Abstract
Pulmonary atresia with intact ventricular septum (PA-IVS) is a rare congenital heart defect defined by membranous or muscular atresia of the right ventricular outflow tract where patients display varying degrees of hypoplasia of the right ventricle. This condition results in cyanosis due to an inability of blood to flow from the right ventricle to the pulmonary arteries, thus requiring immediate surgical intervention after birth. An iPSC line was generated from peripheral blood mononuclear cells of a 11-year-old male patient diagnosed with PA-IVS through Sendai virus-mediated reprogramming. This disease-specific iPSC line was characterized by immunocytochemistry, STR analysis, karyotype analysis, and mycoplasma testing.
1. Resource utility
An iPSC line (Resource Table) was generated from a patient with pulmonary atresia with intact ventricular septum using Sendai virus-mediated epigenetic reprogramming. Differentiated cardiac cells from this PA-IVS-specific iPSC line could serve as clinically relevant cell sources for modelling congenital heart disease and novel drug discovery (Lin et al., 2021).
Resource Table.
| Unique stem cell line identifier | NCHi003-A |
| Alternative name(s) of stem cell line | NCH005 (NCHi003-A) |
| Institution | Center for Cardiovascular Research, Abigail Wexner Research Institute, Nationwide Children’s Hospital, Columbus, OH, 43,215 |
| Contact information of distributor | Dr. Ming-Tao Zhao mingtao.zhao@nationwidechildrens.org |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info required for human ESC or iPSC | Age: 11 years Sex: male Ethnicity: Caucasian |
| Cell Source | Peripheral blood mononuclear cells |
| Clonality | Clonal |
| Method of reprogramming | Sendai virus reprogramming vector including human KLF4, OCT3/4, SOX2, and c-MYC |
| Genetic Modification | NO |
| Type of Genetic Modification | N/A |
| Evidence of the reprogramming transgene loss (including genomic copy if applicable) | N/A |
| Associated disease | Pulmonary Atresia with Intact Ventricular Septum |
| Gene/locus | N/A |
| Date archived/stock date | June 17, 2022 |
| Cell line repository/bank | NCHi003-A (NCH005) is deposited in the iPSC repository of pediatric cardiovascular disease in the Center for Cardiovascular Research at the Abigail Wexner Research Institute at Nationwide Children’s Hospital in Columbus, Ohio, USA. https://hpscreg.eu/cell-line/NCHi003-A |
| Ethical approval | This study was approved by the Internal Review Board (IRB) - STUDY00001788 at Nationwide Children’s Hospital. |
2. Resource details
PA-IVS is a rare congenital heart defect driven primarily by varying degrees of hypoplasia of the right ventricle and membranous or muscular atresia of the right ventricular outflow tract that results in cyanosis (Gorla and Singh, 2022; Rao, 2021; Wright et al., 2019). PA-IVS carries a significant risk of early morbidity and mortality that warrants immediate intervention after birth (Rao, 2021; Wright et al., 2019). The cause of PA-IVS is poorly understood and patient cell-based experimental models are not available (Hall et al., 2022). Here, an iPSC line was generated from the peripheral blood mononuclear cells (PBMCs) of a male patient with PA-IVS and verified to model stages of human development through differentiation into cells from the three germ layers.
An iPSC line was generated from the PMBCs of a patient diagnosed with PA-IVS through Sendai virus vector reprogramming (see Table 1). These PA-IVS iPSCs displayed typical stem cell morphology and colony formation (Fig. 1A). Immunofluorescence staining indicated the expression of pluripotency markers OCT3/4, NANOG, SOX2, TRA-1–60, in the majority of iPSCs (Fig. 1B and D). These iPSCs displayed a normal male karyotype (46, XY) that was confirmed by a whole genome array (Fig. 1C). In vitro germ layer differentiation showed positive staining for layer-specific markers, with endoderm-derived cells expressing FOXA2 and SOX17, mesoderm-derived cells expressing TBX6 and Brachyury, and ectoderm-derived cells expressing PAX6 and OTX2 (Fig. 1E). This PA-IVS iPSC line was tested negative for mycoplasma (Supplementary Fig. 1). In addition, short tandem repeat (STR) analysis confirmed that this iPSC line was originated from the PA-IVS patient (in archive with the journal).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
|
| |||
| Morphology | Photography, bright field | Normal | Fig. 1A |
| Phenotype | Qualitative analysis | Positive expression of OCT3/4, Nanog, Sox2, TRA-1–60 markers | Fig. 1D |
| Quantitative analysis | OCT3/4: 93 % ± 5 % NANOG: 95 % ± 5 % SOX2: 98 % ± 2 % |
Fig. 1B | |
| Genotype | Whole genome array (KaryoStat Assay) | Normal karyotype: 46, XY Resolution 1–2 Mb |
Fig. 1C |
| Identity | Microsatellite PCR (mPCR) OR STR analysis | mPCR not performed | N/A |
| 16 loci tested with matching identity | Submitted in archive with journal | ||
| Mutation analysis (IF APPLICABLE) | Sequencing | N/A | N/A |
| Southern blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Negative result | Supplementary Fig. 1 |
| Differentiation potential | Trilineage in vitro differentiation | Positive staining for three germ layers: Endoderm: SOX17, FOXA2 Mesoderm: Brachyury, TBX6 Ectoderm: PAX6, OTX2 |
Fig. 1E |
| Donor screening | HIV 1 + 2, Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A | |
Fig. 1.
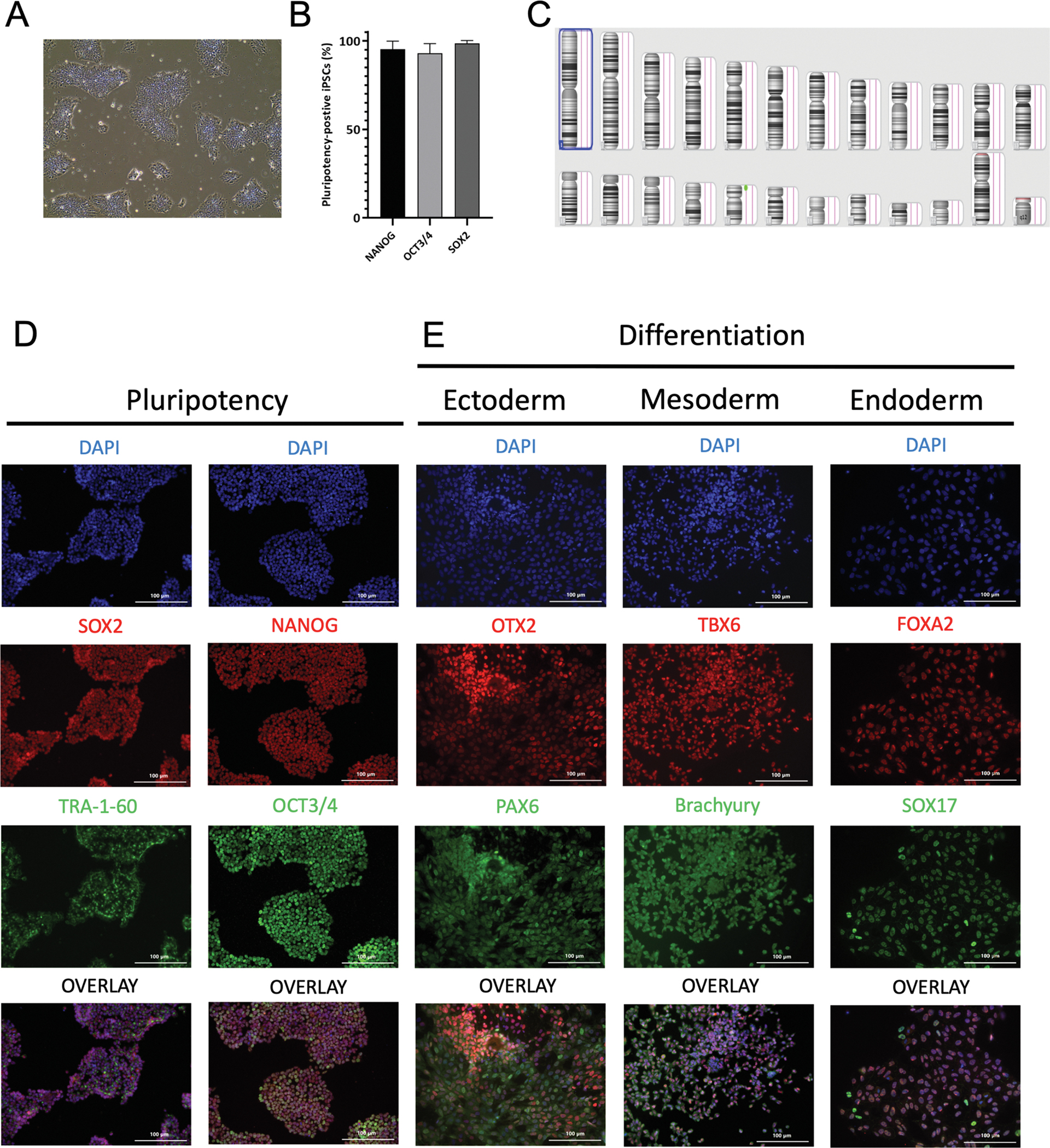
Characterization of an iPSC line derived from a PA-IVS patient.
3. Materials and methods
3.1. Reprogramming and iPSC culture
Patient PBMCs were isolated and incubated for one week in StemPro-34 SFM medium (Thermo Fischer Scientific) that was supplemented with 100 ng/mL SCF (PeproTech), 100 ng/mL FLT3 (Thermo Fischer Scientific), 20 ng/mL IL3 (PeproTech), 20 ng/mL IL6 (Gibco), 20 ng/mL EPO (Thermo Fischer Scientific), and 1X GlutaMAX (Thermo Fischer Scientific). PBMCs were infected using the CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Thermo Fischer Scientific) according to the manufacturer’s protocol. Transduced cells were resuspended in the supplemented StemPro-34 SFM medium and transferred into a Matrigel-coated plate for one week. Medium was then changed to Essential 8 (E8) medium (Thermo Fischer Scientific). After two weeks, emerging iPSC colonies were picked, expanded over multiple passages, and cryopreserved in a liquid nitrogen tank.
iPSCs were maintained in a CO2 incubator at 37 °C and 5 % CO2 in E8 medium. Media was refreshed daily. iPSCs were dissociated using 0.5 mM EDTA and passaged onto a Matrigel-coated plate with E8 medium supplemented with ROCK inhibitor (Y-27632, Selleck Chemicals).
3.2. Immunofluorescent staining
iPSCs (p43) were cultured on Matrigel-coated coverslips in a 24-well plate for 2 days. Cells were fixed with 4 % paraformaldehyde (PFA, Electron Microscopy Sciences) for 15 min. After permeabilization with 0.1 % Triton-X-100 (Sigma) solution for 20 min, cells were blocked with 1X DPBS/0.2 % Bovine Serum Albumin (BSA, Sigma) for 1 h at room temperature. Cells were incubated with primary antibodies diluted in 0.2 % BSA solution overnight at 4 °C (see Table 2). After washing 3 times, cells were then incubated with secondary antibodies for 1 h in the dark. Cells were washed 3 times using 1X DPBS, and counterstained with DAPI solution for 10 min. Coverslips were mounted onto slides with SlowFade Gold Antifade (Thermo Fischer Scientific) and imaged with a fluorescence microscope (Keyence). All steps were performed at room temperature unless otherwise described.
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | ||||
|---|---|---|---|---|
|
|
|
|||
| Antibody | Dilution | Company Cat # | RRID | |
|
| ||||
| Pluripotency Markers | Alexa Fluor 488 Mouse anti-OCT3/4 | 1:2000 | BD Biosciences Cat No: 561,628 | AB_10895977 |
| Pluripotency Markers | Rabbit anti-NANOG | 1:200 | Cell Signaling Technology Cat No: 4903P | AB_10611718 |
| Pluripotency Markers | Mouse anti-TRA-1–60 | 1:200 | Thermo Fischer Scientific Cat No: MA1-023X | AB_2536705 |
| Pluripotency Markers | Rabbit anti-SOX2 | 1:200 | Thermo Fischer Scientific Cat No: PA1-094X | AB_2539863 |
| Endoderm Marker | Goat anti-SOX17 | 1:200 | R&D Systems Cat No: AF1924 | AB_355060 |
| Endoderm Marker | Mouse anti-FOXA2 | 1:200 | Abnova Cat No: H00003170-M10 | AB_534871 |
| Mesoderm Marker | Goat anti-Brachyury | 1:200 | R&D Systems Cat No: AF2085 | AB_2200235 |
| Mesoderm Marker | Rabbit anti-TBX6 | 1:200 | Thermo Fisher Scientific Cat No: PA5-35102 | AB_2552412 |
| Ectoderm Marker | Goat anti-OTX2 | 1:200 | R&D Systems Cat No: AF1979 | AB_2157172 |
| Ectoderm Marker | Rabbit anti-PAX6 | 1:200 | Thermo Fisher Scientific Cat No: 42–6600 | AB_2533534 |
| Secondary Antibody | Goat anti-Mouse IgG (H + L), Alexa Fluor 594 | 1:2000 | Thermo Fisher Scientific Cat# A-11032 | AB_2534091 |
| Secondary Antibody | Goat anti-Mouse IgG (H + L), Alexa Fluor 488 | 1:2000 | Thermo Fisher Scientific Cat# A-11001 | AB_2534069 |
| Secondary Antibody | Goat anti-Rabbit IgG (H + L), Alexa Fluor 594 | 1:2000 | Thermo Fisher Scientific Cat# A-11012 | AB_2534079 |
| Secondary Antibody | Donkey anti-Mouse IgG (H + L), Alexa Fluor 594 | 1:2000 | Thermo Fisher Scientific Cat# R37115 | AB_2556543 |
| Secondary Antibody | Donkey anti-Rabbit IgG (H + L), Alexa Fluor 594 | 1:2000 | Thermo Fisher Scientific Cat# R37119 | AB_2556547 |
| Secondary Antibody | Donkey anti-Goat IgG (H + L), Alexa Fluor Plus 488 | 1:2000 | Thermo Fisher Scientific Cat# A32814 | AB_2762838 |
3.3. Short tandem repeat (STR) analysis
Genomic DNA of iPSCs (p52) and patient’s PBMCs was extracted using Quick-DNA Miniprep Plus Kit (Zymo Research). STRs were amplified using the PowerPlex16 (Promega) system and resolved on a ABI 3730xl Genetic Analyzer (Thermo Fischer Scientific). GeneMapper 5.0 (Thermo Fischer) software was used to call the genotype at each locus and compare between peripheral blood and iPSC samples.
3.4. In vitro differentiation potential
The differentiation potential of iPSCs (p46) was verified through cell differentiation into the three germ layers by measuring the expression of endoderm (OTX2, PAX6), mesoderm (Brachyury, TBX6), and ectoderm (SOX17, FOXA2) markers. Cells of endoderm and ectoderm lineages were generated using a Human Pluripotent Stem Cell Functional Identification Kit (R&D Systems). Cells of mesoderm layers were induced with 6 μM CHIR99021 (Selleck Chemicals) in RPMI 1640 media (Thermo Fischer Scientific) with B27 Supplement minus insulin (Thermo Fischer Scientific) for 2 days. Samples were fixed and stained for their respective markers.
3.5. Karyotype analysis
Human iPSCs (p61) were harvested and sent for KaryoStat+ Assay Service provided by Thermo Fisher Scientific.
3.6. Mycoplasma detection
Supernatant of dense iPSC cultures (p45) was tested using the MycoAlert™ Mycoplasma Detection Kit (Lonza Biosciences).
Supplementary Material
Acknowledgements
This work was partially supported by the NIH/NHLBI grant R01HL155282 (M-T.Z), Diversity Supplement R01HL155282-02S1 (M. A.), Diversity Supplement R01HL155282-02S2 (J.C.), American Heart Association (AHA) Career Development Award 18CDA34110293 (M-T.Z.), Additional Ventures Innovation Fund (AVIF) (V.G. and M-T.Z.), Single Ventricle Research Fund (SVRF) (K.T., V.G., and M-T.Z.), and Tools and Technology Expansion Award (K.T., V.G., and M-T.Z.). The authors would like to acknowledge Dr. Dennis Lewandowski for his assistance in editing this manuscript.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2022.102893.
References
- Gorla SR, Singh AP, 2022. Pulmonary Atresia With Intact Ventricular Septum. StatPearls, StatPearls Publishing Copyright © 2022. StatPearls Publishing LLC., Treasure Island (FL). [PubMed] [Google Scholar]
- Hall B, Alonzo M, Texter K, Garg V, Zhao MT, 2022. Probing single ventricle heart defects with patient-derived induced pluripotent stem cells and emerging technologies. Birth Defects Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, McBride KL, Garg V, Zhao MT, 2021. Decoding Genetics of Congenital Heart Disease Using Patient-Derived Induced Pluripotent Stem Cells (iPSCs). Front. Cell Dev. Biol. 9, 630069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PS, 2021. Single Ventricle—A Comprehensive Review. Children (Basel) 8 (6), 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LK, Knight JH, Thomas AS, Oster ME, St Louis JD, Kochilas LK, 2019. Long-term outcomes after intervention for pulmonary atresia with intact ventricular septum. Heart 105 (13), 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


