Abstract
Objective: Ewing Sarcoma Family of Tumors (ESFT) are a highly aggressive pediatric bone and soft tissue malignancy with poor outcomes in the refractory and recurrent setting. Over 90% of Ewing Sarcoma (ES) tumors are driven by the pathognomonic EWS-ETS chimeric transcripts and their corresponding oncoproteins. It has been suggested that the EWS-ETS oncogenic action can mediate microRNA (miRNA) processing. Importantly, small extracellular vesicles (sEVs), including those frequently referred to as exosomes have been shown to be highly enriched with tumor-derived small RNAs such as miRNAs. We hypothesized that ESFT-specific sEVs are enriched with certain miRNAs which could be utilized toward an exo-miRNA biomarker signature specific to this disease. Methods: We performed miRNAseq to compare both the exo-derived and cell-derived miRNA content from 8 ESFT, 2 osteosarcoma, 2 non-cancerous cell lines, and pediatric plasma samples. Results: We found that sEVs derived from ESFT cells contained nearly 2-fold more number of unique individual miRNAs as compared to non-ESFT samples. Quantitative analysis of the differential enrichment of sEV miRNAs resulted in the identification of 62 sEV-miRNAs (exo-miRNAs) with significant (P < .05) enrichment variation between ESFT and non-ESFT sEV samples. To determine if we could utilize this miRNA signature to diagnose ESFT patients via a liquid biopsy, we analyzed the RNA content of total circulating sEVs isolated from 500 µL plasma from 5 pediatric ESFT patients, 2 pediatric osteosarcoma patients, 2 pediatric rhabdomyosarcoma patients, and 4 non-cancer pediatric controls. Pearson’s clustering of 60 of the 62 candidate exo-miRNAs correctly identified 80% (4 of 5) of pathology confirmed ESFT patients. Importantly, RNAseq analysis of tumor tissue from the 1 outlier, revealed a previously uncharacterized EWS-FLI1 translocation.Conclusions: Taken together, these findings support the development and validation of an exo-miRNA-based liquid biopsy to aid in the diagnosis and monitoring of ESFT.
Keywords: Small extracellular vesicles, exosomes, Ewing Sarcoma, Ewing Sarcoma Family of Tumors, microRNA, exo-miRNAs, biomarkers, liquid biopsy, EWS-FLI1 fusions
Introduction
The Ewing Sarcoma Family of Tumors (ESFT) are the second most common pediatric bone and soft tissue malignancies after osteosarcoma and primarily affect children and young adults in their second decade of life. The disease encompasses a family of tumors believed to derive from the same cell type consisting of osseous Ewing sarcoma, extraosseous Ewing sarcoma, peripheral primitive neuroectodermal tumors (PNET), and Askin tumors.1 Collectively, ESFT is characterized and largely driven by a family of chromosomal translocations which result in a chimeric transcription factor combining the N-terminal, transcriptional activation domain of the EWSR1 gene with the C-terminal, DNA binding domain of one of several members of the ETS family of transcription factors such as FLI1 and ERG.2,3 In over 90% of ESFT cases, the fusion occurs at t(11,22)(q24;q12), resulting in the EWS-FLI1 oncoprotein. An additional form of molecular genetic heterogeneity stems from the location of translocation breakpoints resulting in inclusion of different exon combinations from EWSR1 and FLI1 within the fusion products, with the most commonly occurring Type I followed by Type II.4 The second most abundant translocation after those involving FLI1 occurs at t(21;22)(q22;q12) resulting in the EWS-ERG oncoprotein. A small fraction (<5%) of ESFT patients will have EWSR1 fusions with ETV1, ETV4, or FEV. Detection of translocation presence are typically clinically identified by Florescent In Situ Hybridization (FISH) assay directed at the 3′ and 5′ ends of the EWSR1 gene in samples collected through diagnostic tumor tissue biopsy. Tumor tissue is also used to evaluate the small, round blue cell morphology of ESFT tumors and immunohistology conducted to measure membranous CD99 (MIC2) staining which is nonspecific but highly expressed in ESFT. Given a lack of additional biomarkers, the current methods of disease diagnosis, monitoring, and progression are limited to invasive biopsies and radiation exposure via imaging modalities, such as X-rays, computed tomography scan, magnetic resonance imaging, bone scan, and fluoride-18 fluorodesoxyglucose positron emission tomography. Presently, there remains an unmet need for the development of complementary methodologies toward rapid and specific diagnosis and monitoring of ESFT.
A primary function of the EWS-ETS oncoprotein is transcriptional regulation. In addition to this, a large body of work has demonstrated that EWS-ETS can also act to regulate microRNA (miRNA), synthesis through a combination of aberrant transcription regulation and association with miRNA processing machinery leading to a handful of publications on ESFT cellular miRNA biomarkers.5,6 miRNAs are a class of small, non-coding RNAs between 18 and 24 nucleotides which act as translational repressors by binding to the 3′ untranslated region (UTR) of target gene mRNAs. To date, over 1500 human miRNAs have been identified, each of which can regulate a few to hundreds of mRNAs.7 miRNAs are an essential regulatory mechanism in normal physiology and are frequently dysregulated in pathological states such as cancer. Specifically, miRNAs have been shown to be involved in most recognized pathways of tumorigenesis, including immune suppression, angiogenesis, metastasis, and drug resistance. Importantly, miRNAs can be transferred from cell-to-cell via subsets of sEVs which are from the endosomal system and are commonly referred to as exosomes.8,9 miRNA-sorting into sEVs appears to be a tightly regulated, albeit elusive process with a variety of miRNAs commonly found within sEVs and others almost never enriched into these vesicles.10,11 Although sEVs are produced by nearly all types of healthy cells, sEV production is dramatically elevated in pathological states such as cancer.12 Importantly, these vesicles have been shown to be readily isolated from nearly all forms of bodily fluids including whole blood, serum, and plasma.
Given that ESFT tumors harbor unique miRNA profiles and the fact that some miRNAs can be enriched within sEVs, we hypothesized that miRNA content analysis of ESFT cell lines and their corresponding sEVs may reveal a miRNA signature which could be beneficial toward diagnosis and monitoring of ESFT disease clinical status (regression, progression, and recurrence). To test this hypothesis, we conducted miRNA sequencing of 8 ESFT cell lines and their derived sEVs representing the most clinically relevant EWS-ETS fusions to develop an exo-miRNA signature. We then performed a preliminary evaluation on rare clinical ESFT patient plasma-derived sEVs and were able to identify 4 out of 5 ESFT patients successfully. Interestingly, the single ESFT patient not identified via this exo-miRNA signature carried a rare, previously uncharacterized EWS-FLI1 translocation. Together these data provide the framework for development of a blood-based assay capable of both detecting and monitoring the majority of ESFT tumor types.
Methods
Cell line and culture conditions
Hs919.T, SK-ES-1, RD-ES were purchased from the American Type Culture Collection. TC-32, TC-71, COG-E-352, CHLA-32, CHLA-9, and CHLA-258 cell lines were obtained from the Children’s Oncology Group (COG). F303 was previously generated in the Godwin laboratory.13 U2OS and MG-63 were a kind gift of Dr. Tomoo Iwakuma (University of Kansas Medical Center). All cell lines were cultured at 37°C under a 5% humidified CO2 atmosphere. TC-71, TC-32, CHLA-9, CHLA-32, COG-E-352, and CHLA-258 cell lines were maintained in Iscove’s Modified Dulbecco’s Medium (IMDM), supplemented with 3 mM L-glutamine, 5 mg/ml insulin and transferrin, 5 ng/ml selenium, and 20% exosome free FBS. RD-ES cell line was maintained in RPMI 1640 with L-glutamine, supplemented with 15% exosome free FBS. SK-ES-1 and MG-63 were maintained in McCoy’s 5A with L-glutamine supplemented with 15% exosome free FBS (whole medium). Hs919.T and U2OS cells were maintained in DMEM with high glucose with L-glutamine, supplemented with 20% exosome free FBS. Ten percent penicillin streptomycin was added to all medium.
SEV isolation
sEVs were isolated by differential ultracentrifugation as previously published.14 Briefly, conditioned media was collected every 24 hours while cells were between 60% and 90% confluency. Media was centrifuged at 2500 rpm for 5 minutes to eliminate cellular debris. The media was then ultra-centrifuged using a FIBERLite F40L rotor (Thermo Scientific) at 4°C for 45 minutes at 10 000g to pellet larger microvesicles. The supernatant was collected, transferred to new tubes and ultra-centrifuged for 60 minutes at 110 000g to pellet sEVs. sEVs were resuspended in cold PBS, pooled together, and then spun once more at 110 000g. sEV pellets were resuspended with an appropriate volume of PBS (50-200 µL) and stored at −80°C.
Nanoparticle tracking analysis (NTA)
The NanoSight LM10 (NanoSight Ltd., Minton Park, Amesbury, UK) was used to determine particle count and size distribution of sEV samples by analyzing the Brownian Motion of aliquots of sEVs diluted in PBS. Five measurements were taken for each sample and the mean values were reported.
SDS-PAGE and western blot analysis
Cellular lysates were prepared in RIPA buffer (Thermo Fisher Scientific) and sonicated. Forty microgram of cellular lysates or sEV preparations were separated using Mini-PROTEAN® TGX™ Precast Gels (BioRad) and then transferred to a supported nitrocellulose membrane (BioRad). Primary antibodies for CD81 (mouse monoclonal antibody, clone D5O2Q), CD49 (Mouse monoclonal antibody, D9M81), Flotillin (Rabbit XP monoclonal antibody, clone D2V7J), Alix (Mouse monoclonal antibody, clone 3A9), and CD9 (mouse monoclonal antibody clone D3H4P) were all from Cell Signaling Technology. ß-actin (mouse monoclonal clone AC-15) was from Sigma Aldrich. Anti-rabbit and anti-mouse secondary HRP conjugated antibodies were from Cell Signaling. All antibodies were diluted in either 5% NFM or BSA in TBS according to manufactures recommendations.
RNA isolation
To evaluate miRNAs carried in ESFT-derived EVs (sEV-miRNAs or exo-miRNAs) total RNA was isolated from sEVs obtained from cell culture conditioned media by differential ultracentrifugation (described above). sEVs (~50 µg of total protein) were suspended in a small volume of PBS (<50 µL) and were then lysed using the miRNeasy kit (Qiagen) following the manufacturer’s protocol for total RNA.
For plasma derived sEVs, peripheral blood was collected in ACD tubes. The blood samples were centrifuged @ 1200g for 12 minutes at 4°C. Without disturbing the buffy coat, plasma from 2 ACD tubes (~6 mL) were combined into a 15 mL falcon tube and the samples were centrifuged at 2500g for 15 minutes at 4°C to remove debris and platelets. Once complete, plasma/supernatant, carefully avoiding the pellet, was pipetted into a new 15 mL falcon tube and the samples underwent a final spin at 2500g for 15 minutes at 4°C. All plasma samples were stored at −80°C until sEV purification and RNA isolation. We isolated total RNA from sEVs using the manufacturer protocol with 500 µL plasma and the Qiagen Exo-RNeasy kit. This kit collects all sEVs onto a membrane and directly lyses them resulting in total RNA. All RNA samples were analyzed using the Pico 6000 chip on the Agilent bioanalyzer or the Nanoquant plate (Tecan).
MiRNA next generation sequencing
Isolated miRNA samples were prepared for next generation sequencing using the Bioo Scientific, NEXTflex small RNA Seq v3 kit according to manufacturer’s protocols (Bioo Scientific, Austin, TX). Briefly, 200 ng of isolated miRNA was used for library preparation with 20 cycles of PCR amplification. Size selection was completed using a BluePippin (SAGE Science, Beverly, MA). A 3% gel was used with a selected size range of 135 to 185 base pairs. Samples were pooled for sequencing on an Illumina MiSeq using v3 chemistry (1 × 35 bp read) to a minimum of 700 K reads/sample after trimming.
MiRNA analysis
Single-end sequencing reads were trimmed to remove the NEXTflex adapter and then processed with mirdeep2 v2.0.0.8. First, trimmed reads were collapsed using the mirdeep2 “mapper.pl” script with the following parameters: -e -h -j -m -l 18. Next, the collapsed reads were quantified against miRBase release 22 using the mirdeep2 “quantifier.pl” script with the following parameters: -t hsa -d -W -p hairpin.fa -m mature.fa. Resulting read counts for each miRNA were normalized by total count scaling.15
Availability of data and material: Upon publication all data sets will be made available in GEO, NCBI.
Tet-inducible knock-down
HEK293 T cells were plated in 10 cm dishes and allowed to adhere overnight. The next day cells were transfected according using Lipofectamine 3000 (Thermo Fisher), 1 µg pMD2.G (Addgene #12259), 2 µg/L psPAX.2 (Addgene #12260), 3 µg control shRNA or targeting shRNA plasmid. We used the Tet-plko-puro-Scrambled plasmid as a non-targeting control (Addgene #47541) and the Tet-pLKO-puro plasmid (Addgene #21915) with the previously published FLI1 targeting sequence 5′ CGTCATGTTCTGGTTTGAGAT 3′. Viral particles were collected 2 days post transfection and transferred to 50 mL tubes. Tubes were spun at 200g for 5 minutes, filtered through a 0.4 µm syringe filter, and transferred to 13 mL ultracentrifuge tubes and spun at 120 000g for 1.5 hours. Supernatant was poured off and the viral particles were allowed to dry for 2 minutes before being dissolved in 100 µL of PBS and stored at −80°C.
ESFT cells were treated with 18 µg/ml Polybrene (Thermo Fisher), and 10 µL viral concentrate. Cells were selected using Puromycin in concentrations ranging from 0.5 to 1.0 µg/mL. ESFT cells were treated with Doxycycline 48 hours prior to sEV collection and then allowed to “rest” for 2 days. This cycle was repeated to obtain sEVs under EWS-FLI1 knock-down conditions.
Nanostring
Biological replicates of total RNA were pooled together. We utilized the nCounter Human v3 miRNA panel on the NCOUNTER FLEX following the manufacturers recommended protocol. Around 31 and 100 ng of total RNA were added for sEV and cell lysate preparations respectively. Results were normalized by total count scaling and positive hits were determined for values above the average of 5 individual negative controls.
Availability of data and material: Upon publication all data sets will be made available in GEO, NCBI.
QRT-PCR analysis
Total RNA was reverse transcribed using the TaqMan® Advanced miRNA cDNA synthesis Kit (Invitrogen) according to manufactures instructions. All PCR were performed using TaqMan® Advanced miRNA assays (Invitrogen). qPCR was done utilizing triplicate samples on a BioRad CFX 96 real time instrument (BioRad). Due to a lack of standardized normalization controls for sEV miRNA, we utilized the sequencing data to select 3 miRNAs for normalization. First, we calculated the average counts of each miRNA in each sample set. The samples were then ranked and samples with an average count >1 were selected. From this set we divided the standard deviation by the average to generate a % of variance from the average. Of these, 3 miRNAs were selected to have the highest expression in samples with the least amount of variance from the mean; Let-7a-5p, miR-143-3p, and miR-103a-3p. These miRNAs were used together to normalize all cell-line derived sEV PCR results.
EWSR1 fusion analysis
RNA/DNA next-generation sequencing panel
Genomic DNA and RNA are extracted from manually macro dissected formalin-fixed paraffin-embedded (FFPE) tissue samples using AllPrep DNA/RNA FFPE kit (Qiagen). Following the library preparation by targeted enrichment using a custom Qiagen multimodal panel kit the sample was subjected to NGS to generate FASTQ files (text-based format for storing nucleotide sequences). This assay is a targeted custom NGS Panel that encompasses 486 genes with variable full exon or partial region, 27 MSI loci, and 574 fusions.
Bioinformatics
Data analyses for RNA fusions were performed using QIAGEN’s CLC Genomics Workbench (version 20.0) with manufacturer-recommended settings. The Unique Molecular Index (UMI) reads were trimmed off while retaining the UMI information as an annotation on the read. The trimmed reads were then mapped to the reference transcriptome sequence, and reads were grouped according to their UMI. The fusion events identified were based primarily on the number of fusion crossing reads, and on the number of fusion spanning reads. The fusions detected were then refined. Only the fusions meeting the manufacturer-recommended thresholds were reported: breakpoint distance 10, error rate 0.001, minimum number of supporting reads 2, and maximum P-value .005.
Statistical analysis
Pair wise comparison was performed between groups of interest. In brief, the Bioconductor package “edgeR” was used to preprocess the raw miRNA count data. The data were normalized by library size. For downstream analysis, we only kept genes with >1 cpm in more than or equal to 2 samples out of 47. As a result, a total of 1272 miRNAs were considered for subsequent statistical analysis.
Clinical sample size was limited to the number of pediatric plasma samples available to us and therefore, no power analysis was performed. As an alternative we performed an unsupervised model-based clustering of the 46 miRNAs identified in the clinical samples using a finite mixture of Gaussian distributions and assuming 2 clusters.
Results
ESFT cells release vesicles characterized as small extracellular vesicles
We began by isolating sEVs from 8 ESFT cell lines representing the most common EWS-ETS fusions; CHLA-32, CHLA-9, TC-32, TC-71 (EWS-FLI1 Type I); SK-ES-1, RD-ES (EWS-FLI1 Type II); CHLA-258 (EWS-FLI1 Type III) and COG-E-352 (EWS-ERG); as well as from 2 osteosarcoma cell lines MG-63 and U2OS; a benign osteoid osteoma cell line Hs919.T, and a stromal cell line F303 (Table 1). sEVs were isolated via ultracentrifugation as previously published.16,17 We then subsequently lysed and separated the cellular and sEV proteomic content by SDS page. sEVs were shown to have presence of one or more of common exosomal markers, for example, CD81, Flotillin, ALIX, and CD9 (Supplemental Figure 1).18 A separate aliquot of sEVs were diluted and analyzed for particle size using the NanoSight LX. sEVs consistently averaged <200 nm in diameter with a distribution of ~40 to 300 nm, which is characteristic of this population of small vesicles19 (Supplemental Figure 2). Taken together this result indicates that the vesicles isolated for analysis are consistent with the population of sEVs which contain exosomes.
Table 1.
Experimental cell lines.
| Cell line | Sex | Age | Disease | Phase | Location | Chemo |
|---|---|---|---|---|---|---|
| CHLA-9 | F | 14y | PNET EWS-FLI1 Type I |
Primary | Thoracic | Pre |
| CHLA-32 | F | 8y | PNET EWS-FLI1 Type I |
Primary | Pelvic | Pre |
| TC-32 | F | 17y | PNET EWS-FLI1 Type I |
Primary | Ileum and adjacent soft tissue | Pre |
| TC-71 | M | 22y | ES EWS-FLI1 Type I |
Recurrence/metastatic at diagnosis | Recurrence in Humerus | Post |
| RD-ES | M | 19y | ES EWS-FLI1 Type II |
Primary | Humerus | NA |
| SK-ES-1 | M | 18y | ES EWS-FLI1 Type II |
NA | NA | NA |
| CHLA-258 | F | 14y | PNET EWS-FLI1 Type III |
NA | Lung Metastasis | Post |
| COG-E-352 | M | 17y | PNET EWS-ERG |
Refractory disease | Peripheral blood. Post-mortem (fibula primary) | Post |
| F303 | F | NA | - | - | NA | - |
| Hs919.t | F | 34y | Benign Osteoid Osteoma | - | Bone | - |
| MG-63 | M | 14y | Osteosarcoma | NA | NA | NA |
| U2OS | F | 15y | Osteosarcoma | NA | Tibia | NA |
PNET – Primitive neuroectodermal tumor, ES – Ewing Sarcoma, F-Female, M-Male, NA-Not available, Pre-Prior to Chemotherapy, Post – After Chemotherapy, ‘-’- Not relevant
ESFT sEVs contain a larger, more diverse population of miRNA than non-ESFT sEVs
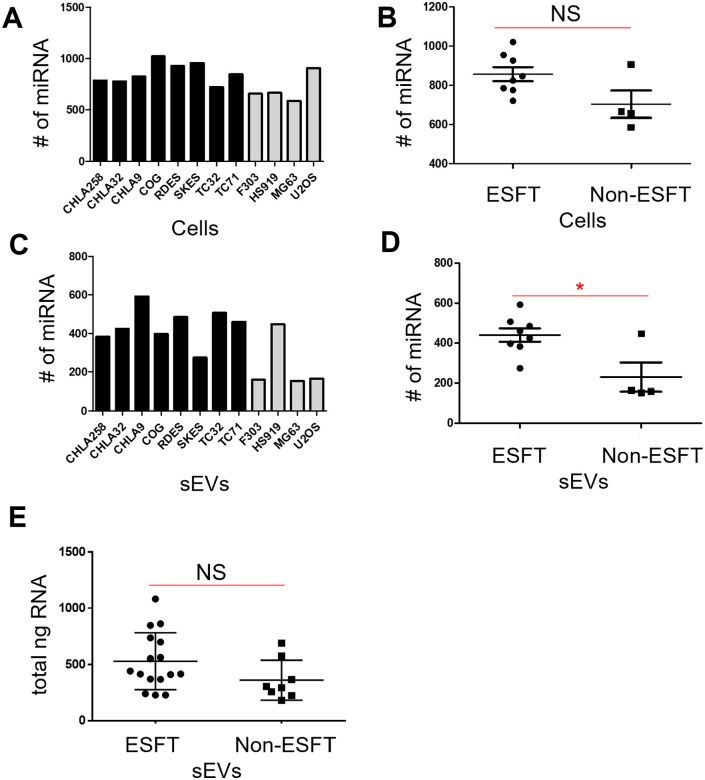
To evaluate miRNA expression in cells and enrichment within sEVs, we isolated total RNA from biological replicates of 8 ESFT and 4 control cell lines and 50 µg of their corresponding sEVs (Table 1). Next generation small RNA sequencing was performed on each sample. In total, we identified a range of 586 to 1021 miRNAs in cells and 151 to 592 miRNAs in their corresponding sEVs (Figure 1A–D, Supplemental Data Files). There was an increase (albeit not significant) in the mean number of individual miRNAs identified between ESFT and non-ESFT cell lines (Figure 1A and B). However, there was a significant increase (>2-fold; P = .0122) in the mean number of miRNAs detected within the ESFT-sEVs versus non-ESFT-sEVs (mean values of 440 and 228 miRNAs, respectively) (Figure 1C and D). Quantification of the total RNA from biological replicates of the 8 ESFT and 4 Non-ESFT sEV samples (50 µg each) indicated there was a slight, but not significant, increase in the amount of total RNA isolated in ESFT-sEV samples as compared to non-ESFT-sEV controls (Table 1 and Figure 1E). These results demonstrate that ESFT-sEVs possess an increased number of individual, unique miRNAs.
Figure 1.
ESFT sEVs are enriched in miRNAs. (A and B) The average number of individual miRNAs found within ESFT cell lines (black bars) was higher, although not significantly, as compared to the Non-ESFT cell lines (gray bars). (C and D) The average number of individual miRNAs was found to be 2-fold, significantly higher, in ESFT sEVs (black bars) compared to Non-ESFT samples (gray bars). (E) Total ng of DNA between ESFT and Non-ESFT sEV samples.
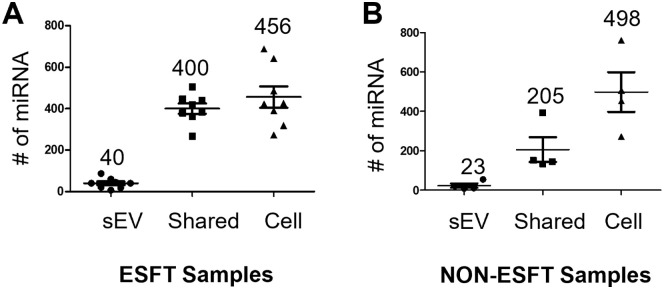
Given the increased number of individual unique miRNAs within the ESFT sEVs, we questioned what percentage of parental cellular miRNAs were being enriched into their corresponding sEVs. To answer this, we quantified the miRNA identified only in cellular lysates, only in sEV lysates, and those shared between cells and sEVs. When comparing the ESFT and non-ESFT cell line samples, a similar number of miRNA (456 and 498, respectively) were detected in cells but not in their corresponding sEVs (Figure 2A and B). In contrast, nearly half (49%), of the miRNA produced in ESFT cells were also found enriched within their corresponding sEVs (440 out of 896 miRNAs), while only a little over a quarter (~30%) of the total non-ESFT miRNAs were identified in their sEVs (228 out of 726 miRNAs) (Figure 2A and B).
Figure 2.
miRNA localization. (A, B) Number of miRNAs identified only within sEVs, cells, or within both cell and corresponding sEVs. (A) 8 ESFT samples. (B) 4 Non-ESFT samples.
Silencing of EWS-FLI1 impacts the enrichment of sEV miRNAs
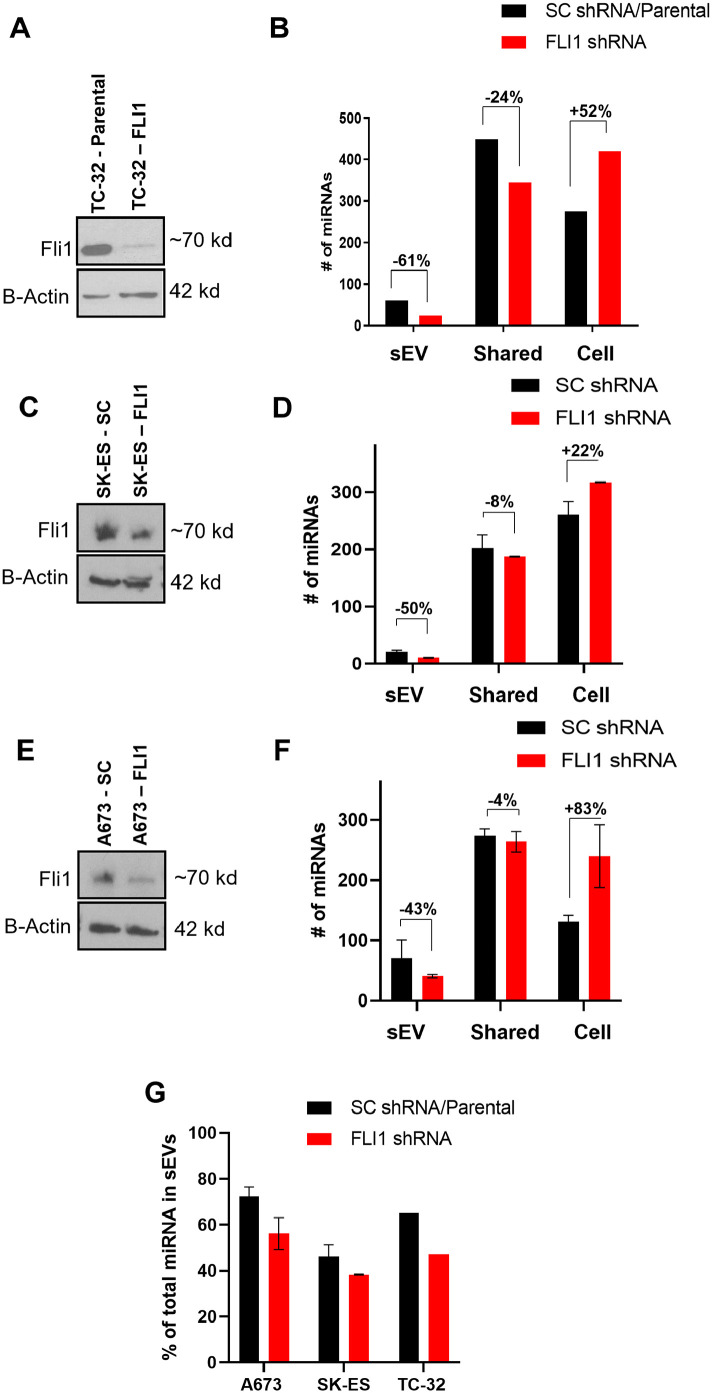
We next questioned whether the EWS-ETS oncoprotein was contributing toward the increased exo-miRNA enrichment in ESFT. To examine this possibility we generated 3 ESFT cell lines with Dox-inducible FLI1 shRNA or a non-targeting shRNA (sc).20 Two cell lines from the original sequencing (TC-32 and SK-ES) representing Types I and II fusions were used. In addition, we utilized the ESFT cell line, A673 which was established from a patient diagnosed with primary rhabdomyosarcoma and later was found to contain the EWS-FLI1 fusion pathognomonic for ESFT.21 Importantly, A673 has been found to be less dependent on the EWS-FLI1 oncoprotein for survival and responds well to EWS-FLI1 knock down. Knock-down of EWS-FLI1 had varying but consistent results on each of the 3 cell lines. In each case, cells were treated with doxycycline in 48-hour intervals with sEVs and cells harvested at the 48-hour mark. Regardless of cell line, the knock-down of cellular EWS-ETS oncoprotein resulted in decreased miRNA enrichment within sEVs and an increased number of miRNAs identified in the cell in comparison to samples treated with a non-targeted shRNA (Figure 3A–F). While preliminary, these data are interesting and suggestive that the EWS-FL1 oncoprotein potentially contributes to miRNA loading and enrichment into sEVs.
Figure 3.
Knock-down of EWS-ETS leads to decreases in sEV-miRNAs. (A–F) Western blots (A, C and E) and corresponding miRNA expression profiles (B, D and F) of 3 ESFT cell lines treated with tet-inducible FLI1 targeting or non-targeting shRNAs. (G) % of total identified miRNAs in sEVs.
Lastly, we asked which miRNAs were produced by all cell lines (ESFT and non-ESFT) but were at levels only detectable in ESFT cells-derived sEVs (Supplemental Figure 3A). We identified 369 miRNAs which were commonly expressed in 75% of cell lines (at least in 3 out of 4 non-ESFT and in 6 out of 8 ESFT cell lines) (Supplemental Figure 3B). We then compared these miRNAs with the list of 125 exo—miRNA which were identified in ESFT-derived but not in non-ESFT-derived sEVs (Supplemental Figure 3B). Of these, 89 exo-miRNA were identified as enriched in ESFT sEVs. Analysis of the expression levels in the parental cells provided some indications as to why these miRNAs are primarily enriched within ESFT sEVs. Around 51 out of the 89 had at least a greater than 2-fold increase in expression within ESFT cells as compared to non-ESFT cells, suggesting that perhaps this group of miRNAs are sorted into sEVs simply because they are more abundantly expressed in their parental cell. Surprisingly, 38 of the miRNAs had similar or even lower (<2-fold) expression within ESFT as compared to non-ESFT, suggestive of a more regulated mechanism of intracellular miRNA sorting.
Identification of an ESFT exo-miRNA signature
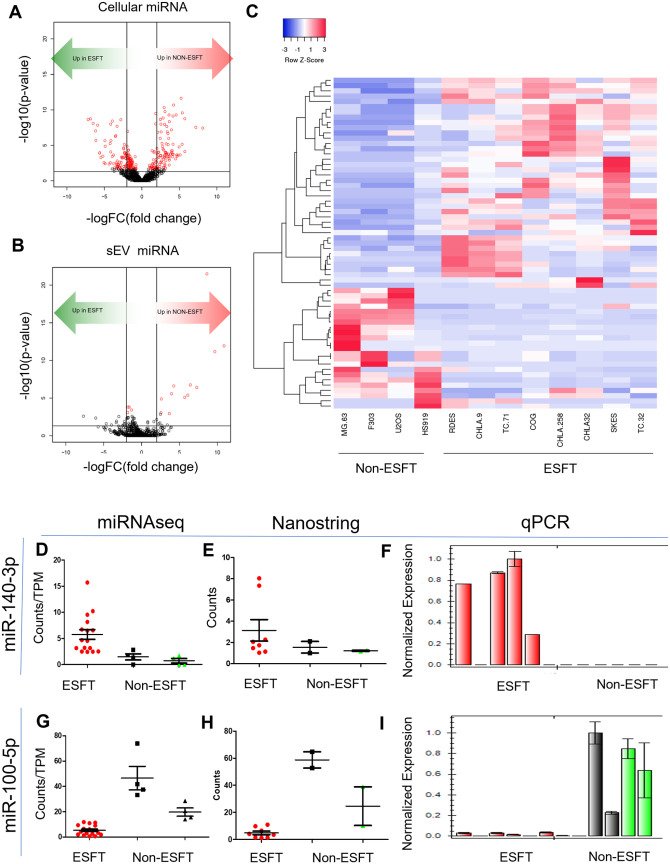
Given the enrichment of miRNA within ESFT sEVs, we next questioned if ESFT sEVs harbored exo-miRNAs which would serve as a liquid-based disease-specific ESFT biomarker panel. To accomplish this, the results from miRNAseq data were evaluated by pair-wise analysis to determine differential enrichment of miRNAs between ESFT sEVs and non-ESFT sEVs. In addition, we also ran the same analysis on the parental cell lines. Of these, we identified exo-miRNAs with an absolute logFC > 2, and an FDR of <0.1 (Figure 4A and B and Supplemental Data Files). In comparing the sEV signatures, we identified 62 exo-miRNAs predicted to be significantly differentially sorted between ESFT and non-ESFT sEV samples (P < .05) (Figure 4B and C). Of these, 41 exo-miRNAs were enriched in ESFT samples and 21 were enriched in non-ESFT samples (Figure 4C). To determine if these results could be validated using different technologies, we evaluated matched samples using the Nanostring miRNA V3 panel which determines counts of 800 miRNAs and by qPCR using pre-designed TaqMan probes. Importantly, we were able to confirm that most of the miRNAs uncovered by NGS were also found using these complementary methods, including miRNAs with the most significant differential enrichment, for example, miR-140-3p and miR-100-5p (Figure 4D–I and Supplemental Figure 4). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (performed by miRPath V.3)22 of the 62 exo-miRNAs, highlighted significant involvement in many tumor-associated pathways known to be involved in ESFT including; PI3K-AKT/mTOR,23 IGF1R,24 and TGF-beta signaling25 (Supplemental Table 1).
Figure 4.
Identification of an ESFT sEV-miRNA signature. (A and B) Volcano plots depicting miRNA with significant enrichment in (A) cells and (B) sEVs. miRNAs above the black horizontal line are significant (uncorrected P < .05), red dots signify genes with false discovery rate of <0.1. The 2 vertical lines denote log fold change of 2 (right) or −2 (left). (C) Heat map of Pearson clustering of the 63 candidate miRNAs. (D–I) Results from matched samples run on the Nanostring Human MiRNA V3 panel (E, H) and biological triplicates run by qPCR (F, I) were compared to miRNAseq results (D, G). Two miRNAs with P < .05 and an FDR of <0.1.
Plasma-derived exo-miRNA can be used to identify ESFT patients from controls
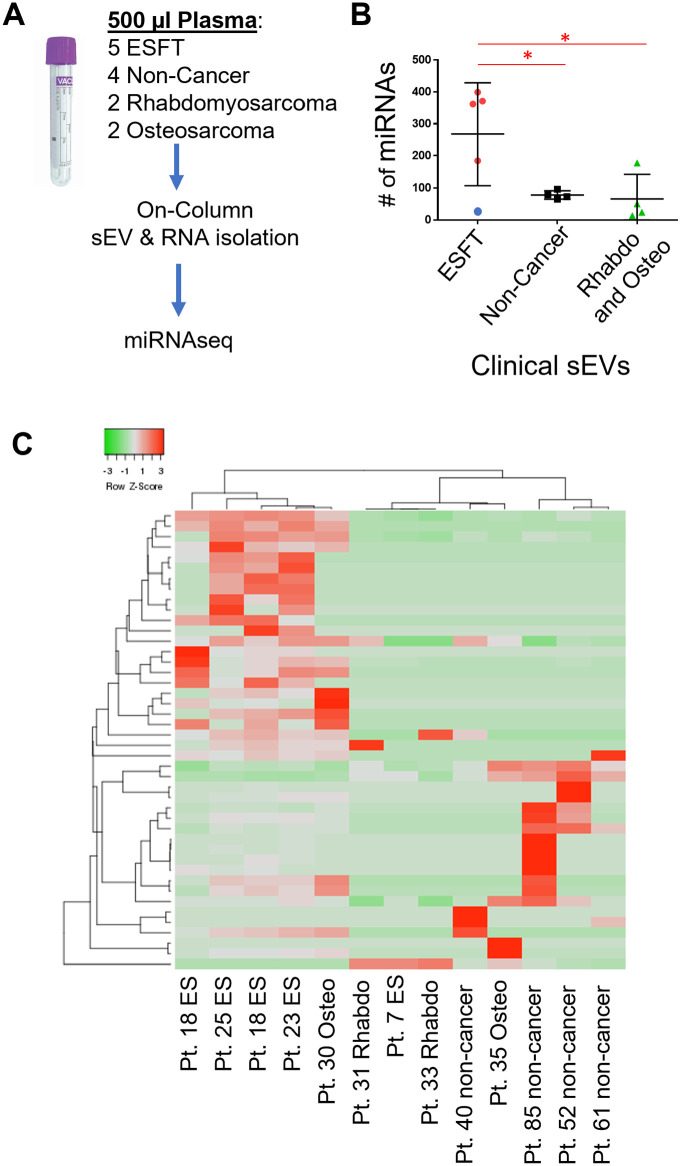
We next evaluated the diagnostic potential of the 62 exo—miRNA panel to distinguish ESFT patients from non-ESFT patients using sEVs isolated from 500 µL of plasma. Total RNA was isolated from plasma-derived sEVs from 5 pediatric ESFT patients (prior to therapy), 4 non-cancer pediatric patients, 2 pediatric rhabdomyosarcoma patients (prior to therapy), and 2 pediatric osteosarcoma patients (prior to therapy) (Figure 5A). The sEV total RNA was sequenced in the same manner as the cell line derived sEVs. Upon quantifying the individual miRNAs identified within these clinical samples we observed that like our in vitro results, the ESFT patients had nearly a 3-fold increase in miRNAs than any of the control groups with an average of 275 exo-miRNAs identified in ESFT pediatric patients and <100 exo-miRNAs identified in pediatric non-cancer, rhabdomyosarcoma, and osteosarcoma samples (Figure 5B). We next evaluated the enrichment pattern of the previously identified 62 exo-miRNAs within these clinical samples. Around 46 out of 62 miRNAs were at detectable levels in plasma-derived sEVs. Using these 46 miRNAs we were able to correctly classify 4 out of the 5 ESFT patient samples using total sEVs from plasma sample (Figure 5C). One osteosarcoma sample clustered more closely with the ESFT samples and was considered a false positive.
Figure 5.
Identification of ESFT patients through sEV miRNA signatures. (A) Schematic of total RNA isolation from plasma sEVs. (B) Number of individual miRNAs identified in clinical plasma sEV samples. (C) Pearson’s clustering of clinical miRNA samples using the 62 miRNA biomarker panel. Box indicates 4 out of the 5 ESFT patients were correctly classified as ESFT.
Given the small sample set of this preliminary study we further tested the 46 exo-miRNAs identified within the clinical samples. We took the 46 miRNAs and performed an unsupervised model-based clustering of the data using a finite mixture of Gaussian distributions and assuming 2 clusters. The results showed that 11/13 samples were correctly clustered. The 2 outliers, PT 7 (ESFT) and PT 30 (osteosarcoma) which was consistent with the results in Figure 5. The rand index, a measure of cluster similarity, was 0.72. The rand index ranges from =0 (no similarity between the true class label and the clustering result) to =1 (the class labels and the clustering result are perfectly in sync) and can be thought of as a measure of accuracy. To further evaluate the predictive power of these 46 exo-miRNAs we next randomly selected 46 miRNAs from the total sequencing results and computed the rand index between the resulting clustering and the true class labels. This process was repeated 1000 times and generated a distribution of rand indices for randomly selected sets of 46 miRNAs. Our results showed that the rand index obtained the identified 46 miRNAs was as large or larger than the rand index obtained from random sets of 46 miRNAs 97.7% of the time. Thus, our exo-miRNA fingerprint leads to a clustering solution that is more reflective of the true underlying status of the sample (ESFT vs non-ESFT) than random miRNA groupings. While these results are intriguing, they will need to be further validated in a collaborative study considering the small sample set given the rarity of the disease. Nevertheless, the finding reported do provide evidence that exo-miRNAs can be used in a liquid-based biopsy assay to help confirm the diagnosis of pediatric ESFT.
Identification of a novel EWS-FLI1 fusion transcript
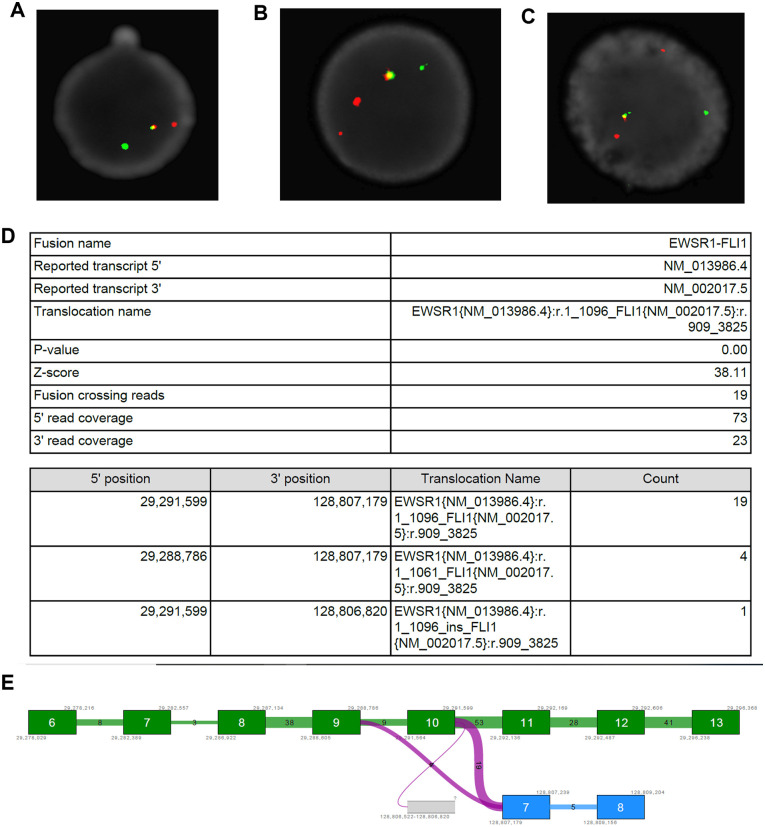
Despite the small number of clinical samples in this evaluation set we observed that one ESFT patient (PT 7) was consistently misidentified. This specific case did not exhibit the increase in the number of miRNA sorted into sEVs (Figure 5B—blue dot) and clustered more strongly with the pediatric rhabdomyosarcoma cases than with pediatric ESFT cases (Figure 5C). Upon further analysis we discovered that this patient’s tumor did not display the high CD99 membranous levels which is a clinical characteristic of ESFT. In fact, in a published study conducted in tandem with this analysis, PT 7 is the only ESFT patient out of 12 samples, to have a CD99 immunohistochemical score of “moderate” by a trained pathologist, while the 11 others tumor samples were ranked as “high.”26 Despite this finding, radiographic imaging including PET, along with both clinical and pathologic findings were all consistent with the diagnosis of localized osseous Ewing Sarcoma of the extremity as well demonstrated presence of EWSR1 breakage as determined by FISH (Figure 6A–C). Given the high promiscuity of EWSR1 in fusion genes and fusion oncoproteins in a variety of malignancies outside of ESFT, we questioned whether this patient may have an alternative EWSR1 binding partner atypical of ESFT. To date, only 12 forms of the EWS-FLI1 transcript have been reported with EWS-FLI1 fusion types I, II, and III being involved in over 90% of cases.27-29 Next generation sequencing using a RNA/DNA multimodal panel of the tumor biopsy for PT 7 revealed a previously undefined EWS-FLI1 genomic rearrangement composed of exons 1 to 10 of the EWSR1 gene and exons 7 to 9 of FLI1, (EWSR1{NM_013986.4}:r.1_1096_FLI1{NM_002017.5}z:r.909_3825) (Figure 6D and E).
Figure 6.
Identification of a previously uncharacterized EWS-FLI1 fusion transcript. (A–C) FISH analysis of the EWSR gene. (D, E) Next generation sequencing results of the EWS-FLI1 fusion depicting a EWS (exon 10) to FLI1 (exon 7) fusion.
Discussion
The ability for both diagnostic identification and monitoring of disease, especially those with arbitrary and non-specific symptoms can be the key toward improved overall outcomes for sarcomas. For pediatric malignancies, this becomes even more imperative given the rarity of disease, the often-vague and non-specific clinical symptoms on initial presentation, and the magnitude of therapy-induced long-term adverse effects. In this study we have developed a miRNA disease signature from ESFT cell line-derived sEVs. We focused on the miRNA content of ESFT sEVs for 2 reasons. As described previously in the introduction, the EWS-ETS oncoprotein has been implicated and shown to be directly involved miRNA regulation in both the nucleus and the cytoplasm. Secondly, previous studies indicate that, while longer RNA fragments can be found within sEVs, most sEV RNA content falls within the small, <40 nt range.30 Importantly, we chose to focus on sEV-derived miRNA due to the protective nature of sEVs themselves and relative ease of isolation from plasma.31
Several studies have shown that ESFT tumor cells contain unique miRNA profiles.32-35 We found this to also be true for ESFT derived sEVs; however, we were surprised by the enrichment of miRNA within correlative ESFT-derived sEVs. In this work we show for the first time that ESFT sEVs contain an increased level and number of individual miRNAs as compared to controls. Surprisingly, this remained true when evaluating ESFT and non-ESFT plasma-derived sEVs. The idea that miRNAs are sorted into sEVs in a controlled manner has been described in several cell models such as breast cancer,36 human embryonic kidney cells,37 colorectal cancer,38 T and B cells, as well as NK cells.39 While the mechanisms of such a regulated process are still elusive and seem to be dependent upon the cellular origin, we observed some evidence of this type of regulated sorting within ESFT cells. Current ideas of the mechanisms of miRNA-sorting include involvement of the neural sphinglmyelinase2 pathway,40 the sumoylated heterogeneous nuclear ribonucleoproteins (hnRN0s) dependent pathway,41 dependence upon uridylation or adenylation of the 3′ ends of miRNAs,42 and an involvement with an Ago2/KRAS-related pathway.43 Of these, there is currently no identifiable direct correlation between ESFT and/or the EWS-ETS oncoprotein and these mechanisms. In these studies, we provided evidence, by way of an inducible EWS-ETS shRNA construct that, in 3 independent ESFT cell lines, there is a decrease in number of miRNAs sorted into sEVs when EWS-FLI1 is silenced. Our previous work has identified the EWS-FLI1 oncoproteins in sEVs such as exosomes.26 We suspect that EWS-FLI1 is important for miRNA enrichment in sEVs however a potential mechanism remains elusive. While this body of work is focused on biomarker development, future work should investigate potential roles of EWS-ETS in the enrichment of miRNAs into sEVs.
Of importance we were able to identify a panel of 62 exo-miRNAs which were differentially enriched into the sEVs of ESFT cells as compared to both benign and non-ESFT controls. This report is, to the best of our knowledge, the first analysis of miRNA content of pediatric ESFT-plasma sEVs. Recent work by Kosela-Paterczyk et al44 has identified significant differences in circulating miRNAs which can differentiate adult Ewing Sarcoma from other adult sarcomas and healthy controls. In these studies, 2 miRNAs were identified with biomarker potential, imiR-142-3p and miR-9-3p. These data corroborate our results given that we also identified miR-142-3p as a lead biomarker candidate with a logFC of 1.82 and P < .004. Interestingly, we did not identify miR- 9-3p, but rather did identify miR-9-5p as a candidate biomarker with a logFC of 1.62 and P = .37. In addition, we found that our ESFT cellular results to agree with work done by Karnuth et al,45. In their work the authors identify a over 30 miRNAs with lower or higher expression levels than mesenchymal stem cells. Of interest, both of our groups identified has-miR-146b-5p to be highly overexpressed and has-miR31 and has-miR-100 to have decreased expression in ESFT cells compared to controls.45 The benefit of using a sEV-based liquid biopsy approach (as compared to circulating RNA or tumor biopsy), is that the basic morphology of the sEV protects delicate cargo, such as miRNAs, and are actively released from viable cells therefore providing a more “real-time” picture of the active tumor. Rapid advancement of microfluidic chip development, especially as it pertains to sEV analysis46-48 further emphasizes the potential for the clinical utility of exo-miRNAs.
In these studies, we were able to correctly classify 80% (4/5) of ESFT patients using our panel of exo-miRNAs. Strikingly, this signature could be evaluated from as little as 500 µL of archival patient plasma acquired by a relatively non-invasive blood draw, and importantly did not require any pre-enrichment for tumor-specific sEVs. Importantly 100% (4/4) of the healthy controls and 75% (3/4) of the non-ESFT sarcoma samples did not show the disease exo-miRNA signature. Regarding the former, the exo-miRNA signature was developed using a panel of ESFT cell lines representative of the most common EWS-ETS gene fusions. Our results lead us to question if the outlier might not be a traditional ESFT tumor. In fact, we discovered a novel and thus rare EWS-FLI1 gene fusion, consistent clinically with ESFT; however, we speculate that the predicted mutant fusion oncoprotein might have different activities related to miRNA processing.
Previous analysis of large cohorts of ESFT samples, such as the Rizzoli Experience, documented 5 distinct EWS-FLI1 fusion transcripts out of 222 ESFT tumor biopsies.29 We did note that this fusion lacks FLI1 exon 6 which most distinctly separates it from the 3 most common EWS-FLI1 fusion transcripts which were also the basis of our discovery sample set. This discovery also reinforces the heterogeneity of this group of tumors, each with distinct features and should continue to be elucidated to identify biomarkers and therapeutic targets for subsets of this tumor type.
Conclusion
Overall, our data demonstrate for the first time the clinical potential of sEV-derived miRNAs to diagnose and monitor disease progression for the most common types of ESFT. While we fully acknowledge that our clinical sample size is not powered to fully demonstrate the potential clinical utility of the exo-miRNA disease signature, we expect that future analyses will require a significantly larger cohort of sample from patients with this rare disease to help validate our exo-miRNA-signature. These future studies will require participating by cooperative groups, for example, Children’s Oncology Group, to improve upon the overall specificity and sensitivity of this early-stage diagnostic ESFT biomarker panel described in the study. With the rapid advancements of microfluidic chip development, it naturally follows that interrogation of plasma-derived sEV miRNAs will become an informative and robust diagnostic tool, especially for patients with tumors that are difficult to biopsy and image, and in cases of monitoring for disease reoccurrence.
Supplemental Material
Supplemental material, sj-docx-1-bmi-10.1177_11772719221132693 for MicroRNA Content of Ewing Sarcoma Derived Extracellular Vesicles Leads to Biomarker Potential and Identification of a Previously Undocumented EWS-FLI1 Translocation by Jennifer Crow, Glenson Samuel, Emily Farrow, Margaret Gibson, Jefferey Johnston, Erin Guest, Neil Miller, Dong Pei, Devin Koestler, Harsh Pathak, Xiaobo Liang, Cooper Mangels and Andrew K Godwin in Biomarker Insights
Supplemental material, sj-xlsx-2-bmi-10.1177_11772719221132693 for MicroRNA Content of Ewing Sarcoma Derived Extracellular Vesicles Leads to Biomarker Potential and Identification of a Previously Undocumented EWS-FLI1 Translocation by Jennifer Crow, Glenson Samuel, Emily Farrow, Margaret Gibson, Jefferey Johnston, Erin Guest, Neil Miller, Dong Pei, Devin Koestler, Harsh Pathak, Xiaobo Liang, Cooper Mangels and Andrew K Godwin in Biomarker Insights
Supplemental material, sj-xlsx-3-bmi-10.1177_11772719221132693 for MicroRNA Content of Ewing Sarcoma Derived Extracellular Vesicles Leads to Biomarker Potential and Identification of a Previously Undocumented EWS-FLI1 Translocation by Jennifer Crow, Glenson Samuel, Emily Farrow, Margaret Gibson, Jefferey Johnston, Erin Guest, Neil Miller, Dong Pei, Devin Koestler, Harsh Pathak, Xiaobo Liang, Cooper Mangels and Andrew K Godwin in Biomarker Insights
Supplemental material, sj-xlsx-4-bmi-10.1177_11772719221132693 for MicroRNA Content of Ewing Sarcoma Derived Extracellular Vesicles Leads to Biomarker Potential and Identification of a Previously Undocumented EWS-FLI1 Translocation by Jennifer Crow, Glenson Samuel, Emily Farrow, Margaret Gibson, Jefferey Johnston, Erin Guest, Neil Miller, Dong Pei, Devin Koestler, Harsh Pathak, Xiaobo Liang, Cooper Mangels and Andrew K Godwin in Biomarker Insights
Acknowledgments
The authors gratefully acknowledge Drs. Katherine Chastain and Joy Fulbright who assisted in the accrual and consenting process of pediatric patients diagnosed at Children’s Mercy-Kansas City onto the pediatric sarcoma biobank protocol (CMH IRB#13010015) and the Biospecimen Repository Core Facility (BRCF) staff at KU Medical Center for processing and banking sample. We would like to thank Kris Laurence who assisted in the accrual, collection, transport, and deidentification of clinical samples from Children’s Mercy to the BRCF. We thank Dr. Tomoo Iwakuma for his gift of osteosarcoma cell lines. We acknowledge the Center for Pediatric Genomic Medicine staff at Children’s Mercy for the miRNAseq analyses of ESFT cell lines. We acknowledge the support of the University of Kansas Cancer Center’s Biospecimen and Biostatistics and Informatics Shared Resources staffs (P30 CA168524); the Massman Family Endowed Fund for Ewing Sarcoma Research, Zachary Durand Endowed Fund for Ewing Sarcoma Research, and Hug Your Kids Foundation. Most importantly, we acknowledge the children and young adults who along with their families consented to enroll and participate in this IRB study at Children’s Mercy-Kansas City by providing clinical specimens.
Footnotes
Supplemental Material: Supplemental material for this article is available online.
Declarations
Ethics Approval and Consent to participate: These studies have been approved by the appropriate institutional research ethics committee and have been performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. As part of a collaboration between Children’s Mercy (CM) in Kansas City, MO and the Biospecimen Repository Core Facility (BRCF) at the KU Medical Center (HSC #5929, Director, A. Godwin), a pediatric sarcoma protocol (CMH IRB#13010015, Glenson Samuel, MD) has been established to obtain tumor and blood specimens from pediatric sarcoma patients undergoing treatment at CM and then subsequently processed and stored in the Pediatric Sarcoma Biobank within the BRCF.
Informed Consent: Written informed consent was obtained from all individual participants included in the study. Pediatric sarcoma patients less than 18 years of age with newly diagnosed sarcomas, that is, ESFT, rhabdomyosarcoma and osteosarcoma, were consented at CM by a study member on the IRB approved study protocol. Copies of consents were given to participant and/or consenting family member. Patients were recruited while either on the inpatient or outpatient setting of Children’s Mercy-Kansas City Department of Hematology Oncology and Bone Marrow Transplantation starting December 2012; recruitment is ongoing. All newly diagnosed and recurrent patients were enrolled; a few patients actively undergoing therapy were also enrolled at the time of initiation of this protocol. Most enrolled patients had not received chemotherapy or radiotherapy prior to study inclusion. Parents of eligible patients aged below 7 years gave written informed consent. Patients 7 years and above were also asked to provide written assent. Samples were collected prior to initiation of therapy, immediately prior to each cycle of therapy and then every 3 months off therapy for the first 2 years only. At enrollment, data on medical history were collected including demographic and clinical details of tumor site, histology, and stage were documented. During the following observation period further blood samples were taken prior to each cycle of chemotherapy or prior to local control (surgery and/or radiation therapy). Blood samples from aged/race matched cancer-free (healthy) individuals were obtained once the volunteer (or legal guardian) provided written, informed consent in accordance with the HSC 5929 umbrella IRB approved protocol. All blood samples, both CM and KUMC/KUCC, were processed and banked by the BRCF staff. De-identified patient specimens, cancer-free controls, and their accompanying clinical data were handled in an anonymous (coded) fashion.
Consent for Publication: Informed consent was obtained to publish research results.
Author contributions: J.C., G.S., and A.K.G. conceived the study and J.C, G.S., A.K.G. helped in the design of the study. J.C., G.S., E.F., M.G., and E.G. developed the methodology. J.C., G.S., M.G., X.L., H.P., C.M., Z.W., and Z.P. carried out experiments and collected data. J.C., J.J., N.M., D.P., and D.K. analyzed, computed, and interpreted the data. J.C. and A.K.G. wrote the manuscript. A.K.G. provided administrative, technical, and material support. All authors reviewed and revised the manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by grants from the NIH (R33 CA214333 to A.K.G.), the NIGMS Kansas Institute for Precision Medicine COBRE (P20 GM130423 to A.K.G.), the MCA Partners Advisory Board from Children’s Mercy Hospital (to G.S. and A.K.G.), the University of Kansas Cancer Center (P30 CA168524 to G.S. and A.K.G.), Alex Lemonade Stand (to G.S.), Braden’s Hope for Childhood Cancer Foundation (to G.S. and A.K.G.), Noah’s Bandage (to G.S. and A.K.G.), Hyundai Hope on Wheels (to G.S), Children’s Mercy-Kansas City (to G.S.), and the Kansas Bioscience Authority Eminent Scholar Program (to A.K.G). A.K.G. is the Chancellors Distinguished Chair in Biomedical Sciences Endowed.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of Data and Material: Upon publication all data sets will be made available in GEO, NCBI.
References
- 1. Maheshwari AV, Cheng EY. Ewing sarcoma family of tumors. J Am Acad Orthop Surg. 2010;18:94-107. [DOI] [PubMed] [Google Scholar]
- 2. Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing’s sarcoma. Oncogene. 2010;29:4504-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sankar S, Bell R, Stephens B, et al. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2013;32:5089-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Alava E, Cajal S. EWS-FLI (ets) fusion transcripts. In: Schwab M, ed. Encyclopedia of Cancer. Springer Berlin Heidelberg; 2011:1352-1355. [Google Scholar]
- 5. Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235-240. [DOI] [PubMed] [Google Scholar]
- 6. Sohn EJ, Park J, Kang SI, Wu YP. Accumulation of pre-let-7g and downregulation of mature let-7g with the depletion of EWS. Biochem Biophys Res Commun. 2012;426:89-93. [DOI] [PubMed] [Google Scholar]
- 7. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351-379. [DOI] [PubMed] [Google Scholar]
- 8. Witwer KW, Théry C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J Extracell Vesicles. 2019;8:1648167-20190801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr. 2007;1:156-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genom Proteom Bioinform. 2015;13:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galardi A, Colletti M, Di Paolo V, et al. Exosomal MiRNAs in pediatric cancers. Int J Mol Sci. 2019;20:4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1-11. [DOI] [PubMed] [Google Scholar]
- 13. Kwon Y, Smith BD, Zhou Y, Kaufman MD, Godwin AK. Effective inhibition of c-MET-mediated signaling, growth and migration of ovarian cancer cells is influenced by the ovarian tissue microenvironment. Oncogene. 2015;34:144-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 15. Tam S, Tsao MS, McPherson JD. Optimization of miRNA-seq data preprocessing. Brief Bioinform. 2015;16:950-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Atay S, Banskota S, Crow J, Sethi G, Rink L, Godwin AK. Oncogenic KIT-containing exosomes increase gastrointestinal stromal tumor cell invasion. Proc Natl Acad Sci USA. 2014;111:711-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crow J, Atay S, Banskota S, Artale B, Schmitt S, Godwin AK. Exosomes as mediators of platinum resistance in ovarian cancer. Oncotarget. 2017;8:11917-11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:E968-E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Willms E, Cabañas C, Mäger I, Wood MJA, Vader P. Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol. 2018;9:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martínez-Ramírez A, Rodríguez-Perales S, Meléndez B, et al. Characterization of the A673 cell line (Ewing tumor) by molecular cytogenetic techniques. Cancer Genet Cytogenet. 2003;141:138-142. [DOI] [PubMed] [Google Scholar]
- 22. Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460-W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zenali MJ, Zhang PL, Bendel AE, Brown RE. Morphoproteomic confirmation of constitutively activated mTOR, ERK, and NF-kappab pathways in Ewing family of tumors. Ann Clin Lab Sci. 2009;39:160-166. [PubMed] [Google Scholar]
- 24. Toretsky JA, Steinberg SM, Thakar M, et al. Insulin-like growth factor type 1 (IGF-1) and IGF binding protein-3 in patients with Ewing sarcoma family of tumors. Cancer. 2001;92:2941-2947. [DOI] [PubMed] [Google Scholar]
- 25. Im YH, Kim HT, Lee C, et al. EWS-FLI1, EWS-ERG, and EWS-ETV1 oncoproteins of Ewing tumor family all suppress transcription of transforming growth factor beta type II receptor gene. Cancer Res. 2000;60:1536-1540. [PubMed] [Google Scholar]
- 26. Samuel G, Crow J, Klein JB, et al. Ewing sarcoma family of tumors-derived small extracellular vesicle proteomics identify potential clinical biomarkers. Oncotarget. 2020;11:2995-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sankar S, Lessnick SL. Promiscuous partnerships in Ewing's sarcoma. Cancer Genet. 2011;204:351-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patócs B, Németh K, Garami M, et al. Multiple splice variants of EWSR1-ETS fusion transcripts co-existing in the Ewing sarcoma family of tumors. Cell Oncol. 2013;36:191-200. [DOI] [PubMed] [Google Scholar]
- 29. Gamberi G, Cocchi S, Benini S, et al. Molecular diagnosis in Ewing family tumors: the Rizzoli experience – 222 consecutive cases in four years. J Mol Diagn. 2011;13:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van den Boorn JG, Dassler J, Coch C, Schlee M, Hartmann G. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev. 2013;65:331-335. [DOI] [PubMed] [Google Scholar]
- 31. O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21:585-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y, Chen G, Liu H, et al. Integrated bioinformatics analysis of miRNA expression in Ewing sarcoma and potential regulatory effects of miR-21 via targeting ALCAM/CD166. Artif Cells Nanomed Biotechnol. 2019;47:2114-2122. [DOI] [PubMed] [Google Scholar]
- 33. Moore C, Parrish JK, Jedlicka P. MiR-193b, downregulated in Ewing sarcoma, targets the ErbB4 oncogene to inhibit anchorage-independent growth. PLoS One. 2017;12:e0178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Satterfield L, Shuck R, Kurenbekova L, et al. MiR-130b directly targets ARHGAP1 to drive activation of a metastatic CDC42-PAK1-AP1 positive feedback loop in Ewing sarcoma. Int J Cancer. 2017;141:2062-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Viera GM, Salomao KB, de Sousa GR, et al. MiRNA signatures in childhood sarcomas and their clinical implications. Clin Transl Oncol. 2019;21:1583-1623. [DOI] [PubMed] [Google Scholar]
- 36. Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, et al. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. eLife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5:e19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen M, Xu R, Rai A, et al. Distinct shed microvesicle and exosome microRNA signatures reveal diagnostic markers for colorectal cancer. PLoS One. 2019;14:e0210003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramanathan S, Shenoda BB, Lin Z, et al. Inflammation potentiates miR-939 expression and packaging into small extracellular vesicles. J Extracell Vesicles. 2019;8:1650595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kosaka N, Iguchi H, Yoshioka Y, Takeshtia F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442-17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649-1658. [DOI] [PubMed] [Google Scholar]
- 43. McKenzie AJ, Hoshino D, Hong NH, et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15:978-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kosela-Paterczyk H, Paziewska A, Kulecka M, et al. Signatures of circulating microRNA in four sarcoma subtypes. Cancer J. 2020;11:874-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Karnuth B, Dedy N, Spieker T, et al. Differentially expressed miRNAs in Ewing sarcoma compared to mesenchymal stem cells: low miR-31 expression with effects on proliferation and invasion. PLoS One. 2014;9:e93067-20140325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14:3773-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang P, Crow J, Lella D, et al. Ultrasensitive quantification of tumor mRNAs in extracellular vesicles with an integrated microfluidic digital analysis chip. Lab Chip. 2018;18:3790-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang P, Samuel G, Crow J, Godwin AK, Zeng Y. Molecular assessment of circulating exosomes towards liquid biopsy diagnosis of Ewing sarcoma family of tumors. Transl Res J Lab Clin Med. 2018;201:136-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-bmi-10.1177_11772719221132693 for MicroRNA Content of Ewing Sarcoma Derived Extracellular Vesicles Leads to Biomarker Potential and Identification of a Previously Undocumented EWS-FLI1 Translocation by Jennifer Crow, Glenson Samuel, Emily Farrow, Margaret Gibson, Jefferey Johnston, Erin Guest, Neil Miller, Dong Pei, Devin Koestler, Harsh Pathak, Xiaobo Liang, Cooper Mangels and Andrew K Godwin in Biomarker Insights
Supplemental material, sj-xlsx-2-bmi-10.1177_11772719221132693 for MicroRNA Content of Ewing Sarcoma Derived Extracellular Vesicles Leads to Biomarker Potential and Identification of a Previously Undocumented EWS-FLI1 Translocation by Jennifer Crow, Glenson Samuel, Emily Farrow, Margaret Gibson, Jefferey Johnston, Erin Guest, Neil Miller, Dong Pei, Devin Koestler, Harsh Pathak, Xiaobo Liang, Cooper Mangels and Andrew K Godwin in Biomarker Insights
Supplemental material, sj-xlsx-3-bmi-10.1177_11772719221132693 for MicroRNA Content of Ewing Sarcoma Derived Extracellular Vesicles Leads to Biomarker Potential and Identification of a Previously Undocumented EWS-FLI1 Translocation by Jennifer Crow, Glenson Samuel, Emily Farrow, Margaret Gibson, Jefferey Johnston, Erin Guest, Neil Miller, Dong Pei, Devin Koestler, Harsh Pathak, Xiaobo Liang, Cooper Mangels and Andrew K Godwin in Biomarker Insights
Supplemental material, sj-xlsx-4-bmi-10.1177_11772719221132693 for MicroRNA Content of Ewing Sarcoma Derived Extracellular Vesicles Leads to Biomarker Potential and Identification of a Previously Undocumented EWS-FLI1 Translocation by Jennifer Crow, Glenson Samuel, Emily Farrow, Margaret Gibson, Jefferey Johnston, Erin Guest, Neil Miller, Dong Pei, Devin Koestler, Harsh Pathak, Xiaobo Liang, Cooper Mangels and Andrew K Godwin in Biomarker Insights