ABSTRACT
Substrates that are selected for degradation by autophagy interact in more complex eukaryotes with Atg8-family proteins via the LC3-interacting region (LIR) that is often preceded by either acidic residues or phosphorylated serine or threonine. These upstream amino acid residues increase the binding affinity of the LIR motif to its binding site on the surface of LC3/GABARAP. It is not fully understood whether or how phosphorylation functionally replaces acidic residues in the LIR-Atg8-family protein interactions. A recent study by Chino et al. discussed in this article analyzed the phosphorylation of two serine residues upstream of the LIR motif in TEX264, a reticulophagy receptor that exhibits a high binding affinity to LC3/GABARAP proteins. The authors found a structural basis for the high-affinity interaction yielded by phosphorylation but not by an acidic residue in place of phosphoserine. Furthermore, finding that phosphorylation of TEX264 generates its high binding affinity to Atg8-family proteins uncovers a mechanistic alternative to that utilized by other reticulophagy receptors when they interact with LC3/GABARAP.
Abbreviations: CSNK2: casein kinase 2; ER: endoplasmic reticulum; IDPR: intrinsically disordered protein region; LIR: LC3-interacting region; p-S: phosphorylated serine
KEYWORDS: Casein kinase 2, crystallography, intrinsically disordered protein region, isothermal titration calorimetry, LC3-Interacting region
One of the indispensable pathways operating in cells under stress conditions is reticulophagy, selective degradation of the endoplasmic reticulum (ER) by autophagy. Maintenance of healthy homeostasis at the ER is essential for the prevention of ER-related diseases, and, therefore, it is not surprising that mammalian cells employ multiple reticulophagy receptors for degradation of their major protein-production site. One of these receptors discovered recently is TEX264 [1,2]. Structure–function analysis of this protein [2] shows that TEX264 has a long intrinsically disordered protein region (IDPR) that functions as a flexible linker bridging the space between the ER and phagophore (amino acid residues 186–313). The C terminus of this IDPR carries the LIR motif (residues 273–276) for the interaction with LC3/GABARAP proteins located at the growing phagophore. Upstream of the IDPR are a GyrI-like domain (residues 41–185) and transmembrane domain (residues 5–27); the latter anchors TEX264 to the ER membrane.
The LIR motif in TEX264 (FEEL) is canonical because it has the consensus sequence Ф0-X1-X2-Ψ3, where Ф0 represents an aromatic amino acid residue (W/F/Y), Ψ represents an aliphatic residue (L/V/I), and X stands for any residue (Figure 1). This short linear motif is typically in a β-strand conformation, and forms an intermolecular β-sheet with the β2-strand of an Atg8-family protein. An earlier study [3] showed that Ф0 and Ψ3 provide major binding forces as they each fit one hydrophobic pocket, the W- and L-site, respectively, on the surface of LC3/GABARAP. Residues upstream of the LIR at positions X−3, X−2, and X−1 as well as those inside the LIR (positions X1 and X2) are often acidic. These residues form electrostatic interactions with various positively charged amino acids in Atg8-family proteins [3]. In some instances, upstream positions X−3-X−1 are occupied by phosphorylated serine or threonine that also contribute to the LIR-Atg8-family protein affinity by either electrostatic forces or hydrogen bonds [4–7]. It is not clear why phosphoserine or phosphothreonine is sometimes in the position of acidic glutamate or aspartate, nor has it been elucidated as to which mechanism, if any, confers a binding advantage for phospho groups at these positions. A recent study by Chino et al. [8], described here, addresses these questions in exploring a phosphorylation-dependent interaction between TEX264 and LC3/GABARAP.
Figure 1.
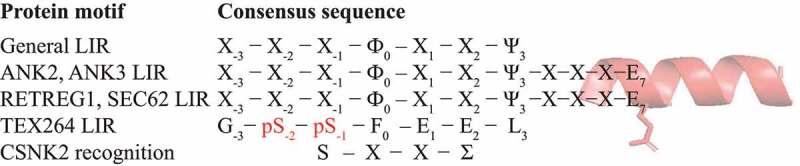
Amino acid sequence alignment of protein motifs and their upstream/downstream sequence features. The C-helix in ankyrins, ANK2 or ANK3, and in reticulophagy receptors, RETREG1 or SEC62, (red ribbon) increases binding affinity of the canonical LIR to LC3/GABARAP. A Glu residue at position 7 is often present in the C-helix. The TEX264 core LIR motif utilizes phosphorylated Ser271 and Ser272 (red) to achieve high-affinity binding to LC3/GABARAP. The TEX264 LIR fulfills the requirement for the CSNK2 recognition motif. Ф, an aromatic amino acid residue (W/Y/F); Ψ, an aliphatic residue (L/V/I); Σ, a negatively charged side chain or group (E/D/p-S/p-Y); X, any amino acid residue.
The researchers performed tandem mass spectrometry (MS/MS) to detect phosphorylated residues in the LIR-containing peptide of TEX264, which exhibits phosphorylation when probed by anti-phosphoserine antibody after immunopurification. The MS/MS analysis revealed four phosphorylated serine residues, in particular, Ser266, Ser269, Ser271, and Ser272 at the positions X−7, X−4, X−2, and X−1, respectively. Mutagenic substitutions to alanine showed that phosphorylation of Ser271 and Ser272 is essential for the interaction of TEX264 with LC3/GABARAP as well as for the function of TEX264 in reticulophagy. By generating a specific antibody against a peptide containing p-S271 and p-S272, Chino et al. showed that TEX264 phosphorylated at these serine residues colocalizes with an autophagosomal cellular fraction, and is a substrate for lysosomal degradation during starvation-induced autophagy.
The sequence of the TEX264 LIR motif with the upstream serine matches a consensus recognition motif of CSNK2 (casein kinase 2) (Figure 1), a constitutively active Ser/Thr kinase that significantly contributes to the generation of the phospho-proteome. Chino et al. found that the treatment of starved HeLa cells with a specific CSNK2 inhibitor, CX4945, suppresses TEX264 phosphorylation, but a CSNK1 inhibitor (D4476) has no effect. Amino acid sequence alignment shows that the two glutamic acids in the TEX264 LIR, E274 and E275, are part of the CSNK2 consensus recognition motif (Figure 1). Alanine substitution of these glutamates probed by anti-phosphoserine antibody shows a significantly decreased level of TEX264 phosphorylation, as is the case with the TEX264S271,272A and TEXS269,271,272A mutants. Together these data clearly reveal that TEX264 is a CSNK2 substrate. Immunostaining and reticulophagy assays with the CSNK2 inhibitor further showed that TEX264 phosphorylation by CSNK2 is required for localization of TEX264 to autophagosomes and for efficient starvation-induced reticulophagy.
Because TEX264 utilizes the LIR peptide to connect with the autophagy machinery via interaction with LC3/GABARAP, Chino et al. investigated how phosphorylation of Ser269, Ser271, and Ser272 affects binding affinity between TEX264 and LC3B or GABARAP and how the phosphorylated peptide compares to its phosphomimetic variants. Probing the TEX264 LIR peptides with isothermal calorimetry (ITC) showed that the phosphorylated versions of the peptide exhibit a strong affinity, in sub-µM Kd values, of TEX264 for LC3B or GABARAP. In comparison, phosphomimetic variants of the TEX264 LIR peptide demonstrate a significantly lower binding affinity to Atg8-family proteins, but higher than unphosphorylated wild type. Immunoprecipitation, immunostaining, and reticulophagy assays confirmed the ITC data, and collectively showed that the efficient function of TEX264 is accompanied by the high-affinity interaction between the receptor and Atg8-family proteins. This interaction can be achieved by phosphorylation of serine residues upstream of the TEX264 LIR motif, but not by Asp or Glu in their place.
To obtain a structural insight into this surprising conclusion, the authors solved the crystal structure of GABARAP in the complex with the phosphorylated TEX264 LIR peptide (residues 269–278 with p-S271 and p-S272; PDB ID: 7VEC) or with the phosphomimetic peptide (residues 271–281 with S272D mutation; PDB ID: 7VED). As expected, the TEX264 LIR motif in both structures interacts with GABARAP in a canonical manner. Specifically, F273 and L276 insert into the hydrophobic W and L pockets, respectively, the motif forms an intermolecular β-sheet with the β2 strand of GABARAP, and Glu274 and Glu275 electrostatically interact with positively charged amino acid residues of GABARAP (Figure 2a). The difference between the two structures is observed in auxiliary interactions. In particular, Leu278, downstream of the phosphorylated LIR, forms hydrophobic contacts with the α3 helix of GABARAP, which is not observed in the phosphomimetic peptide despite having three additional amino acid residues following L278. The upstream region of the phosphorylated LIR creates an extended β-strand that interacts with α1 of GABARAP. The S272D mutation does not allow this extension and interaction, as the LIRS272D sequence is bent upward at D272. The most significant difference between the phosphorylated and phosphomimetic LIR is in the formation of hydrogen bonds. The phosphorylated LIR forms a total of four hydrogen bonds, one between p-Ser272 and Lys46 and three between p-Ser271 and side chains of Tyr5, His9, and Lys48 (Figure 2b). In contrast, the phosphomimetic peptide has only one hydrogen bond between Asp272 and Lys48. Contribution of hydrogen bonds to a high-affinity interaction of TEX264 with GABARAP was confirmed by ITC and in vivo assays with alanine substitutions of Y5, H9, and L48. A search in the PDB database revealed that the phosphorylated LIR in SCOC (short coiled-coil protein), a Golgi protein, interacts with GABARAP in a fashion similar to that of the phosphorylated TEX264 LIR. The phosphorylated serine at position X−2 in the SCOC LIR peptide forms hydrogen bonds with Y5, H9, and L48 of GABARAP (PDB ID: 7AA7). A careful PDB inspection of currently deposited LIR-GABARAP structures showed that triple hydrogen bonds observed with phospho groups are not observed with Asp or Glu. Therefore, Chino et al. conclude that hydrogen bonds mediated by phosphoserines in the upstream sequence of the core LIR provide a specific interaction that dominates over nonspecific salt bridges mediated by negatively charged side chains of Asp or Glu. This mechanism represents a clear example of when a phosphomimetic mutation cannot replace phosphorylation.
Figure 2.
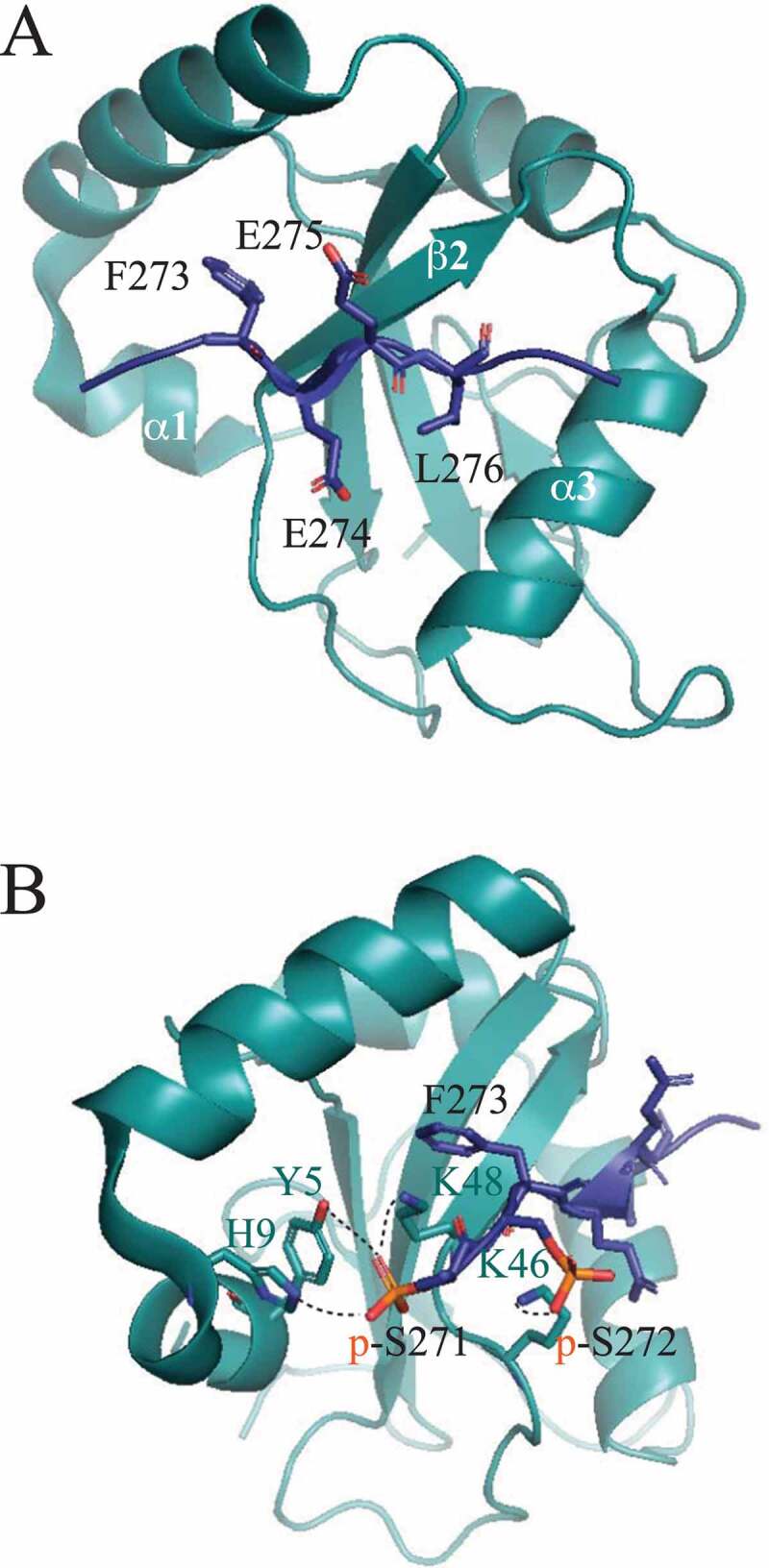
Crystal structure of GABARAP in the complex with the phosphorylated TEX264 peptide (residues 269–278). (A) The core LIR motif in TEX264 is canonical in the interaction with GABARAP. All features of this canonical interface are described in the text. (B) Detailed view of hydrogen bonds (dashed lines) between p-S271 and p-S272 of the TEX264 LIR and Y5, H9, L46, and L48 of GABARAP. GABARAP, dark teal ribbon structure; TEX264 [269-278] peptide, indigo blue.
In general, the LIR sequence is a short linear motif/SLiM located in an IDPR [9–11]. Short linear motifs interact with their binding partners with a low affinity due to a limited binding surface. For a LIR, a typical Kd is in a µM range. If a LIR-containing protein needs to increase this affinity, it needs an additional binding mechanism. Chino et al. showed that phosphorylation of serines upstream of the core LIR motif is one such mechanism. However, there is another mechanism for a high-affinity binding between LIRs and Atg8-family proteins – a C-terminal helix (C-helix) formed by a LIR downstream sequence. Giant ankyrins ANK2 and ANK3 in neuronal axons [12] and reticulophagy receptors (mammalian RETREG1, SEC62, and RTN3 or yeast Atg40) utilize this mechanism [13]. The C-helix in the downstream region achieves sub-µM Kd values, as does p-Ser in the upstream region (Figure 1).
Why do reticulophagy receptors, including TEX264, need a high binding affinity of their LIR to LC3/GABARAP? One possible reason can be the principle of their function, which is the delivery of the fragmented ER to the autophagy machinery for degradation. It has been shown for RETREG1 [14] that the receptor not only connects the ER to the phagophore but also actively bends and deforms the ER lipid bilayer. Strong pulling forces mediated by the LIR-LC3/GABARAP interaction ultimately cause pinching off and fragmentation of the ER, as only ER fragments (i.e., not an intact ER) can be engulfed by the phagophore. It is plausible that the typical low-affinity interaction mediated by the core LIR motif is not sufficient to generate such fragmentation forces.
Future research will hopefully elucidate this phenomenon and answer another puzzling question as to why TEX264 prefers phosphorylation in the LIR upstream sequence over formation of the C-helix in the LIR downstream sequence. At least two possible hypotheses can currently be explored. One comes from the previous study by Chino et al. [2] showing that TEX264 has a long IDPR between the LIR and GyrI-like domain. As mentioned above, this disordered region functions as a linker bridging the space, created by ribosomes, between the ER and phagophore. Shortening of this linker was shown to disrupt the TEX264 function in reticulophagy [2]. Perhaps, if TEX264 utilized a binding-inducible C-helix, the linker in an extended conformation would shorten upon folding and binding to LC3/GABARAP, making the final length of the linker unable to cross the space. The second advantage of phosphorylation/dephosphorylation upstream of the TEX264 LIR can be, as mentioned by the authors [8], employment of phosphoserine as a mechanistic switch between the two TEX264 functions, one in reticulophagy and the other in DNA quality control. If true, this mechanism would elegantly shift a subpopulation of TEX264 to its momentarily needed function without a necessity to synthesize more protein.
Funding Statement
The work was supported by the National Institute of General Medical Sciences [GM131919].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].An H, Ordureau A, Paulo JA, et al. TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol Cell. 2019;74:891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chino H, Hatta T, Natsume T, et al. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol Cell. 2019;74:909. [DOI] [PubMed] [Google Scholar]
- [3].Noda NN, Ohsumi Y, Inagaki F.. Atg8-Family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. [DOI] [PubMed] [Google Scholar]
- [4].Rogov VV, Suzuki H, Fiskin E, et al. Structural basis for phosphorylation-triggered autophagic clearance of salmonella. Biochem J. 2013;454:459–466. [DOI] [PubMed] [Google Scholar]
- [5].Rogov VV, Suzuki H, Marinkovic M, et al. Phosphorylation of the mitochondrial autophagy receptor nix enhances its interaction with LC3 proteins. Sci Rep-UK. 2017;7:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wild P, Farhan H, McEwan DG, et al. Phosphorylation of the autophagy receptor optineurin restricts salmonella growth. Science. 2011;333:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhu YY, Massen S, Terenzio M, et al. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem. 2013;288:1099–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chino H, Yamasaki A, Ode KL, et al. Phosphorylation by casein kinase 2 enhances the interaction between ER-phagy receptor TEX264 and ATG8 proteins. EMBO Rep. 2022;23. 10.15252/embr.202254801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Popelka H, Klionsky DJ. Analysis of the native conformation of the LIR/AIM motif in the Atg8/LC3/GABARAP-binding proteins. Autophagy. 2015;11:2153–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rogov V, Dotsch V, Johansen T, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. [DOI] [PubMed] [Google Scholar]
- [11].Van Roey K, Uyar B, Weatheritt RJ, et al. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem Rev. 2014;114:6733–6778. [DOI] [PubMed] [Google Scholar]
- [12].Li JC, Zhu RC, Chen KY, et al. Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins. Nat Chem Biol. 2018;14:778–787. [DOI] [PubMed] [Google Scholar]
- [13].Mochida K, Yamasaki A, Matoba K, et al. Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes. Nat Commun. 2020;11:3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bhaskara RM, Grumati P, Garcia-Pardo J, et al. Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat Commun. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]


