Abstract
Background:
Savolitinib, a selective MET inhibitor, showed efficacy in patients with non-small cell lung cancer (NSCLC), including pulmonary sarcomatoid carcinoma (PSC), harbouring MET exon 14 skipping alteration (METex14).
Objective:
To analyse post hoc, the association between circulating tumour DNA (ctDNA) biomarkers and clinical outcomes, including resistance, with savolitinib.
Design:
A multicentre, single-arm, open-label phase 2 study.
Methods:
All enrolled patients with baseline plasma samples were included. Outcomes were objective response rate (ORR), progression-free survival (PFS) and overall survival (OS) by baseline METex14 and post-treatment clearance, coexisting gene alterations at baseline and disease progression.
Results:
Among 66 patients with baseline ctDNA sequencing, 46 (70%) had detectable METex14. Frequent coexisting baseline gene alterations included TP53 and POT1 mutations. Patients with detectable baseline METex14 exhibited worse PFS [hazard ratio (HR), 1.77; 95% confidence interval (CI), 0.88–3.57; p = 0.108] and OS (HR, 3.26; 95% CI, 1.35–7.89; p = 0.006) than those without, despite showing a numerically higher ORR. Among 24 patients with baseline detectable METex14 and evaluable postbaseline samples, 13 achieved METex14 clearance post-treatment. Median time to first clearance was 1.3 months (range, 0.7–1.5). METex14 post-treatment clearance was associated with better ORR (92.3%; 95% CI, 64.0–99.8 versus 36.4%; 95% CI, 10.9–69.2; p = 0.0078), PFS (HR, 0.44; 95% CI, 0.2–1.3; p = 0.1225) and OS (HR, 0.31; 95% CI, 0.1–1.0; p = 0.0397) versus non-clearance. Among 22 patients with disease progression, 10 acquired pathway alterations (e.g. in RAS/RAF and PI3K/PTEN) alone or with secondary MET mutations (D1228H/N and Y1230C/H/S).
Conclusion:
ctDNA biomarkers may allow for longitudinal monitoring of clinical outcomes with savolitinib in patients with METex14-positive PSC and other NSCLC subtypes. Specifically, undetectable baseline METex14 or post-treatment clearance may predict favourable clinical outcomes, while secondary MET mutations and other acquired gene alterations may explain resistance to savolitinib.
Registration:
The trial was registered with ClinicalTrials.gov (NCT02897479) on 13 September 2016.
Keywords: circulating tumour DNA, MET exon 14 skipping, non-small cell lung cancer, pulmonary sarcomatoid carcinoma, savolitinib
Introduction
MET exon 14 skipping alteration (METex14) is an emerging biomarker and therapeutic target in non-small cell lung cancer (NSCLC).1 METex14 is present in about 3% of lung adenocarcinoma (LUAD), while it can range from 8 to 32% in pulmonary sarcomatoid carcinoma (PSC),2 which is a rare aggressive NSCLC subtype with relatively poorer prognosis and limited treatment options.3,4 Two selective MET tyrosine kinase inhibitors, namely capmatinib and tepotinib, have been approved by the US Food and Drug Administration for the treatment of patients with NSCLC positive for METex14, based on the GEOMETRY mono-1 and VISION trials, respectively.5–7 However, as most patients enrolled in these trials had LUAD, the activity of selective MET inhibitors in patients with METex14-positive PSC, which is a particularly rare disease, was unclear .6,7
Savolitinib (AZD6094, HMPL-504, volitinib) is a novel, potent and highly selective oral MET tyrosine kinase inhibitor.8 It was evaluated in a recent phase 2 trial, which enrolled a NSCLC cohort containing the largest number of patients with PSC (over one-third of the study cohort) among studies evaluating MET inhibitors to date.9 Savolitinib showed an objective response rate (ORR) of 42.9% and a median progression-free survival (PFS) of 6.8 months, with similar outcomes regardless of NSCLC subtypes.9 To our knowledge, savolitinib is the first MET inhibitor to demonstrate clinical activity in patients with METex14-positive PSC. On the basis of the trial results, savolitinib is currently the only approved therapy for patients with METex14-positive NSCLC (including PSC) in China. Hence, it would be of interest to evaluate the utility of biomarkers in predicting clinical outcomes with savolitinib.
Circulating tumour DNA (ctDNA) allows non-invasive monitoring of genetic alterations over time with tumour evolution and treatment, while tumour biopsies can be limited by tumour inaccessibility, insufficient tissue quantity and quality or intratumoural heterogeneity.10 ctDNA-based next-generation sequencing (NGS) platforms allowing simultaneous screening of multiple genes are widely adopted for the clinical management of NSCLC.11 The validity of ctDNA biomarkers for the longitudinal monitoring of treatment response and resistance towards MET inhibitors remains poorly investigated. Furthermore, genomic profiling studies, particularly those entailing dynamic molecular monitoring over time with treatment, are scarce in PSC.
In this post hoc analysis of the phase 2 savolitinib study, we aimed to assess the association of ctDNA biomarkers, including METex14, with clinical outcomes and development of resistance in patients with METex14-positive NSCLC, including PSC.
Methods
Study design and patients
The multicentre, single-arm, open-label phase 2 study (NCT02897479) of savolitinib in patients with NSCLC, including PSC, was conducted in China. The full study design and eligibility criteria for MET inhibitor-naïve cohort 1 have previously been reported.9 Briefly, eligible patients had histologically diagnosed, unresectable or metastatic METex14-positive PSC or other NSCLC subtypes without EGFR, ALK or ROS1 alterations, and had presented with disease progression or toxicity intolerance towards one or more standard treatments, or were deemed clinically unsuitable for standard treatment. Enrolled patients received either 600 mg (bodyweight ⩾ 50 kg) or 400 mg (bodyweight < 50 kg) oral savolitinib once daily until any discontinuation criterion was met.9 The present study is a post hoc, exploratory analysis of ctDNA biomarkers and their association with clinical outcomes with savolitinib.
Procedures
Plasma samples were prospectively collected at baseline and at each tumour evaluation visit (every 6 weeks within 1 year of the first dose, and every 12 weeks thereafter) until end of treatment. ctDNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (QIAGEN, Hilden, Germany) and quantified by Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA).
Gene alterations were detected by NGS (425-gene panel, Geneseeq Prime; Geneseeq Technology Inc., Nanjing, China).12 Variant calling entailed the use of VarScan2 and ADTEx for the detection of somatic mutations and copy number variations, respectively, in candidate genes. Detectable METex14 is defined by a minimum variant supporting read value of 3. Post-treatment METex14 clearance (molecular response) was assessed using the on-treatment plasma sample that was obtained during the first tumour evaluation visit (at approximately 6 weeks after the first dose). It is defined as undetectable METex14 within the first 6 weeks of savolitinib treatment (i.e. early clearance).
Outcomes
In this post hoc analysis, the end points included ORR (proportion of patients with confirmed complete response or partial response according to Response Evaluation Criteria in Solid Tumours, version 1.1 [RECIST v1.1]), PFS (time from the first dose to disease progression, or death from any cause in the absence of progression) and overall survival (OS) (time from the first dose to death from any cause) according to METex14 status at baseline and post-treatment. In patients with baseline detectable METex14 and postbaseline samples, time to first clearance was determined. Coexisting gene alterations at baseline and those acquired following disease progression were identified.
Statistical analysis
SAS version 9.4 was used for all statistical analyses. The confidence intervals (CIs) for ORR were estimated using the Clopper–Pearson method, and χ2 or Fisher’s exact test was used for the between-group comparison of ORRs. PFS and OS were estimated by the Kaplan–Meier method; 95% CIs of the medians were calculated by the Brookmeyer and Crowley method. PFS was censored at last tumour evaluation, and OS was censored at the last known date of survival if no event occurred. The Cox proportional hazards model was used to estimate the hazard ratios (HRs) for PFS and OS based on METex14 status at baseline and post-treatment; p value was determined using the log-rank test. A multivariate Cox proportional hazards model was constructed using the backward elimination algorithm to account for potential confounding factors affecting survival outcomes in patients with detectable baseline METex14 versus those without; factors with p < 0.1 were included.
Results
Patient characteristics
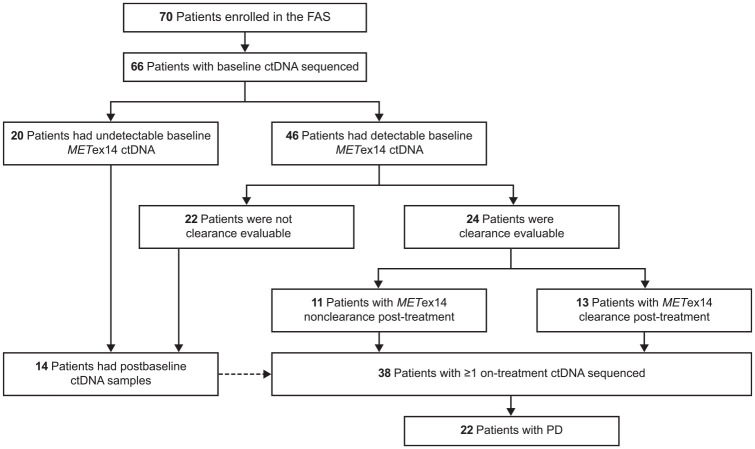
Among 70 patients enrolled in the phase 2 study from 10 February 2017, 66 with baseline plasma samples were included in this post hoc analysis (Figure 1). Of these, 46 (70%) patients had detectable baseline METex14, 24 of whom provided postbaseline samples within the first 6 weeks of savolitinib treatment (i.e. were clearance evaluable). Of the clearance evaluable patients, 13 (54%) achieved METex14 clearance. In addition to the 24 clearance evaluable patients, 14 patients who had undetectable baseline METex14 also provided postbaseline samples – a total of 38 patients had at least one on-treatment ctDNA sequencing. Disease progression occurred in 22 of the 38 patients with both baseline and postbaseline samples, allowing for the analysis of gene alterations associated with savolitinib resistance.
Figure 1.
Patient flow in the post hoc analysis.
ctDNA, circulating tumour DNA; FAS, full analysis set; METex14, MET exon 14 skipping alteration; PD, progressive disease.
Comparing patients with detectable baseline METex14 versus those without, tumour size (assessed by an independent review committee), ctDNA content and blood-based tumour mutational burden (bTMB) were significantly greater, with medians of 95.6 mm versus 51.8 mm, 107.0 ng versus 59.1 ng and 5.29 versus 1.06, respectively. Patients with detectable baseline METex14 were more likely than those without to have PSC (41% versus 15%) and an Eastern Cooperative Oncology Group performance status ⩾1 (91% versus 60%) [Table 1; Supplemental Figure 1(a)–(c)].
Table 1.
Baseline demographics and clinical characteristics in the overall cohort and by ctDNA METex14 status at baseline.
| Characteristic | Total | ctDNA METex14 status at baseline | ||
|---|---|---|---|---|
| N = 66 | Detectable (n = 46) | Undetectable (n = 20) | p Value | |
| Age, median (IQR) (years) | 68.7 (65.4–74.7) | 69.3 (65.6–76.5) | 67.1 (62.0–68.9) | 0.0327 |
| Sex, n (%) | 0.0871 | |||
| Female | 26 (39) | 15 (33) | 11 (55) | |
| Male | 40 (61) | 31 (67) | 9 (45) | |
| ECOG performance status, n (%) | 0.00473 | |||
| 0 | 12 (18) | 4 (9) | 8 (40) | |
| ⩾1 | 54 (82) | 42 (91) | 12 (60) | |
| Time from primary diagnosis to first dosing, median (IQR) (months) | 4.34 (1.28–11.0) | 3.01 (1.25–11.0) | 5.47 (1.31–13.0) | 0.586 |
| Disease stage, n (%) | 0.6348 | |||
| III | 5 (8) | 3 (7) | 2 (10) | |
| IV | 61 (92) | 43 (94) | 18 (90) | |
| Histology, n (%) | 0.0483 | |||
| PSC | 22 (33) | 19 (41) | 3 (15) | |
| Other NSCLC subtypes | 44 (67) | 27 (59) | 17 (85) | |
| Adenocarcinoma | 39 (59) | 24 (52) | 15 (75) | 0.8352 |
| Adenosquamous carcinoma | 1 (2) | 0 | 1 (5) | |
| Squamous cell carcinoma | 3 (5) | 2 (4) | 1 (5) | |
| NSCLC, not otherwise specified | 1 (2) | 1 (2) | 0 | |
| Tumour sites (⩾20% involvement), n (%) | ||||
| Lung | 62 (94) | 44 (96) | 18 (90) | |
| Lymph node | 61 (92) | 43 (94) | 18 (90) | |
| Pleura | 36 (55) | 27 (59) | 9 (45) | |
| Bone | 39 (59) | 31 (67) | 8 (40) | |
| Pleural effusion | 40 (61) | 33 (72) | 7 (35) | |
| Brain | 15 (23) | 11 (24) | 4 (20) | |
| Adrenal gland | 17 (26) | 15 (33) | 2 (10) | |
| Sum of target lesions’ diameters,a median (IQR) (mm) | ||||
| IRC assessment | 86.0 (49.2–118) | 95.6 (56.7–151) | 51.8 (28.3–62.0) | <0.001 |
| Investigators’ assessment | 69.9 (41.1–103) | 89.0 (56.0–121) | 37.8 (20.3–56.0) | <0.001 |
| Prior systemic treatment for advanced disease, n (%) | 0.474 | |||
| 0 | 25 (38) | 18 (39) | 7 (35) | |
| 1 | 31 (47) | 19 (41) | 12 (60) | |
| 2 | 5 (8) | 4 (9) | 1 (5) | |
| 3 | 3 (5) | 3 (7) | 0 | |
| ⩾4 | 2 (3) | 2 (4) | 0 | |
| Type of prior systemic treatment for advanced disease, n (%) | ||||
| Chemotherapy | 39 (59) | 27 (59) | 12 (60) | |
| Immunotherapy | 3 (5) | 2 (4) | 1 (5) | |
| Targeted therapy | 5 (8) | 4 (9) | 1 (5) | |
| Others | 15 (23) | 11 (24) | 4 (20) | |
| Amount of ctDNA, median (IQR) (ng) | 89.4 (52.1–134) | 107 (71.1–150) | 59.1 (49.4–84.3) | 0.00294 |
| bTMB, median (IQR) | 3.17 (1.06–6.34) | 5.29 (2.11–7.40) | 1.06 (0–2.11) | <0.001 |
For investigators’ assessment, five patients without postbaseline tumour evaluation were excluded from the calculation of average sum of target lesions. For IRC assessment, one additional patient assessed to be without target lesion was excluded; six patients in total were excluded from the calculation of average sum of target lesions by the IRC.
bTMB, blood-based tumour mutational burden; ctDNA, circulating tumour DNA; ECOG, Eastern Cooperative Oncology Group; IQR, interquartile range; IRC, independent review committee; METex14, MET exon 14 skipping alteration; NSCLC, non-small cell lung cancer; PSC, pulmonary sarcomatoid carcinoma.
Association of baseline METex14 status with clinical outcomes
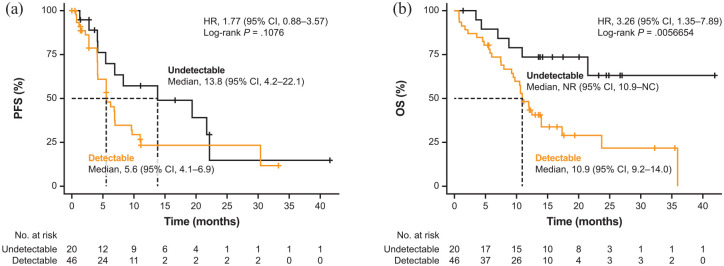
Baseline METex14 status showed association with clinical outcomes. In the detectable baseline METex14 group, ORR was 52.2% (95% CI, 36.9–67.1; 24 of 46 patients) compared with 30.0% (95% CI, 11.9–54.3; 6 of 20 patients) in the undetectable baseline METex14 group (p = 0.096; Supplemental Table 1). Detectable baseline METex14 was associated with a shorter median PFS of 5.6 months (95% CI, 4.14–6.93) compared with 13.8 months (95% CI, 4.17–22.14) in those with undetectable baseline METex14 (HR, 1.77; 95% CI, 0.88–3.57; p = 0.108; Figure 2(a); Supplemental Table 1). Patients with detectable baseline METex14 had significantly shorter median OS of 10.9 months (95% CI, 9.2–13.96), while it was not reached [95% CI, 10.91 months to not calculable (NC)] in patients with undetectable baseline METex14 (HR, 3.26; 95% CI, 1.35–7.89; p = 0.006; Figure 2(b); Supplemental Table 1). Using a multivariate Cox proportional hazards model, which included three covariates (baseline METex14 status, ctDNA content and bTMB levels), the HR for PFS between patients with detectable and undetectable baseline METex14 was 2.68 (95% CI, 1.22–5.91; p = 0.015), while that for OS was 3.72 (95% CI, 1.44–9.59; p = 0.007).
Figure 2.
Kaplan–Meier survival curves for (a) PFS and (b) OS by METex14 ctDNA status at baseline.
CI, confidence interval; ctDNA, circulating tumour DNA; HR, hazard ratio; METex14, MET exon 14 skipping alteration; NC, not calculable; NR, not reached; OS, overall survival; PFS, progression-free survival.
Association of METex14 post-treatment clearance with clinical outcomes
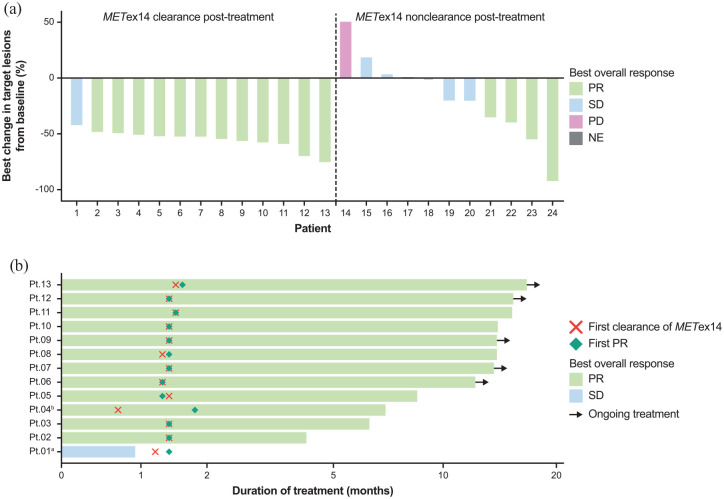
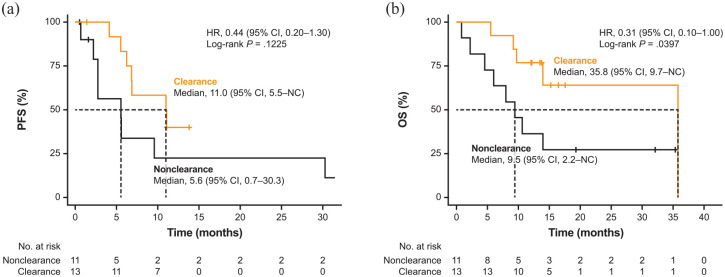
METex14 post-treatment clearance also showed association with clinical outcomes. Patients with METex14 post-treatment clearance exhibited a significantly higher ORR at 92.3% (95% CI, 64.0–99.8; 12 of 13 patients) compared with those with non-clearance at 36.4% (95% CI, 10.9–69.2; 4 of 11 patients; p = 0.0078; Figure 3(a); Supplemental Table 2). Median time to first clearance was 1.3 months (range, 0.7–1.5), which approximately coincided with the time to first partial response [Figure 3(b)]. Owing to a protocol deviation, the plasma sample of one patient was collected before the tumour assessment time point (at 0.7 month), which detected METex14 post-treatment clearance prior to a response. METex14 post-treatment clearance correlated with a longer median PFS versus non-clearance [11.0 months (95% CI: 5.5–NC) versus 5.6 months (95% CI: 0.7–30.3); HR, 0.44; 95% CI, 0.2–1.3; p = 0.1225; Figure 4(a); Supplemental Table 2]. Median OS was significantly longer in patients with METex14 post-treatment clearance than non-clearance [35.8 months (95% CI: 9.7–NC) versus 9.5 months (95% CI: 2.2–NC); HR, 0.31; 95% CI, 0.1–1.0; p = 0.0397; Figure 4(b); Supplemental Table 2].
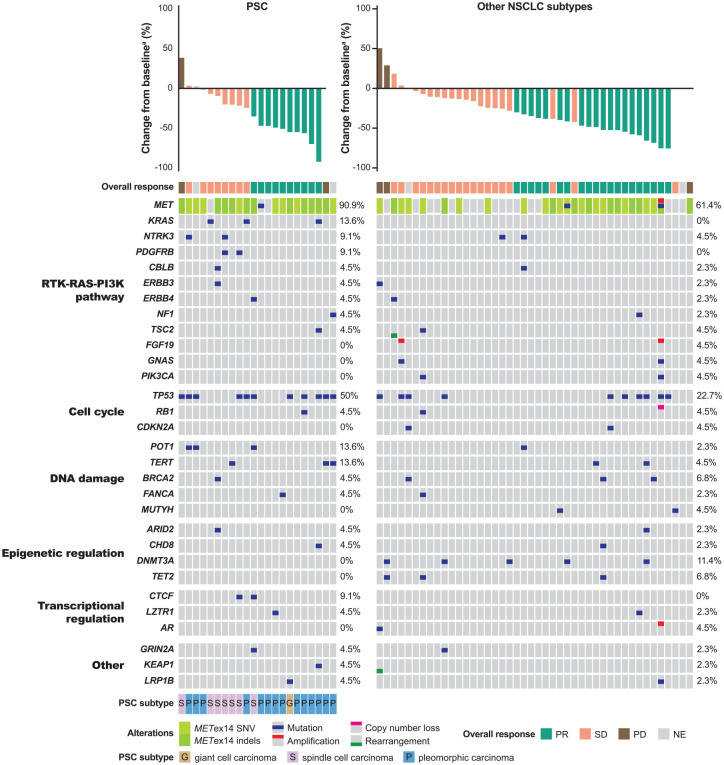
Figure 3.
(a) Best change in target lesions and best overall response by METex14 post-treatment clearance. (b) Time to first METex14 clearance.
aOne patient stopped treatment prior to the tumour assessment time point owing to adverse event; the best overall response for this patient was SD since PR was not confirmed. bOne patient’s sample (Pt 04) was collected prior to the tumour assessment time point (before 6 weeks).
ctDNA, circulating tumour DNA; METex14, MET exon 14 skipping alteration; NE, not evaluable; PD, progressive disease; PR, partial response; Pt, patient; SD, stable disease.
Figure 4.
Kaplan–Meier survival curves for (a) PFS and (b) OS by METex14 post-treatment clearance.
CI, confidence interval; ctDNA, circulating tumour DNA; HR, hazard ratio; METex14, MET exon 14 skipping alteration; NC, not calculable; OS, overall survival; PFS, progression-free survival.
Association of baseline and post-treatment METex14 status with clinical outcomes by NSCLC subtypes
Of the 66 patients in this post hoc analysis cohort, 22 (33.3%) had PSC, while the others had other NSCLC subtypes (mainly LUAD). Patients with PSC had significantly larger tumour sizes and higher ctDNA content than those with other NSCLC subtypes (Supplemental Table 3; Supplemental Figure 1(d)–(f)). Significantly more patients with PSC had detectable baseline METex14 compared with those with other NSCLC subtypes (86% versus 61%; p = 0.0483). Clinical outcomes were analysed by baseline METex14 status in the NSCLC histology subgroups, but it should be noted that subgroup sample sizes were small. Detectable versus undetectable baseline METex14 was associated with significantly higher ORR in patients with other NSCLC subtypes but not in those with PSC. Detectable baseline METex14 status predicted worse PFS and OS outcomes (significant for OS) in patients with other NSCLC subtypes but not in those with PSC (Supplemental Figure 2; Supplemental Table 1). In both subgroups of PSC and other NSCLC subtypes, post-treatment clearance was associated with higher ORR and prolonged PFS and OS compared with non-clearance, although statistical significance was not reached (Supplemental Figure 3; Supplemental Table 2).
Coexisting gene alterations at baseline
bTMB was not significantly different between patients with PSC and other NSCLC subtypes (3.70 versus 3.17; p = 0.995). Baseline coexisting gene alterations were mainly found in the components of the following pathways: receptor tyrosine kinase (RTK)-RAS-phosphoinositide 3-kinase (PI3K), DNA damage, cell cycle and transcriptional and epigenetic regulation. The most frequently coexisting gene alterations with METex14 at baseline were TP53 mutations in 11 (50.0%) of 22 patients with PSC and in 10 (22.7%) of 44 patients with other NSCLC subtypes (Figure 5). Besides TP53 mutations, other frequently altered genes included POT1, TERT and KRAS in three (13.6%) patients each in the PSC subgroup, and DNMT3A in five (11.4%) patients with other NSCLC subtypes (Figure 5). The detectable rates of TP53, POT1, TERT and KRAS mutations in the ctDNA of patients known to have the corresponding gene alterations (from tumour biopsy analysis9) were 64.3, 44.4, 71.4 and 66.7%, respectively.
Figure 5.
Association of baseline METex14 and coexisting gene alterations with best overall response.
aTwo patients with PSC and three with other NSCLC subtypes did not have best change in target lesions data determined by IRC; they either had no target lesion or no tumour assessment during treatment.
IRC, independent review committee; METex14, MET exon 14 skipping alteration; NE, not evaluable; NSCLC, non-small cell lung cancer; PD, progressive disease; PI3K, phosphoinositide 3-kinase; PR, partial response; PSC, pulmonary sarcomatoid carcinoma; RTK, receptor tyrosine kinase; SD, stable disease; SNV, single nucleotide variation.
The number of patients with each coexisting baseline gene alterations detected in the ctDNA was small, which precluded the association with clinical outcomes. The clinical outcomes by TP53 mutations, being the most frequent coexisting gene alteration, were analysed. Patients with mutant-TP53 ctDNA (n = 21) had an ORR 47.6% (95% CI, 25.7–70.2) versus 44.4% (95% CI 29.6–60.0) for patients with wild-type TP53 ctDNA (n = 45; p = 0.8094). PFS was 5.52 months (95% CI, 2.2–11.01) for patients with mutant-TP53 ctDNA versus 6.87 months (95% CI, 5.52–13.8) for patients with wild-type TP53 ctDNA (HR, 0.79; 95% CI, 0.42–1.50; p = 0.4735); OS was 10.91 months (95% CI, 3.61–NC) for patients with mutant-TP53 ctDNA versus 17.31 months (10.61–35.81) for patients with wild-type TP53 ctDNA (HR, 0.71; 95% CI, 0.36–1.43; p = 0.3415).
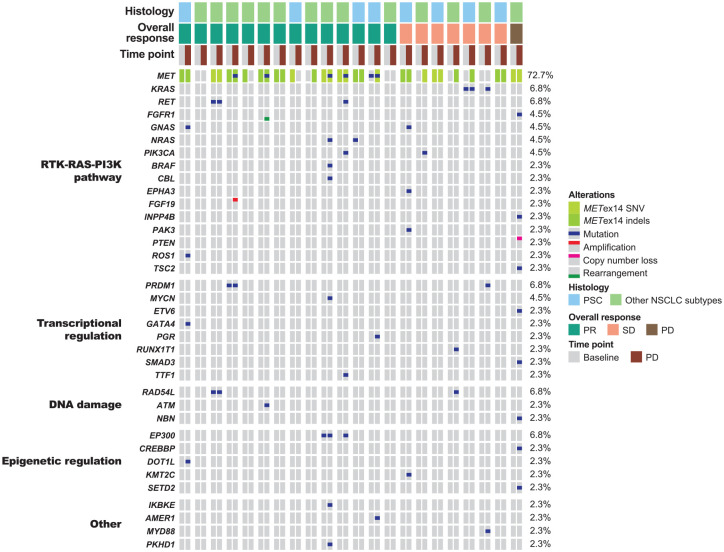
Gene alterations acquired upon disease progression
Upon disease progression, gene alterations were acquired in 10 (3 with PSC and 7 with other NSCLC subtypes) of 22 patients (Figure 6; Supplemental Table 4). These patients mainly acquired gene alterations in the RTK-RAS-PI3K pathway components (e.g. mutations in KRAS, NRAS, BRAF, PIK3CA, TSC2, RET, ROS1, GNAS, EPHA3 and PAK3, PTEN loss, FGF19 amplification and FGFR1 rearrangement), DNA damage pathway components (mutations in RAD54L, ATM and NBN), transcriptional regulators (mutations in MYCN, ETV6, GATA4, PGR, RUNX1T1, SMAD3 and TTF1) and epigenetic modifiers (mutations in CREBBP, DOT1L, KMT2C and SETD2). Secondary MET mutations (D1228H/N and Y1230C/H/S) were acquired in four patients with other NSCLC subtypes, with one patient exhibiting triple MET secondary mutations in trans (Supplemental Figure 4). All secondary MET mutations coexisted with other acquired gene alterations, including mutations in NRAS, BRAF, PIK3CA and ATM, and FGF19 amplification.
Figure 6.
Gene alterations determined from paired ctDNA samples collected at baseline and disease progression (n = 22).
METex14, MET exon 14 skipping alteration; NSCLC, non-small cell lung cancer; PD, progressive disease; PI3K, phosphoinositide 3-kinase; PR, partial response; PSC, pulmonary sarcomatoid carcinoma; RTK, receptor tyrosine kinase; SD, stable disease; SNV, single nucleotide variation.
Discussion
METex14-positive PSC is rare, and the efficacy of MET inhibitors in patients with this NSCLC subtype remains poorly investigated. Our phase 2 study demonstrated favourable clinical outcomes with savolitinib in the largest cohort of patients with METex14-positive PSC to date, as well as in other NSCLC subtypes.9 This post hoc analysis further evaluated ctDNA biomarkers in predicting clinical outcomes. Patients with detectable baseline METex14 showed higher ORR on savolitinib treatment, but had worse survival outcomes, supporting the benefit of early treatment with savolitinib in these patients. METex14 post-treatment clearance was associated with a higher ORR and improved survival outcomes versus non-clearance. Common baseline coexisting gene mutations, although detected at lower frequencies, were similar to those identified from tissue biopsies. Upon disease progression, patients acquired secondary MET mutations and other gene alterations, including those in the RTK-RAS-PI3K pathway, which may be associated with savolitinib resistance. To our knowledge, this is the first comprehensive study of the association between METex14 ctDNA status and survival outcomes with a MET inhibitor in METex14-positive PSC and reports the longitudinal on-treatment ctDNA genomic profiling of PSC in the largest sample size to date.
METex14 ctDNA has been detected in patients with METex14-positive NSCLC identified through tumour biopsy testing. Among patients screened for enrolment using liquid biopsy in the VISION trial, 3.6% had detectable METex14 in their ctDNA,13 consistent with the frequency of such genetic alterations in lung malignancies.2 In our study, 70% of patients with METex14-positive NSCLC had detectable METex14 in their ctDNA at baseline. Thus, plasma-based ctDNA testing can be used to detect METex14,13,14 although some cases may be missed, likely due to low tumour DNA shedding below the detection limit of the assay.15 We observed a significantly higher frequency of detectable baseline METex14 in the PSC subgroup versus in patients with other NSCLC subtypes (86% versus 61%), which may be explained by the larger tumour sizes and higher amount of ctDNA in the former subgroup. Compared with a previous report of crizotinib in a METex14-positive NSCLC cohort (the PROFILE 1001 trial), a higher proportion of patients in the present study had detectable METex14 ctDNA (70% versus 49%),14 possibly reflecting the enrolment of a larger number of patients with PSC and with poor prognoses.
Detectable baseline METex14 ctDNA was associated with a numerically higher ORR but worse survival outcomes (significant for OS) with savolitinib in this study cohort. In particular, the association of detectable baseline METex14 with both poorer PFS and OS reached statistical significance after adjusting for potential confounding factors (ctDNA content and bTMB), supporting the independent predictive value of METex14 ctDNA in patients’ survival. The presence of detectable baseline METex14 ctDNA likely suggests a higher abundance of this driver gene alteration in the tumour tissue, which in turn may entail a higher sensitivity to savolitinib, although the ORR findings warrant further confirmation in a larger sample size for a statistically significant trend. Baseline METex14 detected with liquid biopsy was a criterion used to select patients for tepotinib treatment in the VISION trial.6,16 However, for crizotinib, a nonselective MET inhibitor, similar ORRs were observed regardless of detectable METex14 in ctDNA.14 The association between the abundance of driver gene alteration in the ctDNA and treatment response has been reported for other targeted therapies. Higher plasma mutant EGFR concentrations predicted increased response to EGFR tyrosine kinase inhibitors.17 Detectable baseline METex14 ctDNA (versus undetectable) might also reflect a greater tumour bulk. This is supported by our observation that bTMB was significantly higher in patients with detectable versus undetectable baseline METex14 ctDNA, which potentially explains the worse survival outcomes in the former. According to an earlier MET inhibitor study, PFS was significantly shorter with crizotinib in patients with detectable versus undetectable METex14.14 Through subgroup analyses based on NSCLC histology, we further showed that detectable baseline METex14 was associated with higher ORR and worse survival outcomes in patients with other NSCLC subtypes, consistent with our observation in the overall post hoc ctDNA analysis cohort. This was, however, not observed in the PSC subgroup, likely due to the small number of patients with undetectable METex14 PSC (n = 3) and requires evaluation in a larger subgroup sample size. As detectable baseline METex14 was associated with worse prognosis but predicted response to MET inhibitors, early initiation of treatment in these patients would be beneficial; this was similarly observed with other targeted therapies.17,18–20
METex14 post-treatment clearance potentially predicts radiologic tumour response. In this study, METex14 post-treatment clearance was observed in 54% of evaluable patients, who demonstrated better response and survival outcomes with savolitinib. Similar trends were observed in the NSCLC histology subgroups although statistical significance was not reached possibly due to small sample sizes. To our knowledge, this is the first study to indicate that METex14 post-treatment clearance predicts survival benefit with a MET inhibitor. Although METex14 clearance was also reported to be associated with a higher ORR on tepotinib treatment compared with non-clearance, the association with survival outcomes was not studied.6,16 The clearance of driver gene alterations in the ctDNA was generally correlated with treatment response and better survival outcomes for other targeted therapies.17–19,21
The present post hoc genomic profiling of patients’ ctDNA samples revealed similar types of common baseline coexisting gene alterations as those identified from tumour biopsies.9 Similar to the observations with tumour mutational burden in baseline tissue samples, there was no significant difference in bTMB between patients with PSC and other NSCLC subtypes. From both baseline tumour and ctDNA analyses, TP53 mutations were the most common coexisting gene alterations.9 ctDNA analyses showed that TP53, POT1, TERT and KRAS mutations were more common in the PSC subgroup than in patients with other NSCLC subtypes, consistent with findings from the tumour biopsies.9 Among patients with TP53, POT1, TERT and KRAS mutations in their tumours, the detectable rates of these mutations in the ctDNA ranged from 44.4 to 71.4%.9 In contrast, gene amplifications (e.g. of MDM2 and TERT) observed in baseline tumour tissues were not detected in the ctDNA. These observations suggest that tissue-blood concordance may be present for single nucleotide variations but not copy number variations.
We previously reported that baseline coexisting TP53 and POT1 mutations detected in METex14-positive tumour tissue might adversely affect savolitinib treatment outcomes.9 In this post hoc analysis, patients with baseline coexisting TP53 mutations detected in their ctDNA samples were relatively small, which may explain the lack of significant association observed between TP53-mutant ctDNA and clinical outcomes. Savolitinib treatment outcomes based on POT1-mutant ctDNA were also not determined due to the small number of analysable patients. Concomitant TP53 mutations detected in both tumour and plasma samples were previously shown to be a negative prognostic factor in patients with advanced NSCLC treated with other tyrosine kinase inhibitors,22,23 while the potential prognostic value of POT1 mutations in NSCLC has not been elucidated. The association of common baseline coexisting TP53 and POT1 mutations (detected in tumour biopsies or ctDNA samples) with savolitinib treatment outcomes has to be confirmed in a larger cohort of patients with METex14 NSCLC.
Upon disease progression, acquired genetic alterations detected in the ctDNA can provide insight into the mechanisms of therapeutic resistance. Data on resistance mechanisms of selective MET inhibitors, determined from ctDNA analysis, in METex14-positive NSCLC remain limited and preliminary. In this study, secondary MET mutations (D1228H/N and Y1230C/H/S) were detected in the ctDNA upon disease progression with savolitinib. Multiple secondary MET mutations were detected in trans in one patient, indicative of these mutations being derived from different clones. Several studies in MET inhibitor-treated METex14-positive NSCLC have similarly reported various types of secondary MET mutations, including those at D1228 and Y1230 residues, that were acquired upon disease progression.6,24–27 Mutations involving D1228 and Y1230 residues in the activation loop prevent the binding of type I MET inhibitors, including savolitinib, leading to resistance that can potentially be circumvented by type II MET inhibitors (e.g. cabozantinib, glesatinib and merestinib), supporting the sequential use of type I and II MET inhibitors.28,29
In addition, we observed MET-independent resistance mechanisms in all patients at disease progression. Acquired gene alterations in the RAS/RAF, PI3K/phosphatase and tensin homologue (PTEN) and fibroblast growth factor receptor (FGFR) pathways that may bypass MET signalling were detected in the ctDNA at disease progression. In one patient with METex14 PSC from this study who had tumour biopsy sample at disease progression with savolitinib, amplification of FGFR1, EGFR and KRAS was reported.30 These observations point to the potential of FGFR/EGFR-RAS pathway inhibition in overcoming resistance to savolitinib. Available evidence similarly showed that RAS/RAF/mitogen-activated protein kinase pathway alterations at baseline and upon disease progression were implicated with primary and acquired resistance to other MET inhibitors, respectively, in patients with METex14-positive NSCLC.24–26,31 The inhibition of Src homology 2 domain-containing-phosphatase 2, a common RAS upstream signalling node of multiple oncogenic pathways, was shown to delay and overcome tepotinib resistance in cell lines.32 In the present study, we also observed acquired gene alterations in other components of the RTK-RAS-PI3K pathway, as well as those involved in DNA damage response, and transcriptional and epigenetic regulation. Patients with PSC and other NSCLC subtypes showed acquired gene alterations in similar or related pathways, although this should be interpreted with caution due to the small number of patients with disease progression in this study. Taken together, ctDNA analysis at disease progression can reveal the potential mechanisms of acquired resistance to savolitinib, and hence inform the efficacy of subsequent targeted therapies alone or as an add-on to circumvent savolitinib resistance. Larger prospective trials that investigate the predictive values of these genetic alterations for savolitinib resistance in patients with METex14-positive NSCLC, as well as mechanistic studies to verify the functions of these alterations, are warranted.2
This study has a few limitations. Firstly, as ctDNA dynamic monitoring was a post hoc, exploratory analysis of the trial, the sample size is limited, especially for subgroup analyses by NSCLC subtypes and the study of gene alterations in patients who progressed with savolitinib. The predictive value of these ctDNA biomarkers has to be validated in larger cohorts. Nevertheless, to our knowledge, this study reports the longitudinal on-treatment ctDNA genomic profiling and association of such biomarkers with survival outcomes in the largest sample size of METex14-positive PSC (a particularly rare disease) to date. Secondly, early collection of plasma samples prior to tumour assessment was not planned; hence, it remains to be determined whether METex14 post-treatment clearance precedes radiologic response. Previous evidence has shown that clearance of other targetable alterations, such as EGFR mutations, can predict treatment response and disease progression ahead of radiological results.19,21 Lastly, limitations in liquid biopsy should be considered. Tissue biopsies remain the gold standard in tumour molecular characterization, especially to detect histologic transformation upon acquired therapeutic resistance.11 Both approaches are complementary in clinical practice. Although longitudinal genomic profiling of liquid biopsies was conducted for this cohort, tumour biopsies on-treatment and/or upon disease progression were not available for biomarker analysis.
Conclusion
Baseline and on-treatment ctDNA-based NGS analysis potentially allows for initial prediction and longitudinal monitoring of clinical outcomes with savolitinib in patients with METex14-positive PSC and other NSCLC subtypes. Specifically, undetectable baseline METex14 or post-treatment clearance may predict favourable clinical outcomes. Furthermore, ctDNA-based NGS analysis at disease progression is a non-invasive tool for evaluating potential mechanisms of acquired resistance to savolitinib, which may inform the efficacy of subsequent personalized targeted or combination therapies.
Supplemental Material
Supplemental material, sj-docx-1-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Supplemental material, sj-eps-2-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Supplemental material, sj-eps-3-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Supplemental material, sj-eps-4-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Supplemental material, sj-eps-5-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Acknowledgments
Medical writing support was funded by HUTCHMED and provided by Qing Yun Chong and Henry Chung (Nucleus Global, Shanghai, China), in accordance with Good Publication Practice 3 guidelines.
Footnotes
ORCID iD: Shun Lu  https://orcid.org/0000-0001-8833-7262
https://orcid.org/0000-0001-8833-7262
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Yongfeng Yu, Department of Medical Oncology, Shanghai Chest Hospital, Shanghai Jiaotong University, Shanghai, China.
Yongxin Ren, HUTCHMED, Shanghai, China.
Jian Fang, Peking University Cancer Hospital and Institute, Beijing, China.
Lejie Cao, Anhui Provincial Hospital, The First Affiliated Hospital of University of Science and Technology of China, Hefei, China.
Zongan Liang, West China Hospital of Sichuan University, Chengdu, China.
Qisen Guo, Shandong Cancer Hospital Affiliated to Shandong University, Jinan, China.
Sen Han, Peking University Cancer Hospital and Institute, Beijing, China.
Zimei Ji, Anhui Provincial Hospital, The First Affiliated Hospital of University of Science and Technology of China, Hefei, China.
Ye Wang, West China Hospital of Sichuan University, Chengdu, China.
Yulan Sun, Shandong Cancer Hospital Affiliated to Shandong University, Jinan, China.
Yuan Chen, Tongji Hospital, Huazhong University of Science and Technology, Wuhan, China.
Xingya Li, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China.
Hua Xu, The Second Affiliated Hospital of Nanchang University, Nanchang, China.
Jianying Zhou, The First Affiliated Hospital of Zhejiang University, Hangzhou, China.
Liyan Jiang, Department of Medical Oncology, Shanghai Chest Hospital, Shanghai Jiaotong University, Shanghai, China.
Ying Cheng, Jilin Cancer Hospital, Changchun, China.
Zhigang Han, The Affiliated Cancer Hospital of Xinjiang Medical University, Urumqi, China.
Jianhua Shi, Linyi Cancer Hospital, Linyi, China.
Gongyan Chen, Cancer Hospital of Harbin Medical University, Harbin, China.
Rui Ma, Liaoning Cancer Hospital, Shenyang, China.
Yun Fan, Zhejiang Cancer Hospital, Hangzhou, China.
Sanyuan Sun, Xuzhou Central Hospital, Xuzhou, China.
Longxian Jiao, HUTCHMED, Shanghai, China.
Xiaoyun Jia, HUTCHMED, Shanghai, China.
Linfang Wang, HUTCHMED, Shanghai, China.
Puhan Lu, HUTCHMED, Shanghai, China.
Qian Xu, HUTCHMED, Shanghai, China.
Xian Luo, HUTCHMED, Shanghai, China.
Weiguo Su, HUTCHMED, Shanghai, China.
Shun Lu, Department of Medical Oncology, Shanghai Lung Cancer Center, Shanghai Chest Hospital, Shanghai Jiaotong University, No. 241, Huaihai West Road, Shanghai 200030, China.
Declarations
Ethics approval and consent to participate: This study was conducted in accordance with the Declaration of Helsinki and Guidelines for Good Clinical Practice. The protocol and all amendments were approved by the ethics committees from each participating institute (approval number: LS1626). All patients provided written informed consent prior to enrolment. Patients who participated in this exploratory biomarker post hoc analysis provided additional informed consent.
Consent for publication: Not applicable.
Author contribution(s): Yongfeng Yu: Data curation; Investigation; Resources; Validation; Visualization; Writing – original draft.
Yongxin Ren: Investigation; Supervision; Validation; Visualization; Writing – original draft.
Jian Fang: Data curation; Investigation; Resources; Writing – review & editing.
Lejie Cao: Data curation; Investigation; Resources; Writing – review & editing.
Zongan Liang: Data curation; Investigation; Resources; Writing – review & editing.
Qisen Guo: Data curation; Investigation; Resources; Writing – review & editing.
Sen Han: Data curation; Investigation; Resources; Writing – review & editing.
Zimei Ji: Data curation; Investigation; Resources; Writing – review & editing.
Ye Wang: Data curation; Investigation; Resources; Writing – review & editing.
Yulan Sun: Data curation; Investigation; Resources; Writing – review & editing.
Yuan Chen: Data curation; Investigation; Resources; Writing – review & editing.
Xingya Li: Data curation; Investigation; Resources; Writing – review & editing.
Hua Xu: Data curation; Investigation; Resources; Writing – review & editing.
Jianying Zhou: Data curation; Investigation; Resources; Writing – review & editing.
Liyan Jiang: Data curation; Investigation; Resources; Writing – review & editing.
Ying Cheng: Data curation; Investigation; Resources; Writing – review & editing.
Zhigang Han: Data curation; Investigation; Resources; Writing – review & editing.
Jianhua Shi: Data curation; Investigation; Resources; Writing – review & editing.
Gongyan Chen: Data curation; Investigation; Resources; Writing – review & editing.
Rui Ma: Data curation; Investigation; Resources; Writing – review & editing.
Yun Fan: Data curation; Investigation; Resources; Writing – review & editing.
Sanyuan Sun: Data curation; Investigation; Resources; Writing – review & editing.
Longxian Jiao: Formal analysis; Investigation; Validation; Visualization; Writing – original draft.
Xiaoyun Jia: Investigation; Writing – review & editing.
Linfang Wang: Investigation; Validation; Visualization; Writing – original draft.
Puhan Lu: Investigation; Writing – review & editing.
Qian Xu: Formal analysis; Investigation; Validation; Visualization; Writing – original draft.
Xian Luo: Formal analysis; Investigation; Writing – review & editing.
Weiguo Su: Conceptualization; Funding acquisition; Investigation; Methodology; Project administration; Resources; Supervision; Writing – review & editing.
Shun Lu: Conceptualization; Data curation; Investigation; Methodology; Resources; Supervision; Validation; Visualization; Writing – original draft.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by HUTCHMED (project number: 2016-504-00CH1) and AstraZeneca.
SL reports receiving research support from AstraZeneca, HUTCHMED, Bristol Myers Squibb, Hengrui Therapeutics, BeiGene, Roche and Hansoh; receiving speaker fees from AstraZeneca, Roche, Hansoh and Hengrui Therapeutics and being an advisor and consultant for AstraZeneca, Pfizer, Boehringer Ingelheim, HUTCHMED, Simcere, Zai Lab, GenomiCare, Yuhan Corporation, prIME Oncology, Menarini, InventisBio Co. Ltd and Roche. No disclosures were reported by the other authors.
Availability of data and materials: The trial protocol and statistical analysis plan have been published with the original study (https://www.thelancet.com/journals/lanres/article/PIIS2213-2600(21)00084-9/fulltext). Individual participant data will not be made available to others.
References
- 1. Madison R, Schrock AB, Castellanos E, et al. Retrospective analysis of real-world data to determine clinical outcomes of patients with advanced non-small cell lung cancer following cell-free circulating tumor DNA genomic profiling. Lung Cancer 2020; 148: 69–78. [DOI] [PubMed] [Google Scholar]
- 2. Salgia R, Sattler M, Scheele J, et al. The promise of selective MET inhibitors in non-small cell lung cancer with MET exon 14 skipping. Cancer Treat Rev 2020; 87: 102022. [DOI] [PubMed] [Google Scholar]
- 3. Karim NA, Schuster J, Eldessouki I, et al. Pulmonary sarcomatoid carcinoma: university of cincinnati experience. Oncotarget 2018; 9: 4102–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vieira T, Girard N, Ung M, et al. Efficacy of first-line chemotherapy in patients with advanced lung sarcomatoid carcinoma. J Thorac Oncol 2013; 8: 1574–1577. [DOI] [PubMed] [Google Scholar]
- 5. Hong L, Zhang J, Heymach JV, et al. Current and future treatment options for MET exon 14 skipping alterations in non-small cell lung cancer. Ther Adv Med Oncol 2021; 13: 1758835921992976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paik PK, Felip E, Veillon R, et al. Tepotinib in non-small-cell lung cancer with MET exon 14 skipping mutations. N Engl J Med 2020; 383: 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wolf J, Seto T, Han JY, et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer. N Engl J Med 2020; 383: 944–957. [DOI] [PubMed] [Google Scholar]
- 8. Jia H, Dai G, Weng J, et al. Discovery of (S)-1-(1-(Imidazo[1,2-a]pyridin-6-yl)ethyl)-6-(1-methyl-1H-pyrazol-4-yl)-1H-[1,2,3]triazolo[4,5-b]pyrazine (volitinib) as a highly potent and selective mesenchymal-epithelial transition factor (c-Met) inhibitor in clinical development for treatment of cancer. J Med Chem 2014; 57: 7577–7589. [DOI] [PubMed] [Google Scholar]
- 9. Lu S, Fang J, Li X, et al. Once-daily savolitinib in Chinese patients with pulmonary sarcomatoid carcinomas and other non-small-cell lung cancers harbouring MET exon 14 skipping alterations: a multicentre, single-arm, open-label, phase 2 study. Lancet Respir Med 2021; 9: 1154–1164. [DOI] [PubMed] [Google Scholar]
- 10. Roosan MR, Mambetsariev I, Pharaon R, et al. Utility of circulating tumor DNA in identifying somatic mutations and tracking tumor evolution in patients with non-small cell lung cancer. Chest 2021; 160: 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Akhoundova D, Mosquera Martinez J, Musmann LE, et al. The role of the liquid biopsy in decision-making for patients with non-small cell lung cancer. J Clin Med 2020; 9: 3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang DS, Liu ZX, Lu YX, et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 2019; 68: 1152–1161. [DOI] [PubMed] [Google Scholar]
- 13. Le X, Kowalski D, Cho BC, et al. Liquid biopsy to detect MET exon 14 skipping METex14) and MET amplification in patients with advanced NSCLC: biomarker analysis from VISION study. Cancer Res 2020; 80 (16 Suppl): 3385. [Google Scholar]
- 14. Drilon A, Clark JW, Weiss J, et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med 2020; 26: 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paweletz CP, Lau CJ, Oxnard GR. Does testing error underlie liquid biopsy discordance? JCO Precis Oncol 2019; 3: 1–3. [DOI] [PubMed] [Google Scholar]
- 16. Sakai H, Morise M, Kato M, et al. Tepotinib in patients with NSCLC harbouring MET exon 14 skipping: Japanese subset analysis from the Phase II VISION study. Jpn J Clin Oncol 2021; 51: 1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu YJ, Zhang HB, Liu YH, et al. Estimation of cell-free circulating EGFR mutation concentration predicts outcomes in NSCLC patients treated with EGFR-TKIs. Oncotarget 2017; 8: 13195–13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Madsen AT, Winther-Larsen A, McCulloch T, et al. Genomic profiling of circulating tumor DNA predicts outcome and demonstrates tumor evolution in ALK-positive non-small cell lung cancer patients. Cancers (Basel) 2020; 12: 947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med 2018; 6: 681–690. [DOI] [PubMed] [Google Scholar]
- 20. Papadimitrakopoulou VA, Han JY, Ahn MJ, et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer. Cancer 2020; 126: 373–380. [DOI] [PubMed] [Google Scholar]
- 21. Liao B-C, Hsu W-H, Lee J-H, et al. Serial plasma cell-free circulating tumor DNA tests identify genomic alterations for early prediction of osimertinib treatment outcome in EGFR T790M–positive NSCLC. JTO Clin Res Rep 2021; 2: 100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qin K, Hou H, Liang Y, et al. Prognostic value of TP53 concurrent mutations for EGFR- TKIs and ALK-TKIs based targeted therapy in advanced non-small cell lung cancer: a meta-analysis. BMC Cancer 2020; 20: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iwama E, Sakai K, Azuma K, et al. Exploration of resistance mechanisms for epidermal growth factor receptor-tyrosine kinase inhibitors based on plasma analysis by digital polymerase chain reaction and next-generation sequencing. Cancer Sci 2018; 109: 3921–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Awad MM, Bahcall M, Sholl LM, et al. Mechanisms of acquired resistance to MET tyrosine kinase inhibitors (TKIs) in MET exon 14 (METex14) mutant non-small cell lung cancer (NSCLC). J Clin Oncol 2018; 36 (15 suppl): 9069. [Google Scholar]
- 25. Recondo G, Bahcall M, Spurr LF, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14-mutant NSCLC. Clin Cancer Res 2020; 26: 2615–2625. [DOI] [PubMed] [Google Scholar]
- 26. Rotow JK, Gui P, Wu W, et al. Co-occurring alterations in the RAS-MAPK pathway limit response to MET inhibitor treatment in MET exon 14 skipping mutation-positive lung cancer. Clin Cancer Res 2020; 26: 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dagogo-Jack I, Moonsamy P, Gainor JF, et al. A phase 2 study of capmatinib in patients with MET-altered lung cancer previously treated with a MET Inhibitor. J Thorac Oncol 2021; 16: 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fujino T, Kobayashi Y, Suda K, et al. Sensitivity and resistance of MET exon 14 mutations in lung cancer to eight MET tyrosine kinase inhibitors in vitro. J Thorac Oncol 2019; 14: 1753–1765. [DOI] [PubMed] [Google Scholar]
- 29. Lu X, Peled N, Greer J, et al. MET exon 14 mutation encodes an actionable therapeutic target in lung adenocarcinoma. Cancer Res 2017; 77: 4498–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han S, Fang J, Lu S, et al. Response and acquired resistance to savolitinib in a patient with pulmonary sarcomatoid carcinoma harboring MET exon 14 skipping mutation: a case report. Onco Targets Ther 2019; 12: 7323–7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suzawa K, Offin M, Lu D, et al. Activation of KRAS mediates resistance to targeted therapy in MET exon 14-mutant non-small cell lung cancer. Clin Cancer Res 2019; 25: 1248–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pudelko L, Jaehrling F, Reusch C, et al. SHP2 inhibition influences therapeutic response to tepotinib tumors with MET alterations. iScience 2020; 23: 101832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Supplemental material, sj-eps-2-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Supplemental material, sj-eps-3-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Supplemental material, sj-eps-4-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology
Supplemental material, sj-eps-5-tam-10.1177_17588359221133546 for Circulating tumour DNA biomarkers in savolitinib-treated patients with non-small cell lung cancer harbouring MET exon 14 skipping alterations: a post hoc analysis of a pivotal phase 2 study by Yongfeng Yu, Yongxin Ren, Jian Fang, Lejie Cao, Zongan Liang, Qisen Guo, Sen Han, Zimei Ji, Ye Wang, Yulan Sun, Yuan Chen, Xingya Li, Hua Xu, Jianying Zhou, Liyan Jiang, Ying Cheng, Zhigang Han, Jianhua Shi, Gongyan Chen, Rui Ma, Yun Fan, Sanyuan Sun, Longxian Jiao, Xiaoyun Jia, Linfang Wang, Puhan Lu, Qian Xu, Xian Luo, Weiguo Su and Shun Lu in Therapeutic Advances in Medical Oncology