Abstract
Purpose
Next-generation sequencing (NGS)-based tumor panel testing has been reimbursed by the Korean government since 2017. We evaluated the use of NGS-based tumor panel testing in real-world clinical practice, focusing on molecular profiling (MP)-guided breast cancer treatment.
Methods
A total of 137 breast cancer patients underwent NGS panel testing between December 2017 and July 2020 at Seoul National University Bundang Hospital (SNUBH). Samples from patients were profiled using an in-house SNUBH pan-cancer panel. Sixty-four patients were profiled on SNUBH Pan_Cancer v1.0, targeting 89 genes, while 73 patients were profiled on SNUBH Pan_Cancer v2.0, targeting 546 genes.
Results
Breast cancer subtypes included hormone receptor+/human epidermal growth factor receptor 2 (HER2)− (n = 87), triple-negative (n = 44), and HER2+ (n = 6). Most patients had locally advanced or metastatic cancers (92%). Approximately 92% (126/137) of the patients had significant genomic alterations (tiers I and II), and 62% (85/137) had targetable genomic alterations. The most common targetable genomic alterations were PIK3CA (39%) and ESR1 mutations (9%), followed by ERBB2 (7%), PTEN (7%), BRCA2 (6%), and BRCA1 mutations (4%). Of the 81 patients with locally advanced/metastatic breast cancer with targetable genomic alterations, 6 (7.4%) received MP-guided treatments, including PARP inhibitor (n = 4), ERBB2-directed therapy (n = 1), and PI3K inhibitor (n = 1). Among these 6 patients, 4 participated in clinical trials, 1 underwent treatment at their own expense, and 1 received drugs through an expanded access program. The remaining 66 patients (81%) with targetable genomic alteration did not receive MP-guided treatment due to lack of matched drugs and/or clinical trials, poor performance status, and/or financial burden.
Conclusion
NGS panel testing allowed MP-guided treatment in only 4.7% (6/127) of patients with advanced breast cancer in a real-world setting. The availability of matched drugs is critical for the realistic implementation of personalized treatment.
Keywords: Breast Neoplasms, High-Throughput Nucleotide Sequencing, Precision Medicine
INTRODUCTION
Breast cancer is the second most common cancer worldwide [1], with 22,395 patients newly diagnosed with invasive breast cancer in 2017 in Korea [2]. Although patients with breast cancer show longer survival compared to patients with other cancer types, the 5-year relative survival rate of patients with breast cancer with distant metastasis diagnosed in 2013–2017 was 34.5%. Moreover, breast cancer ranked first (15%) as the leading cause of cancer-related death in women in Korea.
Breast cancer risk assessment and treatment have traditionally been based on hormone receptor (HR) and human epidermal growth factor receptor 2 (HER2) statuses. Advancements in molecular analysis have allowed the further subdivision of breast cancer [3]. In recent years, multigene panels such as Oncotype DX® and MammaPrint® have been commonly used to assess the risk of breast cancer recurrence and the benefits of adjuvant chemotherapy in HR+ breast cancer. Alpelisib and olaparib are approved by the Food and Drug Administration (FDA) for the treatment of patients with HR+/HER2− metastatic breast cancer with PIK3CA and germline BRCA mutations, respectively [4,5]. Pembrolizumab was approved by the FDA for use in the treatment of microsatellite instability-high (MSI-H) solid tumors and tumors with high tumor mutation burden (TMB) [6], whereas larotrectinib and entrectinib were approved for use in the treatment of solid tumors harboring an NTRK gene fusion [7]. In this era of personalized treatment, the need for molecular analysis of breast cancer has become increasingly important.
Next-generation sequencing (NGS) has enabled the identification of novel potential targets for patients with cancer, with targeted NGS panels becoming the most practical genome profiling methods worldwide [8]. NGS panel testing has been reimbursed by the National Health Insurance Service of Korea since March 2017 and has been rapidly adopted in actual clinical practice in Korea. However, data on the clinical application of NGS testing in patients with breast cancer in clinical practice are limited, and the benefits of incorporating it in precision medicine remain controversial.
This study aimed to determine the frequency of targetable genomic alterations using NGS and to assess whether they can be considered targets of molecular profiling (MP)-guided treatments for breast cancer in daily clinical practice.
METHODS
Study population
This study included 137 patients with histologically confirmed breast cancer treated at Seoul National University Bundang Hospital (SNUBH) and who underwent NGS panel testing at the physician’s discretion between December 2017 and July 2020. Tumor histology data, including estrogen receptor (ER), progesterone receptor (PR), HER2, and Ki-67 status, as well as radiological and/or pathological staging, were collected. This study was approved by the Institutional Review Board of SNUBH (B-2010/645-106) and conducted according to the principles of the Declaration of Helsinki. Before NGS panel testing, a consent form for the donation of human materials was completed and submitted in accordance with the Enforcement Regulations of the Bioethics and Safety Act in Korea. The requirement for informed consent was waived by the institutional review board owing to the retrospective study design. Medical records were retrieved from electronic health records, de-identified, and anonymized before the study. No individual-level data were reported.
Analysis of tumor subtypes
Immunohistochemical staining of formalin-fixed paraffin-embedded tissue was performed at the initial diagnosis or at the time of recurrence of metastatic disease. Nuclear tumor cell expression was considered ER- and PR-positive, while membrane staining of tumor cells was considered positive for HER2. The results indicated positive ER and PR expression when ≥ 1% of tumor cells were stained according to the 2010 American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines [9]. HER2 positivity was assessed according to the 2013 ASCO/CAP guidelines [10]. Patients were categorized as having “HR+/HER2− breast cancer,” “HER2+ breast cancer,” or “triple-negative breast cancer (TNBC).”
NGS panel testing and data analysis
Tumor tissues for NGS testing were obtained from archived samples. The samples were profiled on the SNUBH PanCancer panel, which is a targeted sequencing platform in SNUBH, based on the customized Macrogen cancer panel (Seoul, Korea). Sixty-four patients were profiled on SNUBH Pan_Cancer v1.0, which targeted 89 genes, whereas 73 patients were profiled on SNUBH Pan_Cancer v2.0, which targeted 546 genes. MSI and TMB results were reported only for the SNUBH V2 system. Both Pan_Cancer v1.0 and v2.0 were developed by the same manufacturer, with no differences between the 2 panels in terms of sequencing method and results interpretation. Moreover, both panels used HG19 as the human reference genome. A list of the genes included in each panel is provided in Supplementary Table 1.
Samples with coverage < 80% did not meet the quality control standards. Single-nucleotide variants (SNVs) and small insertionå/deletions (indels) were detected using Mutect2, whereas variants were annotated using SnpEff. The variant allele frequency of SNVs/indels was ≥ 2%. We identified copy number variation (CNV) using CNVkit, with an average CN of ≥ 5 defined as a gain (amplification). Gene fusion was determined using LUMPY [11]. For translocations, read counts ≥ 3 were interpreted as positive. MSI was detected using MSI phenotype using NGS (mSINGS) [12]. To calculate TMB, we selected eligible variants that met the following criteria: population DB filter (Exome Aggregation Consortium East Asian [ExAC_EAS] < 1%, gnomAD_EAS < 1%, Korean (in-house DB) < 1%), variant type (nonsynonymous variants only), driver mutation (excluding pathogenic, likely pathogenic variants [Clinvar]), variant allele frequency (≥ 2%), and depth (≥ 200×). TMB was calculated as eligible variants/1.411 MB.
Tiers were classified according to standardized guidelines for the interpretation and reporting of sequence variants in cancer [13]. Somatic variants were classified into 4 tiers based on their level of clinical significance in cancer diagnosis, prognosis, and/or therapeutic, as follows: tier I, variants with strong clinical significance such as FDA-approved, professional guidelines, or well-powered research-based therapy; tier II, variants of potential clinical significance such as FDA-approved treatment for different tumor types or investigational therapies; tier III, variants of unknown clinical significance; and tier IV, benign or likely benign. “Significant genomic alterations” were defined as tier I and II genomic alterations, while “targetable genomic alterations” were defined as genomic alterations with specific targeted therapy. For example, tier II TP53 mutations are significant genomic alterations used in the diagnosis and prognosis of cancer but are not classified as a “targetable genomic alteration” due to the lack of specific targeted therapies. Results of poor quality and suspected errors based on the following criteria were filtered out: variants with < 5% allele frequency, variants with < 100× coverage, and variants in the intron region. The results of the final analysis for each case were reviewed and reported by a professional pathologist.
Statistical analysis
This study aimed to describe the frequency of targetable genomic alterations using NGS and to determine whether they can be used as targets of MP-guided treatments for breast cancer in daily clinical practice. Due to the observational nature of the study, the sample size was not calculated. The variables were presented as median values for continuous variables and percentages (numbers) for categorical variables. Categorical and continuous variables were compared using χ2 and independent samples t-tests, respectively. Progression-free survival (PFS) was calculated using the Kaplan-Meier method. Missing data were not imputed. All tests were 2-sided, and p < 0.05 was considered significant. All analyses were performed using IBM SPSS Statistics for Windows, version 25.0 (IBM, Armonk, NY, USA) and GraphPad Prism 9 (GraphPad Software, Inc., La Jolla, CA, USA).
RESULTS
Patient characteristics and sample information
The clinical characteristics of the patients are summarized in Table 1. The median age was 47 years (range, 30–84 years), and all patients were women. Approximately 88% of the patients (n = 120) had invasive ductal carcinoma, whereas 93% (n = 127) had locally advanced or metastatic breast cancer. HR+/HER2− was the most frequently identified breast cancer subtype (63%, n = 87). Of the 127 patients with locally advanced/metastatic cancer, 41 (32%) had de novo metastatic breast cancer and 86 (68%) had recurrent locally advanced/metastatic cancer. Among the 127 patients with locally advanced/metastatic cancer, 53 (42%) underwent NGS testing at the time of diagnosis of advanced disease, while and (58%) underwent NGS testing during palliative treatment. All 10 patients with resectable breast cancer underwent NGS at the time of diagnosis.
Table 1. Patient characteristics (n = 137).
| Characteristics | No. (%) | ||
|---|---|---|---|
| Age at the time of NGS, median (range) | 51 (34–84) | ||
| Histologic diagnosis | |||
| Invasive ductal carcinoma | 120 (88) | ||
| Invasive lobular carcinoma | 11 (8) | ||
| Others* | 6 (4) | ||
| Subtype | |||
| HR+/HER2− | 87 (63) | ||
| HER2-positive | 6 (4) | ||
| TNBC | 44 (32) | ||
| Stage | |||
| Operable | 10 (7) | ||
| Locally advanced/metastatic | 127 (93) | ||
| NGS panel | |||
| Version 1 | 64 (47) | ||
| Version 2 | 73 (53) | ||
| Timing of NGS | |||
| Operable | |||
| At diagnosis | 10 (100) | ||
| Locally advanced/metastatic | |||
| At diagnosis | 53 (42) | ||
| During palliative treatment | 74 (58) | ||
NGS, next-generation sequencing; HR, hormone receptor; HER2, human epidermal growth factor; TNBC, triple-negative breast cancer.
*Mucinous carcinoma (n = 2), metaplastic carcinoma (n = 2), adenoid cystic carcinoma (n = 1), invasive micropapillary carcinoma (n = 1).
Of the study samples, 82 were obtained by biopsy and 55 were obtained during surgical resection (Table 2). The most common biopsy site was the breast (36%), followed by the liver (19%), lymph nodes (13%), lungs (10%), and skin/soft tissue (7%). Before tissue acquisition for NGS, 61% (n = 84) and 46% (n = 63) of patients received chemotherapy (median lines of treatment: 2; range: 1–7) and endocrine therapy (median: 1; range 1–4), respectively. The median tumor fraction was 70% (range:20%–90%), with 99% of patients showing 100× coverage ≥ 80%.
Table 2. Tissue acquisition methods and quality measures.
| Characteristics | No. (%) | |
|---|---|---|
| Specimen type | ||
| Biopsy | 82 (60) | |
| Resection | 55 (40) | |
| Biopsy site | ||
| Breast | 50 (36) | |
| Liver | 26 (19) | |
| Lymph node | 18 (13) | |
| Lung | 14 (10) | |
| Skin/Soft tissue | 10 (7) | |
| Bone | 8 (6) | |
| Others* | 11 (9) | |
| Prior CT before tissue acquisition | 84 (61) | |
| Anthracycline | 66 (48) | |
| Taxane | 72 (53) | |
| No. of prior lines of CT, median (range) | 2 (1–7) | |
| Prior ET before tissue acquisition | 63 (46) | |
| No. of prior lines of ET, median (range) | 1 (1–4) | |
| Tumor fraction, median (range) | 70 (20–90) | |
| Mean depth, median (range) | 755 (249–2,565) | |
| 100× coverage (%), median (range) | 96.52 (75.46–99.72) | |
| 100× coverage ≥ 95% (pass) | 93 (68) | |
| 95 > 100× coverage ≥ 80% (caution) | 43 (31) | |
| 100× coverage < 80% (fail) | 1 (1) | |
CT, chemotherapy; ET, endocrine therapy.
*Ovary, chest wall (n = 3, respectively), pleura, brain (n = 2, respectively), mediastinum (n = 1).
Mutation landscape
A clinical report containing SNVs, copy number alterations (CNAs), and gene rearrangements detected by NGS and clinical implementation was generated for 137 cases.
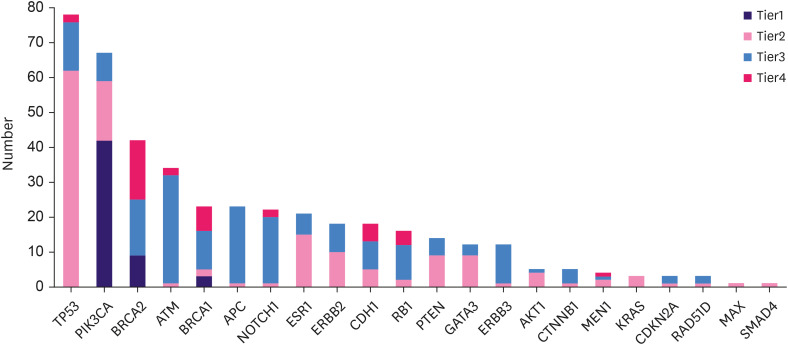
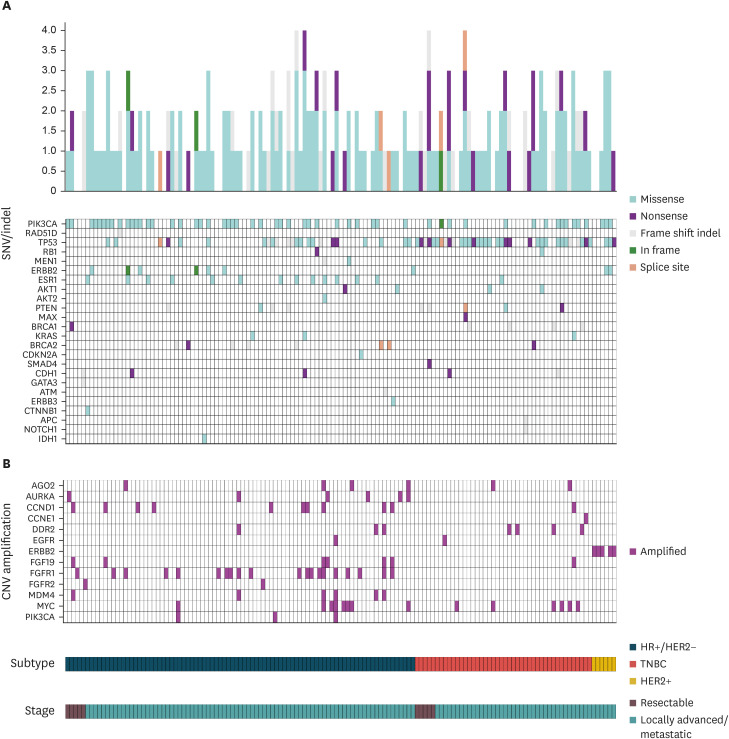
Overall, the cohort revealed 3,620 SNVs/indels. These variants were classified according to the tier system as follows: tier I, 54/3,620 (1.5%); tier II, 141/3,620 (3.9%); tier III, 3,199/3,620 (88.4%); and tier IV, 226/3,620 (6.2%). In tier I, alterations were observed in 3 oncogenes (PIK3CA, BRCA1, and BRCA2), most commonly PIK3CA (77.8% of cases [42/54]), followed by BRCA2 mutations (16.7% [9/54]) and BRCA1 mutations (5.6% [3/54]). Of the 12 patients with tier I BRCA1 and BRCA2 mutations confirmed by NGS testing, 6 (50%) underwent germline BRCA tests, 4 of which were confirmed to have germline BRCA mutations. In tier II, alterations were observed in 20 oncogenes, most frequently TP53 (44.0%, 62/141), PIK3CA (12.1%, 17/141), ESR1 (10.6%, 15/141), ERBB2 (7.1%, 10/141), PTEN (6.4%, 9/141), CDH1 (3.5%, 5/141), and AKT1 (2.8%, 4/141). The details of the frequencies of all tiers I and II gene mutations and whether they were considered targetable genomic alterations are described in Supplementary Table 2. The distribution of SNVs/indels of the selected genes according to tier classification is illustrated in Figure 1. The overall genomic landscape of all the study participants is shown in Figure 2, with Figure 2A showing the number of SNVs/indels of selected genes and the effect of the gene mutation (missense, nonsense, frameshift, in-frame insertion or deletion, and splice site mutation) for each patient.
Figure 1. Distribution of gene alterations (SNVs/indels) (AF ≥ 2%, depth ≥ 100×).
SNV, single-nucleotide variant; indel, insertion or deletion; AF, allele frequency.
Figure 2. Recurrent genes affected by SNVs/indels, copy number alteration. (A) Number and type of SNVs/indels events. (B) Copy number amplifications per sample.
SNV, single-nucleotide variant; CNV, copy-number variation; TNBC, triple-negative breast cancer; HER2, human epidermal growth factor receptor 2.
There were 512 CNAs in the entire cohort. These included 5 tier I alterations (1.0%), 149 tier II alterations (29.1%), and 358 tier III alterations (69.9%). All tier I CNAs exhibited ERBB2 amplification (median CN = 16; range: 8–132). The most common tier II CNA was FGFR1 amplification (n = 20; median CN = 7.5; range: 5–38), followed by MYC amplification (n = 14; median CN = 7; range: 5–37) and CCND1 amplification (n = 11; median CN = 7; range: 5–20). The tier II alterations were DDR2 (n = 7), FGF19 (n = 7), AGO2 (n = 6), AURKA (n = 6), MDM4 (n = 6), PIK3CA (n = 3), EGFR (n = 2), FGFR2 (n = 2), and CCNE1 (n = 1). Figure 2B shows the tier I and II gene amplifications for each patient.
MSI-H tumors and chromosomal translocations were detected in one patient of 73 tested patients (1.4%, MSI-H = 1) and NTRK2-PAN3 fusion in one of 137 tested patients (0.7%).
TMB was evaluated in 72 of the 73 patients tested with SNUBH V2. TMB was not assessed in one patient owing to sequencing quality. The median TMB was 8.5/Mb for all patients (range: 2.8–24.8). TMB did not differ significantly according to age (≤ 50 years vs. > 50 years, 7.1/Mb vs. 9.2/Mb, p = 0.808), subtype (HR+/HER2−, 8.5/Mb; HER2+, 10.3/Mb; TNBC, 8.5/Mb; p = 0.767), or NGS timing (at diagnosis, 8.5/Mb; at recurrence, 9.2/Mb; during palliative treatment, 8.5/Mb; p = 0.434) (Supplementary Figure 1). The median TMB was 8.5/Mb in both groups that had and had not previously received endocrine therapy (p = 0.892). Previous chemotherapy also did not affect TMB status (8.9/Mb for the chemotherapy-naïve group and 8.5/Mb for the chemotherapy-exposed group; p = 0.200).
Clinically significant mutations and application of MP-guided treatment
Approximately 92% (126/137) of patients had significant (tiers I and II) genomic alterations, while 62% (85/137) had targetable genomic alterations.
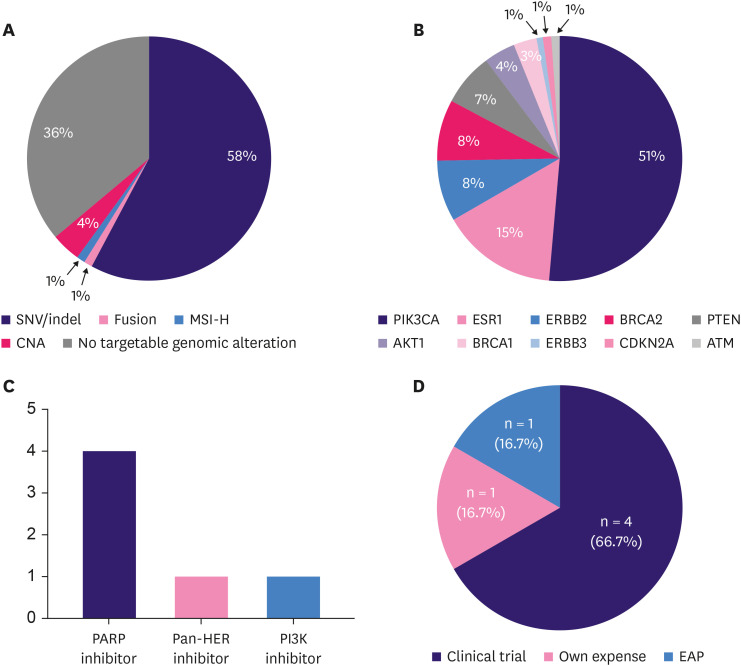
Among patients with locally advanced/metastatic breast cancer, 63.8% (81/127) had targetable genomic alterations, including SNVs/indels (58%, n = 74), CNAs (4%, n = 5), MSI-H (1%, n = 1), and fusion (1%, n = 1) (Figure 3A). The 74 patients with targetable genomic alterations in SNVs/indels had 95 targetable genomic alterations, including 21 patients with more than 2 targetable genomic alterations in SNVs/indels. The most common targetable genomic alterations involving SNVs/indels were PIK3CA mutations (n = 49), followed by ESR1 (n = 14), ERBB2 (n = 8), BRCA2 (n = 8), PTEN (n = 7), AKT1 (n = 4), and BRCA1 (n = 3) mutations (Figure 3B).
Figure 3. MP-guided treatment. (A) Targetable genomic alterations (n = 81) in locally patients with advanced/metastatic breast cancer (n = 127). (B) Composition of targetable genomic alterations of SNVs/indels. (C) Types of MP-guided treatment in patients with advanced breast cancer with targetable genomic alterations. (D) Access to MP-guided treatment.
MP, molecular profiling; NHS, National Health Insurance Service; EAP, expanded access program; SNV, single-nucleotide variant; indel, insertion or deletion; MSI-H, microsatellite instability-high; HER, human epidermal growth factor receptor.
Of the 81 patients with advanced breast cancer with targetable genomic alterations, 7 with ERBB2 alterations received HER2-directed therapy and 2 with BRCA1 mutations received PARP inhibitor therapy. These 9 patients were not categorized as receiving MP-guided treatment because the ERBB2 overexpression and/or amplification and germline BRCA1 mutation status were known before NGS testing and tumor NGS testing did not change the treatment plan for these patients.
Among the 81 patients with advanced breast cancer with targetable genomic alterations, 6 (7.4%) received MP-guided treatment. Four patients with BRCA1 or BRCA2 mutations received a PARP inhibitor, one patient with an ERBB2 p.S310F mutation received neratinib, and one patient with a PIK3CA p.E545K mutation received a PI3K inhibitor (Figure 3C). The details of the 6 patients who received MP-guided treatment are presented in Table 3. The median PFS of MP-guided treatment for all 6 patients was 5.6 months (range, 1.2–9.6 months).
Table 3. Treatment of 6 patients who received molecular-profiling guided treatment.
| Age | Subtype | Timing of NGS | Gene | Genetic alteration | Drug access | Received target therapy | Line of Tx | PFS (mo) |
|---|---|---|---|---|---|---|---|---|
| 69 | HR+/HER2− | During palliative Tx | BRCA1 | p.I1859fs | At own expense | Olaparib | 5th | 6.2 |
| 46 | HR+/HER2− | At initial Dx | BRCA2 | c.8633-2A>T | Clinical trial | Olaparib | 3rd | 2.6 |
| 48 | TNBC | At initial Dx | BRCA1 | p.K608fs | Clinical trial | Olaparib | 2nd | 5.3 |
| 44 | HR+/HER2− | During palliative Tx | BRCA2 | p.R2494* | Clinical trial | PARP/TNK inhibitor | 6th | 1.2 |
| 49 | HR+/HER2− | During palliative Tx | ERBB2 | p.S310F | EAP | Neratinib | 4th | 5.8 |
| 63 | HR+/HER2− | During palliative Tx | PIK3CA | p.E545K | Clinical trial | Alpelisib + fulvestrant | 2nd | 9.6 |
NGS, next-generation sequencing; HER2, human epidermal growth factor; TNBC, triple-negative breast cancer; HR, hormone receptor; Dx, diagnosis; Tx, treatment; EAP, expanded access program.
Of the 4 patients with BRCA mutations, one with a BRCA2 p.I1859fs mutation received olaparib at her own expense did not initially undergo a germline BRCA test because it did not meet the insurance criteria for testing. Germline BRCA gene testing was performed for confirmation only after the NGS test results were reported. This patient remained in the MP-guided treatment group because the tumor NGS test influenced further testing and treatment. A patient with a BRCA2 c.8633-2A>T mutation tested negative for germline BRCA mutations and received olaparib as part of a clinical trial of patients with somatic BRCA mutations. Another patient with a BRCA1 p.K608fs mutation entered the clinical trial and received olaparib. Participants were eligible for this trial if they had homologous recombination repair gene mutations. The clinical assay used in this trial was performed using tumor tissue. Therefore, it was not possible to determine whether the variants were somatic or germline in origin. The patient did not undergo germline BRCA testing. Finally, a patient with a BRCA2 p.R2494 mutation was enrolled in a phase 1 clinical trial of a PARP/TNK inhibitor. Although the patient later tested positive in the germline BRCA test, the decision to enroll in the clinical trial was based on the NGS results. Therefore, this patient remained in the MP-guided treatment group.
Among the 6 patients who received MP-guided treatment, 4 (66.7%) participated in clinical trials, 1 (16.7%) underwent treatment at their own expense, and 1 (16.7%) received drugs through an expanded access program (Figure 3D).
The remaining 66 patients (81%) with targetable genomic alteration did not receive MP-guided treatment. Seventeen patients (25.8%) received endocrine therapy along with CDK4/6 inhibitors as first-line treatment, while 4 patients (6.0%) were followed up without treatment after undergoing palliative resection for oligometastatic disease. The remaining 44 patients (66.7%) were unable to receive MP-guided treatment owing to the lack of matched drugs and/or clinical trials, declining performance status, and/or financial burden.
One patient with an MSI-H tumor also had a germline BRCA1 p.V1833fs mutation and progressed after receiving cytotoxic chemotherapy (paclitaxel and bevacizumab) as first-line treatment. She was included in a clinical trial and was administered eribulin and nivolumab as second-line treatments; however, as the tumor spread to her central nervous system after the first cycle of treatment, she left the trial. The patient subsequently received olaparib and had a PFS of 2.2 months. NTRK2-PAN3 fusion was detected in one patient, but the patient did not receive a TRK inhibitor because of a lack of drug availability and rapid tumor progression.
DISCUSSION
The results of our study demonstrated the usefulness of NGS panel testing for the detection of pathogenic alterations, allowing MP-guided treatment in 4.7% (6/127) of patients with advanced breast cancer and 7.4% (6/81) of patients with advanced breast cancer with targetable genomic alterations. However, NGS panel testing may not always lead to the provision of subsequent therapy because of the deterioration of the patient’s clinical condition, lack of drug availability, difficulty in accessing relevant clinical trials, and financial burden.
NGS panel testing has not only been approved in many countries but has also been covered by health insurance. The FoundationOne®CDx and Oncomine™ Dx Target Test have been approved by the US FDA [14], and the Memorial Sloan Kettering Cancer Center’s Integrated Mutation Profiling of Actionable Cancer Targets NGS assay has received FDA marketing authorization [14]. The Centers for Medicare and Medicaid Services stated that FoundationOne®CDx will receive national coverage for the treatment of all solid tumors in the US [15]. In Korea, most clinical NGS tests are laboratory-developed tests, which have been covered by the National Health Insurance Service since March 2017. Since then, NGS panel testing has been rapidly adopted, with 13,172 tests performed in 2020 [16]. Several studies have reported NGS test results in daily clinical practice [17,18,19,20].
Despite the relatively rapid adaptation of NGS panel testing in clinical practice, the benefits of incorporating NGS in improving PFS and overall survival (OS) remain controversial. Few randomized trials have reported the use of NGS-based treatment approaches. The SHIVA trial, the only precision medicine randomized controlled phase 2 trial, indicated that the use of NGS to match patients to appropriate targeted treatments regardless of cancer type did not improve PFS [21]. However, the NGS-based treatment approach improved the OS of patients with lung cancer [19,22,23]. In oncology, precision medicine studies that evaluated various types of cancers, including breast cancer, showed that MP-matched treatment improved response rate and PFS [8,24]. In our study, patients with advanced breast cancer who received MP-matched treatment had a median PFS of 5.8 months. Although no comparative analysis was performed to evaluate the efficacy of MP-matched and non-matched treatments owing to the heterogeneity of breast cancer subtypes and MP-matched treatment lines, our results suggest that using NGS panel testing to match patients to an appropriate therapy might improve patient outcomes in daily clinical practice.
Another important aspect to consider when using NGS in cancer treatment is the small proportion of sequenced patients with targetable mutations who are eventually treated with sequencing-matched therapies. In the NCI Molecular Analysis for Therapy Choice study, before interim analysis, 5.1% (33 of 645) of patients were eligible for assignment to a sub-protocol arm, of which only 2.5% (16 patients) were enrolled [25]. After the protocol change, the matching rate increased from 5.1% to 25.3%; by July 2017, 12.4% (689 of 5,560) of patients whose tumors were successfully sequenced were finally enrolled in the study and received concordant treatment [26]. In the Molecular Screening for Cancer Treatment Optimization study, which evaluated the clinical benefit of genomic analyses in different types of cancer, 19.2% (199 of 1,035) of patients were finally treated with a matched targeted therapy [27]. In our study, MP-guided treatments were possible in 4.7% (6/127) of patients with advanced breast cancer and 7.4% (6/81) of patients with advanced breast cancer with targetable genomic alterations. Although this MP-guided treatment rate may not seem satisfactory, there is room for improvement, considering that among the 66 patients with targetable genomic alterations without MP-guided treatment, 21 received first-line treatment or were on regular follow-up without treatment. These patients are potential candidates for future MP-guided treatment. Moreover, the PI3K inhibitor alpelisib recently gained approval from the Ministry of Food and Drug Safety in Korea, which will also increase the rate of MP-guided treatment in patients with breast cancer. Of the 66 patients with targetable genomic alterations who did not receive MP-guided treatment in our study, 32 had tier I and II PIK3CA mutations. With the availability of a PI3K inhibitor such as alpelisib, the matching treatment rate increased from 7.4% (6/81) to 58.0% (47/81) when only genetic variation was considered without also considering the patient’s condition or past treatment history.
Regarding the use of NGS in patients with metastatic cancer, the European Society for Medical Oncology suggested that although there is no need to perform tumor NGS in daily practice because the PIK3CA status can be determined by polymerase chain reaction and ERBB2 testing can be performed by immunohistochemistry, molecular screening programs must include patients with advanced breast cancer for clinical trial consideration because a high number of tier II alterations occur in patients with breast cancer [28]. Since daily clinical practice and clinical trials should be viewed as a continuum of treatment, and most patients with advanced breast cancer are treated at, or at least referred to, tertiary hospitals where they have the opportunity to participate in clinical trials, active NGS panel testing of patients with breast cancer in routine clinical practice will provide more opportunities for patient care.
The barriers to MP-guided treatment include access to care options, cost, and insurance coverage [26]. Patients with advanced breast cancer are usually not eligible for clinical trials because of their poor performance status or previous treatment. Outside of clinical trials and approved targeted therapies, patients require costly off-label cancer therapy, which is usually not covered by insurance. In our study, 66.7% of patients with targetable genomic alterations were unable to receive MP-guided treatment for these reasons. The incorporation of NGS tests in the early course of treatment and the availability of matched drugs is critical for the implementation of personalized treatment.
Our study has some limitations. First, NGS panel testing was not routinely performed. Therefore, the patients included in this study may not represent all patients with breast cancer treated at our institution. For instance, HER2+ patients accounted for only 4.3% of the total (6/137), much lower than the actual frequency. Second, different NGS panels were used in the study population owing to updates to the panel during the study period. MSI status and TMB status were determined in only 73 patients profiled using SNUBH V2. These aspects could represent a potential bias in the investigation of genomic profiling in breast cancer. Third, our NGS panel did not detect RNA fusions, which may have led to low linkage rates for MP-guided therapy. Finally, this study was retrospective in nature, and the response rate or PFS/OS was not compared according to MP-guided therapy. In contrast, the strength of our study was the clinical application of NGS in daily practice for the detection of breast cancer and the identification of patients requiring MP-guided treatment.
In a real-world single-institution study, NGS panel testing detected targetable genomic alterations in 59% of all patients with breast cancer and in 63.8% of patients with advanced breast cancer, which led to MP-guided treatments in 4.7% of patients with advanced breast cancer. NGS panel testing during the early disease course and the availability of matched drugs through clinical trials or off-label use are vital for the implementation of personalized treatment.
ACKNOWLEDGMENTS
The authors thank the study participants and their families.
Footnotes
Presentation: This study was presented at Global Breast Cancer Conference 10 and received Best Presentation Award.
Conflict of Interest: Jee Hyun Kim received an honoraria from Roche Korea, Novartis Korea, Pfizer Korea, MSD Korea, Lilly Korea, and Sanofi Korea, and grant/research funding from Ono Korea Ltd. All other authors declare that they have no conflict of interest.
- Conceptualization: Suh KJ, Na HY, Kim IA, Park SY, Kim JH.
- Data curation: Kim SH, Kim YJ, Shin H, Kang E, Lee S, Woo JW, Ahn S, Park SY, Kim JH.
- Formal analysis: Suh KJ, Kim JH.
- Investigation: Suh KJ, Woo JW, Park SY, Kim JH.
- Methodology: Suh KJ, Park SY, Kim JH.
- Project administration: Suh KJ, Park SY, Kim JH.
- Resources: Lee S, Woo JW, Na HY, Ahn S, Park SY.
- Software: Lee S.
- Supervision: Kim SH, Kim YJ, Kim EK, Park SY, Kim JH.
- Visualization: Suh KJ.
- Writing - original draft: Suh KJ, Park SY, Kim JH.
- Writing - review & editing: Suh KJ, Kim SH, Kim YJ, Shin H, Kang E, Kim EK, Lee S, Woo JW, Na HY, Ahn S, Jang BS, Kim IA, Park SY, Kim JH.
SUPPLEMENTARY MATERIALS
Panel gene list
SNV/indel (AF ≥ 2%, depth ≥ 100×) (tier I–II)
Violin plots of tumor mutation burden in clinical subgroup of metastatic breast cancer. (A) All patients, (B) according to subtype, (C) according to timing of NGS.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Information Center. National Cancer Information Center’s homepage. 2021. [Accessed December 12th, 2021]. https://www.cancer.go.kr .
- 3.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Narayan P, Prowell TM, Gao JJ, Fernandes LL, Li E, Jiang X, et al. FDA approval summary: alpelisib plus fulvestrant for patients with HR-positive, HER2-negative, PIK3CA-mutated, advanced or metastatic breast cancer. Clin Cancer Res. 2021;27:1842–1849. doi: 10.1158/1078-0432.CCR-20-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Center for Drug Evaluation and Research. FDA approves olaparib for germline BRCA-mutated metastatic breast cancer. 2018. [Accessed July 18th, 2021]. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-olaparib-germline-brca-mutated-metastatic-breast-cancer .
- 6.Marcus L, Fashoyin-Aje LA, Donoghue M, Yuan M, Rodriguez L, Gallagher PS, et al. FDA approval summary: pembrolizumab for the treatment of tumor mutational burden-high solid tumors. Clin Cancer Res. 2021;27:4685–4689. doi: 10.1158/1078-0432.CCR-21-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Center for Drug Evaluation and Research. FDA approves larotrectinib for solid tumors with NTRK gene fusions. 2019. [Accessed July 18th, 2021]. https://www.fda.gov/drugs/fda-approves-larotrectinib-solid-tumors-ntrk-gene-fusions .
- 8.Stockley TL, Oza AM, Berman HK, Leighl NB, Knox JJ, Shepherd FA, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016;8:109. doi: 10.1186/s13073-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 11.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 13.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlovich CA, Williams PM. Clinical applications of next-generation sequencing in precision oncology. Cancer J. 2019;25:264–271. doi: 10.1097/PPO.0000000000000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Medicare & Medicaid Services. Decision memo for next generation sequencing (NGS) for Medicare beneficiaries with advanced cancer. 2018. [Accessed July 19th, 2021]. https://www.cms.gov/medicare-coverage-database/details/nca-decision-memo.aspx?NCAId=290 .
- 16.Healthcare Bigdata Hub. Health insurance review and assessment service. 2022. [Accessed September 18th, 2021]. http://opendata.hira.or.kr/op/opc/olapDiagBhvInfo.do .
- 17.Lee SH, Lee B, Shim JH, Lee KW, Yun JW, Kim SY, et al. Landscape of actionable genetic alterations profiled from 1,071 tumor samples in Korean cancer patients. Cancer Res Treat. 2019;51:211–222. doi: 10.4143/crt.2018.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon D, Kim B, Shin HC, Kim EJ, Ha SY, Jang KT, et al. Cancer panel assay for precision oncology clinic: results from a 1-year study. Transl Oncol. 2019;12:1488–1495. doi: 10.1016/j.tranon.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, Yoon S, Lee DH, Jang SJ, Chun SM, Kim SW. Real-world utility of next-generation sequencing for targeted gene analysis and its application to treatment in lung adenocarcinoma. Cancer Med. 2021;10:3197–3204. doi: 10.1002/cam4.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park C, Yoon KA, Kim J, Park IH, Park SJ, Kim MK, et al. Integrative molecular profiling identifies a novel cluster of estrogen receptor-positive breast cancer in very young women. Cancer Sci. 2019;110:1760–1770. doi: 10.1111/cas.13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Tourneau C, Delord JP, Gonçalves A, Gavoille C, Dubot C, Isambert N, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 22.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aisner D, Sholl LM, Berry LD, Haura EB, Ramalingam SS, Glisson BS, et al. Effect of expanded genomic testing in lung adenocarcinoma (LUCA) on survival benefit: the Lung Cancer Mutation Consortium II (LCMC II) experience. J Clin Oncol. 2016;34:11510. [Google Scholar]
- 24.Schwaederle M, Parker BA, Schwab RB, Daniels GA, Piccioni DE, Kesari S, et al. Precision oncology: the UC San Diego Moores Cancer Center PREDICT Experience. Mol Cancer Ther. 2016;15:743–752. doi: 10.1158/1535-7163.MCT-15-0795. [DOI] [PubMed] [Google Scholar]
- 25.Flaherty KT, Gray R, Chen A, Li S, Patton D, Hamilton SR, et al. The Molecular Analysis for Therapy Choice (NCI-MATCH) trial: lessons for genomic trial design. J Natl Cancer Inst. 2020;112:1021–1029. doi: 10.1093/jnci/djz245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morash M, Mitchell H, Beltran H, Elemento O, Pathak J. The role of next-generation sequencing in precision medicine: a review of outcomes in oncology. J Pers Med. 2018;8:30. doi: 10.3390/jpm8030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massard C, Michiels S, Ferté C, Le Deley MC, Lacroix L, Hollebecque A, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7:586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 28.Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panel gene list
SNV/indel (AF ≥ 2%, depth ≥ 100×) (tier I–II)
Violin plots of tumor mutation burden in clinical subgroup of metastatic breast cancer. (A) All patients, (B) according to subtype, (C) according to timing of NGS.