Abstract
Cell encapsulation within hydrogel droplets is transforming what is feasible in multiple fields of biomedical science such as tissue engineering and regenerative medicine, in vitro modeling, and cell-based therapies. Recent advances have allowed researchers to miniaturize material encapsulation complexes down to single-cell scales, where each complex, termed a single-cell microgel, contains only one cell surrounded by a hydrogel matrix while remaining <100 μm in size. With this achievement, studies requiring single-cell resolution are now possible, similar to those done using liquid droplet encapsulation. Of particular note, applications involving long-term in vitro cultures, modular bioinks, high-throughput screenings, and formation of 3D cellular microenvironments can be tuned independently to suit the needs of individual cells and experimental goals. In this progress report, an overview of established materials and techniques used to fabricate single-cell microgels, as well as insight into potential alternatives is provided. This focused review is concluded by discussing applications that have already benefited from single-cell microgel technologies, as well as prospective applications on the cusp of achieving important new capabilities.
Keywords: 3D cell culture, cell-based therapies, hydrogels, regenerative medicine, single-cell analysis
1. Introduction
Single-cell techniques have altered the research landscape in the past decade due in large part to advances in instrumentation,[1–5] sample and reagent handling,[6–8] increased computational capabilities,[8] and improved understanding and development of microfluidic-based technologies.[7,9] With these innovations, scientists are now capable of isolating, handling, and assaying individual cells to acquire a deeper understanding of biological heterogeneity across a vast swathe of inquiries.[4] While these discoveries have dramatically transformed efforts in understanding cellular heterogeneity, they have also opened exciting possibilities for single-cell applications in more translational fields like diagnostics and regenerative medicine.
Of these advances, droplet microfluidics has been instrumental in facilitating the growth of single-cell analysis by providing a predictable method for the compartmentalization (i.e., encapsulation) of individual cells into monodisperse, pico-liter-to-nanoliter droplets.[10–14] Perhaps more than any other area, single-cell-omics technologies (e.g., genomics, transcriptomics, proteomics, metabolomics) have benefited from advances in single-cell encapsulation using droplet microfluidics.[15] With these improved technologies and techniques, new insights into the underlying heterogeneity of diseased cells and cell states have lead to developments of novel diagnostic and therapeutic approaches for cancer and microbial infections, with many other areas under study. For example, Azizi et al. generated an immune cell atlas by profiling >45,000 individual cells from primary breast carcinomas and matched, healthy immune cells from breast tissue. Increased diversity in T cell and myeloid lineages was observed in tumors compared to healthy tissue. Additionally, M1 and M2 genetic signatures of tumor-infiltrating macrophages were observed in concert, indicating continuous rather than binary phenotypic states.[16] Another example of this technology revealed intratumor variations in expression in IFN-γ signaling pathway genes and other co-regulatory genes, such as MHCII, for patients with lung adenocarcinoma. This work demonstrated how understanding intratumor gene expression variations can inform appropriate multi-antigen combinatorial therapies for patient-optimized treatments.[17] Analyses with single-cell resolution as demonstrated in these representative works is critical for progressing toward optimized treatment regimens and predicting treatment outcomes for diverse medical applications, including cancer immunotherapies.
Beyond the elucidation of cellular intra- and interpopulation phenotypic heterogeneity,[18,19] single-cell techniques have entered early development for use in therapeutic applications such as cell injection therapies. Additionally, much effort has been aimed at the precise recreation of in vivo cellular microenvironments for in vitro experimental modeling.[20–22] In each of these applications, replication of 3D microenvironmental cues is crucial for eliciting or studying desired cellular behaviors.[23,24] With this in mind, researchers have sought to develop methods that improve upon conventional 2D culturing methods. Of the different solutions proposed, one of particular interest and relevance is the use of hydrogels, defined as 3D crosslinked polymer networks possessing high water content.[23] Using a hydrogel as a cellular encapsulant provides optimal material-to-cell volume ratios and minimal diffusion constraints, with benefits even at the scale of single-cell microgels.[25] Researchers are able to tailor the hydrogel composition to achieve optimal mechanical (e.g., stiffness[26] and stress relaxation[27,28]) and biochemical (e.g., attachment sites[29]) characteristics to suit the target application as well as provide additional protection from external factors (Figure 1).[30–32]
Figure 1.
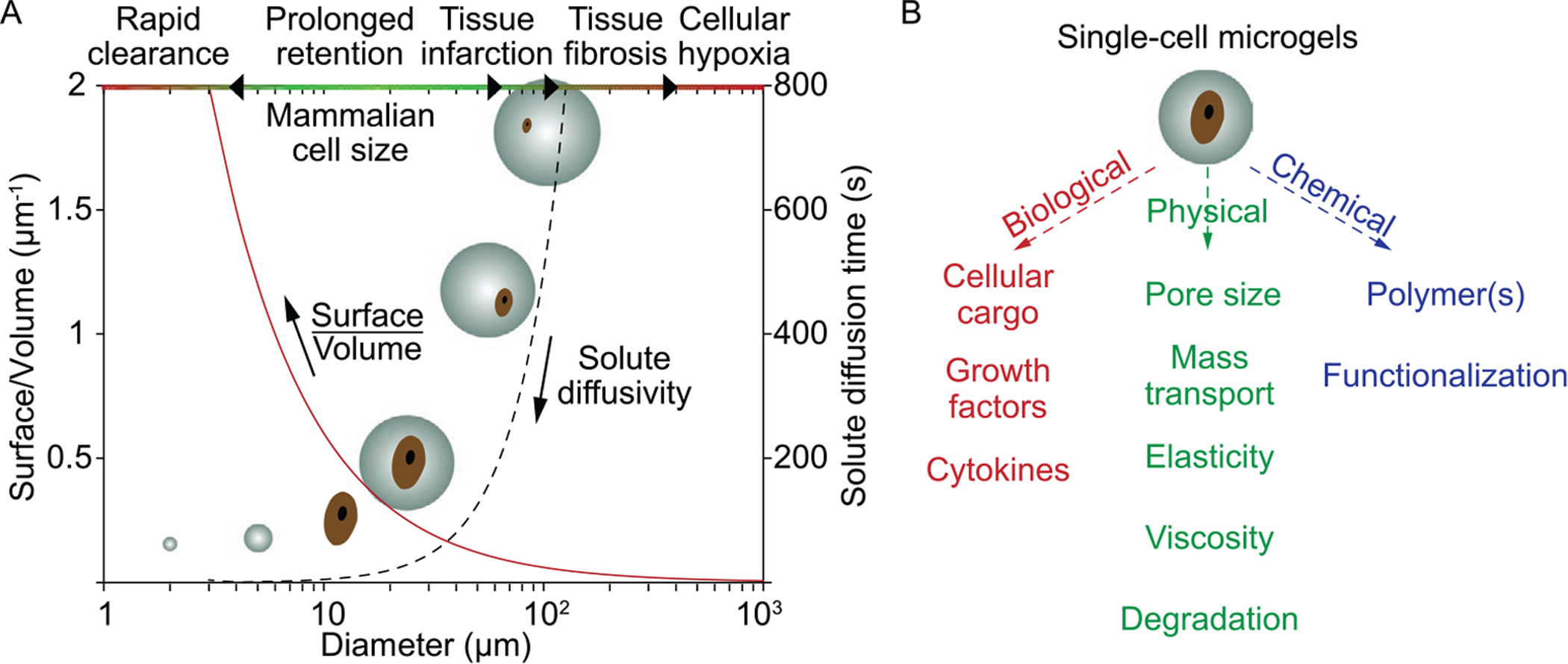
A) Influence of microgel size on physiological response and pharmacokinetics.[21] Cell diameter, Dcell =12 μm, and diffusion coefficient D = 10 μm2 s−1, were used. B) Components of single-cell microgels, with tunable characteristics to suit application needs. Figure (A) was reproduced with permission.[21] Copyright 2018, Elsevier.
In this progress report, we highlight previously demonstrated hydrogel materials for single-cell encapsulation and their target applications, selected fabrication techniques, and applications of cell-laden microgels. While many of the mentioned works involve encapsulation of multiple cells within a hydrogel (i.e., multi-cell microgels), we have inferred how single-cell microgels can benefit potential applications, as well as described possible modifications to current fabrication techniques that are more conducive to single-cell encapsulation. Throughout, we will discuss both the promise and limitations of single-cell microgels as a potentially impactful technology for future diagnostic and therapeutic applications.
2. Materials
A diverse set of natural, synthetic, and composite (i.e., natural and synthetic hybrid) materials has been used to encapsulate cells.[33] However, only a small selection of those materials have been adapted for use within the single-cell paradigm. Choice of hydrogel material and subsequent functionalization is heavily dependent on the intended application. With diverse use cases in cell therapies and tissue engineering, fundamental research, and advanced diagnostics, single-cell microgel material selection requires careful consideration of design criteria including encapsulated cell type, desired cell behavior, time scale of use, external environment, and intended function. These criteria must be matched to critical properties such as chemical and physical biocompatibility, ease of fabrication, and capacity for functionalization.
Here, we provide an overview of the relatively small subset of hydrogel materials that have been used in single-cell encapsulation studies to date (Table 1). The reader is encouraged to survey a more inclusive review of hydrogels used for multi-cell encapsulations, while considering the feasibility for those materials to be applied toward single-cell applications.[33] Additionally, many of the more advanced material modifications used in multi-cell systems to customize the cellular microenvironment (e.g., immunoprotective coatings) have yet to be applied to single-cell microgels.[34] We anticipate continued translation of materials and functionalization techniques from multi-cell studies into the rapidly expanding field of single-cell encapsulation and will highlight a few of the more exciting possibilities in this progress report.
Table 1.
Examples of single-cell microgels previously demonstrated.
| Material | Encapsulated cell type | Functionalization | Crosslinking method | Targeted application | Ref. |
|---|---|---|---|---|---|
| Alginate | MSC,a) OP9,b) HUVECc) | RGD peptides | Ionic, CaCO3 NPs on cell surface | Therapeutic | [46] |
| NIH3T3,d) MSC, HUVEC | RGD peptides | Ionic, Ca2+ freed by acidification | Research | [49] | |
| PCC 6803,e) CC-4532,f) MC3T3-E1g) | N/A | Ionic (CLEX)h) | Research & Therapeutic | [38] | |
| HCT116i) | N/A | Ionic, droplet merging | Research & Diagnostic | [130] | |
| Agarose | CEMj) & K562k) CRL-2265 | N/A | Temperature | Research | [66] |
| GM09947,l) GM09948m) | Reverse primers | Temperature | Research & Diagnostic | [64] | |
| MSC | Fibronectin & fibrinogen | Temperature | Therapeutic | [67] | |
| Dextran | MSC | Tyramine | Enzymatic | Research & Therapeutic | [25] |
| MSC | Tyramine | Enzymatic | Research & Therapeutic | [32] | |
| Hyaluronic acid | MSC | Fibrinogen added | Thiol-ene click chemistry | Research | [40] |
| MSC | Tyramine | Enzymatic | Research & Therapeutic | [32] | |
| Matrigel | RWPE1n) | N/A | Temperature | Research | [36] |
| Collagen-gelatin | 3T3 fibroblasts | N/A | Photopolymerization (riboflavin) | Research | [37] |
| Poly(ethylene glycol) | MSC | TG & MMP | Enzymatic | Research | [123] |
| MSC, bovine chondrocytes | Acrylate | Photopolymerization | Research & Therapeutic | [31] | |
| CHOo) | RGD peptides | Photopolymerization | Research, Diagnostic & Therapeutic | [131] |
Mesenchymal stem cells;
Stromal cell line from bone marrow;
Human umbilical vein endothelial cells;
NIH/Swiss mouse embryonic fibroblast;
Unicellular freshwater cyanobacteria;
Chlamydomonas reinhardtii;
Murine calvarial pre-osteoblasts;
Competitive ligand exchange crosslinking;
Colorectal cell line;
Acute lymphoblastic leukemia;
Chronic myelogenous leukemia;
Female human lymphoid cell line;
Male human lymphoid cell line;
Human epithelial prostate cell line;
Chinese hamster ovary cells.
2.1. Natural Polymers
Natural polymers have seen frequent use in single-cell microgel investigations due to their typical biocompatibility and cell-friendly gelation conditions.[35] The main subtypes of natural polymers used for hydrogels are protein- and polysaccharide-based. However, protein-based gels such as Matrigel[36] and collagen[37] are constrained in their use at the single-cell scale due to a general lack of mechanical stability and fabrication limitations. Polysaccharide-based gels such as alginate[38] and agarose[39] have therefore dominated single-cell microgel research with more limited demonstrations involving dextran[25] and hyaluronic acid.[40]
2.1.1. Alginate
Alginate has emerged as one of the most popular hydrogel materials for single-cell encapsulation due to its favorable biocompatibility, relatively simple mechanism of polymerization,[41] and established laboratory and clinical use.[42] Alginates are a family of natural anionic polysaccharides traditionally derived from brown seaweed, although recent efforts have investigated alginate production via bacterial biosynthesis.[43,44] The structure of alginate consists of chains of (1,4)-linked β-D-mannuronate (M) and α-L-guluronate (G) residues organized into blocks of consecutive M, G, or alternating MG residues.[45] The linear chains can be ionically or covalently crosslinked to form a hydrogel whose properties are influenced by the relative composition of the residue blocks.[45] Alginate of high purity is considered biologically inert, rendering it an attractive material for cell encapsulation for both in vivo and in vitro applications.[41] The reader is directed to multiple well-written references for a more detailed look at the chemical structure, material properties, and applicability of alginate for 3D cell culture.[41,42,45]
One key reason for the early adoption of alginate for single-cell hydrogel encapsulation is its suitability for micro-scale gelation methods. A significant majority of studies involving encapsulation of single cells in alginate microgels employ microfluidic devices to carefully control the introduction of divalent cations into cell-containing droplets of precursor solution. One technique developed by Mao et al. features solid CaCO3 nanoparticles (NPs) either suspended in the alginate precursor solution or pre-adsorbed to the cell surfaces prior to encapsulation (Figure 2A).[46,47] The oil phase is made acidic, usually by the presence of acetic acid, such that the pH of the alginate precursor solution drops upon contact with the oil due to diffusion of H+ ions. Lowered pH stimulates dissolution of the CaCO3 particles and mediates gelation via Ca2+ ion crosslinking. Pre-adsorption of CaCO3 NPs onto the surfaces of cells offers the added benefit that only cell-containing droplets become crosslinked. This selective crosslinking significantly improves single-cell encapsulation efficiency without additional downstream processing (e.g., fluorescence-activated cell/droplet sorting [FACS/FADS]) by reducing production of cell-free microgels.[46] However, the use of CaCO3 as an ion source may lead to structural inhomogeneity due to limited diffusivity of Ca2+ and the rapid kinetics of alginate polymerization.[48] Alternatively, a Ca-EDTA chelate can be included in the alginate precursor solution as a controllable source of divalent cations.[48–50] Contact with an acidified oil phase then similarly triggers release of Ca2+, in this case by dissociating the Ca-EDTA complex. Compared to CaCO3-based methods, Ca-EDTA can be more uniformly dispersed throughout the alginate precursor solution which improves microgel structural homogeneity, although automatic sorting functionality is lost.[48]
Figure 2.
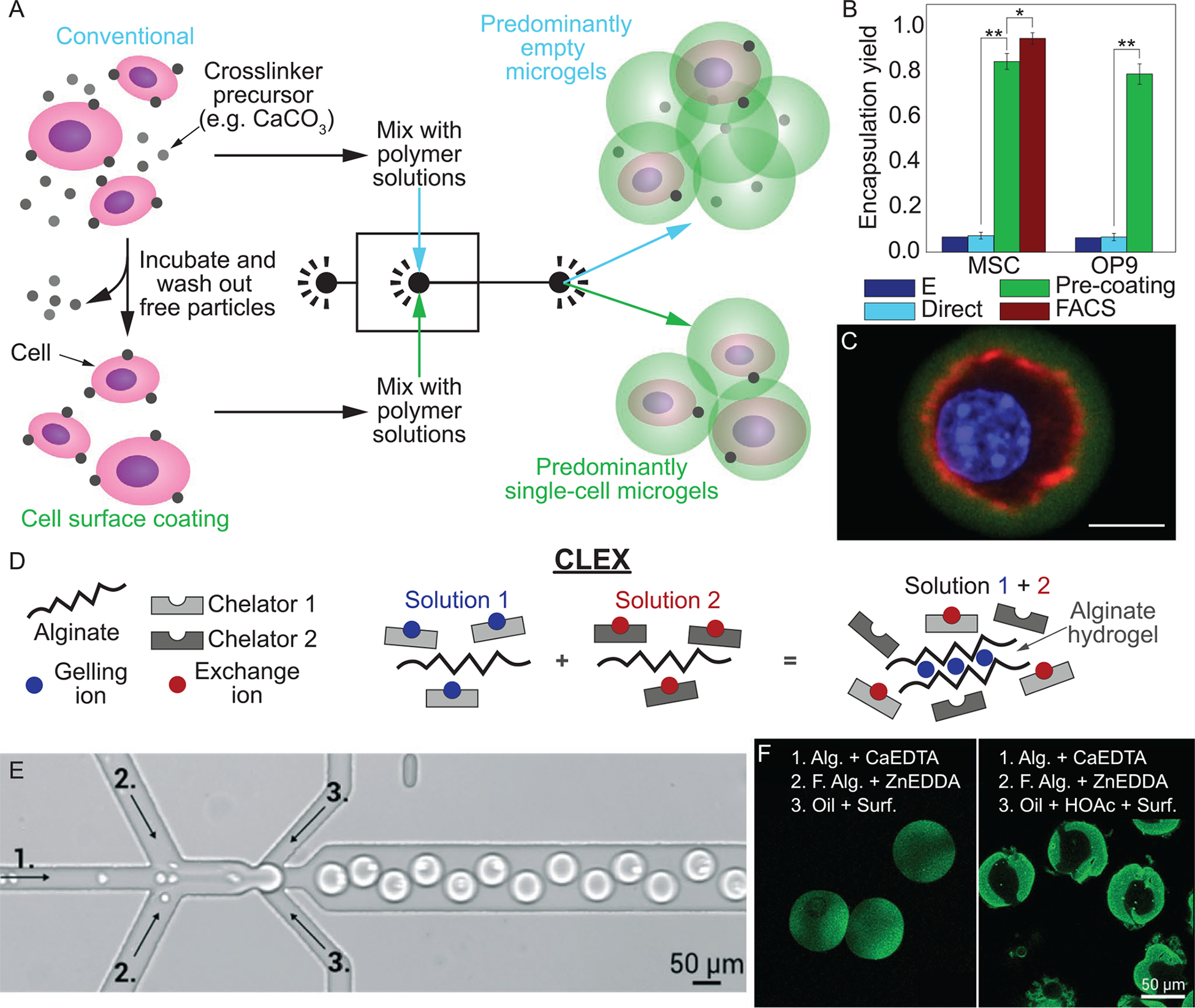
A) Schematic of cell encapsulation using conventional (upper panel) and cell surface coating (lower panel) methods for crosslinking alginate microgels. B) Fraction of alginate microgels containing MSCs and OP9 cells via using conventional (Direct) encapsulation, conventional encapsulation followed by FACS, and cell surface pre-coating. E is theoretical yield for conventional encapsulation. C) Confocal image of encapsulated MSC (green, alginate; red, actin; blue, nucleus), scale bar denotes 10 μm.[46] D) Schematic of CLEX (competitive ligand exchange crosslinking). E) Image of microfluidic device used to fabricate cell-laden microgels using CLEX process, where numbers correspond to solutions in (F). F) Fluorescently labeled cell-free alginate microgels using CLEX (left) and release of Ca2+ from CaEDTA with acidic carrier fluid (right). CLEX microgels are more uniformly stained as a result of the gradual release of Ca2+, compared to non-uniform distribution as a result of rapid gelation. Alg., fluorescent alginate; HOAc, acetic acid; Surf., surfactant.[38] Images (A), (C), and figure (B) are reproduced with permission.[46] Copyright 2016, Springer Nature. Images (D–F) are reproduced with permission.[38] Copyright 2016, Royal Society of Chemistry.
Perhaps the greatest drawback of the above methods is the required exposure of cells to unfavorable pH levels (e.g., pH 4.6) at time scales that are detrimental to cell viability and metabolic activity.[51] Encapsulated cell viability quickly declines as gelation time increases, falling to ≈80% after 2 min and reaching ≈0% after 30 min due to prolonged exposure to acidic conditions.[46,48] To address this limitation, Håti et al. introduced competitive ligand exchange crosslinking (CLEX) as a method for controlling the rate of Ca2+ release, and therefore alginate gelation rate, while maintaining a near physiological pH range (6.0–8.0).[38] An additional chelate (e.g., Zn-EDDA) is introduced through either the continuous phase or a secondary aqueous inlet stream that ultimately mixes with the primary chelate (e.g., Ca-EDTA), as illustrated in Figure 2B. In this specific example, the secondary inorganic cation (Zn2+) has a greater affinity for EDTA than Ca2+, allowing for controlled Ca2+ release dependent on the chelator equilibrium binding constants and solution pH. While requiring optimization of a greater number of experimental parameters, such as the choice of chelates and chelate concentrations, the enhanced control over gelation kinetics, ranging from seconds to minutes, provides a wider range of possible hydrogel architectures and mechanical properties.[52] As a result, the user can achieve improved consistency and versatility in the customization of the single-cell microenvironment, while maintaining high cell viability and minimizing pH-related side effects to cellular metabolic activity. Delayed crosslinking using CLEX may also allow cells to move to the centers of uncrosslinked droplets, which has been identified to reduce incidences of cell egress from microgels.[25]
The above methods use an ionic crosslinking agent (e.g., Ca2+) to form junctions with adjacent guluronate blocks. Such ionically crosslinked gels exhibit the tendency to disintegrate in physiological fluids due to exchange of non-linking Na+ ions with Ca2+.[42] As single-cell alginate microgels move closer to use in long-term in vivo applications, strategies such as the addition of trivalent cations (e.g., Al3+ and Ti3+) or covalent crosslinking of alginate with poly(ethylene glycol) (PEG) diamines can be adapted from established multi-cell approaches.[33,53] Transfer of these techniques for improving gel stability to single-cell methods should be trivial. Mao et al. demonstrated that coating alginate with poly-D-lysine (PDL) to form alginate-PDL-alginate (APA) gels increased in vivo residence time by approximately fivefold compared to untreated gels.[47] While dissolution of a cell’s microgel capsule may be undesirable for certain long-term in vivo applications due to the loss of immunoprotection, this phenomenon can be leveraged in situations where recovery of the cell or other encapsulated materials is required following culture. Controlled disintegration of alginate microgels is possible by exposure to chelating compounds, such as EDTA or phosphate. For example, Zimny et al. used a chelating buffer containing EDTA to easily decrosslink alginate microgels and recover DNA for single-cell genomic analysis following cell lysis.[54]
The most common functionalization of single-cell alginate microgels has been the covalent incorporation of arginylglycylaspartic acid (RGD) adhesion peptides.[46–50] Cellular adhesion to surrounding matrix has been shown to influence cell differentiation,[55] proliferation,[56] and migration.[57] RGD binding sites enable cell attachment via integrins and can be used to elicit a therapeutic phenotype or investigate effects of cellular adhesion with single-cell resolution. Despite the ability of RGD peptides to guide cell adhesion, control over additional material parameters including alginate mechanical properties, soluble factor content, and immunoprotective coatings will likely be necessary for successful implementation of single-cell alginate microgel therapies in clinical use. Considering alginate’s foothold in both multi-cell and single-cell hydrogel encapsulation research, the material is a promising option for both clinical translation of single-cell microgel therapies and first time experimentalists alike.
2.1.2. Agarose
Agarose is a widely popular, natural polysaccharide used in biological research and has received extensive interest for single-cell encapsulation. Extracted from red algae, linear agarose consists of alternating β-D-galactopyranose and 3,6-anhydro-α-L-galactopyranose monomers. Gelation of agarose is temperature mediated, where a structural transition from random coils to double helices occurs below the gelation temperature. The double helices are physically crosslinked by hydrogen bonding between polar groups on the polymer backbone and adjacent water molecules.[58] Molecular weight, concentration, and temperature of gelation are known to strongly influence agarose gel structure and mechanical properties. Specifically, reduced pore size and increased gel stiffness can be achieved by increasing the concentration and/or molecular weight of the agarose or by performing gelation at a lower temperature.[59–61] Similar to alginate, unmodified agarose is not biochemically recognized by mammalian cells and is therefore considered biologically inert.[33]
Agarose is well suited for single-cell encapsulation due to the simplicity and controllability of its thermal gelation mechanism. Most workflows incorporate agarose with low gelation temperatures (<37 °C) to keep the agarose in a liquid state under physiologically relevant temperatures prior to encapsulation.[39,62–70] Gelation then occurs during a brief period of cooling, eliminating the need for potentially cytotoxic crosslinking reagents. Microgels remain crosslinked if raised back to 37 °C due to thermal hysteresis in the gelation behavior of agarose, where the melting point is significantly higher than gelation point.[60] Caution must be taken to minimize exposure of cells to harmful temperatures when fabricating agarose microgels. Many agarose formulations require maintaining the liquid solution above 37 °C, introducing concerns with cell viability[71] or aberrant gene expression.[72] Some strategies attempt to control the temperature of the liquid cell-agarose mixture with feedback-controlled contact heaters[70] or heated air streams,[64] while others wait to combine the cell suspension with hot agarose until immediately before cell encapsulation.[65,66,73] Once crosslinked, agarose is highly stable below its melting point, having been shown to retain gel integrity up to 90 days in vitro[74] and over 100 days in vivo.[75] However, melting can be desirable for select applications, such as in emulsion polymerase chain reaction (ePCR) where gel melting during thermal cycling benefits reagent mixing.[62,64] In situations when heating of microgels to recover contents must be avoided, agarose can be dissolved via enzymatic degradation.[58,70]
For microfluidic applications, additional attention must be given to the relatively high viscosity of liquid agarose solutions, >1 Pa s,[76] since this property can impede throughput or prevent single-cell microgel fabrication due to back pressure limitations. One strategy employed to overcome this limitation is sequential droplet splitting,[77] in which a large agarose droplet is divided N times downstream of initial generation to increase effective throughput by a factor of 2N and produce daughter droplets containing single cells (Figure 3).[65] To maximize the fraction of microgels containing a single cell, cells must be well mixed and suspended at an optimal density in the precursor solution and subsequent parent droplets.
Figure 3.
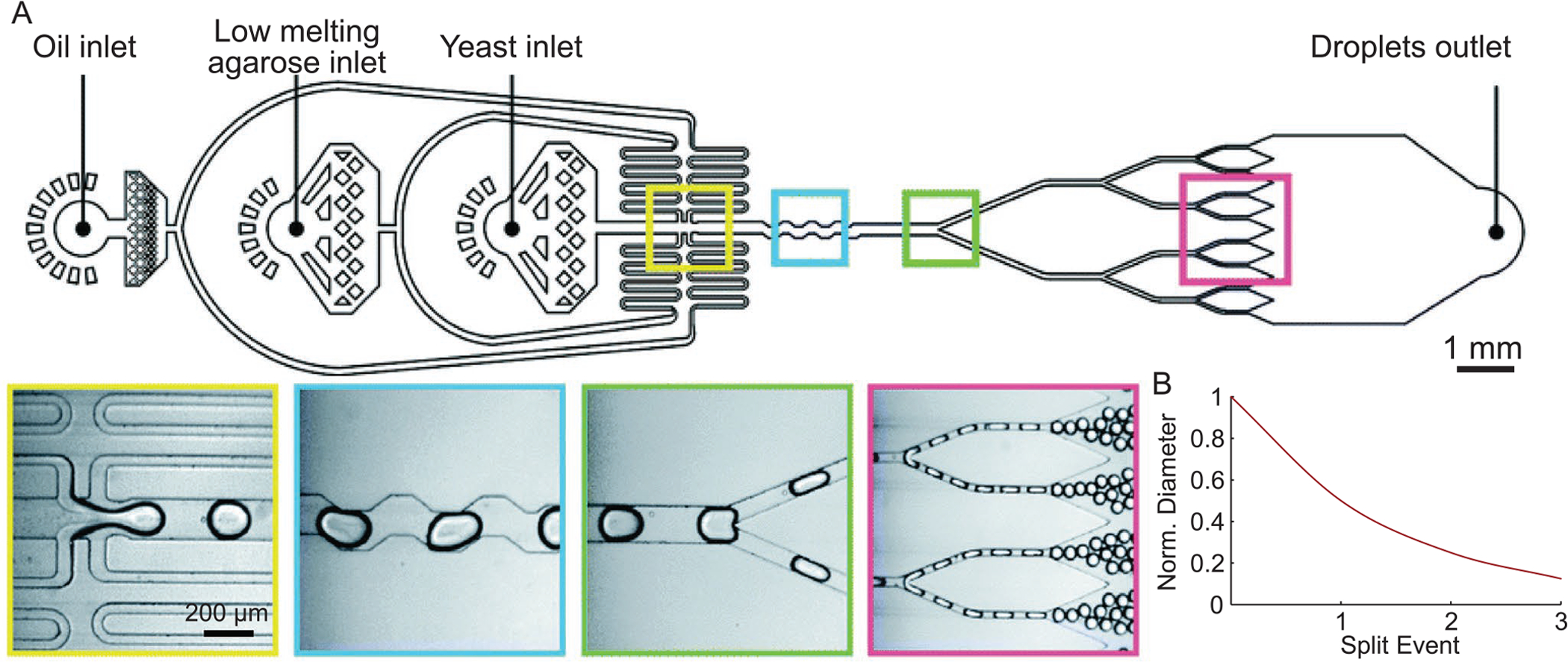
A) Generation of agarose-based microgels. From left to right, images of initial droplet generation event for cell encapsulation, droplet mixer, first droplet splitter, and third droplet splitter. B) Normalized droplet diameter as a result of N splitting events. Images in (A) are reproduced with permission.[65] Copyright 2019, Royal Society of Chemistry.
While single-cell agarose microgels have potential use as in vivo therapeutic cell carriers and in vitro cell culture substrates, they have been most frequently applied in single-cell gene expression assays. Using agarose as the encapsulant allows for facile reagent exchange for ePCR[62–64] and stable compartmentalization of individual cells for extended incubation and subsequent single-cell RNA sequencing (scRNA-seq).[65,66,73] Similar to alginate, agarose microgels require the incorporation of cell-recognizable extracellular matrix (ECM) molecules (e.g., RGD,[78] fibronectin,[67] and fibrinogen[67]) to facilitate direct, cell–material interactions. While, this is not required in many cases due to the brief experimental lifetime of cell-laden microgels, integration of an extended culture period between encapsulation and downstream assays is becoming more common.[65,70] Further tuning of agarose mechanical properties as well as biochemical functionalization will increase the relevance and impact of encapsulation-culture-analysis type studies. Given its established use and compatibility with single-cell encapsulation techniques, agarose is likely to sustain its popularity in single-cell microgel research, particularly for assays in fundamental research and diagnostic applications.
2.1.3. Dextran
Dextran is a natural polymer that has received limited but growing interest for single-cell encapsulation. Dextran is a branched polysaccharide synthesized in bacteria consisting of α-1,6 linked glucopyranoside monomers with α-1,2-, α-1,3-, and/or α-1,4-linked side chains.[79] Similar to alginate, dextran can be ionically or covalently crosslinked. While the crosslinking mechanism is not fully understood, dextran has been observed to form a hydrogel in the presence of K+ ions.[80] However, the K+ concentration required for gelation is >1 M, which is orders of magnitude larger than concentrations normally observed in physiological fluids (e.g., 3–5 mM for extracellular fluid).[81] As a result, the use of covalent crosslinking strategies for most microgel encapsulation applications involving cells is required. Covalent crosslinking of dextran has been performed using a variety of conjugated linking groups, including methacrylate,[82,83] glycidyl methacrylate,[84] hydroxyethyl methacrylate,[85] and tyramine.[25,32] Depending on the chosen crosslinking chemistry, dextran hydrogels can be synthesized with biostable or biodegradable properties.[85] This versatility has afforded dextran relevance for both long-term cell culture and in vivo cell injection therapies, although most existing studies have been limited to multi-cell encapsulation.
The limited adoption of dextran as a single-cell microgel material likely stems in part from the requirement for more specialized gelation strategies compared to other, more popular natural polymers such as alginate and agarose. Much of the work demonstrating encapsulation of single cells in dextran-based microgels has been performed by vanLoo et al.[25,32] In these studies, dextran functionalized with tyramine (Dex-TA) was enzymatically crosslinked by horseradish peroxidase enzyme in the presence of hydrogen peroxide (H2O2) with no negative effects on cell viability. Gelation was physically controlled by diffusion of H2O2 into Dex-TA precursor droplets in a specialized microfluidic device. Tuning of the H2O2 concentration gradient controlled the microgel structure, where lower concentrations resulted in hollow core–shell microgels and increased H2O2 concentrations produced fully crosslinked microgel structures.[32] While the above platform was developed for long-term in vitro culture (≥28 days), the in vivo biocompatibility of dextran suggests potential adaptation of Dex-TA hollow core–shell microgels as protective capsules for therapeutically injected single cells.[33] Similar to the other polysaccha-rides, dextran requires functionalization to enable cell–matrix interactions. Despite limited usage to date, dextran presents another versatile option for single-cell microgel applications.
2.1.4. Hyaluronic Acid
Hyaluronic acid (HA) is a versatile glycosaminoglycan that has been sparsely explored as a single-cell microgel material. Naturally found in all vertebrates as a component of tissue ECM and synovial joint fluid, the structure of HA consists of repeated disaccharide units of alternating D-glucuronic acid and N-aceytl D-glucosamine residues. At physiological pH, the carboxylic acid on the D-glucuronic acid residue is deprotonated and HA behaves as an anionic sodium salt (sodium hyaluronate).[86] The conservation of HA’s structure across species makes it highly bio- and cytocompatible.[87] HA self-aggregation is weak, requiring additional linking groups to form a mechanically stable hydrogel structure. The carboxyl and hydroxyl groups found on the HA backbone are most frequently targeted as sites for chemical functionalization, with a wide variety of crosslinking chemistries and biofunctional moieties already demonstrated. Popular examples of linking groups include PEG-thiol[40,88] and acrylate[89] moieties conjugated to the HA carboxyl by carbodiimide chemistry, each of which allow for cytocompatible gelation conditions.[87]
Similar to dextran, required chemical modifications for specialized gelation mechanisms likely make HA less desirable as a single-cell microgel material compared to alginate and agarose. The potential scope of use for HA microgels for in vivo therapies consists of scenarios in which rapid degradation is desirable.[33,86] Additionally, HA has been shown to promote angiogenesis,[90] which can benefit such in vivo therapies. One of the few examples of HA-based microgels to date was performed by Ma et al., who encapsulated individual mesenchymal stem cells (MSCs) for in vitro culture in thiolated HA (HASH) microgels crosslinked by PEG di-vinyl-sulfone (PEGDVS).[40] HASH was specifically chosen due to its mild gelation conditions and excellent control over the microgel elasticity. The microgels were additionally functionalized with fibrinogen to promote cellular adhesion to the surrounding matrix. A cell suspension in HASH precursor solution with fibrinogen was mixed with PEG-DVS solution in a microfluidic device immediately before encapsulation. Another study performed by vanLoo et al. demonstrated that the tyramine-linked enzymatic crosslinking chemistry used in Dex-TA microgels could also be used to fabricate tyramine-conjugated hyaluronic acid microgels, although cells were not encapsulated.[32] While the requirement for specialized cross-linking functionalization and rapid in vivo degradation rate limits its scope of application, we believe HA microgels have the greatest potential for impact in the area of in vitro, 3D culture of single cells. Its readily functionalizable and highly cytocompatible chemical structure offers fine control over the cellular microenvironment, which can be used to probe cellular behavior with single-cell resolution.
2.1.5. Matrigel
Matrigel is a commercialized extract of ECM proteins which has been extensively used for culturing a wide variety of cell types in 2D and 3D environments. Interest in Matrigel as a single-cell microgel material has thus far been limited. Matrigel is derived from murine Engleberth–Holm–Swarm tumor tissue and contains ECM proteins and growth factors commonly found in the basement membrane.[91] When cultured on or within Matrigel, a variety of cell types including epithelial, endothelial, and stem cells have been observed to differentiate into complex structures. This enables the investigation of certain cellular behaviors which had been impossible to elicit when culturing on single-component materials, such as the formation of acinar structures by epithelial cells and capillary networks by endothelial cells.[91] This emergent behavior is thought to be a result of complex signaling interactions between cells and the surrounding matrix. However, the composition of Matrigel is not well defined and therefore can lead to undesirable variability in experimental results. While the primary components are known to be laminin, collagen IV, and entactin, there have been hundreds of other proteins identified within Matrigel. In particular, a diverse array of growth factors have been detected, which further complicates the interpretation of experimental results.[92] This compositional variability, and inherent complexity with its tumor tissue source, has largely hindered the use of Matrigel for in vivo therapeutic applications and likewise limited its use for some basic science research.
Both the lack of compositional definition and batch-to-batch variability have limited the appeal of Matrigel for single-cell microgel research. However, microgel droplet techniques offer unprecedented throughput and experimental control to previously established Matrigel-based culture assays. Matrigel remains a liquid at 4 °C and spontaneously polymerizes at 37 °C, making it highly compatible with microfluidic encapsulation techniques.[91] To the extent of our knowledge, the only demonstration using Matrigel as a single-cell microgel material was performed by Dolega et al.[36] Human prostatic epithelial cells were individually encapsulated and then observed as they proliferated and self-organized to characterize acinar development, a 3D cell culture assay requiring the complex mixture of basement membrane proteins found in Matrigel (Figure 4). Matrigel-based single-cell microgels may be the ideal candidate in studies that probe the development of complex, 3D cellular structures. Although prior use has been limited, Matrigel is a material which may further diversify the realm of possibility for single-cell microgel investigations.
Figure 4.
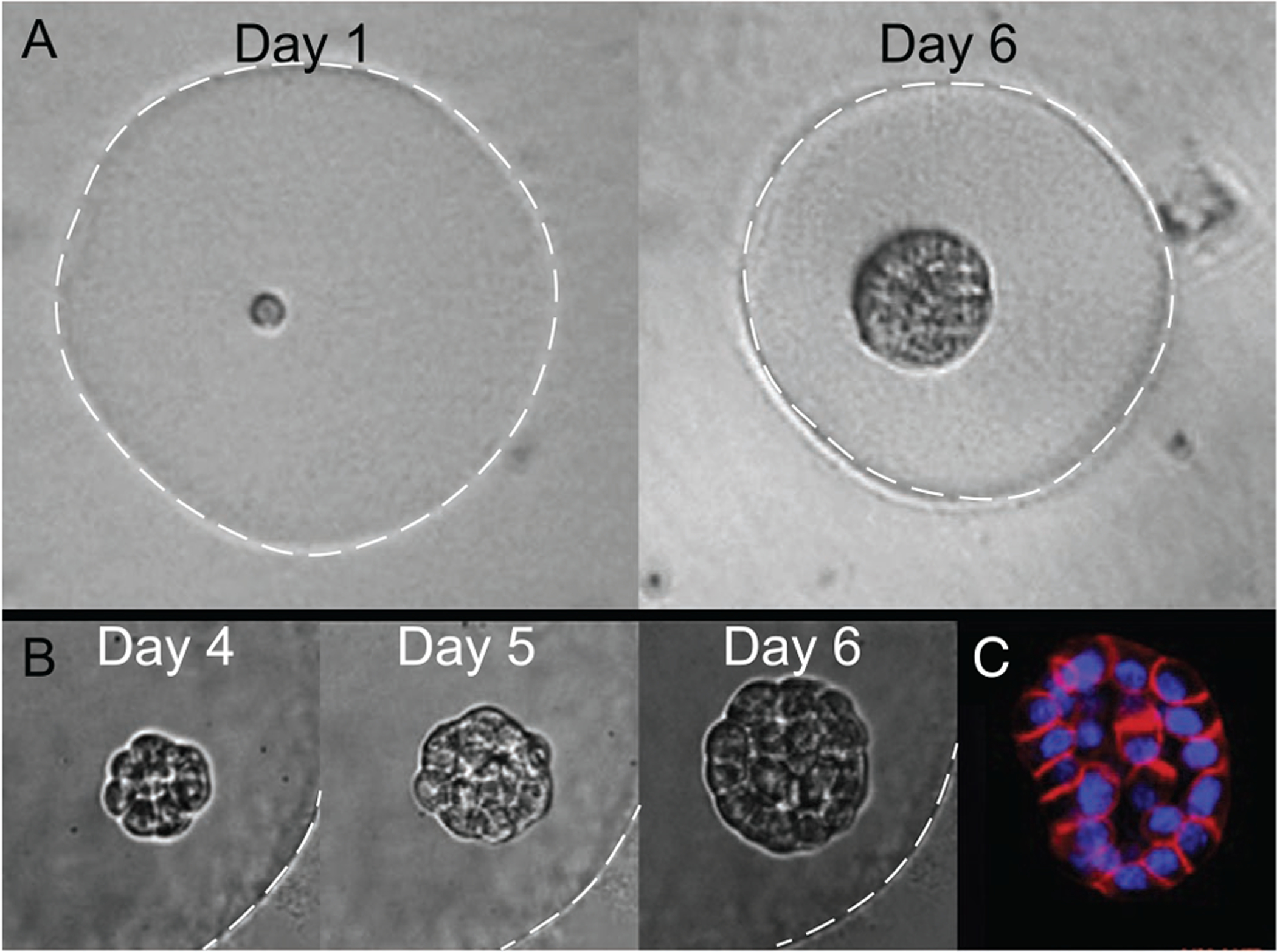
A,B) Phase contrast images of acinus growth within a Matrigel microgel over time starting from a single cell (Day 1). White dashed lines indicate microgel periphery, for clarity. C) Fluorescent staining of actin (red) and nuclei (blue) for visualizing early lumen formation.[36] Reproduced with permission.[36] Copyright 2015, Elsevier.
2.1.6. Collagen
Collagens are a group of ECM proteins which have been frequently adapted for cell culture and also sparsely explored as single-cell microgel materials. The most abundant type of collagen, collagen I, takes the structure of a triple helix and functions as the main structural component of ECM in many tissues.[93] Collagen’s role as an ECM protein confers multiple benefits for single-cell culture, including the natural incorporation of cell adhesive domains and capacity for enzymatic degradation as part of matrix remodeling processes.[93] Collagen I is most frequently extracted from natural sources such as rat tail via an acidic degradation mechanism and undergoes a well-characterized self-aggregation process in vitro when the pH is adjusted to neutral or basic conditions.[94] The characteristics of the resulting hydrogel scaffold can vary widely depending on collagen source, concentration, reaction pH, reaction temperature, and ionic strength of reaction media.[95] High sensitivity to each of these reaction parameters can introduce undesirable variability in the cellular microenvironment, demanding careful attention from researchers to ensure consistency among experiments. Self-aggregated collagen also suffers from low mechanical strength and durability, which may preclude its use for certain in vivo applications. Alternatively, a variety of crosslinking strategies have been implemented to improve the consistency of hydrogel fabrication and customize mechanical properties, such as the use of PEG[96] or riboflavin[37] crosslinkers. Gelatin, a denatured form of collagen I, has been mixed in prescribed ratios with nondenatured collagen to improve control over hydrogel mechanical characteristics (Figure 5).
Figure 5.
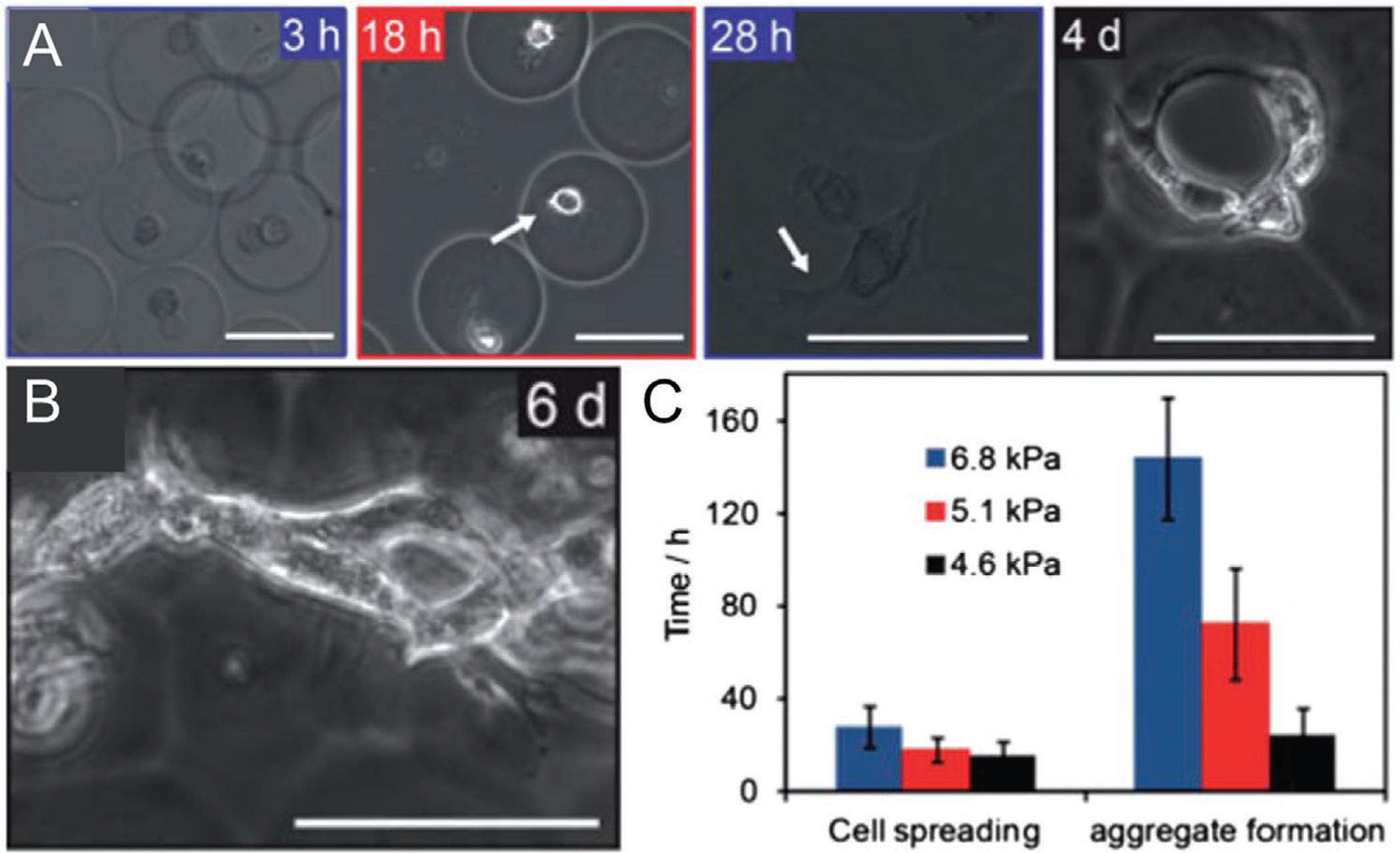
A) Bright field images of 3T3 fibroblasts spreading and invasion in collagen-gelatin microgels over time. B) Fibroblast spreading over aggregated collagen-gelatin microgels, 6 days in culture. C) Time to cell spreading and microgel aggregation for collagen-gelatin microgels of varying stiffnesses. Reproduced with permission.[37] Copyright 2013, Royal Society of Chemistry.
Subdued interest in the use of collagen for single-cell microgels is likely a result of limitations in consistency and durability. However, collagen’s cell-binding domains and biodegradability are attractive for tissue engineering or biological research applications where scaffold degradation is necessary. To the best of our knowledge, the only use of collagen for the synthesis of single-cell microgels was performed by Ma et al. In this study, collagen was mixed with gelatin at varied ratios to overcome the mechanical instability observed in pure collagen microgels.[37] To avoid premature gelation of the precursor solutions, collagen was kept at 4 °C while the gelatin was heated to between 30 and 60 °C. Mixing of the collagen and gelatin precursors was then performed on a microfluidic device immediately prior to cell encapsulation. Crosslinking was achieved using riboflavin, which initiated gelation via radical formation when exposed to blue light. Cell viability was observed to be highly dependent on duration of crosslinking, with an increase from 8 to 16 min of crosslinking causing a reduction from 90% to 20% in cell viability at 24 h. Improved control and durability of collagen formulations will enable expanded use of collagen microgels, particularly when complex mechanical and biochemical ECM cues are required to elicit a desired therapeutic response.
2.2. Synthetic Polymers
Synthetic polymers address some of the shortcomings of natural polymers, including batch-to-batch variations and in vitro/vivo longevity. However, due to less biocompatible crosslinking/gelation conditions, synthetic polymers have experienced a slower adoption for cell encapsulation, and even slower for single-cell encapsulation. Below we discuss the use of PEG, a widely used synthetic polymer in biomedical applications, as a single-cell encapsulant. Additional synthetic polymers with potential applications in single-cell microgels are mentioned at the conclusion of this section.
2.2.1. Poly(ethylene glycol)
PEG is a versatile polymer with applications spanning industrial, commercial, and medical uses. PEG is especially suitable for cell encapsulation because it is biocompatible, water soluble, and bioinert. It can be chemically modified to include bioactive molecules for facilitating cellular recognition and biodegradation. PEG is a polyether consisting of repeated ethylene glycol units.[33] Depending on the functionalization of PEG molecules, the resulting microgels can be crosslinked via photopolymerization[97] or Michael-type addition (MTA) reaction.[98] These cytocompatible crosslinking methods are possible due to the solubility of PEG in water, which is rare for many synthetic polymers. As a result, PEG has been the synthetic polymer of choice for microgel encapsulation of cells.
PEG can be easily modified to allow for cell-friendly on- or off-chip photopolymerization of cell-laden microgels. Similar to many natural polymer-based microgels, fabrication of single-cell PEG microgels can employ microfluidic devices for producing monodisperse microgel droplets. PEG is commonly functionalized with acrylates to form PEG diacrylate (PEGDA),[99] a photopolymerizable PEG derivative. Cells suspended within PEGDA pre-polymer solution containing photoinitiator can be flowed into a microfluidic droplet generator along with a cytocompatible immiscible oil solution to form monodisperse fluid droplets and crosslinked via ultraviolet (UV)-mediated radical generation.[100] Typically, a near-UV wavelength of 365 nm is used for photopolymerization of PEG microgels, which is close to the maximum excitation wavelength of a commonly used photoinitiator, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP). The water solubility of LAP is greater than other photoinitiators such as Irgacure 2959, making LAP more suitable for biological applications. Additionally, exposing cells to wavelengths near 254 nm, which is close to the maximum excitation wavelength of Irgacure 2959, has been shown to have deleterious effects on proliferation rates, viability, and protein characteristics, while no changes were noted following exposure to 365 nm wavelengths.[101] As such, LAP is a preferred photoinitiator for photopolymerizable single-cell microgels. While standard initiation protocols with 365 nm wavelengths have been shown to have insignificant effects on encapsulated cell viability, irradiation times beyond 30 s for PEGDA crosslinking can result in cell death.[97] Alternatively, PEG can be functionalized to PEG norbornene (PEGNB), which has been shown to mitigate deleterious effects of extended UV radiation on the encapsulated cells during photopolymerization.[97,102] Unlike PEGDA polymerization, polymerization of PEGNB is unhindered by the presence of oxygen and can make use of the polymerization-induced reactive oxygen species (ROS).[103] These ROS commonly accumulate during the photopolymerization of PEGDA and negatively affect cell survival by inducing apoptosis.[104] The thiol-ene linkages are formed by step-growth polymerization during PEGNB photopolymerization, while PEGDA is formed via chain-growth polymerization, giving rise to slightly different mechanical properties specifically in response to tangential stresses.[103] Step-growth thiol-ene reactions have been shown to require fewer radicals compared to chain-growth acrylate reactions,[105] along with shorter radical and polymerization times.[106] Overall, PEGNB maintains many of the attributes as PEGDA, while improving on polymerization kinetics and homogeneous polymer network formation.[107] Photopolymerization of PEG molecules permits spatial and temporal control over microgel polymerization. Although, concentration of the photoinitiator must be optimized as it can negatively impact cell viability for both PEGDA and PEGNB systems when used beyond 0.1% weight by volume.[97]
Another cytocompatible method for crosslinking PEG is by MTA reactions, which involve the addition of a carbanion to an acceptor under basic conditions.[108] More specifically, PEG crosslinked via thiol-MTA proceeds between thiolate anions and electron deficient carbon–carbon double bonds.[109] MTA does not rely on cytotoxic free radicals and UV light during the polymerization process, lessening the prevalence of cell death.[110] PEG hydrogels crosslinked in this way have been demonstrated for multiple PEG functionalizations including acrylates,[111] vinyl-sulfone,[112] and maleimides.[110] Similar to photopolymerization, MTA permits microfluidic-based approaches for producing monodisperse cell-laden microgels. Headen et al. demonstrated how multi-cell 4-arm PEG maleimide (PEG-4MAL) microgels could be produced using a microfluidic flow focusing droplet generator. First, cell-laden PEG-4MAL macromer droplets were formed within light mineral oil and surfactant. Immediately following droplet formation, a second oil phase containing an emulsion of dithiothreitol (DTT) was introduced and surrounded the liquid droplet. The DTT rapidly diffused into the droplet, crosslinking the PEG-4MAL.[98] Nucleophilic buffering agents are required for crosslinking via MTA,[113] where unfortunately some buffers (e.g., triethanolamine) can be cytotoxic to cells at high concentrations.[114] As such, cell exposure to nucleophilic buffering agents should be minimized to mitigate potential harm to the encapsulated cells. While MTAs can be applied to different PEG derivatives, reaction kinetics cannot be treated equally due to PEG-4MAL exhibiting faster reaction kinetics and tighter network structures compared to 4-arm PEG-acrylate (PEG-4A) and 4-arm PEG-vinylsulfone (PEG-4VS). Additionally, PEG-4MAL was found to require two orders of magnitude less nucleophilic buffering agent compared to PEG-4A and PEG-4VS, permitting more cytocompatible crosslinking conditions. Phelps et al. achieved crosslinking of PEG-4MAL at lower polymer weight percentages, resulting in a wider range of hydrogel stiffness compared to MTA crosslinked PEG-4A and PEG-4VS and photopolymerized PEG-DA.[110]
PEG hydrogels require additional functionalization to allow for cell-specific bioactivity, for example, adhesion, migration, and biodegradation.[115] Similar to natural polysaccharide-based microgels such as alginate and agarose, RGD adhesion peptides are commonly used to facilitate cell adhesion and migration. In a study focused on β-cell secretion of insulin, many other laminin-derived peptides, such as IKLLI, IKVAV, LRE, PDSGR, RGD, and YIGSR, and the collagen type I sequence, DGEA, were also shown to improve cell viability and preserve cell phenotype.[116] Additionally, controlled degradation of an otherwise stable and inert synthetic polymer like PEG requires further chemical modification. The mechanism of PEGDA degradation remains unclear, with multiple hypotheses existing in the literature.[117–119] Regardless of the means, the time course of degradation can range from a few weeks to many months, depending on crosslinking density and the surrounding environment.[119] Kar et al. synthesized disulfide-modified PEGDA (dPEGDA), which degrades in the presence of encapsulated cells.[120] Cell secreted molecules, such as glutathione (GSH), break down surrounding disulfide bonds in the polymer.[121] Rate of dPEGDA degradation can be tuned by modulating fraction of disulfide moieties, as well as pH, cell type, and local concentration.[120] When considering single-cell microgels, cell number plays a negligible role unless microgels are packed tightly together. Alternatively, matrix-metalloproteinase (MMP)-sensitive sequences can be conjugated to PEG-based hydrogels to allow for cell-mediated degradation of the hydrogel encapsulant. In particular, MMP-sensitive sequences have been introduced into transglutaminase-PEG (TG-PEG) microgels containing single cells.[122] Secretion of MMPs allows for degradation of the surrounding TG-PEG matrix, permitting proliferation, spreading, and migration of encapsulated cells similar to that observed in vivo.[123] Biodegradation is particularly useful for cell transplantation and tissue engineering applications, since the cell-laden microgels can be considered autonomous, requiring no post-encapsulation intervention to initiate hydrogel degradation.
2.2.2. Other
Synthetic polymers other than PEG often require the use of harmful solvents which has limited their use as a cell encapsulant. That being said, a few notable PEG alternatives are available such as polyacrylate derivatives like poly(hydroxyethyl methacrylate-co-methyl methacrylate) (HEMA-MMA)[124] and poly((2-hydroxyethyl)methacrylate-co-(3-aminopropyl)methacrylamide) (P(HEMA-co-APM)).[125] P(HEMA-co-APM), when coupled with RGD-mimicking poly(amidoamine) (PAA) moieties, improves biocompatibility and allows for biodegradation, while maintaining mechanical integrity of the unmodified polymer. NIH3T3 murine embryonic fibroblasts have been encapsulated in P(HEMA-co-APM)/PAA, with no significant effect on cell viability compared to a gelatin methacrylate encapsulated group.[125] Unfortunately, polyacrylate derivatives exhibit low permeability, which has limited further adoption of these polymers.[33]
As opposed to a single-material approach, synthetic polymers have been increasingly used in conjunction with natural polymers to leverage inherent benefits of the two polymer classes. Briefly, natural polymers allow for better cytocompatibility and cell–matrix interactions which are critical for directed cell behavior, while synthetic polymers exhibit long-term mechanical/chemical stability and enable greater control when tuning whole microgel mechanical properties. Many multi-cell encapsulation studies have used PEG in concert with natural polymers like alginate,[126] agarose,[127] hyaluronic acid,[128] or elastin and gelatin.[129] Given the advantages imparted by these material combinations, we believe applications such as long-term in vitro culturing models and cell therapies could benefit from these multi-polymer approaches.
3. Generation of Single-Cell Microgels
Conventional emulsification techniques (e.g., extrusion-based methods) have commonly been adapted over the past 50 years to fit the material and processing requirements for generating cell-laden microgels.[132] This is particularly true for producing multi-cell microgels; however, translation to single-cell microgel production has not been as straightforward. In these applications, primary motivation has been to obtain microgel characteristics that can ensure sufficient nutrient and oxygen transport to maintain high cell viability while also managing host immune response.[133] These characteristics include microgel size,[134] monodispersity,[135] surface smoothness and charge,[136,137] mechanical and chemical properties,[33] and mass transport properties.[31] As a result, new methods capable of generating cell-laden microgels of pL to nL volumes with high monodispersity are being developed (e.g., microfluidic droplet devices). While not obvious, monodispersity plays a key role in producing single-cell microgels of defined diffusion and degradation kinetics while also influencing the biodistribution in certain applications,[138,139] hence an emphasis on fabrication methods capable of producing microgels with good monodispersity. Below, we discuss conventional techniques that have been utilized for producing cell-laden microgels as well as more recent techniques that have been developed to address shortcomings of the original methods.
3.1. Extrusion-Based Single-Cell Microgel Fabrication
Extrusion-based techniques involve dispensing a hydrogel precursor solution containing cells through a narrow orifice (e.g., needle) into a crosslinking environment. Passive (lack of external force) and active (presence of external force) methods have been demonstrated using extrusion-based methodologies with varying successes and limitations. Gravitational extrusion via dripping is the simplest passive extrusion-based technique and relies on competition between the gravitational force on the growing droplet and surface tension of the liquid. Once the droplet reaches a critical size, it is released from the nozzle into a suitable reservoir for microgel crosslinking. However, this passive process results in large microgels (>1 mm diameter),[58] which limits mass transport through the hydrogel matrix to the cell-laden core. Due to the absence of a controllable external force that is capable of manipulating droplet pinch-off, researchers can only vary the hydrogel precursor parameters (e.g., surface tension, viscosity, and density) and the diameter of the nozzle orifice.[140] It is possible to further decrease the overall size of cell-laden microgels by increasing the flow rate of the extruded fluid. This transitions droplet formation from dripping to jet-breaking regimes. By further increasing the flow rate, the spray regime can be achieved.[141] The transition from dripping to jet-breaking is characterized by the minimum average velocity within the orifice (i.e., minimum jet velocity), umin, j[142]
| (1) |
where γ is the surface tension of the liquid, ρ is its density, and dj is the liquid jet radius, which can be approximated to the internal diameter of the nozzle orifice. To prevent collisions between droplets, which can result in droplet coalescence, it is important to limit the velocity of the jet to less than the terminal drop falling velocity.[141] While the jet-breaking or spray regimes allow for production of smaller microgels at higher throughput compared to the dripping regime, the minimum achievable microgel size is still too large in this conventional approach for many single-cell microgel applications.[141]
The integration of tunable secondary forces (e.g., shear and electrostatic) have further decreased the minimal achievable microgel size for extrusion-based techniques. Using a vibrating nozzle, Mazzitelli et al. encapsulated neonatal porcine islets within alginate, achieving microgel sizes below 500 μm with good monodispersity and minimal impact on porcine islet viability, morphology, and functional properties.[143] A nozzle with an internal diameter of 300 mm was used in this study, which dictated an approximate, lower limit on the microgel size. It would be possible to incorporate a smaller nozzle size to produce single-cell microgels with dimensions theoretically more appropriate for optimal mass transport (e.g., <100 μm). Furthermore, early work by Lindblad and Schneider showed that matching the wavelength of the nozzle vibration to between 7 and 14 times the fluid jet diameter produced droplets with high monodispersity, where the droplet diameter, dd can be roughly approximated with dd ≈ 1.64dc, where dc is the inner diameter of the nozzle.[142] This was demonstrated for droplets ranging in diameter from 50 to 700 μm. In the absence of physically changing the nozzle size, the integration of a periodic signal permits controllable droplet size down to levels suitable for single-cell microgels.
Potentially the most promising technique for large-scale microgel production is electrohydrodynamic spraying (i.e., electrospray), which creates a stream of individual droplets from the nozzle. More generally, electrohydrodynamic droplet generation relies on the disruption of the fluid interface with an applied, high electric potential on the extrusion nozzle to reorient ions to the fluid surface, which leads to a repulsion of like charges and reduction in surface tension.[33] This overcomes the surface tension and causes a deformation in the liquid stream and subsequent release of charged droplets with high monodispersity.[144] Different droplet-forming modes are achieved by varying the electrical potential which controls the magnitude of electric stresses at the fluid surface.[145] When a critical electrostatic potential is reached, electrospraying occurs. The electrospray technique has been demonstrated to produce droplets ranging from ≈2 mm down to tens of nm with high monodispersity.[33] Droplet size can be tuned by modulating the electrical potential, inner diameter of the extrusion nozzle, flow rate of the extruding fluid, and the electric (dielectric constant) and rheological (surface tension) characteristics of the extruding fluid.[146,147] Since an increase in electric potential results in the formation of smaller droplets, near a critical electrostatic potential surface tension becomes negligible. The critical electrostatic potential, Uc, can be approximated by
| (2) |
where γ0 is the surface tension of the fluid with zero applied electric potential, and ε0 is the air dielectric constant.[146]
To the best of our knowledge, only one demonstration of single-cell microparticles using the electrospray technique has been reported. Using a design of experiment (DOE) methodology to optimize encapsulation efficiency, microparticle yield, and microparticle size, Esfahani et al. successfully encapsulated NIH3T3 cells within poly(lactic-co-glycolic acid) (PLGA) while only maintaining cell viability at 76%.[148] While this shows the feasibility of using the electrospray technique for generating single-cell microparticle, improvements in cell viability are necessary. This may be challenging given the high shear forces present during droplet formation. Qayyum et al. investigated how electric potential, flow rate, nozzle inner diameter, cell type, and cell density affected microgel size, monodispersity, and cell viability of multi-cell PEG-based microgels. In particular, increases in electrical potential negatively affected cell viability, while flow rate and nozzle inner diameter had no statistically significant impact.[147] While this demonstration produced multi-cell microgels, the relationships between experimental parameters and resulting cell-laden microgels can be translated to the fabrication of single-cell microgels. Cell viability values of >95% have been reported in multi-cell microgels fabricated using electrospray methodologies, which serve as a benchmark for future investigations into the production of single-cell microgels using this technique.[149]
Extrusion-based methods remain a viable option for the production of single-cell microgels, but innate processing limitations may hamper their adaptability. In particular, the excess charge generated on the droplet surface can cause encapsulated cells to migrate toward the droplet interface. While this is advantageous for multi-cell microgels (e.g., improved mass transport), there is a potential for off-centering of encapsulated cells, which has been associated with accelerated cellular egress from microgels.[25] Second, cell-laden droplets generated using extrusion-based methods typically fall into a reservoir or surface for crosslinking. This rapid collision and subsequent deceleration may result in non-spherical microgels,[150] off-centered cell positioning within the microgel, and damaging stresses to the encapsulated cell.[151] One possibility for addressing the non-spherical microgel morphology is to make the droplet curing surface superhydrophobic, which more strongly forces the liquid hydrogel precursor/cell suspension into a spherical orientation.[152] Lastly, extrusion-based techniques can expose cells to high levels of shear, which can be detrimental to cell viability and proliferation.[153,154] While these limitations are inherent to extrusion-based techniques, the negative impact on cell viability and biochemical processes can be minimized by careful consideration and optimization of processing parameters (e.g., flow rate, nozzle diameter).
3.2. Leveraging Microfluidics for Optimizing Monodispersity and Capture Efficiency
Microfluidic-based technologies present a unique platform for manipulating fluid flows and cell behaviors with high precision by integrating device features and force gradients that are of comparable scale to that of cells.[155] The field of microfluidics is vast, but one subset in particular, droplet microfluidics, has seen much growth over the past decades.[156] Droplet microfluidics has improved the consistency of single-cell encapsulation due to its capability to create highly predictable, tunable, and monodisperse populations of microgels with uniform mechanical and chemical properties.[157] Similar to other sections, below we highlight the techniques we believe to be most promising for single-cell microgel production. We encourage readers to refer to other reviews regarding microfluidic-based droplet generation and single-cell encapsulation for more extensive detail.[155,157–161]
3.2.1. Microfluidic-Based Droplet Generation
Droplet microfluidics, although a subcategory of microfluidics, is fairly extensive in its own right. This is due in part to the numerous applications that have benefited from the development of droplet microfluidic platforms, including single-cell -omics,[15] particle synthesis,[162,163] and cell culture.[46] The controlled generation and manipulation of liquid droplets suspended in a fluid medium is fundamental to droplet microfluidics. Similar to conventional methods, microfluidic-based droplet generation can also be classified into two major categories, passive and active. Passive droplet generation is primarily controlled by the flow rates and physical properties of the two immiscible fluids and the device channel geometry.[164] Active methods integrate a controllable external force, such as electrical or acoustic fields, that initiates droplet formation, typically by introducing a pressure gradient.[155] While these two methods vary with respect to the mechanism that initiates droplet generation, the physics behind droplet formation are shared. For brevity, we will refer to the internal, droplet-forming fluid as the dispersed phase (e.g., hydrogel precursor containing cells) and the surrounding fluid as the continuous phase (e.g., fluorinated oil and accompanying surfactant). For the continuous phase, choice of oil and surfactant combination is critical for preserving cell viability and phenotype. Fluorinated oils (e.g., Novec HFE-7500) and fluorosurfactants (e.g., 008-FluoroSurfactant) are a preferred combination for the generation of cell-laden droplets. Fluorinated oils offer high solubility and diffusion of oxygen, thereby reducing the likelihood of exposing cells to hypoxic conditions.[165] Retrieval of newly fabricated single-cell microgels from the continuous phase remains a critical step for maintaining long-term cell viability. Typically, manual washing steps via centrifugation are slow and can expose cells to extended periods of stress, while on-chip isolation methods provide a faster and facile way of retrieving single-cell microgels from the continuous phase.[161]
There are many dimensionless parameters that physically describe microfluidic-based droplet generation (e.g., Reynolds number, Weber number, and Bond number), but for simplicity we focus primarily on the capillary number (Ca). The capillary number is the ratio of viscous force to interfacial tension, Ca =μU/γ, where μ is the fluid dynamic viscosity, U is the fluid velocity, and γ is the interfacial tension between the two fluids. Ca dictates the droplet generation regime, where increasing the Ca will result in a transition from squeezing to dripping, and increasing the Ca further will cause a transition to the jetting regime (Figure 6A).[166,167]
Figure 6.
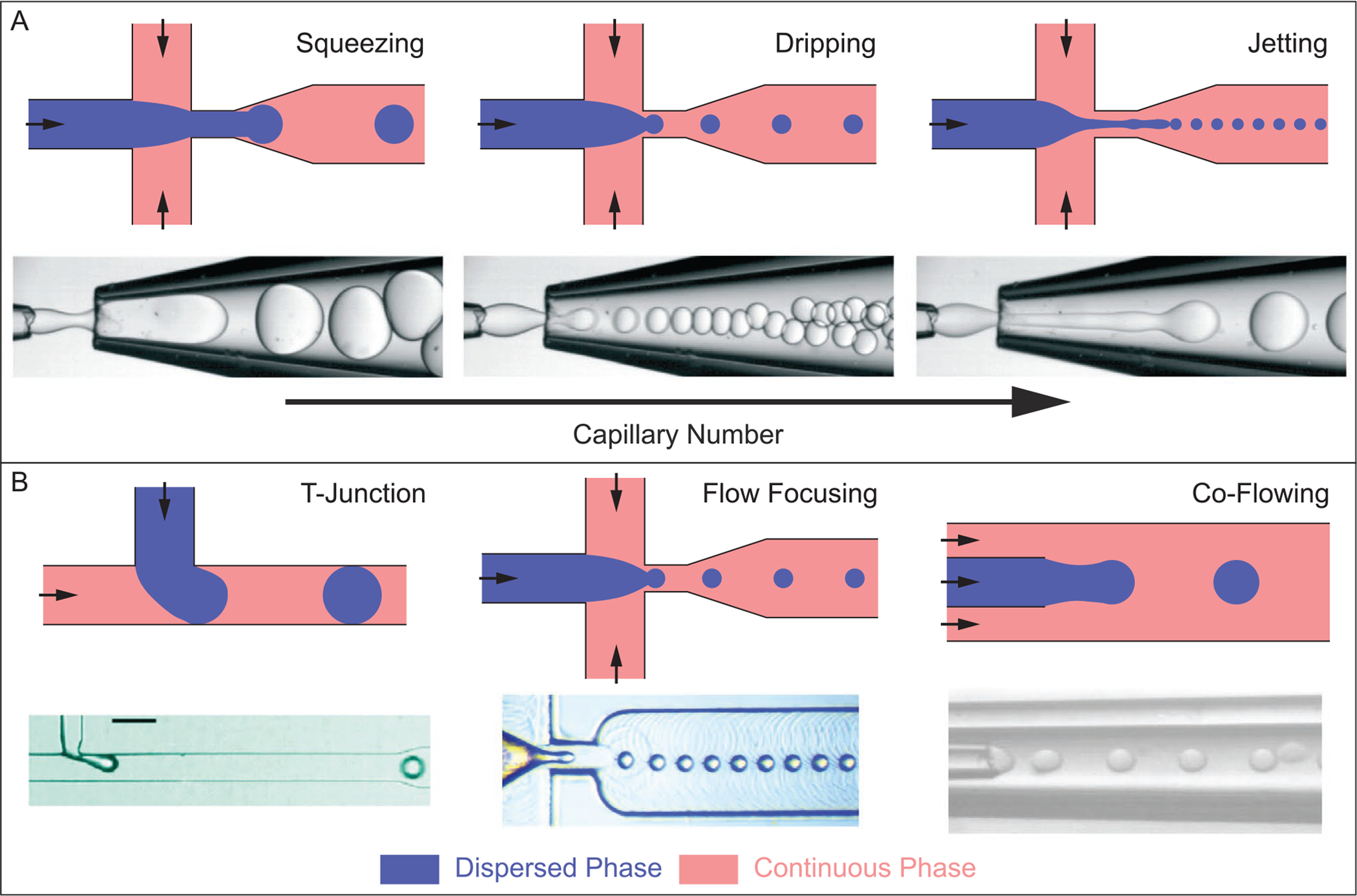
A) Common droplet formation regimes: squeezing, dripping, and jetting. From left to right, increasing the capillary number, Ca, causes a transition from squeezing to dripping to jetting. Increasing Ca typically results in smaller droplet formation, and higher throughput. Channel geometry shown is the flow focusing orientation, but other device geometries follow similar trends for droplet generating regimes. B) Microfluidic-based droplet generator geometries commonly used for producing monodisperse droplets. Images of droplet generation regimes (A) are reproduced with permission.[157] Copyright 2016, Royal Society of Chemistry. Image of T-junction geometry (B) was reproduced with permission.[168] Copyright 2010, American Chemical Society. Image of flow focusing geometry (B) was reproduced with permission.[169] Copyright 2019, Royal Society of Chemistry. Image of co-flowing geometry (B) was reproduced with permission.[170] Copyright 2016, Springer Nature.
In addition to Ca, the geometry of a microfluidic device critically influences droplet formation. Three common device geometries are the T-junction,[168] flow focusing,[171] and co-flowing[172] configurations (Figure 6B). While the designs look quite different, they implement similar physical processes. An interface is formed between the two immiscible fluid streams, where a small volume of dispersed phase collapses into a droplet, effectively separating from the main dispersed stream and becoming surrounded solely by the continuous phase.[155] For the fabrication of single-cell microgels, device channel walls are made hydrophobic through various surface treatment methods.[173,174] Hydrophobicity of channel walls facilitates preferential wetting of the oil continuous phase, ensuring that the aqueous cell and hydrogel precursor droplets do not contact or adhere to the walls of the device. Many microfluidic-based droplet generators are fabricated using standard soft lithography methods with poly(dimethylsiloxane) (PDMS),[175] but devices constructed of coaxial assemblies of glass capillary tubes are also viable options.[176–179] The device landscape for microfluidic-based droplet generators is vast and operating parameters can vary greatly depending on the application, making it a very versatile technology worth adopting for those looking to fabricate highly monodisperse and uniform single-cell microgels.
3.2.2. Integration of Upstream Focusing for Controlled Single-Cell Encapsulation
Droplet-based microfluidics improves upon the size, monodispersity, and shape limitations commonly found in more macro-scale techniques (e.g., extrusion, bulk emulsion). However, continual improvements are needed for encapsulating single cells within their own microgels with high efficiency (i.e., very little empty or multi-cell-laden microgels) and at high throughput (>10 kHz) to minimize the processing time with living cells. Typically, cells suspended within the dispersed phase are randomly spaced within the fluid stream causing the arrival time at the droplet orifice to be random as well. This is not an issue for multi-cell microgels as cell loading can be approximated by a Gaussian distribution.[155] When single-cell encapsulation is desired and multi-cell encapsulation needs to be minimized, cell loading tends to follow the Poisson distribution P(X = k) = λke−λ/(k!).[180] k is the number of cells within the droplet and λ is the average number of cells per droplet, which can be calculated: , where Cc is the cellular concentration in the hydrogel precursor solution and is the average droplet volume. From the Poisson probability distribution equation, we can see that setting the average number of cells per droplet to one, λ = 1, results in an equal quantity of vacant and single-cell-laden droplets (≈37%) and >26% multi-cell-laden droplets. Reducing λ will effectively reduce the probability of multi-cell droplets, while the percent of cell-free droplets will increase as a result. For example, if λ = 0.05, ≈95% of droplets will be empty, and <5% of droplets will contain a single cell. Another way to envision cell loading is by looking at the acceptable percentage of multi-cell droplets (Figure 7C). By establishing the acceptable limit of droplets containing more than one cell, it is possible to calculate λ and the required cell concentration. While this process seems straightforward to implement, Poisson loading of cells in droplets assumes cells are randomly suspended within the dispersed phase. In practice, cell settling within the infusion vessel can occur resulting in inconsistent distribution throughout the encapsulation run. These inconsistencies in the cell suspension prevent researchers from achieving predictable encapsulation efficiencies for a given, predetermined cell concentration. To combat this, density matching reagents such as OptiPrep[9,181] or continual agitation of the cell suspension[182] can be implemented. It should be noted that continual agitation can damage or cause physiological changes to the cells and also has to be carefully managed to avoid affecting downstream fluid behavior. Maintaining a homogeneous suspension of cells prior to encapsulation is necessary for calculating appropriate cell concentrations to achieve desired single-cell encapsulation efficiencies using Poisson loading. By maintaining a consistent cell suspension and targeting a low value for λ, it is possible to minimize the number of multi-cell droplets at the expense of low encapsulation efficiency which demands longer experimental time, more reagents, and additional downstream processing to isolate single-cell droplets.
Figure 7.
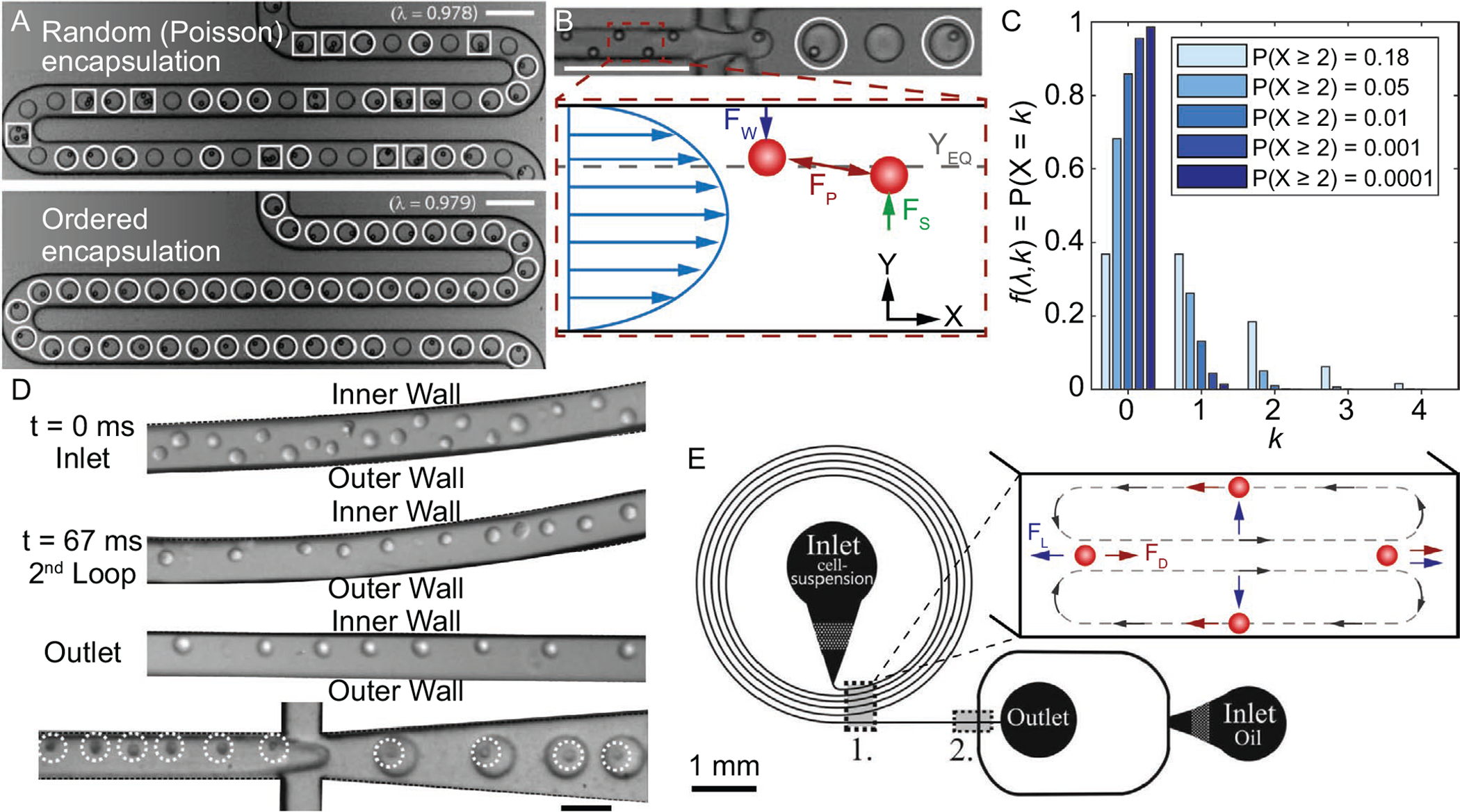
A) (Top) Stochastic (i.e., Poisson) encapsulation of polystyrene microbeads. (Bottom) Inertially ordered encapsulation with similar λ value, 0.98.[183] B) (Top) Inertial ordering of polystyrene microbeads, upstream of droplet formation for ordered encapsulation shown in (A).[183] (Bottom) Schematic of inertial ordering forces within a microchannel, where FW is the wall-induced lift force, FS is the shear-induced lift force, FP is the repulsive force, and YEQ is the equilibrium focusing position. C) Theoretical cell loading density for different probabilities of multi-cell droplet loading events, assuming Poisson distribution. D) Images of cell ordering progression in curved microchannel and subsequent encapsulation.[190] E) Schematic of microfluidic cell encapsulation device with (1) curved microchannel section for focusing cells upstream of (2) droplet formation.[190] (Inset) Schematic of forces present within a curved microchannel, where FD is the Dean force and FL is the net lift force. (A,B) Reproduced with permission.[183] Copyright 2008, Royal Society of Chemistry. C,D) Reproduced with permission.[190] Copyright 2012, Royal Society of Chemistry. Scale bars denote 100 μm for (A) and (B), and 50 μm for (D).
Integrating a method to predictably order and space cells prior to encapsulation should increase encapsulation efficiency and throughput, while also reducing the number of cell-free and multi-cell-laden droplets. The concept of inertial ordering of cells upstream of encapsulation has been demonstrated by Edd et al. (Figure 7A).[183] Polystyrene microparticles and cells were inertially ordered within a rectangular microchannel such that lateral and longitudinal spacing was stable for contiguous particles/cells. Inertial ordering is driven by the presence of two competing forces, termed the shear- and wall-induced lift forces, and the hydrodynamic repulsion effect, as shown in Figure 7B.[155] The shear-induced lift force, which acts down the shear rate gradient toward channel walls, is driven by the shape of the fluid velocity near the particle.[184] The wall-induced lift force, which is directed toward the channel center, depends on the proximity of the nearby wall and the resulting disruption on the axisymmetry of the wake vorticity that is generated on the particle’s surface.[185,186] These two forces act to stabilize the transverse particle focusing, while also providing feedback to the ever increasing longitudinal particle-particle spacing, which is driven by reflected viscous disturbance flows from rotating, neighboring particles.[187] Using this technique, Edd et al. demonstrated single-cell encapsulation efficiency at ≈60% while keeping multi-cell encapsulation events at <5%, with many of the multi-cell events occurring in part due to pre-existing cell aggregation.[183] Following their experimental data trends, it is theoretically possible to achieve ≈97% single-cell encapsulation efficiency, with negligible multi-cell encapsulation events. This implementation would require cells to occupy more than one position in the stream relative to the channel cross section, but it is possible to reduce these terminal transverse positions to a single entity via secondary flows. Secondary flows can be generated within a spiral microchannel due to the presence of centrifugal force that shifts the maximum fluid velocity point toward the concave channel wall.[188,189] These counter-rotating secondary flows, termed Dean vortices, displace cells to a single stream near the convex wall due to an imbalance between the net Dean force and net inertial lift force (i.e., shear- and wall-induced lift forces), shown in Figure 7E.[189] Kemna et al. integrated a spiral microchannel upstream of a flow-focusing droplet generator to pre-align cells to a single transverse location in the stream with deterministic longitudinal ordering (Figure 7D,E).[190] More recently, Li et al. saw a 300% increase in cell utilization (i.e., single-cell and barcoding-bead coencapsulation) when pairing spiral and serpentine microchannels to independently focus cells.[191]
Leveraging inherent fluid behavior at finite Reynolds numbers (Re, ratio of inertial to viscous forces in a fluid) enables predictable encapsulation of single cells within droplets at rates similar to many high-throughput droplet generators (10s of kHz). Re is defined by Re = ρDhUm/μ, where Um is the maximum flow velocity, Dh is the hydraulic diameter of the channel (Dh = 2WcHc/(Wc + Hc), Wc is the width, Hc is the height of the channel, and ρ and μ are the density and viscosity of the fluid, respectively.[187] Typical values for Re in inertial devices are 1 ≤ Re < 400. The examples described above utilized cells that were suspended within a fluid of similar viscosity to water (≈1 mPa s). Most hydrogel precursor solutions are more viscous than water,[21] which when flowed at similar rates and channel dimensions results in reduced values of Re (i.e., reduction in inertial forces relative to viscous forces). When fluid inertial forces are not present, cells cannot be predictably ordered and aligned within a microchannel without applied external fields. One potential solution is to independently flow a low viscosity cell suspension (dispersed phase one) and a hydrogel precursor solution (dispersed phase two) and allow for the two dispersed solutions to mix immediately before droplet formation. While this alternative approach seems straightforward to implement, additional factors would need to be considered such as appropriate mixing of the two dispersed phases for uniform hydrogel structure and suitable Re given a reduction in the individual flow rates of the two dispersed phases.
3.2.3. Downstream Purification and Processing
Another approach to achieving a more purified population of single-cell microgels is to first encapsulate cells with low efficiency (i.e., low average number of cells per droplet, λ ≪ 1) and then sort the resulting microgels downstream to select those containing single cells.[192,193] Microfluidics-based sorting approaches leverage inherent differences in the physical, mechanical, optical, and/or electromagnetic characteristics of particles passing through a microfluidic device.[155,194] Active sorting methods, which function via externally applied forces, have used acoustic fields,[195–199] electric fields,[200,201] magnetic fields,[202,203] and optical traps/forces.[204,205] Passive sorting methods have also emerged as viable options for isolating cells, particles, and droplets.[189,206,207] These approaches have the advantage of typically being higher throughput and easily parallelizable but lack the ability for real-time tuning.[193] Some of the more common passive sorting methods include the previously described straight channel inertial[208] and spiral-induced secondary flow focusing[206] approaches in addition to deterministic lateral displacement.[209] While many of these technologies have been developed for single-phase applications, it is conceivable that they can be adapted for purifying single-cell microgels as well. Already, dielectrophoresis-based microfluidic sorting of fluorescently activated droplets has been used with binary populations of enzymatically activated Escherichia coli encapsulated within liquid droplets at rates ≈2 kHz, with <0.01% and ≈0.1% false positive and negative error rates, respectively.[210] More recently, Caen et al. used similar techniques to sort droplets of varying fluorescence intensity at 200 Hz.[211] Using a gapped divider rather than the standard hard divider for the outlet channel bifurcation, Sciambi and Abate were able to purify an input sample with a starting purity of 6.4% target droplets to 99.3% target droplets at 30 kHz.[212] These examples all used a fluorescent reporter to optically distinguish cell-laden droplets from cell-free droplets, with the cell population being stained prior to encapsulation.
As an alternative to fluorescence, some sorting methods use label-free approaches, which may be advantageous for therapeutic applications requiring unstained or unmodified cells. Using surface acoustic waves,[213] Nam et al. could separate cell-laden microgels based on the number of cells each contained.[214] Density differences between cell-free and cell-laden microgels allowed for sorting selectivity, such that higher density microgels in flow had an increased acoustic contrast factor compared to less dense microgels. Microgels of higher acoustic contrast migrated to standing wave pressure nodes present within the fluid channel at a faster rate than those with lower contrast.[215] This initial study demonstrated that cell loading quantity was distinguishable within an acoustic field, albeit requiring low throughput (≈40 Hz).[214] That said, other microfluidic bulk-acoustic methods have been capable of separating cells of varying size, density, and stiffness at rates >50 MHz,[198,216] so with further development it should be possible to sort cell-free and single-cell microgels at rates exceeding optics-based approaches. Recently, Li et al. demonstrated size-dependent sorting of cell microgels from cell-free microgels in a straight microchannel.[207] Using their specific channel geometry, the authors reported a maximum throughput of ≈1.5 kHz. They noted that further optimization of the sample concentration and Re along with channel parallelization could further increase the throughput. Due to the low complexity of the system, creating a parallel network of these straight channel devices would be simple to develop, with throughput scaling linearly with the number of channels.
Methods initially developed for the separation and purification of cells within single-phase flows can potentially be adapted for the purification of single-cell microgels with high specificity (>90%). Many of the techniques described above provide sufficient performance for many single-cell microgel applications, but additional application-specific optimization could result in better single-cell microgel purity and throughput. It should be noted, trade-offs among throughput, purification, and cell recovery are commonly observed with many sorting methods.[217–219] As such, researchers must prior-itize their chosen metrics. Passive methods tend to be easier to implement, whereas active methods allow for greater control and tuning in real time. To make an informed decision on which microfluidic-based purification approach is appropriate for a specific application, the reader is referred to one of the many review articles discussing these microfluidic-based techniques in more detail.[161,193,220–222]
4. Applications for Single-Cell Microgels
4.1. In Vivo Therapeutics
Cell-based therapies and research strategies involving hydrogel encapsulation have been investigated for the past 40 years, with many applications focusing on diabetes, regenerative medicine, and cancer treatment.[33] Due to their established history, different applications of cell-based therapies vary in maturity. For example, treatment of type I diabetes using microencapsulated islets of insulin-secreting Langerhans cells was initially introduced in 1980.[223] As a result, some of these novel therapies have already reached phase II clinical trials, with phase III trials expected in the near future.[33,224,225] While the majority of research in cell-based therapies has focused on multi-cell microgels, for reasons described below, single-cell approaches are also being explored. To that end, we have focused this review on potential applications that can benefit from single-cell microgels. For additional information on the larger field, the reader is referred to other review articles focusing on applications that use multi-cell microgels.[22,33,226,227]
4.1.1. Tuning Microgel Composition for Improved Protection and Usability of Cellular Cargo
The characteristics of an encapsulating hydrogel are central to its capability to protect its cargo against external factors such as host immune response and mechanical stresses, while also permitting appropriate mass transport for maintaining cell viability. Commonly, researchers tune the hydrogel pore size for controlling permeability, while the appropriate hydrogel stiffness and functionalization can aid in the protection and phenotypic maintenance of the encapsulated cell. These alterations are conceptually simple to implement by modulating the composition percentages of monomer/crosslinker and the polymerization method and conditions.[228] The ability to control these characteristics at cell-relevant scales illustrates the adaptability of hydrogels for encapsulating cells for in vivo therapies.
The precise control of the hydrogel pore size has been shown to be a crucial method of immunological protection of implanted cells (Figure 8).[21,33] Typical hydrogel pore size is ≤0.4 μm, which prevents direct contact between the encapsulated cell and host immune cells. This physical separation prevents direct transplant allorecognition and as a result, investigation into using both allogeneic and xenogeneic sources for transplantation therapies has grown.[33,229–231]
Figure 8.
A) Confocal image of an encapsulated chondrocyte, with cell membrane stained red and nucleus blue. The PEGDA encapsulant (black) prevents fluorescently labeled dextran (green) from reaching cellular cargo; scale bar denotes 10 μm.[31] B) (Left) Off-center cell positioning within a Dex-TA microgel, which leads to early cell egress. (Right) Centered cell positioning within a Dex-TA microgel for prolonged protection from host immune response and long-term in vitro cell culture.[25] C) (Left) Image of PEGDA single-cell (green) microgels integrated into a fibrin macrogel preventing permeation of fluorescently labeled 70 kDa dextran (red) while permitting permeation of BSA (middle and right); scale bar denotes 25 μm.[31] Confocal images in (A) and (C) plots are reproduced with permission.[31] Copyright 2016, Wiley-VCH. Off-center and centered cell positions in microgel images are reproduced with permission.[25] Copyright 2017, Wiley-VCH.
While hydrogel-mediated separation of allogeneic cells from host immune cells can mitigate the immune system’s direct recognition pathway, the problem of indirect recognition still persists.[230] Typically, indirect recognition by the immune system is associated with xenogenic implantation, which is an area of interest due to the shortage of human organs for transplantation. When xenogenic cells are implanted in host animals, they are rejected, even if the animal is immunosuppressed.[229,232] The primary host immune response that is responsible for xenorecognition involves the innate immune system (e.g., natural antibodies, complement, and natural killer cells).[233] The molecules responsible for this pathway (e.g., antibodies ≈150 kDa, complement components ≈10–550 kDa, and cytokines ≈8–80 kDa) are of similar size to nutrients, growth factors, and cellular byproducts.[234] As such a balance must be struck to allow for sufficient mass transport of critical components, while alleviating deleterious molecules.[21] The complex 3D structure of the hydrogel can slow the permeation of complement components enough to hamper cytolytic capabilities.[235] Schneider et al. utilized alginate-based micro-encapsulation of pancreatic islets to extend xenogeneic islet grafts from 4–8 days for nonencapsulated islets to >7 months for encapsulated islets. Encapsulated cells that were retrieved 10 and 36 weeks post-transplantation exhibited >85% viability with minor cellular reactions along the alginate surface, suggesting the root cause for a reduction in glucose clearance rate was pore nutrient and oxygen supply rather than complement-dependent responses.[236] While this study used multi-cell microgels, it demonstrated the importance of cell encapsulation within a selectively permeable substrate for improved in vivo longevity and therapeutic efficacy, which should also be applicable in cases of single-cell microgel transplantation.
The size of a single-cell microgel has been shown to positively correlate with the level of fibrosis around it as well as influence in vivo retention and therapeutic effect.[21] Initial studies using standard microcapsules on the order of ≈1 mm in diameter resulted in a more intense fibrotic response post-implantation compared to microcapsules <350 μm.[237] Additionally, Sakai et al. showed that microcapsules <100 μm in diameter resulted in a 15% decrease in fibrotic overgrowth compared to microcapsules of 387 μm when implanted in the peritoneal cavity.[238] A reduction in overall microgel size benefits the encapsulated system twofold. First, host immune response is decreased; second, better mass transport results from having a thinner hydrogel coating surrounding the cell, permitting freer movement of essential nutrients, therapeutic factors, and waste. While it appears that making microgels as small as possible is beneficial, studies looking at microcapsule retention as a function of size suggests the optimal range is likely application specific.[138,139] Microcapsules of 10 μm in diameter were retained within inflamed, mice joints, while smaller microcapsules escaped.[139] Albeit these two studies used polyester-based microcapsules, the key takeaway of microparticle size affecting retention and biodistribution still applies to hydrogel-based microparticles. While other characteristics, such as surface charge and mechanical properties, also contribute to the retention and biodistribution of microparticles, control over microgel size is made easier with recent developments in fabrication processes. The final size of the microgels should be tailored to the implantation method, terminal location, and projected longevity.
Growth factors[239] and tissue-specific decellularized ECM[240] have been incorporated into microgel matrices to improve viability and direct differentiation and/or protein synthesis of the encapsulated cells.[241] In vitro cultured MSCs, in particular, provide a unique opportunity for cell-based orthopedic therapies due to their ease of isolation, immunomodulatory and immunosuppressive nature, and trophism.[242] MSCs can be differentiated through stimulation with specific growth factors and mechanical environments, including the stiffness of the encapsulating material (e.g., stiff microgel matrix for osteo-genesis or soft for adipogenesis).[243] As shown in Figure 9, MSCs encapsulated in stiffer hydrogel matrices expressed greater levels of alkaline phosphatase (ALP), a common marker used for assessing osteogenesis. The elevated ALP expression observed in the stiffer microgel for these two studies appears agnostic to the hydrogel material, as Mao et al. demonstrated using alginate (Figure 9A), while Lienemann et al. used TG-PEG (Figure 9B).[46,123] Encapsulating stem cells within a 3D hydrogel matrix provides additional advantages beyond a growth scaffold for directed differentiation and phenotypic maintenance.
Figure 9.
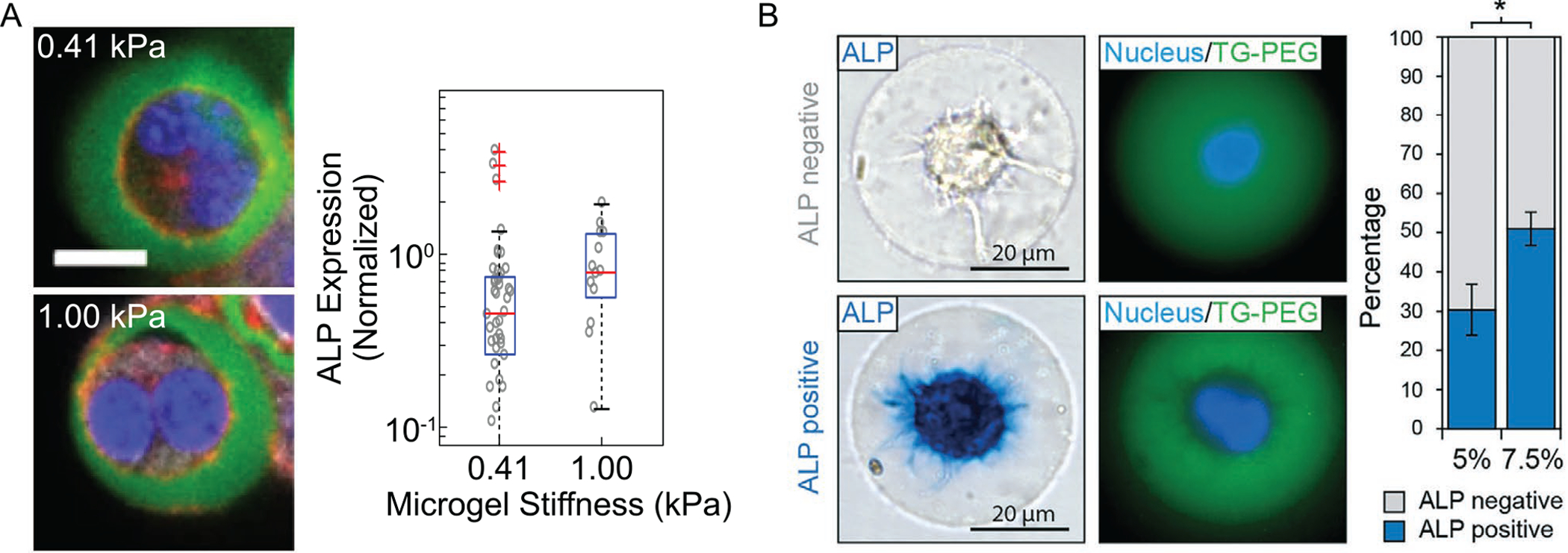
A) Confocal images of MSCs encapsulated within alginate microgels of 0.41 (top) and 1.00 kPa (bottom) stiffness. Blue, nuclei; green, alginate; red, actin; scale bar, 10 μm. ALP expression for cells in the corresponding groups.[46] B) MSCs encapsulated within 5 and 7.5% w/v TG-PEG hydrogels. ALP expression of MSCs in the corresponding groups after culturing 7 days in differentiation medium.[123] Images of single-cell microgels and box plot of ALP expression are reproduced with permission.[46] Copyright 2016, Springer Nature. Images of single-cell microgels and ALP expression percentage plot are reproduced with permission.[123] Copyright 2017, Royal Society of Chemistry.
Integrating targeting molecules into cell-based therapies can provide additional specificity through directed delivery, compared to solely relying on microgel size for appropriate biodistribution and retention. Qi et al. demonstrated enzyme-mediated adhesion of hydrogel cell microcarriers to diseased sites in damaged livers. Leveraging inherent up regulation of transglutaminase (TGase) in necrosing hepatocytes, microcarriers were functionalized with Q- and K-residues which bound to Q- and K-containing proteins present on the liver surface in mice. Functionalized microcarriers resulted in a 100-fold improvement in targeted adhesion compared to free-cell injection, increasing mouse survival rate from 0% to 33%.[244] Using this methodology, it is possible to improve treatment efficacy by directing single-cell microgels to affected sites following systemic distribution from the site of injection.
Controlled degradation of the encapsulating hydrogel matrix is preferred for some applications, such as tissue regeneration, as it allows implanted cells and host tissue to properly integrate at the damaged site at a predictable rate as the hydrogel degrades.[245] Some hydrogel materials can be degraded enzymatically or hydrolytically in vivo. Others are not readily recognized or metabolized by the host, requiring further modifications to provide these characteristics. Degradation rates can be tuned by modulating the molecular weight or composition of the base hydrogel material,[246] chemical modifications such as amide linkages[118] or oxidation,[247] and crosslinking density/mechanism. In vivo temperature and pH have been shown to alter degradation rates, suggesting careful consideration should be made when tailoring the degradation kinetics in vitro.[248] This is even more important in applications where the encapsulated cell is exposed to dynamic in vivo environments.
4.1.2. Outlook of Single-Cell Microgels for In Vivo Therapies
Cell-laden microgels provide a unique opportunity for cell-based therapies in regenerative and personalized medicine by reducing or completely eliminating the need for high dosages of systemic therapeutics, which can cause adverse side effects in otherwise healthy tissues.[249] Traditionally, cell-based therapies rely on the encapsulation of many cells rather than single cells. The use of multi-cell microgels instead of single-cell microgels stems from the former’s ability to regulate cell viability and differentiation through paracrine signaling. Commonly, stem cells are encapsulated in the same microgel with other cell types, such as endothelial cells, to enhance vascularization and recapitulate the cellular microenvironment to promote cell-to-cell interactions.[250] Additionally, the number of cells required for therapeutic effect for many conditions typically exceeds 106 cells per kg, which limits the use of single-cell microgels due to their effective lower packing density. Findings to date suggest that single-cell microgels provide little-to-no benefit over multi-cell microgels for most in vivo therapies. While using only single-cell microgels is impractical and/or sub-optimal for most cell-based therapies, their integration alongside multi-cell microgel-dominated therapies may improve treatment outcomes, and subsequently, the likelihood for clinical adoption.
One application where more compact, single-cell microgels have a unique opportunity for further development is in the area of responsive, circulating cell-based therapeutics. Leveraging cells as the drug releasing agent, therapeutics can be released as needed in cell-mediated quantities as opposed to more conventional, passive drug delivery options in which release is largely unmodulated. As discussed previously, hydrogel encapsulation of the therapeutic cells provides protection from the host immune system, potentially permitting longer circulation times, along with preserving phenotypic integrity and viability prior to administration. Due to their smaller size, single-cell microgels would be preferred over multi-cell as this allows for better circulation potential throughout the vasculature with less chance for entrapment.
While not a direct in vivo application, single-cell microgels have an excellent outlook for use in bioinks. Bioprinting using custom microgels allows for modularity in bottom-up tissue engineering strategies by independently controlling the cellular micro- and macro-environments, prior to and after in vivo implantation.[31] Initially, researchers incorporated porogens[251] or cell-laden microgels[252] into injectables to facilitate directed cell behavior while decoupling inherent properties of the bulk biomaterial, such as porosity and mechanical stiffness. By encapsulating individual cells into their own respective microgels, researchers can tailor the immediate pericellular properties to suit specific requirements for optimal viability and maintaining or directing phenotypic expression (e.g., stiff matrix for osteogenesis, relatively softer for chondrogenesis).[253] Various cell types can be encapsulated independently, providing more control over microenvironmental conditions that influence cell characteristics. These cell type specific microgels can then be cast or printed into a bulk macrogel format that is better suited to the host-specific macroenvironment while replicating the multifunctionality observed in native tissues.[21,31] Additionally, this approach permits tailoring of in vivo therapeutics with single-cell resolution, which may allow for the integration of novel in vivo sensors. One proposed approach is to sparsely incorporate genetically engineered single-cell microgels into the bulk macrogel to serve as real-time sensors for critical microenvironmental conditions, for example, oxygen sensing[254] for real-time subcutaneous monitoring of oxygen content.[255] Ultimately, single-cell microgels can serve as cell type-specific bioinks for diverse tissue printing applications, capitalizing on the stabilization that encapsulation provides to the living cells.
4.2. Enhanced In Vitro Cell Culture
Since the first demonstration of in vitro cell culturing in 1910, researchers have been continuously improving laboratory-based techniques in an attempt to recapitulate cell-specific microenvironments for improved cell viability and directed cell behavior and state.[256] Using conventional cell culturing methods, cells are grown in a sterile tissue-culture grade flask with liquid media specific to the cell type and desired outcome (e.g., proliferation, secretion, differentiation).[257] Culture flasks are placed in an incubator capable of controlling factors such as temperature and ambient gasses, which are some of the numerous parameters critical for cell fate.[256] Depending on the study, researchers can introduce chemical cues and nutrients for directing the cell in bulk culture down a desired pathway. More recently, investigations into modifying the surrounding microenvironment and focal adhesion arrangements have resulted in greater control over cellular differentiation. Earlier efforts looked at recreating and tuning the 2D microenvironment; however, this is often insufficient for recapitulating in vivo conditions.[258–260] Scaffoldless, 3D cell culturing methods (e.g., pellet culture, microwells, and hanging drop) can provide a more accurate representation of the cellular microenvironment, thus permitting more accurate studies into gene and protein expression that control morphogenesis, proliferation, and cell fate.[260] As when used in vivo, encapsulation in a hydrogel offers protection from local mechanical stresses and a means to direct cellular behavior in a 3D microenvironment by tuning hydrogel mechanical properties and incorporating chemical functionalization. Non-negligible shear stresses are present during routine suspension culture,[30] which can affect cell viability, subcellular organization, and genomic stability.[261] As a result, hydrogel encapsulation along with these modifications can improve viability, differentiation, and overall organization of the assembled structure.
Hydrogels can be modified to incorporate various growth factors and cell recognition sites for directing cell differentiation, maintaining cellular morphology and phenotype, and replicating critical cellular adhesion similar to in vivo situations. As previously mentioned, the mechanical properties of the hydrogel can facilitate stem cell differentiation.[40,46,123,253] Coupling this phenomenon with the integration of growth factors into the hydrogel network further replicates in vivo conditions. Incorporating growth factors into the hydrogel matrix improves spatial and temporal control of delivery by tuning diffusion-limited gradients observed in vivo.[23,262] Additionally, enhanced bioactivity of growth factors is frequently observed when encapsulated or attached to hydrogel matrices.[260] Cell proliferation, cytoskeletal reorganization, and other responses necessary for cell survival are strongly dependent on cell-ECM interaction.[56,263] Some hydrogels do not possess cellular binding sites (e.g., alginate and agarose). Many approaches have been developed to append cellular adhesion sites, such as conjugation of peptide motifs and interpenetrating polymer networks (IPN).[260] RGD peptides are one of the most widely and most implemented means to use peptide motifs as a conjugate to polymer chains that can promote proliferation and differentiation and maintain high cell viability.[264,265] IPNs have two or more polymer networks that are at least partially interlaced on a molecular scale but not covalently bonded to each other and cannot be separated without breaking chemical bonds.[266] Researchers can include a material readily recognized by cells, such as collagen, in an IPN along with an inert hydrogel that can form a more favorable cellular microenvironment overall. The desired application and cell type should be considered when deciding which factors and adhesion methods to use.
As with in vivo tissue regeneration, in vitro cell culture can benefit from controlled degradation of the encapsulating hydrogel matrix. Typically, hydrogels degrade via hydrolytic or enzymatic mechanisms.[267] Cell-mediated degradation (e.g., protease sensitivity[123] or disulfide breakdown by GSH secretion[120]) is particularly useful for applications requiring dynamic cell–ECM interaction. This type of degradation can be used to release cell-specific soluble factors, mimicking gradient-dependent responses and the dynamic nature of normal ECM.[37] Controlled hydrogel degradation can enable many cellular processes, such as proliferation and migration, by replicating the responsiveness of native ECM.[123,268] Additionally, some applications require the removal/degradation of the hydrogel to perform downstream assays.
Cells in culture can experience relatively static or prescribed dynamic environments when encapsulated within a hydrogel. The ability to control the surrounding environment allows researchers to preserve the desired cellular phenotype during various handling steps, which is specifically advantageous when the terminal goal is in vivo implantation. As we have discussed, many factors contribute to cell state, phenotype, gene/protein expression, etc.[261] If control over the cellular microenvironment is no longer available during various handling steps, it is difficult to predict the outcome of these subtle fluctuations, which can render studies inaccurate or treatments ineffective. Hydrogel encapsulation of cells allows for a more stable and predictable microenvironment, especially compared to conventional suspension culturing methods.
Lastly, single-cell encapsulation within a hydrogel permits continual or intermittent perfusion of reagents while maintaining isolation of individual cells for studies looking at population heterogeneity. Liquid droplet compartmentalization allows for the isolation of individual cells, but lacks ease of reagent transfer. In one study, mouse embryonic stem cells were individually encapsulated within agarose microgels and continuously imaged for a 68 h period. Microgels were continuously perfused with two different growth media to assess effects on pluripotency with single-cell resolution (Figure 10A).[70] In a separate study, differentiation toward osteogenic and adipogenic lineages of individual MSCs was analyzed following encapsulation within fibrinogen thiolated-HA-PEG (FBNG-HASH-PEG) microgels of varying stiffness. Isolation via hydrogel encapsulation enabled examination of heterogeneity in differentiation response while preventing confounding effects of physical cell-to-cell interactions.[40]
Figure 10.
A) (Top) Bright field images of agarose microgels, with the perimeter traced with red dashed line for clarity, lodged into a microfluidic trapping device. Time-lapse bright field and fluorescence confocal microscopy (GFP expression marking pluripotency) of pluripotent mouse embryonic stem cells continuously perfused with 2i media (middle) and N2B27 media (bottom).[70] B) Workflow of agarose-based droplet microfluidic emulsion PCR.[8] C) Schematic of droplet microfluidics-based Single-Cell Copy Number Variation.[8] Images in (A) are reproduced with permission.[46] Copyright 2018, Springer Nature. Schematics from (B) and (C) are reproduced with permission.[8] Copyright 2019, Wiley-VCH.
4.3. Diagnostics and Screening
Single-cell microgels enable the compartmentalization of individual cells, which can be used to screen and analyze various therapeutics at the single-cell level. Analysis with single-cell resolution allows researchers to investigate cellular heterogeneity, data which are largely obscured in multi-cell methods. This knowledge is becoming more crucial for a broad swathe of biological fields, including antibiotic and cancer treatment screenings.[269,270] While many of the advantages of microgels also exist for the more common liquid droplet cell encapsulation approaches, microgels are unique in their characteristics related to physical protection, controllable mass transport, and replication of ECM. For this reason, we believe microgels provide researchers additional capabilities for future diagnostic and screening applications. In the following sections, we discuss recent works focusing on advanced diagnostics and screening, along with potential future work that can benefit from single-cell microgels, if not already implemented.
4.3.1. High-Throughput Screening
Screening applications have dramatically benefited from advances in next generation sequencing, molecular bar-coding, and the miniaturization and automation of lab-on-a-chip systems. Miniaturized systems typically operate using much smaller volumes compared to conventional bench top techniques, which benefits the system twofold: first, reagent requirements per assay/experiment are dramatically reduced, and second, reactions occur much more rapidly, allowing for high-throughput screens.[271] Droplet microfluidics, in particular, has emerged as an integral tool for rapidly isolating individual cells for downstream, single-cell screening assays. One such application is the screening of monoclonal antibodies, which are the fastest growing class of new drugs with demonstrated treatments for infectious diseases,[272] cancer,[273] and inflammation.[274,275] High-throughput antibody screening using droplet microfluidics was recently demonstrated for identifying ≈450–900 IgG sequences capable of recognizing soluble and membrane bound antigens.[276] While the throughput for this study is modest compared to what is possible with droplet microfluidics, the demonstration of a full and adaptable antibody screening pipeline, which includes encapsulation, sorting, and sequencing of the antigen-binding antibodies, is a step in the right direction for exhibiting the power and feasibility of droplet microfluidics-based approaches.
4.3.2. Single-Cell -Omics
Single-cell sequencing has been a paradigm shift for the development of novel diagnostic and therapeutics in cancer, neurological pathologies, and reproductive and genetic diseases, while also vastly improving our understanding of complex immunological processes and relationships.[277] Single-cell sequencing improves on traditional techniques that miss inherent heterogeneity within a cell population,[278] which has been integral for understanding many biological processes such as tumor formation[279,280] and metastasis.[281] Initial adoption of single-cell techniques was slow due to cost and throughput limitations, but more recent advances in instrumentation and cell handling have addressed many of these concerns. As a result, there have been many efforts aimed at understanding gene/protein expression in heterogeneous biological systems throughout the body to elucidate myriad diseases. One such large-scale endeavor is the Human Cell Atlas project, founded in 2016, which aims to generate comprehensive reference maps for all human cells as a basis for understanding human health and diagnosing, monitoring, and treating disease.[282] This large undertaking is spread among >1700 researchers across >70 countries, each with their own assigned target organ or system. Single-cell -omics techniques, done at scale, will play a crucial role in establishing the reference map by accounting for inherent heterogeneities within cell populations, not just patient-to-patient variation. Given these large collaborative efforts and technological advances, understanding the underlying mechanisms of human disease may be obtainable in the foreseeable future. New data are added continuously, with whole new categories made possible as single-cell technologies add capabilities beyond genomics, transcriptomics, and proteomic characterizations. Below we highlight these three main subcategories but do not discuss other subfields such as metabolomics and epigenomics. We refer the reader to one of the many readily available review articles focusing on the various -omics approaches.[283–286] We conclude this section by providing perspectives on how hydrogel encapsulation may be able to benefit single-cell analyses.
Single-Cell Genomics:
Single-cell genomic studies look at individual cell diversity at the DNA level. As evidenced by the dramatic, global consequences from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), understanding genomic variations in infectious microbes is of utmost importance to inform approaches for targeted interventions, treatments, and prevention of disease.[287] Specifically, phylogenetic classification can be used to evaluate clinical and epidemiological outcomes, as well as designing treatments and eventually vaccines.[288] This genetic traceability applies to non-microbial-derived ailments as well. In particular, cancer research has used single-cell sequencing to map tumor lineage progression by quantifying genomic copy number, as well as mutated variations (Figure 10C).[289] Clinicians can use this phylogenetic information for diagnoses and targeted therapies from standard tumor biopsies.[290]
Understanding tumor heterogeneity is key to the future of precision medicine, with single-cell sequencing being a crucial technology for deciphering this complex ecosystem.[291] Tumor size has been shown to correlate with the degree of cellular heterogeneity.[292] Heterogeneity is also considered a main driver of drug resistance, where cells of differing genomes within the same tumor possess different resistance mechanisms, thus making it much more difficult for effective treatment.[293,294] Analysis of cell heterogeneity within solid tumors is particularly useful for characterizing various cancers but typically requires invasive procedures, which limits its scope in areas of regular treatment monitoring and early stage detection. To address this limitation, researchers began looking at circulating tumor cells (CTCs) when sampling of metastasizing cells from the affected tissues is not feasible. One complication with CTCs is their relatively low abundance within the blood stream, with typical clinical concentrations of ≈10 CTCs per mL or less.[295] Wang et al. demonstrated how CTC tissue of origin could be identified by looking at single nucleotide variants co-occuring within the CTC and primary origin tissue exomes.[296] The ability to trace CTC tissue of origin is important for diagnosis and treatment. Given this ability, CTCs may enable a noninvasive method to monitor therapeutic treatments and early detection of undiagnosed cancers.[290,297,298] While single-cell genomic sequencing provides valuable insight into genetic heterogeneity amongst cell populations, which aids in the development of novel diagnostics and targeted therapeutics, it does not reflect the gene expression differences that influence the broad range of physical, biochemical, and developmental characteristics possible among cells of similar genomes.[299]
Single-Cell Transcriptomics:
Transcriptomics bridges the gap between the cell’s genetic code and the resulting functional molecules that control it.[299] This next level of single-cell analysis gives researchers the ability to determine gene expression, which aids in identifying critical signaling pathways that control cellular responses. In short, single-cell transcriptomics allows for identification of all cell types in a heterogeneous sample and has been used to map the cellular composition of tissues, tumors, and even full organisms.[300,301] Gene expression profiling can be applied in the drug discovery pipeline to interpret small molecule function[302] and illuminate complex underlying biological mechanisms.[303,304] Miyamoto et al. analyzed signaling pathways of 77 prostate CTCs, from 13 patients, when an androgen receptor (AR) inhibitor was administered and compared these data to an untreated group. They observed an increase in noncanonical Wnt signaling, where expression of Wnt5a has shown to reduce the effects of AR inhibitors in prostate cancer cells, while Wnt5a suppression has been shown to restore AR sensitivity.[297] More recently, Shin et al. screened drug candidates in a high-throughput, low cost manner by quantifying cell transcriptomic responses with Drop-Seq and transient transfection with short barcode oligonucleotides.[304] Single-cell transcriptomics performed on ≈37,000 cells revealed novel mucosal ciliated cell states and CD4+ T cell states when looking at healthy and asthmatic lungs.[305] Already, single-cell transcriptomic data from the Human Cell Atlas project is being utilized to identify transmission pathways, degree of susceptibility, and efficient treatment routes for many diseases including the novel SARS-CoV-2.[306,307] CRISPR interference and scRNA-seq was used to identify 664 cis enhancer-gene pairs from the characterization of >250,000 individual cells. Gasperini et al. described this framework as a method for large-scale mapping of enhancer-gene regulatory interactions.[308] Analyzing the transcriptome of diseased cells with single-cell resolution will be critical for the development and optimization of novel therapeutics and early stage diagnostics.
Single-Cell Proteomics:
The next stage of cellular analysis looks at the entire set of proteins that are produced by the cell, i.e., its proteome, which can remove the need to infer protein production from transcriptomic data. Single-cell proteomics is still in its infancy, which up until the last few years seemed to be a distant dream.[309] To make single-cell proteomics a mainstream reality, highly sensitive methods for sample collection, clean-up, and detection are required.[310] Even with these sizeable obstacles, scientists are still optimistic about implementing single-cell proteomics to directly analyze full protein profiles. Unlike transcriptomic studies, no amplification of cellular protein is needed due to its natural, high abundance, even in a single cell. Using mass spectrometry and carrier proteins barcoded with tandem-mass-tags, Specht et al. quantified over 2700 proteins in >1000 individual monocytes and macrophages in ten days of instrument time.[311] While single-cell proteomics is not yet broadly accessible, the future looks bright for this novel technology, given the rate of technological advances over the past decade.
4.3.3. Future Directions in Single-Cell Microgel Diagnostics
Many single-cell diagnostic applications rely on the cell being suspended in fluid, where the fluid can consist of growth/differentiation media, lysis buffer, etc. Traditional single-cell aqueous-in-oil droplets are severely limited for applications that involve static droplet incubation or culturing prior to assaying of encapsulated cells. The limitations stem from difficulties in controlled aqueous reagent transfer to the formed droplet, which typically requires advanced droplet merging[312] techniques or picoinjections.[313] Liquid droplets are also susceptible to coalescence during long-term experiments that expose the droplet to temperature fluctuations and shear. Single-cell microgels are a stable construct which mitigates any potential for coalescence, as well as controllable aqueous reagent transfer through the permeable hydrogel matrix.
As the field of single-cell diagnostics transitions from genomics to transcriptomics to proteomics and beyond, maintaining cellular phenotype and gene/protein expression during an incubation period becomes increasingly more important when characterizing cell states and molecular signatures. In vivo, cells are not typically suspended in fluid, with the exception for blood-borne cell types. Thus, suspending cells within a fluid and restricting critical cellular adhesion may have acute effects on gene and protein expression which can introduce unwanted aberrations during quantification. By encapsulating cells within the appropriate hydrogel matrix, we can better replicate in vivo conditions and observe less perturbed gene/protein expression.
Current sample capture efficiencies for single-cell RNA-seq protocols yield roughly 10–20% of molecules within the cell.[311] While this issue does not considerably impact mammalian cells that contain ≈360,000 mRNA molecules, non-mammalian cells such as yeast typically possess 20,000–60,000 mRNA molecules. As a result, non-mammalian cells may require additional culturing to expand cell numbers and achieve similar levels of genetic material for analysis.[65] Hydrogel encapsulation of individual cells permits the isolated culturing of single-cells such that little to no cell-to-cell influence is observed, preserving heterogeneity of the encapsulated cells. The presence of a physical gel around each cell also removes the possibility of multiple droplets fusing such that more than one cell are contained in an isolated unit.
It is possible to characterize malignancy potential and tumor progression by modulating ECM stiffness and analyzing transcriptomic or proteomic data. Studies show that hydrogel matrix stiffness plays a role in inducing a malignant phenotype.[314,315] Encapsulating individual cancer cells within microgels of differing stiffness and performing transcriptomic or proteomic analyses will give insight into cancer progression pathways. This insight can then be leveraged to inform the field of designer hydrogels for enhanced therapeutic delivery and efficacy.
Similar to fluid-based Drop-Seq methods, hydrogel encapsulation of individual cells compartmentalizes the cell and its byproducts to allow for sequencing with single-cell resolution. The hydrogel material can be functionalized to bind targeted cellular byproducts, which in turn can expedite downstream analysis by essentially filtering these molecules from the overall sample. Lin and Anseth demonstrated affinity binding for controlling the availability of hydrogel-encapsulated proteins in thiol-acrylate photopolymerized PEG.[316] This provides support for the feasibility of designing hydrogels to preferentially select for target molecules, which can be applied to many drug screening applications for observing levels of cell distress in real time.
Lastly, hydrogel encapsulation can preserve the integrity of lysed or excreted cellular components for single-cell analyses. Specifically, Zimny et al. employed single-cell hydrogel encapsulation protocols to expose and preserve long lympho-blast DNA fragments (33 Mbp) to facilitate optical mapping of single-cell genomes.[54] DNA was physically entrapped within the hydrogel encapsulant since the nucleic acid radius of gyration far exceeded hydrogel pore size. The capture and preservation approach also reduced DNA fragmentation, which minimized the additional computations needed for alignment to the reference genome and permitted more experimental testing conditions in the same amount of time.
5. Conclusion
Single-cell microgels have experienced an increase in interest in recent years due to positive developments in micro/nanofabrication, computational abilities, analysis instrumentation, and mechanistic understanding of complex biochemical pathways. In the simplest case, these micron-sized constructs are composed of an individual cell surrounded by hydrogel matrix. The hydrogel can be tailored for specific applications by adjusting microgel thickness, pore size, stiffness, and chemical functionalization. This versatility has opened many doors in the fields of diagnostics and regenerative medicine. While single-cell microgels have their limitations, they still show great promise for use in single-cell analysis and procedures where highly specific cell manipulation/isolation is critical. As technologies continue to develop in the creation and handling of single-cell microgels, we believe their impact in a broad range of applied fields will continue to grow.
Acknowledgements
This work was supported in part by awards from the National Institutes of Health (P30 GM122732) and the Draper Fellows Program. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Draper.
Biographies

Ryan Dubay is a Draper Fellow and Ph.D. candidate at Brown University. He received his bachelor degree in Biomedical Engineering from Western New England University in 2015. Ryan’s current research focuses on high-throughput single-cell mechanophenotyping using microfluidic systems for characterizing and sorting for cell state, disease, and therapeutic potential.

Joseph N. Urban is an Sc.B. candidate in Biomedical Engineering at Brown University. His research experience and current interests include microfluidic tissue-on-chip and organ-on-chip modeling, polymer microparticle drug delivery, and droplet microfluidic processing of single cells for synthetic biology applications.

Eric M. Darling is an associate professor of Medical Science, Orthopaedics, and Engineering at Brown University. He received a B.S. in Engineering from Harvey Mudd College, a Ph.D. in Bioengineering from Rice University, and post-doctoral training in Orthopaedic Research at Duke University. His research involves cell mechanics, mesenchymal stem cells, and cellular heterogeneity. Recent projects have focused on the development of tools and techniques to study these topics in more detail.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Ryan Dubay, Center for Biomedical Engineering, Brown University, 175 Meeting St., Providence, RI 02912, USA; Draper, 555 Technology Sq., Cambridge, MA 02139, USA.
Joseph N. Urban, Center for Biomedical Engineering, Brown University, 175 Meeting St., Providence, RI 02912, USA
Eric M. Darling, Department of Molecular Pharmacology, Physiology, and Biotechnology, Center for Biomedical Engineering, School of Engineering, Department of Orthopaedics, Brown University, 175 Meeting St., Providence, RI 02912, USA
References
- [1].Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER, Cell 2013, 155, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kim ES, Ahn EH, Chung E, Kim DH, Mol. Cells 2013, 36, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mast FD, Ratushny AV, Aitchison JD, J. Cell Biol 2014, 206, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yuan GC, Cai L, Elowitz M, Enver T, Fan G, Guo G, Irizarry R, Kharchenko P, Kim J, Orkin S, Quackenbush J, Saadatpour A, Schroeder T, Shivdasani R, Tirosh I, Genome Biol. 2017, 18, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Garcia HG, Berrocal A, Kim YJ, Martini G, Zhao J, Curr. Top. Dev. Biol 2020, 137, 1. [DOI] [PubMed] [Google Scholar]
- [6].Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA, Nat. Protoc 2013, 8, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhu Z, Yang CJ, Acc. Chem. Res 2017, 50, 22. [DOI] [PubMed] [Google Scholar]
- [8].Matula K, Rivello F, Huck WTS, Adv. Biosyst 2020, 4, 1900188. [DOI] [PubMed] [Google Scholar]
- [9].Headen DM, García JR, García AJ, Microsyst. Nanoeng 2018, 4, 1.31057891 [Google Scholar]
- [10].Kawakatsu T, Kikuchi Y, Nakajima M, Am J. Oil Chem. Soc 1997, 74, 317. [Google Scholar]
- [11].Umbanhowar PB, Prasad V, Weitz DA, Langmuir 2000, 16, 347. [Google Scholar]
- [12].Chabert M, Viovy JL, Proc. Natl. Acad. Sci. U. S. A 2008, 105, 3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kamalakshakurup G, Lee AP, Lab Chip 2017, 17, 4324. [DOI] [PubMed] [Google Scholar]
- [14].Suea-Ngam A, Howes PD, Srisa-Art M, deMello AJ, Chem. Commun 2019, 55, 9895. [DOI] [PubMed] [Google Scholar]
- [15].Wang D, Bodovitz S, Trends Biotechnol. 2010, 28, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, Choi K, Fromme RM, Dao P, McKenney PT, Wasti RC, Kadaveru K, Mazutis L, Rudensky AY, Pe’er D, Cell 2018, 174, 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ma KY, Schonnesen AA, Brock A, Van Den Berg C, Eckhardt SG, Liu Z, Jiang N, JCI Insight 2019, 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gawel DR, Serra-Musach J, Lilja S, Aagesen J, Arenas A, Asking B, Bengner M, Bjorkander J, Biggs S, Ernerudh J, Hjortswang H, Karlsson JE, Kopsen M, Lee EJ, Lentini A, Li X, Magnusson M, Martinez-Enguita D, Matussek A, Nestor CE, Schafer S, Seifert O, Sonmez C, Stjernman H, Tjarnberg A, Wu S, Akesson K, Shalek AK, Stenmarker M, Zhang H, et al. , Genome Med 2019, 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Heath JR, Ribas A, Mischel PS, Nat. Rev. Drug Discov 2016, 15, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jang M, Yang S, Kim P, BioChip J. 2016, 10, 310. [Google Scholar]
- [21].Kamperman T, Karperien M, Le Gac S, Leijten J, Trends Biotechnol. 2018, 36, 850. [DOI] [PubMed] [Google Scholar]
- [22].Daly AC, Riley L, Segura T, Burdick JA, Nat. Rev. Mater 2019, 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tibbitt MW, Anseth KS, Biotechnol. Bioeng 2009, 103, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ, Proc. Natl. Acad. Sci. U. S. A 1992, 89, 9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kamperman T, Henke S, Visser CW, Karperien M, Leijten J, Small 2017, 13, 22. [DOI] [PubMed] [Google Scholar]
- [26].Richardson T, Barner S, Candiello J, Kumta PN, Banerjee I, Acta Biomater. 2016, 35, 153. [DOI] [PubMed] [Google Scholar]
- [27].Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee HP, Lippens E, Duda GN, Mooney DJ, Nat. Mater 2016, 15, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chaudhuri O, Biomater. Sci 2017, 5, 1480. [DOI] [PubMed] [Google Scholar]
- [29].Bian L, Guvendiren M, Mauck RL, Burdick JA, Proc. Natl. Acad. Sci. U. S. A 2013, 110, 10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Horiguchi I, Sakai Y, J. Vis. Exp 2015, 101, e52835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kamperman T, Henke S, van den Berg A, Shin SR, Tamayol A, Khademhosseini A, Karperien M, Leijten J, Adv. Healthcare Mater 2017, 6, 3. [DOI] [PubMed] [Google Scholar]
- [32].van Loo B, Salehi SS, Henke S, Shamloo A, Kamperman T, Karperien M, Leijten J, Mater Today Bio 2020, 6, 100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kupikowska-Stobba B, Lewinska D, Biomater. Sci 2020, 8, 1536. [DOI] [PubMed] [Google Scholar]
- [34].Leung A, Lawrie G, Nielsen LK, Trau M, J. Microencapsul 2008, 25, 387. [DOI] [PubMed] [Google Scholar]
- [35].Gyles DA, Castro LD, Silva JOC, Ribeiro-Costa RM, Eur. Polym. J 2017, 88, 373. [Google Scholar]
- [36].Dolega ME, Abeille F, Picollet-D’hahan N, Gidrol X, Biomaterials 2015, 52, 347. [DOI] [PubMed] [Google Scholar]
- [37].Ma S, Natoli M, Liu X, Neubauer MP, Watt FM, Fery A, Huck WTS, J. Mater. Chem. B 2013, 1, 5128. [DOI] [PubMed] [Google Scholar]
- [38].Hati AG, Bassett DC, Ribe JM, Sikorski P, Weitz DA, Stokke BT, Lab Chip 2016, 16, 3718. [DOI] [PubMed] [Google Scholar]
- [39].Chokkalingam V, Tel J, Wimmers F, Liu X, Semenov S, Thiele J, Figdor CG, Huck WT, Lab Chip 2013, 13, 4740. [DOI] [PubMed] [Google Scholar]
- [40].Ma Y, Neubauer MP, Thiele J, Fery A, Huck WTS, Biomater. Sci 2014, 2, 1661. [DOI] [PubMed] [Google Scholar]
- [41].Andersen T, Auk-Emblem P, Dornish M, Microarrays 2015, 4, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Andersen T, Strand BL, Formo K, Alsberg E, Christensen BE, Chapter 9. Alginates as biomaterials in tissue engineering, Carbohydrate Chemistry, ISBN 978-1-84973-154-6, 2011, pp. 227–258. [Google Scholar]
- [43].Remminghorst U, Rehm BH, Biotechnol. Lett 2006, 28, 1701. [DOI] [PubMed] [Google Scholar]
- [44].Valentine ME, Kirby BD, Withers TR, Johnson SL, Long TE, Hao Y, Lam JS, Niles RM, Yu HD, Microb Biotechnol 2020, 13, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lee KY, Mooney DJ, Prog. Polym. Sci 2012, 37, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mao AS, Shin JW, Utech S, Wang H, Uzun O, Li W, Cooper M, Hu Y, Zhang L, Weitz DA, Mooney DJ, Nat. Mater 2017, 16, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mao AS, Ozkale B, Shah NJ, Vining KH, Descombes T, Zhang L, Tringides CM, Wong SW, Shin JW, Scadden DT, Weitz DA, Mooney DJ, Proc. Natl. Acad. Sci. U. S. A 2019, 116, 15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Utech S, Prodanovic R, Mao AS, Ostafe R, Mooney DJ, Weitz DA, Adv. Healthcare Mater 2015, 4, 1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhang L, Chen K, Zhang H, Pang B, Choi CH, Mao AS, Liao H, Utech S, Mooney DJ, Wang H, Weitz DA, Small 2018, 14, 9. [DOI] [PubMed] [Google Scholar]
- [50].An C, Liu W, Zhang Y, Pang B, Liu H, Zhang Y, Zhang H, Zhang L, Liao H, Ren C, Wang H, Acta Biomater. 2020, 111, 181. [DOI] [PubMed] [Google Scholar]
- [51].Kurkdjian A, Guern J, Annu. Rev. Plant Physiol. Plant Mol. Biol 1989, 40, 271. [Google Scholar]
- [52].Kuo CK, Ma PX, Biomaterials 2001, 22, 511. [DOI] [PubMed] [Google Scholar]
- [53].Eiselt P, Lee KY, Mooney DJ, Macromolecules 1999, 32, 5561. [Google Scholar]
- [54].Zimny P, Juncker D, Reisner W, Biomicrofluidics 2018, 12, 024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dawson E, Mapili G, Erickson K, Taqvi S, Roy K, Adv. Drug Deliv. Rev 2008, 60, 215. [DOI] [PubMed] [Google Scholar]
- [56].Bacakova L, Filova E, Parizek M, Ruml T, Svorcik V, Biotechnol. Adv 2011, 29, 739. [DOI] [PubMed] [Google Scholar]
- [57].Parsons JT, Horwitz AR, Schwartz MA, Nat. Rev. Mol. Cell Biol 2010, 11, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gasperini L, Mano JF, Reis RL, J. R. Soc. Interface 2014, 11, 20140817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pernodet N, Maaloum M, Tinland B, Electrophoresis 1997, 18, 55. [DOI] [PubMed] [Google Scholar]
- [60].Aymard P, Martin DR, Plucknett K, Foster TJ, Clark AH, Norton IT, Biopolymers 2001, 59, 131. [DOI] [PubMed] [Google Scholar]
- [61].Normand V, Lootens DL, Amici E, Plucknett KP, Aymard P, Biomacromolecules 2000, 1, 730. [DOI] [PubMed] [Google Scholar]
- [62].Novak R, Zeng Y, Shuga J, Venugopalan G, Fletcher DA, Smith MT, Mathies RA, Angew. Chem., Int. Ed. Engl 2011, 50, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhu Z, Zhang W, Leng X, Zhang M, Guan Z, Lu J, Yang CJ, Lab Chip 2012, 12, 3907. [DOI] [PubMed] [Google Scholar]
- [64].Geng T, Novak R, Mathies RA, Anal. Chem 2014, 86, 703. [DOI] [PubMed] [Google Scholar]
- [65].Liu L, Dalal CK, Heineike BM, Abate AR, Lab Chip 2019, 19, 1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Delley CL, Abate AR, Lab Chip 2020, 20, 2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Karoubi G, Ormiston ML, Stewart DJ, Courtman DW, Biomaterials 2009, 30, 5445. [DOI] [PubMed] [Google Scholar]
- [68].Zhang H, Jenkins G, Zou Y, Zhu Z, Yang CJ, Anal. Chem 2012, 84, 3599. [DOI] [PubMed] [Google Scholar]
- [69].Bai Y, Weibull E, Joensson HN, Andersson-Svahn H, Sens. Actuators, B 2014, 194, 249. [Google Scholar]
- [70].Kleine-Bruggeney H, van Vliet LD, Mulas C, Gielen F, Agley CC, Silva JCR, Smith A, Chalut K, Hollfelder F, Small 2019, 15, e1804576. [DOI] [PubMed] [Google Scholar]
- [71].Reissis Y, Garcia-Gareta E, Korda M, Blunn GW, Hua J, Stem Cell Res. Ther 2013, 4, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Charlebois DA, Hauser K, Marshall S, Balazsi G, Proc. Natl. Acad. Sci. U. S. A 2018, 115, E10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lan F, Demaree B, Ahmed N, Abate AR, Nat. Biotechnol 2017, 35, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shoichet MS, Li RH, White ML, Winn SR, Biotechnol. Bioeng 1996, 50, 374. [DOI] [PubMed] [Google Scholar]
- [75].Iwata H, Takagi T, Amemiya H, Shimizu H, Yamashita K, Kobayashi K, Akutsu T, J. Biomed. Mater. Res 1992, 26, 967. [DOI] [PubMed] [Google Scholar]
- [76].Fernández E, López D, Mijangos C, Duskova-Smrckova M, Ilavsky M, Dusek K, J. Polym. Sci., Part B: Polym. Phys 2008, 46, 322. [Google Scholar]
- [77].Abate AR, Weitz DA, Lab Chip 2011, 11, 1911. [DOI] [PubMed] [Google Scholar]
- [78].Guaccio A, Borselli C, Oliviero O, Netti PA, Biomaterials 2008, 29, 1484. [DOI] [PubMed] [Google Scholar]
- [79].Robyt JF, Kimble BK, Walseth TF, Arch. Biochem. Biophys 1974, 165, 634. [DOI] [PubMed] [Google Scholar]
- [80].Naji L, Schiller J, Kaufmann J, Stallmach F, Kärger J, Arnold K, Biophys. Chem 2003, 104, 131. [DOI] [PubMed] [Google Scholar]
- [81].Thier SO, Am. J. Med 1986, 80, 3. [DOI] [PubMed] [Google Scholar]
- [82].Huang X, Lowe TL, Biomacromolecules 2005, 6, 2131. [DOI] [PubMed] [Google Scholar]
- [83].Oldenhof S, Mytnyk S, Arranja A, de Puit M, van Esch JH, Sci. Rep 2020, 10, 6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Liu ZQ, Wei Z, Zhu XL, Huang GY, Xu F, Yang JH, Osada Y, Zrinyi M, Li JH, Chen YM, Colloids Surf., B 2015, 128, 140. [DOI] [PubMed] [Google Scholar]
- [85].De Groot C, Biomaterials 2001, 22, 1197. [DOI] [PubMed] [Google Scholar]
- [86].Necas J, Bartosikova L, Brauner P, Kolar J, Vet. Med 2008, 53, 397. [Google Scholar]
- [87].Collins MN, Birkinshaw C, Carbohydr. Polym 2013, 92, 1262. [DOI] [PubMed] [Google Scholar]
- [88].Park J, Lim E, Back S, Na H, Park Y, Sun K, J. Biomed. Mater. Res. A 2010, 93, 1091. [DOI] [PubMed] [Google Scholar]
- [89].Lei Y, Gojgini S, Lam J, Segura T, Biomaterials 2011, 32, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Park D, Kim Y, Kim H, Kim K, Lee YS, Choe J, Hahn JH, Lee H, Jeon J, Choi C, Kim YM, Jeoung D, Mol. Cells 2012, 33, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kleinman HK, Martin GR, Semin. Cancer Biol 2005, 15, 378. [DOI] [PubMed] [Google Scholar]
- [92].Hughes CS, Postovit LM, Lajoie GA, Proteomics 2010, 10, 1886. [DOI] [PubMed] [Google Scholar]
- [93].Parenteau-Bareil R, Gauvin R, Berthod F, Materials 2010, 3, 1863. [Google Scholar]
- [94].Silver FH, Freeman JW, Seehra GP, J. Biomech 2003, 36, 1529. [DOI] [PubMed] [Google Scholar]
- [95].Antoine EE, Vlachos PP, Rylander MN, Tissue Eng., Part B 2014, 20, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Di Caprio N, Bellas E, Adv. Biosyst 2020, 4, 1900286. [DOI] [PubMed] [Google Scholar]
- [97].Jiang Z, Jiang K, McBride R, Oakey JS, Biomed. Mater 2018, 13, 065012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Headen DM, Aubry G, Lu H, García AJ, Adv. Mater 2014, 26, 3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Cruise GM, Hegre OD, Lamberti FV, Hager SR, Hill R, Scharp DS, Hubbell JA, Cell Transplant. 1999, 8, 293. [DOI] [PubMed] [Google Scholar]
- [100].Xia B, Jiang Z, Debroy D, Li D, Oakey J, Biomicrofluidics 2017, 11, 044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ruskowitz ER, DeForest CA, ACS Biomater. Sci. Eng 2019, 5, 2111. [DOI] [PubMed] [Google Scholar]
- [102].Jiang Z, Xia B, McBride R, Oakey J, J. Mater. Chem. B 2017, 5, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Roberts JJ, Bryant SJ, Biomaterials 2013, 34, 9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Xia B, Krutkramelis K, Oakey J, Biomacromolecules 2016, 17, 2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Lin CC, Raza A, Shih H, Biomaterials 2011, 32, 9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].McCall JD, Anseth KS, Biomacromolecules 2012, 13, 2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS, Adv. Mater 2009, 21, 5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Little RD, Masjedizadeh MR, Wallquist O, McLoughlin JI, The Intramolecular Michael Reaction, pp. 315–552, ISBN 0471264180 9780471264187, 2004. [Google Scholar]
- [109].Fu Y, Kao WJ, J. Biomed. Mater. Res. A 2011, 98, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Phelps EA, Enemchukwu NO, Fiore VF, Sy JC, Murthy N, Sulchek TA, Barker TH, Garcia AJ, Adv. Mater 2012, 24, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Elbert DL, Hubbell JA, Biomacromolecules 2001, 2, 430. [DOI] [PubMed] [Google Scholar]
- [112].Lutolf MP, Hubbell JA, Biomacromolecules 2003, 4, 713. [DOI] [PubMed] [Google Scholar]
- [113].Mather BD, Viswanathan K, Miller KM, Long TE, Prog. Polym. Sci 2006, 31, 487. [Google Scholar]
- [114].Shikanov A, Smith RM, Xu M, Woodruff TK, Shea LD, Biomaterials 2011, 32, 2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zhu J, Marchant RE, Expert Rev. Med. Devices 2011, 8, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Weber LM, Hayda KN, Haskins K, Anseth KS, Biomaterials 2007, 28, 3004. [DOI] [PubMed] [Google Scholar]
- [117].Metters A, Anseth KS, Bowman CN, Polymer 2000, 41, 3993. [Google Scholar]
- [118].Browning MB, Cereceres SN, Luong PT, Cosgriff-Hernandez EM, J. Biomed. Mater. Res. A 2014, 102, 4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Reid B, Gibson M, Singh A, Taube J, Furlong C, Murcia M, Elisseeff J, Tissue Eng J. Regen. Med 2015, 9, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kar M, Vernon Shih YR, Velez DO, Cabrales P, Varghese S, Biomaterials 2016, 77, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Forman HJ, Zhang H, Rinna A, Mol. Aspects Med 2009, 30, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ehrbar M, Rizzi SC, Schoenmakers RG, Miguel BS, Hubbell JA, Weber FE, Lutolf MP, Biomacromolecules 2007, 8, 3000. [DOI] [PubMed] [Google Scholar]
- [123].Lienemann PS, Rossow T, Mao AS, Vallmajo-Martin Q, Ehrbar M, Mooney DJ, Lab Chip 2017, 17, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Cheng D, Lo C, Sefton MV, J. Biomed. Mater. Res. A 2008, 87, 321. [DOI] [PubMed] [Google Scholar]
- [125].Gerges I, Tamplenizza M, Rossi E, Tocchio A, Martello F, Recordati C, Kumar D, Forsyth NR, Liu Y, Lenardi C, Macromol. Biosci 2016, 16, 870. [DOI] [PubMed] [Google Scholar]
- [126].Hall KK, Gattas-Asfura KM, Stabler CL, Acta Biomater. 2011, 7, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Rennerfeldt DA, Renth AN, Talata Z, Gehrke SH, Detamore MS, Biomaterials 2013, 34, 8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Jeong CG, Francisco AT, Niu Z, Mancino RL, Craig SL, Setton LA, Acta Biomater. 2014, 10, 3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Cao Y, Lee BH, Irvine SA, Wong YS, Bianco Peled H, Venkatraman S, Polymers 2020, 12, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Sun Y, Cai B, Wei X, Wang Z, Rao L, Meng QF, Liao Q, Liu W, Guo S, Zhao X, Electrophoresis 2019, 40, 961. [DOI] [PubMed] [Google Scholar]
- [131].de Rutte J, Dimatteo R, van Zee M, Damoiseaux R, Di Carlo D 2020, 10.1101/2020.03.09.984245. [DOI]
- [132].Aijaz A, Perera D, Olabisi RM, Polymeric Materials for Cell Microencapsulation, volume 1479 of Methods in Molecular Biology, pp. 79–93, Humana Press, New York, ISBN 978-1-4939-6364-5, 2017. [DOI] [PubMed] [Google Scholar]
- [133].Ling Y, Rubin J, Deng Y, Huang C, Demirci U, Karp JM, Khademhosseini A, Lab Chip 2007, 7, 756. [DOI] [PubMed] [Google Scholar]
- [134].Omer A, Duvivier-Kali V, Fernandes J, Tchipashvili V, Colton CK, Weir GC, Transplantation 2005, 79, 52. [DOI] [PubMed] [Google Scholar]
- [135].Xu Q, Hashimoto M, Dang TT, Hoare T, Kohane DS, Whitesides GM, Langer R, Anderson DG, Small 2009, 5, 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Chen S, Jones JA, Xu Y, Low HY, Anderson JM, Leong KW, Biomaterials 2010, 31, 3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Wen Y, Waltman A, Han H, Collier JH, ACS Nano 2016, 10, 9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Formiga FR, Garbayo E, Diaz-Herraez P, Abizanda G, Simon-Yarza T, Tamayo E, Prosper F, Blanco-Prieto MJ, Eur. J. Pharm. Biopharm 2013, 85, 665. [DOI] [PubMed] [Google Scholar]
- [139].Pradal J, Maudens P, Gabay C, Seemayer CA, Jordan O, Allemann E, Int. J. Pharm 2016, 498, 119. [DOI] [PubMed] [Google Scholar]
- [140].Poncelet D, Neufeld RJ, Goosen MFA, Burgarski B, Babak V, AIChE J. 1999,June 45, 2018. [Google Scholar]
- [141].Bidoret A, Martins E, De Smet BP, Poncelet D, Methods Mol. Biol 2017, 1479, 43. [DOI] [PubMed] [Google Scholar]
- [142].Lindblad NR, Schneide JM, J. Sci. Instrum 1965, 42, 635. [Google Scholar]
- [143].Mazzitelli S, Tosi A, Balestra C, Nastruzzi C, Luca G, Mancuso F, Calafiore R, Calvitti M, J. Biomater. Appl 2008, 23, 123. [DOI] [PubMed] [Google Scholar]
- [144].Naqvi SM, Vedicherla S, Gansau J, McIntyre T, Doherty M, Buckley CT, Adv. Mater 2016, 28, 5662. [DOI] [PubMed] [Google Scholar]
- [145].Ciach T, Int. J. Pharm 2006, 324, 51. [DOI] [PubMed] [Google Scholar]
- [146].Lewinska D, Bukowski J, Kozuchowski M, Kinasiewicz A, Werynski A, Biocybern. Biomed. Eng 2008, 28, 69. [Google Scholar]
- [147].Qayyum AS, Jain E, Kolar G, Kim Y, Sell SA, Zustiak SP, Biofabrication 2017, 9, 025019. [DOI] [PubMed] [Google Scholar]
- [148].Esfahani RR, Jun H, Rahmani S, Miller A, Lahann J, ACS Omega 2017, 2, 2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Xu Y, Peng J, Richards G, Lu S, Eglin D, J. Orthop. Translat 2019, 18, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Gasperini L, Maniglio D, Migliaresi C, J. Bioact. Compat. Polym 2013, 28, 413. [Google Scholar]
- [151].He P, Liu Y, Qiao R, Microfluid. Nanofluid 2014, 18, 569. [Google Scholar]
- [152].Costa AM, Alatorre-Meda M, Oliveira NM, Mano JF, Langmuir 2014, 30, 4535. [DOI] [PubMed] [Google Scholar]
- [153].Blaeser A, Duarte Campos DF, Puster U, Richtering W, Stevens MM, Fischer H, Adv. Healthcare Mater 2016, 5, 326. [DOI] [PubMed] [Google Scholar]
- [154].Shi J, Wu B, Li S, Song J, Song B, Lu WF, Biomed. Phys. Eng. Express 2018, 4, 4. [Google Scholar]
- [155].Collins DJ, Neild A, deMello A, Liu AQ, Ai Y, Lab Chip 2015, 15, 3439. [DOI] [PubMed] [Google Scholar]
- [156].Seemann R, Brinkmann M, Pfohl T, Herminghaus S, Rep. Prog. Phys 2012, 75, 016601. [DOI] [PubMed] [Google Scholar]
- [157].Zhu P, Wang L, Lab Chip 2016, 17, 34. [DOI] [PubMed] [Google Scholar]
- [158].Teh SY, Lin R, Hung LH, Lee AP, Lab Chip 2008, 8, 198. [DOI] [PubMed] [Google Scholar]
- [159].Hummer D, Kurth F, Naredi-Rainer N, Dittrich PS, Lab Chip 2016, 16, 447. [DOI] [PubMed] [Google Scholar]
- [160].Sohrabi S, kassir N, Keshavarz Moraveji M, RSC Adv. 2020, 10, 27560. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [161].Ling SD, Geng Y, Chen A, Du Y, Xu J, Biomicrofluidics 2020, 14, 061508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Chokkalingam V, Weidenhof B, Kramer M, Herminghaus S, Seemann R, Maier WF, Chemphyschem 2010, 11, 2091. [DOI] [PubMed] [Google Scholar]
- [163].Chokkalingam V, Weidenhof B, Kramer M, Maier WF, Herminghaus S, Seemann R, Lab Chip 2010, 10, 1700. [DOI] [PubMed] [Google Scholar]
- [164].Zhao CX, Adv. Drug. Deliv. Rev 2013, 65, 1420. [DOI] [PubMed] [Google Scholar]
- [165].Sklodowska K, Jakiela S, RSC Adv. 2017, 7, 40990. [Google Scholar]
- [166].De Menech M, Garstecki P, Jousse F, Stone HA, J. Fluid Mech 2008, 595, 141. [Google Scholar]
- [167].Nie Z, Seo M, Xu S, Lewis PC, Mok M, Kumacheva E, Whitesides GM, Garstecki P, Stone HA, Microfluid. Nanofluid 2008, 5, 585. [Google Scholar]
- [168].Zagnoni M, Anderson J, Cooper JM, Langmuir 2010, 26, 9416. [DOI] [PubMed] [Google Scholar]
- [169].Lashkaripour A, Rodriguez C, Ortiz L, Densmore D, Lab Chip 2019, 19, 1041. [DOI] [PubMed] [Google Scholar]
- [170].Zhu P, Tang X, Wang L, Microfluid. Nanofluid 2016, 20, 3. [Google Scholar]
- [171].Dubay R, Fiering J, Darling EM, Biomicrofluidics 2020, 14, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Utada AS, Fernandez-Nieves A, Stone HA, Weitz DA, Phys. Rev. Lett 2007, 99, 094502. [DOI] [PubMed] [Google Scholar]
- [173].Abate AR, Lee D, Do T, Holtze C, Weitz DA, Lab Chip 2008, 8, 516. [DOI] [PubMed] [Google Scholar]
- [174].Amstad E, Chemama M, Eggersdorfer M, Arriaga LR, Brenner MP, Weitz DA, Lab Chip 2016, 16, 4163. [DOI] [PubMed] [Google Scholar]
- [175].Xia Y, Whitesides GM, Annu. Rev. Mater. Sci 1998, 28, 153. [Google Scholar]
- [176].Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA, Science 2005, 308, 537. [DOI] [PubMed] [Google Scholar]
- [177].Chu LY, Utada AS, Shah RK, Kim JW, Weitz DA, Angew. Chem., Int. Ed. Engl 2007, 46, 8970. [DOI] [PubMed] [Google Scholar]
- [178].Li W, Zhang L, Ge X, Xu B, Zhang W, Qu L, Choi CH, Xu J, Zhang A, Lee H, Weitz DA, Chem. Soc. Rev 2018, 47, 5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Bandulasena MV, Vladisavljevic GT, Benyahia B, J. Colloid Interface Sci 2019, 542, 23. [DOI] [PubMed] [Google Scholar]
- [180].Moon S, Ceyhan E, Gurkan UA, Demirci U, PLoS One 2011, 6, e21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Liu H, Li M, Wang Y, Piper J, Jiang L, Micromachines 2020, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Sollier E, Amini H, Go DE, Sandoz PA, Owsley K, Di Carlo D, Microfluid. Nanofluid 2015, 19, 53. [Google Scholar]
- [183].Edd JF, Di Carlo D, Humphry KJ, Koster S, Irimia D, Weitz DA, Toner M, Lab Chip 2008, 8, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Stoecklein D, Di Carlo D, Anal. Chem 2019, 91, 296. [DOI] [PubMed] [Google Scholar]
- [185].Zeng L, Balachandar S, Fischer P, J. Fluid Mech 2005, 536, 1. [Google Scholar]
- [186].Di Carlo D, Lab Chip 2009, 9, 3038. [DOI] [PubMed] [Google Scholar]
- [187].Lee W, Amini H, Stone HA, Di Carlo D, Proc. Natl. Acad. Sci. U. S. A 2010, 107, 22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Dean WR, Philos. Mag 2009, 4, 208. [Google Scholar]
- [189].Nivedita N, Papautsky I, Biomicrofluidics 2013, 7, 54101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Kemna EW, Schoeman RM, Wolbers F, Vermes I, Weitz DA, van den Berg A, Lab Chip 2012, 12, 2881. [DOI] [PubMed] [Google Scholar]
- [191].Li L, Wu P, Luo Z, Wang L, Ding W, Wu T, Chen J, He J, He Y, Wang H, Chen Y, Li G, Li Z, He L, ACS Sens. 2019, 4, 1299. [DOI] [PubMed] [Google Scholar]
- [192].Sajeesh P, Sen AK, Microfluid. Nanofluid 2013, 17, 1. [Google Scholar]
- [193].Dalili A, Samiei E, Hoorfar M, Analyst 2018, 144, 87. [DOI] [PubMed] [Google Scholar]
- [194].Darling EM, Di Carlo D, Annu. Rev. Biomed. Eng 2015, 17, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Mueller A, Lever A, Nguyen TV, Comolli J, Fiering J, J. Micromech. Microeng 2013, 23, 12. [Google Scholar]
- [196].Schmid L, Weitz DA, Franke T, Lab Chip 2014, 14, 3710. [DOI] [PubMed] [Google Scholar]
- [197].Augustsson P, Karlsen JT, Su HW, Bruus H, Voldman J, Nat. Commun 2016, 7, 11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Dubay R, Lissandrello C, Swierk P, Moore N, Doty D, Fiering J, Biomicrofluidics 2019, 13, 034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Wu M, Ozcelik A, Rufo J, Wang Z, Fang R, Jun Huang T, Microsyst. Nanoeng 2019, 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Alazzam A, Mathew B, Alhammadi F, J. Sep. Sci 2017, 40, 1193. [DOI] [PubMed] [Google Scholar]
- [201].Song H, Rosano JM, Wang Y, Garson CJ, Prabhakarpandian B, Pant K, Klarmann GJ, Perantoni A, Alvarez LM, Lai E, Lab Chip 2015, 15, 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Pamme N, Wilhelm C, Lab Chip 2006, 6, 974. [DOI] [PubMed] [Google Scholar]
- [203].Myklatun A, Cappetta M, Winklhofer M, Ntziachristos V, Westmeyer GG, Sci. Rep 2017, 7, 6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Zhang H, Liu KK, Soc JR. Interface 2008, 5, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Landenberger B, Hofemann H, Wadle S, Rohrbach A, Lab Chip 2012, 12, 3177. [DOI] [PubMed] [Google Scholar]
- [206].Nivedita N, Ligrani P, Papautsky I, Sci. Rep 2017, 7, 44072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Li M, van Zee M, Goda K, Di Carlo D, Lab Chip 2018, 18, 2575. [DOI] [PubMed] [Google Scholar]
- [208].Di Carlo D, Irimia D, Tompkins RG, Toner M, Proc. Natl. Acad. Sci. U. S. A 2007, 104, 18892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].McGrath J, Jimenez M, Bridle H, Lab Chip 2014, 14, 4139. [DOI] [PubMed] [Google Scholar]
- [210].Baret JC, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Link DR, Weitz DA, Griffiths AD, Lab Chip 2009, 9, 1850. [DOI] [PubMed] [Google Scholar]
- [211].Caen O, Schutz S, Jammalamadaka MSS, Vrignon J, Nizard P, Schneider TM, Baret JC, Taly V, Microsyst. Nanoeng 2018, 4, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Sciambi A, Abate AR, Lab Chip 2015, 15, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Li S, Ding X, Guo F, Chen Y, Lapsley MI, Lin SC, Wang L, McCoy JP, Cameron CE, Huang TJ, Anal. Chem 2013, 85, 5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Nam J, Lim H, Kim C, Yoon Kang J, Shin S, Biomicrofluidics 2012, 6, 24120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Settnes M, Bruus H, Phys. Rev. E: Stat. Nonlinear, Soft Matter Phys 2012, 85, 016327. [DOI] [PubMed] [Google Scholar]
- [216].Adams JD, Ebbesen CL, Barnkob R, Yang AHJ, Soh HT, Bruus H, J. Micromech. Microeng 2012, 22, 7. [Google Scholar]
- [217].Marble HD, Sutermaster BA, Kanthilal M, Fonseca VC, Darling EM, Stem Cell Res. Ther 2014, 5, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Sutermaster BA, Darling EM, Sci. Rep 2019, 9, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Sarnik SA, Sutermaster BA, Darling EM, Cytometry A 2020. [DOI] [PMC free article] [PubMed]
- [220].C. W. t. Shields, Reyes CD, Lopez GP, Lab Chip 2015, 15, 1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Antfolk M, Laurell T, Anal. Chim. Acta 2017, 965, 9. [DOI] [PubMed] [Google Scholar]
- [222].Bayareh M, Chemical Engineering and Processing - Process Intensification 2020, 153, 107984. [Google Scholar]
- [223].Lim F, Sun AM, Science 1980, 210, 908. [DOI] [PubMed] [Google Scholar]
- [224].2012, https://clinicaltrials.gov/ct2/show/NCT01734733?id=NCT01734733&draw=2&rank=1.
- [225].2012, https://clinicaltrials.gov/ct2/show/record/NCT02683629?id=NCT02683629&draw=2&rank=1.
- [226].Foster AA, Marquardt LM, Heilshorn SC, Curr. Opin. Chem. Eng 2017, 15, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Youngblood RL, Truong NF, Segura T, Shea LD, Mol. Ther 2018, 26, 2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Annabi N, Nichol JW, Zhong X, Ji C, Koshy S, Khademhosseini A, Dehghani F, Tissue Eng., Part B 2010, 16, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Brauker J, Martinson LA, Young SK, Johnson RC, Transplantation 1996, 61, 1671. [DOI] [PubMed] [Google Scholar]
- [230].Morris PJ, Trends Biotechnol. 1996, 14, 163. [DOI] [PubMed] [Google Scholar]
- [231].Boardman DA, Jacob J, Smyth LA, Lombardi G, Lechler RI, Curr. Transplant Rep 2016, 3, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Geller RL, Turman MA, Dalmasso AP, Platt JL, J. Am. Soc. Nephrol 1992, 3, 1189. [DOI] [PubMed] [Google Scholar]
- [233].Hoornaert CJ, Le Blon D, Quarta A, Daans J, Goossens H, Berneman Z, Ponsaerts P, Stem Cells Transl. Med 2017, 6, 1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Chan C, Berthiaume F, Nath BD, Tilles AW, Toner M, Yarmush ML, Liver Transpl. 2004, 10, 1331. [DOI] [PubMed] [Google Scholar]
- [235].Hirotani S, Ohgawara H, Cell Transplan. 1998, 7, 407. [DOI] [PubMed] [Google Scholar]
- [236].Schneider S, Feilen PJ, Brunnenmeier F, Minnemann T, Zimmermann H, Zimmermann U, Weber MM, Diabetes 2005, 54, 687. [DOI] [PubMed] [Google Scholar]
- [237].Robitaille R, Pariseau J-F, Leblond FA, Lamoureux M, Lepage Y, Hall J-P, J. Biomed. Mater. Res 1999, 44, 116. [DOI] [PubMed] [Google Scholar]
- [238].Sakai S, Mu C, Kawabata K, Hashimoto I, Kawakami K, J. Biomed. Mater. Res., Part A 2006, 78, 394. [DOI] [PubMed] [Google Scholar]
- [239].Liu G, Pareta RA, Wu R, Shi Y, Zhou X, Liu H, Deng C, Sun X, Atala A, Opara EC, Zhang Y, Biomaterials 2013, 34, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Pathak S, Regmi S, Shrestha P, Choi I, Doh KO, Jeong JH, Small 2019, 15, e1901269. [DOI] [PubMed] [Google Scholar]
- [241].Quinlan E, Lopez-Noriega A, Thompson EM, Hibbitts A, Cryan SA, O’Brien FJ, Tissue Eng J. Regen. Med 2017, 11, 1097. [DOI] [PubMed] [Google Scholar]
- [242].Caplan AI, Tissue Eng., Part B 2009, 15, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Engler AJ, Sen S, Sweeney HL, Discher DE, Cell 2006, 126, 677. [DOI] [PubMed] [Google Scholar]
- [244].Qi C, Li Y, Badger P, Yu H, You Z, Yan X, Liu W, Shi Y, Xia T, Dong J, Huang C, Du Y, Biomaterials 2017, 126, 1. [DOI] [PubMed] [Google Scholar]
- [245].Choe G, Park J, Park H, Lee JY, Polymers 2018, 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Kong HJ, Kaigler D, Kim K, Mooney DJ, Biomacromolecules 2004, 5, 1720. [DOI] [PubMed] [Google Scholar]
- [247].Wright B, De Bank PA, Luetchford KA, Acosta FR, Connon CJ, J. Biomed. Mater. Res. A 2014, 102, 3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Bouhadir KH, Lee KY, Alsberg E, Damm KL, Anderson KW, Mooney DJ, Biotechnol. Prog 2001, 17, 945. [DOI] [PubMed] [Google Scholar]
- [249].Fischbach MA, Bluestone JA, Lim WA, Sci. Transl. Med 2013, 5, 179ps7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Franco CL, Gorenkova N, Hassani Z, Akabawy G, Modo M, West JL, in 34th Annual Meeting of the Society for Biomaterials, Vol. 1, 2010. [Google Scholar]
- [251].Huebsch N, Lippens E, Lee K, Mehta M, Koshy ST, Darnell MC, Desai RM, Madl CM, Xu M, Zhao X, Chaudhuri O, Verbeke C, Kim WS, Alim K, Mammoto A, Ingber DE, Duda GN, Mooney DJ, Nat. Mater 2015, 14, 1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Levato R, Visser J, Planell JA, Engel E, Malda J, Mateos-Timoneda MA, Biofabrication 2014, 6, 035020. [DOI] [PubMed] [Google Scholar]
- [253].Olivares-Navarrete R, Lee EM, Smith K, Hyzy SL, Doroudi M, Williams JK, Gall K, Boyan BD, Schwartz Z, PLoS One 2017, 12, e0170312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Lidsky PV, Lukyanov KA, Misra T, Handke B, Mishin AS, Lehner CF, Development 2018, 145, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Gorocs Z, Rivenson Y, Ceylan Koydemir H, Tseng D, Troy TL, Demas V, Ozcan A, ACS Nano 2016, 10, 8989. [DOI] [PubMed] [Google Scholar]
- [256].Jedrzejczak-Silicka M, History of Cell Culture, Chapter 1, ISBN 978-953-51-3133-5 978-953-51-3134-2, 2017. [Google Scholar]
- [257].Ruijtenberg S, van den Heuvel S, Cell Cycle 2016, 15, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Edmondson R, Broglie JJ, Adcock AF, Yang L, Assay Drug Dev. Technol 2014, 12, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Tabata Y, Lutolf MP, Sci. Rep 2017, 7, 44711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Kang S-M, Lee J-H, Huh YS, Takayama S, ACS Biomater. Sci. Eng 2020, 10.1021/acsbiomaterials.0c00457. [DOI] [PMC free article] [PubMed]
- [261].Hyler AR, Baudoin NC, Brown MS, Stremler MA, Cimini D, Davalos RV, Schmelz EM, PLoS One 2018, 13, e0194170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Ashe HL, Briscoe J, Development 2006, 133, 385. [DOI] [PubMed] [Google Scholar]
- [263].Schwartz MA, Assoian RK, J. Cell Sci 2001, 114, 2553. [DOI] [PubMed] [Google Scholar]
- [264].Jeon O, Alsberg E, Tissue Eng. Part A 2013, 19, 1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Ho SS, Murphy KC, Binder BY, Vissers CB, Leach JK, Stem Cells Transl. Med 2016, 5, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Alemán JV, Chadwick AV, He J, Hess M, Horie K, Jones RG, Kratochvíl P, Meisel I, Mita I, Moad G, Penczek S, Stepto RFT, Pure Appl. Chem 2007, 79, 1801. [Google Scholar]
- [267].Caliari SR, Burdick JA, Nat. Methods 2016, 13, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Kharkar PM, Kiick KL, Kloxin AM, Chem. Soc. Rev 2013, 42, 7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Scheler O, Makuch K, Debski PR, Horka M, Ruszczak A, Pacocha N, Sozanski K, Smolander OP, Postek W, Garstecki P, Sci. Rep 2020, 10, 3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Qian M, Wang DC, Chen H, Cheng Y, Semin. Cell Dev. Biol 2017, 64, 143. [DOI] [PubMed] [Google Scholar]
- [271].Neuzi P, Giselbrecht S, Lange K, Huang TJ, Manz A, Nat. Rev. Drug Discov 2012, 11, 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP, Nature 2014, 514, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Strait RT, Posgai MT, Mahler A, Barasa N, Jacob CO, Kohl J, Ehlers M, Stringer K, Shanmukhappa SK, Witte D, Hossain MM, Khodoun M, Herr AB, Finkelman FD, Nature 2015, 517, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Chan AC, Carter PJ, Nat. Rev. Immunol 2010, 10, 301. [DOI] [PubMed] [Google Scholar]
- [275].Seah YFS, Hu H, Merten CA, Mol. Aspects Med 2018, 59, 47. [DOI] [PubMed] [Google Scholar]
- [276].Gerard A, Woolfe A, Mottet G, Reichen M, Castrillon C, Menrath V, Ellouze S, Poitou A, Doineau R, Briseno-Roa L, Canales-Herrerias P, Mary P, Rose G, Ortega C, Delince M, Essono S, Jia B, Iannascoli B, Richard-Le Goff O, Kumar R, Stewart SN, Pousse Y, Shen B, Grosselin K, Saudemont B, Sautel-Caille A, Godina A, McNamara S, Eyer K, Millot GA, et al. , Nat. Biotechnol 2020, 38, 715. [DOI] [PubMed] [Google Scholar]
- [277].Tang X, Huang Y, Lei J, Luo H, Zhu X, Cell Biosci. 2019, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Wen L, Tang F, Nat. Biotechnol 2018, 36, 408. [DOI] [PubMed] [Google Scholar]
- [279].Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, Riggs M, Eberling Y, Troge J, Grubor V, Levy D, Lundin P, Maner S, Zetterberg A, Hicks J, Wigler M, Genome Res. 2010, 20, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, et al. , Nature 2008, 456, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z, Nat. Cell Biol 2018, 20, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282]. https://www.humancellatlas.org/
- [283].Chappell L, Russell AJC, Voet T, Annu. Rev. Genomics Hum. Genet 2018, 19, 15. [DOI] [PubMed] [Google Scholar]
- [284].Stuart T, Satija R, Nat. Rev. Genet 2019, 20, 257. [DOI] [PubMed] [Google Scholar]
- [285].Hou W, Ji Z, Ji H, Hicks SC, Genome Biol. 2020, 21, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Kumar R, Ghosh M, Kumar S, Prasad M, Front. Microbiol 2020, 11, 1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Andam CP, mSystems 2019, 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Forster P, Forster L, Renfrew C, Forster M, Proc. Natl. Acad. Sci. U. S. A 2020, 117, 9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M, Nature 2011, 472, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Navin N, Hicks J, Genome Med. 2011, 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Schmidt F, Efferth T, Pharmaceuticals 2016, 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, Li Y, Ju YS, Ramakrishna M, Haugland HK, Lilleng PK, Nik-Zainal S, McLaren S, Butler A, Martin S, Glodzik D, Menzies A, Raine K, Hinton J, Jones D, Mudie LJ, Jiang B, Vincent D, Greene-Colozzi A, Adnet PY, Fatima A, et al. , Nat. Med 2015, 21, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Efferth T, Konkimalla VB, Wang YF, Sauerbrey A, Meinhardt S, Zintl F, Mattern J, Volm M, Clin. Cancer Res 2008, 14, 2405. [DOI] [PubMed] [Google Scholar]
- [294].Gonzalez-Silva L, Quevedo L, Varela I, Trends Cancer 2020, 6, 13. [DOI] [PubMed] [Google Scholar]
- [295].Park MH, Reategui E, Li W, Tessier SN, Wong KH, Jensen AE, Thapar V, Ting D, Toner M, Stott SL, Hammond PT, J. Am. Chem. Soc 2017, 139, 2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Wang Y, Guo L, Feng L, Zhang W, Xiao T, Di X, Chen G, Zhang K, Oncol. Rep 2018, 39, 2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, Arora KS, Desai N, Dahl DM, Sequist LV, Smith MR, Kapur R, Wu CL, Shioda T, Ramaswamy S, Ting DT, Toner M, Maheswaran S, Haber DA, Science 2015, 349, 1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, Pazzagli M, Di Leo A, Pinzani P, Mol. Oncol 2015, 9, 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Adams J, Transcriptome: Connecting the genome to gene function 2008, https://www.nature.com/scitable/topicpage/transcriptome-connecting-the-genome-to-gene-function-605/
- [300].Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, Darmanis S, Batson J, Botvinnik O, Chen MB, Chen S, Green F, Jones R, Maynard A, Penland L, Sit RV, Stanley GM, Webber JT, Zanini F, Baghel AS, Bakerman I, Bansal I, Berdnik D, Bilen B, Brownfield D, Cain C, Chen MB, Chen S, Cho M, Cirolia G, Conley SD, Darmanis S, Demers A, Demir K, de Morree A, Divita T, du Bois H, Dulgeroff LBT, Ebadi H, Espinoza FH, Fish M, Gan Q, George BM, Gillich A, Green F, Genetiano G, Gu X, Gulati GS, Hang Y, Hosseinzadeh S, Huang A, Iram T, Isobe T, Ives F, Jones R, Kao KS, Karnam G, Kershner AM, Kiss B, Kong W, Kumar ME, Lam J, Lee DP, Lee SE, Li G, Li Q, Liu L, Lo A, Lu W-J, Manjunath A, May AP, May KL, May OL, Maynard A, McKay M, Metzger RJ, Mignardi M, Min D, Nabhan AN, Neff NF, Ng KM, Noh J, Patkar R, Peng WC, Penland L, Puccinelli R, Rulifson EJ, Schaum N, Sikandar SS, Sinha R, Sit RV, Szade K, Tan W, Tato C, Tellez K, Travaglini KJ, Tropini C, Waldburger L, van Weele LJ, Wosczyna MN, Nature 2018, 562, 367.30283141 [Google Scholar]
- [301].Tabula Muris C, Nature 2020, 583, 590.32669714 [Google Scholar]
- [302].Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH, Cell 2000, 102, 109. [DOI] [PubMed] [Google Scholar]
- [303].Huang Y, de Reynies A, de Leval L, Ghazi B, Martin-Garcia N, Travert M, Bosq J, Briere J, Petit B, Thomas E, Coppo P, Marafioti T, Emile JF, Delfau-Larue MH, Schmitt C, Gaulard P, Blood 2010, 115, 1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Shin D, Lee W, Lee JH, Bang D, Sci. Adv 2019, 5, eaav2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, Brouwer S, Gomes T, Hesse L, Jiang J, Fasouli ES, Efremova M, Vento-Tormo R, Talavera-Lopez C, Jonker MR, Affleck K, Palit S, Strzelecka PM, Firth HV, Mahbubani KT, Cvejic A, Meyer KB, Saeb-Parsy K, Luinge M, Brandsma CA, Timens W, Angelidis I, Strunz M, Koppelman GH, van Oosterhout AJ, et al. , Nat. Med 2019, 25, 1153. [DOI] [PubMed] [Google Scholar]
- [306].Teichmann S, Regev A, Nat. Rev. Mol. Cell Biol 2020, 21, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, Talavera-Lopez C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL, Network HCALB, Nat. Med 2020, 26, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, Trapnell C, Ahituv N, Shendure J, Cell 2019, 176, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Marx V, Nat. Methods 2019, 16, 809. [DOI] [PubMed] [Google Scholar]
- [310].Saha-Shah A, Esmaeili M, Sidoli S, Hwang H, Yang J, Klein PS, Garcia BA, Anal. Chem 2019, 91, 8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Specht H, Emmott E, Petelski AA, Huffman RG, Perlman DH, Serra M, Kharchenko P, Koller A, Slavov N, Genome Biol. 2020, 22, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Teo AJT, Tan SH, Nguyen N-T, Anal. Chem 2019, 92, 1147. [DOI] [PubMed] [Google Scholar]
- [313].Abate AR, Hung T, Mary P, Agresti JJ, Weitz DA, Proc. Natl. Acad. Sci. U. S. A 2010, 107, 19163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM, Cancer Cell 2005, 8, 241. [DOI] [PubMed] [Google Scholar]
- [315].Gu L, Mooney DJ, Nat. Rev. Cancer 2016, 16, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Lin CC, Anseth KS, Adv. Funct. Mater 2009, 19, 2325. [DOI] [PMC free article] [PubMed] [Google Scholar]


