ABSTRACT
At a time when complex diseases affect globally 280 million people and claim 14 million lives every year, there is an urgent need to rapidly increase our knowledge into their underlying etiologies. Though critical in identifying the people at risk, the causal environmental factors (microbiome and/or pollutants) and the affected pathophysiological mechanisms are not well understood. Herein, we consider the variations of autophagy-related (ATG) genes at the heart of mechanisms of increased susceptibility to environmental stress. A comprehensive autophagy genomic resource is presented with 263 single nucleotide polymorphisms (SNPs) for 69 autophagy-related genes associated with 117 autoimmune, inflammatory, infectious, cardiovascular, neurological, respiratory, and endocrine diseases. We thus propose the term ‘autophagopathies’ to group together a class of complex human diseases the etiology of which lies in a genetic defect of the autophagy machinery, whether directly related or not to an abnormal flux in autophagy, LC3-associated phagocytosis, or any associated trafficking. The future of precision medicine for common diseases will lie in our ability to exploit these ATG SNP x environment relationships to develop new polygenetic risk scores, new management guidelines, and optimal therapies for afflicted patients.
Abbreviations: ATG, autophagy‐related; ALS-FTD, amyotrophic lateral sclerosis-frontotemporal dementia; ccRCC, clear cell renal cell carcinoma; CD, Crohn disease; COPD, chronic obstructive pulmonary disease; eQTL, expression quantitative trait loci; HCC, hepatocellular carcinoma; HNSCC, head and neck squamous cell carcinoma; GTEx, genotype-tissue expression; GWAS, genome-wide association studies; LAP, LC3-associated phagocytosis; LC3-II, phosphatidylethanolamine conjugated form of LC3; LD, linkage disequilibrium; LUAD, lung adenocarcinoma; MAF, minor allele frequency; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; NSCLC, non-small cell lung cancer; OS, overall survival; PtdIns3K CIII, class III phosphatidylinositol 3 kinase; PtdIns3P, phosphatidylinositol-3-phosphate; SLE, systemic lupus erythematosus; SNPs, single-nucleotide polymorphisms; mQTL, methylation quantitative trait loci; ULK, unc-51 like autophagy activating kinase; UTRs, untranslated regions; WHO, World Health Organization.
KEYWORDS: Autophagy, cancers, diseases, eQTL, pollutants/exposomics, polymorphism, prognosis, risk, susceptibility, theragnosis
The rising incidence of complex illnesses and their costs have revolutionized basic research needs and approaches, but also patient management, and societal needs. Between 70 to 90% of the risk of developing a disease is due to the air we breathe, the water we drink, the diet we eat, and the surroundings in which we work and live [1]. Visibly polluted, infected, or not, the fact remains that we are now more than ever exposed to environmental risks. Thus, an unhealthy environment can be considered as a pandemic, affecting 280 million people worldwide and claiming 14 million deaths every year from hundreds of diseases, including neurodegenerative, autoimmune, inflammatory illnesses, and cancer [2]. Though critical in identifying the people at risk, the causal environment components (pathogens and/or pollutants) and the compromised physiological mechanisms are not yet well understood. When it comes to environmental stress, we are not equal. However, presented with the same environment, regardless of the pathogens and pollutants we are challenged with, only a small fraction of individuals will develop pathologies. We thus reasoned that the efficiency of our inherited “environmental response machinery” may determine our susceptibility to illness.
Introduction
A. Overview of the autophagy pathway: an immediate cellular response to environmental challenges
Among the many mechanistic pathways, we focus on macroautophagy (hereafter referred to as autophagy), a homeostatic pathway that also provides an immediate adaptive cellular response to environmental injury that ensures cell repair [3]. As suggested by its name (auto = self; phagy = eating), autophagy enables, in all eukaryotic cell types, the turnover of all organelles and most long-lived proteins by a pathway that begins with the formation of a double-membrane compartment, termed a “phagophore,” which captures these components from the cytosol. The phagophore expands into a completed vesicle, an “autophagosome.” Subsequently, the autophagosome rapidly fuses with a lysosome to become an “autolysosome,” in which the content is finally degraded. Successful completion of autophagy requires the coordinated orchestration of more than 69 different autophagy-related (ATG) and other proteins as well as regulators acting at different steps of the process, namely:
ULK (unc-51 like autophagy activating kinase) complex (ULK1, ULK2, ATG13, RB1CC1/FIP200, and ATG101) initiates the induction step.
Once activated, ULK1 phosphorylates BECN1 and ATG14, two components of the class III phosphatidylinositol 3-kinase (PtdIns3K CIII) complex (PIK3C3/VPS34, PIK3R4/VPS15, ATG14, BECN1, and NRBF2), thereby enhancing PIK3C3 activity and phagophore membrane formation.
WIPI1 (WD repeat domain, phosphoinositide interacting 1) and WIPI2 bind phosphatidylinositol-3-phosphate (PtdIns3P), generated by the PtdIns3K CIII; these effectors then recruit ATG16L1 that mediates phagophore expansion through ubiquitination-like reactions.
In the first ubiquitination-like reaction, ATG5 and ATG12 are conjugated to each other in the presence of ATG7 and ATG10. The attachment of the complex containing ATG5, ATG12, and ATG16L1 on the phagophore membrane induces the second complex to covalently conjugate phosphatidylethanolamine to LC3 (LC3-II), which facilitates closure of the phagophore into the autophagosome.
ATG9 (the ATG9-ATG2-WIPI1/Atg18 complex) is another factor essential for expanding the phagophore, which cycles between endosomes, the Golgi, and the phagophore; ATG9 is a lipid scramblase that functions along with ATG2 to transfer lipid components for membrane expansion.
ATG4 first primes LC3 for conjugation, and later removes LC3-II from the outer surface of newly formed autophagosomes; LC3 on the inner surface is eventually degraded when the autophagosome fuses with a lysosome.
The fusion between an autophagosome and a lysosome involves several proteins, including LAMP, RAB7, and the second complex of PtdIns3K CIII (PIK3R4, PIK3C3, UVRAG, and BECN1), resulting in vesicle breakdown and cargo degradation into autolysosomes by lysosomal hydrolases.
Under both baseline conditions and times of stress, several autophagic receptors (including SQSTM1/p62, NBR1, CALCOCO2/NDP52, OPTN, DRAM1, WDFY3/ALFY, and TOLLIP) are recruited to recognize and facilitate the selective elimination of ubiquitinated protein aggregates and damaged/dysfunctional organelles by sequestration within autophagosomes; these aggregates and organelles would otherwise accumulate during the life of the cell (Figure 1).
Figure 1.
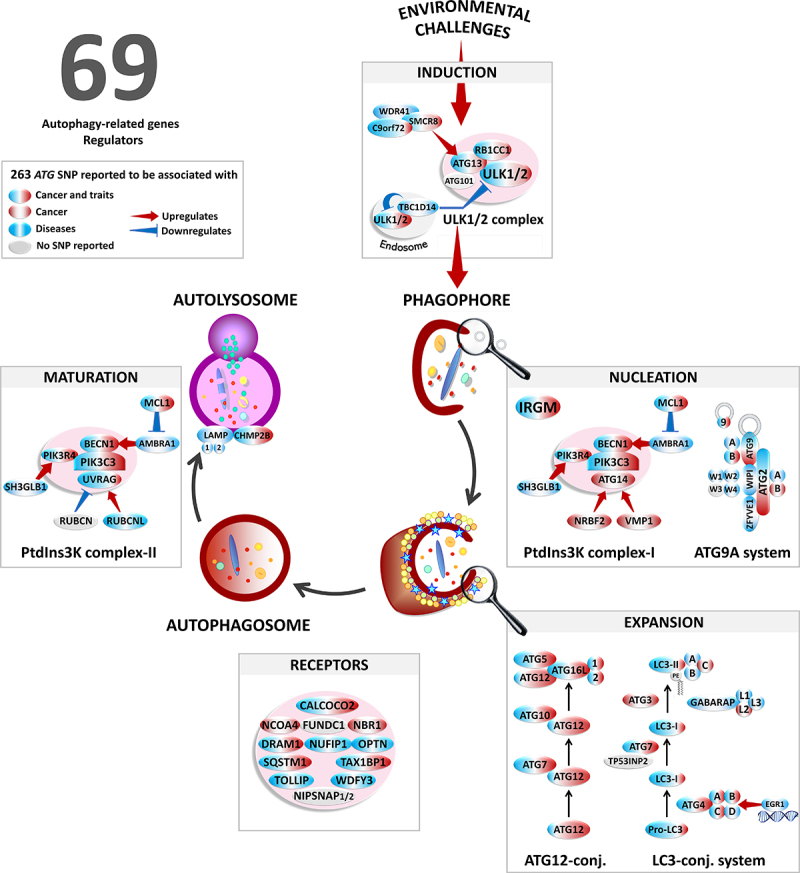
Overview of the autophagy pathway in response to environmental challenges. Schematic illustrating the molecular machinery of autophagy including the major autophagy-related proteins and complexes. Associations of ATG polymorphisms with cancers (red) and non-cancer diseases (blue).
Throughout development and life, autophagy is required for the maintenance, self-renewal, and differentiation of stem-like cells, such as in the hematopoietic system. Likewise, such an intracellular ‘renewal’ (i.e., recycling) process also plays an essential role in determining the homeostasis, functionality, and longevity of post-mitotic cells such as cardiomyocytes and neurons. In a state of emergency, exposure of all cell types to environmental challenges as varied as nutrient starvation, pathogens, and chemical pollutants massively and transiently upregulates the entire autophagy machinery to repair cells and meet their energy needs. Such an intricate interplay between autophagy and the environment is essential for cell/individuals’ adaptation to changing conditions and, when impaired, predisposes to disease.
B. The long and winding road from ATG variations to disease susceptibility
Over the last twenty years, the completion of the human genome project and the remarkable progress of the genome-wide association study (GWAS) have accelerated the identification of hundreds of susceptibility loci for diseases, some of which concern autophagy-related genes. In December 2021, a PubMed search for “autophagy AND (susceptibility OR polymorphism OR SNP OR variant OR variation OR mutation)” yielded 9,076 entries. 243 relevant studies totaling 3,504,075 participants (2 million patients and 1.5 million controls) – 77 are GWAS – have identified 263 common SNPs for 69 ATG genes associated with 185 autoimmune, inflammatory, cardiovascular, neurological, and lung diseases and traits; all are common complex diseases.
We conducted a comprehensive survey of this ATG SNP list using the dbSNP (https://www.ncbi.nlm.nih.gov/snp/) [4], LitVar (https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/LitVar/) [5], HaploReg (https://pubs.Broad-institute.org/mammals/haploreg/haploreg.php, v4.1) [6], and GTEx (Genotype‐Tissue Expression; https://www.gtexportal.org/home/v8-release) [7] public databases:
i) The variety of defects in autophagy that are involved in human pathogenesis resulting in disease initiation, progression, and treatment efficacy is considerable. Strikingly, most diseases are highly polygenic, influenced by many ATG variants, each giving a small effect.
ii) Rather than being a mere inventory of correlations, several ATG variations have pleiotropic effects affecting multiple ‘apparently’ unrelated diseases. Likewise, many of these SNPs have protective effects, allowing them to be maintained in the population. This antagonistic pleiotropy is undoubtedly related to the essential, ubiquitous, and double-edged roles of autophagy in pathologies [8]. These two features, polygenic and pleiotropic inheritances, make it challenging to identify causal SNPs [9] (see Tables S1-S7).
iii) The SNP-disease correlations are even more confusing as 90% of the diseases associated with ATG alleles localize to non-coding genomic regions (intron, intergenic, 5’ and 3’ untranslated regions [UTRs]). Nearly one-third are regulatory SNPs, also called expression quantitative trait loci (eQTL), associated with a difference in transcript abundance. As such, these non-coding regulatory variations can dramatically affect the rate of autophagy flux and how individuals respond to environmental stimuli. However, very few functional genomic studies have so far investigated this ATG SNP x environmental interaction, which represents another critical bottleneck for SNP-disease correlations.
With this in mind, the present review aims to provide the scientific and clinical community with a comprehensive autophagy genomic resource to embrace health risk assessment, precision medicine, and new potential therapeutic opportunities.
Pleiotropy: opening pandora’s box
As autophagy is ubiquitous and essential for the survival of all cell types, its dysfunction can manifest like a ‘constellation’ of symptoms that affect any organ. Within each organ, we will briefly outline here the range of diseases associated with autophagy SNPs (Figures 2–5, Figures S1-S3, and Tables S2-S7). In these figures, panel A will emphasize that diseases can be inflammatory, autoimmune, degenerative (right panel), or associated with cancer (left), and influenced by environmental cues, regardless of their nature. Panel B will help to visualize the autophagic genes, the variation in which may give rise to similar clinical manifestations, i.e., the disease module. In keeping with this, panel C, when included, will present some preclinical mouse models linking a dysfunction in autophagy to the pathogenesis of major complex diseases (see a review [8] for more details on C).
Figure 2.
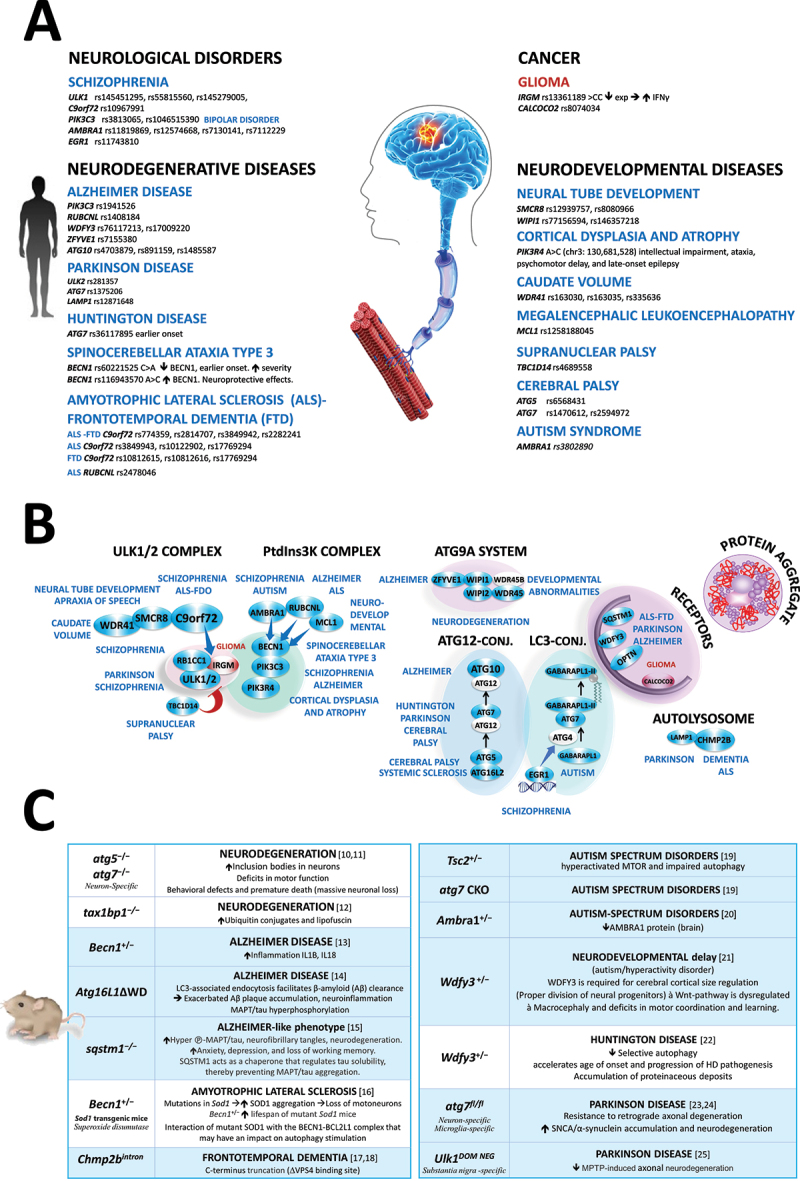
Deficiency in autophagy in human central nervous system diseases. (A) Summary of autophagy-related gene variations. (B) Steps of the autophagy pathway affected by SNPs. (C) Phenotype of autophagy-deficient mousemodels [10–25] -MAPT: phosphorylated-MAPT/tau.
Figure 3.
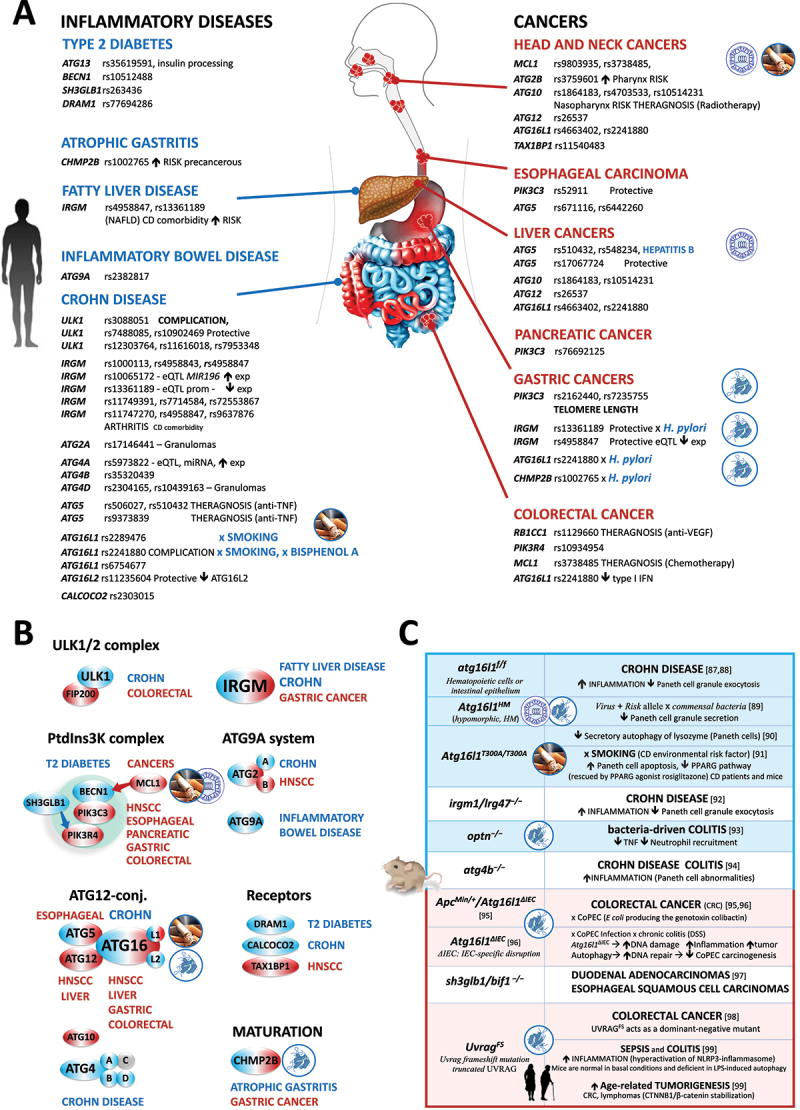
Deficiency in autophagy in human gastrointestinal disorders. (A) Summary of autophagy-related gene variations. (B) Steps of the autophagy pathway affected by SNPs. (C) Phenotypes of autophagy-deficient mouse models [87–93,93–99].
Figure 4.
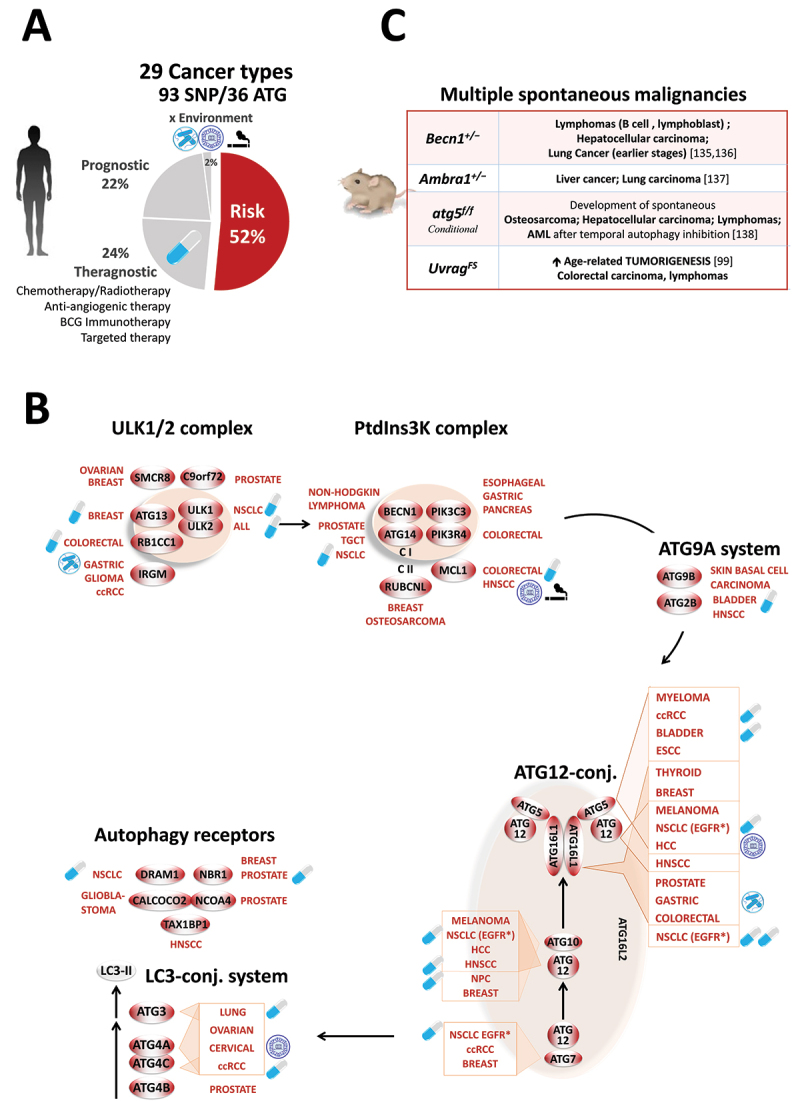
Landscape of ATG polymorphisms in human cancers. (A) Percentage of ATG polymorphisms for 29 cancer types in terms of theragnosis, prognosis, and risk, and relation with environmental factors. (B) Steps of the autophagy pathway affected by SNPs. ESCC, esophageal squamous cell carcinoma. (C) Phenotypes of autophagy-deficient mouse models [99,135–138].
Figure 5.
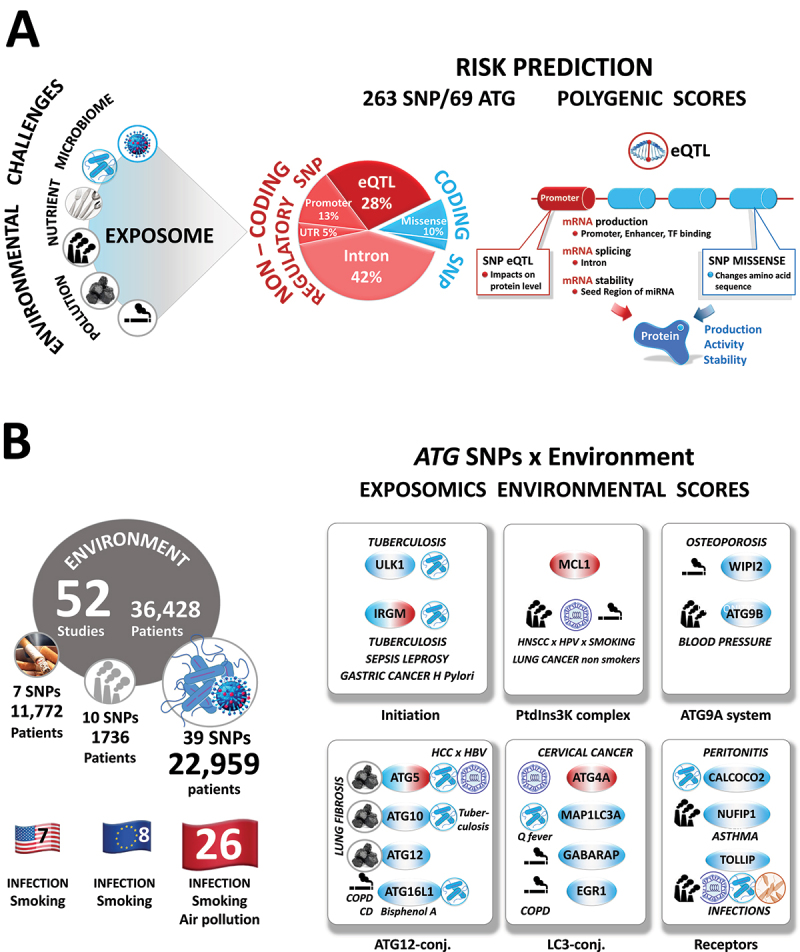
Biomarkers of exposure and, ultimately, of risk assessment and clinical outcome. (A) Right. Distribution (in percentage, left) and predicted functional consequences (right) of 219 coding and non-coding regulatory ATG SNPs. Left, Number of studies, totaling the number of patients, ATG genes, and SNP per Europe, United States, and Asia. Note that whereas 70 to 90% of the risk of developing a disease is due to the environment, only 13% of studies on autophagy gene SNPs have included the environment as a trigger or exacerbating factor. Likewise, air pollution is the fourth most prevalent deadly risk factor worldwide and, so far, most of the studies focused on infection. TF, transcription factor. Related to: (B) The concept of the exposome and atlas of the ‘autophagy gene × environmental’ interactions in the susceptibility of complex human diseases. Because of the long latency period, exposure to a causal agent/mixture typically occurs years to decades before disease diagnosis. The exposome is a unique marker that characterizes the totality of trace chemicals resulting from different routes (internal and external) and times of exposure over the lifetime of an individual.
A. Autophagy and neurodegenerative diseases
Roughly 40 million individuals suffer worldwide from Alzheimer disease and related conditions, a burden that is expected to explode as the population ages. To tackle this public health emergency, a treatment that prevents or delays the development of these devastating diseases is a critical unmet need. Critical for therapeutic intervention, the current consensus is that autophagy is essential to prevent the aging of long-lived post-mitotic neurons. A genetic defect in autophagy may initiate these diseases, alone or with concurrent aggregate-prone mutations [8]. Over the past five decades, we have learned how autophagy degrades selectively damaged organelles and protein aggregates that would otherwise accumulate during life. Beyond these cargos, autophagy is a quality-control system that degrades the aggregate-prone mutated proteins associated with several neurodegenerative diseases (such as Huntington disease, spinocerebellar ataxia, Parkinson disease, amyotrophic lateral sclerosis [ALS], and frontotemporal dementia [FTD]).
Variant discovery. To date, 30 common risk variants have been associated with various neurological disorders, from developmental to neurodegenerative diseases (Figure 2A). The most striking findings include: i) the high polygenic nature of these diseases linked with five to seven ATG variants: For example, Alzheimer disease (PIK3C3 [26], RUBCNL [27], ZFYVE1/DFCP1 [28], WDFY3 [29], and ATG10 [30]), schizophrenia (ULK1 [31], RB1CC1 [32,33], C9orf72 [34], PIK3C3 [35], AMBRA1 [36,37], and EGR1 [38]) and Parkinson disease (ULK2 [39], ATG7 [40], and LAMP1 [41]) among others. ii) Likewise, there is also substantial genetic pleiotropy across various neurological traits: AMBRA1 with schizophrenia [36,37], and autism [42], PIK3C3 with schizophrenia [35] and bipolar disorder [35], ATG7 with cerebral palsy [43], Huntington disease [44], and Parkinson disease [40] (for more details see the Tables S2-S7).
From genetics to biology. Heritability may result from both common and rare genetic variants that affect all steps of the autophagy pathway, from autophagosome formation and substrate sequestration to lysosomal degradation (Figure 2B). For instance, ALS-FTD-linked variations are rare missense, nonsense, and truncating mutations that target the ubiquitin-binding domains of the autophagy receptors SQSTM1 [45], OPTN [46], and UBQLN2 [47], thus compromising the binding and the clearance of ubiquitinated aggregates. Similarly, other mutations are missense or whole-gene deletions of ULK1 [31], WIPI1 [48], WIPI2 [49], WDR45 [50], and GABARAPL1 [51] that impair the formation of autophagosomes. Along these lines, several autophagy-deficient mouse models recapitulate features of neurodegenerative diseases 10-25– (Figure 2C). From a clinical perspective, these loss-of-function mutations are associated with severe neurodegenerative defects that warrant a genetic diagnosis. For the more common, late-onset forms of diseases, 36 ATG variations are frequent SNPs in the promoters, introns, and 3’ UTR that alone confer a small risk. In the absence of a cure, we think identifying the causal ATG variants will be very informative for the early diagnosis and prognosis of these diseases.
B. Autophagy in infectious, autoimmune, and inflammatory diseases
More than ever, we appreciate how autophagy ensures our defense against infection with any pathogen, whether due to bacteria, viruses, parasites, or fungi. In this struggle, the autophagy pathway is essential for our survival as it immediately recognizes, captures, and kills invading pathogens through a selective process called xenophagy or virophagy when referring specifically to viruses. Beyond this innate clearance, autophagy promotes the second wave of adaptive immunity by ensuring antigen presentation to the T cells. All the survival, maturation, and effector properties of recruited troops of immune cells are controlled by autophagy 52. Upon resolution of an infection, autophagy limits the inflammatory response by the degradation of components of inflammasomes (signalphagy [53]). Thus, by orchestrating overall defense, autophagy safeguards the host against infectious, autoimmune, and chronic inflammatory diseases.
Variant Discovery. Several ATG variations confer enhanced susceptibility to bacteria (peritonitis: CALCOCO2 [54]; Buruli ulcer: ATG16L1 [55,56], leprosy: IRGM [57], TOLLIP [58,59]; C. burnetii: ATG5 [60], MAP1LC3A [60]; sepsis: ATG5 [61], IRGM [62], TOLLIP [63]; uropathogenic E. coli: ATG16L1 [64]; and tuberculosis: ULK1 [65,66], ATG10 [67], IRGM [68–73], TOLLIP [74–76]), viral (human papillomavirus: MCL1 [77]; hepatitis B virus: ATG5 [78], ATG16L1 [79], HIV: TOLLIP [80], and rhinovirus: TOLLIP [81,82]), and parasitic pathogens (leishmaniasis: TOLLIP [83]; and malaria: TOLLIP [84]) (Figure 3A). Regarding inflammatory bowel diseases, much has been written about Crohn disease (CD), which provides an excellent paradigm of a complex disease involving an autophagy defect. The first GWAS in 2007 highlighted ATG16L1, and IRGM, as the most robust genetic loci so far described for CD [85,86]. Since then, 8 other genes (including ULK1, ATG2A, ATG4A, ATG4B, ATG4D, ATG5, ATG16L2, and CALCOCO2) with a total of 32 SNPs have been identified. All variations influencing CD are frequent (from 4% to 53% in the general population), and most CD/variant associations are replicated across multiple ethnic groups (Figure 3B).
From genetics to biology. We focus on ATG16L1, which alone recapitulates all features of risk loci of complex diseases. ATG16L1 is part of the ATG12–ATG5-ATG16L1 trimeric complex, which defines the site where LC3 is lipidated on the nascent double-membrane autophagosomes [100]. ATG16L1 is the target of fifteen CD-associated SNPs; all except one are eQTLs located in introns. The most extensively studied SNP is rs2241880, which leads to T300A conversion [85,86]. Despite a massive body of work, how the T300A mutation alters the function of ATG16L1 remains unclear. The existing evidence argues that it has little or no effect on constitutive or starvation-induced autophagy [101–103]. Instead, it impairs a myriad of alternative intracellular trafficking pathways involved in innate immunity, such as the trafficking of secretory vesicles in intestinal Paneth cells [88,104], and the clearance of invading bacteria by xenophagy [101,102,104,105] (Figure 3C). Faced with an emergency, ATG16L1 also mobilizes LC3 and part of the autophagy machinery to pathogen-containing phagosomes to limit infection [106]. Regardless of the pathway involved, i.e., either LC3-associated phagocytosis (LAP), xenophagy or both, the ATG16L1 T300A mutation renders CD patients very vulnerable to bacterial infections.
From one puzzling insight to another, at the molecular level, the function of ATG16L1 in LAP relies on a C-terminal tryptophan-aspartic acid (WD)-repeat domain that is interestingly targeted by a CD-associated mutation (T300A) [107]. Of note, T300A prevents the binding of the WD domain to a transmembrane protein, TMEM59, present on the phagosome, slowing down LC3 lipidation and the LAP route [106–108]. Of interest, upon bacterial challenge, CASP3 (caspase 3) is also found to preferentially cleave the risk allele, decreasing the expression of the full-length protein, leading to impaired xenophagy [104,105]. Even more disturbing, Atg16l1 loss in intestinal epithelial cells exacerbates chronic colitis by increasing apoptosis and/or necroptosis [109–111]. To guide new treatment options, the challenge will be to identify which of these canonical and non-canonical functions of ATG16L1 is turned off in CD.
C. Autophagy and cancer
More than 20,000 articles in PubMed NCBI have addressed the impact of autophagy on tumor development. However, no consensus has yet been reached, as two opposing hypotheses of pro- and anti-tumor autophagy still provoke strong controversy [112]. Indeed, some advocate that this process is harnessed by cancer cells to fuel their metabolism, prevent oxidative stress, and thus promote tumor growth and resistance to anti-tumor therapies. Others propose that autophagy can counteract malignant transformation by degrading signaling proteins, and by limiting chromosomal instability, DNA mutation, and inflammation while promoting immune surveillance, autophagic cell death, and senescence [112]. Reconciling these opposing functions, it was proposed that autophagy may play a critical role in suppressing tumors in the early stages of oncogenesis while sustaining the progression and the resistance of established tumors [112].
Variant discovery. Thirty-six genes throughout the entire autophagy pathway are associated with the risk, theragnostic, and clinical outcome of 30 different cancer types, regardless of the anatomical location or histological type (carcinoma, melanoma, sarcoma, and hematological malignancies) (Figure 4A). Without listing all 93 polymorphisms, we note that the PIK3C3 SNPs tend to be enriched in different gastrointestinal cancers (esophageal [113,114], gastric [115,116], colorectal [117], and pancreatic [118]). Likewise, variations of all members of the ATG12 conjugation system (ATG5, ATG7, ATG10, ATG12, and ATG16L1) are associated with many solid cancer types (head and neck [119–121], breast [122–127], liver [78,128–130], bladder 131, and kidney cancers [132,133], and melanoma [134]), highlighting the pronounced impact of LC3 conversion in tumorigenesis (Figure 4B).
Beyond cancer risk, ATG SNPs emerge as universal predictors for a wide range of anti-tumor treatments as diverse as chemotherapy (anthracycline and/or taxane: ATG5 [124]; cyclophosphamide: ATG13 [139]; ASPG [asparaginase]: ULK2 [140]; and platinum: ULK1, ATG3, ATG14, ATG10, DRAM1 [141]), anti-angiogenic therapy (anti-VEGF: RB1CC1 [142]), immunotherapy (BCG: ATG2B, ATG5 [131]), targeted therapy (pazopanib: ATG4A, ATG4C, ATG5 [132]; gefitinib: ATG5, ATG7, ATG10, ATG16L2 [143]), and radiotherapy (ATG10 [121]; ATG12, ATG16L2 [144]; and NBR1 [145]).
From genetics to biology. For patient management, the autophagy SNPs are confusingly protective or risk-enhancing. Located in non-coding genomic regions, these associations remain devoid of any molecular hypothesis. A few studies have so far documented a slight increase or decrease in ATG expression, but whether this is enough to have an impact on the autophagy flux remains elusive. Identifying the causal variants and understanding the underlying molecular mechanisms represent important challenges to clarify their contributions to tumorigenesis. We hope that the characterization of the cancer-associated ATG SNPs will help to estimate the cancer risk and guide the use of anti-cancer molecules.
D. Presumed guilty by association
Overall, by piecing together information from 243 studies, we observed that 67 ATG variants over the 263 are associated with several pairs of diseases; many are recognized comorbidities.
● Several are shared across multiple inflammatory or autoimmune manifestations that target the gastrointestinal tract, lung, heart, or multiple miscellaneous tissues: such as CD and asthma (ULK1, and ATG12), CD and rheumatoid arthritis (IRGM, ATG5, ATG16L1, and ATG16L2), CD and ankylosing spondylitis (IRGM), CD and cardiovascular diseases (ATG4C, ATG4D, and ATG16L1), CD and systemic lupus erythematosus (SLE; IRGM, ATG5, and ATG16L2), CD, SLE and Grave disease (IRGM) and asthma and SLE (ATG5), among others. This agrees with the co-occurrence of inflammatory and autoimmune diseases and the long-recognized roles of autophagy in immunity and inflammation [52, 132].
● Equally anticipated from the role of inflammation in carcinogenesis, a growing number of studies have highlighted the association of 13 ATG variants with one or more inflammatory diseases to one or more cancers: such as CD and NSCLC (ULK1), CD and cervical cancers (ATG4A), CD, asthma and cancers (ATG5), CD and gastric cancer glioma (IRGM), CD, arthritis and gastric cancer (IRGM), CD, chronic obstructive pulmonary disease (COPD), rheumatoid arthritis and cancers (ATG16L1), CD, rheumatoid arthritis, SLE and NSCLC (ATG16L2), rheumatoid arthritis and multiple myeloma (ATG5), SLE and cancers (ATG5), and gastritis and gastric cancer (CHMP2B).
● Similarly, thirteen ATGs are associated with two to eight cancers (ATG2B, ATG4A, ATG5, ATG7, ATG10, ATG12, ATG14, ATG16L1, ATG16L2, IRGM, MCL1, NBR1, and VMP1), indicating extensive pleiotropy. It may seem also intuitive that ATG variations could predispose carriers to multiple independent primary cancers, a neglected hypothesis that deserves attention given the high morbidity of this malignancy. Along this line, mice with a mono-allelic deletion of Becn1 show multi-site cancers (lymphoma, hepatocellular carcinoma and lung carcinoma) [135,136]. This was further supported by the deletion of Ambra1 (liver and Lung cancers) [137] or atg5 (osteosarcoma, hepatocellular carcinoma and lymphoma) [138] and the expression of the UvragFS mutant (colorectal carcinoma, and lymphomas) [99] (Figure 4C).
● Intriguingly, Pandora’s box is also opened with ATG variants affecting ‘apparently’ unrelated phenotypes, such as degenerative diseases and cancer. For instance, several ATG7 alleles are common to ccRCC and Parkinson disease. ATG10 SNPs are a risk for breast cancer and Alzheimer disease, whereas C9orf72 variants are related to prostate cancer and ALS-FTD, ALS, or schizophrenia. The same issue applies to cancers and cardiovascular diseases (ATG9B, ATG7, ATG16L1, and ATG16L2), (see for references Tables S2-S7).
● Even more remarkable is that 20 ATG SNPs across 6 autophagy genes (IRGM, ATG4A, ATG5, ATG10, ATG16L1, and ATG16L2) are associated with a cluster of diseases from autoimmune, inflammatory and infectious to degenerative diseases, and cancers. The most notable example is here again the ATG16L1 T300A SNP that alone confers a pleiotropic systemic risk for 18 distinct illnesses. These include, beyond CD and the intestine boundaries, the following: cardiovascular [146], infectious [55,56,64,79], inflammatory diseases (COPD [147], rheumatoid arthritis [148], psoriasis [149], and Paget disease of bone [150]), and eight cancers (breast [126], head and neck [119], lung [151,152], thyroid [153], gastric [154,155], colorectal [156,157], and hepatocellular [129] cancers, and melanoma [134]).
From genetics to biology. Such a disease network agrees with the myriad of roles of autophagy in cancer, inflammation, infection, and neurodegeneration, thus pointing to an autophagy defect as the shared liability for these common diseases. However, caution is warranted when interpreting the data, because most arise from independent studies, and only three studies have associated an ATG SNP with two comorbidities in the same cohort (ATG16L1: HCC and cirrhosis [158], IRGM: Crohn disease and non-alcoholic fatty liver disease [159], and CD and arthritis [160]). Understanding such a high level of pleiotropy might be an important step towards developing new autophagy-modulating drugs that might benefit multiple conditions.
Translating ATG SNP pleiotropy into the clinic
A. Proof of principle and limitations of transgenic mice
In the quest for this demonstration, mouse models have been instrumental in establishing the liability of an autophagy defect in cancer, neurodegenerative, inflammatory, autoimmune, and cardiovascular diseases [8]. A significant cornerstone is the demonstration in 2005 that the loss of autophagy in the central nervous system is sufficient to recapitulate the accumulation of aggregates and neurodegeneration in mice [10,11]. Thereafter, it was elegantly demonstrated that pharmacological activation of autophagy reduces, whereas inhibition of autophagy increases, the formation and the neurotoxicity of aggregate-prone aggregates (mutant HTT [huntingtin], mutant SNCA/α-synuclein, and mutant MAPT/tau [161–163]). Along these lines, the mono-allelic loss of several autophagy genes (Atg16l1, Irgm1, Atg4c, Atg5, Becn1, Uvrag, Ambra1, etc.) was demonstrated in mice to predispose to neurodegeneration, inflammatory disease, age-related cardiac injury, and cancer (see for references Figures 2C-5C, S1C-S3C).
Although these elegant preclinical models have undoubtedly linked defects in autophagy to pathologies, murine models do not sufficiently reflect the natural history of human diseases. The limitations include: i) essential differences between human and mouse physiologies, and hence, the functions of autophagy may be more or less different, and ii) deletions/loss-of-function ATG mutations performed in transgenic mice are not typical of common variations identified in complex human diseases (see Tables S2-S7). Far from the polygenic nature of human diseases, these mouse models explore the homozygous deletion of one gene with critical lethal phenotypes. Lastly, while pollutants and microbes are major environmental variables for human pathogenesis, laboratory mice are housed in highly controlled, sanitized, and ventilated cages. Only a few studies have emphasized the impact of tobacco and pathogens on the development of inflammatory diseases such as CD and COPD in Atg16l1 [89,91], egr1 [164], and map1lc3b [165] murine models. This difference severely limits the translation of findings from mouse models to the bedside of patients.
B. From the noise to risk threshold and polygenetic score
Fourteen years after the first GWAS on CD in 2007, 76 GWAS and 242 genetic studies later, we are still surprised to find a colossal gap between ATG SNP and disease risk correlations. The considerable investment and the subsequent information have not yet improved patient outcome, either for risk-stratification or response to therapy. The mutational landscape of autophagy can be viewed as an iceberg. The small visible portion of the iceberg, herein presented, represents the hotspots of missense, or frequently reported autophagy variations. In contrast, the larger and submerged portion of the iceberg is silent/non-coding mutations that have been overlooked, never reported, or insufficiently characterized.
Approximately 90% of the ATG SNPs are located in non-coding regions (Figure 5A). Given that these regions contain regulatory sequences (promoters, enhancers, introns, and 3ʹUTR), we assume that these non-coding variations may control mRNA abundance by influencing the transcription, splicing, and stability through binding of a transcription factor, splicing machinery, or microRNA. Therefore, any variation in these non-coding sequences might alter gene expression, affecting the autophagy flux, and disease susceptibility and severity.
One such example is the IRGM rs10065172 (c.313C>T), one of the most significant risk SNPs for CD. However, its discovery, a decade ago, has met some skepticism [166]. This was because this synonymous SNP does not change the sequence of the IRGM protein and was thus considered silent. Likewise, its high prevalence in 10% of unaffected populations argues against a pathogenic effect. Of interest, we showed that this type of synonymous SNP is of clinical relevance [167]. Indeed, this SNP changes the seed region of MIR196, a microRNA overexpressed in the inflamed intestinal epithelia of CD patients. Disrupting IRGM mRNA/miRNA regulation was sufficient to have an impact on IRGM expression and impair downstream clearance of bacteria by autophagy [167].
A few studies have reported the consequences of other non-coding ATG SNPs on the binding of upstream regulators, the activity of the promoters or 3ʹUTR by luciferase assays, and the downstream mRNA expression. Of interest, one of the biggest surprises to emerge from our analysis was that the variants linked to a disease tended to be active in the specific tissues that are relevant to this trait. To give the reader an overview of autophagic defects, we have thus completed the tables with the SNP annotations, the impact on transcription factor or miRNA binding, methylation, and expression quantitative trait loci (mQTL and eQTL) as much as possible from the literature or our analyses. We would like to encourage, through this review, collaborative effort to gain insight into the variant-regulator-phenotype ‘trio’ for an improved understanding of the regulation of the autophagy network that is linked to pathogenesis.
One hypothesis that emerges from these observations is that the rate of the autophagy flux is exquisitely sensitive to any change in the protein level of any autophagy member. Most ATG variants are non-coding and regulatory, with typically a slight effect on the level of ATG proteins. This infers that each variant alone is not pathogenic. As they do not interfere with the reproduction of their carriers, these SNPs escape negative selection. Some alleles such as ATG16L1T300A are even positively selected as they offer a selective advantage against some pathogens (Mycobacterium ulcerans [56], S. typhimurium [101], Uropathogenic Escherichia coli [64]) or tumor development (non-small cell lung cancers [151], thyroid cancer [153], colorectal cancer [156], and gastric cancer [168]). Likewise, given their high frequency, the co-occurrence of multi-allelic combinations is common in a large number of individuals.
Thus, we propose that: i) the synergy of all ATG alleles may become strong enough to reach a particular ‘threshold’ of susceptibility to the disease. ii) Exceeding such a risk threshold is dependent on multiple alleles, age, and exposure to an environmental factor. As a result, each variant alone has a limited predictive power, and when combined into a polygenetic risk score, the association may be more robust. With the increasing use of molecular profiling, we regret that there is no autophagy gene panel-based testing when identifying patients at risk for complex diseases. Although relevant for a pathway, no study has brought together the effects of all ATG variants (i.e., for the 69 autophagy members) into a polygenic risk score despite the importance it could have in estimating the risk, the genetic overlap between traits, and phenotype severity. Further studies are thus urgently needed to develop pan-autophagy polygenetic scores and validate their clinical utility in routine clinical practice.
C. The missing environmental link
The significant variability of disease expression by affected or unaffected ‘healthy’ carriers advocates for the intervention of environmental factors. Neither exposure to environmental challenges nor the presence of multiple genetic autophagy variants are individually the direct cause of a disease. Both combined genetic and environmental liabilities interact to push the individual over the threshold leading to disease. This notion presents considerable challenges and opportunities for the management of human diseases. We hope that understanding how autophagy gene-environment interactions affect disease outcome will guide more efficient and personalized treatment strategies. Limiting exposure to environmental factors or manipulating the host autophagy repair machinery should delay the onset of these diseases.
So far, the effects of ATG SNP-environment interactions have been primarily detected through the susceptibility to infection (Figure 5B). Likewise, approximately 15% of cancers are associated with hepatitis B virus (HBV), human papillomavirus, and Helicobacter pylori. While these infections are widespread, most infected people will not develop cancer. Of interest, people carrying variations in several autophagy genes have a greater risk of developing gastric (H. pylori: ATG16L1 T300A [154,155], IRGM [155] and CHMP2B [169]), head and neck (HPV16 x smoking: MCL1 [77]), hepatocellular (HBV: ATG16L1 T300A [129], and ATG5 [78,130]), and cervical (HPV: ATG4A [170]) cancers in response to infection.
As a point of entry, the lungs are constantly exposed to microbes, irritants, allergens, or pollutants (Figure S1). At play, autophagy is immediately upregulated to protect the host from these environmental insults. This might explain why the respiratory system is particularly vulnerable to a spectrum of alterations of autophagy in asthma (ULK1 [171], MAP1LC3B [171], ATG5 [171–174], ATG7 [173] and NUFIP1 [175]), pulmonary fibrosis (ATG5, ATG10, and ATG12 [176], TOLLIP [177–179]), chronic obstructive pulmonary disease (COPD: ATG16L1 [147], and EGR1 [147,164]), tuberculosis (ULK1 [65,66], IRGM [68–73], ATG4C [180], and TOLLIP [74–76]), and lung cancers (NSCLC: ULK1, ATG3, ATG14, DRAM1 [141], ATG4A [181], ATG5 182, ATG10 [141,151,183], ATG12 [144,151], ATG16L1 [151], ATG16L2 [144]; NSCLC EGFR*: ATG5, ATG7, ATG10, ATG12, ATG16L1, ATG16L2 [143]). Of global growing concern, COPD and lung cancers are the third most common cause of death and the leading cause of cancer-related death, respectively. Both result from chronic exposure to cigarette smoke and air pollution, a “silent killer” claiming five million lives worldwide every year (WHO, 2021 [2]). However, despite the magnitude of the health effects, only a few ATG variants have been associated with exposure to air pollutants, such as pulmonary fibrosis (coal: ATG5, ATG10, and ATG12 [176]), asthma (diisocyanate: NUFIP1 [175]), acute respiratory infections (AIR quality x rhinovirus: TOLLIP [81,82]), or lung cancer (non-smokers: MCL1 [184]).
From genetics to biology. We acknowledge that the promising start to studies associating specific ATG SNPs with disease suffers from several limitations. So far, the ATG SNP-environment interactions remain elusive. From 243 studies (3.5 million participants), 52 studies (totaling 36,428 patients and 22,800 controls) have documented ‘autophagy gene × environmental’ interactions in conditions of infection (bacteria: 28 studies, 19,043 patients; virus: 9 studies, 2966 patients; and parasite: 2 studies, 950 patients), tobacco usage (7 studies, 11,772 patients), exposure to coal (1 study, 705 patients), bisphenol A (1 study, 200 patients), diisocyanate (1 study, 88 patients), and air pollution (3 studies, 854 patients). Of these studies, four were African, eight European, seven American, and twenty-six Asian. However, due to differences in allele frequencies, the predictive power of these findings in relation to European populations is limited. Thus, for future precision medicine we encourage new studies that broaden the ethnic diversity.
Attention should also be paid to gene regulation as most disease-associated SNPs are cis-eQTL, the expression of which is plastic and highly specific for a particular environmental cue. We now appreciate that pollutants are endocrine disrupters of signaling pathways and lead to abnormal gene expression. However, previous efforts have underestimated the impact of pollutants on the expression of ATG eQTL variants. Several arguments can be put forward to explain this huge gap: i) There is no assessment of the type of environmental exposure (external and internal levels, latency periods) in the investigated population-case cohorts. ii) Rather than a single-environment stress paradigm, it should be emphasized that we are exposed in the ‘real world’ to low but chronic exposure to a large variety of chemicals and stressors. Our modern lifestyle risk factors include pollutants, medication, pathogens, smoking, alcohol abuse, and diets rich in processed foods. iii) This intricate interaction is further complicated by the long latency period, with exposure to a causal agent typically occurring years to decades before disease diagnosis [185]. Thus, during our lifetime, our body accumulates long-lived and hydrophobic chemicals resulting from different routes and times of exposure. The totality of the pollutants and their metabolites in the blood and tissues is called the exposome [185,186]. iv) However, only a very limited number of widespread pollutants are currently monitored by the agencies that measure the quality of air, water, and food. As a result, it is challenging, if ever possible, to identify from ‘short’ monitoring the causal pollutants and stressors that affect the expression of ATG eQTL and thereby the onset of disease.
To tackle the complex mixture of contaminants, we propose to stimulate interdisciplinary research to develop, test, and validate internal and external biomarkers that could provide more accurate estimates of environmental exposure relevant to chronic environmental diseases. The impact of the environment on human health should then be reconstructed through a combination of blood-based autophagy polygenetic and exposomics risk scores (i.e., we propose to call this the ‘genexposomic’ score). Identifying autophagy genexposomics biomarkers will provide both prevention programs to delay the onset of these devastating diseases in the at-risk population and the rationale of the manipulation of autophagy in new therapeutic opportunities.
Impact of autophagy SNP in the era of precision medicine: a place for autophagopathies
Taken together, the compilation of the ATG variations presented herein is a major step forward in understanding these common and untreatable diseases. However, as yet there are no clinical practice guidelines harnessing this genetic information, most likely because of the small effect of each variant. If these associations are confirmed, detecting the ATG risk alleles from blood samples will be critical for risk stratification, patient diagnosis, and treatment decisions. Such ATG SNP tests may identify CD patients with an increased risk of cancer, thus enabling diagnosis of cancer at an early stage when they have the best chance of being cured. This new information may also limit treatment choices, as immunosuppressive drugs for CD may further increase the risk of cancer. Although relatively rare, clinicians dealing with CD should also be aware of several IRGM SNPs linked to the increased risk of opportunistic tuberculosis. Similarly, we should remember that some ATG SNPs are associated with cancer and neurological diseases. Therefore, a critical challenge will be to recognize these ATG SNP carriers to help disease screening programs, prophylaxis, and precision care to promptly relieve complications of comorbidity. We thus propose the term ‘autophagopathy’ to group together a class of genetic diseases the etiology of which concerns a defect in the autophagy machinery, whether directly related to an abnormal autophagic flux, LC3-associated phagocytosis, or any associated trafficking. This model assumes that neither genetic autophagy variants nor exposure to environmental challenges alone directly cause disease. However, SNPs that lead to low-autophagy impairment may alter the cell’s ability to detoxify damaged organelles when challenged by an environmental factor. As a corollary, any ATG SNP would predispose individuals to develop this wide variety of diseases (such as cancer, infection, and neurodegenerative, cardiovascular, metabolic, and inflammatory diseases), only upon exposure to this particular environmental risk; the nature of which will determine the organ affected, and the diseases.
Thus, through this comprehensive atlas, we aim to bring autophagy into precision medicine. In the near future, physicians will offer patients the option to have their genome sequenced as a routine diagnostic procedure in their health care regimen. The autophagy-targeted SNP panel and the related polygenetic and exposomics risk scores will help predict an individual’s risk of developing an autophagopathy. Because it will provide a diagnosis, and early treatment options, the implementation of this approach is a major global health issue.
Supplementary Material
Acknowledgments
This work was supported by grants from “Association pour la Recherche contre le Cancer” (BR MAD IG fellows; GENEXPOSOMICS Canc’air), “Agence régionale santé Provence Alpes Côte d’Azur and Direction régionale de l’Environnement, de l’aménagement et du logement, Région SUD (IG: Région SUD, plan régional santé environnement, DREAL PACA, ARS PACA, PRSE PACA), “Cancéropole PACA”, French Government “Institut National de la Santé et de la Recherche Médicale”, French national research agency (“Investments for the Future” LABEX SIGNALIFE # ANR-11-LABX-0028-01), Children’s Medical Safety Research Institute (BM and RK G, CMSRI, Vaccinophagy project R17033DJA), INCA Plan Cancer, and NIH (DJK: NIGMS GM131919). Authors would like to thank Christiane Brahimi-Horn for helpful comments.
Funding Statement
This work was supported by the Association pour la Recherche contre le Cancer [GENEXPOSOMICS Canc’air]; Cancéropole PACA [Immunoquest]; French national research agency [ANR-11-LABX-0028-01]; NIH [NIGMS GM131919]; Région SUD [GENEXPOSOMICS]; DREAL PACA, ARS PACA, PRSE PACA [GENEXPOSOMICS]; INCA Plan Cancer [Plan Cancer]; Children’s Medical Safety Research Institute [Vaccinophagy R17033DJA].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Rappaport SM, Smith MT.. Epidemiology. Environment and disease risks. Science. 2010. Oct 22;330(6003):460–461. doi: 10.1126/science.1192603. PubMed PMID: 20966241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].https://www.who.int/news-room/fact-sheets/detail/ambient-(outdoor)-air-quality-and-health.
- [3].Yin Z, Pascual C, Klionsky DJ.. Autophagy: machinery and regulation. Microb Cell. 2016. Dec 5;3(12):588–596. doi: 10.15698/mic2016.12.546. PubMed PMID: 28357331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Smigielski EM, Sirotkin K, Ward M, et al. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000. Jan 1;28(1):352–355. doi: 10.1093/nar/28.1.352. PubMed PMID: 10592272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Allot A, Peng Y, Wei C-H, et al. LitVar: a semantic search engine for linking genomic variant data in PubMed and PMC. Nucleic Acids Res. 2018. Jul 2;46(W1):W530–W536. doi: 10.1093/nar/gky355. PubMed PMID: 29762787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ward LD, Kellis M.. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016. Jan 4;44(D1):D877–81. doi: 10.1093/nar/gkv1340. PubMed PMID: 26657631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carithers LJ, Ardlie K, Barcus M, et al. A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv Biobank. 2015. Oct 20;13(5):311–319. doi: 10.1089/bio.2015.0032. PubMed PMID: 26484571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. EMBO J. 2021. Oct 1;40(19):e108863. doi: 10.15252/embj.2021108863. PubMed PMID: 34459017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schaid DJ, Chen W, Larson NB.. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet. 2018. Aug;19(8):491–504. doi: 10.1038/s41576-018-0016-z. PubMed PMID: 29844615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006. Jun 15;441(7095):885–889. doi: 10.1038/nature04724. PubMed PMID: 16625204. [DOI] [PubMed] [Google Scholar]
- [11].Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006. Jun 15;441(7095):880–884. doi: 10.1038/nature04723. PubMed PMID: 16625205. [DOI] [PubMed] [Google Scholar]
- [12].Sarraf SA, V. Shah H, Kanfer G, et al. Loss of TAX1BP1-directed autophagy results in protein aggregate accumulation in the brain. Mol Cell. 2020. Dec 3;80(5):779–795.e10. doi: 10.1016/j.molcel.2020.10.041. PubMed PMID: 33207181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Houtman J, Freitag K, Gimber N, et al. Beclin1‐driven autophagy modulates the inflammatory response of microglia via NLRP3. EMBO J. 2019. Feb 15;38(4):1–15. doi: 10.15252/embj.201899430. PubMed PMID: 30617086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Heckmann BL, Teubner BJW, Boada-Romero E, et al. Noncanonical function of an autophagy protein prevents spontaneous Alzheimer’s disease. Sci Adv. 2020. Aug 14;6(33):eabb9036. doi: 10.1126/sciadv.abb9036. PubMed PMID: 32851186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ramesh Babu J, Lamar Seibenhener M, Peng J, et al. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008. Jul;106(1):107–120. doi: 10.1111/j.1471-4159.2008.05340.x. PubMed PMID: 18346206. [DOI] [PubMed] [Google Scholar]
- [16].Nassif M, Valenzuela V, Rojas-Rivera D, et al. Pathogenic role of BECN1/Beclin 1 in the development of amyotrophic lateral sclerosis. Autophagy. 2014. Jul;10(7):1256–1271. doi: 10.4161/auto.28784. PubMed PMID: 24905722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Krasniak CS, Ahmad ST.. The role of CHMP2BIntron5 in autophagy and frontotemporal dementia. Brain Res. 2016. Oct 15;151–157. doi: 10.1016/j.brainres.2016.02.051. PubMed PMID: 26972529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ghazi-Noori S, Froud KE, Mizielinska S, et al. Progressive neuronal inclusion formation and axonal degeneration in CHMP2B mutant transgenic mice. Brain. 2012. Mar;819–832. doi: 10.1093/brain/aws006. PubMed PMID: 22366797. [DOI] [PubMed] [Google Scholar]
- [19].Tang G, Gudsnuk K, Kuo S-H, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014. Sep 3;83(5):1131–1143. doi: 10.1016/j.neuron.2014.07.040. PubMed PMID: 25155956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dere E, Dahm L, Lu D, et al. Heterozygous ambra1 deficiency in mice: a genetic trait with autism-like behavior restricted to the female gender. Front Behav Neurosci. 2014. May 16;181. doi: 10.3389/fnbeh.2014.00181. PubMed PMID: 24904333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Le Duc D, Giulivi C, Hiatt SM, et al. Pathogenic WDFY3 variants cause neurodevelopmental disorders and opposing effects on brain size. Brain. 2019. Sep 1;142(9):2617–2630. doi: 10.1093/brain/awz198. PubMed PMID: 31327001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fox LM, Kim K, Johnson CW, et al. Huntington’s disease pathogenesis is modified in vivo by Alfy/Wdfy3 and selective macroautophagy. Neuron. 2020. Mar 4;105(5):813–821.e6. doi: 10.1016/j.neuron.2019.12.003. PubMed PMID: 31899071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Choi I, Zhang Y, Seegobin SP, et al. Microglia clear neuron-released α-synuclein via selective autophagy and prevent neurodegeneration. Nat Commun. 2020. Mar 13;11(1):1386. doi: 10.1038/s41467-020-15119-w. PubMed PMID: 32170061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cheng H-C, Kim SR, Oo TF, et al. Akt suppresses retrograde degeneration of dopaminergic axons by inhibition of macroautophagy. J Neurosci. 2011. Feb 9;31(6):2125–2135. doi: 10.1523/JNEUROSCI.5519-10.2011. PubMed PMID: 21307249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Balke D, Tatenhorst L, Dambeck V, et al. AAV-mediated expression of dominant-negative ULK1 increases neuronal survival and enhances motor performance in the MPTP mouse model of Parkinson’s disease. Mol Neurobiol. 2020. Feb 24;57(2):685–697. doi: 10.1007/s12035-019-01744-0. PubMed PMID: 31446549. [DOI] [PubMed] [Google Scholar]
- [26].Shulman JM, Chipendo P, Chibnik LB, et al. Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am J Hum Genet. 2011. Feb 11;88(2):232–238. doi: 10.1016/j.ajhg.2011.01.006. PubMed PMID: 21295279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gómez-Ramos A, Podlesniy P, Soriano E, et al. Distinct X-chromosome SNVs from some sporadic AD samples. Sci Rep. 2015. Dec 9;5(1):18012. doi: 10.1038/srep18012. PubMed PMID: 26648445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lee JH, Cheng R, Graff-Radford N, et al. Analyses of the National Institute on Aging Late-Onset Alzheimer’s Disease Family Study: implication of additional loci. Arch Neurol. 2008. Nov;65(11):1518–1526. doi: 10.1001/archneur.65.11.1518. PubMed PMID: 19001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li QS, Parrado AR, Samtani MN, et al. Variations in the FRA10AC1 fragile site and 15q21 are associated with cerebrospinal fluid Aβ1-42 level. PLoS One. 2015. Aug 7;10(8):e0134000. doi: 10.1371/journal.pone.0134000. PubMed PMID: 26252872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Briones N, Dinu V.. Data mining of high density genomic variant data for prediction of Alzheimer’s disease risk. BMC Med Genet. 2012. Jan 25;13(1):7. doi: 10.1186/1471-2350-13-7. PubMed PMID: 22273362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Al Eissa MM, Fiorentino A, Sharp SI, et al. Exome sequence analysis and follow up genotyping implicates rare ULK1 variants to be involved in susceptibility to schizophrenia. Ann Hum Genet. 2018. Mar 17;82(2):88–92. doi: 10.1111/ahg.12226. PubMed PMID: 29148569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Errichiello E, Giorda R, Gambale A, et al. RB1CC1 duplication and aberrant overexpression in a patient with schizophrenia: further phenotype delineation and proposal of a pathogenetic mechanism. Mol Genet Genomic Med. 2021. Jan 19;9(1):e1561. doi: 10.1002/mgg3.1561. PubMed PMID: 33340270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Degenhardt F, Priebe L, Meier S, et al. Duplications in RB1CC1 are associated with schizophrenia; identification in large European sample sets. Transl Psychiatry. 2013. Nov 26;e326. doi: 10.1038/tp.2013.101. PubMed PMID: 26151896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wason JMS, Dudbridge F.. Comparison of multimarker logistic regression models, with application to a genomewide scan of schizophrenia. BMC Genet. 2010. Dec 9;11(1):80. doi: 10.1186/1471-2156-11-80. PubMed PMID: 20828390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stopkova P, Saito T, Papolos DF, et al. Identification of PIK3C3 promoter variant associated with bipolar disorder and schizophrenia. Biol Psychiatry. 2004. May 15;55(10):981–988. doi: 10.1016/j.biopsych.2004.01.014. PubMed PMID: 15121481. [DOI] [PubMed] [Google Scholar]
- [36].Rietschel M, Mattheisen M, Degenhardt F, et al. Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol Psychiatry. 2012. Sep;17(9):906–917. doi: 10.1038/mp.2011.80. PubMed PMID: 21747397. [DOI] [PubMed] [Google Scholar]
- [37].Li Z, Chen J, Yu H, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017. Nov 1;49(11):1576–1583. doi: 10.1038/ng.3973. PubMed PMID: 28991256. [DOI] [PubMed] [Google Scholar]
- [38].T-M Hu, Chen S-J, Hsu S-H, et al. Functional analyses and effect of DNA methylation on the EGR1 gene in patients with schizophrenia. Psychiatry Res. 2019. May;276–282. doi: 10.1016/j.psychres.2019.03.044. PubMed PMID: 30952071. [DOI] [PubMed] [Google Scholar]
- [39].Fung H-C, Scholz S, Matarin M, et al. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006. Nov;5(11):911–916. doi: 10.1016/S1474-4422(06)70578-6. PubMed PMID: 17052657. [DOI] [PubMed] [Google Scholar]
- [40].Zhao X, Chen Y, Wang L, et al. Associations of ATG7 rs1375206 polymorphism and elevated plasma ATG7 levels with late-onset sporadic Parkinson’s disease in a cohort of Han Chinese from southern China. Int J Neurosci. 2020. Dec;130(12):1206–1214. doi: 10.1080/00207454.2020.1731507. PubMed PMID: 32065549. [DOI] [PubMed] [Google Scholar]
- [41].Pankratz N, Wilk JB, Latourelle JC, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009. Jan;124(6):593–605. doi: 10.1007/s00439-008-0582-9. PubMed PMID: 18985386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mitjans M, Begemann M, Ju A, et al. Sexual dimorphism of AMBRA1-related autistic features in human and mouse. Transl Psychiatry. 2017 Oct 10;7(10):e1247. doi: 10.1038/tp.2017.213. PubMed PMID: 28994820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Xu J, Xia L, Shang Q, et al. A variant of the autophagy-related 5 gene is associated with child cerebral palsy. Front Cell Neurosci. 2017 Dec 18;407. doi: 10.3389/fncel.2017.00407. PubMed PMID: 29326554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Metzger S, Saukko M, Van Che H, et al. Age at onset in Huntington’s disease is modified by the autophagy pathway: implication of the V471A polymorphism in Atg7. Hum Genet. 2010. Oct;128(4):453–459. doi: 10.1007/s00439-010-0873-9. PubMed PMID: 20697744. [DOI] [PubMed] [Google Scholar]
- [45].Morgan S, Shatunov A, Sproviero W, et al. A comprehensive analysis of rare genetic variation in amyotrophic lateral sclerosis in the UK. Brain. 2017. Jun 1;140(6):1611–1618. doi: 10.1093/brain/awx082. PubMed PMID: 28430856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Guo Q, Wang J, Weng Q.. The diverse role of optineurin in pathogenesis of disease. Biochem Pharmacol. 2020. Oct;114157. doi: 10.1016/j.bcp.2020.114157. PubMed PMID: 32687832. [DOI] [PubMed] [Google Scholar]
- [47].Zhang K, Wang A, Zhong K, et al. UBQLN2-HSP70 axis reduces poly-Gly-Ala aggregates and alleviates behavioral defects in the C9ORF72 animal model. Neuron. 2021. Jun 16;109(12):1949–1962.e6. doi: 10.1016/j.neuron.2021.04.023. PubMed PMID: 33991504. [DOI] [PubMed] [Google Scholar]
- [48].Wang L, Ren A, Tian T, et al. Whole-exome sequencing identifies damaging de novo variants in anencephalic cases. Front Neurosci. 2019. Nov 29; 1285. doi: 10.3389/fnins.2019.01285. PubMed PMID: 31849593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jelani M, Dooley HC, Gubas A, et al. A mutation in the major autophagy gene, WIPI2, associated with global developmental abnormalities. Brain. 2019. May 1;142(5):1242–1254. doi: 10.1093/brain/awz075. PubMed PMID: 30968111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kruer MC, Boddaert N.. Neurodegeneration with brain iron accumulation: a diagnostic algorithm. Semin Pediatr Neurol. 2012. Jun;19(2):67–74. doi: 10.1016/j.spen.2012.04.001. PubMed PMID: 22704259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Griswold AJ, Ma D, Cukier HN, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet. 2012. Aug 1;21(15):3513–3523. doi: 10.1093/hmg/dds164. PubMed PMID: 22543975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity. 2021. Mar 9;54(3):437–453. doi: 10.1016/j.immuni.2021.01.018. PubMed PMID: 33691134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Belaid A, Ndiaye PD, Klionsky DJ, et al. Signalphagy: scheduled signal termination by macroautophagy. Autophagy. 2013. Oct 25;9(10):1629–1630. doi: 10.4161/auto.25880. PubMed PMID: 24004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lutz P, Krämer B, Kaczmarek DJ, et al. A variant in the nuclear dot protein 52kDa gene increases the risk for spontaneous bacterial peritonitis in patients with alcoholic liver cirrhosis. Dig Liver Dis. 2016. Jan;48(1):62–68. doi: 10.1016/j.dld.2015.09.011. PubMed PMID: 26493630. [DOI] [PubMed] [Google Scholar]
- [55].Manry J, Vincent QB, Johnson C, et al. Genome-wide association study of Buruli ulcer in rural Benin highlights role of two LncRNAs and the autophagy pathway. Commun Biol. 2020. Apr 20;3(1):177. doi: 10.1038/s42003-020-0920-6. PubMed PMID: 32313116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Capela C, Dossou AD, Silva-Gomes R, et al. Genetic variation in autophagy-related genes influences the risk and phenotype of Buruli ulcer. PLoS Negl Trop Dis. 2016 Apr 29;10(4):e0004671. doi: 10.1371/journal.pntd.0004671. PubMed PMID: 27128681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yang D, Chen J, Shi C, et al. Autophagy gene polymorphism is associated with susceptibility to leprosy by affecting inflammatory cytokines. Inflammation. 2014. Apr 22;37(2):593–598. doi: 10.1007/s10753-013-9773-1. PubMed PMID: 24264476. [DOI] [PubMed] [Google Scholar]
- [58].Montoya-Buelna M, Fafutis-Morris M, Tovar-Cuevas AJ, et al. Role of toll-interacting protein gene polymorphisms in leprosy Mexican patients. Biomed Res Int. 2013. Nov 4;459169. doi: 10.1155/2013/459169. PubMed PMID: 24294608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shah JA, Berrington WR, Vary JC, et al. Genetic variation in Toll-interacting protein is associated with leprosy susceptibility and cutaneous expression of interleukin 1 receptor antagonist. J Infect Dis. 2016. Apr 1;213(7):1189–1197. doi: 10.1093/infdis/jiv570. PubMed PMID: 26610735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jansen AFM, Schoffelen T, Bleeker-Rovers CP, et al. Genetic variations in innate immunity genes affect response to Coxiella burnetii and are associated with susceptibility to chronic Q fever. Clin Microbiol Infect. 2019. May;25(5):631.e11-e15. doi: 10.1016/j.cmi.2018.08.011. PubMed PMID: 30616015. [DOI] [PubMed] [Google Scholar]
- [61].Shao Y, Chen F, Chen Y, et al. Association between genetic polymorphisms in the autophagy-related 5 gene promoter and the risk of sepsis. Sci Rep. 2017. Aug 24;7(1):9399. doi: 10.1038/s41598-017-09978-5. PubMed PMID: 28839236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kimura T, Watanabe E, Sakamoto T, et al. Autophagy-related IRGM polymorphism is associated with mortality of patients with severe sepsis. Salluh JIF, editor. PLoS One. 2014. Mar 13;9(3):e91522. doi: 10.1371/journal.pone.0091522. PubMed PMID: 24626347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Song Z, Yin J, Yao C, et al. Variants in the Toll-interacting protein gene are associated with susceptibility to sepsis in the Chinese Han population. Crit Care. 2011. Jan 10;15(1):R12. doi: 10.1186/cc9413. PubMed PMID: 21219635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang C, Bauckman KA, Ross ASB, et al. A non-canonical autophagy-dependent role of the ATG16L1 T300A variant in urothelial vesicular trafficking and uropathogenic Escherichia coli persistence. Autophagy. 2019. Mar;15(3):527–542. doi: 10.1080/15548627.2018.1535290. PubMed PMID: 30335568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang R-R, Liang L, Chen W-W, et al. ULK1 polymorphisms confer susceptibility to pulmonary tuberculosis in a Chinese population. Int J Tuberc Lung Dis. 2019. Feb 1;23(2):265–271. doi: 10.5588/ijtld.18.0174. PubMed PMID: 30808462. [DOI] [PubMed] [Google Scholar]
- [66].Horne DJ, Graustein AD, Shah JA, et al. Human ULK1 variation and susceptibility to Mycobacterium tuberculosis infection. J Infect Dis. 2016. Oct 15;214(8):1260–1267. doi: 10.1093/infdis/jiw347. PubMed PMID: 27485354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Songane M, Kleinnijenhuis J, Alisjahbana B, et al. Polymorphisms in autophagy genes and susceptibility to tuberculosis. Carvalho LH, editor. PLoS One. 2012 Aug 6;7(8):e41618. doi: 10.1371/journal.pone.0041618. PubMed PMID: 22879892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bahari G, Hashemi M, Taheri M, et al. Association of IRGM polymorphisms and susceptibility to pulmonary tuberculosis in Zahedan, Southeast Iran. ScientificWorldJournal. 2012. Sep 23;950801. doi: 10.1100/2012/950801. PubMed PMID: 23049477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Intemann CD, Thye T, Niemann S, et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009. Sep 11;5(9):e1000577. doi: 10.1371/journal.ppat.1000577. PubMed PMID: 19750224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].King KY, Lew JD, Ha NP, et al. Polymorphic allele of human IRGM1 is associated with susceptibility to tuberculosis in African Americans. PLoS One. 2011. Jan 21;6(1):e16317. doi: 10.1371/journal.pone.0016317. PubMed PMID: 21283700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lu Y, Li Q, Peng J, et al. Association of autophagy-related IRGM polymorphisms with latent versus active tuberculosis infection in a Chinese population. Tuberculosis (Edinb). 2016. Mar;47–51. doi: 10.1016/j.tube.2016.01.001. PubMed PMID: 26980495. [DOI] [PubMed] [Google Scholar]
- [72].Xie H, Li C, Zhang M, et al. Association between IRGM polymorphisms and tuberculosis risk: A meta-analysis. Medicine (Baltimore). 2017. Oct;96(43):e8189. doi: 10.1097/MD.0000000000008189. PubMed PMID: 29068986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yao Q, Zhu Y, Wang W, et al. Polymorphisms in autophagy-related gene IRGM are associated with susceptibility to autoimmune thyroid diseases. Biomed Res Int. 2018. Jun 11;2018(1):7959707. doi: 10.1155/2018/7959707. PubMed PMID: 29992164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wu S, Huang W, Wang D, et al. Evaluation of TLR2, TLR4, and TOLLIP polymorphisms for their role in tuberculosis susceptibility. APMIS. 2018. Jun 20;126(6):501–508. doi: 10.1111/apm.12855. PubMed PMID: 29924447. [DOI] [PubMed] [Google Scholar]
- [75].Shah JA, Vary JC, Chau TTH, et al. Human TOLLIP regulates TLR2 and TLR4 signaling and its polymorphisms are associated with susceptibility to tuberculosis. J Immunol. 2012. Aug 15;189(4):1737–1746. doi: 10.4049/jimmunol.1103541. PubMed PMID: 22778396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Shah JA, Musvosvi M, Shey M, et al. A functional Toll-interacting protein variant is associated with bacillus Calmette-Guérin-specific immune responses and tuberculosis. Am J Respir Crit Care Med. 2017. Aug 15;196(4):502–511. doi: 10.1164/rccm.201611-2346OC. PubMed PMID: 28463648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhou Z, Sturgis EM, Liu Z, et al. Genetic variants of NOXA and MCL1 modify the risk of HPV16-associated squamous cell carcinoma of the head and neck. BMC Cancer. 2012 May 1;12(1):159. doi: 10.1186/1471-2407-12-159. PubMed PMID: 22548841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li N, Fan X, Wang X, et al. Genetic association of polymorphisms at the intergenic region between PRDM1 and ATG5 with hepatitis B virus infection in Han Chinese patients. J Med Virol. 2020. Aug 1;92(8):1198–1205. doi: 10.1002/jmv.25629. PubMed PMID: 31729038. [DOI] [PubMed] [Google Scholar]
- [79].Sharma A, Kaur S, Duseja A, et al. The autophagy gene ATG16L1 (T300A) variant is associated with the risk and progression of HBV infection. Infect Genet Evol. 2020. Oct;104404. doi: 10.1016/j.meegid.2020.104404. PubMed PMID: 32526369. [DOI] [PubMed] [Google Scholar]
- [80].Wang M-G, Wang J, J-Q He. Genetic association of TOLLIP gene polymorphisms and HIV infection: a case-control study. BMC Infect Dis. 2021. Jun 21;21(1):590. doi: 10.1186/s12879-021-06303-4. PubMed PMID: 34154540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Rodrigues AF, Santos AM, Ferreira AM, et al. Year-long rhinovirus infection is influenced by atmospheric conditions, outdoor air virus presence, and immune system-related genetic polymorphisms. Food Environ Virol. 2019. Dec;11(4):340–349. doi: 10.1007/s12560-019-09397-x. PubMed PMID: 31350695. [DOI] [PubMed] [Google Scholar]
- [82].Huang C, Jiang D, Francisco D, et al. Tollip SNP rs5743899 modulates human airway epithelial responses to rhinovirus infection. Clin Exp Allergy. 2016. Dec;46(12):1549–1563. doi: 10.1111/cea.12793. PubMed PMID: 27513438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].de Araujo FJ, da Silva LD, Mesquita TG, et al. Polymorphisms in the TOLLIP gene influence susceptibility to cutaneous leishmaniasis caused by Leishmania guyanensis in the Amazonas state of Brazil. PLoS Negl Trop Dis. 2015 Jun 24;9(6):e0003875. doi: 10.1371/journal.pntd.0003875. PubMed PMID: 26107286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Brasil LW, Barbosa LRA, de Araujo FJ, et al. TOLLIP gene variant is associated with Plasmodium vivax malaria in the Brazilian Amazon. Malar J. 2017. Mar 13;16(1):116. doi: 10.1186/s12936-017-1754-7. PubMed PMID: 28288644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007. Feb 1;39(2):207–211. doi: 10.1038/ng1954. PubMed PMID: 17200669. [DOI] [PubMed] [Google Scholar]
- [86].Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007. May;39(5):596–604. doi: 10.1038/ng2032. PubMed PMID: 17435756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008. Nov 13;456(7219):264–268. doi: 10.1038/nature07383. PubMed PMID: 18849965. [DOI] [PubMed] [Google Scholar]
- [88].Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008. Nov 13;456(7219):259–263. doi: 10.1038/nature07416. PubMed PMID: 18849966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010. Jun 25;141(7):1135–1145. doi: 10.1016/j.cell.2010.05.009. PubMed PMID: 20602997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Bel S, Pendse M, Wang Y, et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017. Sep 8;357(6355):1047–1052. doi: 10.1126/science.aal4677. PubMed PMID: 28751470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liu T, Kern JT, VanDussen KL, et al. Interaction between smoking and ATG16L1T300A triggers Paneth cell defects in Crohn’s disease. J Clin Invest. 2018. Nov 1;128(11):5110–5122. doi: 10.1172/JCI120453. PubMed PMID: 30137026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Liu B, Gulati AS, Cantillana V, et al. Irgm1-deficient mice exhibit paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am J Physiol - Gastrointest Liver Physiol. 2013. Oct 15;305(8):573–584. doi: 10.1152/ajpgi.00071.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chew TS, O’Shea NR, Sewell GW, et al. Optineurin deficiency in mice contributes to impaired cytokine secretion and neutrophil recruitment in bacteria-driven colitis. Dis Model Mech. 2015 Aug 1;8(8):817–829. doi: 10.1242/dmm.020362. PubMed PMID: 26044960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Cabrera S, ÁF Fernández, Mariño G, et al. ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy. 2013. Aug 14;9(8):1188–1200. doi: 10.4161/auto.24797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lucas C, Salesse L, Hoang MHT, et al. Autophagy of intestinal epithelial cells inhibits colorectal carcinogenesis induced by colibactin-producing Escherichia coli in ApcMin/+ mice. Gastroenterology. 2020. Apr;158(5):1373–1388. doi: 10.1053/j.gastro.2019.12.026. PubMed PMID: 31917256. [DOI] [PubMed] [Google Scholar]
- [96].Salesse L, Lucas C, Hoang MHT, et al. Colibactin-producing Escherichia coli induce the formation of invasive carcinomas in a chronic inflammation-associated mouse model. Cancers (Basel). 2021. Apr 24;13(9):2060. doi: 10.3390/cancers13092060. PubMed PMID: 33923277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007. Oct;9(10):1142–1151. doi: 10.1038/ncb1634. PubMed PMID: 17891140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].He S, Zhao Z, Yang Y, et al. Truncating mutation in the autophagy gene UVRAG confers oncogenic properties and chemosensitivity in colorectal cancers. Nat Commun. 2015. Nov 3;6(1):7839. doi: 10.1038/ncomms8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Quach C, Song Y, Guo H, et al. A truncating mutation in the autophagy gene UVRAG drives inflammation and tumorigenesis in mice. Nat Commun. 2019 Dec 12;10(1):5681. doi: 10.1038/s41467-019-13475-w. PubMed PMID: 31831743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Fujita N, Itoh T, Omori H, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008 May 1;19(5):2092–2100. doi: 10.1091/mbc.e07-12-1257. PubMed PMID: 18321988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Messer JS, Murphy SF, Logsdon MF, et al. The Crohn’s disease: associated ATG16L1 variant and Salmonella invasion. BMJ Open. 2013. Jun 20;3(6):e002790. doi: 10.1136/bmjopen-2013-002790. PubMed PMID: 23794574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kuballa P, Huett A, Rioux JD, et al. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One. 2008. Oct 13;3(10):e3391. doi: 10.1371/journal.pone.0003391. PubMed PMID: 18852889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Fujita N, Saitoh T, Kageyama S, et al. Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem. 2009. Nov 20;284(47):32602–32609. doi: 10.1074/jbc.M109.037671. PubMed PMID: 19783656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lassen KG, Kuballa P, Conway KL, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A. 2014. May 27;111(21):7741–7746. doi: 10.1073/pnas.1407001111. PubMed PMID: 24821797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Murthy A, Li Y, Peng I, et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014. Feb 27;506(7489):456–462. doi: 10.1038/nature13044. PubMed PMID: 24553140. [DOI] [PubMed] [Google Scholar]
- [106].Boada-Romero E, Letek M, Fleischer A, et al. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 2013. Feb 20;32(4):566–582. doi: 10.1038/emboj.2013.8. PubMed PMID: 23376921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Fletcher K, Ulferts R, Jacquin E, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018. Feb 15;37(4):e97840. doi: 10.15252/embj.201797840. PubMed PMID: 29317426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Boada-Romero E, Serramito-Gómez I, Sacristán MP, et al. The T300A Crohn’s disease risk polymorphism impairs function of the WD40 domain of ATG16L1. Nat Commun. 2016. Jun 8;11821. doi: 10.1038/ncomms11821. PubMed PMID: 27273576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Matsuzawa-Ishimoto Y, Hine A, Shono Y, et al. An intestinal organoid-based platform that recreates susceptibility to T-cell-mediated tissue injury. Blood. 2020. Jun 25;135(26):2388–2401. doi: 10.1182/blood.2019004116. PubMed PMID: 32232483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med. 2017. Dec 4;214(12):3687–3705. doi: 10.1084/jem.20170558. PubMed PMID: 29089374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Slowicka K, Serramito-Gómez I, Boada-Romero E, et al. Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat Commun. 2019. Apr 23;10(1):1834. doi: 10.1038/s41467-019-09667-z. PubMed PMID: 31015422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Belaid A, Roméo B, Filippakis H, et al. Autophagy-driven cancer drug development. Autophagy Cardiometabolic Dis. Elsevier; 2018. p. 255–275. doi: 10.1016/B978-0-12-805253-2.00021-3. [DOI] [Google Scholar]
- [113].Ng D, Hu N, Hu Y, et al. Replication of a genome-wide case-control study of esophageal squamous cell carcinoma. Int J cancer. 2008 Oct 1;123(7):1610–1615. doi: 10.1002/ijc.23682. PubMed PMID: 18649358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Hu N, Wang C, Hu Y, et al. Genome-wide association study in esophageal cancer using GeneChip mapping 10K array. Cancer Res. 2005. Apr 1;65(7):2542–2546. doi: 10.1158/0008-5472.CAN-04-3247. PubMed PMID: 15805246. [DOI] [PubMed] [Google Scholar]
- [115].Mangino M, Richards JB, Soranzo N, et al. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J Med Genet. 2009. Jul 1;46(7):451–454. doi: 10.1136/jmg.2008.064956. PubMed PMID: 19359265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhang N, Zheng Y, Liu J, et al. Genetic variations associated with telomere length confer risk of gastric cardia adenocarcinoma. Gastric Cancer. 2019 Nov 21;22(6):1089–1099. doi: 10.1007/s10120-019-00954-8. PubMed PMID: 30900102. [DOI] [PubMed] [Google Scholar]
- [117].Abulí A, Fernández-Rozadilla C, Giráldez MD, et al. A two-phase case-control study for colorectal cancer genetic susceptibility: candidate genes from chromosomal regions 9q22 and 3q22. Br J Cancer. 2011. Sep 6;105(6):870–875. doi: 10.1038/bjc.2011.296. PubMed PMID: 21811255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhao L-L, Liu H-L, Luo S, et al. Associations of novel variants in PIK3C3, INSR and MAP3K4 of the ATM pathway genes with pancreatic cancer risk. Am J Cancer Res. 2020. Jul 1;10(7):2128–2144. PubMed PMID: 32775006. [PMC free article] [PubMed] [Google Scholar]
- [119].Fernández-Mateos J, Seijas-Tamayo R, Klain JCA, et al. Analysis of autophagy gene polymorphisms in Spanish patients with head and neck squamous cell carcinoma. Sci Rep. 2017. Jul 31;7(1):6887. doi: 10.1038/s41598-017-07270-0. PubMed PMID: 28761177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Song X, Yuan Z, Yuan H, et al. ATG12 expression quantitative trait loci associated with head and neck squamous cell carcinoma risk in a Chinese Han population. Mol Carcinog. 2018. Aug;57(8):1030–1037. doi: 10.1002/mc.22823. PubMed PMID: 29637616. [DOI] [PubMed] [Google Scholar]
- [121].Yang Z, Liu Z.. Potentially functional variants of autophagy‐related genes are associated with the efficacy and toxicity of radiotherapy in patients with nasopharyngeal carcinoma. Mol Genet Genomic Med. 2019. Dec 8;7(12):e1030. doi: 10.1002/mgg3.1030. PubMed PMID: 31692259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Zhou J, Hang D, Jiang Y, et al. Evaluation of genetic variants in autophagy pathway genes as prognostic biomarkers for breast cancer. Gene. 2017. Sep 5;549–555. doi: 10.1016/j.gene.2017.06.053. PubMed PMID: 28669927. [DOI] [PubMed] [Google Scholar]
- [123].Qin Z, Xue J, He Y, et al. Potentially functional polymorphisms in ATG10 are associated with risk of breast cancer in a Chinese population. Gene. 2013. Sep 25;527(2):491–495. doi: 10.1016/j.gene.2013.06.067. PubMed PMID: 23850577. [DOI] [PubMed] [Google Scholar]
- [124].Li M, Ma F, Wang J, et al. Genetic polymorphisms of autophagy-related gene 5 (ATG5) rs473543 predict different disease-free survivals of triple-negative breast cancer patients receiving anthracycline- and/or taxane-based adjuvant chemotherapy. Chin J Cancer. 2018. Dec 31;37(1):4. doi: 10.1186/s40880-018-0268-1. PubMed PMID: 29382381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Guo X, Lin W, Bao J, et al. A comprehensive cis-eQTL analysis revealed target genes in breast cancer susceptibility loci identified in genome-wide association studies. Am J Hum Genet. 2018. May 3;102(5):890–903. doi: 10.1016/j.ajhg.2018.03.016. PubMed PMID: 29727689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].El-Amir MI, Wahman MM, Khaled HA, et al. Role of ATG16L1 (rs2241880) and Interleukin 10 (rs1800872) polymorphisms in breast cancer among Egyptian patients. Egypt J Immunol. 2020. Jan;27(1):65–76. PubMed PMID: 33180389. [PubMed] [Google Scholar]
- [127].Dorling L, Kar S, Michailidou K, et al. The relationship between common genetic markers of breast cancer risk and chemotherapy-induced toxicity: a case-control study. PLoS One. 2016. Jul 8;11(7):e0158984. doi: 10.1371/journal.pone.0158984. PubMed PMID: 27392074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Shen M, Lin L.. Functional variants of autophagy-related genes are associated with the development of hepatocellular carcinoma. Life Sci. 2019. Oct 15;235(4):116675. doi: 10.1016/j.lfs.2019.116675. PubMed PMID: 31340167. [DOI] [PubMed] [Google Scholar]
- [129].Wisetsathorn S, Tantithavorn V, Hirankarn N, et al. Gene polymorphisms of autophagy machinery and the risk of hepatitis B virus-related hepatocellular carcinoma in a Thai population. ScienceAsia. 2017. Dec;43(6):362. doi: 10.2306/scienceasia1513-1874.2017.43.362. [DOI] [Google Scholar]
- [130].Li N, Fan X, Wang X, et al. Autophagy-related 5 gene rs510432 polymorphism is associated with hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Immunol Invest. 2019. May;48(4):378–391. doi: 10.1080/08820139.2019.1567532. PubMed PMID: 30907204. [DOI] [PubMed] [Google Scholar]
- [131].Buffen K, Oosting M, Quintin J, et al. Autophagy controls BCG-induced trained immunity and the response to intravesical BCG therapy for bladder cancer. PLoS Pathog. 2014 Oct 30;10(10):e1004485. doi: 10.1371/journal.ppat.1004485. PubMed PMID: 25356988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Santoni M, Piva F, De Giorgi U, et al. Autophagic gene polymorphisms in liquid biopsies and outcome of patients with metastatic clear cell renal cell carcinoma. Anticancer Res. 2018. Oct 1;38(10):5773–5782. doi: 10.21873/anticanres.12916. PubMed PMID: 30275199. [DOI] [PubMed] [Google Scholar]
- [133].Wang Z, Tao L, Xue Y, et al. Association of ATG7 Polymorphisms and Clear Cell Renal Cell Carcinoma Risk. Curr Mol Med. 2019. Jan;19(1):40–47. doi: 10.2174/1566524019666190227202003. PubMed PMID: 30827239. [DOI] [PubMed] [Google Scholar]
- [134].White KAM, Luo L, Thompson TA, et al. Variants in autophagy-related genes and clinical characteristics in melanoma: a population-based study. Cancer Med. 2016. Nov 22;5(11):3336–3345. doi: 10.1002/cam4.929. PubMed PMID: 27748080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Yue Z, Jin S, Yang C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003. Dec 9;100(25):15077–15082. doi: 10.1073/pnas.2436255100. PubMed PMID: 14657337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003. Dec 15;112(12):1809–1820. doi: 10.1172/JCI20039. PubMed PMID: 14638851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Cianfanelli V, Fuoco C, Lorente M, et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015. Jan;17(1):20–30. doi: 10.1038/ncb3072. PubMed PMID: 25438055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Cassidy LD, Young ARJ, Young CNJ, et al. Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat Commun. 2020. Dec 16;11(1):307. doi: 10.1038/s41467-019-14187-x. PubMed PMID: 31949142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Liu B, An T, Li M, et al. The association between early-onset cardiac events caused by neoadjuvant or adjuvant chemotherapy in triple-negative breast cancer patients and some novel autophagy-related polymorphisms in their genomic DNA: a real-world study. Cancer Commun. 2018. Dec 4;38(1):71. doi: 10.1186/s40880-018-0343-7. PubMed PMID: 30514381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Wolthers BO, Frandsen TL, Baruchel A, et al. Asparaginase-associated pancreatitis in childhood acute lymphoblastic leukaemia: an observational Ponte di Legno Toxicity Working Group study. Lancet Oncol. 2017. Sep;18(9):1238–1248. doi: 10.1016/S1470-2045(17)30424-2. PubMed PMID: 28736188. [DOI] [PubMed] [Google Scholar]
- [141].Wang S, Song X, Zhao X, et al. Association between polymorphisms of autophagy pathway and responses in non-small cell lung cancer patients treated with platinum-based chemotherapy. Yi chuan. 2017. Mar 20;39(3):250–262. doi: 10.16288/j.yczz.16-294. PubMed PMID: 28420621. [DOI] [PubMed] [Google Scholar]
- [142].Berger MD, Yamauchi S, Cao S, et al. Autophagy-related polymorphisms predict hypertension in patients with metastatic colorectal cancer treated with FOLFIRI and bevacizumab: Results from TRIBE and FIRE-3 trials. Eur J Cancer. 2017. May;13–20. doi: 10.1016/j.ejca.2017.02.020. PubMed PMID: 28347919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Yuan J, Zhang N, Yin L, et al. Clinical implications of the autophagy core gene variations in advanced lung adenocarcinoma treated with gefitinib. Sci Rep. 2017. Dec 19;7(1):17814. doi: 10.1038/s41598-017-18165-5. PubMed PMID: 29259263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Wen J, Liu H, Wang L, et al. Potentially functional variants of ATG16L2 predict radiation pneumonitis and outcomes in patients with non-small cell lung cancer after definitive radiotherapy. J Thorac Oncol. 2018. May;13(5):660–675. doi: 10.1016/j.jtho.2018.01.028. PubMed PMID: 29454863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Sanchez A, Schoenfeld JD, Nguyen PL, et al. Common variation in BRCA1 may have a role in progression to lethal prostate cancer after radiation treatment. Prostate Cancer Prostatic Dis. 2016. Jun;19(2):197–201. doi: 10.1038/pcan.2016.4. PubMed PMID: 26926928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Orsatti CL, Sobreira ML, Sandrim VC, et al. Autophagy-related 16-like 1gene polymorphism, risk factors for cardiovascular disease and associated carotid intima-media thickness in postmenopausal women. Clin Biochem. 2018. Nov;12–17. doi: 10.1016/j.clinbiochem.2018.09.006. PubMed PMID: 30236831. [DOI] [PubMed] [Google Scholar]
- [147].Chen C-Z, C-Y Ou, Wang R-H, et al. Association of Egr-1 and autophagy-related gene polymorphism in men with chronic obstructive pulmonary disease. J Formos Med Assoc. 2015. Aug;114(8):750–755. doi: 10.1016/j.jfma.2013.07.015. PubMed PMID: 24012056. [DOI] [PubMed] [Google Scholar]
- [148].Mo JJ, Zhang W, Wen QW, et al. Genetic association analysis of ATG16L1 rs2241880, rs6758317 and ATG16L2 rs11235604 polymorphisms with rheumatoid arthritis in a Chinese population. Int Immunopharmacol. 2021. Apr;107378. doi: 10.1016/j.intimp.2021.107378. PubMed PMID: 33529915. [DOI] [PubMed] [Google Scholar]
- [149].Douroudis K, Kingo K, Traks T, et al. Polymorphisms in the ATG16L1 gene are associated with psoriasis vulgaris. Acta Derm Venereol. 2012. Jan;92(1):85–87. doi: 10.2340/00015555-1183. PubMed PMID: 21879234. [DOI] [PubMed] [Google Scholar]
- [150].Usategui-Martín R, García-Aparicio J, Corral-Gudino L, et al. Polymorphisms in autophagy genes are associated with paget disease of bone. PLoS One. 2015 Jun 1;10(6):e0128984. doi: 10.1371/journal.pone.0128984. PubMed PMID: 26030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Li Q, Zhou X, Huang T, et al. The Thr300Ala variant of ATG16L1 is associated with decreased risk of brain metastasis in patients with non-small cell lung cancer. Autophagy. 2017. Jun 3;13(6):1053–1063. doi: 10.1080/15548627.2017.1308997. PubMed PMID: 28441070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Al-Ali R, Fernández-Mateos J, González-Sarmiento R.. Association of autophagy gene polymorphisms with lung cancer. Gene Reports. 2017. Jun;74–77. doi: 10.1016/j.genrep.2017.02.001. [DOI] [Google Scholar]
- [153].Huijbers A, Plantinga TS, Joosten LAB, et al. The effect of the ATG16L1 Thr300Ala polymorphism on susceptibility and outcome of patients with epithelial cell-derived thyroid carcinoma. Endocr Relat Cancer. 2012. May 3;19(3):L15–8. doi: 10.1530/ERC-11-0302. PubMed PMID: 22302078. [DOI] [PubMed] [Google Scholar]
- [154].Burada F, Ciurea ME, Nicoli R, et al. ATG16L1 T300A polymorphism is correlated with gastric cancer susceptibility. Pathol Oncol Res. 2016. Apr;22(2):317–322. doi: 10.1007/s12253-015-0006-9. PubMed PMID: 26547861. [DOI] [PubMed] [Google Scholar]
- [155].Castaño-Rodríguez N, Kaakoush NO, Goh K-L, et al. Autophagy in Helicobacter pylori infection and related gastric cancer. Helicobacter. 2015. Oct;20(5):353–369. doi: 10.1111/hel.12211. PubMed PMID: 25664588. [DOI] [PubMed] [Google Scholar]
- [156].Grimm WA, Messer JS, Murphy SF, et al. The Thr300Ala variant in ATG16L1 is associated with improved survival in human colorectal cancer and enhanced production of type I interferon. Gut. 2016. Mar;65(3):456–464. doi: 10.1136/gutjnl-2014-308735. PubMed PMID: 25645662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Nicoli ER, Dumitrescu T, Uscatu CD, et al. Determination of autophagy gene ATG16L1 polymorphism in human colorectal cancer. Rom J Morphol Embryol. 2014;55(1):57–62. PubMed PMID: 24715166. [PubMed] [Google Scholar]
- [158].Reuken PA, Lutz P, Casper M, et al. The ATG16L1 gene variant rs2241880 (p.T300A) is associated with susceptibility to HCC in patients with cirrhosis. Liver Int. 2019. Dec;39(12):2360–2367. doi: 10.1111/liv.14239. PubMed PMID: 31484215. [DOI] [PubMed] [Google Scholar]
- [159].Simon TG, Van Der Sloot KWJ, Chin SB, et al. IRGM gene variants modify the relationship between visceral adipose tissue and NAFLD in patients with Crohn’s disease. Inflamm Bowel Dis. 2018. Sep 15;24(10):2247–2257. doi: 10.1093/ibd/izy128. PubMed PMID: 29788077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Kee BP, Ng JG, Ng CC, et al. Genetic polymorphisms of ATG16L1 and IRGM genes in Malaysian patients with Crohn’s disease. J Dig Dis. 2020. Jan 13;21(1):29–37. doi: 10.1111/1751-2980.12829. PubMed PMID: 31654602. [DOI] [PubMed] [Google Scholar]
- [161].Ravikumar B, Acevedo-Arozena A, Imarisio S, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005 Jul 26;37(7):771–776. doi: 10.1038/ng1591. PubMed PMID: 15980862. [DOI] [PubMed] [Google Scholar]
- [162].Rubinsztein DC, Gestwicki JE, Murphy LO, et al. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007. Apr;6(4):304–312. doi: 10.1038/nrd2272. PubMed PMID: 17396135. [DOI] [PubMed] [Google Scholar]
- [163].Jia K, Hart AC, Levine B.. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 2007. Jan 8;3(1):21–25. doi: 10.4161/auto.3528. PubMed PMID: 17172799. [DOI] [PubMed] [Google Scholar]
- [164].Chen Z-H, Kim HP, Sciurba FC, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One. 2008. Oct 2;3(10):e3316. doi: 10.1371/journal.pone.0003316. PubMed PMID: 18830406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Chen Z-H, Lam HC, Jin Y, et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci U S A. 2010. Nov 2;107(44):18880–18885. doi: 10.1073/pnas.1005574107. PubMed PMID: 20956295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007. Jul 6;39(7):830–832. doi: 10.1038/ng2061. PubMed PMID: 17554261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011. Mar;43(3):242–245. doi: 10.1038/ng.762. PubMed PMID: 21278745. [DOI] [PubMed] [Google Scholar]
- [168].Ma C, Storer CE, Chandran U, et al. Crohn’s disease-associated ATG16L1 T300A genotype is associated with improved survival in gastric cancer. EBioMedicine. 2021. May;103347. doi: 10.1016/j.ebiom.2021.103347. PubMed PMID: 33906066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Xu Q, Wu Y, Li Y, et al. SNP-SNP interactions of three new pri-miRNAs with the target gene PGC and multidimensional analysis of H. pylori in the gastric cancer/atrophic gastritis risk in a Chinese population. Oncotarget. 2016. Apr 26;7(17):23700–23714. doi: 10.18632/oncotarget.8057. PubMed PMID: 26988755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Mao J, Wu L, Wang W, et al. Nucleotide variation in ATG4A and susceptibility to cervical cancer in Southwestern Chinese women. Oncol Lett. 2018. Mar 20;15(3):2992–3000. doi: 10.3892/ol.2017.7663. PubMed PMID: 29435029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Poon AH, Chouiali F, Tse SM, et al. Genetic and histologic evidence for autophagy in asthma pathogenesis. J Allergy Clin Immunol. 2012. Feb;129(2):569–571. doi: 10.1016/j.jaci.2011.09.035. PubMed PMID: 22040902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Martin LJ, Gupta J, Jyothula SSSK, et al. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PLoS One. 2012. Apr 20;7(4):e33454. doi: 10.1371/journal.pone.0033454. PubMed PMID: 22536318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Pham DL, Kim S-H, Losol P, et al. Association of autophagy related gene polymorphisms with neutrophilic airway inflammation in adult asthma. Korean J Intern Med. 2016. Mar 1;31(2):375–385. doi: 10.3904/kjim.2014.390. PubMed PMID: 26701229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Zeki AA, Yeganeh B, Kenyon NJ, et al. Autophagy in airway diseases: a new frontier in human asthma? Allergy. 2016. Jan;71(1):5–14. doi: 10.1111/all.12761. PubMed PMID: 26335713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Yucesoy B, Kaufman KM, Lummus ZL, et al. Genome-wide association study identifies novel loci associated with diisocyanate-induced occupational asthma. Toxicol Sci. 2015. Jul;146(1):192–201. doi: 10.1093/toxsci/kfv084. PubMed PMID: 25918132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Yuan J, Han R, Esther A, et al. Polymorphisms in autophagy related genes and the coal workers’ pneumoconiosis in a Chinese population. Gene. 2017. Oct 20;36–42. doi: 10.1016/j.gene.2017.08.017. PubMed PMID: 28844669. [DOI] [PubMed] [Google Scholar]
- [177].Bonella F, Campo I, Zorzetto M, et al. Potential clinical utility of MUC5B und TOLLIP single nucleotide polymorphisms (SNPs) in the management of patients with IPF. Orphanet J Rare Dis. 2021. Feb 27;16(1):111. doi: 10.1186/s13023-021-01750-3. PubMed PMID: 33639995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Fingerlin TE, Murphy E, Zhang W, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013. Jun;45(6):613–620. doi: 10.1038/ng.2609. PubMed PMID: 23583980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Noth I, Zhang Y, Ma S, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013. Jun;1(4):309–317. doi: 10.1016/S2213-2600(13)70045-6. PubMed PMID: 24429156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Daya M, van der Merwe L, van Helden PD, et al. Investigating the role of gene-gene interactions in TB susceptibility. PLoS One. 2014. Apr 28;10(4):e0123970. doi: 10.1371/journal.pone.0123970. PubMed PMID: 25919455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].He Q, Lu Y, Hu S, et al. An intron SNP rs807185 in ATG4A decreases the risk of lung cancer in a southwest Chinese population. Eur J Cancer Prev. 2016. Jul;25(4):255–258. doi: 10.1097/CEJ.0000000000000174. PubMed PMID: 26061994. [DOI] [PubMed] [Google Scholar]
- [182].Nikseresht M, Shahverdi M, Dehghani M, et al. Association of single nucleotide autophagy-related protein 5 gene polymorphism rs2245214 with susceptibility to non-small cell lung cancer. J Cell Biochem. 2018. Sep 22;120(2):1924–1931. doi: 10.1002/jcb.27467. PubMed PMID: 30242869. [DOI] [PubMed] [Google Scholar]
- [183].Xie K, Liang C, Li Q, et al. Role of ATG10 expression quantitative trait loci in non-small cell lung cancer survival. Int J cancer. 2016. Oct 1;139(7):1564–1573. doi: 10.1002/ijc.30205. PubMed PMID: 27225307. [DOI] [PubMed] [Google Scholar]
- [184].Jiang Y, Wang W, Wang J, et al. Functional regulatory variants of MCL1 contribute to enhanced promoter activity and reduced risk of lung cancer in nonsmokers: implications for context-dependent phenotype of an antiapoptotic and antiproliferative gene in solid tumor. Cancer. 2012. Apr 15;118(8):2085–2095. doi: 10.1002/cncr.26502. PubMed PMID: 21887682. [DOI] [PubMed] [Google Scholar]
- [185].Wild CP, Scalbert A, Herceg Z.. Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk. Environ Mol Mutagen. 2013. Aug 14;54(7):480–499. doi: 10.1002/em.21777. PubMed PMID: 23681765. [DOI] [PubMed] [Google Scholar]
- [186].Wild CP. The exposome: from concept to utility. Int J Epidemiol. 2012. Feb;41(1):24–32. doi: 10.1093/ije/dyr236. PubMed PMID: 22296988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


