Abstract
Germline mutations leading to aneuploidy are rare, and their tumor-promoting properties are mostly unknown at the molecular level. We report here novel germline biallelic mutations in MAD1L1, encoding the spindle assembly checkpoint (SAC) protein MAD1, in a 36-year-old female with a dozen of neoplasias. Functional studies demonstrated lack of full-length protein and deficient SAC response, resulting in ~30 to 40% of aneuploid blood cells. Single-cell RNA analysis identified mitochondrial stress accompanied by systemic inflammation with enhanced interferon and NFκB signaling both in aneuploid and euploid cells, suggesting a non–cell autonomous response. MAD1L1 mutations resulted in specific clonal expansions of γδ T cells with chromosome 18 gains and enhanced cytotoxic profile as well as intermediate B cells with chromosome 12 gains and transcriptomic signatures characteristic of leukemia cells. These data point to MAD1L1 mutations as the cause of a new variant of mosaic variegated aneuploidy with systemic inflammation and unprecedented tumor susceptibility.
MAD1L1 mutations found in an individual with high tumor susceptibility and a systemic inflammation response to aneuploidy.
INTRODUCTION
Aneuploidy is a frequent finding in spontaneous abortions and a near-universal characteristic of tumor cells (1). However, germline mutations leading to chromosomal instability are rare in humans and lead to clinical and genetically heterogeneous syndromes (2). The hallmark of these patients is the presence of mosaic aneuploidies, mainly trisomies and, more rarely, monosomies, and a variegated distribution of affected chromosomes in different cells and tissues throughout the body.
Human diseases characterized by chromosomal instability are typically associated to mutations in genes related to cell division. Mutations in genes associated with centrosome and microtubule dynamics, chromosome morphology, and kinetochore functions affect the majority of patients with primary microcephaly (3), and alteration of different cohesin subunits and associated regulators results in a variety of pathologies known as cohesinopathies (4). Aneuploidy, however, is only occasionally observed in these pathologies (4). On the other hand, mosaic variegated aneuploidy (MVA) typically correlates with mutations in components of the spindle assembly checkpoint (SAC), a multigene network that, together with other biological processes, controls the accurate chromosome segregation into daughter cells during cell division and permits ordered cell cycle progression from metaphase to anaphase (5, 6). The SAC monitors the complete bipolar attachment of kinetochores on chromosomes to microtubules of the mitotic spindle and delays anaphase entry in the presence of unattached kinetochores. This is achieved through the formation of the mitotic checkpoint complex (MCC), composed of BUBR1, mitotic arrest deficiency 2 (MAD2), BUB3 and CDC20, and the subsequent inhibition of the anaphase-promoting complex/cyclosome (APC/C). SAC deficiency results in chromosome segregation in the presence of unattached chromosomes, generating daughter cells with abnormal chromosome numbers. In patients, these alterations are associated with a heterogeneous and nonspecific phenotype that typically involves pre- and postnatal growth retardation, eye and facial anomalies, variable developmental delay, and intellectual disability (7–9).
Despite the widespread presence of aneuploidy in human tumors, to what extent chromosomal instability plays oncogenic or tumor suppressor roles is unclear (1, 10). Cancer predisposition is not a common feature in patients with microcephaly or cohesinopathies. MVA is associated with a higher increase in some childhood malignancies, such as rhabdomyosarcoma, Wilms tumor, and leukemia, although these neoplasias are only observed in a subset of patients with MVA, and specific MVA-associated mutations have been suggested to impair tumor progression (9, 11). Common features of patients with MVA are therefore intellectual disability and growth retardation, with or without microcephaly and tumors.
We describe here the first germline biallelic mutations in MAD1L1 as a novel cause of aneuploidy in an individual with no intellectual disability and an unprecedented number of neoplasias. The MAD1L1 product MAD1 is an essential component of the mitotic checkpoint originally identified as a protein required for mitotic arrest in the presence of mitotic poisons (12, 13). MAD1 recruits the MCC component MAD2 to unattached kinetochores, thus promoting the formation of the MCC and APC/C inhibition (14–16). Lack of Mad1l1 is lethal in the mouse during early development (17), and no germline pathogenic mutations in the MAD1L1 gene have been described in humans. The proband displays biallelic mutations that are present in her parents in a heterozygous manner. The proband developed 12 neoplasias including five malignant tumors before the age of 36 years. Cellular studies suggested defective SAC function associated to high levels of aneuploidy (30 to 40%) in peripheral blood cells. Single-cell transcriptomic studies allowed the first analysis of the molecular consequences of aneuploidy in an individual with germline chromosomal instability. These studies suggested the presence of a non–cell autonomous inflammatory response to aneuploidy and identified small populations of cells with selected aneuploidies and specific functional or premalignant properties of physiological or clinical relevance.
RESULTS
Identification of germline biallelic mutations in MAD1L1
The proband, a female, was born in 1986 after an uncomplicated term pregnancy of a 26-year-old mother and a 30-year old father, both healthy and not consanguineous. Apgar was 9 of 10. Birth weight was 2510 g (3rd < P < 10th centile), with a length of 48 cm (10th < P < 25th centile). Developmental delay and slightly psychomotor retardation were apparent during the first months of life (Table 1). Several café-au-lait spots were noted before the 6 months of life. At the age of 2 years, she had a stage III embryonal rhabdomyosarcoma of the left auditory canal that was treated with chemotherapy and radiotherapy. The impairment of her growth curve was interpreted as a consequence of the radiotherapy treatment, and growth hormone therapy was indicated. In 2001, several bone masses suggesting enchondromatosis were seen in the femur, humerus, and ulna. In addition, in 2001, a stage IB clear cell cervical carcinoma with ectocervix and endocervix involvement was diagnosed, without evidences of human papillomavirus infection, and treated with hysterectomy, bilateral adnexectomy, brachytherapy, and external radiotherapy. The proband had not been exposed to diethylstilbestrol in utero. In 2006, a pleomorphic adenoma of left parotid gland was surgically removed. One year later, she suffered a left mastoidectomy with left parotidectomy due to a low-grade fusiform cell sarcoma. During the period 2006–2010, several dysplastic nevi, a mammary lipoma, and a pilomatrixoma were removed. In 2010, a hemithyroidectomy was performed by a multinodular goiter. In 2012, a polypectomy of one adenoma of the colon harboring an intramucosal adenocarcinoma was carried out, and 2 years later, a pT3N0M0 rectal adenocarcinoma was resected. Another tubular adenoma was removed in the control colonoscopy in 2014.
Table 1. A comparison of phenotypes in individuals with germline aneuploidy.
CRC, colorectal carcinoma; ERMS, embryonic rhabdomyosarcoma; WT, Wilms tumor.
| Gene | N of individuals | Major phenotype | Age* | Aneuploidy† | Cancer | Other neoplasias |
Representative
references |
| BUB1B | 24 | Intellectual disability, growth retardation, short stature, microcephaly, cataracts, facial dysmorphism, skin alterations, Dandy-Walker |
68 (0.4–47) | 4.5–78% | ERMS, WT, ampulla Vater carcinoma, CRC, gastric cancer, rhabdomyosarcoma (two patients with both ERMS and WT) |
GI adenomas, myelodysplasia |
(7, 33, 58) |
| CEP57 | 12 | Intellectual disability, microcephaly, growth retardation, facial dysmorphism, skull abnormalities, rhizomelic shortening |
29 | 12–22% | No | No | (26, 59, 60) |
| TRIP13 | 6 | Microcephaly, seizures, growth retardation, skin alterations |
43 (10) | 10–76% | WT | No | (27) |
| CENATAC | 2 | Microcephaly, developmental delay, maculopathy |
47 | 7.3–8.5% | No | No | (29) |
| BUB1 | 2 | Intellectual disability, microcephaly, growth retardation, microcephaly, facial dysmorphism, skin alterations |
17 | 35–42% in one of the individuals |
No | No | (28) |
| MAD1L1 | 1 | Developmental delay, microcephaly, growth retardation, facial dysmorphism, skin alterations |
36 | 39–43% | Rhabdomyosarcoma, cervical carcinoma, fusiform cell sarcoma, and colon and rectal adenocarcinomas (all in the same patient) |
Multiple dysplastic nevi, parotid gland adenoma, a mammary lipoma, colon adenomas, enchondromas, pilomatrixoma |
This work |
*Oldest age known (or death) in years.
†Aneuploidy levels are difficult to compare due to the use of different technologies, protocols, and cell types analyzed.
Physical examination in October 2017 revealed a weight of 44 kg (third centile), a height of 146 cm (P < 3), and an occipitofrontal circumference of 50 (P < 3). She had bitemporal narrowing, deeply set eyes, a midface hypoplasia, micrognathia, low-set and posteriorly rotated ears, nystagmus, bilateral pes cavus, and bilateral hammer toes (fig. S1, A to C). She showed very notable cutaneous features with multiple widespread hyperpigmented lesions of variable tone. Hyperpigmented lesions in the anterior part of the trunk had a quadrant/blaschkoid wide band distribution, affecting both sides of the body, and millimeter clear pigmented macular lesions on the back of the hands and feet, right axillae, and right iliac fossa (fig. S1, C to E). Alternating with pigmented areas, she had patchy areas of hypopigmented and healthy skin. The hypopigmentation was blaschkoid in narrow band in both upper extremities and the left pretibial region and with more erratic/phyllodes distribution in the rest of the body. She showed hyperkeratotic plaques with furfuraceous desquamation on her knees and compact hyperkeratosis in the soles of the feet in support areas (fig. S1, C to E).
We performed biopsies of the hyper, hypopigmented, and healthy skin and the hyperkeratotic areas of the knee. Optical microscopy with hematoxylin-eosin staining showed basal pigmentation in the hyperpigmented and healthy skin, more notable in the hyperpigmented skin. In the samples of healthy and hypopigmented skin, scattered melanocytes were observed. Immunohistochemistry with Melan-A technique evidenced an increase in basal melanocytes in hyperpigmented and normal skin, with virtually no pigment in the basal hypopigmented skin. The biopsy of the hyperkeratotic zone showed a subacute spongiosis dermatitis, with irregular descending hyperplasia of the epidermis with foci of spongiosis with vesiculation and slight lymphocyte exocytosis and superficial perivascular lymphocytic infiltrate in the dermis. The radiographic study of the upper and lower extremities revealed the existence of lesions suggestive of enchondromas in the left femur and in the right arm (fig. S1, F and G). There were no evidences of mental retardation, hypotonia, seizures, progeroid traits, or immunodeficiency.
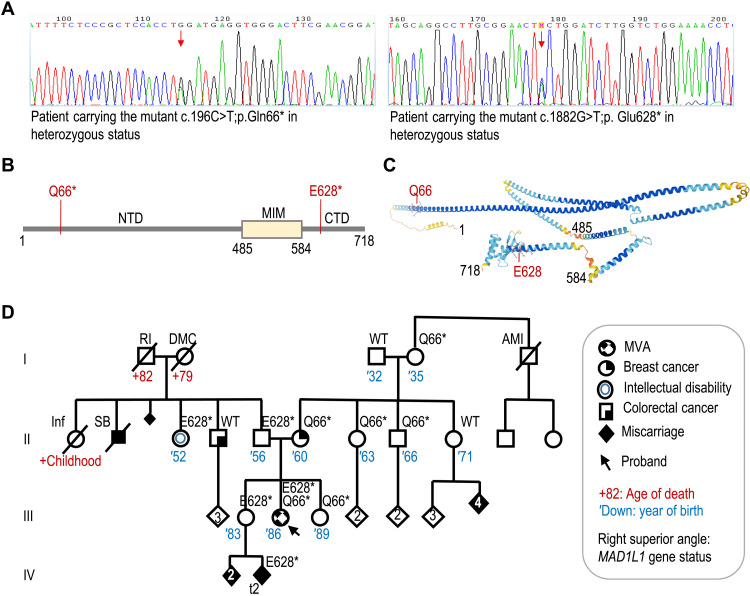
We initially ruled out mutations in MLH1, MSH2, MSH6, PMS2, NF1, and TP53 by focused DNA sequencing to discard alterations in genes related to constitutional mismatch repair deficiency, neurofibromatosis, and Li-Fraumeni syndromes. Furthermore, the colorectal and endometrial tumors of the proband were microsatellite stable, and the tumor immunohistochemistry analysis results in conserved expression of the four mismatch repair proteins. Whole-exome sequencing (WES) of peripheral blood samples from the proband identified two different stop-gained mutations in MAD1L1: c.196C>T; p.Q66* in exon 4 and c.1882G>T; p.E628* in exon 18 (Fig. 1A). The first one, affecting most of the protein, has been described in a European individual [Exome Aggregation Consortium (ExAC); minor allele frequency (MAF) of 8.304 × 10−6]. The second one is not present in ExAC, Genome Aggregation Database (gnomAD), Catalogue of Somatic Mutations in Cancer (COSMIC), or The Cancer Genome Database. This mutation prevents the expression of the globular head of the C-terminal domain (CTD; Fig. 1, B and C), a region critical for binding to MAD2 or CDC20 and for SAC activity (18, 19). By Sanger analysis, we determined that the proband’s mother and other maternal relatives carry the first mutation, while the second one had a paternal origin (Fig. 1D and fig. S1H). The older sister of the proband (III-4) carried the paternal mutation, and the younger sister (III-6) carried the maternal one. At the age of 45 years, the proband’s mother was diagnosed with a grade III invasive ductal carcinoma on the left breast, estrogen and progesterone receptors positive, and HER2 negative. No alterations were found in BRCA1, BRCA2, PALB2, CHEK2, ATM, BRIP1, RAD51C, and RAD51D genes in the peripheral blood of the mother. The older sister of the proband, carrier of the paternal MAD1L1 mutant allele, was diagnosed with type 1 diabetes at the age of 17 years. She had three early miscarriages on May and September of 2017 and on January 2018. A maternal sister (II-9), carrier of MAD1L1 wild-type alleles, had four consecutive miscarriages of unknown cause. Paternal grandmother had seven pregnancies, two of them finished in spontaneous abortion, and one was a stillborn male without detectable congenital abnormalities. Paternal aunt (II-3) has an intellectual disability due to an infection during early childhood. She is carrier of the MAD1L1 p.E628* allele. Another brother of the father (II-4), also carrying MAD1L1 wild-type alleles, had colorectal cancer at the age of 55 years (Fig. 1D).
Fig. 1. Biallelic loss-of-function mutations in MAD1L1.
(A) Chromograms showing the two heterozygous mutations in the MAD1L1 gene in the proband. (B) Schematic representation of the MAD1 protein and the mutations found in the proband. NTD, N-terminal domain; MIM, MAD2 interaction motif. (C) Predicted structure of MAD1 (AlphaFold) showing critical residues limiting the different domains and the position of the Q66 and E628 residues mutated. (D) Pedigree of the family with the status of MAD1L1 gene shown on the right superior angle of each individual. AMI, acute myocardial infarction; DMC, diabetes mellitus complications; Inf, infection; RI, renal insufficiency; SB, stillbirth; t2, trisomy chromosome 2.
MAD1L1 mutations induce germline and tumor aneuploidy
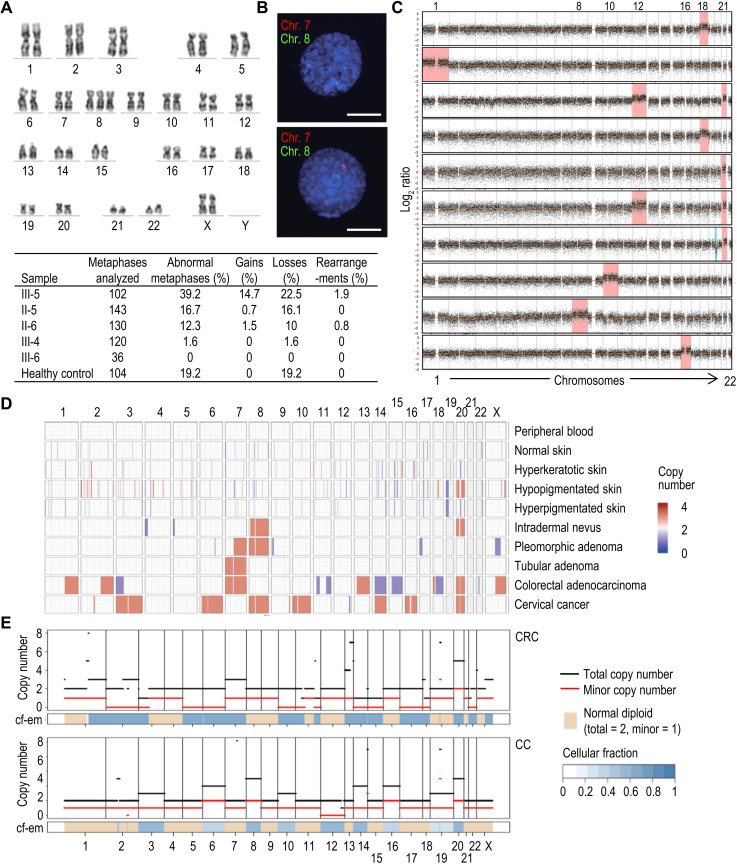
Metaphase spreads and G-banding in peripheral blood mononuclear cells (PBMCs) identified 39% of aneuploid metaphases in the proband (Fig. 2A). The number of chromosomal gains found in the parents was low (0.7% in the father and 1.5% in the mother), and these alterations were not found in the sisters or healthy controls. Analysis of specific aberrations in chromosomes 7, 8, 20, and X using fluorescence in situ hybridization (FISH) confirmed that copy number gains in these chromosomes were more frequent than losses (Fig. 2B and fig. S2A). Further studies using shallow whole-genome sequencing in single cells identified whole chromosome aneuploidies in 10 of 23 (43.5%) proband cells but not in controls (Fig. 2C and fig. S2B). Trisomy of chromosome 21 was the most frequent alteration in addition to recurrent gains of chromosomes 12 and 18. Monosomies were not observed using shallow whole-genome sequencing, and del(20p) was observed in one of the cells (Fig. 2C).
Fig. 2. Analysis of aneuploidy.
(A) Representative example of the analysis of metaphase spreads from peripheral blood cells. The image shows an example of trisomy of chromosome 8 (Chr. 8) in the proband. The table at the bottom of the panel shows the summary of cytogenetic analysis of G-banding karyotype of family members proband (III-5), father (II-5), mother (II-6), older sister (III-4), younger sister (III-6), and a 46,XX healthy control. (B) Examples of two-color FISH with centromeric chromosomes 7 (red) and 8 (green) DNA probes. Representative chromosome 8 trisomic nucleus with two signals for chromosome 7 and three signals for chromosome 8 (top nucleus) and representative chromosome 7 trisomic nucleus with three signals for chromosome 7 and two signals for chromosome 8 (bottom nucleus). Scale bars, 5 μm. See fig. S2A for quantitative data. (C) Detection of aneuploidies using single-cell DNA sequencing. Ten aneuploid cells of the 23 cells analyzed in the proband are shown (see also fig. S2B). Red and blue indicates significant increase or decrease in copy number, respectively. (D) CNV plot calculated from the array comparative genomic hybridization data of 10 III-5 samples including the peripheral blood, skin, and different types of cancer (blue, copy number loss; red, copy number gain). Note that only clonal aneuploidies, and not variegated aneuploidies in which each chromosome gain or loss affects a small percentage of cells, can be detected using this technique. (E) FACETS view representing copy number inferred from WES data in different neoplasias of the proband. CRC, rectal adenocarcinoma; CC, cervical carcinoma.
We also performed an analysis of apparently normal and pathological tissues of the proband by different platforms. Comparative genome hybridization (CGH) analysis of normal and pathological tissues identified chromosomal gains in most pathological conditions, whereas losses were only observed in colorectal carcinoma (CRC). The progressive acquisition of copy number variants (CNV) is evident from tubular adenoma, with gain of chromosome 7 as unique alteration, to CRC with many additional alterations (Fig. 2D).
We further analyzed genomic changes in neoplasias developed by the proband by WES. FACETS (fraction and allele-specific copy number estimates from tumor sequencing) (20) analysis suggested that, in agreement with the previous observations, gains of one or several complete chromosomes were relatively frequent, while losses of complete chromosomes were observed only in CRC (Fig. 2E and fig. S3A). Total copy number changes were higher in more severe neoplasias [CRC and cervical carcinoma (CC)], being remarkable the high proportion of uniparental alterations (partial or complete disomies, trisomies, or tetrasomies) in the CRC. However, these uniparental alterations were infrequent in CC, only involving chromosome 12. The somatic variants found in the proband’s neoplastic tissues corresponded to canonical tumorigenesis pathways in these pathologies, including mutations in APC (p.R1255X), TP53 (p.V25F), RET (p.S922Y), and KRAS (p.G12A) in interval colorectal cancer (ICRC) and CRC (fig. S3B). None of these mutations was identified in tubular adenoma (TA), neither in other tissues sequenced. CTNNB1 mutations and a frameshift mutation in POLE were identified in pleomorphic adenoma (PA). Missense mutations in RECQL4 and FAT4 were the most common alterations in CC (fig. S3B).
CGH analysis of the third abortion in the older sister (III-4) identified a trisomy in chromosome 2 (fig. S3C). CGH analysis of the ductal breast cancer of the proband’s mother showed mainly partial chromosome gains and losses, with reduced whole chromosome copy number alterations, as usually observed in sporadic breast cancer (fig. S3, D and E). WES analysis of this tumor identified the heterozygous MAD1L1 p.(Q66*) mutation in addition to new mutations in TP53 and PIK3CA (fig. S3B).
Mutations in MAD1L1 result in defective SAC function
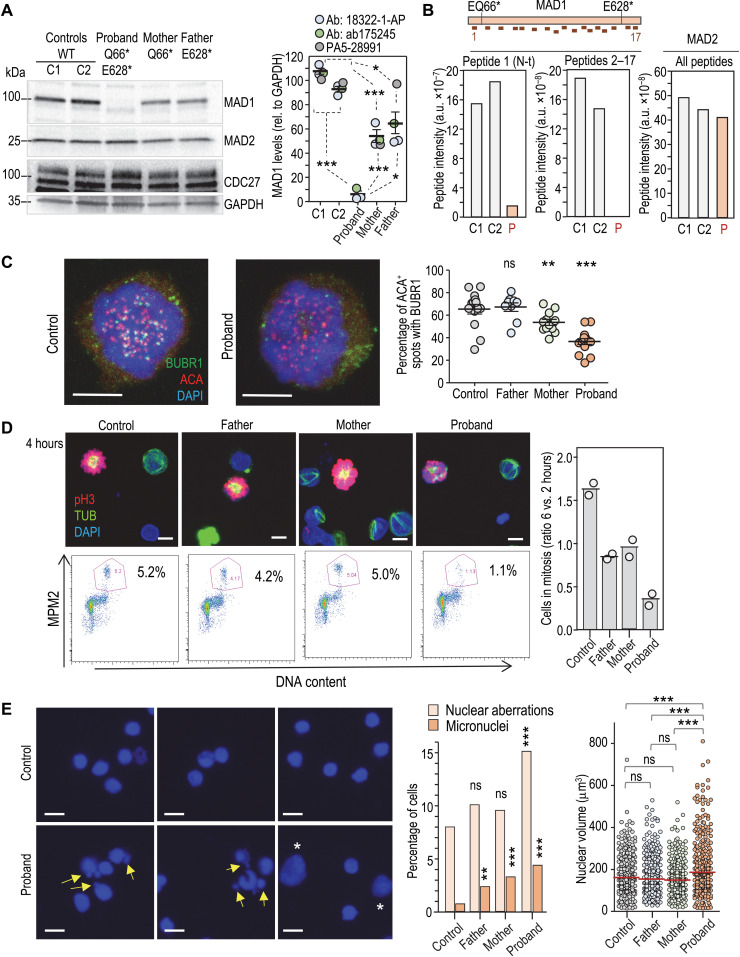
To understand the functional effect of MAD1L1 mutations in chromosome segregation, we established cultures of lymphocytes from the peripheral blood of the proband, parents, and additional wild-type controls. MAD1L1 mutations resulted in lack of full-length MAD1 protein in peripheral lymphocytes from the proband and reduced levels in lymphocytes from the heterozygous parents (Fig. 3A). Reduced levels of putative shorter peptides were observed as a consequence of the MAD1L1 mutations (fig. S4A). Mass spectrometry analysis of total protein lysates from these cells found abundant signal for MAD1 and MAD2 peptides in control cells. However, only a highly reduced signal was found for MAD1 peptide 1 (N-terminal domain upstream of the Q66* mutation), and all the other peptides (2 to 17) were not detected in cells from the proband (Fig. 3B and fig. S4B). MAD2 peptides were readily detected in the proband, suggesting the specific absence of peptides representing most of the MAD1 protein in proband lymphocytes or their presence at levels below the threshold of detection by mass spectrometry. Unexpectedly, mutant lymphocytes proliferated normally after mitogenic stimuli (fig. S4C), and no obvious defects were observed in mitotic figures from early-passage cultures. However, proband and, to a lesser extent, mother cells displayed reduced load of BUBR1, the protein encoded by the BUB1B gene mutated in MVA1, in kinetochores lacking attached microtubules in the presence of taxol (Fig. 3C). In addition, the taxol-induced mitotic arrest was deficient in lymphocytes from the proband (Fig. 3D), and accordingly, a larger amount of aberrant nuclei with abnormal shape and lobuli, as well as interphasic cells with micronuclei, and increased nuclear volume were observed, suggesting premature exit and/or abnormal recovery from taxol in the presence of a misfunctional SAC (Fig. 3E).
Fig. 3. Defective chromosome segregation in MAD1L1 mutant lymphocytes.
(A) Representative immunoblot using anti-MAD1 18322-1-AP in peripheral blood cells. The quantification using three different antibodies is shown to the right. See also fig. S4A. Ab, antibody. (B) Mass spectrometry analysis in blood cells, with separate plots for the total intensity of MAD1 peptide 1 (N terminus; see schematic representation above indicating the relative positions of the Q66* and E628* mutations), MAD1 peptides 2 to 17, and MAD2 peptides. a.u., arbitrary units. (C) Immunofluorescence of BUBR1 and ACAs in lymphocytes treated with taxol. Scale bars, 5 μm. Signals were quantified from mitotic cells from (n = 14), proband (n = 15), mother (n = 12), and father (n = 9) cells. (D) Representative micrographs (top) and fluorescence-activated cell sorting analysis (bottom) of lymphocytes in the presence of taxol. Top panels show immunofluorescence for phosphorylated histone H3 (pH3; red), α-tubulin (green), and DAPI (blue). A minimum of 5000 cells per condition was scored for MPM2 staining (two independent experiments with >300 cells per condition). Scale bars, 5 μm. The percentage of mitotic cells after 6 hours in taxol normalized versus the number of cells that entered mitosis at 2 hours is plotted to the right (n = 2 independent experiments). (E) Representative example (two independent experiments) of interphase cells after treatment with taxol showing aberrant cells (abnormal shape and fragmented lobuli) and micronuclei (arrows), as well as cells large nuclei (asterisks). Scale bars, 10 μm. Plots show the percentage of nuclear aberrations and micronuclei (>300 cells per condition), as well as nuclear volume (>500 cells per condition). In (A), (C), and (E), ns, not significant; **P < 0.01; ***P < 0.001; Student’s t test with Welch’s correction (A), one-way analysis of variance with Bonferroni correction (C and E, nuclear volume), or chi-square, two-coiled Fisher exact test (E, nuclear aberrations and micronuclei).
A systemic inflammation response to aneuploidy
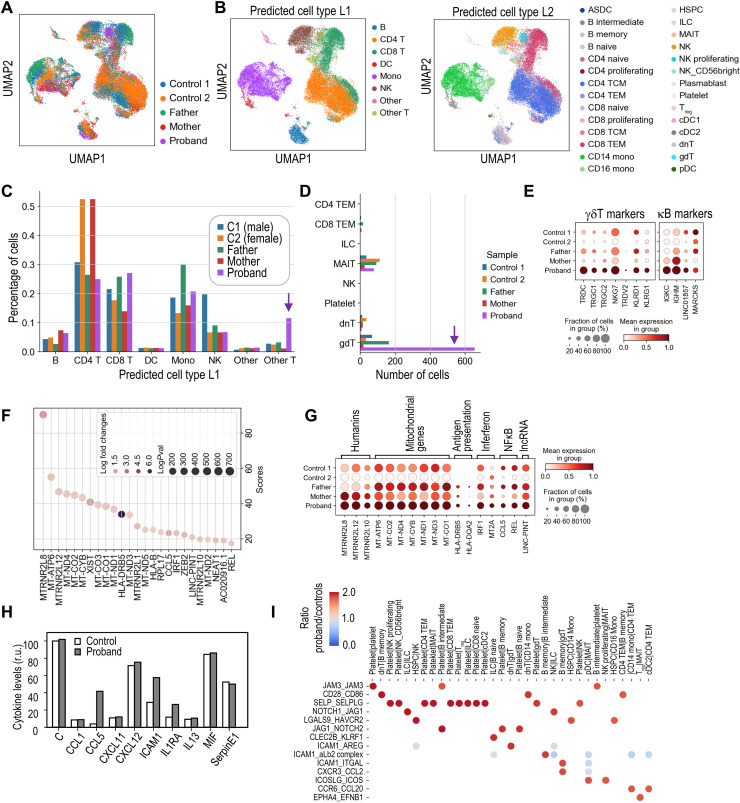
To understand the consequences of aneuploidy in vivo, we sequenced a total of 33,604 individual PBMCs from the proband, both heterozygous parents and two healthy donors (Fig. 4A and fig. S5A). We assigned each cell using available algorithms for PBMC classification (21) to 8 (L1), 28 (L2), or 48 (L3) different cell types (Fig. 4B and fig. S5, B and C). The relative abundance of the different cell types in all samples suggested the expansion of T cells expressing the δ and γ chains of the T cell receptor (TRD and TRG genes; γδ T cells) and, to a lesser extent, B cells expressing the κ light chain (IGKC) in the proband (Fig. 4, C to E, and fig. S5D).
Fig. 4. Transcriptomic analysis of peripheral blood cells with biallelic MAD1L1 mutations.
(A) Uniform manifold approximation and projection (UMAP) of cells from the indicated samples. (B) UMAP distribution of cell types (L1 and L2 classifications following the Azimuth algorithms). DC, dendritic cell; Treg, regulatory T cell. ASDC, AXL(+) dendritic cells; TCM, Central memory T cells; TEM, T effector memory; HSPC, Hematopoietic stem and progenitor cells; ILC, Innate lymphoid cells; MAIT, Mucosal-associated invariant T cells. (C) Percentage of L1 cell types in the indicated samples. (D) Number of cells in the specific cell types included in the L1 “other T” classification showing an overrepresentation of γδ T cells in the proband. (E) Expression of specific markers of γδ T cells and B cells expressing the κ chain. (F) List and scores (Wilcoxon rank sum test) of the top up-regulated genes in proband versus control PBMCs. (G) Aggregated expression of the set of top genes up-regulated in proband cells in the different individuals analyzed. lncRNA, long noncoding RNA. (H) Relative levels of a panel of cytokines in the plasma of controls and the proband. r.u., relative units. (I) Top ligand-receptor interaction pairs overrepresented in the proband compared to the controls. The scale represents the ratio between the significant means for each ligand-receptor interaction pair (y axis) and pair of interacting cell types (x axis) in the proband versus controls.
Analysis of the 25 top genes up-regulated in proband PBMCs versus control cells identified multiple mitochondrial genes (MT-ATP6, MT-ND4, MT-CO2, MT-CYB, etc.), as well as several members of the humanin family, being the humanin-like MTRNR2L8, the top-scoring up-regulated gene in proband cells (Fig. 4, F and G). Humanins have been shown to be released to the extracellular milieu upon mitochondrial stress and to act as mitokynes (mitochondrial cytokines) with cytoprotective effects (22). Among the most up-regulated genes, we could also detect the chemokine CCL5 and members of the interferon (IRF1 and MT2A) and nuclear factor κB (NFκB) (REL) signaling pathways. In accordance, CCL5 up-regulation was also detected in the plasma of the proband together with intercellular adhesion molecule–1 (Fig. 4H). A specific analysis of ligand-receptor interactions (23) suggested increased signals involved in platelet activation as well as molecules involved in antigen presentation and interferon signaling, natural killer (NK) and γδ T cell activation, recruitment of leukocytes to circulation sites of inflammation, and other general inflammatory signals (Fig. 4I). The strongest differences between proband and control ligand-receptor interactions were found in molecules expressed in platelets, such as the junctional adhesion molecule 3 or the selectin P. Other ligand-receptor interaction pairs overrepresented in proband cells included molecules involved in antigen presentation and interferon signaling, NK and γδ T cell activation, recruitment of leukocytes to circulation sites of inflammation, and other general inflammatory signals (Fig. 4I).
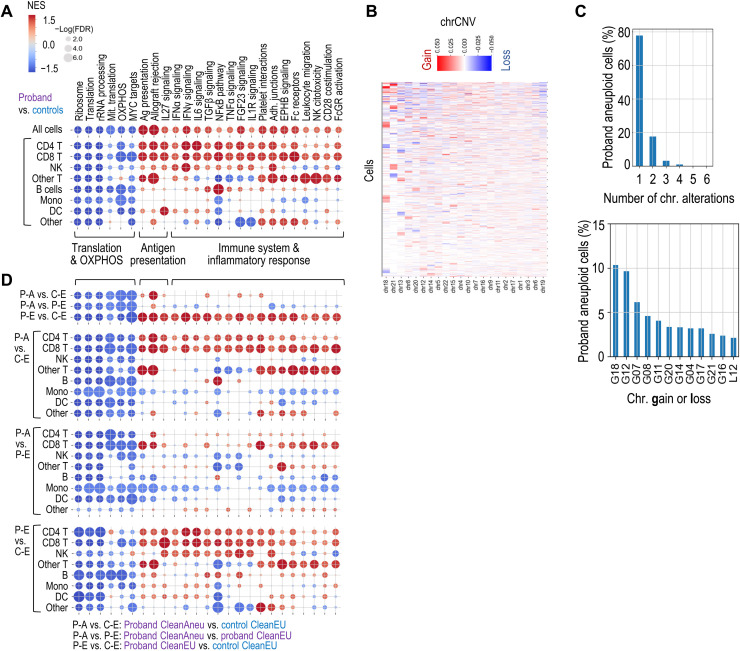
Pathway enrichment analysis suggested significant down-regulation of pathways related to protein synthesis (ribosome biogenesis, translation, and RNA processing), along down-regulation of oxidative phosphorylation (OXPHOS) and MYC targets (Fig. 5A and fig. S6, A and B). On the other hand, proband cells displayed enhanced expression of genes related to inflammatory response, including interferon and NFκB signaling pathways. Whereas down-regulation of ribosome and OXPHOS was shared by most cell types, inflammatory pathways were most evident in T cells (Fig. 5A and fig. S6, A to C). Pathways related with antigen presentation were also significantly up-regulated in proband’s cells, with major histocompatibility complex class II molecules between the most up-regulated genes in the proband, especially in professional antigen-presenting cells (dendritic cells, monocytes, and B cells) and in the expanded population of γδ T cells (Fig. 5A and fig. S6, C and D).
Fig. 5. Pathway analysis of aneuploid and euploid MAD1L1 mutant cells.
(A) Gene set enrichment analysis (GSEA) used preranked differential expression of genes in the indicated comparisons. The list of pathways represents a selection of differentially regulated pathways in the different conditions. (B) Plot showing the average CNV score per chromosome (chrCNV score). Chromosomes are ordered by the number of extreme values in the chrCNV score, i.e., from chromosomes with higher monosomy or trisomy scores (left) to chromosomes with higher representation or disomies (right). (C) Distribution of cells with numerical alterations in one or more chromosomes in the proband (top) and percentage of cells with the indicated aneuploidies in the proband (bottom). G, gain; L, loss of the indicated chromosomes. (D) Deregulation of the selected pathways in proband aneuploid cells (P-A) versus control euploid (C-E) cells, proband aneuploid cells versus proband euploid cells (P-A versus P-E), and proband euploid cells versus control euploid cells (P-E versus C-E). Scale as in (A). FDR, false discovery rate; rRNA, ribosomal RNA; IFNα, interferon-α; TGFβ, transforming growth factor–β; TNFα, tumor necrosis factor–α. NES, normalized enrichment score.
To estimate chromosomal gains and losses, we made use of the inferCNV algorithm (24), which infers DNA copy number changes from the gene expression data, using matched control cell types for normalization (see Materials and Methods for details). To classify every single cell as aneuploid (i.e., harboring at least one chromosomal gain or loss) or euploid (all disomic chromosomes), we first calculated the average CNV score per chromosome for each cell (chrCNV) and established a threshold for classifying each chromosome as monosomic, disomic, or trisomic based on the kernel distribution of the CNV score in the proband and controls (fig. S7, A and B). The estimation of aneuploidies from using single-cell RNA sequencing (scRNA-seq) data suggested a wide spectrum of chromosomal changes in different cell types (Fig. 5B and fig. S7A). The lowest ratios of aneuploidy were found in progenitor cells, whereas 67.9% of intermediate (or transitional) B cells were aneuploid in the proband (fig. S7C). Most aneuploid cells contained a single chromosomal gain or loss (77.8%), and gains of chromosomes 18 and 12 were the most abundant alterations (10.3 and 9.7% of all aberrations; Fig. 5C). Cells with massive gains and losses were not abundant (less than 5% of aneuploid cells carried more than two events), suggesting that these events were infrequent or abnormal cells were eliminated rapidly.
We next compared the transcriptomic profile of aneuploid cells by using a more stringent threshold to define “clean euploid” (CleanEU) and “clean aneuploid” (CleanAneu) cells (fig. S7, B and C), thus avoiding the noise in the inferCNV approach. Only cells with at least one mono- or trisomy with the stringent thresholds were considered as CleanAneu (13.6% in the proband versus 4.0% in controls), and only cells with all disomic chromosomes using these stringent conditions were classified as CleanEU. Analysis of the 25-gene signature overexpressed in the proband suggested that these genes were significantly overexpressed not only in aneuploid cells but also, to a lesser extent, in CleanEU cells from the proband (fig. S7D). Pathway analysis detected the down-regulation of translation and OXPHOS and up-regulation of antigen presentation–related pathways in CleanAneu cells from the proband when compared to CleanEU cells from either the proband or controls. However, aneuploid and euploid cells from the proband did not show differences in immune pathways, whereas this inflammatory response was evident when comparing CleanEU cells from the proband with CleanEU cells from the controls (Fig. 5D). The p53 pathways were not significantly deregulated in these comparisons [false discovery rate of >0.25 for all the L1 cell types when using HALLMARK_p53_PATHWAY from gene set enrichment analysis (GSEA)]. Together, all these data suggest that aneuploidy induces a cell-autonomous response characterized by increased antigen presentation and down-regulation of translation and OXPHOS, as well as a systemic non–cell autonomous inflammatory response.
Clonal expansion of specific B and T cell subsets
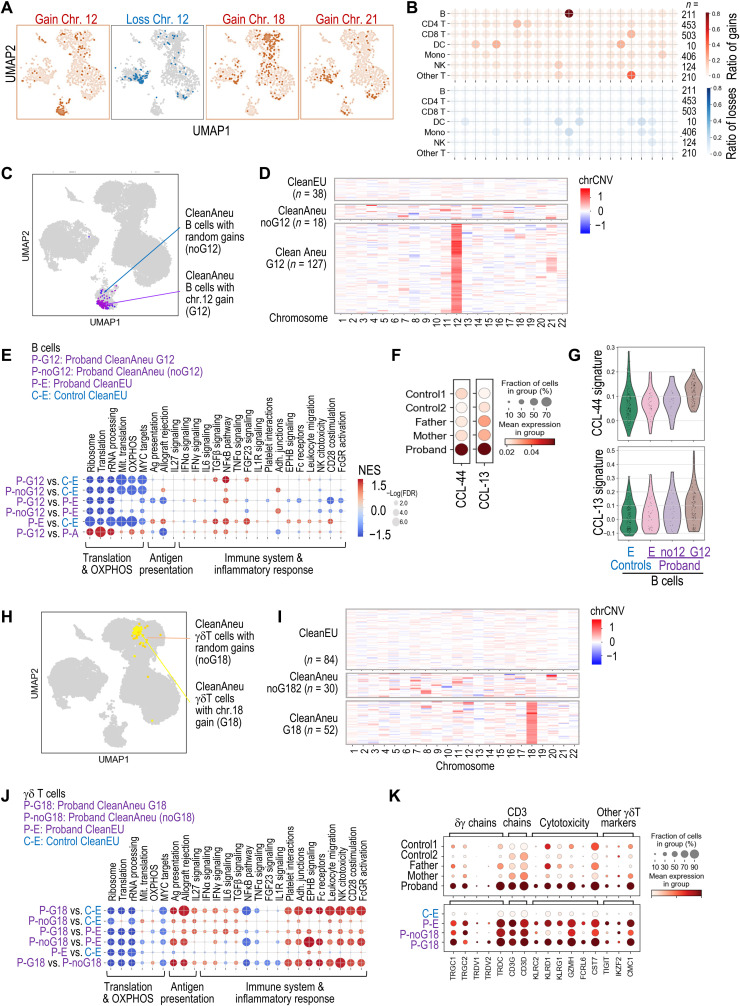
Whereas random aneuploidies affected most cell types, we observed a preference for chromosome 18 gain in γδ T cells and chromosome 12 gain in intermediate B cells expressing the κ light chain subunit and, to a lesser extent, the λ subunit, frequently accompanied of concomitant gain of chromosome 21 (Fig. 6, A and B, and fig. S8, A and B). Both intermediate B cells and γδ T cells were expanded in the proband (Fig. 6B and fig. S5D), suggesting clonal or subclonal expansion of aneuploid cells.
Fig. 6. Transcriptomic analysis of random versus clonally selected aneuploidies.
(A) Distribution of aneuploid cells with gains (red) or losses (blue) of the indicated chromosomes. (B) Ratio of cells with gain (red) or loss (blue) of each chromosome among proband aneuploid cells, showing the selection of chromosome 12 or 18 gains in B cells or other T cells, respectively. The number of cells with gain or loss was divided by the total number of aneuploid cells for each cell type. (C) UMAP representation of aneuploid B cells with gain of chromosome 12 (G12) or random aneuploidies. (D) Heatmap of the chrCNV score in B cells classified as CleanEU, random aneuploidies with no G12 (noG12), or aneuploidies with G12. chrCNV values are scaled from −1 (loss of one chromosome) to +1 (gain of a copy). (E) GSEA of the pathways selected in Fig. 5 in comparisons between control euploid B cells (C-E), proband euploid B cells (P-E), and proband aneuploid B cells with G12 (P-G12) or random aneuploidies (P-noG12). (F) Violin and aggregated dot plots showing the expression of a 44- or 13-gene signature typical of chronic lymphocytic leukemia (CLL) in the indicated samples. (G) Violin plots of the 44- and 13-gene CLL signatures. (H) UMAP representation of aneuploid γδ T cells with G18 or random aneuploidies (noG18). (I) ChrCNV heatmap in γδ T cells classified as CleanEU, random aneuploidies with noG18, or aneuploidies with G18. (J) GSEA in the indicated comparisons between control euploid γδ T cells (C-E), proband euploid γδ T cells (P-E), and proband aneuploid γδ T cells with G12 (P-G12) or random aneuploidies (P-noG12). (K) Aggregated expression of γδ T cell markers in the indicated samples.
Because aneuploidy is frequently detrimental for cell proliferation (1), we next asked whether the clonal selection of chromosome 12 gains (G12) in B cells could provide specific benefits. Transcriptional analysis of proband (P) aneuploid cells with G12 (Fig. 6, C and D) versus control euploid cells (C-E) showed inhibition of translation and OXPHOS, similar to other aneuploid cells, but no evidences for immune alteration in these cells (P-G12 versus C-E; Fig. 6E). Actually, these P-G12 aneuploid cells displayed reduced inflammatory-related pathways when compared to euploid cells from the proband (P-G12 versus P-E). A direct comparison between proband aneuploid B cells with G12 versus B cells with random aneuploidies confirmed that G12 cells displayed reduced inflammatory-related pathways (P-G12 versus P-noG12; Fig. 6E and fig. S8, C and D), suggesting that these clones were protected from the negative effects of aneuploidy. In addition, these B cells displayed increased expression of two gene signatures that define chronic lymphocytic leukemia (CLL) (Fig. 6F) (25). This effect was specifically due to aneuploid cells with G12 and was not observed in aneuploid B cells with random aneuploidies (Fig. 6G).
Notably, a similar analysis of δγ T cell clones with G18 (Fig. 6, H and I) indicated that the inflammatory-like response was enhanced in aneuploid γδ T cells with G18 (Fig. 6J and fig. S8, E and F). These G18 clones displayed increased expression of cytotoxicity markers such as cell lectin-like receptors (KLRC, KLRD, and KLRG transcripts) as well as granzyme H (GZMH), Fc receptor–like 6 (FCRL6), or cystatin F (CST7) (Fig. 6K). Only 30% of the expanded γδ T cell population was aneuploid (Fig. 6I and fig. S8B), suggesting that the G18 clone arose secondary to an expansion of highly cytotoxic γδ T cells within the immune response against the aneuploidy-associated stress.
DISCUSSION
Germline aneuploidy syndromes group heterogeneous entities with mutations in genes involved in cell division. Previous studies have identified mutations in four regulators of the SAC, namely, BUB1B [MVA1; Online Mendelian Inheritance in Man (OMIM) #257300] (7), CEP57 (MVA2; OMIM #614114) (26), TRIP13 (MVA3; #617598) (27), and BUB1 (28), in addition to CENATAC, encoding a spliceosome factor involved in the expression of mitotic regulators (29). Common phenotypic alterations in these patients include developmental delay, facial dimorphism, and intellectual disability, with a more diverse phenotype in other characteristics such as microcephaly or tumor susceptibility (Table 1).
The biallelic loss-of-function mutations in MAD1L1, described here, result in lack of full-length protein, defective SAC, and a wide spectrum of phenotypic alterations. Whether truncated MAD1 peptides are expressed in this individual is unclear, although whole lysate mass spectrometry analysis only identified reduced amounts of a peptide belonging to the N terminus of the protein, upstream of the Q66 residue, in proband cells. These peptides are unlikely to be functional, as the two mutations in the proband prevent expression of the C-terminal globular domain of MAD1, known to be critical for its function in the SAC (18, 19, 30, 31). Although the proband developed some abnormalities typical of other syndromes (e.g., café-au-lait spots in cohesinopathies), the presence of high levels of mosaic aneuploidy suggests her inclusion within the MVA syndromes. Differential characteristics of this individual versus other patients with MVA are long survival and no intellectual disability among others. The proband shows a notable high cancer susceptibility to a wide spectrum of 12 different neoplasias since early age, including five malignancies by the age of 36 years. Less than 50% of patients with MVA develop tumors, and the number of neoplasias per patient is typically reduced to one, with only two MVA1 individuals reported to develop both rhabdomyosarcoma and Wilms tumors (Table 1) (32, 33). Individuals carrying heterozygous MAD1L1 mutations did not display significant levels of aneuploidy, despite the reduction in protein levels (Fig. 3A). However, the finding of multiple abortions and cancer in some family members suggests the need of clinical monitoring of these individuals.
Whereas germline mutations in SAC components are typically linked to specific cognitive and medical features (9), the cellular and systemic consequences of chromosomal instability have not been explored in patients. In agreement with previous studies in cellular or mouse models (34–38), our single-cell transcriptomic analysis of proband cells suggested defects in translation and ribosome biogenesis, along activation of interferon and NFκB signaling. In Drosophila, the reduction in ribosomal proteins associated to aneuploidy has been proposed to trigger the clearance of aneuploid cells from tissues through cell competition (39), although whether these mechanisms exist in humans remains unclear. Single-cell analysis allows the comparison of aneuploid and euploid cells after inferring chromosomal gains and losses from gene expression data. Our data suggest that the inflammatory response is present in euploid cells from the individual with aneuploidy and leads to increased levels of specific cytokines and inflammatory molecules in the blood plasma. Whereas we cannot rule out that inflammation is secondary to other pathologies, the last neoplasias in the proband were surgically removed in 2014 (without radio- or chemotherapy), and she has been free of other relevant pathologies until the recollection of the blood samples in 2021.
Nuclear humanins are among the top transcripts up-regulated in the MAD1L1 mutant individual. Humanins are short mitochondrial and nuclear-encoded peptides released to the extracellular environment upon mitochondrial stress (22, 40). The up-regulation of humanins and mitochondrial genes, likely as a consequence of OXPHOS defects, might suggest the existence of mitochondrial stress or dysfunction in the proband, which could be a secondary effect of the chronic inflammation (41, 42). Humanins are also overexpressed in fibroblasts from patients with Down (trisomy 21) syndrome (43, 44), in which mitochondrial dysfunction is a prominent feature (45). The possible value of humanins as a biomarker in genome imbalance conditions and its role during the response to aneuploidy remain to be explored. In addition, multiple mitochondrial components have been shown to act as damage-associated molecular patterns (46), suggesting that humanins and other mitochondrial proteins up-regulated in the proband could contribute to the systemic inflammatory response observed.
In addition to random aneuploidies, scRNA-seq analysis identified small cell populations with clonal or subclonal gains. B cells with G12 displayed increased expression of genes that characterize malignant CLL cells (25), suggesting possible pathogenic properties. Accordingly, trisomy 12 is seen in approximately 16% of cases of CLL and is associated with poor prognosis (47, 48). The majority of patients with CLL have been shown to carry prediagnostic B cell clones 6 months to 6 years before the development of clinically recognized leukemia, suggesting the relevance of close selection in this disease (49). Whether these B cells with G12 represent a premalignant state is uncertain, but the detection of these clones may call for frequent monitoring for expected pathologies at early stages.
On the other hand, the expanded γδ T cells with G18 display properties of activated cytotoxic cells with increased expression of granzymes, or killer lectin-like receptors such as KLRG1, and with T cell markers commonly up-regulated in patients with chronic inflammation (42, 50). The expression of some of these molecules is shared with NK cells, which, using in vitro coculture assays, were previously proposed to mediate the immune response against aneuploidy (51). γδ T cells play a major role in both innate and adaptive immune responses not only against pathogens but also against own tumor and stressed cells (52, 53). However, their specific links with aneuploidy have not been established yet.
In summary, our study reports biallelic MAD1L1 mutations in a novel variant of mosaic aneuploidy accompanied by unprecedented high susceptibility to a wide spectrum of tumors, some of them were never reported in patients with MVA (Table 1). In addition to its mitotic role, MAD1 has been reported to participate in additional cellular structures or functions such as nuclear pore complex (54), nucleoplasm organization and chromatin regulation (55), integrin secretion and cell migration (56), and p53 destabilization (57). Whereas we did not find evidences for deregulation of the p53 pathway in proband or aneuploid cells, we cannot rule out that some of these functions may contribute to the phenotype of MAD1L1 mutant individuals. However, our data suggest that the up-regulation of pathways involved in systemic inflammation (also observed in cellular models) (35, 36) and clonal expansion of specific cell populations may be present in other MVA individuals and may require clinical follow-up. Exploiting the enhanced immune response associated to aneuploidy could provide new opportunities for the clinical management of these patients.
MATERIALS AND METHODS
Study design
We obtained peripheral blood samples from the proband, parents, and other family members or controls; from biopsies of the hyper, hypopigmented, and healthy skin of the proband; and from hyperkeratotic areas of the knee. Paraffin-embedded tissues were obtained from intradermal and compound nevi, pilomatrixoma, pleomorphic adenoma and tubular adenoma of the proband, and also from colorectal and endometrial carcinomas of the proband. We also obtained fresh sample of the third miscarriage of the older sister of the proband and paraffin-embedded tissue from the maternal breast carcinoma. Informed consent was obtained from all family members, and the study was approved by The Committee for Ethical Research of the Hospital Universitario de Fuenlabrada (Madrid, Spain; reference number 18/26).
Microsatellite instability and DNA repair defects
For the evaluation of microsatellite instability, five quasi-monomorphic markers (NR-21, BAT-26, NR-22, NR-24, and BAT-25) were amplified in the proband’s tumor DNA and in a DNA sample from peripheral blood leukocytes by two Multiplex polymerase chain reaction (PCR). Tumor DNA was extracted from a 30-μm-thick formalin-fixed, paraffin-embedded tumor sample using the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN GmbH, Hilden, Germany). For DNA sequencing, selected genes were analyzed using the OncoPlus-GeneSGKit capture kit (Sistemas Genómicos S.L.) and the MiSeq platform (Illumina). DNA from peripheral blood leukocytes was extracted using the Maxwell RSC automatic extractor (Promega, Madison, Wisconsin, WI, USA), following the manufacturer’s instructions. Each Multiplex PCR was performed using the QIAGEN Multiplex PCR Kit. Forward primers were end-labeled with 6-carboxyfluorescein and hexachlorofluorescein fluorescent marker according to the amplification size. Primer sequences and PCR conditions can be provided upon request. The PCR products were separated by size on an ABI 3730XL DNA analyzer (Applied Biosystems, Waltham, MA, USA). The results obtained were analyzed with Peak Scanner v1.0 software.
Detection of DNA mismatch repair deficiencies by immunohistochemistry was performed using a Dako autostainer or a Leica bond autostainer. The antibody clones used were ES05 (1:50) for MLH1, FE11 (1:50) for MSH2, clone EP49 (1:50) for MSH6, and clone A16-4 (1:200) for PMS2.
Exome sequencing
For WES, we used SureSelect Target Enrichment System v6 (Agilent Technologies), following the manufacturer’s protocols. Briefly, 3 μg of high-quality genomic DNA was fragmented using Covaris technology, and DNA fragments were end-repaired and ligated to adapters before hybridization. Sequencing was performed using a 100-bp paired-end on a HiSeq 2000 (Illumina Inc., USA). Reads were mapped against hg19 using Burrows-Wheeler Aligner (BWA) and in-house scripts. Germline variant calling was performed by Genome Analysis Toolkit (GATK) (https://gatk.broadinstitute.org/hc/en-us) and VarScan (http://varscan.sourceforge.net/) algorithms, whereas MuTect, Strelka, and VarScan were used and combined to detect somatic variants. Repeated variants in all samples were filtered out as a step of quality control, besides variants with an allele frequency of ≤0.3 or a read depth of ≤10 were also discarded. The variants were annotated with an in-house annotation pipeline gathering information from different sources to help its interpretation: germline population datasets (gnomAD; https://gnomad.broadinstitute.org/), variants genetic variation [COSMIC, International Cancer Genome Consortium (ICGC); https://cancer.sanger.ac.uk/cosmic], clinical relevance (ClinVar; www.ncbi.nlm.nih.gov/clinvar), genotype-phenotype relationships (OMIM; www.omim.org), gene databases on oncogenic role [Tumor Suppresor Gene Database (TSGene; https://bioinfo.uth.edu/TSGene/)], and in-house database on genes involved in hereditary cancer. For prioritization, variants with a MAF of ≥0.1% were filtered out. All noncoding, intergenic, intronic, 3′ and 5′ untranslated regions (UTRs), and synonymous nonsplice site variants were also removed. Remaining variants were selected according to a recessive mode of inheritance. Thus, we selected genes carrying homozygous or compound heterozygous variants.
Metaphase spreads
For karyotyping analysis, peripheral blood cells were stimulated with phytohemagglutinin-l (PHA) and interleukin-2 (IL-2), exposed to colcemid (0.5 μg/ml; Life Technologies) for 1 hour at 37°C to enrich cells at metaphases stage, and harvested routinely. Briefly, cells were pelleted and exposed to hypotonic treatment (75 mM KCl solution), fixed in cold Carnoy’s solution [methanol:acetic acid (3:1)], and spread onto clean glass slides. The classical G-banding protocol was performed by treating the slides with trypsin at 37°C and staining the chromosomes using Leishman solution. Metaphase images were digitally acquired with a charge-coupled device (CCD) camera (Photometrics SenSys) connected to a Zeiss Axioplan2 microscope using a 100× objective and using Metapher 4 software v3.1.125. Last, chromosomes were manually classified using Ikaros software v5.0 according to the International System for Human Cytogenetic Nomenclature (2013).
Comparative genome hybridization
CGH was performed using the Agilent SurePrint G3 Human CGH 60 K microarray (Agilent Technologies) by spanning the entire human genome at a median resolution of 41 kb. For the hybridization, 500 ng of genomic DNA from the test samples and human reference DNA (Agilent Technologies) were differentially labeled with Cy5–2′-deoxycytidine 5′-triphosphate (dCTP) and Cy3-dCTP by random priming. An Agilent DNA Microarray scanner G2565CA (Agilent Technologies) was used to scan the arrays, and data were extracted and visualized using Feature Extraction software v10.7. Copy number–altered regions were detected with Agilent Genomic Workbench software v7.0 (Agilent Technologies) using the Aberration Detection Method 2 algorithm set as 6, with a minimum number of three consecutive probes.
Fluorescence in situ hybridization
FISH analyses were performed according to the manufacturers’ instructions on peripheral blood cells treated with colcemid for 1 hour, exposed to hypotonic treatment (75 mM KCl solution), fixed in cold Carnoy’s solution [methanol:acetic acid (3:1)], and spread onto clean glass slides. After dehydration (70, 80, and 100% ethanol), the samples were denatured in the presence of the specific probe at 75°C for 5 min and left overnight for hybridization at 37°C in a hybridizer machine (Dako). The probes used detect copy number alterations in chromosomes 7 and 8 (KBI-20031; Kreatech), 20 (KBI-10203; Kreatech), and X (KBI-20030; Kreatech). Last, the samples were washed with 20× SSC buffer with Tween 20 at 63°C and stained with 4′,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml)/antifade solution (Palex Medical). FISH images were captured using a Leica DM5500B microscope with a CCD camera (Photometrics SenSys) connected to a PC running the CytoVision software 7.2 image analysis system (Applied Imaging). Images were blinded analyzed to score for the number of FISH signals.
Variant calling
Variants were annotated with an in-house annotation pipeline gathering information from different sources to help its interpretation: germline population datasets (gnomAD), somatic variants genetic variation (COSMIC, ICGC), clinical relevance (ClinVar), genotype-phenotype relationships (OMIM), gene databases on oncogenic role such as Tumor Suppresor Gene Database (TSGene), and an in-house database on genes involved in hereditary cancer prepared from literature searching candidates. Variants with a population MAF higher than 0.1% were filtered out. All noncoding, intergenic, intronic, 3′ and 5′ UTRs, and synonymous nonsplice site variants were also removed. Remaining variants were studied considering a recessive model of inheritance, thus selecting genes carrying homozygous or compound heterozygous variants. This approach led to 12 genes with at least two non-outfiltered suggestive variants (29 remaining variants), with a single highly suggestive candidate gene (MAD1L1), included at the TSGene Database and carrying two protein-truncating variants.
Single-cell DNA sequencing
For single-cell DNA sequencing, genomic DNA was amplified with the GenomePlex Single Cell Whole Genome Amplification Kit (Sigma-Aldrich). Amplified DNA was purified, barcoded and pooled, and sequenced on an Illumina Hiseq2000. Sequencing reads were trimmed to 40 nucleotides and aligned to reference sequence hg19/GRCh37 human genome reference using BWA (0.6.1). QDNAseq (v1.30.0) was used to estimate gene copy number using 100-kb bins and to generate segmentation plots (https://bioconductor.org/packages/release/bioc/html/QDNAseq.html).
Analysis of SAC function
PBMCs were extracted from blood samples by centrifugation on a Ficoll cushion (Cytiva) for 30 min at 1800 rpm at room temperature (RT) and washed several times in phosphate-buffered saline (PBS). After 30 min of adhesion step at 37°C in RPMI 1640, nonadherent cells were cultured in RPMI 1640 in the presence of PHA (5 μg/ml; Sigma-Aldrich) to induce T lymphocyte proliferation. After 2 days, PHA was washed, and IL-2 (50 U/ml; STEMCELL Technologies) was added to the medium and replaced every 2 days for a time period of 5 days.
To study mitotic progression, PBMCs were synchronized in G2 by overnight incubation with 5 μM RO-3306 (Selleck Chemicals, S7747). Cell cycle arrest was released by washing cells twice in PBS, and 1 μM taxol (Selleck Chemicals, S1150) was subsequently added to arrest cells in mitosis. Flow cytometry analysis was performed on PBMCs fixed with 4% paraformaldehyde (PFA), permeabilized with 0.5% Triton X-100–PBS, and stained with anti-MPM2 (Millipore, 05-368). Cells were washed with PBS and incubated with a secondary antibody conjugated to Alexa Fluor 488 (Molecular Probes). Secondary antibody was washed with PBS, and DNA was stained with DAPI (2 mg/ml) for 30 min at 37°C.
Protein analysis
For immunofluorescence, PBMCs were attached for 30 min to coverslips previously treated with 0.01% poly-l-lysine (Sigma-Aldrich, P6282). Cells were fixed with 4% PFA, permeabilized with 0.05% Triton X-100–PBS, and stained overnight with primary antibodies against the following: phospho–histone-3 (Cell Signaling Technology, 9701), α-tubulin (Sigma-Aldrich, T9026), BUBR1 (provided by S. Taylor, University of Manchester, UK), or anti-centromeric antigen (ACA) (Antibodies Incorporated, 15-235). Coverslips were washed twice in PBS and incubated with secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 647 (Molecular Probes) and DAPI. Secondary antibodies were washed three times in PBS, and coverslips were mounted in Fluoromount (Bionova).
For Western blot analysis, cultured PBMCs were lysed in radioimmunoprecipitation assay [25 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% Na deoxycholate, and complete protease inhibitor cocktail] for 15 min at 4°C and centrifuged for 10 min at 14000g, and the supernatant was saved for analysis of proteins. Protein extracts were quantified with Pierce bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific), mixed with loading buffer [350 mM tris-HCl (pH 6.8), 30% glycerol, 10% SDS, 0.6 M dithiothreitol, and 0.1% bromophenol blue], boiled for 5 min, and subjected to electrophoresis using the standard SDS–polyacrylamide gel electrophoresis method. Proteins were then transferred to a nitrocellulose membrane (Bio-Rad), blocked for 1 hour at RT in tris-buffered saline (TBS) with 0.1% Tween 20 containing 5% bovine serum albumin (BSA), and incubated overnight at 4°C with specific primary antibodies. Membranes were washed 10 min in TBS–Tween 20 and incubated with peroxidase-conjugated secondary antibodies (Dako) for 45 min at RT. Last, the membranes were washed three times for 5 min and developed using enhanced chemiluminescence reagent (Western Lightning Plus-ECL; PerkinElmer). The following primary antibodies were used: anti-MAD1 (ProteinTech, 18322-1-AP; Abcam, ab175245; and Thermo Fisher Scientific, PA5-28991), anti-MAD2 (Abcam, ab10691), anti-CDC27 (Abcam, ab10538), anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling Technology, 5174).
For mass spectrometry studies, protein concentration from lysates was determined by micro-BCA using BSA as standard. Then, 200 μg of each sample was digested by means of the ProtiFi S-Trap Mini Spin Column Digestion Protocol. Samples were labeled using tandem mass tag (TMT) reagent 11-plex following the manufacturer’s instructions. Samples were mixed in 1:1 ratios based on the total peptide amount, and the final mixture was desalted using a Sep-Pak C18 cartridge (Waters Corporation) and dried prior high pH reverse phase high-performance liquid chromatography (HPLC) prefractionation. Peptides were prefractionated offline by means of high pH reverse phase chromatography using an Ultimate 3000 HPLC system equipped with a sample collector. Raw files were processed with MaxQuant (v1.6.0.16) using the standard settings against a human protein database (UniProtKB/TrEMBL) supplemented with contaminants. A global normalization of log2-transformed intensities across samples was performed using the LOESS function.
Cytokines were analyzed from plasma samples using the Proteome Profiler Human Cytokine Array Kit (R&D Systems, ARY005B) following the manufacturer’s instructions. Relative expression levels of detected cytokines were quantified using Fiji (https://imagej.net/software/fiji/).
Single-cell transcriptomics
PBMCs were isolated from blood samples by centrifugation on a Ficoll cushion (Cytiva) for 30 min at 1800 rpm and washed in PBS. Fc receptors were blocked using FcR Blocking Reagent (Miltenyi Biotec, 130-059-901) according to manufacturer’s instructions. Dead cells were removed with a Dead Cell Removal kit (Miltenyi Biotec, 130-090-101). Viability was assessed (>90%), and cells were subjected to single-cell isolation. Generation of gel beads in emulsion (GEMs), barcoding, GEM-RT clean-up, cDNA amplification, and library construction were all performed as recommended by the manufacturer. Samples were loaded onto a 10× Chromium single cell controller chip B (10x Genomics) for a targeted recovery of ~10,000 cells per condition, as described in the manufacturer’s protocol (Chromium Single Cell 3′ GEM, Library and Gel Bead Kit v3, PN-1000075). Data were processed using CellRanger (10x Genomics) and Scanpy (https://scanpy.readthedocs.io/en/stable/). All the three classifications, L1 (7 cell types), L2 (30 cell types), and L3 (57 cell types), were used for more general or specific definitions of cell types (https://azimuth.hubmapconsortium.org/references/#Human%20-%20PBMC). Gene enrichment and pathway analysis were performed with Metascape (https://metascape.org/gp/index.html), GSEA (www.gsea-msigdb.org/gsea/index.jsp), Enrichr (https://maayanlab.cloud/Enrichr/), and their Python wrap GSEApy (https://gseapy.readthedocs.io/en/latest/). Ligand-receptor interactions were investigated using CellPhoneDB (23).
Inference of aneuploidy from scRNA-seq data
The prediction of aneuploidies was performed using inferCNV in their R (https://github.com/broadinstitute/infercnv) and Python (https://icbi-lab.github.io/infercnvpy/infercnv.html) implementations and using a window of 250 genes and cell types predicted in the L3 classification in Azimuth. To estimate chromosomal gains and losses in every individual cell, estimated DNA copy number changes from the gene expression data using matched control cell types for normalization (fig. S6A). To classify every single cell as aneuploid (i.e., harboring at least one chromosomal gain or loss) or euploid (all disomic chromosomes), we first calculated the average CNV score per chromosome for each cell (chrCNV; Fig. 5B) and established a threshold for classifying each chromosome as monosomic, disomic, or trisomic based on the kernel distribution of the CNV score in the proband and controls (fig. S6B). We also used more stringent thresholds (fig. S6B) to define “clean” disomies (CleanEU) and either mono- or trisomies (CleanAneu) that may allow a better confidence in the transcriptional comparison between these groups. Only cells with at least one mono- or trisomy with the stringent thresholds were considered as CleanAneu (13.6% in the proband versus 4.0% in controls), and only cells with all disomic chromosomes using these stringent conditions were classified as CleanEU.
Statistical analysis
Statistical comparisons were performed using Prism 8 (GraphPad), using the specific tests indicated in each figure legend. P values were corrected for multiple testing where appropriate using Bonferroni corrections unless otherwise indicated. Genomic analysis was performed using Python 3.8, R 4.1.3, and the packages indicated in the corresponding section.
Acknowledgments
We thank S. Taylor (University of Manchester, UK) and K. Wassmann (CNRS, Paris) for antibodies and suggestions and P. X. de Embún, F. García, and E. Zarzuela (Proteomics Unit, CNIO) for mass spectrometry analysis.
Funding: This work is supported by Spanish Ministry of Science, Juan de la Cierva programme (C.V.-B.); Spanish Ministry of Science and Innovation, Agencia Estatal de Investigación (MCI-AEI), BIO2017-91272-EXP (S.R.-P.); Spanish National Research and Development Plan, ISCIII, and FEDER, PI17/02303, PI20/01837, and DTS19/00111 (S.R.-P.); Spanish National Research and Development Plan, ISCIII, and FEDER, PI21/01641 (R.T.-R.); Fundación Científica de la Asociación Española contra el Cancer, LABAE20049RODR (S.R.-P.); MCI-AEI/FEDER, RTI2018-095582-B-I00, and RED2018-102723-T (M.M.); Comunidad de Madrid iLUNG and scCANCER programmes, B2017/BMD-3884 and Y2020/BIO-6519 (M.M.); and MCI-AEI, Severo Ochoa Center of Excellence, CEX2019-000891-S (S.R.-P., M.M., and M.U.).
Author contributions: Conceptualization: M.M. and M.U. Methodology: C.V.-B., A.G.-C., A.H.-N., M.U., S.R.-P., M.M., and M.U. Investigation: C.V.-B., M.T., A.G.-C., A.H.-N., D.G.-S., D.R., B.H., J.P., A.O., F.M., R.T.-R., S.R.-P., M.M., and M.U. Visualization: C.V.-B., A.S.-B., B.P., M.M., and M.U. Funding acquisition: J.P., S.R.-P., M.M., and M.U. Project administration: M.M. and M.U. Supervision: M.M. and M.U. Writing: C.V.-B., M.M., and M.U.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: scRNA-seq data are available at the GEO repository under the accession code GSE197267. Proteomics data are available at ProteomeXchange (project accession: PXD035765). R and Python notebooks used in this work are accessible at Zenodo (DOI: 10.5281/zenodo.6960105) and GitHub (https://github.com/malumbreslab). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
REFERENCES AND NOTES
- 1.Ben-David U., Amon A., Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 21, 44–62 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Nagaoka S. I., Hassold T. J., Hunt P. A., Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan T. P., Holland A. J., Time is of the essence: The molecular mechanisms of primary microcephaly. Genes Dev. 35, 1551–1578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remeseiro S., Cuadrado A., Losada A., Cohesin in development and disease. Development 140, 3715–3718 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Musacchio A., Salmon E. D., The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Lara-Gonzalez P., Pines J., Desai A., Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell Dev. Biol. 117, 86–98 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D., Kidd A., Mehes K., Nash R., Robin N., Shannon N., Tolmie J., Swansbury J., Irrthum A., Douglas J., Rahman N., Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 36, 1159–1161 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Micale M. A., Schran D., Emch S., Kurczynski T. W., Rahman N., Van Dyke D. L., Mosaic variegated aneuploidy without microcephaly: Implications for cytogenetic diagnosis. Am. J. Med. Genet. A 143A, 1890–1893 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Castillo H., Vasquez-Velasquez A. I., Rivera H., Barros-Nunez P., Clinical and genetic heterogeneity in patients with mosaic variegated aneuploidy: Delineation of clinical subtypes. Am. J. Med. Genet. A 146A, 1687–1695 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Vasudevan A., Schukken K. M., Sausville E. L., Girish V., Adebambo O. A., Sheltzer J. M., Aneuploidy as a promoter and suppressor of malignant growth. Nat. Rev. Cancer 21, 89–103 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Aziz K., Sieben C. J., Jeganathan K. B., Hamada M., Davies B. A., Velasco R. O. F., Rahman N., Katzmann D. J., van Deursen J. M., Mosaic-variegated aneuploidy syndrome mutation or haploinsufficiency in Cep57 impairs tumor suppression. J. Clin. Invest. 128, 3517–3534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R., Murray A. W., Feedback control of mitosis in budding yeast. Cell 66, 519–531 (1991). [DOI] [PubMed] [Google Scholar]
- 13.Hardwick K. G., Murray A. W., Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 131, 709–720 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfieri C., Chang L., Zhang Z., Yang J., Maslen S., Skehel M., Barford D., Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature 536, 431–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faesen A. C., Thanasoula M., Maffini S., Breit C., Muller F., van Gerwen S., Bange T., Musacchio A., Basis of catalytic assembly of the mitotic checkpoint complex. Nature 542, 498–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer E. S., Yu C. W. H., Bellini D., McLaughlin S. H., Orr C. M., Wagner A., Freund S. M. V., Barford D., Molecular mechanism of Mad1 kinetochore targeting by phosphorylated Bub1. EMBO Rep. 22, e52242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwanaga Y., Chi Y. H., Miyazato A., Sheleg S., Haller K., Peloponese J. M. Jr., Li Y., Ward J. M., Benezra R., Jeang K. T., Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 67, 160–166 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Kim S., Sun H., Tomchick D. R., Yu H., Luo X., Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc. Natl. Acad. Sci. U.S.A. 109, 6549–6554 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji W., Luo Y., Ahmad E., Liu S. T., Direct interactions of mitotic arrest deficient 1 (MAD1) domains with each other and MAD2 conformers are required for mitotic checkpoint signaling. J. Biol. Chem. 293, 484–496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen R., Seshan V. E., FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai H., Liu Y., Men H., Zheng Y., Protective mechanism of humanin against oxidative stress in aging-related cardiovascular diseases. Front Endocrinol (Lausanne) 12, 683151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efremova M., Vento-Tormo M., Teichmann S. A., Vento-Tormo R., CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Patel A. P., Tirosh I., Trombetta J. J., Shalek A. K., Gillespie S. M., Wakimoto H., Cahill D. P., Nahed B. V., Curry W. T., Martuza R. L., Louis D. N., Rozenblatt-Rosen O., Suva M. L., Regev A., Bernstein B. E., Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornet E., Debliquis A., Rimelen V., Civic N., Docquier M., Troussard X., Drenou B., Matthes T., Developing molecular signatures for chronic lymphocytic leukemia. PLOS ONE 10, e0128990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snape K., Hanks S., Ruark E., Barros-Nunez P., Elliott A., Murray A., Lane A. H., Shannon N., Callier P., Chitayat D., Clayton-Smith J., Fitzpatrick D. R., Gisselsson D., Jacquemont S., Asakura-Hay K., Micale M. A., Tolmie J., Turnpenny P. D., Wright M., Douglas J., Rahman N., Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat. Genet. 43, 527–529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yost S., de Wolf B., Hanks S., Zachariou A., Marcozzi C., Clarke M., de Voer R., Etemad B., Uijttewaal E., Ramsay E., Wylie H., Elliott A., Picton S., Smith A., Smithson S., Seal S., Ruark E., Houge G., Pines J., Kops G., Rahman N., Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nat. Genet. 49, 1148–1151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalhal S., Bader I., Rooimans M. A., Oostra A. B., Balk J. A., Feichtinger R. G., Beichler C., Speicher M. R., van Hagen J. M., Waisfisz Q., van Haelst M., Bruijn M., Tavares A., Mayr J. A., Wolthuis R. M. F., Oliveira R. A., de Lange J., Biallelic BUB1 mutations cause microcephaly, developmental delay, and variable effects on cohesion and chromosome segregation. Sci. Adv. 8, eabk0114 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Wolf B., Oghabian A., Akinyi M. V., Hanks S., Tromer E. C., van Hooff J. J. E., van Voorthuijsen L., van Rooijen L. E., Verbeeren J., Uijttewaal E. C. H., Baltissen M. P. A., Yost S., Piloquet P., Vermeulen M., Snel B., Isidor B., Rahman N., Frilander M. J., Kops G., Chromosomal instability by mutations in the novel minor spliceosome component CENATAC. EMBO J. 40, e106536 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinrich S., Sewart K., Windecker H., Langegger M., Schmidt N., Hustedt N., Hauf S., Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO Rep. 15, 291–298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruse T., Larsen M. S., Sedgwick G. G., Sigurdsson J. O., Streicher W., Olsen J. V., Nilsson J., A direct role of Mad1 in the spindle assembly checkpoint beyond Mad2 kinetochore recruitment. EMBO Rep. 15, 282–290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganmore I., Smooha G., Izraeli S., Constitutional aneuploidy and cancer predisposition. Hum. Mol. Genet. 18, R84–R93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuura S., Matsumoto Y., Morishima K., Izumi H., Matsumoto H., Ito E., Tsutsui K., Kobayashi J., Tauchi H., Kajiwara Y., Hama S., Kurisu K., Tahara H., Oshimura M., Komatsu K., Ikeuchi T., Kajii T., Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am. J. Med. Genet. A 140, 358–367 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Williams B. R., Prabhu V. R., Hunter K. E., Glazier C. M., Whittaker C. A., Housman D. E., Amon A., Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheltzer J. M., Torres E. M., Dunham M. J., Amon A., Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. U.S.A. 109, 12644–12649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stingele S., Stoehr G., Peplowska K., Cox J., Mann M., Storchova Z., Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8, 608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davoli T., Uno H., Wooten E. C., Elledge S. J., Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chunduri N. K., Storchova Z., The diverse consequences of aneuploidy. Nat. Cell Biol. 21, 54–62 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Ji Z., Chuen J., Kiparaki M., Baker N., Cell competition removes segmental aneuploid cells from Drosophila imaginal disc-derived tissues based on ribosomal protein gene dose. eLife 10, e61172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wijenayake S., Storey K. B., The role of humanin in natural stress tolerance: An underexplored therapeutic avenue. Biochim. Biophys. Acta Gen. Subj. 1866, 130022 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Missiroli S., Genovese I., Perrone M., Vezzani B., Vitto V. A. M., Giorgi C., The role of mitochondria in inflammation: From cancer to neurodegenerative disorders. J. Clin. Med. 9, 740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y., Shao Q., Peng G., Exhaustion and senescence: Two crucial dysfunctional states of T cells in the tumor microenvironment. Cell. Mol. Immunol. 17, 27–35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bik-Multanowski M., Pietrzyk J. J., Midro A., MTRNR2L12: A candidate blood marker of early Alzheimer’s disease-like dementia in adults with Down syndrome. J. Alzheimers Dis. 46, 145–150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salemi M., Ridolfo F., Salluzzo M. G., Cannarrella R., Giambirtone M., Caniglia S., Tirolo C., Ferri R., Romano C., Humanin gene expression in fibroblast of Down syndrome subjects. Int. J. Med. Sci. 17, 320–324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dierssen M., Fructuoso M., de Lagran M. M., Perluigi M., Barone E., Down syndrome is a metabolic disease: Altered insulin signaling mediates peripheral and brain dysfunctions. Front. Neurosci. 14, 670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grazioli S., Pugin J., Mitochondrial damage-associated molecular patterns: From inflammatory signaling to human diseases. Front. Immunol. 9, 832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Que T. H., Marco J. G., Ellis J., Matutes E., Babapulle V. B., Boyle S., Catovsky D., Trisomy 12 in chronic lymphocytic leukemia detected by fluorescence in situ hybridization: Analysis by stage, immunophenotype, and morphology. Blood 82, 571–575 (1993). [PubMed] [Google Scholar]
- 48.Balatti V., Bottoni A., Palamarchuk A., Alder H., Rassenti L. Z., Kipps T. J., Pekarsky Y., Croce C. M., NOTCH1 mutations in CLL associated with trisomy 12. Blood 119, 329–331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landgren O., Albitar M., Ma W., Abbasi F., Hayes R. B., Ghia P., Marti G. E., Caporaso N. E., B-cell clones as early markers for chronic lymphocytic leukemia. N. Engl. J. Med. 360, 659–667 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akbar A. N., Henson S. M., Are senescence and exhaustion intertwined or unrelated processes that compromise immunity? Nat. Rev. Immunol. 11, 289–295 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Wang R. W., Vigano S., Ben-David U., Amon A., Santaguida S., Aneuploid senescent cells activate NF-kappaB to promote their immune clearance by NK cells. EMBO Rep. 22, e52032 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribot J. C., Lopes N., Silva-Santos B., γδ T cells in tissue physiology and surveillance. Nat. Rev. Immunol. 21, 221–232 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Bonneville M., O’Brien R. L., Born W. K., γδ T cell effector functions: A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10, 467–478 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Bravo V., Maciejowski J., Corona J., Buch H. K., Collin P., Kanemaki M. T., Shah J. V., Jallepalli P. V., Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 156, 1017–1031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raich N., Mahmoudi S., Emre D., Karess R. E., Mad1 influences interphase nucleoplasm organization and chromatin regulation in Drosophila. Open Biol. 8, 180166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan J., Zhu F., Zasadil L. M., Yu J., Wang L., Johnson A., Berthier E., Beebe D. J., Audhya A., Weaver B. A., A Golgi-localized pool of the mitotic checkpoint component Mad1 controls integrin secretion and cell migration. Curr. Biol. 24, 2687–2692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wan J., Block S., Scribano C. M., Thiry R., Esbona K., Audhya A., Weaver B. A., Mad1 destabilizes p53 by preventing PML from sequestering MDM2. Nat. Commun. 10, 1540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rio Frio T., Lavoie J., Hamel N., Geyer F. C., Kushner Y. B., Novak D. J., Wark L., Capelli C., Reis-Filho J. S., Mai S., Pastinen T., Tischkowitz M. D., Marcus V. A., Foulkes W. D., Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia. N. Engl. J. Med. 363, 2628–2637 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Pinson L., Mannini L., Willems M., Cucco F., Sirvent N., Frebourg T., Quarantotti V., Collet C., Schneider A., Sarda P., Genevieve D., Puechberty J., Lefort G., Musio A., CEP57 mutation in a girl with mosaic variegated aneuploidy syndrome. Am. J. Med. Genet. A 164A, 177–181 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Santos-Simarro F., Pacio M., Cueto-Gonzalez A. M., Mansilla E., Valenzuela-Palafoll M. I., Lopez-Grondona F., Lledin M. D., Schuffelmann C., Del Pozo A., Solis M., Vallcorba P., Lapunzina P., Menendez Suso J. J., Siccha S. M., Montejo J. M., Mena R., Jimenez-Rodriguez C., Garcia-Minaur S., Palomares-Bralo M., Mosaic variegated aneuploidy syndrome 2 caused by biallelic variants in CEP57, two new cases and review of the phenotype. Eur. J. Med. Genet. 64, 104338 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8