Abstract
Objectives
Clinical phenotyping and predicting treatment responses in SLE patients is challenging. Extensive blood transcriptional profiling has identified various gene modules that are promising for stratification of SLE patients. We aimed to translate existing transcriptomic data into simpler gene signatures suitable for daily clinical practice.
Methods
Real-time PCR of multiple genes from the IFN M1.2, IFN M5.12, neutrophil (NPh) and plasma cell (PLC) modules, followed by a principle component analysis, was used to identify indicator genes per gene signature. Gene signatures were measured in longitudinal samples from two childhood-onset SLE cohorts (n = 101 and n = 34, respectively), and associations with clinical features were assessed. Disease activity was measured using Safety of Estrogen in Lupus National Assessment (SELENA)-SLEDAI. Cluster analysis subdivided patients into three mutually exclusive fingerprint-groups termed (1) all-signatures-low, (2) only IFN high (M1.2 and/or M5.12) and (3) high NPh and/or PLC.
Results
All gene signatures were significantly associated with disease activity in cross-sectionally collected samples. The PLC-signature showed the highest association with disease activity. Interestingly, in longitudinally collected samples, the PLC-signature was associated with disease activity and showed a decrease over time. When patients were divided into fingerprints, the highest disease activity was observed in the high NPh and/or PLC group. The lowest disease activity was observed in the all-signatures-low group. The same distribution was reproduced in samples from an independent SLE cohort.
Conclusions
The identified gene signatures were associated with disease activity and were indicated to be suitable tools for stratifying SLE patients into groups with similar activated immune pathways that may guide future treatment choices.
Keywords: childhood-onset SLE, interferon, neutrophils, plasma cells, gene signatures, biomarkers, disease activity, clustering analysis
Rheumatology key messages.
Gene signatures are associated with disease activity and change over time in childhood-onset SLE.
A combination of gene signatures into fingerprints is an easy and reliable clustering strategy.
Fingerprints are robust tools that can stratify patients into groups with similar biological disease profiles.
Introduction
SLE is an autoimmune disease characterized by its heterogeneity at the clinical, cellular and molecular levels [1]. This often poses a challenge for clinicians seeking to reliably divide patients into homogeneous disease subgroups. Additionally, patients with distinct clinical disease phenotypes respond to the same medication and vice versa, underlining the fact that solely using clinical phenotype to decide which treatment to start is not enough. Identification of tools for clustering patients into homogeneous groups with similar underlying aberrantly activated immune pathways, to guide treatment for blocking these pathways, is an important research topic. Transcriptional profiling resulted in the identification of so-called ‘gene signatures’. A gene signature is a group of simultaneously upregulated genes caused by a change in the cell’s biological processes. In SLE, multiple gene signatures with correlations to unique clinical features have been identified. However, application in clinical practice is challenging, due to a lack of consensus regarding the genes representing a signature, and the lack of feasibility of implementing transcriptional profiling on individual patients in the clinical setting.
The most well-known gene signature in SLE is the type I (IFN-I) gene signature, which is present in >50% of the patients and has been correlated with disease activity in several cross-sectional studies [2–6]. Transcriptomic data for SLE blood has revealed three different upregulated IFN-annotated modules, respectively, called M1.2, M3.4 and M5.12 [7]. The M1.2 module is induced by IFN-I, while both the M3.4 and M5.12 modules are induced by a combination of IFN-I and type II IFN (IFN-II). When studied over time in SLE patients, each module displayed a different dynamic pattern, with the highest variation in the M5.12 module. These fluctuations indicated that the IFN signature could be used as biomarker for disease activity. However, the few studies that have investigated the parallel change in disease activity with change in IFN gene signatures over time, have shown a lack of association [8, 9]. This implicates the involvement of immune pathways other than the IFN route.
When focusing on pathways that have already been shown to correlate with SLE disease manifestations and/or disease activity, two other gene signatures stand out: the neutrophil (NPh) and plasma cell (PLC) signatures [8–12]. Neutrophils and plasma cells are increased in SLE patients and play a role in disease pathogenesis [13, 14]. Neutrophils of SLE patients are more active, have lower phagocytic capacity and are prone to spontaneously release Neutrophil Extracellular Traps [15]. Plasma cells are the source of pathogenic auto-antibodies in SLE. The NPh signature was associated with lupus nephritis and vascular inflammation, while the PLC signature was correlated with disease activity [8–10, 16]. Moreover, in studies with extensive transcriptional profiling, these and other gene signatures were used to divide patients into subgroups with similar biological disease profiles [12, 17].
Here, we investigated whether we could translate complex transcriptomic data, reflecting multiple different immune pathways, into simple gene signatures suitable for introduction into clinical practice. Additionally, we studied their associations with disease activity and clinical outcome using clinical data and blood samples prospectively collected over time from two childhood-onset SLE (cSLE) cohorts. cSLE is an excellent disease model to study, as cSLE represents the more severe clinical phenotype, has a higher genetic component and children with SLE lack the comorbidities common in adult-onset SLE and that may confound translational studies.
Methods
Patient recruitment
Patients fulfilled the SLICC classification criteria or the 2019 EULAR/ACR criteria for SLE [18, 19]. Blood specimens, demographics and clinical characteristics were prospectively collected. Disease activity was assessed by the Safety of Estrogen in Lupus National Assessment (SELENA)-SLEDAI at each visit [20]. Disease flares were indicated by an increase of >3 or >12 points from the previous visit for a mild/moderate or severe flare, respectively. Disease domains were derived from the SELENA-SLEDAI items.
Additionally, 51 healthy controls (HCs), without symptoms of underlying viral infections or the use of any medications, were included. Written informed consent was obtained from all participants in compliance with the Declaration of Helsinki. The study was approved by the medical ethics review committee of the Erasmus Medical Center, Rotterdam, the Netherlands (MEC-2019–0412).
Clustering strategy
A semi-manual and an automated clustering strategy was performed (Supplementary Fig. S1, available at Rheumatology online). For the semi-manual clustering strategy, the combination of a positive or negative score per gene signature was used to identify 16 clusters. These clusters were subsequently divided into three so-called fingerprints consisting of patients with mutually exclusive combinations of positive and negative gene signatures. For the automated clustering strategy, an unsupervised hierarchical clustering method was used to identify clusters that were enriched in the cSLE cohort.
Supplementary methods
Details on blood collection and real-time PCR, gene selection and signature definitions, ultrasensitive IFN-α Simoa and statistics are described in the Supplementary Material, available at Rheumatology online.
Results
Cohort description
Between March 2013 and January 2021, 101 cSLE patients with median disease duration of 0.5 (0–8.2) years at enrolment (Cohort-I, Table 1) were prospectively recruited at the outpatient clinic of three academic hospitals in the Netherlands and one in the Czech Republic. Fifty-one HCs were included in the study. For 73/101 patients, blood samples from 2–4 longitudinal time points with a median follow-up time of 344 (29–1542) days were available. As a replication cohort, 34 adults with cSLE with median disease duration of 15.8 (3.8–40) years were included (Cohort-II, Table 1).
Table 1.
Patient and healthy control characteristics
| COHORT-I |
COHORT-II |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Category | Total cSLE cohort (n = 101) | EMC cohort (n = 49) | AUMC cohort (n = 35) | TCH cohort (n = 12) | RUMC cohort (n = 5) | Adult cSLE cohort (n = 34) | HCs (n = 51) | ||
| Demographics | |||||||||
| Gender | Female | 84 (83.2%) | 41 (83.7%) | 30 (85.7%) | 8 (66.7%) | 5 (100%) | 32 (94.1%) | 40 (78.4%) | |
| Male | 17 (16.8%) | 8 (16.3%) | 5 (14.3%) | 4 (33.3%) | 0 (0%) | 2 (5.9%) | 11(21.6%) | ||
| Ethnicity | White | 43 (42.6%) | 25 (51.0%) | 15 (42.9%) | 11 (91.7%) | 2 (40%) | 26 (76.5%) | 45 (88.2%) | |
| Non-white | 58 (57.4%) | 24 (49.0%) | 20 (57.1%) | 1 (8.3%) | 3 (60%) | 8 (23.5%) | 6 (11.8%) | ||
| Age at enrolment (years) | 15.6 (5.1–23) | 15.2 (5.2–18.1) | 16.7 (11.8–23) | 14.4 (5.1–17.2) | 16.1 (15.5–17.6) | 32.4 (18.5–56.4) | 29 (20–65) | ||
| Disease duration at enrolment (years) | 0.51 (0–8.2) | 0.18 (0–6.8) | 1.11 (0–8.2) | 0.04 (0–1.8) | 0.59 (0–4.8) | 15.8 (3.8–40) | |||
| SELENA-SLEDAI at enrolment | 4 (0–27) | 4 (0–18) | 4 (0–27) | 4 (0–15) | 4 (0–10) | 4 (0–14) | |||
| ≤4 | 53 (52.5%) | 25 (51.0%) | 18 (51.4%) | 7 (58.3%) | 3 (60%) | 23 (67.6%) | |||
| 5–7 | 12 (11.9%) | 9 (18.4%) | 2 (5.7%) | 0 (0%) | 1 (20%) | 8 (23.5%) | |||
| ≥8 | 36 (35.6%) | 15 (30.6%) | 15 (42.9%) | 5 (41.7%) | 1 (20%) | 3 (8.9%) | |||
| Longitudinal samples | 73 (72.2%) | 43 (87.8%) | 18 (51.4%) | 9 (75%) | 3 (60%) | 0 | 0 | ||
| Number of visits (median) | 2 (1–4) | 2 (1–4) | 2 (1–4) | 2 (1–4) | 2 (1–3) | ||||
| Follow-up time (days) | 344 (29–1542) | 267 (51–1542) | 511 (98–1059) | 91 (29–182) | 120.5 (105–182) | ||||
| Flare | Mild/moderate | 14 (19.2%) | 7 (14.2%) | 6 (17.1%) | 1 (8.3%) | 0 (0%) | |||
| Severe | 7 (9.6%) | 1 (2.0%) | 6 (17.1%) | 0 (0%) | 0 (0%) | ||||
| Treatment at enrolment | |||||||||
| None | 27 (26.7%) | 15 (30.6%) | 8 (22.9%) | 3 (25%) | 0 (0%) | 0 (0%) | 51 (100%) | ||
| HCQ | 67 (66.3%) | 33 (67.3%) | 24 (68.6%) | 6 (50%) | 4 (80%) | 31 (91.2%) | |||
| MMF | 28 (27.7%) | 16 (32.7%) | 9 (25.7%) | 0 (0%) | 3 (60%) | 14 (41.2%) | |||
| MTX | 3 (3.0%) | 2 (4.1%) | 0 (0%) | 0 (0%) | 1 (20%) | 2 (5.9%) | |||
| AZA | 8 (7.9%) | 2 (4.1%) | 5 (14.3%) | 1 (8.3%) | 0 (0%) | 16 (47.1%) | |||
| CYC | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 8 (23.5%) | |||
| Prednisone | 29 (28.7%) | 15 (30.6%) | 8 (22.9%) | 4 (33.3%) | 2 (40%) | 31 (91.2%) | |||
| Rituximab | 4 (4.0%) | 0 (0%) | 4 (11.4%) | 0 (0%) | 0 (0%) | 1 (2.9%) | |||
| Belumimab | 1 (1.0%) | 0 (0%) | 1 (2.9%) | 0 (0%) | 0 (0%) | 0 (0%) | |||
Data are presented as median (range) or as number (% of total). Non-white ethnicity = Hindu, Suriname, Hispanic, Asian, African American, Mixed. AUMC: Amsterdam University Medical Center; cSLE: childhood-onset SLE; EMC: Erasmus Medical Center; HC: healthy control; RUMC: Radboud University Medical Center; SELENA: Safety of Estrogen in Lupus National Assessment; TCH: Czech Republic Palacky University Olomouc.
Four dynamic gene signatures were present in SLE patients
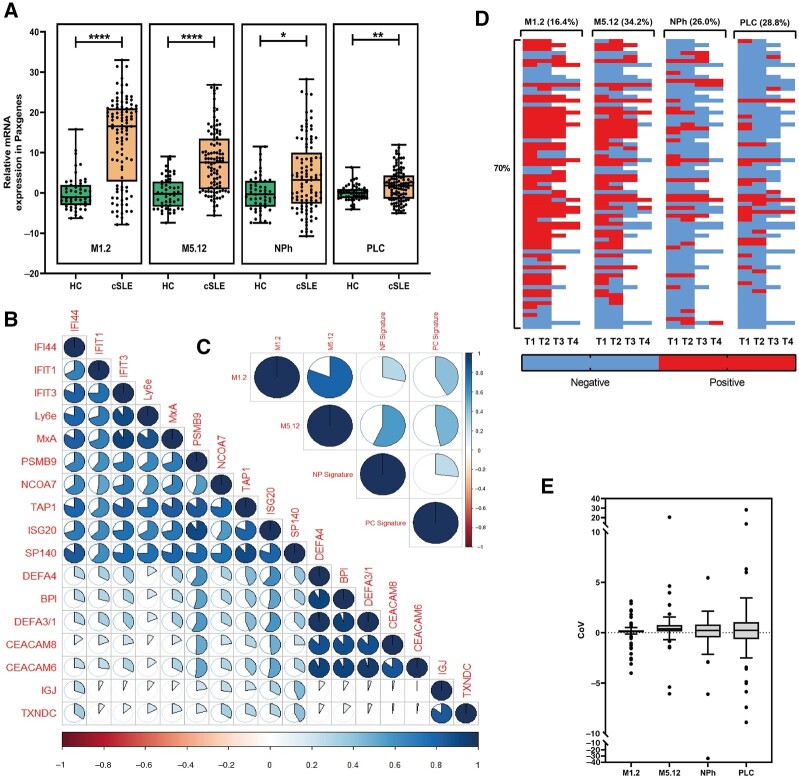
Four gene signatures were assessed based on indicator genes in 51 HCs and 101 cSLE patients (Supplementary Fig. S1 and Table S1, available at Rheumatology online). All four gene signatures were significantly higher expressed in patients compared to HCs (Fig. 1A). High positive associations were present between the genes representing each gene signature, while poor associations were found between genes of different gene modules (Fig. 1B). The M1.2 and M5.12 IFN signatures had the highest association, followed by the M5.12 and NPh signature, while poor associations were observed between the other signatures (Fig. 1C).
Fig. 1.
Four dynamic gene signatures are present in cSLE patients
(A) Relative expression of four gene signature scores in HCs (N = 51) vs cSLE patients (N = 101). (B) Correlation matrix between gene signature–associated genes based on relative expression. (C) Correlation matrix between each individual gene signature. (D) Heatmap indicating a positive or negative gene signature score over time in 73 cSLE patients. Each row represents the same patient. Horizontal percentages indicate the number of patients who showed a dynamic signature over time. The vertical percentage indicates the number of patients with at least one dynamic gene signature. (E) Coefficient of variation (CoV) per gene signature indicating the intra-individual difference per gene signature. Each dot represents the CoV of one patient. Lines indicate the mean (s.d.). The Mann–Whitney U-test was used to compare two groups; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. For correlations, Spearman’s rho was used. A full circle represents a rho of 1. cSLE: childhood-onset SLE; HC: healthy control; NPh: neutrophil signature; PLC: plasma cell signature; T: time point.
In 73 cSLE patients, each gene signature was determined at a second time point, and in 42 cSLE patients at a third and/or fourth time point (Fig. 1D). In 51 out of the 73 patients (70%), at least one or more signatures changed from positive to negative or vice versa (Fig. 1D). The M5.12 IFN signature showed the highest variability within individual patients [coefficient of variation (CoV) ± S.D. 0.66 ± 2.74], followed by the PLC- [CoV 0.54 ± 4.39], NPh- [CoV −0.35 ± 4.23] and M1.2 IFN [CoV 0.05 ± 1.14] signatures (Fig. 1E). These findings imply that gene signatures are driven by different pathways and have a dynamic character over time, which makes them potential biomarkers for changes in disease activity.
Gene signatures were associated with disease activity
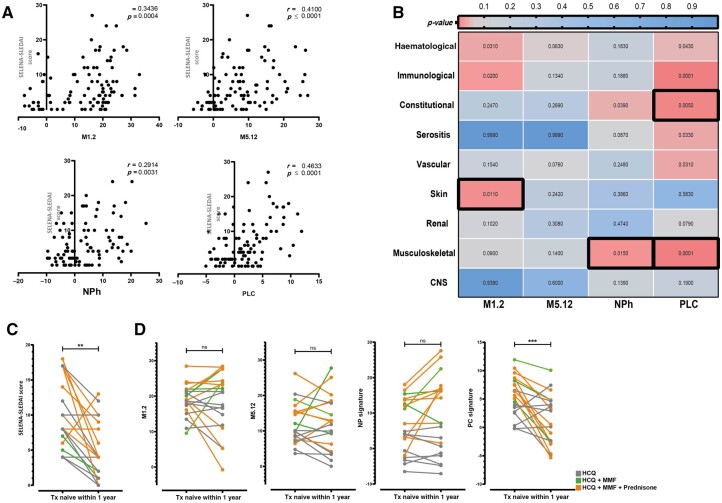
To investigate associations with disease characteristics, we stratified patients into groups based on a low or high gene signature, using the mean + 2 × SDHC per score as a threshold. A high M1.2 IFN, M5.12 IFN, NPh or PLC signature was more prevalent in patients with a higher SELENA-SLEDAI (Supplementary Fig. S2A–D, available at Rheumatology online). The highest association was found for the PLC signature (P < 0.0001, r = 0.473) (Fig. 2A).
Fig. 2.
Gene signatures are associated with disease activity
(A) Correlation between the SELENA-SLEDAI and gene signature scores. (B) Heatmap indicating the correlation between a high signature and a specific disease domain derived from the SELENA-SLEDAI. Numbers indicate the P-value based on univariate analysis. Black outlined boxes indicate domains that were significant in the multivariate model. (C) Longitudinal SELENA-SLEDAI and (D) gene signature scores from 20 Txnaive cSLE patients at the first and second time points (median time between two samples = 62.5 days). The Mann–Whitney U-test was used to compare two groups; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. For correlations, Spearman’s rho was used. Fisher’s exact test was used to compare categorical data. cSLE: childhood-onset SLE; NPh: neutrophil signature;ns: not significant; PLC: plasma cell signature; SELENA: Safety of Estrogen in Lupus National Assessment; Txnaive: treatment naïve.
Furthermore, we investigated the association of disease domains, derived from the SELENA-SLEDAI, with the different gene signatures (Fig. 2B). Univariate analysis showed skin domain, haematological domain, and immunological domain involvement to be associated with a high M1.2 IFN signature. No domain was associated with a high M5.12 IFN signature. Constitutional and musculoskeletal domain involvement showed an association with a high NPh signature. Additionally, all domains except for the skin, renal and CNS were associated with a high PLC signature. In the multivariate model, skin domain involvement was associated with a high M1.2 IFN signature, musculoskeletal domain involvement was associated with a high NPh and PLC signature, and the constitutional domain was associated with a high PLC signature (Fig. 2B).
Next, we determined whether changes over time in disease activity were accompanied by changes in gene signatures. For this purpose, 20 treatment-naïve (Txnaive) children with at least one subsequent sample after start of treatment (median time between samples: 62.5 days) were chosen as the optimal patient group for investigating this. Decrease in disease activity over time was accompanied by a significant reduction in the PLC signature, but was not mirrored by changes in the M1.2 IFN, M5.12 IFN and NPh signatures (Fig. 2C, D). Interestingly, when investigating the effect of medication, the additional use of MMF and/or prednisone to HCQ treatment also led to a decrease in the PLC signature (P = 0.0005), whereas the addition of prednisone increased the NPh signature (P = 0.0195) (Fig. 2D). In addition, the IFNa2 levels, neutrophil counts and anti-dsDNA levels were measured in the same samples in order to study factors other than medication use that may influence the signatures. Neutrophil counts and anti-dsDNA levels showed the same trend as the NPh and PLC signatures, while the IFNa2 levels in general decreased, in contrast with varying activation of the M1.2 and M5.12 IFN signatures (Supplementary Fig. S3A–C, available at Rheumatology online). Together, these data indicate that the gene signatures were associated with disease activity and were influenced by medication use and cell compositions. However, the correlation coefficients (Fig. 2A) were low, indicating that testing individual gene signatures will not be sufficient for identifying homogeneous subgroups of patients.
Gene fingerprints identified SLE patients with similar disease activity
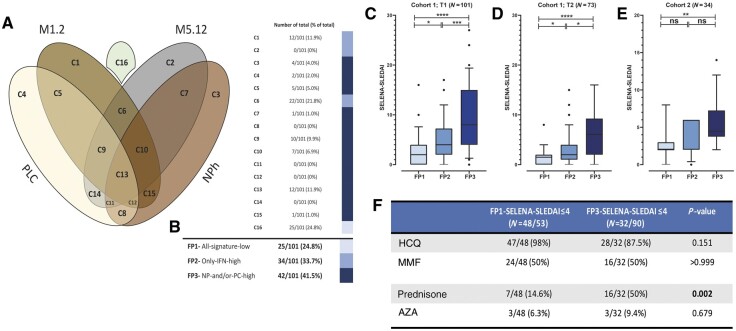
In our search to find homogeneous subgroups of patients, we first used a semi-manual clustering strategy. Based on the four described gene signatures, patients were allocated into 16 unique clusters (Fig. 3A). A cluster represented a combination of either a positive or negative gene signature score. To reduce data complexity, the clusters were distributed over three mutually exclusive groups with matching underlying activated immune pathways, forming a so-called fingerprint. Fingerprint-1 was described as ‘all-signatures-low’, which indicated patients with a low score in all four gene signatures. Fingerprint-1 consisted of patients in cluster 16 (n = 25; 24.8%). Fingerprint-2 represented patients with high IFNs, i.e. a high M1.2 and/or high M5.12 score, and consisted of patients in clusters 1, 2 and 6 (n = 34; 33.7%). Finally, fingerprint-3 defined patients with high NPh and/or PLC signature, independent of the IFN signatures, and included patients in clusters 3, 4, 5, 7–15 (n = 42; 41.5%). Notably, the majority of these patients had a positive M1.2 and/or M512 gene signature (36/42) (Fig. 3A, B).
Fig. 3.
Gene fingerprints identify SLE patients with similar disease activity
(A) Venn diagram of gene clusters. Overlap between the ellipses in the Venn diagram indicates a positive gene score for the involved gene signatures. (B) Patient distribution over three fingerprint groups. Coloured bars represent the clusters that are involved in each fingerprint. (C) SELENA-SLEDAI distribution per fingerprint group in cSLE cohort-I; first time point (n = 101). (D) SELENA-SLEDAI distribution per fingerprint group in cSLE cohort-I; second time point (n = 73). (E) SELENA-SLEDAI distribution per fingerprint group in cSLE cohort-II; replication cohort (n = 34). (F) Medication use in patients with FP1 and FP3 with a SELENA-SLEDAI of ≤4. Dots represent individual patients. The Mann–Whitney U-test was used to compare the two groups; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. cSLE: childhood-onset SLE; FP: fingerprint; NPh: neutrophil signature; ns: not significant; PLC: plasma cell signature; SELENA: Safety of Estrogen in Lupus National Assessment; T: time point.
As a first step, we cross-sectionally analysed data from samples taken at entry into the study (Fig. 3C). Disease activity differed significantly between the three fingerprint groups. Patients with fingerprint-1 (median SELENA-SLEDAI = 2) had the lowest disease activity, while patients with fingerprint-3 had the highest disease activity (median SELENA-SLEDAI = 8). Interestingly, patients with fingerprint-2 formed an intermediate group, indicating that these patients had immunological activation and clinical disease activity, but which was less prominent than patients with fingerprint-3 (Fig. 3C). In the fingerprint-3 group, the highest number of patients was treatment naïve (Txnaive;N = 20/42) and recently diagnosed (median 10 days, Supplementary Fig. S4A and B, available at Rheumatology online). Only 8 patients in this group had a disease duration of >1 year, and of these, 3 had a disease flare.
To filter out the component of high disease activity and lack of immunosuppressive medication in Txnaive patients (n = 27/101) at the first time point, we analysed 73 samples taken at a subsequent second time point. This confirmed the observation from the first time point: patients within fingerprint-1 had the lowest disease activity, while patients with fingerprint-3 had the highest disease activity (Fig. 3D).
To further address whether disease duration influenced our findings, the fingerprints were determined in a cohort of 34 adults with cSLE with a median disease duration of 15.8 years (cohort-II, Table 1). The distribution of patients within cohort-II, based on fingerprints, showed an identical association with disease activity. This excluded disease duration from being a factor influencing the fingerprints (Fig. 3E). Moreover, to investigate why a selection of patients with low disease activity (SELENA-SLEDAI ≤4) had fingerprint-3, we compared the use of medication in those patients with a SELENA-SLEDAI of ≤4 who were in the fingerprint-1 group with that of those who were in the fingerprint-3 group (Fig. 3F). Patients in the fingerprint-3 group with a SELENA-SLEDAI of ≤4 were more often on prednisone (P < 0.002). This indicated that these patients, despite having low disease activity, still had activation of the underlying immune pathways, leading to high NPh and/or PLC gene signature expression.
Gene fingerprints and clinical phenotype
We performed univariate and multivariate logistic regression analyses to test for the involvement of specific organ domains in patients in the fingerprint groups: fingerprint-1 was associated with significantly less skin involvement in the multivariate model, and fingerprint-3 was associated with involvement of the musculoskeletal, constitutional and immunological organ domains (Supplementary Table S2, available at Rheumatology online).
Auto-antibody profiling of the patients revealed that patients with fingerprint-3 had higher anti-dsDNA levels than patients with fingerprints-1 and 2, reflecting the higher disease activity found in patients with fingerprint-3 (Supplementary Fig. S4C, D, available at Rheumatology online). Interestingly, patients with fingerprint-3 were also more often anti-dsDNA positive than the other patients. Anti-SSA antibodies were primarily present in patients with fingerprint-2 and 3, while anti-SM and anti-RNP did not differ between the fingerprint groups (Supplementary Fig. S4D).
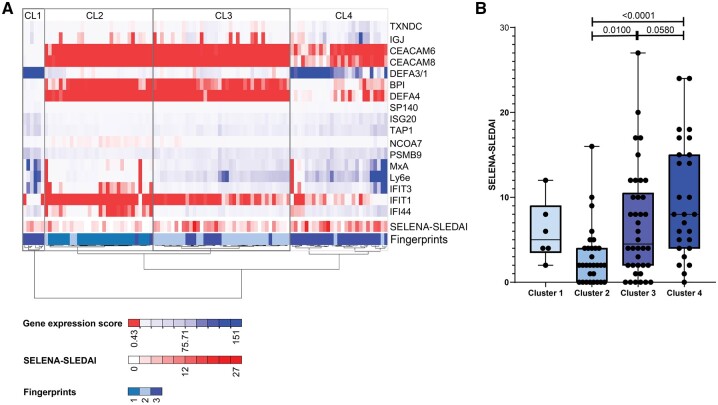
Hierarchical clustering identified gene fingerprints
To investigate the robustness of the identified gene fingerprints, we additionally performed an automated clustering strategy to assess clusters that were enriched in our patient cohort. Unsupervised hierarchical clustering identified four major clusters that paralleled the identified gene fingerprints (Fig. 4A). Cluster 1 represented a small group of patients, all with fingerprint-3. Cluster 2 represented patients with fingerprint-1. Cluster 3 represented patients with primarily fingerprint-2, while cluster 4 represented patients with primarily fingerprint-3. As in the respective fingerprint groups, patients with cluster 2 had the lowest disease activity, while patients with cluster 4 had the highest disease activity (Fig. 4B). Interestingly, within cluster 3, patients who had a fingerprint-3 had a higher disease activity. These data indicate that gene fingerprints are robust tools that match with an automated clustering method and correctly identify patients with similar disease activity.
Fig. 4.
Hierarchical clustering parallels the identified gene fingerprints
(A) Unsupervised hierarchical clustering using Ward’s agglomerative method, passing the Euclidean distance between samples, identifying Clusters 1, 2, 3 and 4. Fingerprints 1, 2, 3 and SELENA-SLEDAI are depicted in the lowest two rows of the heatmap. Each column represents one patient. (B) Association between SELENA-SLEDAI and clusters. Dots represent individual patients. The Mann–Whitney U-test was used to compare two groups (4B); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Red to white colour indicates the magnitude of gene expression described as 2−ΔCt. SELENA: Safety of Estrogen in Lupus National Assessment.
Discussion
We studied four gene signatures in SLE, derived from previously described transcriptomic data, to develop a method that can easily be applied in clinical practice. These gene signatures were associated with disease activity in general and with disease domains derived from the SELENA-SLEDAI. Upon subgrouping of patients into fingerprints, we found a significant difference in disease activity between the fingerprint groups. Fingerprint-1 was associated with low disease activity, while fingerprint-3 represented the patients with high disease activity. We replicated these findings in samples collected over time within the same patient cohort, as well as in cross-sectional samples of a replication cohort. Hierarchical clustering identified similar gene fingerprints, indicating the robustness of our strategy.
Transcriptional profiling is elaborate and costly, resulting in signatures that consist of large gene sets that are simultaneously upregulated [12]. Translation of these data into signatures of a restricted number of genes would facilitate introduction into clinical practice. Therefore, we used a principal components analysis (PCA) approach to identify genes that explained >95% of the total variance in the gene groups. Between the indicator genes that describe a specific gene signature, we observed a high association, while genes from different gene signatures lacked this association. These results are in line with previous studies, in which indicator genes from each individual gene signature were found to be driven by their own unique pathway [12, 21]. Previously, Chiche et al. described the intra-individual variation of the M1.2 and M5.12 IFN gene signatures obtained from transcriptomics in longitudinal samples from 29 SLE patients [7]. We were able to reproduce these findings for the M1.2 and M5.12 IFN gene signatures obtained via the PCA approach, in our longitudinal cohort of 73 patients. Interestingly, demographic and clinical differences between the two cohorts didn’t affect these observations. We further demonstrated, for the first time, that the NPh and PLC signatures, showed a dynamic character over time. Importantly, the presence of the described signatures in our cohort was in line with previous findings obtained by microarray analysis by Banchereau and colleagues [12]. This indicated that the signatures were a reliable approach that could substitute transcriptomic analysis.
Confirming previous data, disease activity was associated with each individual gene signature [5, 7, 10, 12]. In line with the findings of Banchereau et al., the PLC signature showed the highest level of association with disease activity in a cross-sectional cohort [12]. Additionally, we showed in our longitudinal cohort of Tx-naïve patients that the PLC signature was also significantly aligned with disease activity. Yet, the correlation coefficients were rather low. Disease activity is measured by scoring the involvement of various disease domains. Here we showed that each gene signature was linked to specific disease domains. The M1.2 IFN signature was associated with the skin domain, while the NPh and PLC signatures were significantly associated with the musculoskeletal and constitutional domains. In contrast to previous findings, the NPh signature was not associated with renal involvement [10, 12, 16]. As shown by Banchereau et al. and Wither et al., the NPh signature was mostly increased during the active phase of lupus nephritis [10, 12]. The contrast between the results of our study and previous published results might be due to the relatively low number of patients who had active lupus nephritis at the time of sample collection. Nevertheless, these data indicate that a low correlation of individual gene signatures with disease activity could be a consequence of the effect of these signatures on different disease domains.
Our longitudinal data indicated that there might be an association between prednisone use and a positive NPh signature. This observation was in line with previous data showing that neutrophil numbers are increased in individuals using CSs [12, 22, 23]. Our study is the first longitudinal study that has confirmed the previous findings by Banchereau et al. that CSs influence the NPh signature. In addition, the finding that the PLC signature in longitudinal cSLE samples is sensitive to changes in disease activity and is affected by the use of prednisone and MMF is in line with previously described results [12]. Studies in the MRL/lpr mouse model for SLE showed that prednisone treatment was associated with a significant decrease in plasma cell numbers [24], which in turn was linked to a decrease in BLIMP-1, which regulates plasma cell formation [25]. Interestingly, BLIMP-1 is correlated with increased plasma cell numbers and disease activity in SLE patients [25, 26]. Moreover, the neutrophil count and anti-dsDNA represented the NPh and PLC signatures, indicating that cell compositions were drivers of the gene signatures. Regarding the IFNa2 levels, we showed that the IFN gene signatures did not parallel changes in disease activity, while ultrasensitive analysis of IFNa2 did. This was in line with previous findings [27]. This finding indicated that, in contrast to the NPh and PLC signatures, the IFN gene signatures were not influenced by the use of medication and potentially had a biological role in disease manifestation. Future longitudinal studies in SLE patients are needed to confirm our observations.
We identified three fingerprints by cluster analysis of four different gene signatures. These fingerprints were able to discriminate between patients who were in remission (fingerprint-1- ‘all-signatures-low’) and those who had high disease activity (fingerprint-3- ‘high NPh and/or PLC’) in two different cohorts. This observation was in line with previous work showing that adult SLE patients with a low IFN-I signature had significantly lower disease activity compared with patients with a high IFN-I and a high NPh signature [23]. Moreover, the demonstration that unsupervised hierarchical clustering analysis paralleled the identification of the gene fingerprint groups showed the robustness of this novel approach.
Our logistic regression model indicated that fingerprints were associated with different organ domains. The identified associations highly reflected the drivers of each fingerprint group. Fingerprint-1 was particularly driven by the M1.2 IFN gene signature, as this signature was associated with skin involvement. Fingerprint-2 seemed to be driven by the M5.12 gene signature, as this signature was not associated with any organ domain. Finally, fingerprint-3 was driven by the NPh and PLC signatures, as these signatures were associated with the musculoskeletal and constitutional domains. Moreover, our results illustrated that patients’ autoantibody profiles differed between patients within the various fingerprint groups, adding to our knowledge of the relationships between autoantibody profile, disease activity and disease phenotype [28, 29], yet also highlighting the gaps in our knowledge and underlining the fact that mere autoantibody profiles are not enough for subtyping SLE patients.
Previous data showed that the presence of an IFN signature is associated with an increased chance of disease flare in 5 years [30], supporting a role in disease pathogenesis. Also, higher baseline serum IFN-alpha levels measured by Simoa during SLE remission identified patients at risk of relapse [31]. Interestingly, in our cohorts, the patients with only high IFN scores were clustered together in fingerprint-2, and they had intermediate DASs when compared with patients with fingerprint-1 and fingerprint-3. Further studies will be needed to show whether these patients may be particularly at risk of developing a disease flare. Recently, Northcott et al. showed that high expression of IFN-I is associated with limited efficacy of glucocorticoids in SLE patients, suggesting that IFN gene signatures can predict treatment efficacy and therefore are candidates for improving individualized treatment choices in the future [32].
This study has several strengths. This is a study measuring four different gene signatures in a longitudinal multicentre cohort of SLE patients with ±25% being Txnaive. Moreover, the reproducibility of our observations in an independent replication cohort and unsupervised clustering strategy with similar findings shows the robustness of this approach. Our study also has limitations. With the current cohort of 101 patients, we were underpowered to study specific disease phenotypes such as lupus nephritis and neuropsychiatric lupus. Furthermore, disease activity was assessed by the SELENA-SLEDAI. A disadvantage of this scoring system is that it does not consider improvement or worsening of disease items. Therefore, the SELENA-SLEDAI is less sensitive to changes in disease activity compared with other measurement scales [33]. Finally, it is important to mention that gene signatures and fingerprints, especially the NPh and PLC signatures, may be influenced by medication use. Therefore, medication use should always be considered as a confounding factor before the results are interpreted based on fingerprints alone.
In conclusion, this study showed that using PCA to identify indicator genes for gene groups is a successful method for translating existing transcriptomic data into a tool that can be applied in clinical practice. We confirmed the activation of four gene signatures previously identified by transcriptomics and reproduced these data in an independent replication cohort. Moreover, combining the gene signatures into so-called fingerprints enabled us to stratify patients into subgroups with similar activated immune pathways that were associated with disease activity over time in our longitudinal cohort study. The heterogeneity of SLE is reflected in the variability of drug responsiveness between patients. This is expected to increase, given the current focus on the development of new biologics that target specific molecules or immune pathways. We have identified a molecular tool for stratifying patients into groups with similar biological disease profiles, which has the potential to guide individualized treatment choices and to improve upon the trial-and-error treatment approach of the present time. Our findings should be confirmed in large longitudinal studies to elucidate the applicability of this tool for prediction of responses to treatments interfering with the aberrantly activated immune pathways.
Supplementary Material
Acknowledgements
The research for this study was performed within the framework of the Erasmus Postgraduate School of Molecular Medicine. The authors thank all the cSLE patients and HCs for taking part in this study. The authors thank I. Bodewes, S. Bergkamp, D. Timmermans, K. van Rijswijk and L. van den Berg for assistance with sample collection and data acquisition. Furthermore, the authors thank Dr. J. Göpfert, head of Applied Biomarkers and Immunoassays at Natural and Medical Sciences Institute (NMI) for the measurement of the IFNa2 with the Simoa. All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. S.K. and M.A.V. contributed to the study conception and design. M.J.W., C.G.v.H.-M., S.T. and M.A.V. contributed to the experimental work. M.J.W., D.S.-M., C.G.v.H.-M., S.J.v.T., N.G., E.J.H.S., E.P.A.H.H., P.C.E.H.M., D.M.C.B., D.D., M.V., J.M.v.d.B., K.B., S.K. and M.A.V. contributed to the acquisiton of data. M.J.W., D.S.-M., S.J.v.T., S.K. and M.A.V. contributed to the analysis and interpretation of the data. M.J.W., S.K. and M.A.V. contributed to the writing of the draft manuscript. M.J.W., D.S.-M., C.G.v.H.-M., S.J.v.T., N.G., E.J.H.S., E.P.A.H.H., P.C.E.H.M., D.M.C.B., D.D., M.V., J.M.v.d.B., K.B., S.K. and M.A.V. contributed to the rewriting of the manuscript.
Funding: This work was made possible by the support of the Sophia Children’s Hospital Fund [B18-04], NVLE (Dutch patient organization for Lupus, APS, Scleroderma and MCTD) [BP12-1–261] and the Dutch Arthritis Society [CO-19–001].
Disclosure statement: The authors have declared no conflicts of interest.
Contributor Information
M Javad Wahadat, Department of Immunology, Erasmus MC; Department of Paediatric Rheumatology, Erasmus MC—Sophia Children’s hospital, University Medical Center Rotterdam, Rotterdam.
Dieneke Schonenberg-Meinema, Department of Pediatric Immunology, Rheumatology and Infectious diseases, Emma Children's Hospital, Amsterdam University Medical Centres, University of Amsterdam, Amsterdam.
Cornelia G van Helden-Meeuwsen, Department of Immunology, Erasmus MC.
Sander J van Tilburg, Department of Immunology, Erasmus MC.
Noortje Groot, Department of Paediatric Rheumatology, Erasmus MC—Sophia Children’s hospital, University Medical Center Rotterdam, Rotterdam.
Ellen J H Schatorjé, Department of Paediatric Rheumatology, Amalia Children’s Hospital, Radboudumc; Department of Paediatric Rheumatology, St. Maartenskliniek, Nijmegen.
Esther P A H Hoppenreijs, Department of Paediatric Rheumatology, Amalia Children’s Hospital, Radboudumc; Department of Paediatric Rheumatology, St. Maartenskliniek, Nijmegen.
Petra C E Hissink Muller, Department of Pediatrics, Division of Pediatric Rheumatology, Willem Alexander Children’s Hospital, Leiden University Medical Center, Leiden, The Netherlands.
Danielle M C Brinkman, Department of Pediatrics, Division of Pediatric Rheumatology, Willem Alexander Children’s Hospital, Leiden University Medical Center, Leiden, The Netherlands.
Denis Dvorak, Paediatric Rheumatology, Department of Paediatrics, Faculty of Medicine and Dentistry, Palacky University Olomouc and University Hospital, Olomouc, Czech Republic.
Marleen Verkaaik, Department of Paediatric Rheumatology, Erasmus MC—Sophia Children’s hospital, University Medical Center Rotterdam, Rotterdam.
J Merlijn van den Berg, Department of Pediatric Immunology, Rheumatology and Infectious diseases, Emma Children's Hospital, Amsterdam University Medical Centres, University of Amsterdam, Amsterdam.
Kateřina Bouchalova, Paediatric Rheumatology, Department of Paediatrics, Faculty of Medicine and Dentistry, Palacky University Olomouc and University Hospital, Olomouc, Czech Republic.
Sylvia Kamphuis, Department of Paediatric Rheumatology, Erasmus MC—Sophia Children’s hospital, University Medical Center Rotterdam, Rotterdam.
Marjan A Versnel, Department of Immunology, Erasmus MC.
Data availability statement
The data underlying this article are available in the article and in its online supplementary material, available at Rheumatology online.
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1. Mohan C, Putterman C.. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat Rev Nephrol 2015;11:329–41. [DOI] [PubMed] [Google Scholar]
- 2. Baechler EC, Batliwalla FM, Karypis G. et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 2003;100:2610–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennett L, Palucka AK, Arce E. et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med 2003;197:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kirou KA, Lee C, George S. et al. Coordinate overexpression of interferon-alpha–induced genes in systemic lupus erythematosus. Arthritis Rheum 2004;50:3958–67. [DOI] [PubMed] [Google Scholar]
- 5. Feng X, Wu H, Grossman JM. et al. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum 2006;54:2951–62. [DOI] [PubMed] [Google Scholar]
- 6. Chaussabel D, Quinn C, Shen J. et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 2008;29:150–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiche L, Jourde-Chiche N, Whalen E. et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol 2014;66:1583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Villanueva E, Yalavarthi S, Berthier CC. et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol 2011;187:538–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Streicher K, Morehouse CA, Groves CJ. et al. The plasma cell signature in autoimmune disease. Arthritis Rheumatol 2014;66:173–84. [DOI] [PubMed] [Google Scholar]
- 10. Wither JE, Prokopec SD, Noamani B. et al. Identification of a neutrophil-related gene expression signature that is enriched in adult systemic lupus erythematosus patients with active nephritis: clinical/pathologic associations and etiologic mechanisms. PLoS One 2018;13:e0196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carlucci PM, Purmalek MM, Dey AK.. Neutrophil subsets and their gene signature associate with vascular inflammation and coronary atherosclerosis in lupus. JCI Insight 2018;3:e99276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banchereau R, Hong S, Cantarel B. et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 2016;165:551–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Odendahl M, Jacobi A, Hansen A. et al. Disturbed peripheral B lymphocyte homeostasis in systemic lupus erythematosus. J Immunol 2000;165:5970–9. [DOI] [PubMed] [Google Scholar]
- 14. Kaplan MJ. Neutrophils in the pathogenesis and manifestations of SLE. Nat Rev Rheumatol 2011;7:691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wirestam L, Arve S, Linge P, Bengtsson AA.. Neutrophils—important communicators in systemic lupus erythematosus and antiphospholipid syndrome. Front Immunol 2019;10:2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jourde-Chiche N, Whalen E, Gondouin B. et al. Modular transcriptional repertoire analyses identify a blood neutrophil signature as a candidate biomarker for lupus nephritis. Rheumatology (Oxford) 2017;56:477–87. [DOI] [PubMed] [Google Scholar]
- 17. Toro-Dominguez D, Martorell-Marugan J, Goldman D. et al. Stratification of systemic lupus erythematosus patients into three groups of disease activity progression according to longitudinal gene expression. Arthritis Rheumatol 2018;70:2025–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petri M, Orbai AM, Alarcon GS. et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum 2012;64:2677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aringer M, Costenbader K, Daikh D. et al. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Arthritis Rheumatol 2019;71:1400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petri M, Kim MY, Kalunian KC. et al. ; OC-SELENA Trial. Combined oral contraceptives in women with systemic lupus erythematosus. N Engl J Med 2005;353:2550–8. [DOI] [PubMed] [Google Scholar]
- 21. Bodewes ILA, Al-Ali S, van Helden-Meeuwsen CG. et al. ; UK Primary Sjögren’s Syndrome registry. Systemic interferon type I and type II signatures in primary Sjögren’s syndrome reveal differences in biological disease activity. Rheumatology (Oxford) 2018;57:921–30. [DOI] [PubMed] [Google Scholar]
- 22. Shoenfeld Y, Gurewich Y, Gallant LA, Pinkhas J.. Prednisone-induced leukocytosis. Influence of dosage, method and duration of administration on the degree of leukocytosis. Am J Med 1981;71:773–8. [DOI] [PubMed] [Google Scholar]
- 23. Chasset F, Ribi C, Trendelenburg M. et al. ; for the Swiss SLE Cohort Study (SSCS). Identification of highly active systemic lupus erythematosus by combined type I interferon and neutrophil gene scores vs classical serologic markers. Rheumatology (Oxford) 2020;59:3468–78. [DOI] [PubMed] [Google Scholar]
- 24. Yan SX, Deng XM, Wang QT, Sun XJ, Wei W.. Prednisone treatment inhibits the differentiation of B lymphocytes into plasma cells in MRL/MpSlac-lpr mice. Acta Pharmacol Sin 2015;36:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo J, Niu X, Liu H. et al. Up-regulation of transcription factor Blimp1 in systemic lupus erythematosus. Mol Immunol 2013;56:574–82. [DOI] [PubMed] [Google Scholar]
- 26. Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ. et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 2003;19:607–20. [DOI] [PubMed] [Google Scholar]
- 27. Mathian A, Mouries-Martin S, Dorgham K. et al. Monitoring disease activity in systemic lupus erythematosus with single-molecule array digital enzyme-linked immunosorbent assay quantification of serum interferon-alpha. Arthritis Rheumatol 2019;71:756–65. [DOI] [PubMed] [Google Scholar]
- 28. To CH, Petri M.. Is antibody clustering predictive of clinical subsets and damage in systemic lupus erythematosus? Arthritis Rheum 2005;52:4003–10. [DOI] [PubMed] [Google Scholar]
- 29. Jurencak R, Fritzler M, Tyrrell P. et al. Autoantibodies in pediatric systemic lupus erythematosus: ethnic grouping, cluster analysis, and clinical correlations. J Rheumatol 2009;36:416–21. [DOI] [PubMed] [Google Scholar]
- 30. Mai L, Asaduzzaman A, Noamani B. et al. The baseline interferon signature predicts disease severity over the subsequent 5 years in systemic lupus erythematosus. Arthritis Res Ther 2021;23:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathian A, Mouries-Martin S, Dorgham K. et al. Ultrasensitive serum interferon-alpha quantification during SLE remission identifies patients at risk for relapse. Ann Rheum Dis 2019;78:1669–76. [DOI] [PubMed] [Google Scholar]
- 32. Northcott M, Gearing LJ, Nim HT. et al. Glucocorticoid gene signatures in systemic lupus erythematosus and the effects of type I interferon: a cross-sectional and in-vitro study. Lancet Rheumatol 2021;3:e357–70. [DOI] [PubMed] [Google Scholar]
- 33. Mikdashi J, Nived O.. Measuring disease activity in adults with systemic lupus erythematosus: the challenges of administrative burden and responsiveness to patient concerns in clinical research. Arthritis Res Ther 2015;17:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material, available at Rheumatology online.