Abstract
Background
Selected molecular biomarkers were incorporated into the US cancer registry reporting for patients with brain tumors beginning in 2018. We investigated the completeness and validity of these variables and described the epidemiology of molecularly defined brain tumor types.
Methods
Brain tumor patients with histopathologically confirmed diagnosis in 2018 were identified within the Central Brain Tumor Registry of the United States and NCI’s Surveillance, Epidemiology, and End Results Incidence databases. The brain molecular markers (BMM) site-specific data item was assessed for coding completeness and validity. 1p/19q status, MGMT promoter methylation, WHO grade data items, and new ICD-O-3 codes were additionally evaluated. These data were used to profile the characteristics and age-adjusted incidence rates per 100 000 population of molecularly defined brain tumors with 95% confidence intervals (95% CI).
Results
BMM completeness across the applicable tumor types was 75%-92% and demonstrated favorable coding validity. IDH-wildtype glioblastomas’ incidence rate was 1.74 (95% CI: 1.69-1.78), as compared to 0.14 for WHO grade 2 (95% CI: 0.12-0.15), 0.15 for grade 3 (95% CI: 0.14-0.16), and 0.07 for grade 4 (95% CI: 0.06-0.08) IDH-mutant astrocytomas. Irrespective of WHO grade, IDH mutation prevalence was highest in adolescent and young adult patients, and IDH-mutant astrocytomas were more frequently MGMT promoter methylated. Among pediatric-type tumors, the incidence rate was 0.06 for H3K27M-mutant diffuse midline gliomas (95% CI: 0.05-0.07), 0.03 for SHH-activated/TP53-wildtype medulloblastomas (95% CI: 0.02-0.03), and <0.01 for both C19MC-altered embryonal tumor with multilayered rosettes and RELA-fusion ependymomas.
Conclusions
Our findings illustrate the success of developing a dedicated, integrated diagnosis variable, which provides critical molecular information about brain tumors related to accurate diagnosis.
Keywords: biomarkers, brain tumor, CBTRUS, IDH, molecular epidemiology
Key Points.
Critical brain tumor molecular data are now available from US cancer registries.
There was a high level of reporting completeness for new brain molecular data items.
We report the first epidemiological estimates for molecularly defined brain tumors.
Importance of the Study.
In 2018, CBTRUS—with associated cancer registry stakeholders—implemented new integrated diagnosis ICD-O-3 codes and molecular site-specific data items for patients with brain tumors, including a brain molecular markers (BMM) data item that reported: (1) IDH and 1p/19q statuses for adult-type diffuse gliomas, (2) SHH-activated and TP53-wildtype status for medulloblastomas, and (3) C19MC alteration for embryonal tumor with multilayered rosettes. We demonstrate a high level of completeness (75%-92%) and validity for BMM coding. By leveraging the new BMM, MGMT promoter methylation, and WHO grade data items, we generated the first national incidence and epidemiology estimates for adult-type diffuse gliomas (by IDH and MGMT statuses), H3 K27M-mutant diffuse midline gliomas, molecular subgroups of medulloblastomas, C19MC-altered embryonal tumor with multilayered rosettes, and RELA-fusion ependymomas. Our findings underscore the value of developing dedicated integrated diagnosis data items for cancer reporting and illustrate their utility in helping to advance our understanding of patients’ brain tumors at the population level.
Before 2016, the classification of central nervous system (CNS) tumors has historically been defined by histological findings and ancillary immunohistochemical and/or ultrastructural examination. Molecular investigation of brain tumors has led to the identification of biomarkers with important diagnostic, prognostic, and predictive value.1 For instance, in adult-type diffuse gliomas, isocitrate dehydrogenase 1/2 (IDH) mutations and whole-arm codeletion of 1p and 19q are associated with favorable prognosis—as well as with improved survival in patients treated with radiotherapy or alkylating chemotherapy.2–7 Additionally, O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation is predictive of a survival benefit from temozolomide treatment among glioblastoma patients.8–10 Beyond diffuse gliomas, molecular subtyping has also been shown to have prognostic value in such tumor types as medulloblastomas and ependymomas.11–13 Reflecting their clinicopathologic utility, many of these molecular biomarkers were integrated into the revised 4th edition of the WHO Classification of Tumors of the CNS (WHO-CNS4) released in 2016, detailed in the practical recommendations from the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy (ie, cIMPACT-NOW), and implemented in the clinical practice guidelines from the National Comprehensive Cancer Network.14–22 The 5th edition of the WHO CNS (WHO-CNS5) builds on the transformative changes of the WHO-CNS4, restructuring both CNS tumor taxonomy and nomenclature into a more parsimonious classification system.23
Starting in 2010, site-specific factors were implemented in the US cancer registry reporting for WHO grade, MGMT promoter methylation status, chromosome arm 1p loss of heterozygosity, and chromosome arm 19q loss of heterozygosity; however, a majority of the molecular biomarkers required for accurate brain tumor classification according to the WHO-CNS4 remained unreported and several of the tumor types lacked unique ICD-O-3 codes.24,25 Therefore, in 2017—in close collaboration with the North American Association of Central Cancer Registries (NAACCR)’s Site-Specific Data Items Task Force, Centers for Disease Control and Prevention’s (CDC) National Program of Cancer Registries, NCI’s Surveillance, Epidemiology and End Results (SEER), and other stakeholders—the Central Brain Tumor Registry of the United States (CBTRUS) spearheaded the design of a new “brain molecular markers” (BMM) site-specific data item that integrated IDH mutational status for adult-type diffuse gliomas (and 1p/19q codeletion for oligodendrogliomas), SHH-activated and TP53-wildtype status for medulloblastomas, and C19MC alteration for embryonal tumor with multilayered rosettes (Table 1).26 Registry reporting of the BMM data item, as well as updated site-specific data items for clinical and pathological grade, began in all US central cancer registries on January 1, 2018. Concurrently, updated site-specific data items for MGMT promoter methylation, 1p and 19q loss of heterozygosity statuses were implemented by central cancer registries included in the SEER system. Furthermore, in concert with the NAACCR’s “International Classification of Diseases for Oncology, third edition (ICD-O-3) Update Implementation Committee,” several updated ICD-O-3 histological/behavior codes for WHO-CNS4 entities (Supplementary Table 1) were incorporated into the collection practices of cancer registries effective January 1, 2018—including codes for molecularly defined diagnoses, such as “diffuse midline glioma, H3 K27M-mutant” (grouped among “diffuse midline glioma, H3 K27-altered” in WHO-CNS5), “ependymoma, RELA fusion-positive” (“supratentorial ependymoma, ZFTA fusion-positive” in WHO-CNS5), and others. We have previously detailed the history of these important efforts.26
Table 1.
Coding Definitions for the Brain Molecular Markers (BMM) Data Items
| Code | Description |
|---|---|
| 01 | Diffuse astrocytoma, IDH-mutant (9400/3) |
| 02 | Diffuse astrocytoma, IDH-wildtype (9400/3) |
| 03 | Anaplastic astrocytoma, IDH-mutant (9401/3) |
| 04 | Anaplastic astrocytoma, IDH-wildtype (9401/3) |
| 05 | Glioblastoma, IDH-wildtype (9440/3) |
| 06 | Oligodendroglioma, IDH-mutant and 1p/19q codeleted (9450/3) |
| 07 | Anaplastic oligodendroglioma, IDH-mutant and 1p/19q codeleted (9451/3) |
| 08 | Medulloblastoma, SHH-activated, and TP53-wildtype (9471/3) |
| 09 | Embryonal tumor with multilayered rosettes, C19MC-altered (9478/3) |
| 85 | Not applicable: Histology not 9400/3, 9401/3, 9440/3, 9450/3, 9451/3, 9471/3, 9478/3 |
| 86 | Benign or borderline tumor |
| 87 | Test ordered, results not in chart |
| 88 | Not applicable: Information not collected for this case (If this item is required by your standard setter, use of code 88 will result in an edit error.) |
| 99 | Not documented in patient record; No microscopic confirmation; Brain molecular markers not assessed or unknown if assessed |
NAACCR Data Item #3816. This data item applies only to ICD-O-3 histology codes: 9400/3, 9401/3, 9440/3, 9450/3, 9451/3, 9471/3, and 9478/3. If a histology is not included in this list, code 85 was assigned. Only one code is applicable for each tumor.
The incorporation of molecular biomarkers and WHO-CNS4-5 entities into registry reporting for patients with brain tumors represents a significant step forward in realizing precision neuro-oncology. In order to inform the widespread implementation of these new brain tumor-specific variables, we first evaluated their completeness and validity in the CBTRUS database, which provides population-based data of patients with brain tumors. We then leveraged the CBTRUS data to provide the initial national estimates of the epidemiology of molecularly defined brain tumor types in the United States. Importantly, we analyzed the molecular biomarker data both in the context of the revised WHO-CNS4—upon which the new variables and codes were designed—and in the context of the WHO-CNS5, so as to facilitate their clinical relevance.
Materials and Methods
Data Sources and Variable Design
CBTRUS—in collaboration with the CDC’s National Program of Cancer Registries and the NCI’s SEER programs—is the largest population-based registry focused on primary malignant and non-malignant brain and other CNS tumors in the United States, covering the entire US population.27–30 The NAACCR Item #3816 “Brain Molecular Markers” (BMM) site-specific data item was developed using the revised WHO-CNS4 criteria and was implemented by cancer registries for patients diagnosed starting on January 1, 2018; with the intent to capture clinically important brain cancer subtypes identified by molecular markers that, at the time, were not defined in the ICD-O-3. Concomitantly, new brain site-specific data items were implemented for “chromosome 1p: loss of heterozygosity (LOH)” (NAACCR Item #3801), “chromosome 19q: loss of heterozygosity LOH” (NAACCR Item #3802), “Methylation of O6-Methylguanine-Methyltransferase (MGMT)” (NAACCR Item #3889), and “Grade Pathological” (ie, WHO CNS grade, NAACCR Item #3844). While BMM was available for all 51 central cancer registries, data on 1p/19q loss of heterozygosity and MGMT promoter methylation were available for SEER18 registries only (a subset of CBTRUS data, including ~28% of the US population).30 The brain site-specific data items’ code definitions are detailed in the Supplementary Note. Therefore, patients diagnosed with a histopathologically confirmed brain tumor from January 1, 2018 to December 31, 2018 (the last date of available data for this CBTRUS data release) were identified from the CBTRUS analytic database and a special dataset provided by SEER databases using ICD-O-3 codes. Cases reported as death certificate only or diagnosed by autopsy were excluded. This study was approved as part of an exempt protocol by the institutional review board of Duke University Health System.
Completeness and Frequency of Brain Molecular Biomarker Site-Specific Data Items
For the BMM data item, completeness was defined as a BMM code 1-9 being reported for a case with an applicable ICD-O-3 histology, which were “diffuse” astrocytoma (9400/3), “anaplastic” astrocytoma (9401/3), glioblastoma (9440/3), “oligodendroglioma, NOS” (9450/3), “anaplastic” oligodendroglioma (9541/3), desmoplastic/nodular medulloblastoma (9471/3), and embryonal tumor with multilayered rosettes (9478/3); whereas BMM codes 87, 88, and 99 were classified as incomplete. Cases were considered erroneously coded if they had an applicable ICD-O-3 histology but an inappropriate BMM code 1-9 for that histology, or were coded as 85 (ie, that the ICD-O-3 was not applicable) or 86 (ie, that the tumor was benign or borderline). For analyses of adult-type diffuse gliomas, molecular statuses were categorized as IDH-mutant astrocytoma (BMM codes 1 and 3), IDH-wildtype astrocytoma and glioblastoma (BMM codes 2, 4, and 5), and IDH-mutant and 1p/19q-codeleted oligodendroglioma (BMM codes 6 and 7).
The chromosome 1p and 19q data items were considered complete if a case had a code 0 (deletion not present) or 1 (deletion present) reported for both 1p and 19q. Cases were considered incomplete if either 1p or 19q statuses were unknown (codes 5-9). Completeness of MGMT promoter methylation was defined by codes 0-3; and otherwise incomplete if reported as codes 6-9. The completeness of 1p and 19q loss of heterozygosity and of MGMT promoter methylation was assessed across infiltrative gliomas as defined by both ICD-O-3 codes and BMM tumor types. Among adult-type diffuse gliomas, the internal validity was defined as the concordance between BMM and 1p/19q loss of heterozygosity coding. Pathological grade was considered as available if reported as a code corresponding to WHO CNS grades 1-4, low-grade NOS, or high-grade NOS; and otherwise considered as unavailable (codes A-D, 9). Completeness was assessed using data from CBTRUS for BMM and from the SEER18 Incidence database for 1p/19q, MGMT promoter methylation, and pathological grade.
Validation of BMM Coding From Medical Records
To assess the external validity of BMM coding, the institutional data submitted to cancer registries for brain tumor patients histopathologically diagnosed in 2018-2019 at Brigham and Women’s Hospital, Dana-Farber Cancer Institute, and Massachusetts General Hospital were reviewed and compared to corresponding integrated diagnosis pathology data from the medical record. The positive predictive values and specificities of BMM coding were calculated. 2018 and 2019 were selected to ensure sufficient sample size for the validation of BMM and because coding practices were unchanged from 2018 to 2019. This analysis was approved by the Mass General Brigham Institutional Review Board (#2019P000950).
Epidemiology of Brain Tumors With Molecular Biomarkers
Using data provided by CBTRUS, annual age-adjusted incidence rates and demographic statistics—including age, sex, and race/ethnicity—were evaluated across molecularly defined brain tumor types and subtypes. In addition to molecular tumor types identified by the BMM data item, novel ICD-O-3 brain tumor types and codes (implemented for patients diagnosed starting on January 1, 2018) were also evaluated. Incidence rates were generated per 100 000 population and age adjusted to the 2000 US Standard Population, with corresponding 95% confidence intervals (CIs) calculated as described previously.31 Tumor types were analyzed by both their WHO-CNS4 classification and—when possible—their WHO-CNS5 classification.
For adult-type diffuse gliomas, tumors were further stratified by WHO CNS grade using the pathological grade data item. Because the WHO-CNS5 grading criteria had not yet been incorporated into registry reporting, the grades reported here were based on the revised WHO-CNS4 grading criteria. Cases reported as grade 1 or “low-grade, NOS” were combined into the grade 2 category because there no longer are grade 1 adult-type diffuse gliomas in the WHO-CNS5 classification. Cases reported with a new “glioblastoma, IDH-mutant” ICD-O-3 code (9445/3) were grouped with BMM-defined IDH-mutant astrocytomas. Additionally, the distribution of IDH mutation was investigated among adult-type diffuse astrocytic gliomas, as stratified by sex, age (pediatric <15 years old, adolescent and young adult 15- to 39 years old, adult ≥40 years old), and MGMT promoter methylation status. IDH mutation distributions were analyzed using both the revised WHO-CNS4 classification—from which the BMM data item was developed, and which includes the former “diffuse” and “anaplastic” terminology—and the WHO-CNS5 classification (by stratifying tumors according to their WHO pathological grade). Patient race/ethnicity was categorized as White non-Hispanic, Black non-Hispanic, Hispanic, Asian/Pacific Islander, other, or unknown.
Data Analysis
The BMM data item was the primary focus of our analyses, with the 1p/19q, MGMT promoter methylation, and WHO CNS grade data items additionally evaluated as a secondary aim. Cells containing either <16 cases, or which could be used to back-calculate a cell value of <16 cases, were suppressed in accordance with CBTRUS data use agreements with CDC and NCI. All frequencies and figures were generated in R v4.1.1.32 Incidence rates were calculated using SEER*Stat v8.3.9.33
Results and Discussion
Brain Molecular Marker Completeness in Registry Data
Because 2018 was the first year that the new site-specific data items were implemented by cancer registries, we first examined the completeness of the BMM data item in CBTRUS data (BMM codes are defined in Table 1). We identified 15 079 patients who were histopathologically confirmed with BMM-applicable ICD-O-3 codes for adult-type diffuse glioma and medulloblastoma (and <16 with embryonal tumor with multilayered rosettes) in 2018. Overall, a BMM molecular status was reported for 79% of cases, ranging from 78% of adult-type diffuse astrocytic gliomas (with IDH status) to 90% of oligodendrogliomas (with IDH and 1p/19q statuses) and 81% of desmoplastic/nodular medulloblastomas (with SHH-activated and TP53-wildtype statuses) (Table 2). Although <16 embryonal tumor with multilayered rosettes were identified, all were reported to be C19MC-altered. This initial level of BMM completeness compared favorably to that of other molecular variables in the first year of their introduction: for instance, in 2010 <50% of oligodendrogliomas had complete reporting for 1p and 19q loss of heterozygosity variables.24,34
Table 2.
Completeness and Frequency of the Reported Brain Molecular Markers (BMM) Site-Specific Data Item by US Central Cancer Registries for Diagnosis Year 2018
| Total, n | BMM Coding, % | Erroneously Codeda (85, 86), % | Incomplete coding (87 88 99)b, % |
|||||
|---|---|---|---|---|---|---|---|---|
| IDHmut (1, 3) | IDHwt (2, 4, 5) | IDHmut 1p/19q codel (6, 7) | SHH-activated + TP53wt (8) | C19MC- altered (9) | ||||
| Adult-type diffuse astrocytic glioma | 13 999 | 8% | 70% | 0 | 0 | 0 | <1% | 22% |
| 9400/3: “Diffuse” astrocytoma | 1304 | 43% | 31% | 0 | 0 | 0 | <1% | 25% |
| 9401/3: “Anaplastic” astrocytoma | 1363 | 43% | 43% | 0 | 0 | 0 | <1% | 15% |
| 9440/3: Glioblastoma | 11 332 | 0 | 78% | 0 | 0 | 0 | <1% | 22% |
| Adult-type diffuse oligodendroglial glioma | 976 | 0 | 0 | 90% | 0 | 0 | 0 | 10% |
| 9450/3: “Oligodendroglioma, NOS” | 655 | 0 | 0 | 89% | 0 | 0 | 0 | 11% |
| 9451/3: “Anaplastic” oligodendroglioma | 321 | 0 | 0 | 92% | 0 | 0 | 0 | 8% |
| 9471/3: Desmoplastic/ nodular medulloblastoma | 104 | 0 | 0 | 0 | 81% | 0 | 0 | 19% |
| 9478/3: ETMR | <16 | 0 | 0 | 0 | 0 | 100% | 0 | 0 |
Abbreviations: BMM, brain molecular markers; ETMR, embryonal tumor with multilayered rosettes; mut, mutant; wt, wildtype.
Data provided by CDC’s National Program of Cancer Registries and NCI’s Surveillance, Epidemiology and End Results Program, November 2020 submissions.
aNo cases were erroneously reported as BMM code 85 (ie, not applicable histology), and n < 16 cases were erroneously reported as BMM code 86 (ie, benign or borderline tumor).
bIn <0.5% of cases (n = 54), BMM testing was reported as ordered, but the result was not available in the chart (BMM code 87). No cases were reported as BMM code 88 (ie, the registry did not collect information about that case). BMM code 99 (n = 3104) can indicate a lack of molecular testing, lack of testing documentation in the patient’s record, or unknown if molecular status was assessed (including testing was done, but not reported by the registry).
Incomplete BMM coding was highest among cases reported as “diffuse” astrocytomas (9400/3; likely representing grade 2 astrocytomas in WHO-CNS5)—with 25% of such cases missing an IDH mutation status in the registry data. Of note, the molecular site-specific data items use a single code to indicate a lack of testing, lack of testing documentation in the patient’s record, and unknown if molecular status was assessed—all of which were considered as incomplete data in this study. If required IDH and/or 1p/19q testing was truly infeasible in a subset of cases (eg, testing failed due to insufficient tissue), such cases could be classified as “Not Otherwise Specified (NOS).” 14 Across all BMM-applicable ICD-O-3 diagnoses, <1% received an improper BMM code (eg, a glioblastoma being erroneously encoded as a benign/borderline tumor type) and none received a BMM code that was specified for a different tumor type (eg, a glioblastoma being erroneously encoded as a SHH-activated and TP53-wildtype medulloblastoma).
Validation of Brain Molecular Marker Coding Using Multi-Institutional Data
To help validate BMM coding in the real-world setting, we compared the BMM codes submitted to cancer registries for 950 patients who were histopathologically diagnosed with brain tumors between 2018 and 2019 at three hospitals to the molecular diagnoses from their corresponding pathology reports (Supplementary Table 3). Among BMM-coded IDH-mutant astrocytomas (n = 65), the positive predictive value was 95.4%, with the erroneously encoded cases representing an IDH-wildtype astrocytoma, BRAF-mutant astrocytoma, and H3 K27-altered diffuse midline glioma (each 1.5%). Among BMM-coded IDH-wildtype astrocytomas and glioblastomas (n = 429), the positive predictive value was 98.1%, with 0.9% (n = 4) actually IDH-mutant and 0.9% (n = 4) representing H3 K27-altered diffuse midline gliomas. The specificity of BMM coding for adult-type diffuse gliomas ranged from 93.7% to 99.9%. For the BMM codes for SHH-activated and TP53-wildtype medulloblastoma and embryonal tumor with multilayered rosettes C19MC-altered, the positive predictive values were both 100%, although only five and two patients, respectively, were identified in this cohort. Although it is possible that this limited cohort derived from academic institutional data may not be wholly representative of the registry coding practices nationwide, these results provide reassurance regarding the validity of BMM coding.
Completeness and Concordance of Ancillary Brain Molecular Biomarker Site-Specific Data Items
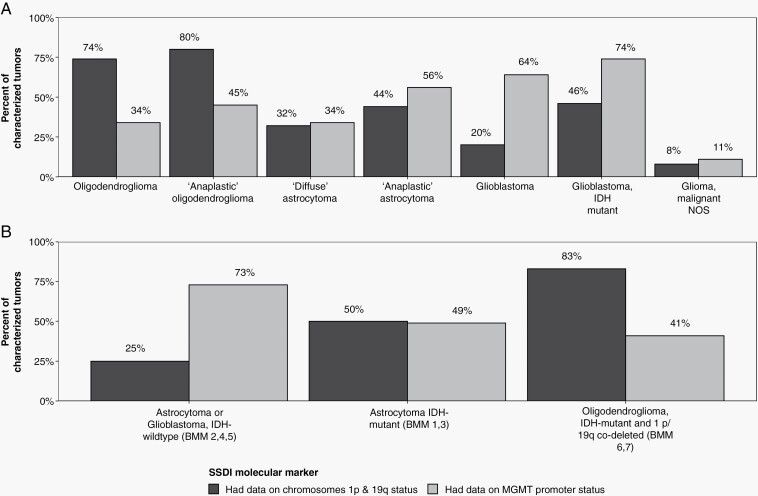
In addition to BMM being introduced in 2018, several site-specific data items were also newly designed to replace the prior brain Site-Specific Factors—including chromosome 1p and 19q loss of heterozygosity, and MGMT promoter methylation, as well as pathological grade (ie, WHO CNS grade). Having previously described the completeness of those retired versions,24,25 we evaluated the completeness of registry coding for the new site-specific data item versions (Figure 1; Supplementary Tables 1 and 2)—using data derived from SEER, a subset of the larger CBTRUS dataset. The completeness of the 2018 1p and 19q data items surpassed that of their former versions: whereas in 2012 only 53%-54% of ICD-O-3-coded oligodendrogliomas had complete 1p and 19q data,24 74%-80% did for the 1p and 19q in 2018 (Figure 1A). Notably, the completeness of 1p and 19q data items for grade 2 and “anaplastic” oligodendroglioma cases was 74% and 80%, respectively. However, given the diagnostic necessity of 1p/19q status for diagnosing an oligodendroglioma in 2018, incomplete reporting likely represents suboptimal collection by registries. For example, even among cases reported by BMM coding as IDH-mutant and 1p/19q-codeleted oligodendrogliomas—for which 1p/19q codeletion identification is required—only 82% also had complete data for 1p and 19q data items. By comparison, 25% of IDH-wildtype astrocytomas and glioblastomas and 50% of IDH-mutant astrocytomas were accompanied by complete 1p/19q data items (Figure 1B), likely reflecting that in the former case, 1p/19q testing is not indicated for IDH-wildtype gliomas, and in the latter case, that an IDH-mutant diffuse astrocytic glioma with clear ATRX loss and/or strong p53 positivity is sufficient for an astrocytoma diagnosis even in the absence of 1p/19q data.15
Figure 1.
Completeness of 1p/19q and MGMT promoter methylation statuses in adult-type diffuse gliomas from US central cancer registries for diagnosis year 2018. Completeness of 1p/19q loss of heterozygosity (black) data was defined as cases having a “present” or “not present” code for both 1p and 19q loss of heterozygosity site-specific data items. Completeness of MGMT promoter methylation (gray) data was defined as cases with codes 0-3. Other codes were considered as incomplete reporting. Completeness was assessed for adult-type diffuse gliomas, categorized both by (A) histology-based ICD-O-3 coding (WHO-CNS4) and (B) brain molecular marker-based integrated diagnosis classification (WHO-CNS5). Data provided by NCI’s SEER Program, November 2020 submissions.
Because 1p/19q status for adult-type diffuse gliomas was reported in both the BMM and separate 1p and 19q data items, we examined the internal concordance between these variables (Supplementary Table 4). Of the BMM-coded IDH-mutant and 1p/19q-codeleted oligodendrogliomas with complete data for BMM and 1p/19q variables (n = 273), 85.3% also were coded as 1p and 19q loss of heterozygosity, but 14.7% had a retained 1p, 19q, or both. Conversely, of the BMM-coded IDH-mutant and IDH-wildtype astrocytic gliomas with complete data (n = 205 and n = 879), 88.8%-90.1% were also encoded as having a retained 1p, 19q, or both; however, 9.9%-11.2% cases were reported to have both 1p and 19q loss of heterozygosity. Given that in our institutional cohort, only 2.3% (n = 1) of the 44 BMM-coded IDH-mutant and 1p/19q-codeleted oligodendrogliomas was in fact 1p/19q-retained and none of the BMM-coded IDH-mutant or IDH-wildtype astrocytic gliomas were actually 1p/19q-codeleted, together these data suggest that integrated diagnosis site-specific data items that are based on WHO-defined tumor types may provide more accurate reporting than separate variables for each individual mutation status (eg, separate data items for 1p and 19q statuses).
Based on the striking capability of MGMT promoter methylation status to predict the survival associated with temozolomide, MGMT promoter methylation testing was part of the standard-of-care workup for patients with glioblastoma in 2018.21 Herein, we found that 73% of BMM-coded IDH-wildtype astrocytomas and glioblastomas also had complete data about MGMT promoter status (Figure 1); in contrast to just 38% of ICD-O-3-coded glioblastomas from 2010 to 2016.24,35
Furthermore, a new pathological grade site-specific data item was implemented in 2018, which was defined using the WHO grade from the resection specimen—including the new inclusion of codes for “low-grade, NOS” and “high-grade, NOS,” which will enable the reporting of cases where a definitive grade could not be assigned. Completeness of pathological and clinical grade coding varied by tumor type (Supplementary Tables 1 and 2), including 66%-87% of adult-type diffuse gliomas, 79%-84% of ependymal tumors, and 72%-87% of meningiomas. These levels were similar to those reported for the prior version of the WHO grade variable.24,25 Particularly for adult-type diffuse gliomas, we have previously shown that the dedicated WHO grade variable provided superior information regarding tumor behavior, as compared to the behavior defined by ICD-O-3 coding alone.36 Herein, for instance, only 73% of cases reported as “diffuse” astrocytoma (ie, WHO grade 2 astrocytoma in WHO-CNS4; 9400/3) were in fact coded as grade 2, whereas 20% were grades 3-4. These data underscore the advantages of WHO-CNS5’s move from “diffuse” and “anaplastic” terminology to numeric grading within tumor types.
Epidemiology of Adult-Type Diffuse Gliomas by IDH Status
The favorable completeness and validity of the BMM data item were then leveraged to characterize the epidemiology of molecularly defined brain tumors in the United States—including the annual age-adjusted incidence rate, sex, age at diagnosis, and race/ethnicity. These analyses supplement the histology-based CBTRUS 2021 report on brain tumor epidemiology.27 Adult-type diffuse gliomas were classified using BMM into IDH-mutant astrocytoma, IDH-wildtype astrocytoma and glioblastoma, or IDH-mutant and 1p/19q-codeleted oligodendroglioma and then stratified by WHO grade (Table 3). Grading was reported in registry data using the WHO-CNS4 criteria. Of note, whereas the WHO-CNS5 recognizes IDH-wildtype glioblastoma (grade 4), it does not recognize IDH-wildtype astrocytomas of grades 2-3. Such histologic grades 2-3 IDH-wildtype diffuse gliomas without molecular features of glioblastoma (EGFR amplification, polysomy 7/monosomy 10, or TERT promoter mutation—which are not captured by registries) should now be rendered a descriptive diagnosis, with the recognition that this classification will evolve over time.
Table 3.
Annual Age-Adjusted Incidence, Median Age, Sex, and Race/Ethnicity of Molecularly Defined Brain Tumors From US Central Cancer Registries for Diagnosis Year 2018
| Tumor Type | ICD-O-3 Histology Codes | Gradea | Total Cases | Age-Adjusted Incidence per 100 000 (95% CI) | Age (Median, Interquartile Range) | Sexg | Race/Ethnicityf | ||
|---|---|---|---|---|---|---|---|---|---|
| Female (%) | White Non- Hispanic (%) | Black Non- Hispanic (%) | Hispanic (%) | ||||||
| Adult-type diffuse glioma | |||||||||
| IDHmut astrocytoma (BMM 1, 3) | 9400/3 9401/3 9445/3 | 2 | 425 | 0.14 (0.12-0.15) | 34 (27, 45) | 41.6% | 80.5% | 5.0% | 10.2% |
| 3 | 465 | 0.15 (0.14-0.16) | 37 (29, 48) | 44.7% | 82.2% | 5.4% | 9.5% | ||
| 4 | 241 | 0.07 (0.06-0.08) | 47 (36, 60) | 42.3% | 80.1% | 8.5% | 9.3% | ||
| IDHwt astrocytoma and glioblastomab (BMM 2, 4, 5) | 9400/3 9401/3 9440/3 | 2 | 190 | 0.05 (0.05-0.06) | 54 (32, 66) | 47.9% | 78.2% | -- | -- |
| 3 | 369 | 0.10 (0.09-0.11) | 59 (47, 70) | 45.5% | 84.4% | 6.6% | 5.7% | ||
| 4 | 6878 | 1.73 (1.69-1.78) | 65 (56, 72) | 40.2% | 83.2% | 6.0% | 8.3% | ||
| IDHmut and 1p/19q-codeleted oligodendroglioma (BMM 6,7) | 9450/3 9451/3 |
2 | 437 | 0.14 (0.13-0.15) | 42 (33, 54) | 47.6% | 76.0% | 5.6% | 14.0% |
| 3 | 274 | 0.08 (0.07-0.09) | 48 (37, 57) | 46.0% | 76.4% | -- | -- | ||
| Medulloblastoma | |||||||||
| SHH-activated and TP53wt (BMM 8) | 9471/3 | 76 | 0.03 (0.02-0.03) | 21 (6, 29) | 36.8% | 60.3% | -- | -- | |
| SHH-activated and TP53mut | 9476/3 | <16 | n/ac | -- | -- | -- | -- | -- | |
| WNT-activated | 9475/3 | <16 | n/ac | -- | -- | -- | -- | -- | |
| Non-WNT/non-SHH | 9477/3 | 47 | n/ac | 7 (4, 10) | 52.3% | 53.2% | -- | 36.4% | |
| Other tumor typesd,e | |||||||||
| Diffuse midline glioma, H3 K27M- mutant | 9385/3 | 144 | 0.05 (0.04-0.06) | 14 (7, 28) | 53.5% | 58.2% | -- | 22.7% | |
| ETMR C19MC- altered (BMM 9) | 9478/3 | <16 | <0.01 | -- | -- | -- | -- | -- | |
| RELA-fusion ependymoma | 9396/3 | <16 | <0.01 | -- | -- | -- | -- | -- |
Abbreviations: BMM, brain molecular markers variable; CI, confidence interval; ETMR, embryonal tumor with multilayered rosettes; mut, mutant; wt, wildtype.
Data provided by CDC’s National Program of Cancer Registries and NCI’s Surveillance, Epidemiology and End Results Program, November 2020 submissions.
-- Suppressed if case counts are <16.
aAdult-type diffuse glioma cases reported as WHO grade 1 or “low-grade, NOS” were grouped with WHO grade 2.
bIn WHO-CNS5, all IDH-wildtype adult-type diffuse astrocytic gliomas are classified as glioblastoma, IDH-wildtype, WHO CNS grade 4, without separate grades 2 or 3.
cBoth histologically defined and new molecularly defined ICD-O-3 codes for medulloblastomas were reported in the registry data; however, only a single ICD-O-3 diagnosis can be reported per case. As a result, the national incidence rates could only be estimated for BMM-coded SHH-activated and TP53-wildtype subtypes.
dThe implementation of updated ICD-O-3 codes in 2018 also included 9509/1, which groups together three distinct glioneuronal and neuronal tumor types: papillary glioneuronal tumor, rosette-forming glioneuronal tumor, diffuse leptomeningeal glioneuronal tumor. Altogether, these three diagnoses were reported in 51 patients and associated with an AAIR of 0.02 (95% CI: 0.01-0.02), median age at diagnosis of 24 years old (interquartile range 13-39), with 52.9% in females.
eIn 2018, a separate ICD-O-3 code was introduced for pilomyxoid astrocytomas (9425/3)—a subtype of pilocytic astrocytomas that is thought to be more aggressive in behavior. Cases presented at a median age of 3 years old (interquartile range 1-7; n = 29).
fFor race/ethnicity, patients with Asian/Pacific Islander, other, or unknown races/ethnicities were suppressed due to low cell counts.
g% males = 100%−% females.
Among adult-type diffuse gliomas, IDH-wildtype glioblastomas (WHO grade 4) exhibited the highest age-adjusted incidence rates (1.74, 95% CI: 1.69-1.78). In contrast, the incidence of IDH-mutant astrocytomas was markedly lower: 0.14 for WHO grade 2 (95% CI: 0.12-0.15), 0.15 for grade 3 (95% CI: 0.14-0.16), and 0.07 for grade 4 (95% CI: 0.06-0.08) cases. Although the incidence rates of WHO grade 2 IDH-mutant and 1p/19q-codeleted oligodendrogliomas (0.14, 95% CI: 0.13-0.14) were comparable to their IDH-mutant astrocytoma counterparts, the grade 3 oligodendrogliomas presented with a lower incidence rate of 0.08 (95% CI: 0.07-0.09). There was a male predominance across all adult-type diffuse gliomas, regardless of IDH status, peaking at 59.8% of IDH-wildtype glioblastomas.
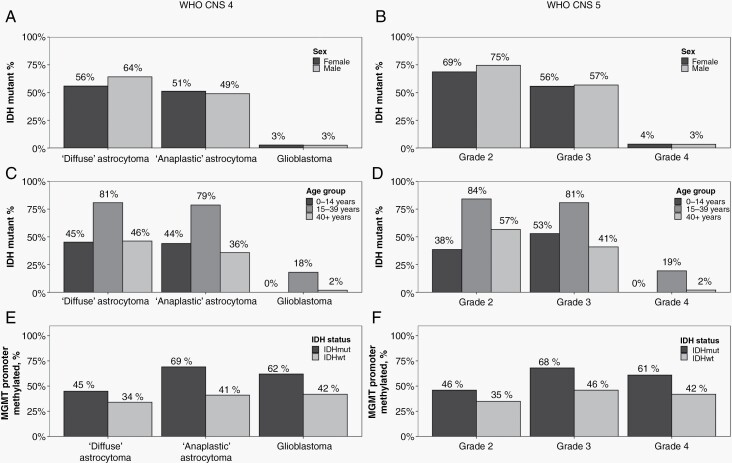
To further investigate how the IDH status of diffuse astrocytic gliomas was distributed by age and sex, we examined those cases with complete IDH data (by either BMM or ICD-O-3 coding) using both the revised WHO-CNS4 and WHO-CNS5 schema (Figure 2). Whereas WHO grade 2 “diffuse” astrocytomas were more likely IDH-mutant in males (ie, 64% compared to 56% of females using WHO-CNS4, Figure 2A; or 75% compared to 69% of females using WHO-CNS, Figure 2B), the IDH status was similar between the sexes for cases reported as grade 3 “anaplastic” astrocytomas or grade 4 glioblastomas (Figure 2A and B). Notably, the prevalence of IDH mutation was strikingly highest among astrocytomas in adolescent and young adult (15- to 39 years old) patients (Figure 2C and D), regardless of WHO grade. For instance, 81% of grade 3 adolescent and young adult tumors were IDH-mutant, compared to only 41% of adult (≥40 years old) and 53% of pediatric (<15 years old) tumors, respectively. Overall, adult-type diffuse astrocytic gliomas were rare in the pediatric population, with an age-adjusted incidence rate of 0.06 for IDH-mutant and 0.15 for IDH-wildtype astrocytic gliomas (inclusive of all grades, with no grade 4 IDH-mutant astrocytomas observed).
Figure 2.
IDH mutation and MGMT promoter methylation statuses of adult-type diffuse astrocytic gliomas from US central cancer registries for diagnosis year 2018. The percentage of IDH mutation among adult-type diffuse astrocytic gliomas reported in CBTRUS data for 2018, as stratified by patients’ sex (A, B) and age (C, D), was determined using the brain molecular markers (BMM) site-specific data item. Additionally, the percentage of MGMT promoter methylation among cases, as stratified by IDH status (E, F), was assessed using the BMM and MGMT promoter methylation data items (SEER data only). Results are shown for categorization of tumor types by both the WHO-CNS4 (using histology-defined ICD-O-3 codes, upon which the BMM was developed; A, C, E) into “diffuse astrocytoma” (9400/3), “anaplastic astrocytoma” (9401/3), or “glioblastoma” (9440/3, 9445/3); and by the WHO-CNS5 (B, D, F). Because the WHO-CNS5 grading criteria had not yet been incorporated into registry reporting, the grades reported here were based on the revised WHO-CNS4 grading criteria and categorized using clinically relevant WHO pathological grades into WHO CNS grades 2, 3, or 4. In accordance with WHO-CNS5, cases reported as “grade 1” or “low-grade” were integrated into “WHO CNS grade 2.” Data provided by CDC’s National Program of Cancer Registries and NCI’s SEER Programs (2020).
Moreover, across all WHO CNS grades of adult-type diffuse astrocytic gliomas, IDH-mutant tumors were more commonly MGMT promoter methylated than IDH-wildtype ones (Figure 2E and F). In particular, 61% of IDH-mutant grade 4 astrocytomas were characterized by MGMT promoter methylation, compared to 42% of IDH-wildtype grade 4 glioblastomas. Irrespective of IDH status, grade 2 astrocytomas were least likely to be methylated (46% of IDH-mutant and 35% of IDH-wildtype), whereas grade 3 astrocytomas were most likely to be methylated (68% of IDH-mutant and 46% of IDH-wildtype).
WHO-CNS4 discouraged the use of “oligoastrocytoma” (9382/3) given that such mixed histology gliomas could be distinctly classified by molecular profiling as oligodendrogliomas or astrocytomas, and—moreover—recognized “gliomatosis cerebri” (9381/3) as a general growth pattern rather than a unique diagnostic entity.22 Reflecting these changes, “oligoastrocytoma” (n = 23) and “gliomatosis cerebri” (n < 16) were only rarely reported in the 2018 registry data.
Epidemiology of Molecularly Defined Pediatric-Type Brain Tumors
The WHO-CNS5 draws on the growing molecular dissection of gliomas in the pediatric population to clearly distinguish pediatric-type high-grade diffuse gliomas from their adult-type counterparts. However, among pediatric-type high-grade glioma types, only H3 K27M-mutant diffuse midline glioma (n = 144; which would be included among H3 K27-altered diffuse midline gliomas in WHO-CNS5) was distinctly reported in 2018, for which the age-adjusted incidence rate was 0.06 (95% CI: 0.05-0.07; Table 3)—a comparable rate to WHO grade 4 IDH-mutant astrocytomas. Nationally, H3 K27M-mutant diffuse midline gliomas presented at a median age of 14 years old (interquartile range 7-28) and exhibited a female predominance (53.8% of cases). In the subset of patients with MGMT status, 96% were reported to be promoter unmethylated.
Beyond the molecular classification of diffuse gliomas, 2018—importantly—also saw the implementation of molecular encoding for medulloblastomas, including a distinct BMM code for SHH-activated and TP53-wildtype medulloblastomas (among 9471/3 desmoplastic/nodular medulloblastomas) and new molecularly defined ICD-O-3 codes for SHH-activated and TP53-mutant (9476/3), WNT-activated (9475/3), and non-WNT/non-SHH (9477/3) subgroups. How the clinical characteristics, molecular underpinnings, and outcomes differ between these subgroups have been extensively reviewed elsewhere.37,38 Overall, the CBTRUS 2021 report estimated that the age-adjusted incidence rates of medulloblastomas ranged from 0.49 (95% CI: 0.46-0.51) in pediatrics to 0.02 (95% CI: 0.02-0.03) in adults.22
WHO-CNS4 and WHO-CNS5 maintained both histological and molecular classifications of medulloblastomas. Thus, cancer registries retained the original histologically defined ICD-O-3 codes for medulloblastomas alongside the new molecularly defined codes; however, only a single ICD-O-3 diagnosis can be reported per case and in 2018 the majority (83%) of medulloblastomas continued to be reported using the histologically defined codes (eg, nationally, n < 16 WNT-activated [9475/3] and n < 16 SHH-activated and TP53-mutant [9476/3] molecularly coded medulloblastomas were reported; whereas n = 211 “medulloblastoma, NOS” [9470/3] cases were reported without molecular classification). As a result, the national incidence rates could only be estimated for SHH-activated and TP53-wildtype medulloblastomas (BMM code 8; n = 77): with an incidence rate of 0.03 (95% CI: 0.02-0.03) cases per 100 000 across all ages, a median age at diagnosis of 20 years old (interquartile range 6-30), and a marked male predominance (62.3%) (Table 3). For non-WNT/non-SHH medulloblastomas (9477/3; ie, group 3 or group 4 medulloblastomas; n = 47), although incidence could not be assessed, patients presented at younger ages (median 8 years old, interquartile range 4-10) and 57.4% were male.
Practically, the medulloblastoma results further suggest that, whenever possible, the incorporation of molecular characteristics into registry collection protocols should be attempted using dedicated integrated diagnosis site-specific data items (eg, the BMM site-specific data item) or exclusive integrated diagnosis ICD-O-3 codes (eg, 9385/3 for H3 K27M-mutant diffuse midline glioma), instead of adding new molecularly defined ICD-O-3 codes on top of existing histologically defined ICD-O-3 codes. Among other molecularly defined pediatric tumor types, <16 C19MC-altered embryonal tumor with multilayered rosettes and <16 RELA-fusion ependymoma cases were reported in CBTRUS data from 2018 (representing an age-adjusted incidence rate <0.01 per 100 000), precluding a detailed assessment of their epidemiology.
Limitations
CBTRUS data, while covering 99% of the US population in 2018, are still constrained by common registry-related limitations. First, although the new molecularly defined site-specific data items and ICD-O-3s provide invaluable information for the study of brain tumors, the systematic and complicated nature of cancer registry reporting standards means that reported data may lag behind contemporary classification schema (eg, using WHO-CNS4-based definitions for BMM). As the understanding of the molecular drivers of cancer grows, so will the need for sophisticated, versatile, integrated diagnosis variables to capture those data. Similarly, because WHO-CNS5 grading criteria (particularly regarding molecular markers) had not yet been incorporated into registry reporting, we analyzed tumor types by both WHO-CNS4 and WHO-CNS5 classifications but the grading was based on WHO-CNS4 criteria due to this limitation of the registries.
Second, molecular site-specific data items combine a lack of testing, a lack of testing documentation in the patient record, and unknown testing status as a single code—thus we could not differentiate between molecular testing that was truly not conducted and testing that was done but not captured by cancer registry abstraction. Given the diagnosis-defining, standard-of-care role of IDH status for adult-type diffuse gliomas, it suggests that BMM incompleteness for 2018 cases may largely represent suboptimal cancer registry reporting and/or documentation of molecular results in the medical record. Furthermore, there remains sizeable incompleteness of the pathological grade data item even among tumor types where WHO CNS grading is essential—likely including many patients that had biopsy only, which would instead be reported in the clinical grade data item. To overcome these issues across the various variables, the NAACCR, CDC, and NCI all perform regular evaluations of data completeness and quality; implement edits; and provide feedback to the central cancer registries for improving data collection efforts.
Third, because the age-adjusted incidence rates of adult-type diffuse glioma types were estimated based on cases with complete BMM (ie, IDH and 1p/19q) data and 10%-22% of diffuse gliomas had incomplete BMM data, it is likely that the incidence rates of IDH-mutant astrocytomas, IDH-wildtype astrocytomas and glioblastomas, and IDH-mutant and 1p/19q-codeleted oligodendrogliomas were underestimated. Likewise, incidence rates of BMM-encoded SHH-activated and TP53-wildtype medulloblastomas are likely underestimated. Fourth, due to immature follow-up data, CDC and NCI presently suppress overall survival data for patients diagnosed in 2018. These survival data will be available in future years.
Conclusions
This is the first year for which brain tumor molecular data are available from the US cancer registries and CBTRUS (for diagnosis year 2018 in the United States). We found a high level of reporting completeness for new molecular site-specific data items among relevant brain tumor histologies, including the BMM, MGMT promoter methylation, and 1p/19q data items; along with demonstrating the favorable coding validity of the novel BMM data item. By leveraging these new variables, alongside the newly implemented integrated diagnosis ICD-O-3 codes, we were able to generate the first national incidence and epidemiology estimates for adult-type diffuse gliomas (by IDH and 1p/19q, MGMT promoter methylation, and WHO CNS grade status), for H3 K27M-mutant diffuse midline gliomas, and for molecular subgroups of medulloblastomas. To facilitate their use by both research and clinical stakeholders, we conducted these analyses in the context of both WHO-CNS4 (from which the codes are defined) and WHO-CNS5 (representing the current clinically relevant classification). Together our findings underscore the value of developing dedicated site-specific data items for the encoding of integrated diagnoses and illustrate their utility in helping to advance our understanding of patients’ brain tumors at the population level.
Supplementary Material
Acknowledgments
The authors thank Reda Wilson and Mary Elizabeth O’Neil from the CDC and Ingrid Stendhal and the cancer registrars at Brigham and Women’s Hospital, Dana-Farber Cancer Institute, and Massachusetts General Hospital for their invaluable assistance. The CBTRUS data presented in this report were provided through an agreement with the CDC’s National Program of Cancer Registries. In addition, CBTRUS used data from the research data files of the NCI’s SEER Program. CBTRUS acknowledges and appreciates these contributions to this report and to cancer surveillance in general.
Conflict of interest statement. There are no conflicts of interest relevant to this manuscript to report.
Authorship statement. Conceptualization and supervision: J.B.I., C.K., J.S.B.-S., and Q.T.O.; methodology: J.B.I., C.S., C.N., G.C., and Q.T.O.; data collection: J.B.I. and C.G.; formal analysis and investigation: J.B.I., C.S., and C.N.; critical interpretation of results and writing: J.B.I., C.S., C.N., G.C., and Q.T.O.; read and approved final version: all authors.
Contributor Information
J Bryan Iorgulescu, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA; Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA.
Chuxuan Sun, Department of Biostatistics, Duke University School of Medicine, Durham, NC, USA.
Corey Neff, Central Brain Tumor Registry of the United States, Hinsdale, IL, USA; Department of Neurosurgery, Duke University School of Medicine, Durham, NC, USA.
Gino Cioffi, Central Brain Tumor Registry of the United States, Hinsdale, IL, USA; Trans Divisional Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, USA.
Catherine Gutierrez, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA; Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA.
Carol Kruchko, Central Brain Tumor Registry of the United States, Hinsdale, IL, USA.
Jennifer Ruhl, Surveillance, Epidemiology, and End Results program, National Cancer Institute, Bethesda, MD, USA.
Kristin A Waite, Trans Divisional Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, USA.
Serban Negoita, Surveillance, Epidemiology, and End Results program, National Cancer Institute, Bethesda, MD, USA.
Jim Hofferkamp, North American Association of Central Cancer Registries, Springfield, IL, USA.
Tarik Tihan, Division of Neuropathology, Department of Pathology, University of California San Francisco, San Francisco, CA, USA.
Roger McLendon, Department of Pathology, Duke University School of Medicine, Durham, NC, USA; Duke Cancer Institute, Duke University Medical Center, Durham, NC, USA.
Daniel J Brat, Department of Pathology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Quinn T Ostrom, Central Brain Tumor Registry of the United States, Hinsdale, IL, USA; Department of Neurosurgery, Duke University School of Medicine, Durham, NC, USA; The Preston Robert Tisch Brain Tumor Center, Duke University School of Medicine, Durham, NC, USA; Duke Cancer Institute, Duke University Medical Center, Durham, NC, USA.
Jill S Barnholtz-Sloan, Central Brain Tumor Registry of the United States, Hinsdale, IL, USA; Center for Biomedical Informatics & Information Technology, National Cancer Institute, Bethesda, MD, USA.
Funding
CBTRUS has been funded by the Centers for Disease Control and Prevention under Contract No. 75D30119C06056 Amendment/Modification No: 00002, the American Brain Tumor Association, Novocure, the Musella Foundation, The Sontag Foundation, National Brain Tumor Society, the Pediatric Brain Tumor Foundation, the Uncle Kory Foundation, and the Zelda Dorin Tetenbaum Memorial Fund, and private and in-kind donations. J.B.I. acknowledges support from the National Cancer Institute (NCI; K12CA090354) and Conquer Cancer Foundation/Sontag Foundation. The research services of J.S.B.-S., K.W., and G.C. were provided by the Division of Cancer Epidemiology and Genetics of the NCI. Contents are solely the responsibility of the authors and do not necessarily represent the official views of either the CDC or the NCI.
References
- 1. Horbinski C, Ligon KL, Brastianos P, et al. The medical necessity of advanced molecular testing in the diagnosis and treatment of brain tumor patients. Neuro Oncol. 2019;21(12):1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–9861. [DOI] [PubMed] [Google Scholar]
- 3. van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. [DOI] [PubMed] [Google Scholar]
- 4. Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hartmann C, Hentschel B, Wick W, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120(6):707–718. [DOI] [PubMed] [Google Scholar]
- 6. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hegi ME, Diserens A-C, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 9. Malmström A, Grønberg BH, Marosi C, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. [DOI] [PubMed] [Google Scholar]
- 10. Wick W, Platten M, Meisner C, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. [DOI] [PubMed] [Google Scholar]
- 11. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhukova N, Ramaswamy V, Remke M, et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J Clin Oncol. 2013;31(23):2927–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123(4):473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Louis DN, Wesseling P, Paulus W, et al. cIMPACT-NOW update 1: not Otherwise Specified (NOS) and Not Elsewhere Classified (NEC). Acta Neuropathol. 2018;135(3):481–484. [DOI] [PubMed] [Google Scholar]
- 15. Louis DN, Giannini C, Capper D, et al. cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018;135(4):639–642. [DOI] [PubMed] [Google Scholar]
- 16. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136(5):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellison DW, Hawkins C, Jones DTW, et al. cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600E mutation. Acta Neuropathol. 2019;137(4):683–687. [DOI] [PubMed] [Google Scholar]
- 18. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020;139(3):603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Louis DN, Wesseling P, Aldape K, et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020;30(4):844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellison DW, Aldape KD, Capper D, et al. cIMPACT-NOW update 7: advancing the molecular classification of ependymal tumors. Brain Pathol. 2020;30(5):863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nabors LB, Portnow J, Ammirati M, et al. NCCN guidelines insights: central nervous system cancers, version 1.2017. J Natl Compr Canc Netw. 2017;15(11):1331–1345. [DOI] [PubMed] [Google Scholar]
- 22. Louis D, Ohgaki H, Cavenee W, eds. WHO Classification of Tumours. Vol. 1, revised 4th ed.Lyon, France: International Agency for Research on Cancer; 2016. [Google Scholar]
- 23. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ostrom QT, Gittleman H, Kruchko C, et al. Completeness of required site-specific factors for brain and CNS tumors in the Surveillance, Epidemiology and End Results (SEER) 18 database (2004-2012, varying). J Neurooncol. 2016;130(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lym RL, Ostrom QT, Kruchko C, et al. Completeness and concordancy of WHO grade assignment for brain and central nervous system tumors in the United States, 2004-2011. J Neurooncol. 2015;123(1):43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kruchko C, Gittleman H, Ruhl J, et al. Cancer collection efforts in the United States provide clinically relevant data on all primary brain and other CNS tumors. Neurooncol. Pract. 2019;6(5):330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ostrom QT, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2014-2018. Neuro Oncol. 2021;23(Supplement_3):iii1–iii105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kruchko C, Ostrom QT, Gittleman H, Barnholtz-Sloan JS. The CBTRUS story: providing accurate population-based statistics on brain and other central nervous system tumors for everyone. Neuro Oncol. 2018;20(3):295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. National Program of Cancer Registries and Surveillance EaER. SEER*Stat Database: NPCR and SEER Incidence – U.S. Cancer Statistics 2001–2018 Public Use Research Database, 2020 submission (2000–2018), United States Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute.2021. www.cdc.gov/cancer/uscs/public-use
- 30. Surveillance Epidemiology and End Results (SEER) Program. SEER*Stat Database: incidence - SEER Research Data, 21 Registries, Nov 2020 Sub (2000-2018) - Linked To County Attributes - Time Dependent (1990-2018) Income/Rurality, 1969-2018 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2021 based on the November 2020 submission. 2021.
- 31. Tiwari RC, Clegg LX, Zou Z. Efficient interval estimation for age-adjusted cancer rates. Stat Methods Med Res. 2006;15(6):547–569. [DOI] [PubMed] [Google Scholar]
- 32. R Core Team. R: A Language and Environment for Statistical Computing. 2021. http://www.R-project.org/. Accessed April 20, 2021.
- 33. Surveillance Epidemiology and End Results (SEER) Program. SEER*Stat software version 8.3.9. 2021. www.seer.cancer.gov/seerstat. Accessed July 21, 2021.
- 34. Charlton ME, Karlitz JJ, Schlichting JA, Chen VW, Lynch CF. Factors associated with guideline-recommended KRAS testing in colorectal cancer patients: a population-based study. Am J Clin Oncol. 2017;40(5):498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lamba N, Chukwueke UN, Smith TR, et al. Socioeconomic disparities associated with MGMT promoter methylation testing for patients with glioblastoma. JAMA Oncol. 2020;6(12):1972–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iorgulescu JB, Torre M, Harary M, et al. The misclassification of diffuse gliomas: rates and outcomes. Clin Cancer Res. 2019;25(8):2656–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hovestadt V, Ayrault O, Swartling FJ, Robinson GW, Pfister SM, Northcott PA. Medulloblastomics revisited: biological and clinical insights from thousands of patients. Nat Rev Cancer. 2020;20(1):42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.