Overexpression of the AtpD subunit of the chloroplast ATP synthase in rice increases the abundance and activity of the complex and stimulates photosynthetic electron transport at high light and high CO2.
Keywords: ATP synthase, CO2 assimilation, electron transport, photosynthesis, proton motive force, thylakoid membrane
Abstract
ATP, produced by the light reactions of photosynthesis, acts as the universal cellular energy cofactor fuelling all life processes. Chloroplast ATP synthase produces ATP using the proton motive force created by solar energy-driven thylakoid electron transport reactions. Here we investigate how increasing abundance of ATP synthase affects leaf photosynthesis and growth of rice, Oryza sativa variety Kitaake. We show that overexpression of AtpD, the nuclear-encoded subunit of the chloroplast ATP synthase, stimulates both abundance of the complex, confirmed by immunodetection of thylakoid complexes separated by Blue Native-PAGE, and ATP synthase activity, detected as higher proton conductivity of the thylakoid membrane. Plants with increased AtpD content had higher CO2 assimilation rates when a stepwise increase in CO2 partial pressure was imposed on leaves at high irradiance. Fitting of the CO2 response curves of assimilation revealed that plants overexpressing AtpD had a higher electron transport rate (J) at high CO2, despite having wild-type-like abundance of the cytochrome b6f complex. A higher maximum carboxylation rate (Vcmax) and lower cyclic electron flow detected in transgenic plants both pointed to an increased ATP production compared with wild-type plants. Our results present evidence that the activity of ATP synthase modulates the rate of electron transport at high CO2 and high irradiance.
Introduction
The efficiency of light interception is one of the major factors affecting crop yield (Zhu et al., 2010), and its improvement presents a promising route for increasing plant productivity (Evans, 2013; Long et al., 2015; Walter and Kromdijk, 2022). Absorption of light and conversion of light into chemical energy are performed by the light reactions of photosynthesis which are localized to the thylakoid membranes of chloroplasts. Using the energy of light, electrons originating from water split by PSII are transferred by the chain of cofactors to PSI to reduce NADP+. This process is known as linear electron flow (LEF). Between the two photosystems, the cytochrome b6f complex (Cytb6f) links electron transport with the translocation of protons across the thylakoid membrane, from stroma to lumen, which establishes the electrochemical proton gradient termed the proton motive force (pmf). The latter is used by the ATP synthase complex to produce ATP from ADP and Pi. During the so-called dark reactions of photosynthesis, CO2 is fixed into sugars by enzymes of the photosynthetic carbon reduction cycle which requires a minimum of 3 ATP and 2 NADPH to fix 1 CO2. Since the chloroplast ATP synthase needs 4.7 protons to produce 1 ATP (von Ballmoos et al., 2008), LEF only results in production of 2.6 ATP molecules per 2 NADPH. To achieve a ratio of 3 ATP to 2 NADPH and to supply ATP for other metabolic processes, plants run cyclic electron flow (CEF) around PSI that establishes additional pmf and thus increases ATP production (Joliot and Joliot, 2005).
The capacity of the photosynthetic light reactions must be closely adjusted to their metabolic consumption by the photosynthetic carbon reduction cycle and other anabolic pathways (Cruz et al., 2005; Walker et al., 2020). Therefore, ATP synthase activity is tightly controlled by multilevel regulatory systems providing feedback from both light and dark reactions (Kanazawa et al., 2017). Redox modulation of ATP synthase through the thioredoxin system stimulates its activity under low light and in response to dark–light transition (Carrillo et al., 2016; Kohzuma et al., 2017). At low CO2, the activity of ATP synthase is rapidly inhibited (Kanazawa and Kramer, 2002), which allows a build-up of the transmembrane proton gradient, a major component of pmf, that triggers non-photochemical quenching (NPQ) (Takizawa et al., 2007). The latter is a suite of photoprotective reactions aimed at reducing the excitation energy reaching the reaction centres of PSII by dissipating a part of absorbed light as heat in the PSII antennae (Malnoë, 2018). PROTON GRADIENT REGULATION5 (PGR5), mediating one of the CEF routes, cooperates with ATP synthase in building up the proton gradient to up-regulate NPQ (Ruban, 2016; Yamori and Shikanai, 2016).
ATP synthase is comprised of nine subunits organized in two major subcomplexes, CF0 and CF1. The membrane-integral CF0 subcomplex consists of four subunits (a, b, bʹ, and c) and the extrinsic CF1 subcomplex is made up of five subunits (α, β, γ, δ, and ε) (Capaldi and Aggeler, 2002; Hahn et al., 2018). ATP synthesis in CF1 is powered by the CF0 rotary motor in the membrane. In Arabidopsis thaliana (Arabidopsis), the bʹ, δ, and γ subunits are encoded by the nuclear genes atpG, atpD, and atpC1/C2, respectively. The δ subunit (hereafter AtpD) plays an important role in stabilizing the structure of the complex and regulating ATP synthesis because it forms a peripheral stalk together with subunits b and bʹ that acts as a stator to prevent unproductive rotation of CF1 with CF0 (Maiwald et al., 2003; Hahn et al., 2018). Plants with reduced AtpD abundance show decreased LEF and increased NPQ (Price et al., 1995; Yamori et al., 2011).
Rice (Oryza sativa) is one of the world’s staple crops, and substantial efforts are being made towards improving rice productivity. The Kitaake variety is often used as a model crop because it is smaller and has a shorter life cycle compared with indica varieties, and transformation techniques have been established. Here we test whether overexpression of the AtpD subunit of ATP synthase affects the complex formation and photosynthesis in rice. We show that overexpressing AtpD results in enhanced abundance and activity of ATP synthase and has the potential to be used for photosynthesis improvement.
Materials and methods
Construct assembly and transformation
The coding sequence of O. sativa atpD (OsKitaake02g334900.1, Phytozome, https://phytozome.jgi.doe.gov/) was codon optimized for rice and the Golden Gate cloning system (Engler et al., 2014) using the IDT online tool (https://sg.idtdna.com/CodonOpt) and translationally fused at the 3ʹ end with the glycine linker and the Myc-tag-coding sequence (EQKLISEEDL). The resulting sequence was assembled with the Zea mays Ubiquitin1 promoter and the bacterial Nos terminator into the second expression module of the pAGM4723 binary vector. The first module of the binary vector contained the coding sequence of the hygromycin phosphotransferase gene (hpt) combined with the O. sativa Actin1 promoter and Nos terminator. The construct was transformed into O. sativa ssp. japonica variety Kitaake using Agrobacterium tumefaciens strain AGL1 following the procedure described in Ermakova et al. (2021). T0 plants resistant to hygromycin were transferred to soil and analysed for the presence of AtpD-Myc by immunoblotting with Myc antibodies and for the hpt copy number by digital PCR (iDNA Genetics, Norwich, UK). Lines 2, 9, and 15 were selected for further analysis based on a stronger AtpD-Myc signal from immunoblots per transgene insertion number and the availability of seeds. Homozygous T2 seeds were obtained by selfing T0 and T1 plants and then used in all experiments unless stated otherwise. Wild-type (WT) plants were used as control in all experiments.
Plant growth conditions
Plants were grown in a controlled-environment chamber (Model PGC Flex, Conviron, Winnipeg, Canada) under ambient CO2 partial pressure (pCO2), 16 h photoperiod, 28 °C day, 22 °C night, and 60% humidity. Irradiance at 400 μmol photons m−2 s−1 was supplied by a mixture of Pentron Hg 4 ft fluorescent tubes (54 W 4100 K cool white, Sylvania, Wilmington, MA, USA) and halogen incandescent globes (42 W 2800 K warm white clear glass 630 lumens, CLA, Brookvale, Australia). Plants were grown in 1.2 litre pots in a potting mix composed of 80% peat/10% perlite/10% vermiculite (pH 5.6–5.8) mixed with 5 g l−1 of slow-release fertilizer (Osmocote, Evergreen Garden Care, Australia). All pots were kept at field water capacity. All measurements were performed on the mid-distal leaf blade portions of the youngest fully expanded leaves from the central stem of 4-week-old plants.
Gas exchange
Gas exchange and fluorescence analyses were performed using a LI-6800 (LI-COR Biosciences, Lincoln, NE, USA) equipped with a fluorometer head 6800-01A (LI-COR Biosciences). Leaves were first equilibrated at 381 µbar pCO2 in the reference side, leaf temperature 25 °C, 60% humidity, flow rate of 500 µmol s−1, and 1500 µmol photons m−2 s−1 (90% red/10% blue actinic light). For the CO2 response curves, a stepwise increase of pCO2 from 0 to 1525 µbar was imposed at 3 min intervals. The maximum carboxylation rate of Rubisco (Vcmax), the rate of electron transport (J), and triose phosphate usage (TPU) were obtained using the fitting routine of Sharkey et al. (2007). Leaf mesophyll conductance to CO2 of 0.67 µmol CO2 m−2 s−1 bar−1 previously determined for rice (von Caemmerer and Evans, 2015) was used in the fitting routine. The CO2 compensation point was calculated from the CO2 response curves recorded at different O2 partial pressures (pO2). Light–response curves were measured during a stepwise decrease of irradiance from 2000 µmol m−2 s−1 to 0 µmol m−2 s−1 at 3 min intervals and at 381 µbar pCO2 in the reference side. The quantum yield of PSII was calculated at different irradiances and pCO2 upon the application of a multiphase saturating pulse (8000 µmol m−2 s−1) according to Genty et al. (1989).
Protein isolation and western blotting
Total protein extracts were isolated from 0.5 cm2 frozen leaf discs and separated by SDS–PAGE according to Ermakova et al. (2019). Proteins were transferred to a nitrocellulose membrane and probed with antibodies against various photosynthetic proteins in dilutions recommended by the manufacturer: Rieske (AS08 330, Agrisera, Vännäs, Sweden), AtpH (Agrisera, AS09591), Myc-tag (ab9132, Abcam, Cambridge, UK), whole ATP synthase (Agrisera, AS08370), D1 (Agrisera, AS10 704), and PGR5 (Agrisera, AS163985). Quantification of immunoblots was performed with Image Lab software (Biorad, Hercules, CA, USA).
Thylakoid isolation and Blue Native-PAGE
Thylakoid membranes from the mid portions of the three youngest fully expanded leaves collected from one plant were ground in 100 ml of ice-cold grinding buffer (50 mM HEPES-NaOH, pH 7.5, 330 mM sorbitol, 5 mM MgCl2) in an Omni Mixer (Thermo Fisher Scientific, Tewksbury, MA, USA). The homogenate was passed through a double layer of Miracloth (Merck Millipore, Burlington, MA, USA) and centrifuged at 6000 rpm, 4 °C for 5 min. Pellets were first resuspended in ice-cold shock buffer (50 mM HEPES-NaOH, pH 7.5, 5 mM MgCl2) and centrifuged again. The resulting pellets were resuspended in ice-cold storage buffer (50 mM HEPES-NaOH, pH 7.5, 100 mM sorbitol, 10 mM MgCl2) and centrifuged again. Finally, pellets were resuspended in an equal aliquot of the storage buffer, snap-frozen in liquid N2, and stored at −80 °C. Preparation of the samples and Blue Native-PAGE followed Rantala et al. (2018). The gel was scanned, then incubated for 30 min in the transfer buffer (25 mM Tris, 25 mM glycine, 20% methanol, 0.1% SDS) and blotted to a nitrocellulose membrane. Western blotting was then performed as usual.
Electron transport and electrochromic shift analyses
To obtain PSII parameters (PhiPSII, the effective quantum yield; PhiNPQ, the yield of non-photochemical quenching; PhiNO, the yield of non-regulated non-photochemical quenching) and PSI parameters (PhiPSI, the effective quantum yield; PhiND, the non-photochemical yield caused by donor side limitation; PhiNA, the non-photochemical yield caused by the acceptor side limitation), fluorescence analysis was performed simultaneously with the spectroscopic measurements at 820 nm using the Dual-PAM-100 (Heinz Walz, Effeltrich, Germany). Measurements were done using red actinic light and 300 ms saturating pulses of 10 000 µmol m−2 s−1. Leaves were dark-adapted for 30 min to record the minimal and maximal levels of fluorescence in the dark. Then a saturating pulse was given after pre-illumination with far-red light to record the maximal and minimal oxidation levels of P700 (the reaction centre of PSI). To allow for a brief photoactivation, the leaves were next illuminated for 8 min with actinic light of 378 µmol m−2 s−1 and briefly dark-adapted again for 2 min. After that, photosynthetic parameters were assessed over a range of irradiances from 0 to 2043 µmol m−2 s−1 at 2 min intervals by applying a saturating pulse at the end of each illumination period. The parameters were calculated according to Kramer et al. (2004) and Klughammer and Schreiber (2008). The kinetics of P700 oxidation upon the change of light intensity presented in Fig. 5 were extracted from these measurements.
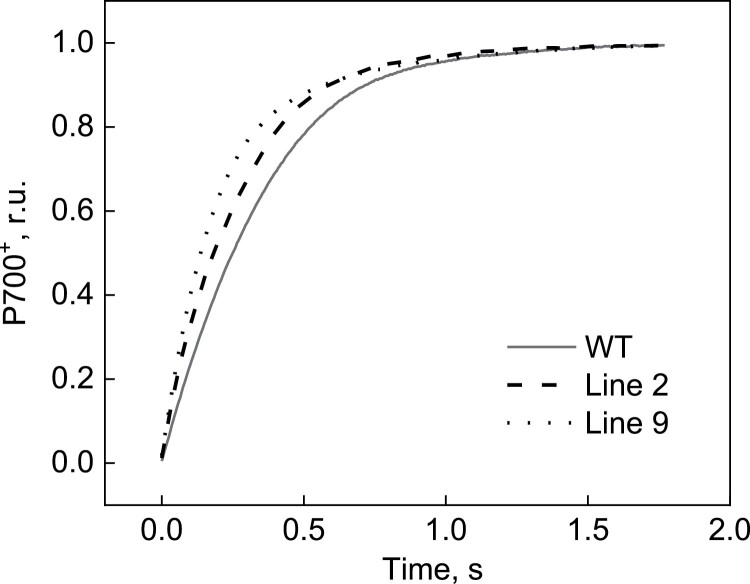
Fig. 5.
Oxidation kinetics of the reaction centres of PSI (P700) in WT O. sativa and transgenic lines overexpressing AtpD during an increase of irradiance from 218 µmol m−2 s−1 to 417 µmol m−2 s−1. Curves were normalized to the same amplitude to facilitate comparison of the kinetics and present an average of three biological replicates.
The electrochromic shift (ECS) signal was monitored as the absorbance change at 515–550 nm using the Dual-PAM-100 equipped with the P515/535 emitter–detector module (Heinz Walz). Leaves were first dark adapted for 40 min, and the absorbance change induced by a single turnover flash was measured. Dark interval relaxation of the ECS signal was recorded after 3 min of illumination with red actinic light of increasing irradiance. Proton conductivity of the thylakoid membrane through the ATP synthase was calculated as an inverse of the time constant obtained by fitting the first-order ECS relaxation (Sacksteder and Kramer, 2000). Total pmf was estimated from the amplitude of the rapid decay of the ECS signal normalized for ECS signal change induced by the single turnover flash.
Chlorophyll, protein, and Rubisco active sites
Frozen leaf discs were ground using the Qiagen TissueLyser II (Qiagen, Venlo, The Netherlands) and total Chl (a+b) was extracted in 80% acetone buffered with 25 mM HEPES-KOH and measured according to Porra et al. (1989). The amount of Rubisco sites was assayed by [14C]carboxyarabinitol bisphosphate binding as described in Ruuska et al. (2000). Total protein content was measured from the same samples by Coomassie Plus protein assay reagent (Thermo Fisher Scientific).
Statistical analysis
For all measurements, the relationship between mean values of transgenic and WT plants was tested using heteroscedastic Student’s t-tests.
Results
Overexpression of AtpD increases abundance of ATP synthase
The gene construct for AtpD overexpression (AtpD-OE hereafter) was transformed into rice calli, and T0 plants resistant to hygromycin were selected and transferred to soil. T0 plants were analysed for the presence of AtpD-Myc protein and for gene insertions based on the hpt copy number (Fig. 1). Out of 26 T0 plants, 19 plants showed detectable levels of AtpD-Myc. Lines 2, 9, and 15 were selected for further analysis, and homozygous T2 plants were studied in all experiments.
Fig. 1.
Selection of T0 rice plants transformed with the construct for AtpD overexpression (AtpD-OE). Immunodetection of AtpD-Myc in leaf protein extracts, 15 µg of protein loaded for each sample. The copy number of the hygromycin phosphotransferase gene (hpt) detected by digital PCR was used to estimate the number of construct insertions. *Lines 2, 9, and 15 were selected for further experiments.
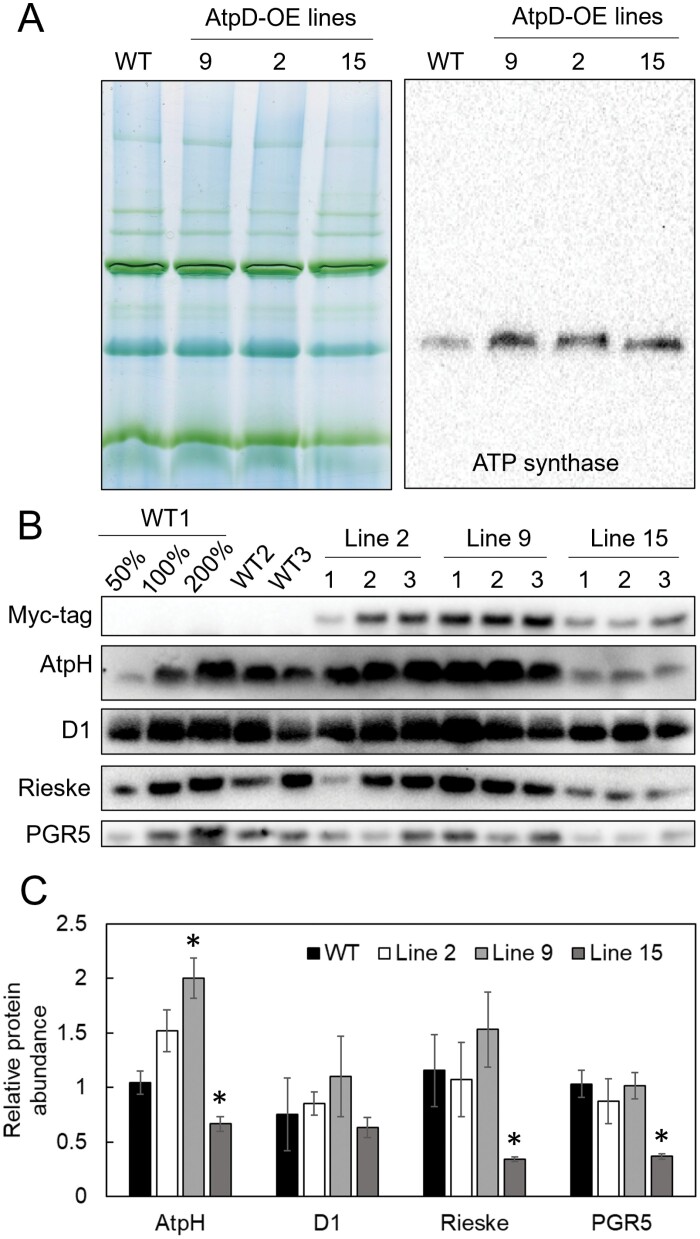
The three selected AtpD-OE lines demonstrated increased abundance of the whole ATP synthase complex in their thylakoid membranes, compared with WT plants, when the samples were normalized on a Chl (a+b) basis (Fig. 2A). When total leaf protein extracts were loaded on a leaf area basis, lines 2 and 9 had more AtpD-Myc than line 15 and also increased abundance of AtpH, the c subunit of the F0 complex, compared with WT plants (Fig. 2B). Abundances of D1 (PSII core subunit), Rieske (Cytb6f core subunit), and PGR5 were unaltered in transgenic lines 2 and 9, whilst line 15 had significantly less AtpH, Rieske, and PGR5, compared with the WT (Fig. 2C). Line 9 had increased leaf Chl content compared with the WT (Table 1). Despite having an increased abundance of ATP synthase on a chlorophyll basis, line 15 contained significantly less chlorophyll per leaf area (Table 1) and was excluded from further physiological analysis.
Fig. 2.
Immunodetection of photosynthetic proteins in WT O. sativa and transgenic lines overexpressing AtpD. (A) Thylakoid protein complexes separated by Blue Native-PAGE and probed with antibodies against whole ATP synthase; 10 µg of Chl (a+b) loaded in each lane. (B) Leaf protein samples loaded on a leaf area basis and probed with antibodies against Myc-tag, AtpH (subunit c of ATP synthase), D1 subunit of PSII, Rieske subunit of Cyt b6f, and PGR5 (PROTON GRADIENT REGULATION5). (C) Relative quantification of immunoblots. Mean ±SE, n=3 biological replicates. Asterisks indicate statistically significant differences between transgenic lines and the WT (t-test, P<0.05).
Table 1.
Gas exchange and fluorescence parameters of wild-type (WT) and AtpD-OE rice plants
| Parameter | WT | Line 2 | Line 9 | Line 15 |
|---|---|---|---|---|
| Chl (a+b), mmol m−2 | 0.66 ± 0.06 | 0.72 ± 0.03 | 0.86 ± 0.02* | 0.40 ± 0.03* |
| Chl a/b | 4.44 ± 0.04 | 4.40 ± 0.06 | 4.50 ± 0.06 | 4.59 ± 0.05* |
| Soluble protein, g m−2 | 8.49 ± 0.78 | 10.23 ± 0.37 | 12.45 ± 0.35* | 5.84 ± 0.58* |
| F V/FM | 0.806 ± 0.003 | 0.799 ± 0.005 | 0.804 ± 0.003 | 0.791 ± 0.009 |
| V cmax, µmol CO2 m−2 s−1 | 106.8 ± 8.3 | 133.8 ± 14.5 | 157.4 ± 12.6* | N/A |
| J (LEF), µmol e− m−2 s−1 | 125.1 ± 8.0 | 153.0 ± 1.7* | 166.8 ± 5.1* | N/A |
| J/Vcmax | 1.17 ± 0.03 | 1.17 ± 0.13 | 1.09 ± 0.11 | N/A |
| TPU, µmol CO2 m−2 s−1 | 9.02 ± 0.39 | 10.65 ± 0.08* | 11.24 ± 0.40* | N/A |
| R d, µmol CO2 m−2 s−1 | 1.52 ± 0.12 | 1.32 ± 0.07 | 1.40 ± 0.13 | N/A |
| R d/ Vcmax | 0.0142 ± 0.0005 | 0.0100 ± 0.0010* | 0.0090 ± 0.0006* | N/A |
| Rubisco sites, µmol m−2 | 34.1 ± 1.7 | 35.0 ± 1.9 | 36.9 ± 1.4 | N/A |
| Rubisco sites/soluble protein | 4.02 | 3.42 | 2.97 | N/A |
| LMA, g(DW) m−2 | 54.2 ± 2.6 | 50.5 ± 3.6 | 53.2 ± 1.4 | 51 ± 2.2 |
F V/FM, the maximum quantum efficiency of PSII; Vcmax, maximum carboxylation rate allowed by Rubisco; J, the rate of photosynthetic electron transport based on NADPH requirement; TPU, triose phosphate use; Rd, dark respiration rate. Mean ±SE, n=4 biological replicates. Asterisks indicate statistically significant differences between the WT and transgenic plants (t-test, P<0.05). N/A, not assessed.
Increased ATP synthase activity in AtpD-OE plants
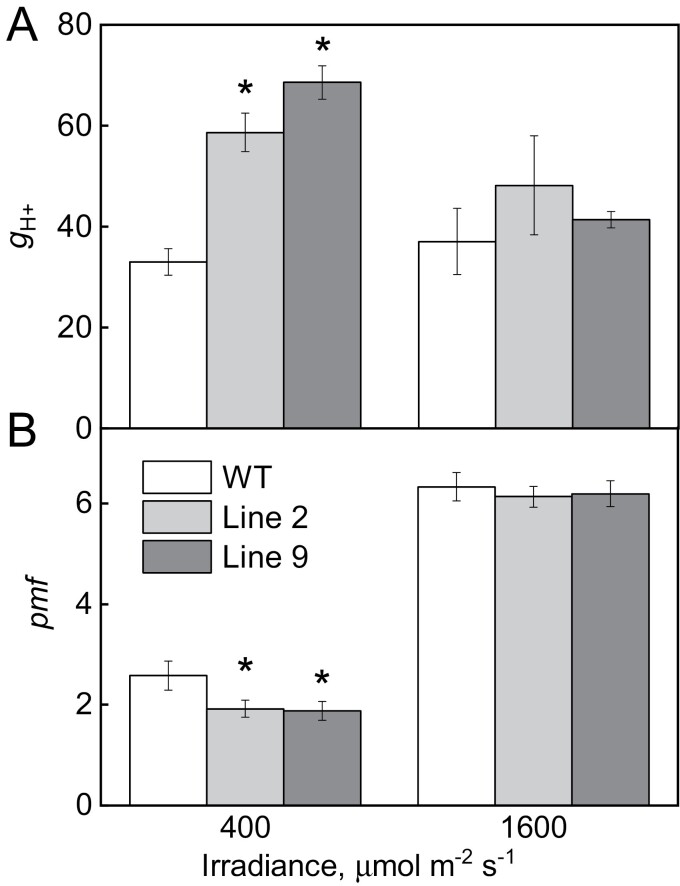
Proton conductivity of the thylakoid membrane and pmf in leaves of WT and AtpD-OE plants were estimated from the speed and amplitude of ECS relaxation upon the light–dark transition. Lines 2 and 9 showed significantly increased thylakoid proton conductivity at 400 µmol m−2 s−1, indicating a higher activity of ATP synthase compared with the WT (Fig. 3). When measured at 1600 µmol m−2 s−1, no significant differences in proton conductivity were detected between the genotypes. In line with that, the amplitude of the fast ECS decay, representing a balance between the build-up and dissipation of pmf, was significantly decreased in both AtpD-OE lines at 400 µmol m−2 s−1 but not at 1600 µmol m−2 s−1, compared with WT plants (Fig. 3).
Fig. 3.
Proton conductivity of the thylakoid membrane (gH+) and proton motive force (pmf) estimated from the dark interval relaxation kinetics of ECS in WT O. sativa and transgenic lines overexpressing AtpD. Mean ±SE, n=3-4 biological replicates. Asterisks indicate statistically significant differences between transgenic lines and the WT (t-test, P<0.05).
Electron transport properties of AtpD-OE plants
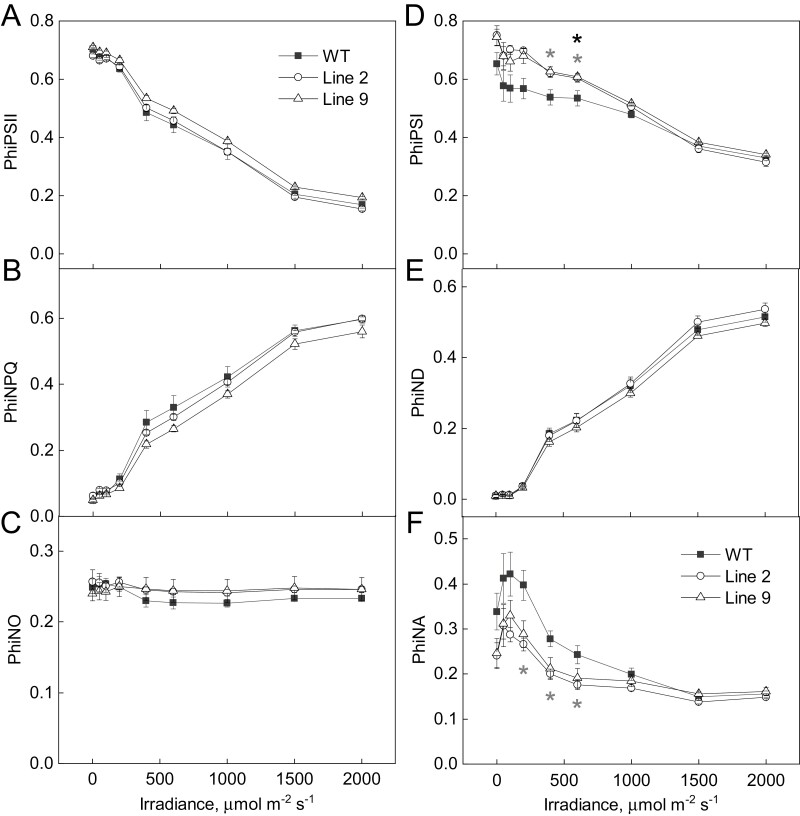
Fluorescence analysis of WT and AtpD-OE plants demonstrated that the maximum quantum efficiency of PSII (FV/FM) did not differ between genotypes (Table 1), and neither did the PSII electron transport parameters PhiPSII, PhiNPQ, and PhiNO when measured at different irradiances and ambient pCO2 (Fig. 4, left panels). Spectroscopic analysis of the redox state of P700, the reaction centre of PSI, at different irradiances (Fig. 4, right panels) showed that the quantum yield of PSI (PhiPSI) was increased in AtpD-OE lines (significant for line 9 at 548 µmol m−2 s−1 and for line 2 at 417 and 548 µmol m−2 s−1). The detected increase in PhiPSI in AtpD-OE plants could be attributed to the lower PSI acceptor side limitation (PhiNA), which was significant in line 2 between 218 µmol m−2 s−1 and 548 µmol m−2 s−1. The donor side limitation of PSI (PhiND) did not differ between the WT and AtpD-OE plants at any irradiance (Fig. 4, right panels). Further comparison of the kinetics of P700 oxidation during the increase of irradiance from 218 µmol m−2 s−1 to 417 µmol m−2 s−1 revealed faster oxidation in AtpD-OE lines, compared with the WT (Fig. 5). This result suggested that PhiNA in AtpD-OE plants was probably reduced due to an increased electron sink capacity downstream of PSI and not due to up-regulated CEF (Joliot and Johnson, 2011).
Fig. 4.
Electron transport properties of WT O. sativa and transgenic lines overexpressing AtpD at different irradiances. PhiPSII, quantum yield of PSII; PhiNPQ, quantum yield of non-photochemical quenching; PhiNO, quantum yield of non-regulated non-photochemical quenching; PhiPSI, quantum yield of PSI; PhiND, non-photochemical loss due to the oxidized PSI donors; PhiNA, non-photochemical loss due to the reduced PSI acceptors. Mean ±SE, n=3 biological replicates. Grey asterisks indicate statistically significant differences between line 2 and the WT, black asterisks—between line 9 and the WT (t-test, P<0.05).
Gas exchange analysis of plants with increased ATP synthase abundance
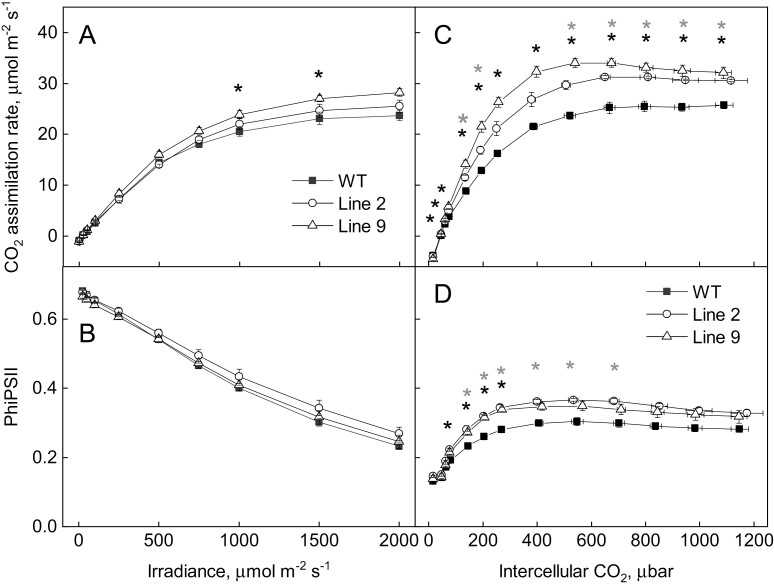
CO2 assimilation rates and PhiPSII were assayed in WT and AtpD-OE plants at different irradiances and pCO2 (Fig. 6). When the light response of photosynthesis was analysed at 381 µbar pCO2 and high light, line 9 showed significantly increased assimilation rates between 1000 µmol m−2 s−1 and 1500 µmol m−2 s−1, compared with the WT (Fig. 6, left panels). The response of CO2 assimilation to intercellular pCO2 (ACi curves) was measured at a constant irradiance of 1500 µmol m−2 s−1 (Fig. 6, right panels). Line 9 had significantly increased assimilation rates at all intercellular pCO2 except the lowest one, compared with the WT. Line 2 also had significantly increased assimilation rates at ambient and high intercellular pCO2. Both AtpD-OE lines showed increased PhiPSII, compared with WT plants (significant for line 2 at 133–650 µbar and for line 9 at 74–257 µbar).
Fig. 6.
Gas exchange and fluorescence analysis of WT O. sativa and transgenic lines overexpressing AtpD. Mean ±SE, n=4 biological replicates. Grey asterisks indicate statistically significant differences between line 2 and the WT, black asterisks—between line 9 and the WT (t-test, P<0.05).
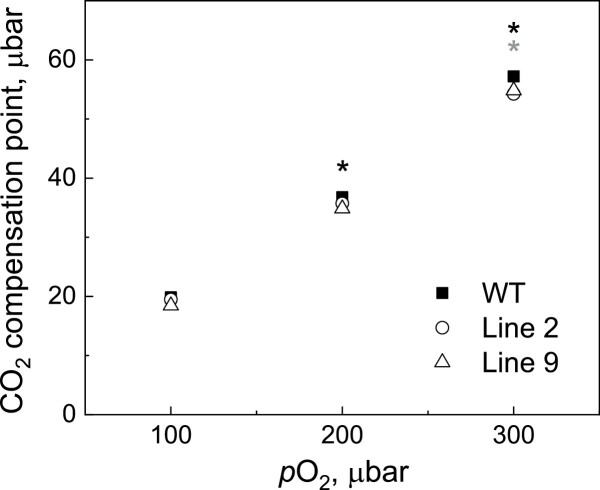
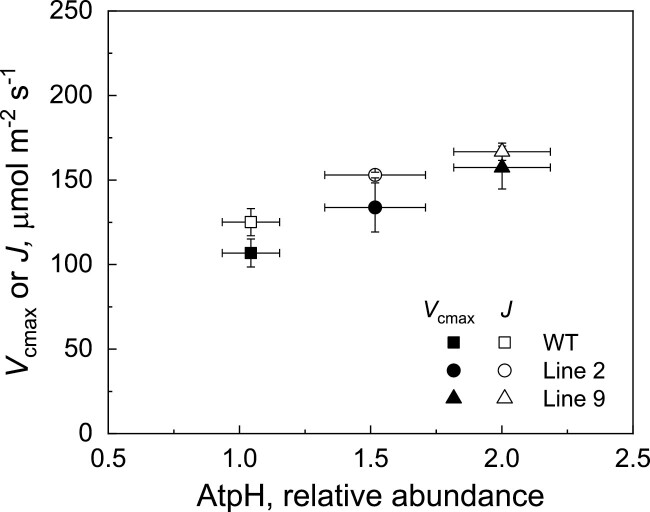
Fitting of the ACi curves revealed significantly increased J, the rate of photosynthetic electron transport, and TPU, the triose phosphate use, in AtpD-OE lines 2 and 9, compared with the WT (Table 1). Line 9 also had significantly increased Vcmax, the maximum carboxylation rate allowed by Rubisco (P=0.18 for line 2). The J/Vcmax ratio and the rate of respiration in the dark (Rd) did not differ between WT and AtpD-OE plants, whilst the Rd/Vcmax ratio was significantly lower than in the WT in lines 2 and 9. In line with the observed decrease in Rd/Vcmax, AtpD-OE plants also showed a lower CO2 compensation point, significant for line 9 at ambient and high pO2 and for line 2 at high pO2 (Fig. 7). Overall, a positive correlation was observed between both Vcmax and J values and ATP synthase abundance (estimated as the relative abundance of the AtpH subunit from immunoblots on Fig. 2) for all three genotypes (Fig. 8). The total abundance of Rubisco active centres measured in the leaves subjected to gas exchange analysis did not differ between WT and AtpD-OE plants, but the total soluble protein content was significantly increased in line 9, compared with the WT (Table 1).
Fig. 7.
Leaf CO2 compensation point measured at different O2 partial pressures (pO2). Mean ±SE, n=3 biological replicates; SE is smaller than the symbols. Grey asterisks indicate statistically significant differences between line 2 and the WT, black asterisks—between line 9 and the WT (t-test, P<0.05).
Fig. 8.
Gas exchange parameters obtained by fitting the CO2 response curves of assimilation versus relative protein abundance of AtpH from the immunoblots in Fig. 2.
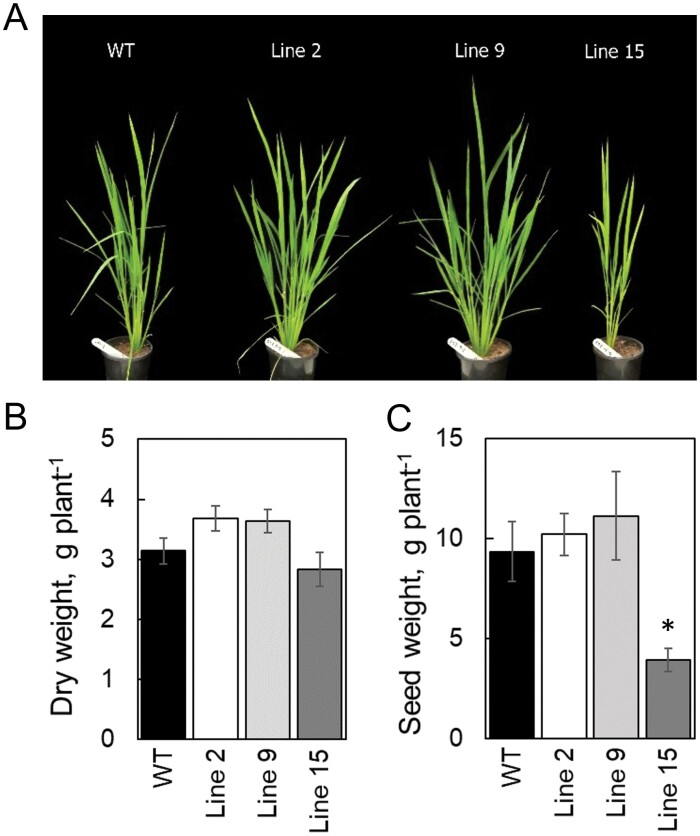
Biomass and grain yield of AtpD-OE plants
Leaf mass per area in AtpD-OE plants was similar to that of the WT (Table 1). During the mid-tillering stage, AtpD-OE plants of lines 2 and 9 were slightly larger than WT plants: P=0.097 for line 2 and P=0.111 for line 9 (Fig. 9). The total weight of seeds produced by AtpD-OE lines 2 and 9 was similar to that of the WT, whilst plants of line 15 produced significantly fewer seeds (Fig. 9C).
Fig. 9.
Biomass and seed yield of WT O. sativa and transgenic lines overexpressing AtpD. (A) Phenotype of plants during the mid-tillering stage, 4 weeks after germination. (B) Dry weight of the plants harvested at mid-tillering stage, mean ±SE, n=8 biological replicates. (C) Total weight of seeds produced by the plants, mean ±SE, n=4 biological replicates. Asterisks indicate statistically significant differences between transgenic lines and the WT (t-test, P<0.05).
Discussion
Increasing the rate of electron transfer during the light reactions of photosynthesis is predicted to be of benefit for enhancing crop yield (Simkin et al., 2019). Promising strategies for accelerating electron transport include up-regulating the abundance of Cytb6f by overexpressing the Rieske subunit to alleviate rate limitation at the step of plastoquinol oxidation (Simkin et al., 2017; Ermakova et al., 2019), speeding up the delivery of electrons to PSI by engineering plants with algal cytochrome c6 (Chida et al., 2007; López-Calcagno et al., 2020), and facilitating faster relaxation of NPQ (Kromdijk et al., 2016). Although the low productivity of the chloroplast ATP synthase with the high H+/ATP ratio of 4.67 was suggested to be favourable for avoiding photodamage (Davis and Kramer, 2020), we were interested to test the effects of increased ATP synthase activity on electron transport. Although the activity of ATP synthase correlates with transcript and protein levels of its subunits (Kohzuma et al., 2009), multiple additional levels of regulation make this complex a regulatory hub collecting signals from light-harvesting reactions, the photosynthetic carbon reduction cycle, and central metabolism (Schöttler and Tóth, 2014).
Rice plants overexpressing the AtpD subunit of ATP synthase demonstrated increased abundance of the whole complex, and two AtpD-OE lines had increased ATP synthase activity detected as higher proton conductivity of the thylakoid membrane (Figs 2, 3). Moreover, two AtpD-OE lines showed a proportional increase in whole-chain electron transport and CO2 assimilation rates at high pCO2 (Table 1; Figs 6, 8). This is in line with the C3 photosynthesis model which predicts electron transport limitations at high irradiance and high pCO2 as well as at lower irradiance (Farquhar and von Caemmerer, 1981; von Caemmerer and Farquhar, 1981). Studies on tobacco plants with reduced atpD transcript levels have previously shown a close correlation between CO2 assimilation rate and AtpD abundance (Price et al., 1995; Yamori et al., 2011). Given that there are equally close correlations between Rieske content and chloroplast electron transport and CO2 assimilation rates, previous works suggested that Cytb6f and ATP synthase co-limit electron transport (Price et al., 1995; Yamori et al., 2011, 2016). Our results indicate that, at high irradiance and high CO2, electron transport is primarily limited by ATP synthase, and the limitation at Cytb6f could be overcome by increasing AtpD content. At ambient pCO2, a significant increase of thylakoid proton conductivity was only detected in AtpD-OE plants at growth irradiance but not at high irradiance (Fig. 3), supporting a down-regulation of ATP synthase in conditions when the light reactions are limited by Rubisco activity and metabolic regeneration of NADP+, ADP, and Pi (Kohzuma et al., 2017).
Curiously, we also observed an increase in in vivo Rubisco activity (Vcmax) in two AtpD-OE lines, despite similar amounts of Rubisco in the leaves (Table 1), which matched similar abundances of Rieske and other electron transport components (Fig. 2; Table 1). The lower CO2 compensation point detected in two AtpD-OE lines was also in line with the increased Vcmax and lower Rd/Vcmax (Table 1; Fig. 7) (Azcon-Bieto and Osmond, 1983; von Caemmerer, 2000). The higher Vcmax was probably due to the higher activation state of Rubisco in plants overexpressing AtpD. The active sites of Rubisco become inactivated by binding sugar phosphates and require Rubisco activase to restore their activity (Salvucci et al., 1985). The activase is strongly dependent on ATP as it is both regulated by the ATP/ADP ratio and uses ATP for the reaction (Streusand and Portis, 1987; Robinson and Portis, 1988). Higher thylakoid proton conductivity in AtpD-OE plants could provide more ATP for Rubisco activase and sustain Rubisco carboxylation activity in conditions promoting the deactivation, for instance upon exposure to low CO2 (von Caemmerer and Edmondson, 1986). Since CEF is strongly inhibited by ATP (Fisher et al., 2019), a lower CEF, seen as a lower PhiNA and faster P700 oxidation kinetics (Figs 4, 5), indicated an increased ATP production in AtpD-OE plants. Because activation of Rubisco is one of the promising traits for improving crop photosynthesis (Parry et al., 2013; Carmo-Silva et al., 2015; Taylor et al., 2022), exploring the relationship between the thylakoid proton conductivity and Rubisco activation could be of great interest for future research.
Importantly, we have developed a method for increasing the abundance and activity of ATP synthase by overexpressing one subunit of the complex. It was previously shown that AtpD is critical for stabilizing the complex and that its abundance correlates with the electron transport rate (Engelbrecht et al., 1989; Price et al., 1995). AtpD is one of the two nuclear gene-encoded subunits of chloroplast ATP synthase. Interestingly, despite the transfer of organellar genes to the nucleus being favourable for the cell energy budget, only a limited number of genes encoding subunits of the thylakoid complexes were translocated to the nucleus; proteins with the highest abundance are more likely to be retained in an organellar genome (Kelly, 2021). This suggests that nuclear-encoded subunits limit the assembly of thylakoid complexes and are involved in retrograde–anterograde signalling pathways integrating chloroplast light reactions with cellular metabolism (Lyska et al., 2013; Pribil et al., 2014). Taking advantage of these signalling pathways provides an opportunity for increasing the abundance of complexes by overexpressing single subunits, as previously reported for Cytb6f (Simkin et al., 2017; Ermakova et al., 2019) and as shown here for ATP synthase. However, whilst overexpression of Rieske in Arabidopsis allowed higher electron transport and CO2 assimilation rates, this was accompanied by the increased abundance of not only Cytb6f but also ATP synthase subunits, as well as PSII and PSI (Simkin et al., 2017). In contrast, two rice AtpD-OE lines did not display increases in thylakoid membrane complexes other than the ATP synthase and showed a lower pmf, suggesting that ATP synthase activity exceeded Cytb6f activity. Therefore, increasing Cytb6f abundance could be complementary to AtpD overexpression to further boost the electron transport rate and stimulate photosynthesis.
Conclusion
Photosynthetic light reactions are tightly regulated to prevent photodamage, but in some conditions overcoming or reducing the regulation of photosynthesis could be of benefit for plant productivity (Kromdijk et al., 2016). Here we show that overexpressing the AtpD subunit of the chloroplast ATP synthase was sufficient to increase abundance of the whole complex. Moreover, two AtpD-OE lines showed increased ATP synthase activity, resulting in higher electron transport rates at high pCO2 and high irradiance, as well as higher assimilation rates. The gas exchange properties of AtpD-OE plants suggested that increased biomass and seed yield could be expected when plants are grown at high light and high pCO2. Further experiments including additional lines and targeted overexpression in leaves will clarify whether increasing the AtpD content presents a novel route for enhancing crop yield.
Acknowledgements
We thank Aleu Mani George for [14C]carboxyarabinitol bisphosphate binding assays, and the Australian Plant Phenomics Facility supported under the National Collaborative Research Infrastructure Strategy of the Australian Government for plant growth facilities.
Glossary
Abbreviations
- CEF
cyclic electron flow
- Cytb6f
cytochrome b6f complex
- ECS
electrochromic shift
- LEF
linear electron flow
- NPQ
non-photochemical quenching
- P700
reaction centre of PSI
- pCO2
CO2 partial pressure
- PGR5
PROTON GRADIENT REGULATION5
- PhiNA
non-photochemical yield of PSI caused by acceptor side limitation
- PhiND
non-photochemical yield of PSI caused by donor side limitation
- PhiNO
yield of non-regulated non-photochemical reactions in PSII
- PhiNPQ
yield of non-photochemical quenching
- PhiPSI
effective quantum yield of PSI
- PhiPSII
effective quantum yield of PSII
- pmf
proton motive force
- WT
wild type
Contributor Information
Maria Ermakova, Centre of Excellence for Translational Photosynthesis, Division of Plant Science, Research School of Biology, The Australian National University, Canberra, Australian Capital Territory, Australia.
Eiri Heyno, Centre of Excellence for Translational Photosynthesis, Division of Plant Science, Research School of Biology, The Australian National University, Canberra, Australian Capital Territory, Australia.
Russell Woodford, Centre of Excellence for Translational Photosynthesis, Division of Plant Science, Research School of Biology, The Australian National University, Canberra, Australian Capital Territory, Australia.
Baxter Massey, Centre of Excellence for Translational Photosynthesis, Division of Plant Science, Research School of Biology, The Australian National University, Canberra, Australian Capital Territory, Australia.
Hannah Birke, Centre of Excellence for Translational Photosynthesis, Division of Plant Science, Research School of Biology, The Australian National University, Canberra, Australian Capital Territory, Australia.
Susanne von Caemmerer, Centre of Excellence for Translational Photosynthesis, Division of Plant Science, Research School of Biology, The Australian National University, Canberra, Australian Capital Territory, Australia.
Elizabete Carmo-Silva, Lancaster University, UK.
Author contributions
SvC, HB, and ME: design; ME, EH, RW, BM, and HB: performing the research and data analysis; ME and SvC: writing.
Conflict of interest
The authors declare no conflict of interest.
Funding
This research was supported by the Australian Research Council Centre of Excellence for Translational Photosynthesis (CE140100015).
Data availability
Raw data and materials are available from the corresponding author upon request.
References
- Azcon-Bieto J, Osmond CB.. 1983. Relationship between photosynthesis and respiration. Plant Physiology 71, 574–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi RA, Aggeler R.. 2002. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends in Biochemical Sciences 27, 154–160. [DOI] [PubMed] [Google Scholar]
- Carmo-Silva E, Scales JC, Madgwick PJ, Parry MAJ.. 2015. Optimizing Rubisco and its regulation for greater resource use efficiency. Plant, Cell & Environment 38, 1817–1832. [DOI] [PubMed] [Google Scholar]
- Carrillo LR, Froehlich JE, Cruz JA, Savage LJ, Kramer DM.. 2016. Multi-level regulation of the chloroplast ATP synthase: the chloroplast NADPH thioredoxin reductase C (NTRC) is required for redox modulation specifically under low irradiance. The Plant Journal 87, 654–663. [DOI] [PubMed] [Google Scholar]
- Chida H, Nakazawa A, Akazaki H, et al. 2007. Expression of the algal cytochrome c6 gene in Arabidopsis enhances photosynthesis and growth. Plant & Cell Physiology 48, 948–957. [DOI] [PubMed] [Google Scholar]
- Cruz JA, Avenson TJ, Kanazawa A, Takizawa K, Edwards GE, Kramer DM.. 2005. Plasticity in light reactions of photosynthesis for energy production and photoprotection. Journal of Experimental Botany 56, 395–406. [DOI] [PubMed] [Google Scholar]
- Davis GA, Kramer DM.. 2020. Optimization of ATP synthase c-rings for oxygenic photosynthesis. Frontiers in Plant Science 10, 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrecht S, Schurmann K, Junge W.. 1989. Chloroplast ATP synthase contains one single copy of subunit δ that is indispensable for photophosphorylation. European Journal of Biochemistry 179, 117–122. [DOI] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S.. 2014. A Golden Gate modular cloning toolbox for plants. ACS Synthetic Biology 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Ermakova M, Arrivault S, Giuliani R, et al. 2021. Installation of C4 photosynthetic pathway enzymes in rice using a single construct. Plant Biotechnology Journal 19, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova M, Lopez-Calcagno PE, Raines CA, Furbank RT, von Caemmerer S.. 2019. Overexpression of the Rieske FeS protein of the cytochrome b6f complex increases C4 photosynthesis in Setaria viridis. Communications Biology 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR. 2013. Improving photosynthesis. Plant Physiology 162, 1780–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S.. 1981. Electron transport limitations on the CO2 assimilation rate of leaves: a model and some observations in Phaseolus vulgaris L. In: Akoyunoglou G, ed. Proceedings of the Fifth International Congress on Photosynthesis, Vol. IV. Philadelphia: Balaban, 163–175. [Google Scholar]
- Fisher N, Bricker TM, Kramer DM.. 2019. Regulation of photosynthetic cyclic electron flow pathways by adenylate status in higher plant chloroplasts. Biochimica et Biophysica Acta 1860, 148081. [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR.. 1989. The relationship between the quantum yield of photosynthetic electron-transport and quenching of chlorophyll fluorescence. Biochimica et Biophysica Acta 990, 87–92. [Google Scholar]
- Hahn A, Vonck J, Mills DJ, Meier T, Kühlbrandt W.. 2018. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360, eaat4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Johnson GN.. 2011. Regulation of cyclic and linear electron flow in higher plants. Proceedings of the National Academy of Sciences, USA 108, 13317–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Joliot A.. 2005. Quantification of cyclic and linear flows in plants. Proceedings of the National Academy of Sciences, USA 102, 4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa A, Kramer DM.. 2002. In vivo modulation of nonphotochemical exciton quenching (NPQ) by regulation of the chloroplast ATP synthase. Proceedings of the National Academy of Sciences, USA 99, 12789–12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa A, Ostendorf E, Kohzuma K, et al. 2017. Chloroplast ATP synthase modulation of the thylakoid proton motive force: implications for Photosystem I and Photosystem II photoprotection. Frontiers in Plant Science 8, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. 2021. The economics of organellar gene loss and endosymbiotic gene transfer. Genome Biology 22, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer C, Schreiber U.. 2008. Saturation pulse method for assessment of energy conversion in PS I. PAM Application Notes 1, 3. [Google Scholar]
- Kohzuma K, Cruz JA, Akashi K, Hoshiyasu S, Munekage YN, Yokota A, Kramer DM.. 2009. The long-term responses of the photosynthetic proton circuit to drought. Plant, Cell & Environment 32, 209–219. [DOI] [PubMed] [Google Scholar]
- Kohzuma K, Froehlich JE, Davis GA, Temple JA, Minhas D, Dhingra A, Cruz JA, Kramer DM.. 2017. The role of light–dark regulation of the chloroplast ATP synthase. Frontiers in Plant Science 8, 1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer D, Johnson G, Kiirats O, Edwards G.. 2004. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynthesis Research 79, 209–218. [DOI] [PubMed] [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP.. 2016. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354, 857–861. [DOI] [PubMed] [Google Scholar]
- Long SP, Marshall-Colon A, Zhu X-G.. 2015. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66. [DOI] [PubMed] [Google Scholar]
- López-Calcagno PE, Brown KL, Simkin AJ, Fisk SJ, Vialet-Chabrand S, Lawson T, Raines CA.. 2020. Stimulating photosynthetic processes increases productivity and water-use efficiency in the field. Nature Plants 6, 1054–1063. [DOI] [PubMed] [Google Scholar]
- Lyska D, Meierhoff K, Westhoff P.. 2013. How to build functional thylakoid membranes: from plastid transcription to protein complex assembly. Planta 237, 413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiwald D, Dietzmann A, Jahns P, Pesaresi P, Joliot P, Joliot A, Levin JZ, Salamini F, Leister D.. 2003. Knock-out of the genes coding for the Rieske protein and the ATP-synthase delta-subunit of Arabidopsis. Effects on photosynthesis, thylakoid protein composition, and nuclear chloroplast gene expression. Plant Physiology 133, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoë A. 2018. Photoinhibition or photoprotection of photosynthesis? Update on the (newly termed) sustained quenching component qH. Environmental and Experimental Botany 154, 123–133. [Google Scholar]
- Parry MAJ, Andralojc PJ, Scales JC, Salvucci ME, Carmo-Silva AE, Alonso H, Whitney SM.. 2013. Rubisco activity and regulation as targets for crop improvement. Journal of Experimental Botany 64, 717–730. [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE.. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta 975, 384–394. [Google Scholar]
- Pribil M, Labs M, Leister D.. 2014. Structure and dynamics of thylakoids in land plants. Journal of Experimental Botany 65, 1955–1972. [DOI] [PubMed] [Google Scholar]
- Price G, Yu J, Caemmerer S, Evans J, Chow W, Anderson J, Hurry V, Badger M.. 1995. Chloroplast cytochrome b6f and ATP synthase complexes in tobacco: transformation with antisense RNA against nuclear-encoded transcripts for the Rieske FeS and ATPD polypeptides. Functional Plant Biology 22, 285–297. [Google Scholar]
- Rantala M, Paakkarinen V, Aro E-M.. 2018. Separation of thylakoid protein complexes with two-dimensional Native-PAGE. Bio-protocol 8, e2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SP, Portis AR Jr. 1988. Involvement of stromal ATP in the light activation of ribulose-1,5-bisphosphate carboxylase/oxygenase in intact isolated chloroplasts. Plant Physiology 86, 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV. 2016. Nonphotochemical chlorophyll fluorescence quenching: mechanism and effectiveness in protecting plants from photodamage. Plant Physiology 170, 1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Andrews TJ, Badger MR, Price GD, von Caemmerer S.. 2000. The role of chloroplast electron transport and metabolites in modulating Rubisco activity in tobacco. Insights from transgenic plants with reduced amounts of cytochrome b/f complex or glyceraldehyde 3-phosphate dehydrogenase. Plant Physiology 122, 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder CA, Kramer DM.. 2000. Dark-interval relaxation kinetics (DIRK) of absorbance changes as a quantitative probe of steady-state electron transfer. Photosynthesis Research 66, 145–158. [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Portis AR, Ogren WL.. 1985. A soluble chloroplast protein catalyzes ribulosebisphosphate carboxylase/oxygenase activation in vivo. Photosynthesis Research 7, 193–201. [DOI] [PubMed] [Google Scholar]
- Schöttler MA, Tóth SZ.. 2014. Photosynthetic complex stoichiometry dynamics in higher plants: environmental acclimation and photosynthetic flux control. Frontiers in Plant Science 5, 188–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey TD, Bernacchi CJ, Farquhar GD, Singsaas EL.. 2007. Fitting photosynthetic carbon dioxide response curves for C3 leaves. Plant, Cell & Environment 30, 1035–1040. [DOI] [PubMed] [Google Scholar]
- Simkin AJ, López-Calcagno PE, Raines CA.. 2019. Feeding the world: improving photosynthetic efficiency for sustainable crop production. Journal of Experimental Botany 70, 1119–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, McAusland L, Lawson T, Raines CA.. 2017. Overexpression of the RieskeFeS protein increases electron transport rates and biomass yield. Plant Physiology 175, 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streusand VJ, Portis AR Jr. 1987. Rubisco activase mediates ATP-dependent activation of ribulose bisphosphate carboxylase 1. Plant Physiology 85, 152–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa K, Cruz JA, Kanazawa A, Kramer DM.. 2007. The thylakoid proton motive force in vivo. Quantitative, non-invasive probes, energetics, and regulatory consequences of light-induced pmf. Biochimica et Biophysica Acta 1767, 1233–1244. [DOI] [PubMed] [Google Scholar]
- Taylor SH, Gonzalez-Escobar E, Page R, Parry MAJ, Long SP, Carmo-Silva E.. 2022. Faster than expected Rubisco deactivation in shade reduces cowpea photosynthetic potential in variable light conditions. Nature Plants 8, 118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ballmoos C, Cook GM, Dimroth P.. 2008. Unique rotary ATP synthase and its biological diversity. Annual Review of Biophysics 37, 43–64. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S. 2000. Biochemical models of leaf photosynthesis. Collingwood: CSIRO Publishing. [Google Scholar]
- von Caemmerer S, Edmondson DL.. 1986. Relationship between steady-state gas-exchange, in vivo ribulose bisphosphate carboxylase activity and some carbon-reduction cycle intermediates in Raphanus sativus. Australian Journal of Plant Physiology 13, 669–688. [Google Scholar]
- von Caemmerer S, Evans JR.. 2015. Temperature responses of mesophyll conductance differ greatly between species. Plant, Cell & Environment 38, 629–637. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD.. 1981. Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153, 376–387. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Kramer DM, Fisher N, Fu X.. 2020. Flexibility in the energy balancing network of photosynthesis enables safe operation under changing environmental conditions. Plants 9, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Kromdijk J.. 2022. Here comes the sun: how optimization of photosynthetic light reactions can boost crop yields. Journal of Integrative Plant Biology 64, 564–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W, Kondo E, Sugiura D, Terashima I, Suzuki Y, Makino A.. 2016. Enhanced leaf photosynthesis as a target to increase grain yield: insights from transgenic rice lines with variable Rieske FeS protein content in the cytochrome b6/f complex. Plant, Cell & Environment 39, 80–87. [DOI] [PubMed] [Google Scholar]
- Yamori W, Shikanai T.. 2016. Physiological functions of cyclic electron transport around Photosystem I in sustaining photosynthesis and plant growth. Annual Review of Plant Biology 67, 81–106. [DOI] [PubMed] [Google Scholar]
- Yamori W, Takahashi S, Makino A, Price GD, Badger MR, von Caemmerer S.. 2011. The roles of ATP synthase and the Cytochrome b6f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiology 155, 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X-G, Long SP, Ort DR.. 2010. Improving photosynthetic efficiency for greater yield. Annual Review of Plant Biology 61, 235–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data and materials are available from the corresponding author upon request.