photosynthetic GAPA1, GAPA-2, and GAPB, plastidic glycolytic GAPCp1 and GAPCp2, cytosolic glycolytic GAPC1 and GAPC2, and the NADP-dependent non-phosphorylating cytosolic ALDH11A3) [23] and three PGKs (plastidial PGK1, PGK2 and cytosolic PGK3) [24] in Arabidopsis.
Here we showed that PA suppresses the formation of autophagosomes through interacting with cytosolic glycolytic GAPDHs (GAPC1 and GAPC2) and PGK3, the key members of glycolysis that localizes at the endoplasmic reticulum. PA promoted the interactions of GAPCs-ATG3 and PGK3-ATG6 proteins which further inhibited the function of ATG8e or VPS34 by competitive interactions.
Results
PA suppresses autophagy in Arabidopsis
To examine the role and functional mechanism of PA in autophagosome formation, Arabidopsis seedlings expressing GFP-ATG8e fusion protein, which has been widely used as a marker of autophagic structures [25], were treated with PA (10 μM), 1-butanol (inhibitor of phospholipase D, 0.4%) or PLDζ inhibitor (200 nM) for 12 h in the absence or presence of concanamycin A (Con A, a V-ATPase inhibitor). Observation of GFP fluorescence in root meristem zone (MZ) and elongation zone (EZ) showed that in contrast to control, GFP-ATG8e–labeled puncta and ring-like structures (Figure 1A) were significantly increased under 1-butanol or PLDζ inhibitor treatment (Figures 1B, S1A), while decreased upon PA treatment (Figure 1B).
Figure 1.
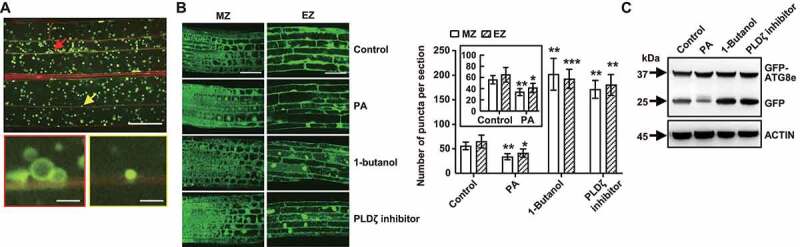
Phosphatidic acid (PA) suppresses autophagosome formation in Arabidopsis roots. (A) GFP-ATG8e–labeled dots and ring-like structures in elongation zone (EZ) of Arabidopsis roots after 1-butanol treatment. Arabidopsis seedlings expressing GFP-ATG8e fusion protein were grown on 1/2 MS medium for 5 days, then transferred to 1/2 MS medium containing 1-butanol (0.4%) for 12 h in the presence of concanamycin A (Con A, 0.5 μM) and visualized by confocal microscopy. Representative images were shown (upper, bar: 50 μm). The GFP-ATG8e dots or ring-like structures are indicated by arrows and enlarged (bottom, bar: 5 μm). (B) Observation (left, bars = 50 μm) and measurement (right) of puncta per root section of GFP-ATG8e–labeled dots and ring-like structures in the meristem zone (MZ) and EZ of Arabidopsis roots under treatment with phosphatidic acid (PA), 1-butanol or PLDζ inhibitor. Arabidopsis seedlings expressing GFP-ATG8e fusion protein were grown on 1/2 MS medium for 5 days, then transferred to 1/2 MS medium containing PA (10 μM), 1-butanol (0.4%) or PLDζ inhibitor (200 nM) for 12 h in the presence of Con A (0.5 μM) and visualized by confocal microscopy. Representative images were shown. Experiments were biologically repeated for three times and data were shown as mean ± SD (n = 15). Statistical significance was determined by Student's t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001, compared with control). (C) Immunodetection of free GFP release during autophagy-mediated vacuolar degradation of GFP-ATG8e under PA, 1-butanol or PLDζ inhibitor treatment. Arabidopsis seedlings expressing GFP-ATG8e fusion protein were grown on 1/2 MS medium for 7 days, then transferred to 1/2 MS medium containing PA (10 μM), 1-butanol (0.4%) or PLDζ inhibitor (200 nM) for 12 h in the presence of Con A (0.5 μM), followed by protein extraction and immunoblotting analyses. The experiments were biologically repeated for three times and ACTIN was used as a protein loading control.
Upon induction of autophagy, GFP-ATG8e recruits other autophagy-associated proteins to form autophagosomes, which subsequently fuse with the tonoplast to deliver the contents of autophagic bodies into vacuole. Within the vacuole lumen, GFP-ATG8e is sheared into ATG8e fragments and free GFP by acid hydrolase [26], and detection of free GFP is widely used to monitor the extent of autophagy. Consistent with the confocal observations, the ratio of free GFP to GFP-ATG8e was substantially decreased after PA treatment, whereas largely increased under 1-butanol or PLDζ inhibitor treatment (Figure 1C). To further confirm this, Arabidopsis seedlings expressing GFP-ATG8e were exposed to 1-butanol (0.4%) or PLDζ inhibitor (200 nM) in the absence or presence of PA (10 μM). Observations revealed that after 1-butanol or PLDζ inhibitor treatment, formation of GFP–labeled puncta and ring-like structures was obviously increased in root cells, however, such accumulation was moderately attenuated in the presence of PA (Figure S1B). These results indicated that PA negatively regulates autophagy in Arabidopsis.
PLD deficiency resulted in the accumulation of autophagic structures
PA is mainly produced by PLD that hydrolyzes the structural phospholipids. To investigate the role of PA in autophagy, we treated 5-day-old wild type (WT), pldα1, pldζ1, pldβ1, pldδ (knock-out T-DNA insertion mutants of 4 representative PLD genes) and PLDδ-OE seedlings with Con A, and then monodansylcadaverine (MDC) staining was performed to observe the autophagosomes. Confocal observations indicated that the MDC-stained vesicles were significantly accumulated in pldα1, pldζ1, pldβ1, and pldδ root cells (Figure 2A), whereas those were reduced in PLDδ-OE root cells (Figure 2B), compared with those of WT. To confirm the altered accumulation of MDC-labeled vesicles that representing the changed autophagosomes, we used transmission electron microscopy (TEM) to monitor the autophagic activity. Analyses of the ultrastructure of Arabidopsis leaves revealed that in compared to WT, autophagy-related structures were increased in pldα1, pldζ1, pldβ1, and pldδ leaves (Figure 2C), while decreased in PLDδ-OE plants (Figure 2D), confirming that PLD deficiency causes the accumulated autophagic structures.
Figure 2.
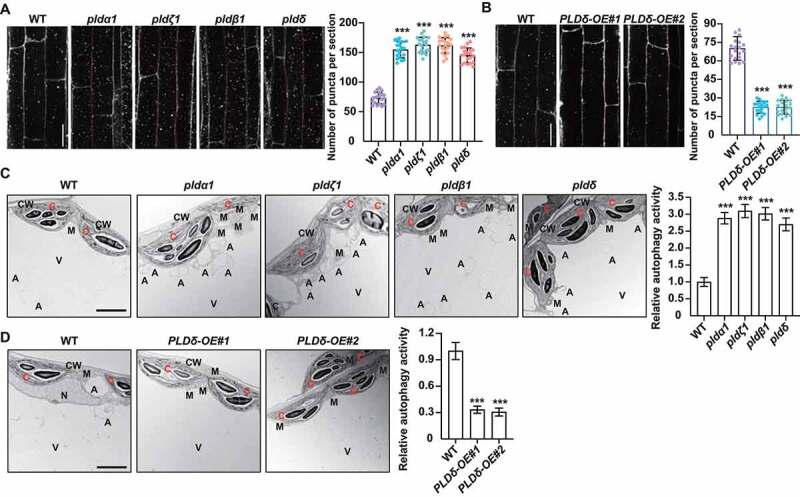
Deficiencies of PLDs result in promoted autophagy. (A and B) MDC staining (left) and quantification of puncta per root section (right) of WT, pldα1, pldζ1, pldβ1, pldδ (A) and PLDδ-OE (B). Five-day-old seedlings were treated with Con A (0.5 μM), followed by staining with MDC. The labeled autophagosomes of root cells were visualized by fluorescence confocal microscopy using a DAPI-specific filter and representative images were shown (bars: 50 μm). Numbers of puncta per root section were measured and data were shown as mean ± SD (n = 20). Statistical significance was determined by Student's t-test (***, P < 0.001, compared to WT). (C and D) TEM analyses (left, bars = 50 μm) and quantification of autophagic structures (right) in WT and pldα1, pldζ1, pldβ1, pldδ seedlings (C), and seedlings overexpressing PLDδ (D). Three-week-old seedlings were treated with Con A (0.5 μM) and mesophyll cells were observed. “A” indicates the autophagic structures. CW, cell wall; C, chloroplast; M, mitochondrion; V, vacuole. Experiments were biologically repeated for three times and data were shown as mean ± SD (n = 10). Statistical significance was determined by Student's t-test (***, P < 0.001, compared to WT).
Deficiencies of ATG genes lead to premature leaf senescence and hypersensitivity to nutrient deprivation [27–29]. Studies showed that suppression of PLDα1 and PLDδ attenuates abscisic acid-promoted leaf senescence [30] and overexpressing acyl-CoA binding proteins 3 (ACBP3-OE) leads to PA accumulation and promoted leaf senescence [31] in Arabidopsis. Indeed, analyses of seedling growth showed that when grown under nutrient-rich conditions, pld mutants and PLDδ-OE plants did not display obvious growth differences compared to WT, while upon the carbon or nitrogen starvation, pld mutants displayed more tolerance, as reflected by the delayed yellowing of seedlings and increased chlorophyll contents (Figure S1C). By contrast, the PLDδ-OE lines were more sensitive to carbon and nitrogen starvation than WT (Figure S1D), confirming that PLD negatively regulates the autophagy-associated response to nutrient starvation in Arabidopsis.
PA binds to GAPCs or PGK3
To explore the mechanism of autophagy inhibition by PA, candidate PA-binding proteins were searched and analyzed, including glycerol-3-phosphate dehydrogenase (GAPDH) and phosphoglycerate kinase (PGK) [32] that involve in the glycolysis pathway. Since PA and autophagosome are mainly localized in the cytosol, we focused on cytosolic glycolytic GAPCs (GAPC1 and GAPC2) and cytosolic PGK3. Consistent with previous study showing that PA binds to GAPC [32], lipid-protein blot overlay assays showed that purified Arabidopsis GAPCs and PGK3 (recombinant expressed in Escherichia coli) bound to PA, as well as various PtdIns monophosphates and PtdIns bisphosphates (Figure S2A). Liposomal binding assay was further performed (liposomes were produced with a mixture of dioleoyl PC and dioleoyl PA) and western blotting using anti-His antibodies showed that bound GAPCs or PGK3 (pellet with liposomes) increased and non-binding protein (supernatant with liposomes) decreased with increasing PA concentration, while no significant change in pellet and supernatant with increasing PC (Figure 3A). These results confirmed the specific binding of PA to GAPCs and PGK3.
Figure 3.
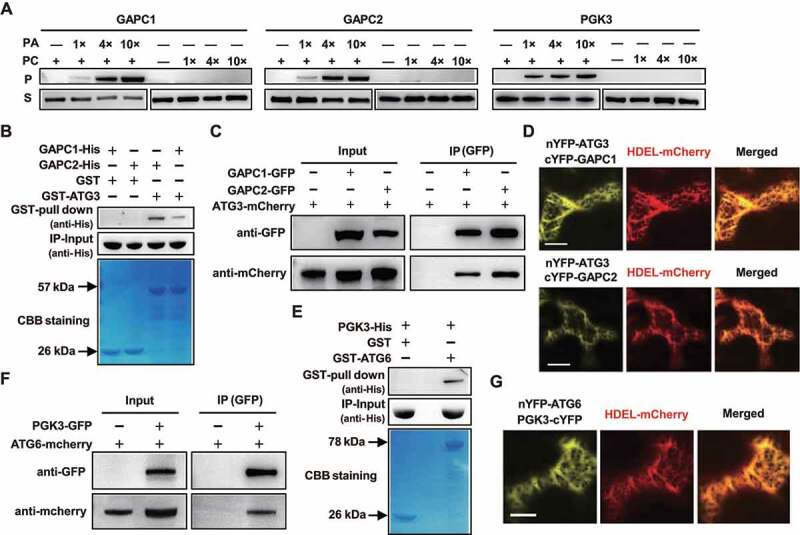
GAPCs interacted with ATG3, and PGK3 interacted with ATG6, at endoplasmic reticulum (ER). (A) Liposome binding assay showed the correlation between bound GAPCs or PGK3 and PA. Purified His-tagged GAPCs or PGK3 were incubated with liposomes containing di18:1-PC only or di18:1-PA/PC (1/3 mole ratio). 1×, 4×, and 10× refer to the concentration gradient of PC or PA/PC liposomes used. Bound (pellet with liposomes, P) GAPCs or PGK3 and non-binding protein (supernatant with liposomes, S) were detected by immunoblotting. (B) GST pull-down assays revealed that GAPC1-His and GAPC2-His directly interact with GST-ATG3. Recombinantly expressed (from E. coli) GST or GST-ATG3 proteins were incubated with GAPC1-His or GAPC2-His and analyzed by immunoblot assays using an anti-His antibody (upper panel). Input of His-tagged proteins (middle panel) and equal aliquots of glutathione beads loaded with GST-ATG3 or GST (separated by SDS-PAGE and stained with Coomassie blue, bottom panel) were shown. (C) Co-IP assay demonstrated the interaction between ATG3-mCherry and GAPC1-GFP or GAPC2-GFP in vivo. GFP-tagged GAPCs was transiently co-expressed with mCherry-tagged ATG3 in N. benthamiana leaves and immunoprecipitated by GFP affinity magnetic beads. Resultant immunoprecipitation (IP) and cell lysate were analyzed by immunoblotting using anti-GFP or anti-mCherry antibodies. (D) Fluorescence observation of N. benthamiana leaf epidermal cells co-expressing GAPC1-cYFP/GAPC2-cYFP and nYFP-ATG3 with ER marker HDEL-mCherry. Representative images were shown (bar = 50 μm). (E) GST pull-down assays revealed that PGK3-His directly interacts with GST-ATG6. Recombinantly expressed (from E. coli) GST or GST-ATG6 proteins were incubated with PGK3-His and analyzed by immunoblot assays using an anti-His antibody (upper panel). Input of His-tagged proteins (middle panel) and equal aliquots of glutathione beads loaded with GST-ATG6 or GST (separated by SDS-PAGE and stained with Coomassie blue, bottom panel) were shown. (F) Co-IP assay demonstrated the interaction between ATG6-mCherry and PGK3-GFP in vivo. GFP-tagged PGK3 was transiently co-expressed with mCherry-tagged ATG6 in N. benthamiana leaves and immunoprecipitated by GFP affinity magnetic beads. Resultant immunoprecipitation (IP) and cell lysate were analyzed by immunoblotting using anti-GFP or anti-mCherry antibodies. (G) Fluorescence observation of N. benthamiana leaf epidermal cells co-expressing PGK3-cYFP and nYFP-ATG6 with ER marker HDEL-mCherry. Representative images were shown (bar = 50 μm).
Interaction of GAPCs-ATG3 or PGK3-ATG6 at the ER
PA participates in various signaling pathways through binding with and affecting the activity, subcellular localization, protein interaction or complex assembly of target proteins [33]. Considering that PA-GAPCs interaction did not affect the activity and subcellular localization of GAPCs in Arabidopsis [32] and GAPCs interact with ATG3 to suppress autophagy in Nicotiana benthamiana [34], it is speculated that PA may affect the interaction of GAPCs with interacting proteins by binding. Confocal observations revealed that GAPC1-GFP and GAPC2-GFP colocalized with ATG3-mCherry in tobacco leaves (Figure S3A), and an in vitro GST (glutathione S-transferase) affinity-isolation assay showed that GAPC1-His or GAPC2-His was only detected upon incubation with GST-ATG3 (Figure 3B), indicating that Arabidopsis GAPCs directly interact with ATG3 in vitro. A co-immunoprecipitation (co-IP) analyses demonstrated the interaction of GAPCs-GFP with ATG3-mCherry in vivo (Figure 3C), which is further confirmed by bimolecular fluorescence complementation (BiFC) studies showing the detectable YFP fluorescence under co-expressed nYFP-ATG3 and GAPCs-cYFP in N. benthamiana leaves (nYFP-OsHAL3 and OsHAL3-cYFP served as a positive control [35], Figure S3B). Previous analyses showed that GAPCs and PGK3 locate at endoplasmic reticulum (ER), the major source of autophagosomal membranes [36], and consistently, observation of N. benthamiana co-expressing nYFP-ATG3/GAPCs-cYFP with ER marker HDEL-mCherry revealed that GAPCs interact with ATG3 at ER (Figure 3D).
To search for the PGK3-associated autophagy components, we used yeast two-hybrid (Y2H) analyses to examine the possible interactions of PGK3 with known ATG proteins. Results showed that PGK3 specifically interacted with ATG6 (Figure S3C). GST pull-down assay (Figure 3E) and co-immunoprecipitation (co-IP) analyses (Figure 3F) demonstrated the interaction of PGK3 with ATG6 in vitro and in vivo, which is further confirmed by BiFC studies using N. benthamiana leaves (Figure S3D). Similar as GAPCs-ATG3, observation of the co-expressed nYFP-ATG6/PGK3-cYFP with HDEL-mCherry showed that PGK3-ATG6 complex also localizes at ER (Figure 3G). These results indicate that GAPCs-ATG3 or PGK3-ATG6 complex may participate in the early regulation of autophagy at the ER.
PA promotes the GAPCs-ATG3 or PGK3-ATG6 interaction through direct binding
Based on the binding of GAPCs or PGK3 with PA, whether PA specifically affects the interaction of GAPCs-ATG3 or PGK3-ATG6 to participate in autophagy regulation through direct binding is examined. GST pull-down assay were performed and results showed the interaction of GAPCs-ATG3 or PGK3-ATG6 was significantly enhanced in the presence of PA (Figure 4A,B), whereas there was no significant difference in the presence of PC (Figure S2B, C). A co-immunoprecipitation (co-IP) analyses further revealed that the interaction of GAPCs-ATG3 or PGK3-ATG6 was significantly enhanced in the presence of PA in vivo, but not PC (Figure 4C,D). These suggested that PA specifically promotes the GAPCs-ATG3 or PGK3-ATG6 interaction in vitro and in vivo.
Figure 4.
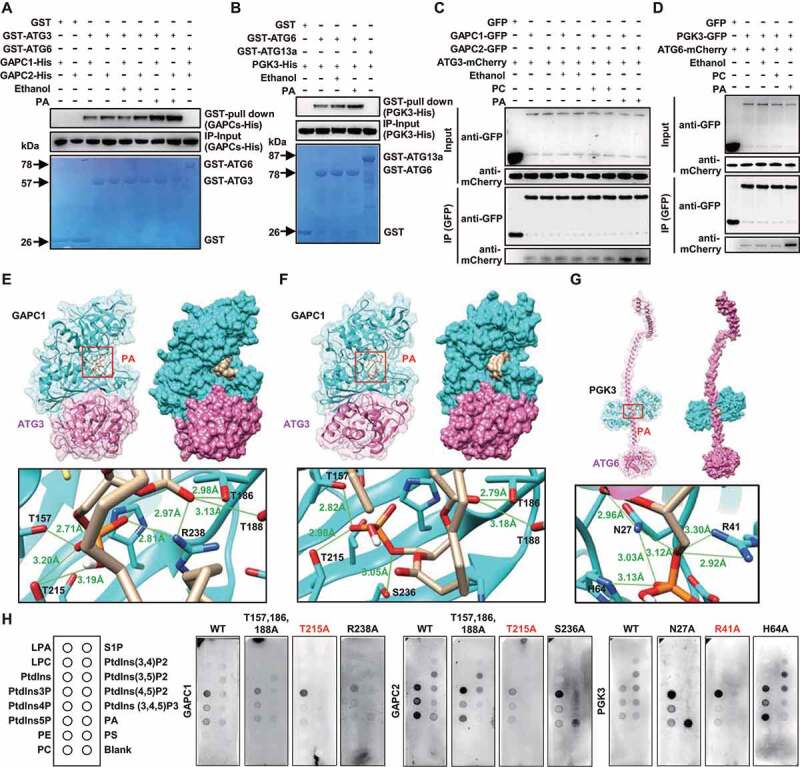
PA binds to GAPCs or PGK3 and promotes the interaction of GAPCs-ATG3 or PGK3-ATG6 in vivo and in vitro. (A and B) GAPCs-His (A) or PGK3-His (B) was precipitated by GST-ATG3 (A, GST-ATG6 was used as negative control) or GST-ATG6 (B, GST-ATG13a was used as negative control) using agarose beads in the absence or presence of PA (10 μM). The pellet fraction was eluted and analyzed by immunoblotting using anti-His antibodies (upper panel). Input of His-tagged proteins (middle panel) and equal aliquots of glutathione beads loaded with GST-ATG3 or GST-ATG6 or GST (separated by SDS-PAGE and stained with Coomassie blue, bottom panel) were shown. (C and D) PA promotes the formation of GAPCs-ATG3 (C) or PGK3-ATG6 (D) complexes in vivo. Cell lysate from Arabidopsis seedlings expressing GAPCs-GFP and ATG3-mCherry or PGK3-GFP and ATG6-mCherry was subjected to a GFP trap assay. Resultant immunoprecipitation (IP) and cell lysate were analyzed by immunoblotting using anti-GFP or anti-mCherry antibodies. (E-G) Structural diagram of the PA-GAPCs-ATG3 (E, F) or PA-PGK3-ATG6 (G) complex based on the flexible docking model. Proteins are shown in the cartoon and surface model, with GAPCs or PGK3 colored in cyan, ATG3 or ATG6 colored in pink and 18:1 PA colored in yellow. In the zoom graph, the amino acids of GAPCs or PGK3 protein that could establish hydrogen bonds with PA are predicted based on the docking model. Green font and lines indicate the hydrogen bonds and distances. (H) Lipid-protein blot overlay assays of the PA binding capacity of GAPCs and PGK3 (WT and variants containing various amino acid mutation). Recombinant expressed GAPCs, PGK3, and variants (fused with His-tag) were incubated with PIP strips and detected using anti-His antibodies. LPA, lysophosphatidic acid; LPC, lysophosphocholine; PtdIns, phosphatidylinositol; PtdIns3P, PtdIns-3-phosphate; PtdIns4P, PtdIns-4-phosphate; PtdIns5P, PtdIns-5-phosphate; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PtdIns(3,4)P2, PtdIns-3,4-bisphosphate; PtdIns(3,5)P2, PtdIns-3,5-bisphosphate; PtdIns(4,5)P2, PtdIns-4,5-bisphosphate; PtdIns(3,4,5)P3, PtdIns-3,4,5-trisphosphate; PA, phosphatidic acid; PS, phosphatidylserine.
Fluorescence resonance energy transfer with fluorescence lifetime imaging microscopy (FRET-FLIM), a technique determining resonance energy transfer by fluorescence lifetime imaging, has the advantage of less interference factors and becomes a common technique for studying the conformational changes of biomolecules and dynamic interactions between molecules in living cells [37]. Analyses by FRET-FLIM assay revealed a reduced fluorescence lifetime of GAPCs-GFP or PGK3-GFP when co-expressing ATG3-mCherry or ATG6-mCherry, which is much decreased under PA treatment (Figure S4A, B), indicating that PA enhances the interactions of ATG3-GAPCs and PGK3-ATG6, which was further confirmed by BiFC assay using N. benthamiana leaves expressing nYFP-ATG3 or nYFP-ATG6 with GAPCs-cYFP or PGK3-cYFP in the presence of PA (Figure S4C, with similar protein levels, Figure S4D, E).
Templated-based modeling approach together with protein-small molecule and protein-protein docking were used to analyze the overall structure of PA-GAPCs-ATG3 or PA-PGK3-ATG6 and amino acids of GAPCs or PGK3 protein that could establish hydrogen bonds with PA. Structural models of 18:1 PA, GAPC1 (PDB code 6QUQ) and ATG3 (PDB code 3VX8) were obtained from ChemSpider or Protein Data Bank (PDB). Using homologous modeling of Swiss-Model approach, we obtained a highly confident structural model for the studied proteins (Figure S5). The binding interfaces of the trimolecular complex composed of PA-GAPCs-ATG3 or PA-PGK3-ATG6 were predicted by semi flexible docking using Autodock Vina [38] and a hybrid docking method using the HDOCK algorithm [39]. A cavity inside the GAPCs or PGK3 protein for binding PA molecules is adjacent to the binding interfaces of the GAPCs-ATG3 or PGK3-ATG6 complex (Figure 4E–G). Detailed analyses showed that the amino acids in GAPCs or PGK3 proteins can establish hydrogen bonds with PA (bottom panel, Figure 4E–G).
Further, the predicted interaction sites of GAPCs (Threonine, T, position 215) and PGK3 (Arginine, R, position 41) were mutated to A (Alanine, A, due to its neutral nature, short side chain and no charge, which is used to study the effect of mutated amino acid), and used to examine the effects of these residues on PA binding capacity. Analyses by using lipid-protein blot overlay assays showed that binding of PA with the mutated form of GAPC (GAPCsT215A) or PGK3 (PGK3R41A) was substantially decreased (Figure 4H) compared with normal GAPC or PGK3, demonstrating the crucial role of T215 of GAPCs and R41 of PGK3 in PA binding and indicating that GAPCs and PGK3 proteins bound to PA to facilitate the formation of GAPCs-ATG3 or PGK3-ATG6 complexes.
PA attenuates ATG3-ATG8e and ATG6-VPS34 interactions through binding to GAPCs or PGK3
The mechanism of how enhanced GAPC-ATG3 or PGK3-ATG6 interactions regulate autophagy was further investigated. ATG3 is an E2-like conjugating enzyme that catalyzes the formation of ATG8-PE conjugate [26], and ATG6 activates PtdIns3K/VPS34 activity to induce PtdIns3P production and recruits other autophagy-related proteins [10], during the autophagy process. Interactions of ATG3-ATG8e and ATG6-VPS34 are required for autophagosome formation. We therefore examined whether the enhanced GAPCs-ATG3 or PGK3-ATG6 interactions interrupt the ATG3-ATG8e or ATG6-VPS34 interactions to inhibit autophagy.
Consistent with previous studies [1,40], an affinity-isolation assay showed that His-ATG8e is pulled down by GST-ATG3 (Figure 5A) and VPS34-His is pulled down by GST-ATG6 (Figure 5B). Interestingly, the interaction of ATG3-ATG8e or ATG6-VPS34 was reduced under added GAPCs-His or PGK3-His (Figure 5A,B), indicating that GAPCs or PGK3 represses the ATG3-ATG8e or ATG6-VPS34 interaction in vitro. Furthermore, by transiently co-expressing GFP-ATG8e, ATG3-mCherry, and GAPCs-Flag, or ATG6-GFP, VPS34-mCherry, and PGK3-Flag, in N. benthamiana leaves, analyses showed that ATG3-mCherry or VPS34-mCherry co-immunoprecipitated with GFP-ATG8e or ATG6-GFP; however, as expected, the interactions were reduced upon co-expression of GAPCs-Flag (Figure 5C) or PGK3-Flag (Figure 5D), and the ATG8e-ATG3 or ATG6-VPS34 interaction was further suppressed under exogenous PA (Figure 5C,D). These results indicated that GAPCs or PGK3 interfered with the ATG8e-ATG3 or ATG6-VPS34 interactions to inhibit autophagy, and PA enhances the inhibitory effects of GAPCs or PGK3.
Figure 5.
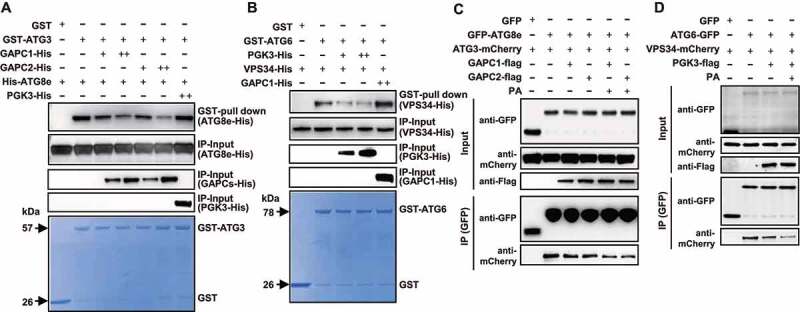
PA attenuates the interaction of ATG3-ATG8e or ATG6-VPS34 by binding to GAPCs or PGK3. (A and B) GST affinity‐isolation assay showed the suppressed ATG3-ATG8e association by GAPCs (A, PGK3-His was used as a negative control) or ATG6-VPS34 association by PGK3 (B, GAPC1-His was used as a negative control). The pellet fraction was eluted and analyzed by immunoblotting using anti-His antibodies (upper panel). Input of GAPCs-His or PGK3-His (middle panel) and equal aliquots of glutathione beads loaded with GST-ATG3 or GST-ATG6 or GST (separated by SDS-PAGE and stained with Coomassie Brilliant Blue, bottom panel) were shown. The experiments were biologically repeated for three times and representative images were shown. (C) GAPCs-Flag and PA inhibit the interaction of ATG3-mCherry and GFP-ATG8e in vivo. GAPCs-Flag or GFP were transiently co-expressed in N. benthamiana leaves with ATG3-mCherry and GFP-ATG8e. After immunoprecipitated by GFP affinity magnetic beads, ATG3-mCherry, GFP-ATG8e, and GAPCs-Flag were detected by immunoblot analyses with anti-mCherry, anti-GFP, and anti-Flag antibody, respectively. (D) PGK3-Flag and PA inhibit the interaction of VPS34-mCherry and ATG6-GFP in vivo. PGK3-Flag or GFP were transiently expressed in N. benthamiana leaves with VPS34-mCherry and ATG6-GFP. After immunoprecipitated by GFP affinity magnetic beads, VPS34-mCherry, ATG6-GFP, and PGK3-Flag were detected by immunoblot analyses with anti-mCherry, anti-GFP, and anti-Flag antibody, respectively.
FRET-FLIM assays were further performed to investigate the PA effect on ATG3-ATG8e and ATG6-VPS34 interactions. By using GFP-ATG8e/ATG3-mCherry, ATG6-GFP/VPS34-mChery, GFP-ATG8e and ATG6-GFP, observations showed that the fluorescence lifetime of GFP-ATG8e or ATG6-GFP was reduced when co-expressing with ATG3-mCherry or VPS34-mCherry, whereas such reduction was strongly inhibited under PA treatment (Figure S6A, B), indicating the interactions of ATG3-ATG8e and ATG6-VPS34 is attenuated by PA. Being consistent, overexpression of ATG3 or VPS34 obviously induce the autophagic body accumulation in vacuole, whereas such accumulation was strongly reduced under PA treatment (Figure S6C, D).
Templated-based modeling approach together with protein-protein docking were used to analyze the conservation of the overall structure of individual subunits and their respective binding interface. The binding interfaces of the heterodimer composed of GAPCs-ATG3, ATG3-ATG8e, PGK3-ATG6 and ATG6-VPS34 were predicted by a hybrid docking method using the HDOCK algorithm [39]. Results showed that GAPC1, GAPC2 or ATG8e interact with ATG3 at the same binding interface, i.e. via 6, 8 and 4 hydrogen bonds respectively (Figure 6A–C). Analyses of the amino acids involved in the formation of hydrogen bonds revealed that either GAPCs or ATG8e are linked to ATG3 via Tyrosine (Y) at position 205 (Figure 6A–C), suggesting that GAPCs or ATG8e may competitively bind ATG3 at this site or binding interface. Further, Y205 site was mutated, and analyses of the GAPCs-ATG3 or ATG3-ATG8e interaction by using GST pull-down assay showed the substantially decreased interaction with GAPCs (Figure 6D) or ATG8e (Figure 6E) of the mutated form of ATG3 (ATG3Y205A) compared with normal ATG3, demonstrating the crucial role of Y205 of ATG3 in GAPCs-ATG3 or ATG3-ATG8e interactions.
Figure 6.
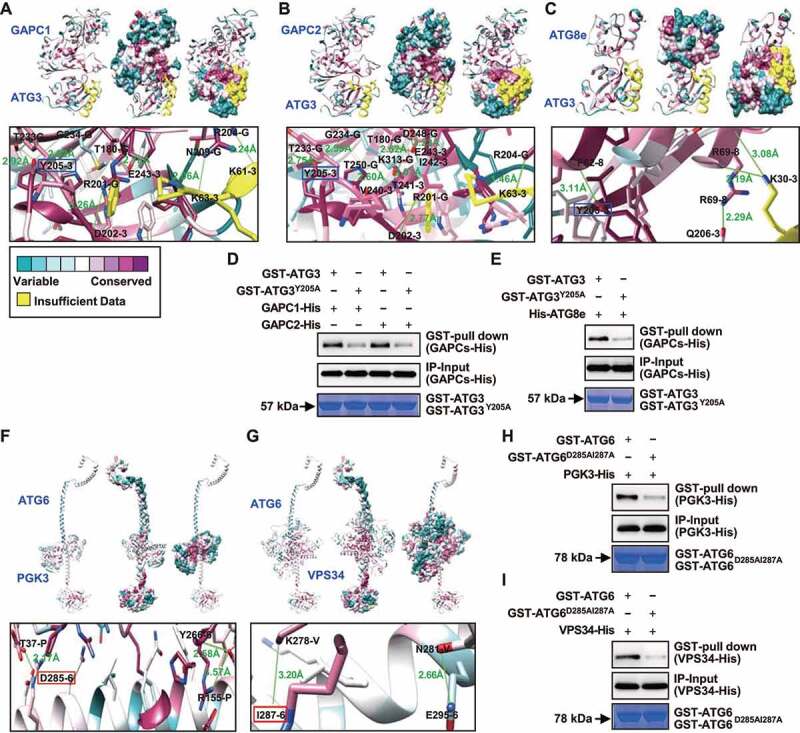
Template-based modeling and protein docking indicate that GAPCs or PGK3 interacts with ATG3 or ATG6 competitively with ATG8e or VPS34. (A-C) Heterodimer formed by GAPCs (A, B) or ATG8e (C) with ATG3 was calculated by a hybrid docking approach using the HDOCK algorithm. Analyses of conserved amino acid residues was performed by the Consurf server and mapped on the solvent-excluded surface of each protein. In the zoom graph, the amino acids in GAPCs or ATG8e protein that could establish hydrogen bonds with ATG3 are predicted based on the docking model. Green font and lines indicate the hydrogen bonds and distances. Amino acids followed with G, 3 or 8 indicate GAPCs, ATG3 or ATG8e. GAPCs or ATG8e are linked to ATG3 through Y205 (highlighted by blue box). (D and E) GST affinity-isolation assays revealed that residue Y205 in ATG3 is important for binding with GAPCs (D) or ATG8e (E). Fusion proteins GAPCs-His or His-ATG8e were incubated with glutathione agarose-bound GST-ATG3 or GST-ATG3Y205A, and analyzed by immunoblot assays using an anti-His antibody (upper panel). Input of His-tagged proteins (middle panel) and equal aliquots of glutathione beads loaded with GST-ATG3 or GST-ATG3Y205A (separated by SDS-PAGE and stained with Coomassie blue, bottom panel) were shown. (F and G) Heterodimer formed by PGK3 (F) or VPS34 (G) with ATG6 was calculated by a hybrid docking approach using the HDOCK algorithm. Analyses of conserved amino acid residues was performed by the Consurf server and mapped on the solvent-excluded surface of each protein. In the zoom graph, the amino acids in PGK3 or VPS34 protein that could establish hydrogen bonds with ATG6 are predicted based on the docking model. Green font and lines indicate the hydrogen bonds and distances. Amino acids followed by P, V, and 6 indicate PGK3, VPS34, and ATG6. PGK3 or VPS34 are coupled to ATG6 via D285 or I287 (highlighted by red box), respectively, which are located in close proximity to each other. (H and I) GST pull-down assays revealed that residues D285 and I287 in ATG6 is important for binding with PGK3 (H) or VPS34 (I). Fusion proteins PGK3-His or VPS34-His were incubated with glutathione agarose-bound GST-ATG6 or GST-ATG6D285AI287A and analyzed by immunoblot assays using an anti-His antibody (upper panel). Input of His-tagged proteins (middle panel) and equal aliquots of glutathione beads loaded with GST-ATG6 or GST- GST-ATG6D285AI287A (separated by SDS-PAGE and stained with Coomassie blue, bottom panel) were shown.
In addition, structural analyses showed that PGK3 or VPS34 interact with ATG6 at the same binding interface, i.e. via 3 and 2 hydrogen bonds respectively (Figure 6F,G), and PGK3 or VPS34 are linked to ATG6 via Aspartic acid (D) at position 285 or Isoleucine (I) at position 287, which are in close proximity to each other, suggesting that PGK3 or VPS34 may competitively bind ATG6 at these sites or binding interfaces. Similarly, D285 and I287 sites were mutated, and analyses of the PGK3-ATG6 or ATG6-VPS34 interaction by using GST affinity-isolation assay showed the substantially decreased interaction with PGK3 (Figure 6H) or VPS34 (Figure 6I) of the mutated form of ATG6 (ATG6D285AI287A) compared with normal ATG6, demonstrating the crucial role of D285 and I287 of ATG6 in PGK3-ATG6 or ATG6-VPS34 interactions. Interestingly, these sites or binding interfaces are formed by highly conserved amino acids (Figure 6), suggesting that they are structurally important and supporting their key roles in the formation of heterodimers.
GAPCs and PGK3 are required for the PA-suppressed autophagy
Previous studies showed that the numbers of autophagic puncta in gapc1 leaves were increased compared with WT seedlings [41]. To assess the role of GAPCs and PGK3 in autophagy, we treated 5-day-old WT, gapc1, gapc2, gapc1 gapc2 and pgk3 seedlings with Con A, and then MDC staining was performed to observe the autophagosomes. Analyses indicated that the MDC-stained vesicles were significantly accumulated in gapc1, gapc2, gapc1 gapc2 and pgk3 root cells (Figure S7A, B). By crossing gapc1, gapc2, gapc1 gapc2 and pgk3 mutants with seedlings expressing GFP-ATG8e, observation further showed that in contrast to the root cells of WT, there were more GFP-ATG8e–labeled dots and ring-like structures in various mutants (Figure 7A,B). Being consistent, accumulation of free GFP was obviously increased in gapc1, gapc2, gapc1 gapc2 and pgk3 mutants (Figure 7C,D). Seedling overexpressing GAPCs or PGK3 were further generated and observation of the 5-day-old WT, GAPC1-OE, GAPC2-OE, and PGK3-OE seedlings, which were treated with Con A and then stained with MDC, showed that the MDC-stained vesicles were reduced in GAPC1-OE, GAPC2-OE, and PGK3-OE root cells (Figure S7C, D). In addition, by crossing GAPC1-OE, GAPC2-OE, and PGK3-OE with seedlings expressing GFP-ATG8e, observations showed that in contrast to those of WT, there were less GFP-ATG8e–labeled dots and ring-like structures in GAPC1-OE, GAPC2-OE, and PGK3-OE root cells (Figure 7E,F), confirming the inhibitory effects of GAPCs and PGK3 on autophagy.
Figure 7.
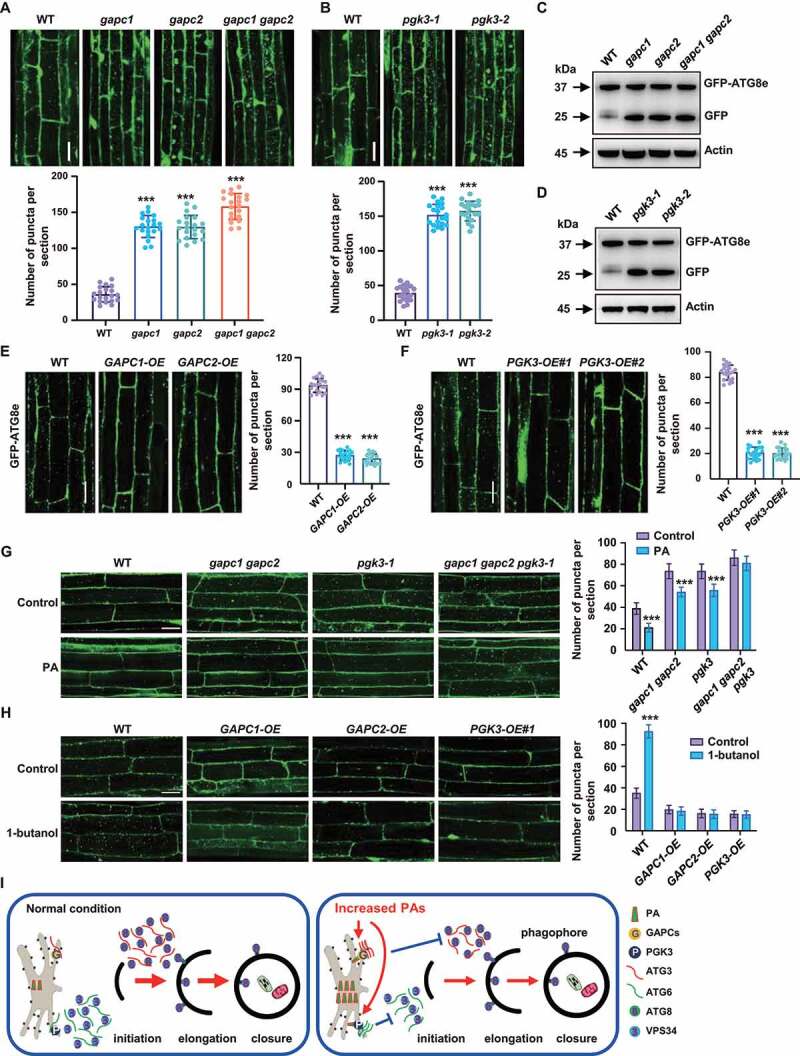
GAPCs and PGK3 inhibit autophagy in Arabidopsis. (A and B) Fluorescence observation (upper, bars: 50 μm) and quantification of puncta per root section (bottom) of WT, gapc (A) and pgk3 seedlings (B) expressing GFP-ATG8e. Five-day-old seedlings were treated with Con A (0.5 mM, 2 h) and visualized by confocal microscopy. Numbers of puncta per root section were measured and data were shown as mean ± SD (n = 20). Experiments were biologically repeated for three times. Statistical significance was determined by Student's t-test (***, P < 0.001, compared to WT). (C and D) Immunodetection of the free GFP release during autophagy-mediated vacuolar degradation of GFP-ATG8e in WT, gapc (C) or pgk3 (D) expressing GFP-ATG8e. One-week-old seedlings grown on 1/2 MS medium were transferred to 1/2 MS medium containing Con A (0.5 μM), followed by protein extraction and immunoblotting analyses. The experiments were biologically repeated for three times and ACTIN was used as a protein loading control. (E and F) Fluorescence observation (upper, bars = 50 μm) and quantification of puncta per root section (bottom) of WT, GAPCs-OE (E) and PGK3-OE (F) lines expressing GFP-ATG8e. Five-day-old seedlings were treated with Con A (0.5 mM, 4 h) and visualized by confocal microscopy. Numbers of puncta per root section were measured and data were shown as mean ± SD (n = 20). Experiments were biologically repeated for three times. Statistical significance was determined by Student's t-test (***, P < 0.001, compared to WT). (G) Observation (left) and quantitation (right) of GFP-ATG8e–labeled dots and ring-like structures in WT, gapc1 gapc2, pgk3, and gapc1 gapc2 pgk3 roots in the absence or presence of PA. Arabidopsis seedlings expressing GFP-ATG8e fusion protein were grown on 1/2 MS medium for 5 days, then transferred to 1/2 MS medium containing Con A (0.5 μM) for 8 h in the absence or presence of PA (10 μM) and visualized by confocal microscopy. Representative images were shown (bars = 50 μm) and numbers of puncta per root section were measured. Experiments were biologically repeated for three times and data were shown as mean ± SD (n = 20). Statistical significance was determined by Student's t-test (***, P < 0.001). (H) Observation (left) and quantitation (right) of GFP-ATG8e–labeled dots and ring-like structures in WT, GAPCs-OE, and PGK3-OE roots in the absence or presence of PLD inhibitor 1-butanol. Arabidopsis seedlings expressing GFP-ATG8e fusion protein were grown on 1/2 MS medium for 5 days, then transferred to 1/2 MS medium containing Con A (0.5 μM) for 8 h in the absence or presence of 1-butanol (0.4%) and visualized by confocal microscopy. Representative images were shown (bars = 50 μm) and numbers of puncta per root section were measured. Experiments were biologically repeated for three times and data were shown as mean ± SD (n = 20). Statistical significance was determined by Student's t-test (***, P < 0.001). (I) A hypothetical model illustrating how PA regulates autophagy in Arabidopsis. Under normal conditions, ATG6 activates VPS34 activity to form the PtdIns3P-enriched pre-autophagosomal structures (PAS), which organizes the site of autophagosome formation, and ATG3 catalyzes the formation of ATG8-PE conjugate to regulate the phagophore elongation. Under nitrogen or carbon starvation or other environmental stimuli, biosynthesis of phospholipids were rapidly promoted and increased phosphatidic acid (PA) enhances the interaction of glycolytic proteins GAPCs with ATG3 or PGK3 with ATG6 through direct binding GAPCs and PGK3, leading to the attenuated interactions of ATG3-ATG8e or ATG6-VPS34 thus suppressed autophagy.
We further phenotypically analyzed the mutants (gapc1, gapc2, gapc1 gapc2, pgk3-1, and pgk3-2) and overexpression lines (GAPC1-OE, GAPC2-OE, PGK3-OE#1, and PGK3-OE#2) under nutrient starvation conditions. When grown under normal conditions, mutants and overexpression lines are similar to WT; while upon nitrogen or carbon starvation, the mutants are more tolerant than WT, as reflected by the delayed yellowing of seedlings and increased chlorophyll contents (Figure S7E), whereas the overexpression lines were more sensitive to carbon and nitrogen starvation (Figure S7F). These results indicated that GAPCs and PGK3 negatively regulate the autophagy-associated response to nutrient starvation in Arabidopsis.
To demonstrate that GAPCs and PGK3 are required for the inhibited autophagy by PA, GFP-ATG8e, gapc1 gapc2 GFP-ATG8e, pgk3 GFP-ATG8e, and gapc1 gapc2 pgk3 GFP-ATG8e seedlings were treated with Con A in the absence or presence of PA. Analyses indicted that after PA treatment, the formation of GFP–labeled puncta and ring-like structures were decreased in WT, gapc1 gapc2, and pgk3 root cells, however, such decrease was not evident in the gapc1 gapc2 pgk3 background (Figure 7G). Further, Arabidopsis seedlings expressing GFP-ATG8e fusion protein in WT, GAPC1-OE, GAPC2-OE, PGK3-OE background were treated with Con A in the absence or presence of 1-butanol and analyses revealed that after 1-butanol treatment, the formation of GFP–labeled puncta and ring-like structures were obviously increased in WT root cells, while such increase was not observed in GAPC1-OE, GAPC2-OE, PGK3-OE background (Figure 7H). Taken together, these results indicated that GAPCs and PGK3 are involved in and necessary for the PA-mediated inhibition of autophagy.
Discussion
Eukaryotes have evolved a variety of sophisticated pathways to recycle the intracellular components [42,43]. In addition to the ubiquitin-26S proteasome system, which degrades the ubiquitinated proteins, mainly the short-lived proteins, another more vital pathway is the autophagy, which involves double-membrane vesicles engulfing and delivering protein aggregates, lipid bodies, long-lived proteins and damaged organelles to the vacuole for breakdown [44,45]. Previous study has illuminated how cells spatially tune synthesis and flux of phospholipids for autophagosome biogenesis during autophagy [46]. Here, we demonstrated that PA inhibits autophagy through binding glycolytic proteins GAPCs and PGK3 to promote their interactions with ATG3 and ATG6, which attenuates the interactions of ATG3-ATG8e or ATG6-VPS34 (Figure 7I). Our studies greatly expand the understanding of regulation of autophagy and provide evidence on the crucial roles of PA through binding to distinct target protein.
TRAF1a and TRAF1b regulate autophagy dynamics by modulating the ubiquitination and stability of ATG6 [47]. We demonstrated that PGK3 interacts with ATG6 to disrupt the ATG6-VPS34 interaction (Figure 5B), hence negatively regulates autophagy in Arabidopsis, providing new insights into the regulatory mechanisms of ATG6. NbGAPCs-NbATG3 interaction negatively regulates autophagy in N. benthamiana [34] and Cotton curlyleaf mudan virus βC1 protein induces autophagy by disrupting the NbGAPCs-NbATG3 interaction [48], our studies showed that GAPCs negatively regulate autophagy by interacting with ATG3 hence to disrupt the ATG3-ATG8e interaction in Arabidopsis (Figure 5A), revealing the molecular mechanism of how GAPCs-ATG3 interaction inhibits autophagy. Consistent with previous studies [1,49], structural analyses revealed that ATG3 binding to ATG8 was expected to be independent of both the LIR/AIM docking site (LDS) and ubiquitin-interacting motif (UDS), and binding of VPS34 to a ATG6 region encompassing the CC2 domain. In addition, GAPCs or ATG8e are linked to ATG3 via Y205, and PGK3 or VPS34 are linked to ATG6 via D285 and I287 (Figure 6), providing a structural basis for the mechanism of competitive inhibition.
As a cone-shaped lipid with a small and highly charged head-group, PA is not only the main precursor for phospholipids synthesis, but also an important second messenger molecule involving in a variety of signal transduction pathways through binding distinct target proteins [50,51]. In Arabidopsis, PA binds to PP2AA1 to promote its plasma membrane localization [52], interacts with WER to regulate its localization and function [53], binds to the core clock regulators LHY and CCA1 to modulate their functions [51], binds to PINOID to enhance its phosphorylation activity [54], and interacts with the ABI1-PP2C complex or the Gα subunit of heterotrimeric G protein to involve in the ABA-promoted stomatal closure [55]. We demonstrated that PA binds glycolytic proteins GAPCs and PGK3 (Figure 3A) to promote their interactions with ATG proteins. Structural analyses revealed that PA binds to a cavity inside the GAPCs or PGK3 protein. The amino acids in GAPCs or PGK3 proteins that can establish hydrogen bonds with PA are different from those in ATG3 or ATG6, which implies that GAPCs and PGK3 proteins bound to PA are more tightly structured, facilitating the formation of GAPCs-ATG3 or PGK3-ATG6 complexes and hence suppressing the ATG3-ATG8e or ATG6-VPS34 interactions. These provide clues of phospholipid molecules on autophagy and expand the mechanism of how PA regulates target proteins.
In animals, PLD1 produces distinct specie of PA that competitively binds to the FRB domain of MTOR, thus activating the MTOR kinase by displacing DEPTOR, an endogenous inhibitor of MTOR, leading to the inhibition of autophagy [20], which is consistent with that treatment with inhibitors and knockdown of PLD1 resulted in the accumulation of autophagic structures [56]. Our studies confirmed that autophagy of plant cells was significantly induced under 1-butanol or PLDζ inhibitor treatment, while suppressed upon PA treatment (Figures 1B, S1A). However, the effect of PLD inhibitors is moderately counteracted by the addition of PA (Figure S1B), suggesting that in addition to PA production, PLD inhibitors may affect autophagy through other pathways.
Consistent with previous study showing that PA inhibits autophagy by reducing the affinity of ATG6 towards the VPS34 complex [57], our studies demonstrated that PA binds PGK3 to promote its interactions with ATG6, which attenuates the interactions of ATG6-VPS34. Considering the conservation of PA production and autophagosome formation, these results provide informative clues for how PA reduces the affinity of ATG6 and VPS34 complexes in animals. In addition, regarding to the relatively conserved functions and regulatory mechanism of phospholipids in mammal and plant cells, PA inhibition of autophagy by binding glycolytic proteins further help to elucidate the underlying mechanism of how phospholipids regulate autophagy.
Under various conditions, particularly environmental stimuli and nutritional deficiency, PA level increases through hydrolyzing structural phospholipids by PLD. Considering the induced autophagy under stress conditions and nutritional deficiency, suppression of autophagy by PA through binding to GAPCs and PGK3 proteins forms a regulatory feedback loop that reduces the promoted autophagic activity, thus serving as a protective mechanism to maintain the autophagy at proper levels.
Materials and methods
Plant materials, growth condition and treatments
Arabidopsis thaliana lines used in this study were all with Colombia-0 (Col-0) background. The pldα1 and pldζ1 mutants, plants expressing 35S:GFP-ATG8e and PLDδ-overexpression lines were previously described [5,53,54,58]. Mutants pldβ1, pldδ, gapc1, gapc2, gapc1 gapc2, gapc3-1 and pgk3-2 were obtained from the Arabidopsis Information Resources Centre (ABRC) and verified by PCR. The gapc1 gapc2 and pgk3 mutants were crossed to generate the gapc1 gapc2 pgk3 triple mutants.
Seeds were surface sterilized, vernalized at 4°C for 3 days, and sown on half-strength Murashige and Skoog (MS) (PhytoTechnology Laboratories, M519) medium plate and vertically grown at 22°C under 16-h light/8-h dark photoperiod. For nitrogen starvation, 1-week-old seedlings were transferred to MS or nitrogen-deficient MS medium (PhytoTechnology Laboratories, M531) and grown under normal conditions for 5 days. For carbon starvation, 1-week-old seedlings were transferred to MS or sucrose-deficient MS medium and grown under continuous darkness for 7 days.
Constructs and recombinant protein expression
Most constructs used in this study were generated using Hieff Clone Plus One Step Cloning Kit (Yeasen Biotechnology, 10911ES20) according to the manufacturer<apos;>s instructions. Gene-specific primers with 15-bp extensions homologous to the corresponding vectors were listed in supplemental table 1. For transformation, full-length cDNAs of GAPC1, GAPC2, and PGK3 were amplified and subcloned into BglII/SpeI-digested pCambia1302 (Abcam, ab275760) to generate 35S:GAPC1-GFP, 35S:GAPC2-GFP and 35S:PGK3-GFP; full-length cDNAs of ATG3 and ATG6 were amplified and subcloned into BamHI/EcoRI-digested pCambia1306 to generate 35S:ATG3-mCherry and 35S:ATG6-mCherry. For yeast two-hybrid (Y2H) assay, full-length cDNAs of PGK3, ATG1a, ATG3, ATG5, ATG6, ATG7, ATG8e, ATG13a, ATG18a, and VPS34 were amplified and subcloned into EcoRI/BamHI-digested pGADT7 and pGBKT7. For the BiFC study, cDNAs of ATG3 and ATG6 were amplified and subcloned into SmaI/SalI-digested pCambia1300S-YN to generate the YN-protein fusion construct. cDNAs of GAPC1, GAPC2, and PGK3 were amplified and subcloned into SmaI/SpeI-digested pCambia2300S-YC to generate the protein-YC fusion constructs.
For protein expression, full-length cDNAs of ATG3 and ATG6 were amplified and subcloned into BamHI/EcoRI-digested pGEX-4 T-1 vector for expression of GST-tagged fusion proteins in Escherichia coli. Full-length cDNAs of GAPC1, GAPC2 and PGK3 were amplified and subcloned into BamHI/EcoRI-digested pET-28a for expression of the His-tagged fusion proteins in E. coli.
Measurement of chlorophyll contents
Chlorophyll contents were measured according to Xiao et al. [31]. Arabidopsis leaves were extracted by immersion in 1 mL of N,N-dimethylformamide (Sigma, 33120) for 48 h in dark at 4°C. Absorbance was determined at 664 and 647 nm, and total chlorophyll content was measured and normalized to gram fresh weight per sample.
Detection of autophagy
To determine the effect of PA on autophagosome formation, 5-day-old seedlings expressing eGFP-ATG8e grown on 1/2 MS medium (PhytoTechnology Laboratories, M519) were transferred to 1/2 MS medium containing PA (10 μM; Avanti Polar Lipids, 840875), 1-butanol (0.4%; Sigma, 281549), or PLDζ inhibitor (200 nM; Avanti Polar Lipids, VU0285655-1), in the absence or presence of concanamycin A (Con A, 0.5 μM; Sangon Biotech, A601179) and grown under normal conditions. Primary root cells were observed using a Leica SP8 laser scanning confocal microscope (Leica, Germany), and the excitation and emission wavelengths were 488 and 507 nm, respectively.
For monodansylcadaverine (MDC) staining, seedlings were treated for the indicated times and subsequently incubated in 0.05 mM MDC (Sigma, D4008) in phosphate-buffered saline (PBS; Ambion, AM9625) for 10 min, followed by three times washes with PBS at room temperature. Primary root cells were observed using a Leica SP8 laser scanning confocal microscope with a DAPI-specific filter. The excitation and emission wavelengths for MDC were 345 and 455 nm, respectively.
Transmission electron microscopy (TEM) analyses
Leaf sections (1 mm2) cut from 3-week-old Arabidopsis seedlings were vacuum-infiltrated and fixed immediately with 2.5% glutaraldehyde (Sigma, G5882) in 0.1 M PBS overnight at 4°C. Samples were washed three times with PBS, post-fixed with 1% OsO4 (Sigma, O5500) rinsed three times with PBS again, dehydrated in a graded ethanol series, replaced ethanol with acetone, and embedded in SPI-PON 812 resin (SPI Science, 90529–77-4). The ultrathin sections stained with 2% (w/v) uranyl acetate (Polysciences, 21447–25) and 2.6% (w:v) lead citrate (Sigma, 15326). Observation and capturing of images were performed by using a transmission electron microscope (TEM, Hitachi H-7650, Japan) at 80 kV.
Protein extraction and western blot analyses
Total proteins were extracted from N. benthamiana leaves using an ice-cold RIPA lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100 [Sigma, X100], 1% sodium deoxycholate [Sigma, D6750], 0.1% SDS) supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF; Roche, 10837091001). The homogenates were placed on ice for 30 min, then centrifuged (12,000 g) at 4°C for 15 min. The supernatant was transferred to a new microfuge tube before electrophoresis. For immunoblot analyses, total proteins were separated by 12% SDS–PAGE, electroblotted to a nitrocellulose membrane (Biosharp, 68100230) and analyzed using anti-GFP (Abcam, ab290) antibodies.
Yeast two-hybrid (Y2H), bimolecular fluorescence complimentation (BiFC) and subcellular localization analyses
Gal4-based yeast two-hybrid assays were performed according to the manufacturer<apos;>s instructions (Clontech, PT4084-1). Vectors pGADT7 and pGBKT7 were used to generate the prey and bait constructs, respectively. Yeast strain AH109 containing different pairs of constructs were grown on synthetic dropout (SD) medium absence of tryptophan, leucine, and histidine.
For BiFC assay, cDNAs of ATG3 and ATG6 or GAPC1, GAPC2 and PGK3 were fused to the N terminus of YFP or C terminus of YFP, transformed to A. tumefaciens strain GV3101 and infiltrated the leaves of 5-week-old N. benthamiana plants. Infiltrated plants were grown in dark for 48 h, and fluorescence signals were observed using Leica SP8 laser scanning confocal microscope (Leica, Germany). For PA treatment, the leaves were infiltrated with PA (10 mM) for 8 h before observations.
For subcellular localization analyses, cDNAs of ATG3 or ATG6 was fused to mCherry, cDNAs of GAPC1, GAPC2 or PGK3 was fused to GFP, transformed to A. tumefaciens strain GV3101 and infiltrated the leaves of 5-week-old N. benthamiana plants. For concanamycin A (Con A) treatment, the samples were additionally infiltrated with Con A (0.5 μM) for 8 h before observation. ER marker HDEL-mCherry was transformed to investigate whether the proteins interact with each other at ER.
Protein expression, purification, GST affinity isolation and Co-IP assays
GST, GST-ATG3, GST-ATG6, GAPC1-His, GAPC2-His, PGK3-His, His-ATG3-mCherry, His-ATG6-mCherry, and SNAP-His-ATG8e were recombinant expressed using E. coli strain BL21 (DE3). Expression of protein was induced by supplementing isopropyl β-d-1-thiogalactopyranoside (IPTG, 0.5 mM; Sangon Biotech, A600168) at 28°C for 4 h. Cell lysates were used for affinity purification using Ni-NTA His-binding resin according to the manufacturer<apos;>s instructions (Yeasen, 20502ES60). Eluted proteins were further purified using a HiLoad 26/60 Superdex 200 PG column (GE Healthcare, 17–1071-01). Resultant proteins were flash-frozen and stored at −80°C.
GST-binding resin (100 μl; Yeasen, 20508ES10) were used to pull down protein complexes in binding buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, 1× protease cocktail inhibitors). After binding at 4°C for 3 h, beads were washed five times with washing buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 1 mM EDTA), boiled in 2× SDS sample buffer, and analyzed by immunoblot using anti-His or anti-GST antibody.
For protein extraction and immunoprecipitation, total cell lysates were prepared in lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA, 0.5% Triton X-100) containing a protease inhibitor cocktail (Sigma, P8340), then incubated with anti-GFP magnetic microbeads overnight at 4°C. Samples were washed three times in wash buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA with 1× Complete Protease Inhibitor Cocktail) and then eluted by boiling in 2× SDS sample buffer. Samples were separated by SDS-PAGE and analyzed by immunoblot using appropriate antibodies.
Lipid–protein binding and liposomal binding
Lipid–protein blot overlay assays were performed using PIP strips or PIP arrays. Proteins were recombinant expressed in E. coli and purified. Nitrocellulose membranes containing immobilized lipids (PIP strips or PIP arrays; Thermo Fisher Scientific, P23751) were incubated in blocking solution (1× TBST [50 mM Tris-HCl, 150 mM NaCl, 0.1% [v:v] Tween-20 [Sigma, P9416], pH 7.6, 3% [w:v] BSA [Sangon Biotech, A602448) for 1 h. Membranes were incubated overnight in 10 ml of 1× TBST containing 60 μg of recombinant proteins. After washing (3× for 10 min) using 1× TBST, the membranes were incubated at room temperature with 1:3000 anti-His antibodies (Abmart, M30111) for 2 h and rinsed three times with 10 ml 1× TBST. The signals were detected following the ECL Plus immunoblot method.
Liposomal binding was performed according to previously described [53] with slight modifications. Briefly, dioleoyl PC and dioleoyl PA were mixed in the molar ratio of 3:1 and then dried with nitrogen gas. Lipids were rehydrated for 1 h in extrusion buffer (250 mM raffinose [Sigma, R0250], 25 mM Tris-HCl, pH 7.5, 1 mM DTT). The extrusion buffer was extruded by the liposome extruder through the polycarbonate membrane (0.2 μm pore size) to produce an optically clear suspension of small unilamellar liposomes. The small unilamellar liposomes was resuspended in binding buffer (25 mM Tris-HCl, pH 7.5, 1.25 mM potassium chloride, 0.5 mM EDTA, 1 mM DTT). Different concentrations of liposomes were incubated with the purified proteins for 1 h and the liposomes containing proteins were centrifuged (14,000 g) for 30 min. The pellets were washed twice with binding buffer and the supernatant transferred to a new tube. The pellets and supernatant resuspended in SDS-PAGE sample buffer for immunoblotting.
Fluorescence resonance energy transfer with fluorescence lifetime imaging microscopy (FRET-FLIM) assays
FRET-FLIM experiments were carried out by using a Leica STELLARIS laser scanning confocal microscope (Leica, Germany) assembly of white laser and the fluorescence lifetime system. The GAPC1-GFP, GAPC2-GFP, PGK3-GFP, GFP-ATG8e, and ATG6-GFP alone were used as a donor and whose fluorescence lifetime were measured as the negative control. The excitation and emission wavelengths were 488 and 507 nm, respectively. Detailed experimental principle and conditions of FRET-FLIM are previously described [59]. All measurements were taken from whole field images expressing fluorescent protein at similar levels.
Structure modeling
To model a structure of individual complex subunits in this study, we used threading algorithms, namely Swiss-Model [60]. For subsequent protein-small molecule or protein-protein docking and analyzes, we used the models calculated using the Swiss-model program as the obtained models had the best ProSA score [61]. The binding interfaces of the complex in this study were predicted by semi flexible docking using Autodock Vina [38] or a hybrid docking method using the HDOCK algorithm [39]. To analyze amino acid conservation, we employed the Consurf server with the default parameters [62]. The structures were visualized in the ChimeraX or PyMOL program.
Statistical analyses
The significance of difference between two groups was determined by using a Student's t-test. The level of statistical significance is indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Supplementary Material
Acknowledgments
We thank Prof. Wen-Hua Zhang (Nanjing Agricultural University, China) and Prof. Jin-Xing Lin (Beijing Forestry University, China) for providing plda1 mutant and PLDδ-overexpression line, Dr. Peng-Chao Hao for help on western blot analyses, and Dr. Lin-Liang Yin and Dr. Wen-Jun Cai (engineer of Olympus) for their help on laser confocal microscope observation.
Funding Statement
The study was supported by National Natural Science Foundation of China [NSFC, 91954206, 31721001], “Ten-Thousand Talent Program” and Collaborative Innovation Center of Crop Stress Biology, Henan Province.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Marshall RS, Hua Z, Mali S, et al. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 2019;177(3):766–781. DOI: 10.1016/j.cell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Marshall RS, Vierstra RD.. Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol. 2018;69:173–208. [DOI] [PubMed] [Google Scholar]
- [3].Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. [DOI] [PubMed] [Google Scholar]
- [4].Chen L, Liao B, Qi H, et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy. 2015;11(12):2233–2246. DOI: 10.1080/15548627.2015.1112483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guan B, Lin Z, Liu D, et al. Effect of waterlogging-induced autophagy on programmed cell death in Arabidopsis roots. Front Plant Sci. 2019;10:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hanaoka H, Noda T, Shirano Y, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129(3):1181–1193. DOI: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5(7):954–963. [DOI] [PubMed] [Google Scholar]
- [8].Xiong Y, Contento AL, Bassham DC. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy. 2007;3(3):257–258. [DOI] [PubMed] [Google Scholar]
- [9].Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. [DOI] [PubMed] [Google Scholar]
- [10].Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Popelka H, Klionsky DJ. Multiple structural rearrangements mediated by high-plasticity regions in Atg3 are key for efficient conjugation of Atg8 to PE during autophagy. Autophagy. 2021;17(8):1805–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Noda NN, Fujioka Y, Hanada T, et al. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 2013;14(2):206–211. DOI: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davis S, Wang J, Ferro-Novick S. Crosstalk between the secretory and autophagy pathways regulates autophagosome formation. Dev Cell. 2017;41(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Obara K, Sekito T, Niimi K, et al. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283(35):23972–23980. DOI: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li J, Yu F, Guo H, et al. Crystal structure of plant PLDα1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D. Cell Res. 2020;30(1):61–69. DOI: 10.1038/s41422-019-0244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kolesnikov YS, Nokhrina KP, Kretynin SV, et al. Molecular structure of phospholipase D and regulatory mechanisms of its activity in plant and animal cells. Biochemistry. 2012;77(1):1–14. DOI: 10.1134/S0006297912010014. [DOI] [PubMed] [Google Scholar]
- [17].Lin DL, Yao HY, Jia LH, et al. Phospholipase D-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN-FORMED2. New Phytol. 2020;226(1):142–155. DOI: 10.1111/nph.16330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yao HY, Xue HW. Phosphatidic acid plays key roles regulating plant development and stress responses. J Integr Plant Biol. 2018;60(9):851–863. [DOI] [PubMed] [Google Scholar]
- [19].Fang Y, Vilella-Bach M, Bachmann R, et al. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294(5548):1942–1945. DOI: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- [20].Yoon MS, Rosenberger CL, Wu C, et al. Rapid mitogenic regulation of the mTORC1 inhibitor, DEPTOR, by phosphatidic acid. Mol Cell. 2015;58(3):549–556. DOI: 10.1016/j.molcel.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Plaxton WC. The organization and regulation of plant glycolysis. Ann Rev Plant Physiol Plant Mol Biol. 1996;47:185–214. [DOI] [PubMed] [Google Scholar]
- [22].Rosa-Téllez S, Anoman AD, Flores-Tornero M, et al. Phosphoglycerate kinases are co-regulated to adjust metabolism and to optimize growth. Plant Physiol. 2018;176(2):1182–1198. DOI: 10.1104/pp.17.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zaffagnini M, Fermani S, Costa A, et al. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci. 2013. 12;4:450. DOI: 10.3389/fpls.2013.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li R, Qiu Z, Wang X, et al. Pooled CRISPR/Cas9 reveals redundant roles of plastidial phosphoglycerate kinases in carbon fixation and metabolism. Plant J. 2019;98(6):1078–1089. DOI: 10.1111/tpj.14303. [DOI] [PubMed] [Google Scholar]
- [25].Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;42(4):598–608. [DOI] [PubMed] [Google Scholar]
- [26].Ichimura Y, Imamura Y, Emoto K, et al. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279(39):40584–40592. DOI: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- [27].Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010;62(3):483–493. [DOI] [PubMed] [Google Scholar]
- [28].Doelling JH, Walker JM, Friedman EM, et al. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277(36):33105–33114. DOI: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- [29].Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;42(4):535–546. [DOI] [PubMed] [Google Scholar]
- [30].Fan L, Zheng S, Wang X. Antisense suppression of phospholipase D alpha retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell. 1997;9(12):2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xiao S, Gao W, Chen QF, et al. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 2010;22(5):1463–1482. DOI: 10.1105/tpc.110.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim SC, Guo L, Wang X. Phosphatidic acid binds to cytosolic glyceraldehyde-3-phosphate dehydrogenase and promotes its cleavage in Arabidopsis. J Biol Chem. 2013;288(17):11834–11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Testerink C, Dekker HL, Lim ZY, et al. Isolation and identification of phosphatidic acid targets from plants. Plant J. 2004;39(4):527–536. DOI: 10.1111/j.1365-313X.2004.02152.x. [DOI] [PubMed] [Google Scholar]
- [34].Han S, Wang Y, Zheng X, et al. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell. 2015;27(4):1316–1331. DOI: 10.1105/tpc.114.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sun SY, Chao DY, Li XM, et al. OsHAL3 mediates a new pathway in the light-regulated growth of rice. Nat Cell Biol. 2009;11(7):845–851. DOI: 10.1038/ncb1892. [DOI] [PubMed] [Google Scholar]
- [36].Zhuang X, Chung KP, Luo M, et al. Autophagosome biogenesis and the endoplasmic reticulum: a plant perspective. Trends Plant Sci. 2018;23(8):677–692. DOI: 10.1016/j.tplants.2018.05.002. [DOI] [PubMed] [Google Scholar]
- [37].Long Y, Stahl Y, Weidtkamp-Peters S, et al. In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature. 2017;548(7665):97–102. DOI: 10.1038/nature23317. [DOI] [PubMed] [Google Scholar]
- [38].Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yan Y, Tao H, He J, et al. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020;15(5):1829–1852. DOI: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- [40].Liu F, Hu W, Li F, et al. AUTOPHAGY-RELATED14 and its associated phosphatidylinositol 3-kinase complex promote autophagy in Arabidopsis. Plant Cell. 2020;32(12):3939–3960. DOI: 10.1105/tpc.20.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Henry E, Fung N, Liu J, et al. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 2015;11(4):e1005199. DOI: 10.1371/journal.pgen.1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bassham DC. Function and regulation of macroautophagy in plants. Biochim Biophys Acta. 2009;1793(9):1397–1403. [DOI] [PubMed] [Google Scholar]
- [43].Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10(6):385–397. [DOI] [PubMed] [Google Scholar]
- [44].Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17(9):526–537. [DOI] [PubMed] [Google Scholar]
- [45].Masclaux-Daubresse C, Chen Q, Havé M. Regulation of nutrient recycling via autophagy. Curr Opin Plant Biol. 2017;39:8–17. [DOI] [PubMed] [Google Scholar]
- [46].Schütter M, Giavalisco P, Brodesser S, et al. Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell. 2020;180(1):135–149. DOI: 10.1016/j.cell.2019.12.005. [DOI] [PubMed] [Google Scholar]
- [47].Qi H, Xia FN, Xie LJ, et al. TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell. 2017;29(4):890–911. DOI: 10.1105/tpc.17.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ismayil A, Yang M, Haxim Y, et al. Cotton leaf curl multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with glyceraldehyde-3-phosphate dehydrogenases. Plant Cell. 2020;32(4):1124–1135. DOI: 10.1105/tpc.19.00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rostislavleva K, Soler N, Ohashi Y, et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350(6257):7365. DOI: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang X, Devaiah SP, Zhang W, et al. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45(3):250–278. DOI: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- [51].Kim SC, Nusinow DA, Sorkin ML, et al. Interaction and regulation between lipid mediator phosphatidic acid and circadian clock regulators. Plant Cell. 2019;31(2):399–416. DOI: 10.1105/tpc.18.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gao HB, Chu YJ, Xue HW. Phosphatidic acid (PA) binds PP2AA1 to regulate PP2A activity and PIN1 polar localization. Mol Plant. 2013;6(5):1692–1702. [DOI] [PubMed] [Google Scholar]
- [53].Yao H, Wang G, Guo L, et al. Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis. Plant Cell. 2013;25(12):5030–5042. DOI: 10.1105/tpc.113.120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang P, Shen L, Guo J, et al. Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED2 auxin efflux transporter in response to salt stress. Plant Cell. 2019;31(1):250–271. DOI: 10.1105/tpc.18.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mishra G, Zhang W, Deng F, et al. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science. 2006;312(5771):264–266. DOI: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- [56].Bae EJ, Lee HJ, Jang YH, et al. Phospholipase D1 regulates autophagic flux and clearance of α-synuclein aggregates. Cell Death Differ. 2014;21(7):1132–1141. DOI: 10.1038/cdd.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jang YH, Choi KY, Min DS. Phospholipase D-mediated autophagic regulation is a potential target for cancer therapy. Cell Death Differ. 2014;21(4):533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Xing J, Li X, Wang X, et al. Secretion of Phospholipase Dδ Functions as a Regulatory Mechanism in Plant Innate Immunity. Plant Cell. 2019;31(12):3015–3032. DOI: 10.1105/tpc.19.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Alvarez LAJ, Widzgowski B, Ossato G, et al. SP8 FALCON: A novel concept in fluorescence lifetime imaging enabling video-rate confocal FLIM. Nat Methods. 2019;16:1069–1071.31619775 [Google Scholar]
- [60].Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ashkenazy H, Abadi S, Martz E, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


