Abstract
Post-traumatic stress disorder (PTSD) can develop following severe trauma, but the extent to which genetic and environmental risk factors contribute to individual clinical outcomes is unknown. Here, we compared transcriptional responses to hydrocortisone exposure in human induced pluripotent stem cell (hiPSC)-derived glutamatergic neurons and peripheral blood mononuclear cells (PBMCs) from combat veterans with PTSD (n = 19 hiPSC and n = 20 PBMC donors) and controls (n = 20 hiPSC and n = 20 PBMC donors). In neurons only, we observed diagnosis-specific glucocorticoid-induced changes in gene expression corresponding with PTSD-specific transcriptomic patterns found in human postmortem brains. We observed glucocorticoid hypersensitivity in PTSD neurons, and identified genes that contribute to this PTSD-dependent glucocorticoid response. We find evidence of a coregulated network of transcription factors that mediates glucocorticoid hyper-responsivity in PTSD. These findings suggest that induced neurons represent a platform for examining the molecular mechanisms underlying PTSD, identifying biomarkers of stress response, and conducting drug screening to identify new therapeutics.
Subject terms: Gene expression profiling, Post-traumatic stress disorder
The authors generated stem cell-derived neurons from combat veterans with and without PTSD and found PTSD-dependent gene expression changes in response to glucocorticoids. This highlights how stress response may be altered in individuals with PTSD.
Main
Although unequivocally precipitated by environmental events, PTSD develops in only a minority of trauma survivors1, prompting a search for risk factors that increase the probability of developing this condition following trauma exposure. Convergent lines of evidence are consistent with a heritable component to PTSD risk. There is a concordance between PTSD diagnosis in monozygotic and dizygotic twins2. Genome-wide association studies (GWAS) estimate single nucleotide polymorphism (SNP)-based heritability from 5% to 30%3–5 and identify loci significantly associated with PTSD3,5, although loci vary with ancestry, sex and type of trauma. This highlights the necessity of developing paradigms that examine the impact of PTSD genetic risk factors in the context of exposure to an environmental stressor.
Insights into the gene by environment (G×E) interactions underlying the psychiatric symptoms of PTSD remain poorly resolved, reflecting the lack of a cohesive neurobiological framework to investigate these mechanisms. To date, most studies of PTSD pathophysiology have focused on PBMCs, with PTSD patients showing lower ambient cortisol and heightened glucocorticoid sensitivity relative to healthy controls6,7 coupled with increased expression of innate immune genes8,9. Toward resolving the relative contributions of genetic risk and environmental stress to PTSD pathophysiology, it is critical to deconvolve the impact of stress in a cell-specific manner, including determinants of stress responses across brain and blood cells. Glucocorticoid receptor signaling is the most overlapping pathway between murine blood and brain and is highly associated with individual differences in response to trauma exposure10. The degree to which PTSD symptoms arise as a result of elevated/sustained peripheral response to stress6,10 and/or abnormal cellular or circuit response to peripheral stress cues11–13 remains unresolved. In either case, preclinical data suggests that trauma-induced perturbations in glucocorticoid signaling result in glutamate-induced excitotoxicity, leading to decreased glutamatergic neural activity, dendritic retraction and reduced synaptic density14. Drugs that modulate the glucocorticoid15,16 and glutamatergic17 systems represent potential avenues for pharmacologic intervention.
An improved understanding of the pathophysiology of PTSD requires the development of appropriate human-specific models. Understanding the extent to which the dysregulated stress response reflects cell-type-specific preexisting genetic vulnerabilities will improve genetic-based diagnosis and potentially identify new therapeutic targets to prevent or reverse PTSD. hiPSC-based models have the potential to address some of the main limitations of clinical and animal studies, evaluating human glutamatergic neurons that are genetically identical to those of their donors (reviewed in Fernando et al.18). Although there have been a handful of studies of glucocorticoid response in live human neurons19,20, none have contrasted neurons derived from trauma-exposed controls and PTSD cases.
Here, we describe a hiPSC-based study of PTSD, comparing glutamatergic neurons from combat veterans with (n = 19 donors) and without (n = 20 donors) PTSD, to test the hypothesis that PTSD risk variants and glucocorticoid response (for example, to dexamethasone (DEX) and hydrocortisone (HCort)21,22) interact in a cell-type-specific manner. Transcriptional profiles of hiPSC neurons were compared with a well-matched and largely overlapping PBMC study of combat veterans with (n = 20 donors) and without (n = 20 donors) PTSD. First, glucocorticoid-induced blood and neuronal responses were significantly enriched for immune response, brain development and neurodevelopmental disorder genes, with specific upregulation of disorder genes in neurons only. Second, a baseline PTSD diagnosis-specific signature was undetectable in either human neurons or PBMCs. Third, glucocorticoid hypersensitivity occurred in samples from PTSD cases, with diagnosis-specific effects greatest at low doses, and significantly more robust in neurons than PBMCs. This PTSD-specific neuronal glucocorticoid-response signature was enriched for transcriptomic patterns observed in postmortem (PM) brain tissue from PTSD cases. Critical aspects of glucocorticoid response are encoded in patient genetics, consistent with a clear genetic predisposition to PTSD. These findings suggest possible therapeutic strategies to minimize the likelihood of PTSD following trauma exposure, a particularly valuable outcome for military and first-responder personnel.
Results
To study how glucocorticoids influence gene expression in donor-specific brain and blood cells, skin and blood samples were collected from well-matched and largely overlapping (30 shared individuals) hiPSC-derived glutamatergic neuron and PBMC cohorts respectively, comprised of combat veterans with (n = 19 hiPSC donors and n = 20 PBMC donors) and without (n = 20 hiPSC donors and n = 20 PBMC donors)) PTSD (Supplementary Table 1). For technical reasons, glucocorticoid treatment of NGN2-neurons (batch 1 n = 15 versus 15, batch 2 n = 4 versus 5) and PBMCs (batch A n = 10 versus 10, batch B n = 10 versus 10 (ref. 23)) were completed independently and then meta-analyzed together to adjust for batch effects (Supplementary Table 2).
Dexamethasone-stimulated transcriptional responses in PBMCs
Treatment of PBMCs in vitro with the synthetic glucocorticoid DEX is a promising method to explore molecular responses to stress hormones in PTSD23. To continue these earlier findings, identical methods were applied to expand this study of combat-exposed veterans to (n = 20) and without (n = 20) PTSD (Fig. 1a) (Supplementary Tables 1 and 2).
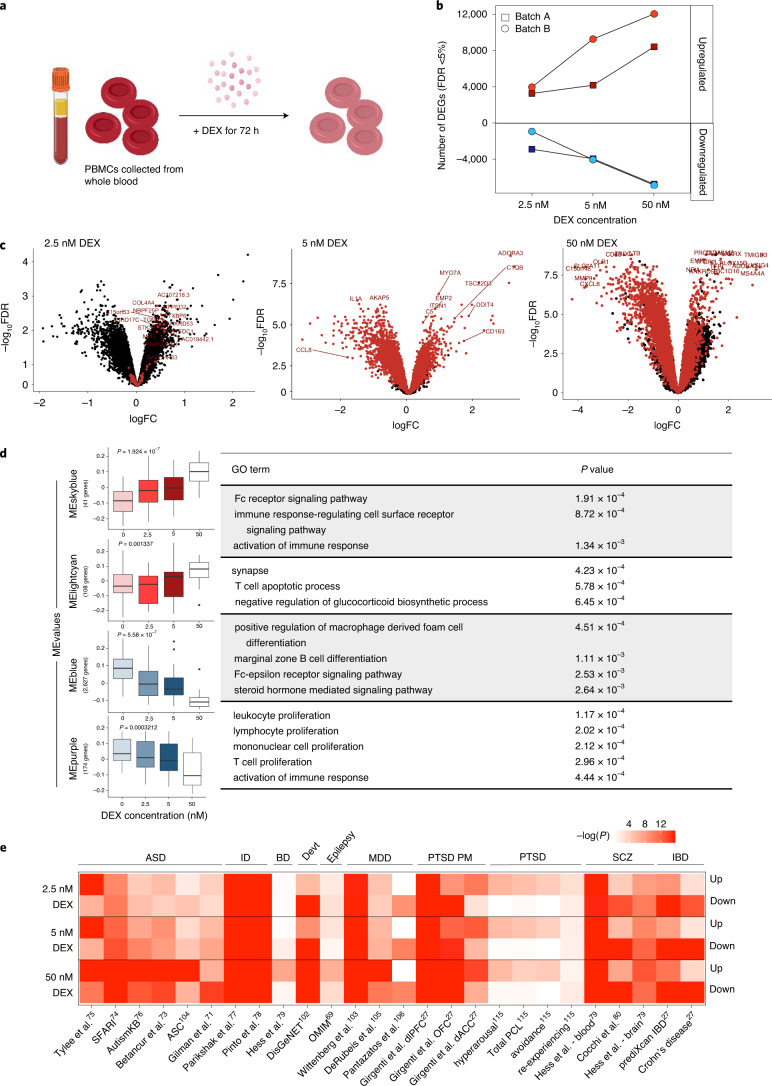
Fig. 1. Transcriptional response to DEX in PBMCs.
a, PBMCs from 20 PTSD cases and 20 combat-exposed controls were treated with DEX for 72 h and RNA-seq was performed. b, The number of DEGs observed in batch A (n = 10 versus 10) (squares) and batch B (n = 10 versus 10) (circles) are upregulated and downregulated (y axis) across three different concentrations of DEX conditioning (x axis) at a Bonferroni-corrected P value threshold. c, Meta-analysis of expression logFC (differences observed between vehicle and DEX exposure) was plotted against –log(FDR) for each gene. Red points indicate significantly DEGs in the meta-analysis. Sample-size-based meta-analysis was conducted using METAL with a Cochran’s Q test for heterogeneity. Benjamini–Hochberg adjusted P values are reported to correct for multiple testing. d, ME values from modules identified by WGCNA were correlated with increasing DEX concentrations for all DEX-treated samples (n = 40 individuals). Top correlated modules with DEX concentration are shown here (P values are labeled above each boxplot). Correlation P values were derived from a two-tailed t distribution. Data are presented as minimum, first quartile, median, third quartile and maximum. Each module was subjected to GO enrichment analysis and the topmost significant enrichment terms and their associated Benjamini–Hochberg adjusted P values are displayed. e, Gene set enrichment of DEX-dependent DEGs across psychiatric disorder and neurodevelopmental gene sets27. Devt, genesets involved in neurodevelopmental disorders.
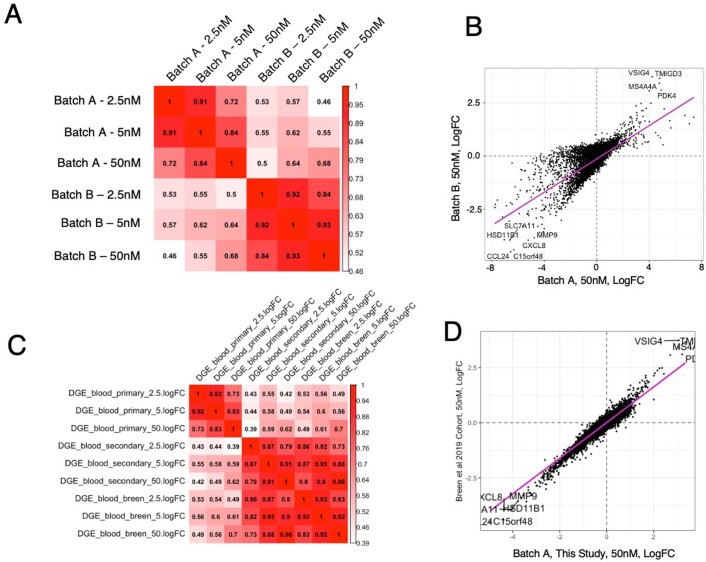
RNA-sequencing (RNA-seq) was generated from cultured PBMCs treated for 72 h with three concentrations of DEX (2.5 nM, 5 nM and 50 nM), and analyzed relative to baseline samples. To identify reliable markers of glucocorticoid activation independent of PTSD, diagnosis as well as other confounds were adjusted for (Methods). Incubation with increasing concentrations of DEX (2.5 nM, 5 nM and 50 nM, respectively) identified 6,153, 8,114 and 15,128 differentially expressed genes (DEGs) in batch A, and 4,880, 13,297 and 18,856 DEGs in batch B, respectively (Q value < 0.05) (Fig. 1b and Supplementary Table 4). Overall transcriptome-wide concordance of DEX-induced fold changes (FC) relative to vehicle between the two batches was exceedingly high (average r = 0.738) (Extended Data Fig. 5a), and was highly concordant with reported responses to DEX in PBMCs23 (r = 0.7 in the 50 nM dose) (Extended Data Fig. 5c,d). These findings demonstrate strong conservation of transcriptional changes in PBMCs in response to DEX that are independent of PTSD and other potentially confounding factors.
Extended Data Fig. 5. Comparison of PBMC batches.
a, Pairwise correlations between PBMC batches. b, Transcriptome-wide correlation between batches at 50 nM DEX dose. c, Pairwise correlations between PBMC batches and Breen et al23. d, Transcriptome-wide correlation between our study and Breen et al.23 at the 50 nM DEX dose.
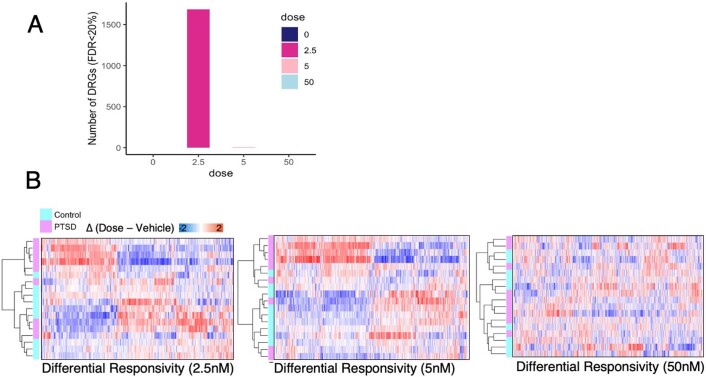
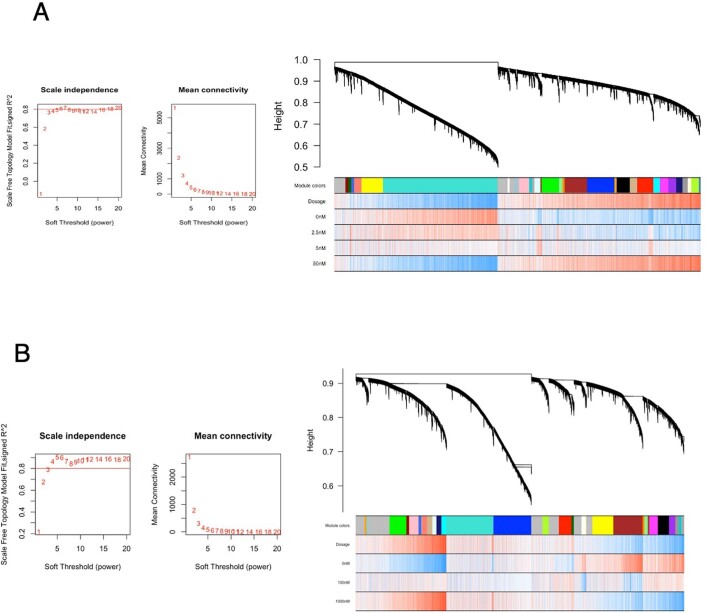
Gene coexpression modules were calculated to probe functional consequences of DEX exposure in PBMCs. Of 36 significant coexpression modules (M), 24 were significantly dynamically regulated in response to increasing concentrations of DEX (Supplementary Table 5). Gene ontology (GO) analysis found enrichment in neuronal regulation terms such as synapse (module eigengene (ME) lightcyan, P = 4.23 × 10–4) and immune regulation terms such as activation of immune response (ME skyblue, P = 1.34×10–3) and lymphocyte proliferation (ME purple, P = 2.02 × 10–4), and glucocorticoid signaling terms such as steroid hormone mediated signaling pathway (ME blue, P = 2.64 × 10–3) and Fc receptor signaling pathway (ME skyblue, P = 1.91 × 10–4) (Fig. 1d). These enrichments are representative of shared regulation of genes involved in cell migration, cytoskeletal and cell-adhesion properties, which are broadly shared by immune and neural cells, through common processes such as cell migration, degranulation and cellular outgrowth, which may explain the neural term represented in this module. Together, these enrichments are consistent with known glucocorticoid response impacts on cell signaling24, neurogenesis25 and immunosuppression26 (Fig. 1d). Gene set enrichment for psychiatric disorder gene sets27 revealed enrichment of DEX-upregulated PBMC DEGs broadly across neuropsychiatric disorder risk genes, but not across PTSD-specific signatures (Fig. 1e). This may indicate that. after adjusting for diagnosis, glucocorticoid treatment of PBMCs alone is insufficient to recapitulate PTSD signatures. Although glucocorticoid treatment of PBMCs (batch A) was highly correlated to the DEX-induced gene responses previously reported (batch B)23 (Extended Data Fig. 5a), the larger meta-analysis did not identify PTSD-specific DEX-induced differential response genes (PBMC-DRGs) at a false discovery rate (FDR)-corrected threshold (Extended Data Fig. 9).
Extended Data Fig. 9. PTSD(+) specific responses to DEX in PBMCs.
a, Genes that differ in their response to DEX in PTSD(+) donors compared to PTSD(-) donors, here termed differential response genes (DRGs), at an FDR threshold of 20% (nonsignificant) b, Unsupervised clustering of nominally significant PTSD DRGs.
HCort-induced transcriptional responses in neurons
We28–31 and others32–39 have demonstrated that NGN2-neurons are greater than 95% pure glutamatergic neurons, robustly express glutamatergic genes, release neurotransmitters, produce spontaneous synaptic activity and recapitulate the impact of psychiatric disease-associated genes. Well-matched to the PBMCs described above, 21-day-old NGN2-neurons from combat-exposed veterans with (n = 19) and without (n = 20) PTSD (Supplementary Table 1) were treated with two to three distinct concentrations of HCort (100 nM, 1,000 nM or 2,500 nM) and an untreated baseline condition for 24 h before RNA-seq (Fig. 2a).
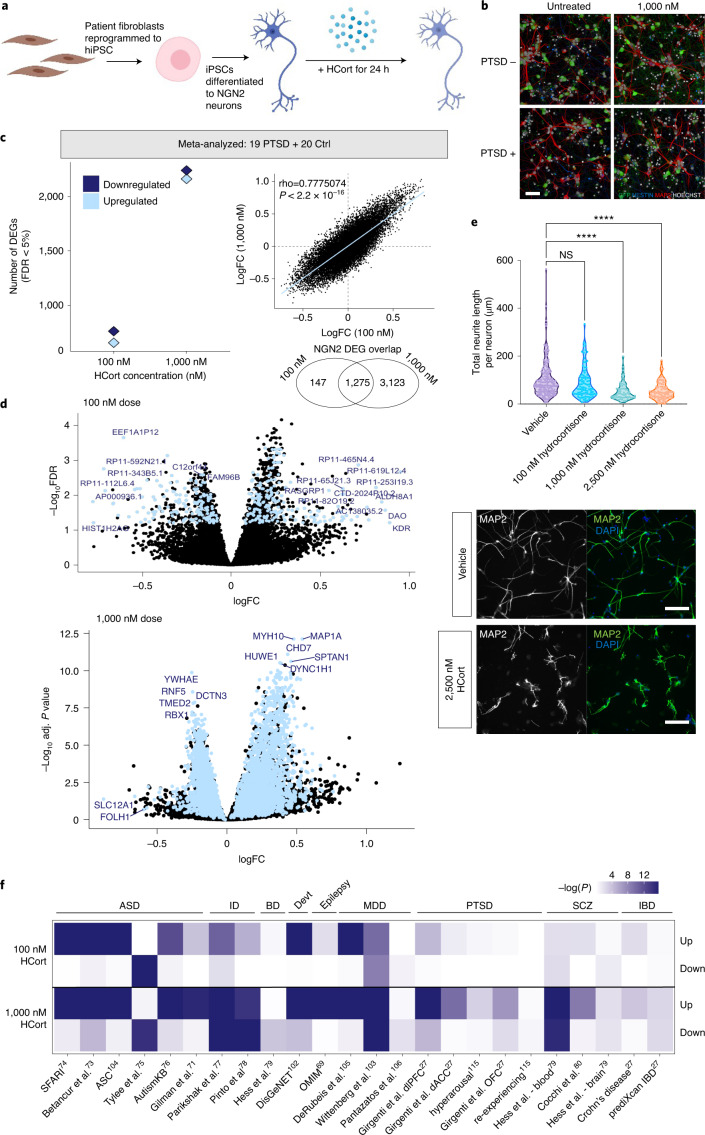
Fig. 2. Gene expression changes to HCort in hiPSC-derived neurons.
a, hiPSC-derived NGN2-neurons were treated with HCort for 24 h and RNA-seq performed. b, NGN2-neurons stained for neuronal markers NESTIN and MAP2, nucleic marker HOECHST and green fluorescent protein to confirm neuronal identity and morphology across all conditions. c, Meta-analyzed DEGs in response to increasing concentrations of HCort shows robust changes in NGN2-neurons. A comparative analysis of transcriptome-wide log2FC in response to different concentrations of HCort in NGN2-neurons shows similar responses, indicating a conserved response across all donors to HCort in NGN2-neurons. Sample-size-based meta-analysis was conducted using METAL with a Cochran’s Q test for heterogeneity. Benjamini–Hochberg adjusted (adj.) P values are reported to correct for multiple testing. d, Meta-analysis of expression LogFC (differences observed between vehicle and HCort exposure) was plotted against –log(P value) for each gene. Blue points indicate significantly DEGs in the meta-analysis. e, Morphological analysis of neurite outgrowth in day 7 NGN2-neurons showing dose-dependent decrease in neurite outgrowth with HCort exposure. A Kruskal–Wallis test with a post hoc Dunn’s multiple comparisons test was performed on data for neurite length per neuron. **** P < <0.0001. NS, nonsignificant. P value for the NS comparison was 0.2121. Representative images of neurite morphology to HCort exposure shown below. f, Gene set enrichment of HCort-dependent DEGs across psychiatric disorder and neurodevelopmental gene sets27.
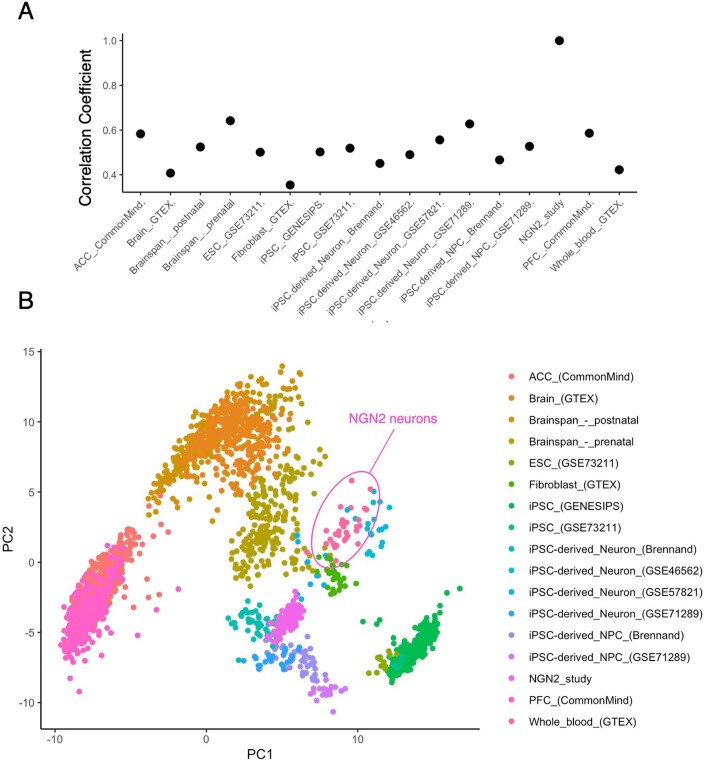
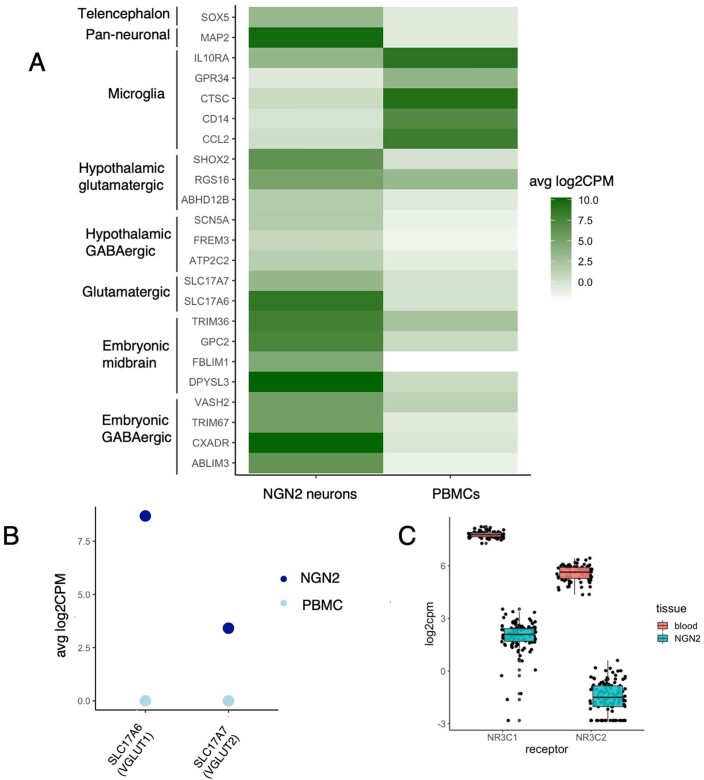
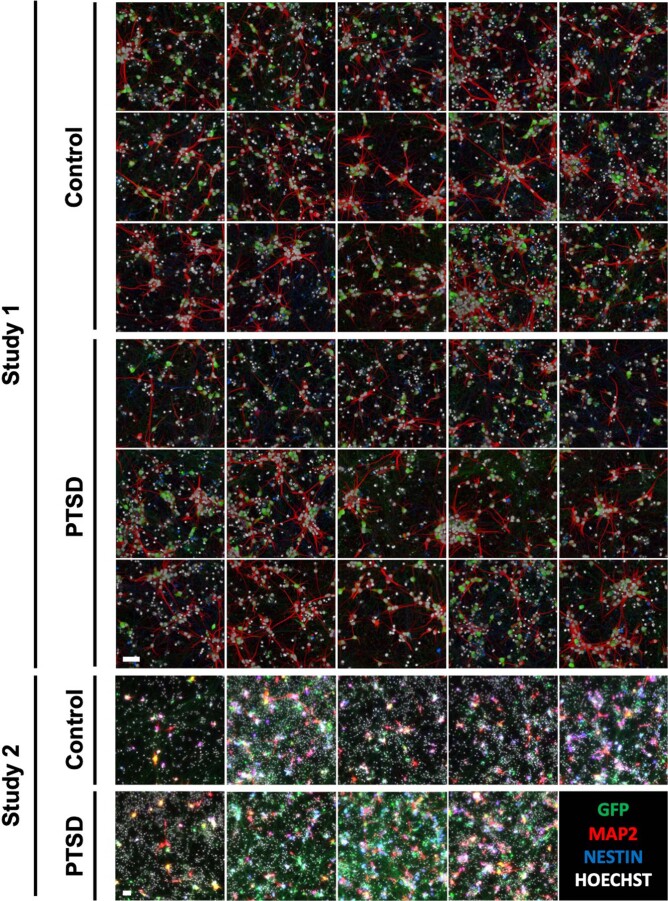
To confirm the neuronal identity and developmental specificity of these data, a large comparative and integrative analysis was performed across 16 existing transcriptome datasets including 2,716 independent samples from a range of hiPSC-derived neurons and developmentally distinct PM brain tissue40. A high degree of transcriptomic convergence was observed for our hiPSC-derived neurons with fetal brain tissue and hiPSC-derived neurons described in previous reports, confirming their early developmental gene expression profiles (Extended Data Fig. 3). Neuronal fate was further confirmed by demonstrated expression of pan-neuronal and synaptic genes36 in hiPSC-derived neurons but not in PBMCs (Extended Data Fig. 4a) and VGLUT expression in NGN2-neurons (Extended Data Fig. 4b). Glucocorticoid and mineralocorticoid receptor expression was additionally confirmed for each cell type (Extended Data Fig. 4c), with PBMCs demonstrating higher expression of both receptors, consistent with heightened glucocorticoid transcriptional response in PBMCs. Immunostaining of a parallel well demonstrated consistent cell number (4,307 ± 1,313) and neuronal marker expression (78.5 ± 6% microtubule-associated protein 2 (MAP2)-positive and 0.4 ± 0.6% NESTIN-positive) (Fig. 2b and Extended Data Fig. 1) that did not differ significantly by diagnosis or glucocorticoid treatment.
Extended Data Fig. 3. Developmental specificity analysis.
a, Pairwise correlation between NGN2s from our study with cell types across 16 independent studies. b, PCA analysis of cell types within all 16 studies with our NGN2-neurons.
Extended Data Fig. 4. Neuronal fate specificity analysis.
a, Expression of hallmark pan-neuronal and neuronal subtype specific genes in NGN2-neurons and PBMCs. b, Average log2CPM expression of VGLUT1 and VGLUT2 in NGN2-neurons and PBMCs. c, Expression of GR and MR in NGN2-neurons (from n = 39 individuals) and PBMCs (from n = 40 individuals). Boxplot data are presented as minimum, 1st quartile, median, 3rd quartile and maximum.
Extended Data Fig. 1. Immunostaining of hiPSC-derived NGN2-neurons.
Immunostaining of Hoechst (white), GFP (green), MAP2 (red) and NESTIN (blue) across all participants prior to HCort treatment. Imaging was repeated after treatment with the 100 and 1000 HCort dosages. Study 1 and 2: Scale bar = 50 µm.
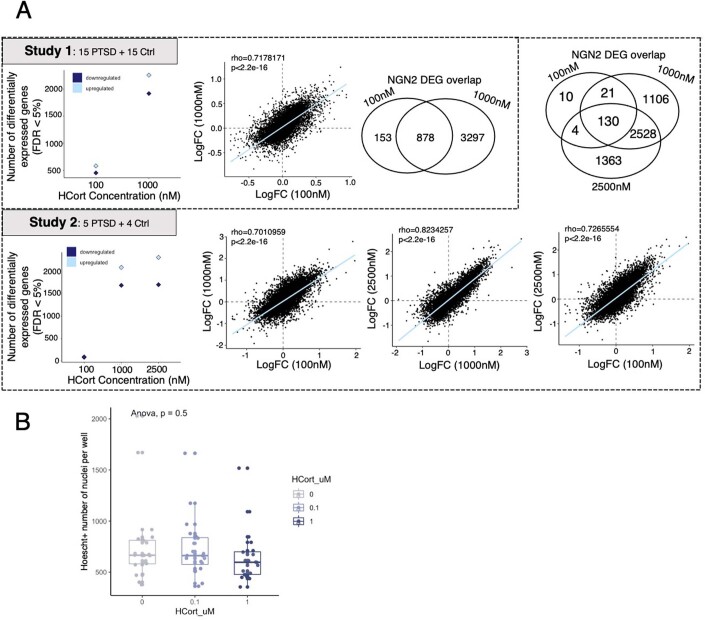
HCort dose-dependent transcriptional responses were resolved relative to those in baseline untreated NGN2-neurons. To identify reliable markers of glucocorticoid activation independent of PTSD, diagnosis as well as other confounds were adjusted for. Incubation with increasing HCort concentrations (batch 1: 0 nM, 100 nM and 1,000 nM; batch 2: 0 nM, 100 nM, 1,000 nM and 2,500 nM) resulted in 1,031 and 4,175 DEGs in batch 1, and 165, 3,785 and 4,025 DEGs in batch 2, respectively (Q value < 0.05) (Fig. 2c and Extended Data Fig. 6a). Transcriptome-wide concordance was examined using HCort-associated log2 FC and HCort responses between doses was highly concordant and preserved across increasing concentrations of HCort in both batches (Extended Data Fig. 6a). Meta-analysis of differential expression summary statistics found 1,837 significant response genes in the 100 nM dose and 5,956 in the 1,000 nM dose (Fig. 2d). To assess whether this large transcriptional response to HCort elicited nonspecific cell toxicity, we measured cell density of untreated and HCort-treated neurons by quantifying the number of Hoescht positive neurons in untreated and treated neurons. We observed no significant differences in cell density between doses (Extended Data Fig. 6b). This suggests that HCort treatment in our neurons did not induce significant cell toxicity. As with PBMCs, gene set enrichment for psychiatric disorder gene sets27 revealed enrichment of HCort upregulated NGN2-neuron DEGs broadly across neuropsychiatric disorder risk genes, but not across PTSD-specific signatures (Fig. 2f), again suggesting that glucocorticoid treatment of NGN2-neurons, without considering PTSD diagnosis, is insufficient to recapitulate PTSD signatures.
Extended Data Fig. 6. Comparison of NGN2 batches.
a, Hcort-responsive DEGs across independent batches. Transcriptome-wide concordance is plotted between dosages for each batch. b, Quantification of cell number with HCort treatment showing no significant cell density between doses. One-way ANOVA was performed in n = 39 biologically independent individuals treated with each dose. Data are presented as minimum, 1st quartile, median, 3rd quartile and maximum.
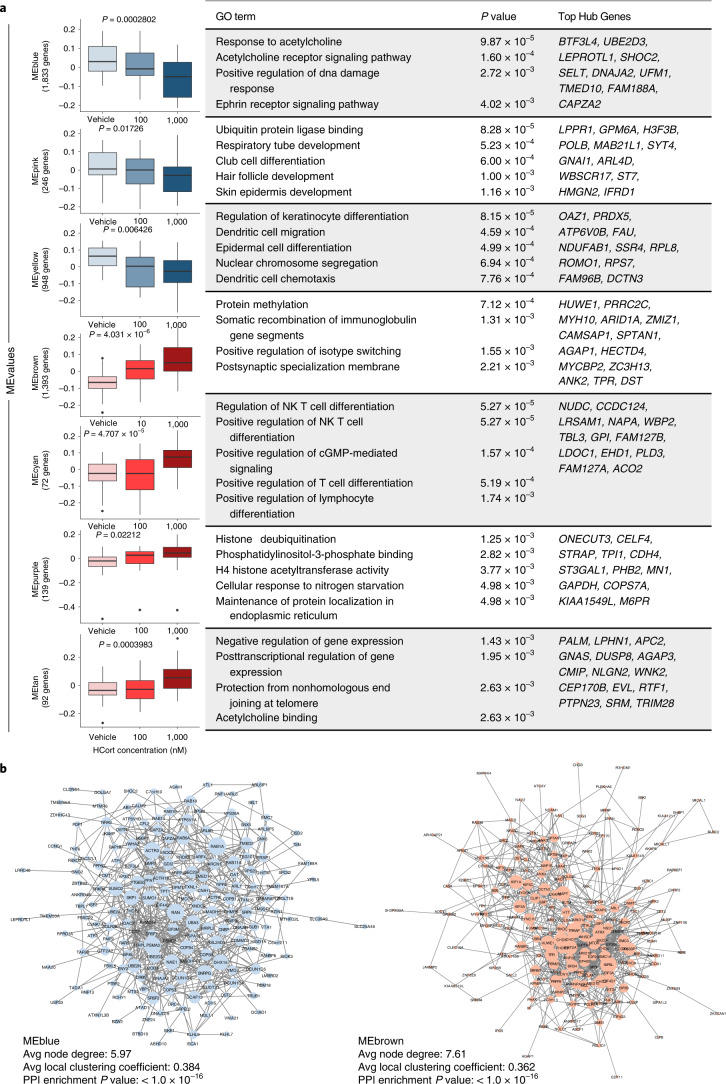
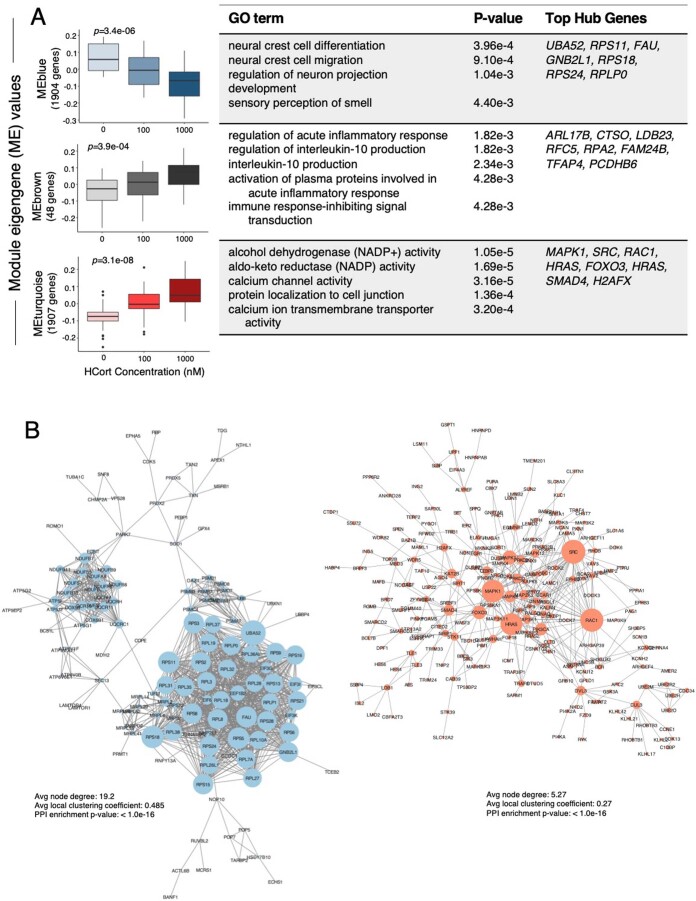
Weighted gene coexpression modules were calculated from the meta-analyzed studies to better understand the functional aspects of the HCort-induced transcriptional changes in NGN2-neurons (Extended Data Fig. 7). Seven significant coexpression modules (M1–7) were identified that were dynamically regulated in response to increasing concentrations of HCort in our signed analysis (Fig. 3a). ME values for M1–3 were downregulated with increasing concentrations of HCort and enriched for biological processes related to acetylcholine signaling, such as acetylcholine receptor signaling pathway (P = 1.60 × 10–4), protein degradation such as ubiquitin protein ligase binding (P = 8.28 × 10–5) and skin regulation such as regulation of keratinocyte differentiation (P = 8.15 × 10–5). These signatures are consistent with known glucocorticoid inhibition of acetylcholine signaling41 and role in skin atrophy42. Glucocorticoid-acetylcholine signaling interactions occur in pathways affecting memory consolidation43, and alter glutamatergic synapses and synaptic stability44, suggesting that glucocorticoid exposure alters acetylcholine signaling to impact glutamatergic synaptic biology. Remaining ME values for modules M4–7 significantly increased with HCort treatment. Many terms within these modules enriched for immune processes such as positive regulation of isotype switching (P = 1.55 × 10–3), regulation of NK T cell differentiation (P = 5.27 × 10–5), and somatic recombination of immunoglobulin gene segments (P = 1.31 × 10–3), processes that are well-established markers of glucocorticoid activation8,9. Finally, enrichments for hallmark glucocorticoid processes of histone acetylation45, such as H4 histone acetyltransferase activity (P = 3.77 × 10–3) and transcriptional suppression (negative regulation of gene expression (P = 1.43 × 10–3)), were enriched. Unsigned modules significantly associated with neural projection terms, such as neural crest cell differentiation (P = 3.96 × 10–4) and regulation of neuron projection development (P = 1.04 × 10–3), and immune terms, such as regulation of acute inflammatory response (P = 1.83 × 10–3) (Extended Data Fig. 8). To visualize module connectivity, protein–protein interactions (PPIs) of genes within modules were mapped, finding a significant observed number of edges in all modules (P < 1.0 × 10–16) (Fig. 3b). Overall, HCort treatment of NGN2-neurons, independent of diagnosis, resulted in robust downregulation of acetylcholine signaling and skin development, and upregulation of inflammation-modulating and cell-signaling genes.
Extended Data Fig. 7. Weighted gene co-expression network analysis of (A) PBMCs and (B) NGN2-neurons.
The β-power required to satisfy scale-free topology (SFT) and corresponding mean connectivity for gene co-expression network construction. Hierarchical gene cluster tree and module structure and gene-treatment color bands. The first color band underneath the tree indicates the detected modules and subsequent bands indicate treatment correlation, when red indicates a strong relationship and blue indicates a strong negative relationship.
Fig. 3. HCort stimulated coexpression modules in NGN2-neurons.
a, WGCNA identified three groups of coregulated gene modules. Pearson correlation was used to assess changes in ME values with increasing concentration of HCort (P values are labeled above each boxplot). Correlation P values were derived from a two-tailed t distribution. Data are presented as minimum, first quartile, median, third quartile and maximum. Each module was subjected to GO enrichment analysis and the topmost significant enrichment terms and their associated Benjamini–Hochberg adjusted P values are displayed. Top hub genes (kME > 0.6) within each module are displayed for quick interpretation of GR-stimulated gene coexpression modules and candidate individual genes. b, Network visualization of PPIs within modules indicating clusters and network hubs. STRING analysis was used to identify the observed number of edges. Enrichment of observed edges was assessed against expected edges to determine a PPI P value of < 1.0 × 10–16 for each network. Avg, average.
Extended Data Fig. 8. Unsigned WGCNA Network.
a, Unsigned Weighted gene co-expression network analysis (WGCNA) identified coregulated gene modules. Pearson correlation was used to assess changes in module eigengene (ME) values with increasing concentration of HCort (p-values are labeled above each boxplot). Correlation p-values were derived from a two-tailed t-distribution. Data are presented as minimum, 1st quartile, median, 3rd quartile and maximum. Each module was subjected to gene ontology enrichment analysis and the topmost significant enrichment terms and their associated Benjamini–Hochberg adjusted P-values are displayed. Top hub genes (kME > 0.6) within each module are displayed for quick interpretation of GR-stimulated gene co-expression modules and candidate individual genes. b, Network visualization of protein-protein interactions within modules indicating clusters and network hubs. STRING analysis was used to identify the observed number of edges. Enrichment of observed edges was assessed against expected edges to determine a Protein Protein Interaction (PPI) p-value of p < 1.0e-16 for each network.
As transcriptomic responses to HCort exposure enriched in neuronal projection and signaling terms, we sought to validate these functional measures of HCort exposure in NGN2-neurons. To visualize the effects of HCort exposure on neurite outgrowth, we performed neurite tracing analysis on NGN2-neurons treated with HCort, finding a dose-dependent decrease in neurite length per neuron after stimulation with 100 nM, 1,000 nM and 2,500 nM of HCort (Fig. 2e). This finding is concordant with previously described dendritic retraction phenotypes in response to trauma-related glucocorticoid signaling46.
PTSD diagnosis-dependent differences in human neurons
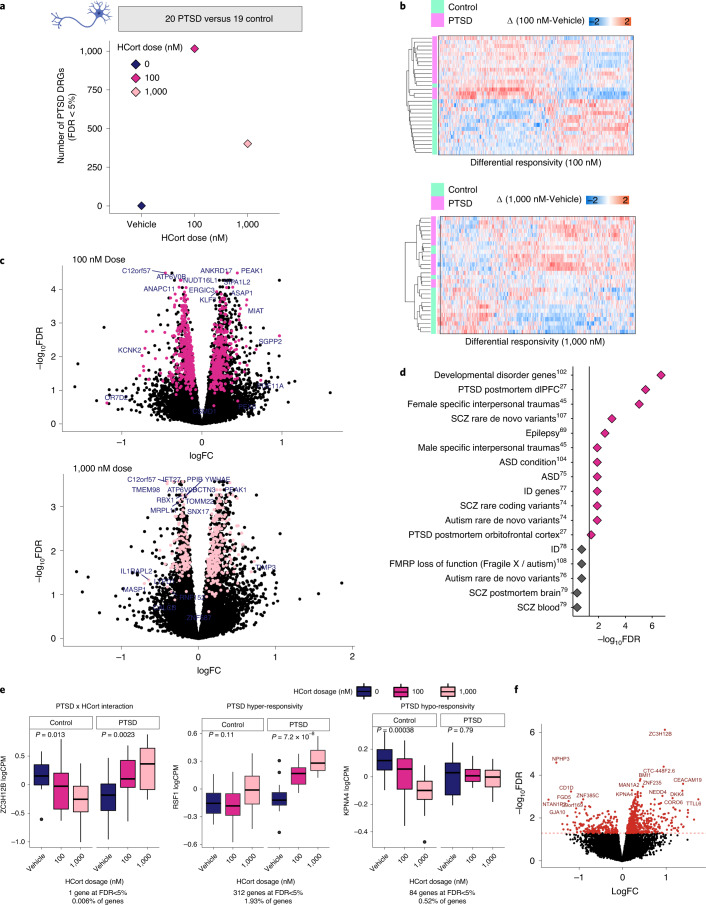
PTSD-specific transcriptional responses to glucocorticoid exposure were examined in human neurons. Baseline gene expression profiles (vehicle; 0 nM HCort) between PTSD(+) and PTSD(–) groups were compared, with no significant differences in gene expression observed (Q value < 0.05) (Fig. 4a). Linear contrast analysis was used to examine the extent to which genes respond differently to HCort, relative to baseline, in PTSD(+) relative to PTSD(–) combat veterans, here termed differential response genes (DRGs); 1,016 and 402 PTSD-specific DRGs were identified in NGN2-neurons following treatment with 100 nM and 1,000 nM HCort, respectively (Fig. 4a and Supplementary Table 6). The significant DRGs in NGN2-neurons predicted PTSD; for each individual, unsupervised classification revealed a clear pattern of HCort response dysregulation that correctly classified NGN2-neurons from PTSD(+) and PTSD(–) groups (Fig. 4b). Meta-analysis revealed shared DRGs across batches (Fig. 4c), demonstrating the validity of this PTSD signature. A gene set enrichment analysis of our DRGs against 17 psychiatric disorder gene sets revealed significant enrichment in PM PTSD dorsolateral prefrontal cortex (dlPFC)27 (P = 2.30 × 10–7), and orbitofrontal cortex (OFC) (P = 0.016) as well as female and male specific interpersonal traumas (P = 1.03 × 10–6, P = 0.005, respectively) (Fig. 4d). Altogether, PTSD-specific neuronal DEGs are not detectable at baseline, are most significant in response to low (100 nM) glucocorticoid exposure and enriched for PM PTSD transcriptomic signatures.
Fig. 4. PTSD(+) specific responses to HCort in NGN2-neurons.
a, Genes that differ in their response to HCort in PTSD(+) donors compared with PTSD(–) donors, here termed DRGs, were detected in both the 100 nM and 1,000 nM dose, indicating PTSD diagnosis-specific responses to HCort. b, Significant NGN2-DRGs correctly classify PTSD(+) from PTSD(–) participants using an unsupervised approach. c, Meta-analysis of expression LogFC DRGs (differences observed between PTSD(+) and PTSD(–)) were plotted against –log(P value). Pink points indicate significantly DEGs in the meta-analysis, representing PTSD case-specific response genes to HCort. Sample-size-based meta-analysis was conducted using METAL with a Cochran’s Q test for heterogeneity. Benjamini–Hochberg adjusted P values are reported to correct for multiple testing. d, Gene set enrichment of significant DRGs across psychiatric disorder gene sets (epilepsy69, developmental delay70, ASD71–76, ID77,78, SCZ79,80 and FMRP targets81). e, The interactive effect of PTSD diagnosis and HCort exposure on gene expression are modeled, and three main observed patterns of direction of effect in significantly interactive genes are represented here. Interaction statistics were derived from the linear model with a diagnosis by HCort interaction coefficient. From among the 740 genes with significant Benjamini–Hochberg-adjusted P values <0.05, genes were identified that responded significantly to HCort in either cases or controls by performing one-way ANOVA on log2CPM normalized expression to increasing HCort exposure separately in PTSD cases and controls. We denote genes with a significant ANOVA P value in controls but not in PTSD cases as ‘PTSD hypo-responsive’, genes with a significant ANOVA P value in both PTSD cases and controls but with opposite directions of effect as ‘interactive’ and genes with a significant ANOVA P value to HCort in PTSD cases but not in controls as ‘PTSD hyper-responsive’. Data are presented as minimum, first quartile, median, third quartile and maximum. The patterns of three representative genes that indicate patterns of PTSD by HCort interaction, PTSD hyper-responsivity and PTSD hypo-responsivity are plotted, demonstrating examples of three biologically meaningful patterns of diagnosis by HCort interaction. f, logFC of all significantly interactive diagnosis by HCort genes are plotted against the P value of their interaction term, with most significant genes representing those with most significant interactive effects.
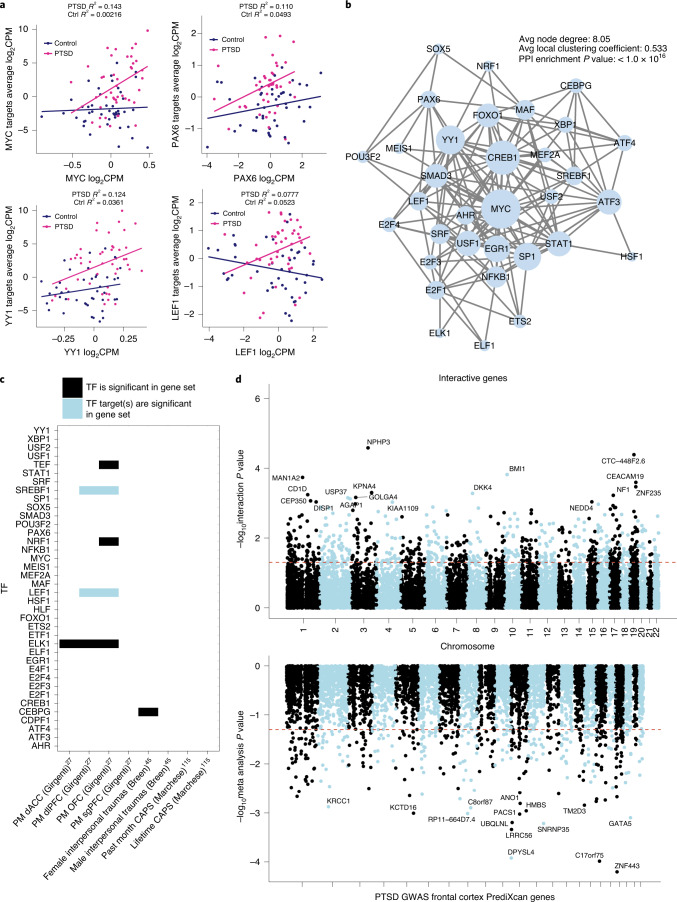
The dose-dependent impact of HCort on PTSD case/control status was tested, revealing 740 genes with a significant diagnosis by HCort exposure interactive effect (FDR < 5%) (Fig. 4f). From this, three gene categories of interest were evaluated: (1) PTSD by HCort interaction, where expression effect with increasing HCort exposure was significant and in opposite directions in cases and controls (1 gene at FDR < 5%); (2) PTSD hypo-responsivity, where increasing HCort exposure only caused a significant expression change in controls, but not cases (84 genes at FDR < 5%) and (3) PTSD hyper-responsivity, where increasing HCort exposure caused a significant expression change in cases, but not controls (312 genes at FDR < 5%) (Fig. 4e). To assess spatial association of significantly interactive genes with common variants associated in PTSD, we mapped imputed expression of PTSD GWAS3 variants against the significance of the HCort by PTSD interaction term (Fig. 5d). Shared peaks were visible in chromosomes 10, 17 and 19.
Fig. 5. TFs driving PTSD hyper-responsivity.
a, PTSD hyper-responsive genes were shown to be enriched for several TF targets. Four of the enriched TFs driving hyper-responsivity in PTSD cases are shown here. Normalized expression of the TF is graphed on the x axis with average expression of the TF targets on the y axis in PTSD case and control NGN2-neurons stimulated with HCort, demonstrating differential regulation in stimulated cases versus controls. b, Network visualization of PPIs amongst identified TFs mediating PTSD hyper-responsivity. STRING analysis was used to identify the observed number of edges. Enrichment of observed edges was assessed against expected edges to determine a PPI P value < 1.0 × 10–16. c, Overlap of TFs (black) and their targets (blue) identified in our study with significantly DEGs in other PTSD studies. dACC, dorsal anterior cingulate cortex; sgPFC, subgenual prefrontal cortex. d, Manhattan plot of significantly interactive genes in our study compared with Manhattan plot of imputed expression from PTSD GWAS indicating spatial orientation of significantly interactive genes.
Transcription factor regulation of PTSD hyper-responsivity
Given previous reports of glucocorticoid hyper-responsivity in PTSD6, putative neuronal driver genes that may mediate this hyper-responsivity were predicted. The FUMA pipeline was used to perform gene set enrichment for transcription factor (TF) targets, finding 38 significantly enriched TFs for which our hyper-responsive genes were targets. To examine differences in regulatory function of these TFs within our study, the relationship between TF expression and target expression was investigated in PTSD cases and controls treated with HCort (Fig. 5a). In a subset of putative driver genes, increased TF levels more tightly corresponded to increased expression of target genes in neurons derived from PTSD cases than controls. These different patterns of TF coregulation in PTSD suggest a significant role for these TFs in driving PTSD-specific transcriptional response. The coregulation network was mapped by visualizing PPIs (Fig. 5b), finding a significant observed number of edges (P < 1.0 × 10–16), showing that linked expression of genes in this network translate to a shared effector regulation. Four of these 38 TFs, and two targets of identified TFs, were noted to be differentially expressed in previously reported PM27 and/or blood47 studies of PTSD (P value of 0.011, binomial test with an α of 0.05) (Fig. 5c). Altogether, coordinated transcriptional regulation and proteomic interaction of a network of TFs with shared effector regulation may mediate glucocorticoid hyper-responsiveness in PTSD.
Discussion
Here, an hiPSC-based model was used to explore the cell-type-specific and diagnosis-dependent glucocorticoid treatment responses of PBMCs and NGN2-glutamatergic neurons derived from male combat-exposed PTSD cases (n = 19 hiPSC donors and n = 20 PBMC donors) and controls (n = 20 hiPSC donors and n = 20 PBMC donors). Both blood and neuronal glucocorticoid responses were significantly enriched for immune response genes; neuronal glucocorticoid responses were also associated with brain development and neurodevelopmental disorder genes. Although a PTSD diagnosis-specific signature was not detectable at baseline in either cell type, glucocorticoid hypersensitivity in PTSD was observed, with diagnosis-specific effects greatest in neurons at low HCort doses.
These findings are consistent with the glucocorticoid hypersensitivity hypothesis; for example, patients with PTSD have altered blood sensitivity to glucocorticoids6 and perturbations in glucocorticoid receptor signaling have been shown for PTSD in PM brain tissue10. That the PTSD-relevant gene set enrichment in HCort neuronal response was significant only if the effect of PTSD diagnosis was included in our model (compare Fig. 4d and Fig. 2e) suggests that environmental stress alone, independent of genetic risk, is insufficient to recapitulate PTSD biology. Further studies into the genetic risk elements imparting PTSD-specific HCort response, such as detecting HCort-responsive expression quantitative trait loci, may fine-map PTSD-associated risk loci. This will allow for more formal studies of colocalization, expanding on Fig. 5d, to spatially associate noncoding risk elements in PTSD with functional consequences. These findings emphasize the crucial role of stress response in PTSD risk but cannot rule out the potential impact of subsequent stress recovery; future work will be necessary to resolve the relative contributions of these alternatives.
The cell type(s) of origin in PTSD remain unresolved. It will be critical to explore diagnosis-dependent glucocorticoid responses across additional subtypes of brain cells to establish more physiologically relevant models. Although robust upregulation of immune response gene modules was observed in both cell types, the cell-type specific differences were confounded by cohort composition as well as experimental differences in the methods of glucocorticoid treatment used in each cell type (PBMCs: 72 h with 2.5 nM, 5 nM or 50 nM DEX; NGN2-neurons: 24 h with 100 nM, 1,000 nM or 2,500 nM HCort). Are blood-specific glucocorticoid changes a critical part of disease risk, initiation and/or progression, resulting in the release of additional cytokines and inflammatory molecules that impact brain function? Or are they simply an incomplete biomarker of causal events occurring in the brain? Within the brain, are neuron-specific responses adequate to interpret G×E interactions? Certainly, future experiments should investigate cell-autonomous and noncell-autonomous effects across a larger variety of neuronal, astroglial and microglial cell types. Moreover, an increasing amount of direct brain imaging48–50 and recent genomic analyses implicate specific subtypes of inhibitory GABAergic neurons in PTSD27,51. Transcriptome-wide association studies using PTSD GWAS52, as well as two independent transcriptomic studies of PM brain tissue from PTSD cases and controls27,51, associated GABAergic signaling with PTSD risk and illness state. This suggests that decreases in GABAergic activity confer susceptibility to traumatic stress and may be involved in the pathophysiology of PTSD17. Therefore, inclusion of GABAergic neurons, alone and in coculture with glutamatergic neurons, would be of particular interest in subsequent studies of neuronal glucocorticoid response.
Given the similarity of hiPSC-derived neurons to fetal brain cells53–57, our hiPSC-based model of PTSD may best reflect the G×E impact of developmental stress exposure, as demonstrated by robust enrichments for expression of autism spectrum disorder (ASD) risk genes, as observed here and previously58. Consistent with this, glucocorticoid treatment increases the proliferation of hiPSC-derived neural progenitor cells59, and impairs neuronal differentiation and maturation in hiPSC-derived neurons20,60 and primary mouse neurons61. Nonetheless, it would be an oversimplification to speculate that glucocorticoid response in hiPSC-derived neurons represents the ‘pretrauma state’, and that analyses of patient PBMCs and brain tissue reflect the ‘post-trauma state’. Stress impacts risk to psychiatric disorders throughout the lifespan—across prenatal development62,63, childhood52 and adulthood52,64,65. Moreover, the significant and positive relationship between our observed associations in NGN2-neurons and previously demonstrated transcriptional PTSD signatures in PM brains suggests that some impacts of glucocorticoid exposure may persist through to adulthood. Notably, glucocorticoid stimulation is not a specific model for PTSD; stress response is comorbid in many psychiatric disorders. HCort-responsive genes therefore likely represent aspects of stress response shared across PTSD and other neuropsychiatric disorders, such as the shared impact on social cognition between PTSD and ASD66,67. Combined analysis of context-dependent hiPSC models with cross-lifespan datasets, such as PM brains, may uncover long-term glucocorticoid-dependent PTSD signatures with which to refine hallmarks of PTSD susceptibility following combat exposure.
Because trauma exposure, even in the absence of PTSD, can impact glucocorticoid response68, future studies may usefully include donors with no trauma exposure. Toward this exploration, hCort response in NGN2-neurons was examined from one additional control without a reported trauma exposure, observing 13% overlap (805 of 6,190) of our HCort-responsive neuronal genes (exact binomial test 0.1300485). Likewise, glucocorticoid response in hiPSC-derived cerebral organoids from healthy and presumably trauma-free controls20 induced 16% (989 of 6,190) of the HCort-responsive genes here detected in NGN2-neurons (exact binomial test 0.1597738, P < 2.2 × 10–16).
hiPSC-derived neurons represent a promising platform with which to screen for genetic and/or pharmacological interventions that can prevent, ameliorate or reverse acute or chronic neuronal responses to stress. While glucocorticoid-responsive transcriptomic changes are sufficient to stratify trauma-exposed PTSD cases and controls, they are not yet clinically informative. Clinical utility is limited by variability of traumatic exposures, complexity of involved cell types, impracticality of individualized hiPSC culture and transcriptomic analysis, poor understanding of the specific genetic loci mediating differential HCort response, and lack of in-depth characterization of how in vitro phenotypes correspond to clinical phenotypes or treatment susceptibility. To translate our findings into clinically meaningful risk-stratification, it is imperative that future studies test whether our signatures relate to clinical severity and/or treatment responsiveness, and thereby uncover new therapeutic target(s) for PTSD. The current work indicates that if the biological mediators of environmental risk can be predicted, then hiPSC-based models can be used to test genotype-dependent and cell-type-specific environmental responses. Thus, given the critical importance of gene by stress (and G×E more broadly), we believe that, moving forward, functional genomics approaches must integrate stem cell models and genome engineering to resolve the impact of patient-specific variants across cell types and environmental stressors. Our hope is that dissecting how disease risk variants interact with the environment across the diverse cell types that comprise the human brain will ultimately improve diagnostics, predict clinical trajectories and identify pathways that could serve as presymptomatic targets of therapeutic intervention.
Methods
Participants
A total of 49 individuals were recruited to yield well-matched and largely overlapping (30 shared individuals) hiPSC and PBMC cohorts, comprised of combat veterans with (n = 19 hiPSC donors and n = 20 PBMC donors) and without (n = 20 hiPSC donors and n = 20 PBMC donors) PTSD. Detailed information about donor breakdown in the hiPSC and PBMC studies, by experimental batch, is presented in Supplementary Table 2. Participants in this cross-sectional study were combat-exposed Operation Enduring Freedom (OEF), Operation Iraqi Freedom (OIF) and Operation New Dawn (OND) veterans with and without PTSD who provided written informed consent (VA HS no. YEH-16-03 and ISMMS HS no. 15-00886) and from whom a viable blood and/or fibroblast sample was provided and sufficient RNA for genome-wide expression analyses was extracted. Participants were compensated $300 for involvement in this study. Eligibility for participation was determined as previously described23. Participants were included serially in the order in which they consented until the enrollment targets were attained. All participants reported a DSM-IV criterion A combat trauma82; all experienced deployment to active military combat zones and experienced one or more significant combat-related traumas. Individuals with and without PTSD did not have significant differences in childhood or predeployment trauma, deployment number or cumulative duration. Participants underwent psychological evaluation using the Structured Clinical Interview for DSM-5 (SCID)83 and the Clinician Administered PTSD Scale (CAPS)84 for determination of PTSD diagnosis and severity. Eligibility criteria and thresholds were based on CAPS for DSM-IV; PTSD(+) had a current CAPS-IV total score above 40 (frequency plus intensity), whereas PTSD(–) participants were combat-exposed veterans with a total score below 40. Although DSM-IV criteria for PTSD were used for inclusion, note that PTSD(+) participants also met criteria for PTSD based on DSM-5. Diagnostic and clinical exclusions included: (1) presence of moderate or severe substance use disorder within the past 6 months, (2) lifetime history of primary psychotic or bipolar I disorders (BD), (3) self-reported history of moderate or severe traumatic brain injury, (4) neurological disorder or main systemic illness, (5) treatment with systemic steroids and, for PTSD(−) only, (6) current or recurrent major depressive disorder (MDD). Psychotropic medication was permitted, but dosage stabilization for at least 2 weeks was required. Around 20% of individuals across both groups are currently treated with psychiatric medications. Current oral steroid treatment was an exclusion based on the impact of systemic steroids on the hypothalamic-pituitary-adrenal axis. Given the small sample size, there was no additional matching performed for clinical characteristics such as index trauma types, duration of the disorder, comorbidities and psychiatric medications at time of recruitment. Available clinical information is summarized in Supplementary Table 1 and presented in detail in Supplementary Table 2.
Biopsy collection and generation/validation of hiPSCs
Biological samples (blood and fibroblast) were collected from eligible participants at the James J. Peters (JJP) Veterans Affairs Medical Center (VAMC). Blood processing occurred at the JJP VAMC and fibroblasts were transferred to the New York Stem Cell Foundation Research Institute (NYSCF). All hiPSCs were reprogrammed together in randomized batches as fibroblast biopsies were obtained over time.
Skin fibroblasts of PTSD and non-PTSD participants were collected via skin punch biopsy taken from the upper buttocks. Upon collection, biopsies were placed immediately in Biopsy Collection Medium (Cascade Biologics Medium 106 (Life Technologies, catalog no. M-106-500), 1× Antibiotic-Antimycotic (Life Technologies, catalog no. 15240-062) and LSGS (Life Technologies, catalog no. S-003-10)) and stored at 4 °C for a maximum of 24 h. Biopsies were dissected into pieces of around 1 mm3 and plated in Biopsy Plating Medium (Knockout-DMEM (Life Technologies, catalog no. 10829-018), 10% fetal bovine serum (Life Technologies, catalog no. 100821-147), 2 mM GlutaMAX (Life Technologies, catalog no. 35050-061), 0.1 mM MEM nonessential amino acids (Life Technologies, catalog no. 11140-050), 1× Antibiotic-Antimycotic (Life Technologies, catalog no. 15240-062) and 0.1 mM 2-mercaptoethanol (Life Technologies, catalog no. 21985-023)). Upon the outgrowth of fibroblasts, samples were entered into an automated pipeline producing vials of cells for both hiPSC reprogramming and backup stock. A cell pellet was collected, with DNA isolated using an EPMotion and the ReliaPrepTM 96 gDNA Miniprep HT System (Promega, catalog no. A2671). This DNA was used to confirm sample identity throughout the reprogramming process. Fibroblasts were reprogrammed using modified mRNA (Reprocell, catalog no. 000076) and enriched using antifibroblast meads (Miltentyi Biotec, catalog no. 130-050-601) in automated procedures. Reprogrammed hiPSCs were then expanded using PSC Feeder Free Medium (Thermo Fisher Scientific, catalog no. A14577SA) and grown on Cultrex-coated plates (R&D Systems, catalog no. 3434-010-02). Cells were routinely passaged using our automated platform in the presence of Accutase (Thermo Fisher, catalog no. A11105-01). Following passage, cells were maintained in PSC medium supplemented with 1 µM thiazovivin (Sigma-Aldrich, catalog no. SML1045-25MG). All cells were frozen in Synthafreeze (Thermo Fisher, catalog no. A12542-01) into master stocks.
As part of the hiPSC validation process, all samples were tested for the absence of Mycoplasma (Lonza, catalog no. LT07-710) and confirmed to be sterile (Hardy Diagnostics, catalog no. K82). Samples were confirmed to key karyotypically normal using the Illumina Core Exome Genotyping Chip (Illumina, catalog no. 20030770) and cnvPartition v.3.2.0 (Illumina, Genome Studio). No cell lines displayed karyotypic abnormalities (no reported copy number variations ⫺2.5 MB in size). All reported copy number variations are shown in each certificate of analysis. hiPSC lines were confirmed to be viable post-thaw, achieving a minimum of 50% confluency within 10 days). Sample identity testing was performed using the SNPTrace assay, confirming correct sample association between parental fibroblast and hiPSC line. Gene expression analysis was using a custom Nanostring panel85 to confirm expression of pluripotency markers such as POU5F1, NANOG and SOX2, and lack of expression of early differentiation markers such as AFP (Mesoderm), SOX17 (Endoderm) and NR2F2 (Ectoderm). A scorecard panel was used to confirm propensity to differentiate85. All hiPSC lines used in this study passed the above qualify control and have a certificate of analysis.
PBMC isolation and culture
Isolation of PBMCs, cell culture and incubation with DEX were performed as described23 (Fig. 1a). PBMC data from n = 10 combat veterans with PTSD and n = 10 without PTSD is first reported here (batch A) and n = 10 combat veterans with PTSD and n = 10 without PTSD was reported previously23 (batch B) (Supplementary Table 2). In brief, blood was collected in EDTA-containing vacutainer tubes (VWR) and PBMCs were isolated by density gradient centrifugation using Ficoll-Paque (GE Healthcare) and washed twice in HBSS (Thermo Fisher, 14175). Mononuclear cells were then counted manually using a Cellometer disposable counting chamber (Nexcelom Bioscience LLC). The cells were resuspended in complete RPMI, containing RPMI-1640 (Gibco), 10% fetal bovine serum, 50 U ml–1 penicillin–streptomycin mixture (Gibco) at a density of 1.75–2.00 × 106 cells ml–1 of the medium. PBMCs were prepared at 2.5 × 106 cells ml–1 in complete RPMI for DEX treatment experiments. Following incubation at 37 °C, 5% (v/v) CO2 for 72 h, the plates were centrifuged at 900g for 15 min at 4 °C and supernatant was collected and pooled from each DEX concentration well. The cell pellet on the bottom of each well was resuspended in TRIzol reagent. Cell lysates for each DEX dose were pooled, aliquoted and stored at −80 °C until RNA isolation.
Automated generation of hiPSC-derived NGN2-neurons
Glutamatergic NGN2-neurons were generated from hiPSCs, using high-throughput automated differentiations, in two batches (batch 1 n = 15 versus 15; batch 2 n = 4 versus 5) (Supplementary Table 2), as previously described with some modifications36. hiPSCs were single cell passaged after a 20-min dissociation with Accutase (STEMCELL Technologies) at 37 °C and 5% CO2. A total of 1 million cells per well were plated in 12-well Cultrex-coated (R&D Systems, catalog no. 3434-010-02) tissue culture plates (Corning Costar) in PSC Feeder Free Medium (Thermo Fisher Scientific, catalog no. A14577SA) with 1 µM thiazovivin (Sigma-Aldrich, catalog no. SML1045). Lentivirus (generated by ALSTEM) carrying pLV-TetO-hNGN2-eGFP-Puro (Addgene, catalog no. 79823) and FUdeltaGW-rtTA (Addgene, catalog no. 19780) was diluted to a multiplicity of infection of one each (1 million genome counts of each vector per transduction) in 100 µl DPBS, no calcium, no magnesium (Thermo Fisher Scientific) and added directly after cell seeding. After 24 h, the medium was exchanged with Neural Induction Medium (NIM) comprising a 50:50 mix of DMEM/F12 and Neurobasal, with 1× B27 plus vitamin A, 1× N2, 1× Glutamax (Thermo Fisher Scientific) and 1 µM doxycycline hyclate (Sigma-Aldrich). After 24 h, the medium was removed and NIM was added with doxycycline plus 5 µg ml–1 puromycin. A full medium exchange was performed the next day with NIM plus doxycycline and puromycin; 24 h later, cells were passaged by incubating with Accutase for 30 min at 37 °C and 5% CO2. A series of 96-well plates (PerkinElmer CellCarrier Ultra) were coated with 0.1% polyethylenimine (Sigma-Aldrich, catalog no. 408727) in 0.1 M borate buffer pH 8.4 for 30 min at room temperature, washed five times with water and prefilled with 100 µl per well of neural coating medium comprising Brainphys medium (STEMCELL Technologies) with 1× B27 plus vitamin A, 1 µM thiazovivin, 5 µg ml–1 puromycin, 250 µM dibutyryl cAMP (dbcAMP, Sigma-Aldrich), 40 ng ml–1 brain-derived neurotrophic factor (BDNF, R&D Systems), 40 ng ml–1 glial cell line-derived neurotrophic factor (GDNF, R&D Systems), 200 µM ascorbic acid (Sigma-Aldrich) and 10 µg ml–1 natural mouse laminin (Sigma-Aldrich). A sample of cells were stained with 10 µg ml–1 Hoechst plus 1:500 acridine orange/propidium iodide solution and counted on a Celigo imager (Nexcelom Bioscience). A total of 50,000 cells per well were seeded into neural coating medium-filled 96-well plates in 100 µl per well of neural medium comprising Brainphys medium with 1× B27 plus vitamin A, 1 µM thiazovivin, 5 µg ml–1 puromycin, 250 µM dbcAMP, 40 ng ml–1 BDNF, 40 ng ml–1 GDNF, 200 µM ascorbic acid and 1 µg ml–1 natural mouse laminin. At 24 h after seeding, medium was exchanged for neural selection medium comprising Brainphys medium with 1× B27 plus vitamin A, 250 µM dbcAMP, 40 ng ml–1 BDNF, 40 ng ml–1 GDNF, 200 µM ascorbic acid, 1 µg ml–1 natural mouse laminin and 2 µM arabinosylcytosine (Ara-C, Sigma-Aldrich). After 48 h, the neural selection medium was fully exchanged and after a further 48 h the medium was fully exchanged with neural maintenance medium (NMM) comprising Brainphys medium with 1× B27 plus vitamin A, 250 µM dbcAMP, 40 ng ml–1 BDNF, 40 ng ml–1 GDNF, 200 µM ascorbic acid and 1 µg ml–1 natural mouse laminin. Thereafter, every 48 h, half the medium was exchanged with NMM until day 21 posttransduction passage. In Batch 1, all medium exchanges were performed using a Hamilton Star liquid handler set to 5 µl s–1 for aspirate and dispense as part of the NYSCF Global Stem Cell Array® Team86. Passages were fully automated and performed on a robotic cluster comprising a Thermo Fisher Scientific C24 Cytomat incubator, a Hamilton Star liquid handler, an Agilent microplate centrifuge, a Precise Automation PreciseFlex 400 Sample Handler and a PerkinElmer Opera Phenix. In Batch 2, although medium exchanges were fully automated, passages were performed manually.
At harvest, medium was removed using the Bluewasher (BlueCatBio) and cells were lysed for 5 min using RLT plus buffer (Qiagen), snap frozen on dry ice and stored at −80 °C. A replicate plate was fixed for immunofluorescence analysis by adding 32% paraformaldehyde (Electron Microscopy Sciences) directly to medium to a final concentration of 4% and incubated at room temp for 15 min. Cells were washed three times with HBSS (Thermo Fisher Scientific), stained overnight with mouse anti-Nestin 1:3,000 (Millipore, catalog no. 09-0024), chicken anti-MAP2 1:3,000 (Abcam, catalog no. 09-0006) in 5% normal goat serum (Jackson ImmunoResearch) in 0.1% Triton X-100 (Thermo Fisher Scientific) in HBSS. Primary antibodies were counterstained with goat anti-mouse Alexa Fluor 555 and goat anti-chicken Alexa Fluor 647 and 10 µg ml–1 Hoechst for 1 h at room temp. Cells were washed three times with HBSS. Batch 1: nine fields (×40 magnification) were imaged per well (one well per condition per line) using the PerkinElmer Opera Phenix microscope. Batch 2: nine fields (×20 magnification) were imaged per well (two wells per hiPSC line) using the ArrayScan automated microscope (Thermo Fisher Scientific). Interwell variability in neuronal identity and maturity was assessed using automated image analyses. NESTIN-positive neural progenitor cells were demarcated from MAP2-positive postmitotic neurons (Extended Data Fig. 1). Variation between batches may reflect discrepancies in imaging methods, rather than biological differences in neuronal morphology or maturity.
Glucocorticoid treatment
Preliminary studies were conducted to identify optimal culture and glucocorticoid stimulation conditions. These pilot studies sought to evaluate the transcriptional effects of HCort and DEX on NGN2-neurons, and optimize the length of glucocorticoid treatment and concentrations. Neither quantitative PCR for six glucocorticoid regulatory genes (covering ten concentrations of DEX) nor RNA-seq (covering three concentrations of DEX) revealed significant gene expression differences following 72 h of exposure87, consistent with minimal upregulation of FKBP5 mRNA expression following DEX treatment of hiPSC neurons19. This is consistent with previous reports of DEX treatment of primary cultures, which found neurons to be significantly less responsive than astrocytes88. Together, our preliminary data uphold the view that astrocyte88 or endothelial89 response, rather than neuronal response, mediates brain-level effects of DEX treatment. As our preliminary experiments found a strong genome-wide effect of HCort treatment, we used HCort treatment for neuronal glucocorticoid exposure. Serum measurements of cortisol in patients with and without PTSD have been found to average around 492.52 nM90, intermediate between our 100 and 1,000 nM doses. HCort treatment medium was prepared by first dissolving HCort (Sigma-Aldrich, catalog no. H0888) in ethanol to make a 2.8 mM stock. HCort ethanol stock was then diluted to 0.2 mM in HBSS. Ethanol was equalized to 15 µM in control and all treatment media. DEX treatment medium was prepared by dissolving DEX (Sigma-Aldrich, catalog no. D1156) in HBSS6. The final treatment medium was prepared by diluting HCort or DEX stocks into NMM/RPMI, before applying to cells by fully exchanging medium. Neurons were treated with HCort for 24 h (baseline, 100 nM, 1,000 nM and 2,500 nM), PBMCs with DEX for 72 h (baseline, 2.5 nM, 5 nM and 50 nM). Baseline medium conditions are estimated to contain 58 nM of corticosterone from neuronal supplement B27 (Thermo Fisher), which may predispose neurons to a higher effective concentration for glucocorticoid stimulation91, and may bias responsive genes toward those that respond to severe stressors, rather than homeostatic or regulatory changes, such as circadian rhythms.
RNA extraction and quality control
For NGN2-neurons, RNA was harvested with an RNeasy Plus Micro Kit (Qiagen). For PBMCs, RNA was extracted from TRIzol-lysed PBMCs using the miRNAeasy Mini Kit (Qiagen). Following each extraction, RNA quantity was measured using a Nano Drop 2000 Spectrophotometer (Thermo Scientific) and RNA quality and integrity was measured with an Agilent 2100 Bioanalyzer (Agilent). All RNA integrity numbers (RINs) in the current study were high: NGN2-neurons (8.8 ± 0.53) and PBMCs (7.5 ± 0.95).
RNA-seq data generation
A low-input RNA-seq protocol was applied for the generation of RNA-seq data from NGN2-neurons. Specifically, polyA enriched RNAs were subjected to RNA-seq library preparation using the SMART-Seq v.4 Kit (SSv.4; Takara) and sequenced using a paired-end 150 bp configuration with 30 M supporting reads per sample. For RNA-seq data generation from PBMCs, Ribo-zero rRNA deleted RNAs were subjected to RNA-sequencing library preparation using the Illumina TruSeq Stranded Total RNA kit (Illumina) and sequenced using a paired-end 150 bp configuration with 20 M supporting reads per sample.
RNA-seq data preprocessing
All RNA-seq FASTQ files underwent matching analytical procedures, as described previously. In brief, resulting short reads with Illumina adapters were trimmed and low-quality reads were filtered using TrimGalore92 (–illumina option). All high-quality reads were then processed for alignment using the hg38 reference and the ultrafast universal RNA-seq aligner STAR with default parameters93. Mapped bam files were sorted using Samtools and short read data were quantified using featureCounts94 with the following parameters: -T 5, -t exon and -g gene_id. Subsequently, all read counts were exported and all downstream analyses were performed in the R statistical computing environment. Raw count data was subjected to nonspecific filtering to remove low-expressed genes that did not meet the requirement of a minimum of two counts per million (CPM) in at least around 40% of samples. This filtering threshold was applied to NGN2-neurons and PBMCs separately. All expression values were converted to log2 reads per kilobase of transcript, per million mapped reads (RPKM) and subjected to unsupervised principal component analysis (PCA) to identify and remove outlier samples that lay outside 95% confidence intervals from the grand averages. This identified two outlier samples in NGN2-neuron Batch 1 and one outlier sample in Batch 2 that were excluded from our analysis.
Developmental specificity analysis
RNA-seq datasets from existing PM brain tissue and hiPSC models were integrated to validate the developmental specificity of our samples using a previously described approach40. In brief, a total of 16 independent studies were collected covering 2,716 independent samples and 12,140 genes. These samples cover a broad range of hiPSCs, neural progenitor cells, mature neurons, prenatal and postnatal brain tissues. All expression values were converted to log2 RPKM and collectively normalized using quantile normalization using the limma R package. Subsequently, for each independent sample present in our hiPSC neuron dataset, we performed pairwise correlation analysis (using Pearson’s correlation coefficients) across all independent samples and subsequently aggregated the correlation coefficients for each external study and/or cell type. Next, and as a complementary approach, all datasets were jointly analyzed and integrated using PCA (Extended Data Fig. 3).
Differential gene expression analyses
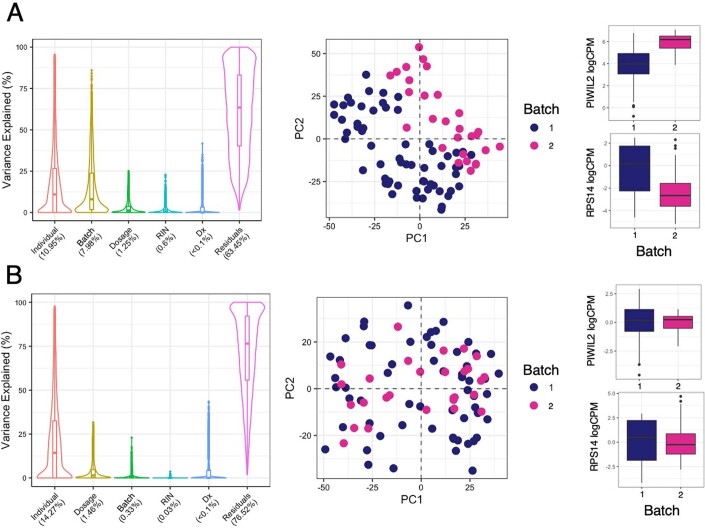
Transcriptional signatures were generated using a strategy adapted from Hoffman et al.40, using scripts available at www.synapse.org/hiPSC_COS. Gene expression values were normalized using VOOM95. Confounders explaining a significant proportion of variance in gene expression were identified using variancePartition96 (that is, experimental batch, treatment, individual as a repeated measure (that is, interdonor effects), PTSD diagnosis and RIN) (Extended Data Fig. 2a). A significant batch effect was observed (Extended Data Fig. 2a–c), which was corrected for by constructing a linear model of batch and extracting the residuals (Extended Data Fig. 2d–f). Next, differential gene expression analyses were conducted using a moderated t test from the R package limma95. Models examining the HCort-dependent, PTSD-independent effect (Figs. 1–3) included adjustment for the possible confounding influence of PSTD diagnosis and RIN, while PTSD-dependent models (Figs. 4 and 5) included diagnosis as a main outcome. Due to the repeated measures study design, where individuals are represented by multiple independent technical replicates, the duplicateCorrelation function was applied in the limma analysis and gene level significance values were adjusted for multiple testing using the Benjamini and Hochberg method to control the FDR. Genes with FDR < 5% were considered significantly differentially expressed. To integrate differential gene expression results between batches, summary statistics from differential gene expression analyses to both glucocorticoid-dependent and PTSD-dependent responses in each batch were meta-analyzed using METAL97. Significant results across both studies were identified using a Benjamini–Hochberg-adjusted P value threshold (FDR < 0.05).
Extended Data Fig. 2. Adjustment of Batch effects.
a, VariancePartition and PCA analysis on uncorrected data from two batches (batch 1, n = 30, batch 2, n = 9 biologically independent individuals) indicating large batch effect. Boxplot data presented are minimum, 1st quartile, median, 3rd quartile and maximum. Example of gene with large batch effect across batch 1 (n = 30) and batch 2 (n = 9). b, VariancePartition, PCA, and example gene after batch correction for same batches as above.
Neurite outgrowth analysis
To determine functional and mechanistic consequences of HCort treatment, NGN2-neurons were seeded as 1.5 × 104 cells per well in a 96-well plate coated with 4× Matrigel at day 5. At day 6, NGN2-neurons were treated with HCort for 24 h (0 nM (vehicle), 100 nM, 1,000 nM and 2,500 nM) as in previous experiments. At day 7, cultures were fixed using 4% formaldehyde/sucrose in PBS with Ca2+ and Mg2+ for 10 min at room temperature (RT). Fixed cultures were washed twice in PBS and permeabilized and blocked using 0.1% Triton/2% normal donkey serum (NDS) in PBS for 2 h. Cultures were then incubated with primary antibody solution (1:1,000 MAP2 anti-chicken (Abcam, catalog no. ab5392) in PBS with 2% NDS) overnight at 4 °C. Cultures were then washed three times with PBS and incubated with secondary antibody solution (1:500 donkey anti-chicken Alexa 647 (Life technologies, catalog no. A10042) in PBS with 2% NDS) for 1 h at RT. Cultures were washed a further three times with PBS with the second wash containing 1 μg ml–1 4,6-diamidino-2-phenylindole. Fixed cultures were then imaged on a CellInsight CX7 HCS Platform with a ×20 objective (0.4 numerical aperture) and neurite tracing analysis performed using the neurite tracing module in the Thermo Scientific HCS Studio v.4.0 Cell Analysis Software. A total of 12–24 wells were imaged per condition across a minimum of two independent cell lines, with nine images acquired per well for neurite tracing analysis. A Kruskal–Wallis test with a post hoc Dunn’s multiple comparisons test was performed on data for neurite length per neuron using Graphpad Prism. This analysis was performed in day 7 neurons to ensure optimal density to observe neurite outgrowth phenotypes, which are less quantifiable by these methods in more mature neurons.
Weighted gene coexpression network analysis and functional annotation
Signed coexpression networks were built for PBMCs and NGN2-neurons using WGCNA98. To construct a global weighted network for each cell type, a total of 20,101 and 16,146 postquality-control genes were used in PBMCs and NGN2-neurons respectively. The absolute values of Biweight midcorrelation coefficients (optimal for small sample sizes) were calculated for all possible gene pairs within each cell type, and resulting values were transformed using a β-power (β = 4 for NGN2-neurons and β = 7 for PBMCs) so that the final correlation matrices followed an approximate scale-free topology (Extended Data Fig. 6). To determine which modules, and corresponding biological processes, were most associated with HCort/DEX, singular value decomposition of each module’s expression matrix was used; the resulting ME), equivalent to the first principal component, represented the overall expression profiles for each module. Each module was enriched for GO biological processes, molecular factors, cellular components and molecular pathways using ToppGene99. All genes passing nonspecific filtering in the current dataset were used as a genomic background. Only gene sets that passed a multiple test adjustment using the Benjamini–Hochberg procedure (adj. P < 0.05) were deemed significant. ME values were correlated with dosage by Pearson correlation. Protein–protein association networks were constructed using STRING100. For protein–protein association analysis of WGCNA modules, genes with high module membership (>0.8) were selected for STRING analysis and computation of PPI enrichment P values. Networks were visualized by cytoscape101.
Gene coexpression module preservation analysis
Gene coexpression modules that were disrupted or created in response to glucocorticoids across NGN2-neurons and PBMCs, a permutation-based preservation statistic (Zsummary)2 with 200 random permutations was used to measure the (dis)similarity in correlation patterns for the genes within these gene sets: Zsummary > 10 indicates strong evidence of preservation, 2 < Zsummary < 10 indicates weak-to-moderate evidence of preservation and Zsummary < 2 indicates minimal-to-no evidence of preservation. For this analysis, we specifically focused on dynamically regulated, glucocorticoid-responsive functional modules that were identified in either NGN2-neurons or PBMCs, respectively.
Integration of disease-associated genes and gene sets
Concordance of observed PBMC and neuron transcriptomic signatures to glucocorticoid stimulation (Figs. 1e and 2e), or between PTSD cases and controls (Fig. 4d) and previously reported psychiatric disorder and neurodevelopmental expression patterns102–108 was determined (Extended Data Fig. 10). Psychiatric disorder enrichment was determined using genetic and genomic disease-related gene lists for PTSD, MDD, schizophrenia (SCZ), BD, ASD, alcohol use disorder, and inflammatory bowel disease (IBD)27. Glucocorticoid dysregulation of neurodevelopmental genes was examined using risk genes associated with epilepsy69, developmental delay70, autism spectrum disorder71–76, intellectual disability (ID)77,78, SCZ79,80 and fragile X mental retardation protein (FMRP) target81 genes. All gene sets were assessed by overrepresentation analysis109 using WebGestaltR110, with significant enrichment calculated using hypergeometric distribution. All P values from all gene sets were adjusted for multiple testing using the Benjamini–Hochberg procedure, using a P value < 0.05 threshold to determine significance. These gene lists are available in Supplementary Table 3.
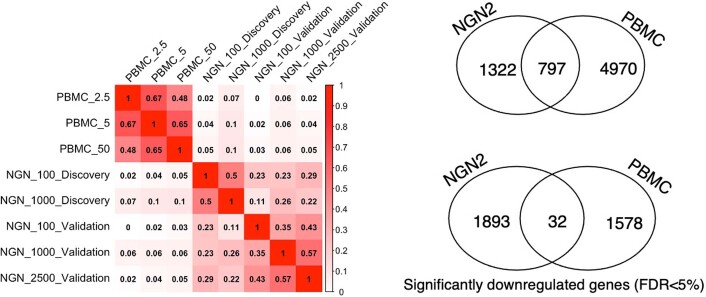
Extended Data Fig. 10. Concordance of PBMC signature with NGN2 signature.
Pairwise Pearson-correlations between transcriptome-wide signatures of PBMC and NGN2 batches.
Clustering of PTSD expression patterns
To assess whether significant gene expression patterns could predict PTSD, the differential log2CPM of 1,016 and 402 PTSD-specific DRGs in NGN2-neurons following 100 nM and 1,000 nM of HCort, respectively, were plotted using pheatmap111. K-means clustering was applied with Euclidian distance using average linkage clustering and tested for observed clustering of PTSD(+) and PTSD(–) gene signatures (Fig. 4b).
Interaction analysis
To test the interaction of HCort concentration with PTSD diagnosis, the linear effect of the HCort by PTSD interaction term was modeled using limma, adjusting for RIN and donor as a repeated measure. Of the 740 genes with significant Benjamini–Hochberg-adjusted P values <0.05, genes were identified that responded significantly to HCort in either cases or controls by performing one-way analysis of variance (ANOVA) on log2CPM normalized expression to increasing HCort exposure separately in PTSD cases and controls. Genes with a significant ANOVA P value in controls but not in PTSD cases as ‘PTSD hypo-responsive’, genes with a significant ANOVA P value in both PTSD cases and controls, but with opposite directions of effect as ‘interactive’ and genes with a significant ANOVA P value to HCort in PTSD cases but not in controls were labeled as ‘PTSD hyper-responsive’ (Fig. 4e). We selected these three categories to maximize biological interpretability, and due to previous reports of glucocorticoid hypersensitivity in PTSD, suggesting hyper- and hypo-sensitivity of targets as physiologically relevant to PTSD. To assess spatial association of genes with the most significant interactive effects with known common variant effectors involved in PTSD, expression of PTSD GWAS3 variants was imputed using PrediXcan112 and mapped against the significance of the HCort by PTSD interaction term in our study (Fig. 5d).
To identify upstream drivers of the PTSD-specific HCort response signature, enrichment for TF targets, defined based on MSigDB C3 gene set groupings113, was performed using the FUMA pipeline114. TFs were tested for their overlap with genes previously reported in PM brains27 of individuals with PTSD and genes associated with CAPS scores in whole blood115. The significance of the number of TFs enriched in PTSD gene sets was tested using a binomial test.
Statistics and reproducibility
Sample sizes for this study were chosen based on comparisons with other idiopathic designs40,116,117. Using an isogenic within-donor design, we were adequately powered to resolve Hcort specific effects. No statistical method was used to predetermine sample size for case–control comparisons. Data was assumed to be normal due to log2-transformation of all expression data. RNA-seq sample data was excluded based on PCA of genome-wide log2RPKM values to identify outlier samples that lay outside 95% confidence intervals from the grand averages. This identified two outlier samples in NGN2-neuron Batch 1 and one outlier sample in Batch 2 that were excluded from our analysis. All experimentation and quality control on hiPSC lines was performed with blinding to disorder status. Sorting of samples occurred with HCort treatment conditions laid out with increasing dosages from top to bottom for both sample groups. Control and case samples were randomized across plates. RNA-seq from samples were randomized for sequencing.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41593-022-01161-y.
Supplementary information
Supplementary Table 1. Comparison of clinical characteristics of veterans with and without PTSD. Supplementary Table 2. a, Demographic information and PTSD severity of individuals included within this dataset. b, Data dictionary of terms used in demographic feature table. Supplementary Table 3. Curated disorder-associated gene sets. Supplementary Table 4. Results from meta-analysis of differential expression by batch for PBMCs and NGN2-neurons treated with HCort.-based meta-analysis was conducted using METAL with a Cochran’s Q test for heterogeneity. a, Meta-analysis results for PBMCs treated with 2.5 nM DEX. b, Meta-analysis results for PBMCs treated with 5 nM DEX. c, Meta-analysis results for PBMCs treated with 50 nM DEX. d, Meta-analysis results for NGN2-neurons treated with 100 nM HCort. e, Meta-analysis results for NGN2-neurons treated with 1,000 nM HCort. Supplementary Table 5. a,b, GO enrichment results of coregulated expression modules that vary significantly in PBMCs treated with DEX (a), and NGN2-neurons treated with HCort (b). Supplementary Table 6. a,b, Results from meta-analysis of PTSD-effect differential expression by batch for NGN2-neurons treated with 100 nM (a) and 1,000 nM of HCort (b). Sample size-based meta-analysis was conducted using METAL with a Cochran’s Q-test for heterogeneity.
Acknowledgements
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the USAMRMC BAA for Extramural Medical Research under Award No. W81XWH-15-1-0706. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. This material is the result of work supported with resources and the use of facilities at the James J. Peters VAMC in Bronx, New York. The contents of this manuscript do not represent the views of the United States Department of Veterans Affairs or the United States Government. Further, this work was supported by the New York Stem Cell Foundation Research Institute (NYSCF). Funding for this work was received from DOD W81XWH-15-1-0706 (R.Y.), R01ES033630 (K.J.B, L.H.), R01MH124839 (L.H.) and R01MH118278 (L.H.). We dedicate this work to the memory of our beloved founding CEO Susan L. Solomon, in honor of her pioneering impact on stem cell research and technologies and her unwavering commitment to accelerating better treatments and cures for patients.
Extended data
Author contributions
R.Y. and K.J.B conceived of the study. Clinical characterization, patient recruitment, and skin biopsies were overseen by H.N.B., F.D., I.M., J.D.F., L.M.B., M.S. and R.Y. Automated hiPSC reprogramming, neuronal induction and hCort treatment was completed T.R., C.J.H., B.M., J.G., D.P., S.A.N. and the NYSCF Global Stem Cell Array® Team; manual neuronal differentiation and treatment by M.C. Bioinformatic analysis was conducted by M.S.B., C.S., C.X. and L.M.H. Phenotypic analysis was conducted by P.J.M.D. The manuscript was written by K.J.B., R.Y., M.S.B., C.S., H.N.B.,T.R. and D.P., although all authors contributed to data interpretation and revision of the manuscript. All authors approved the final manuscript.
Peer review
Peer review information
Nature Neuroscience thanks Onno Meijer, Eric Vermetten, Jun Yao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Owing to constraints reflecting the original patient consents, the hiPSCs will be made available by the authors upon reasonable request and IRB approval. RNA-seq files will be uploaded to the gene expression omnibus and will release following manuscript publication.
Code availability
All computational code is available at GitHub: https://github.com/BreenMS.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Carina Seah, Michael S. Breen, Tom Rusielewicz, Heather N. Bader.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Daniel Paull, Email: dpaull@nyscf.org.
Kristen J. Brennand, Email: kristen.brennand@yale.edu
Rachel Yehuda, Email: Rachel.Yehuda@va.gov.
NYSCF Global Stem Cell Array® Team:
Lauren Bauer, Katie Brenner, Geoff Buckley-Herd, Sean DesMarteau, Patrick Fenton, Peter Ferrarotto, Jenna Hall, Selwyn Jacob, Travis Kroeker, Gregory Lallos, Hector Martinez, Paul McCoy, Frederick J. Monsma, Dorota Moroziewicz, Reid Otto, Kathryn Reggio, Bruce Sun, Rebecca Tibbets, Dong Woo Shin, Hongyan Zhou, and Matthew Zimmer
Extended data
are available for this paper at 10.1038/s41593-022-01161-y.
Supplementary information
The online version contains supplementary material available at 10.1038/s41593-022-01161-y.
References
- 1.Yehuda R, et al. Post-traumatic stress disorder. Nat. Rev. Dis. Primers. 2015;1:15057. doi: 10.1038/nrdp.2015.57. [DOI] [PubMed] [Google Scholar]
- 2.Kremen WS, Koenen KC, Afari N, Lyons MJ. Twin studies of posttraumatic stress disorder: differentiating vulnerability factors from sequelae. Neuropharmacology. 2012;62:647–653. doi: 10.1016/j.neuropharm.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nievergelt CM, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat. Commun. 2019;10:4558. doi: 10.1038/s41467-019-12576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan LE, et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol. Psychiatry. 2018;23:666–673. doi: 10.1038/mp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gelernter J, et al. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat. Neurosci. 2019;22:1394–1401. doi: 10.1038/s41593-019-0447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yehuda R, Golier JA, Yang RK, Tischler L. Enhanced sensitivity to glucocorticoids in peripheral mononuclear leukocytes in posttraumatic stress disorder. Biol. Psychiatry. 2004;55:1110–1116. doi: 10.1016/j.biopsych.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Yehuda R, et al. The cortisol and glucocorticoid receptor response to low dose dexamethasone administration in aging combat veterans and holocaust survivors with and without posttraumatic stress disorder. Biol. Psychiatry. 2002;52:393–403. doi: 10.1016/s0006-3223(02)01357-4. [DOI] [PubMed] [Google Scholar]
- 8.Somvanshi PR, et al. Role of enhanced glucocorticoid receptor sensitivity in inflammation in PTSD: insights from computational model for circadian-neuroendocrine-immune interactions. Am. J. Physiol. Endocrinol. Metab. 2020;319:E48–E66. doi: 10.1152/ajpendo.00398.2019. [DOI] [PubMed] [Google Scholar]
- 9.Breen MS, et al. Gene networks specific for innate immunity define post-traumatic stress disorder. Mol. Psychiatry. 2015;20:1538–1545. doi: 10.1038/mp.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daskalakis NP, Cohen H, Cai G, Buxbaum JD, Yehuda R. Expression profiling associates blood and brain glucocorticoid receptor signaling with trauma-related individual differences in both sexes. Proc. Natl Acad. Sci. USA. 2014;111:13529–13534. doi: 10.1073/pnas.1401660111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yehuda R, et al. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol. Psychiatry. 2015;77:356–364. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Cathomas F, Murrough JW, Nestler EJ, Han MH, Russo SJ. Neurobiology of resilience: interface between mind and body. Biol. Psychiatry. 2019;86:410–420. doi: 10.1016/j.biopsych.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorsch ZS, et al. Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat. Neurosci. 2019;22:1413–1423. doi: 10.1038/s41593-019-0462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popoli M, Yan Z, McEwen BS, Sanacora G. The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat. Rev. Neurosci. 2011;13:22–37. doi: 10.1038/nrn3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehrner A, et al. A randomized, double-blind, placebo-controlled trial of hydrocortisone augmentation of prolonged exposure for PTSD in US combat veterans. Behav. Res. Ther. 2021;144:103924. doi: 10.1016/j.brat.2021.103924. [DOI] [PubMed] [Google Scholar]
- 16.Golier JA, et al. A pilot study of mifepristone in combat-related PTSD. Depress. Res. Treat. 2012;2012:393251. doi: 10.1155/2012/393251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Averill LA, et al. Glutamate dysregulation and glutamatergic therapeutics for PTSD: evidence from human studies. Neurosci. Lett. 2017;649:147–155. doi: 10.1016/j.neulet.2016.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernando MB, Ahfeldt T, Brennand KJ. Modeling the complex genetic architectures of brain disease. Nat. Genet. 2020;52:363–369. doi: 10.1038/s41588-020-0596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieberman R, Kranzler HR, Levine ES, Covault J. Examining FKBP5 mRNA expression in human iPSC-derived neural cells. Psychiatry Res. 2017;247:172–181. doi: 10.1016/j.psychres.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruceanu C, et al. Cell-type-specific impact of glucocorticoid receptor activation on the developing brain: a cerebral organoid study. Am. J. Psychiatry. 2022;179:375–387. doi: 10.1176/appi.ajp.2021.21010095. [DOI] [PubMed] [Google Scholar]
- 21.Aden P, et al. Glucocorticoids dexamethasone and hydrocortisone inhibit proliferation and accelerate maturation of chicken cerebellar granule neurons. Brain Res. 2011;1418:32–41. doi: 10.1016/j.brainres.2011.08.053. [DOI] [PubMed] [Google Scholar]
- 22.Levone B,R, et al. Adult-born neurons from the dorsal, intermediate, and ventral regions of the longitudinal axis of the hippocampus exhibit differential sensitivity to glucocorticoids. Mol. Psychiatry. 2021;26:3240–3252. doi: 10.1038/s41380-020-0848-8. [DOI] [PubMed] [Google Scholar]
- 23.Breen MS, et al. Differential transcriptional response following glucocorticoid activation in cultured blood immune cells: a novel approach to PTSD biomarker development. Transl. Psychiatry. 2019;9:201. doi: 10.1038/s41398-019-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crabtree GR, Munck A, Smith KA. Glucocorticoids inhibit expression of Fc receptors on the human granulocytic cell line HL-60. Nature. 1979;279:338–339. doi: 10.1038/279338a0. [DOI] [PubMed] [Google Scholar]
- 25.Odaka H, Adachi N, Numakawa T. Impact of glucocorticoid on neurogenesis. Neural Regen. Res. 2017;12:1028–1035. doi: 10.4103/1673-5374.211174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cain DW, Cidlowski JA. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 2017;17:233–247. doi: 10.1038/nri.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girgenti MJ, et al. Transcriptomic organization of the human brain in post-traumatic stress disorder. Nat. Neurosci. 2021;24:24–33. doi: 10.1038/s41593-020-00748-7. [DOI] [PubMed] [Google Scholar]
- 28.Schrode N, et al. Synergistic effects of common schizophrenia risk variants. Nat. Genet. 2019;51:1475–1485. doi: 10.1038/s41588-019-0497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, et al. Transformative network modeling of multi-omics data reveals detailed circuits, key regulators, and potential therapeutics for Alzheimer’s disease. Neuron. 2021;109:257–272 e214. doi: 10.1016/j.neuron.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flaherty E, et al. Neuronal impact of patient-specific aberrant NRXN1alpha splicing. Nat. Genet. 2019;51:1679–1690. doi: 10.1038/s41588-019-0539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho SM, et al. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods. 2016;101:113–124. doi: 10.1016/j.ymeth.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pak C, et al. Cross-platform validation of neurotransmitter release impairments in schizophrenia patient-derived NRXN1-mutant neurons. Proc. Natl Acad. Sci. USA. 2021;118:e2025598118. doi: 10.1073/pnas.2025598118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marro SG, et al. Neuroligin-4 regulates excitatory synaptic transmission in human neurons. Neuron. 2019;103:617–626 e616. doi: 10.1016/j.neuron.2019.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. The fragile X mutation impairs homeostatic plasticity in human neurons by blocking synaptic retinoic acid signaling. Sci. Transl. Med. 2018;10:eaar4338. doi: 10.1126/scitranslmed.aar4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi F, et al. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science. 2016;352:aaf2669. doi: 10.1126/science.aaf2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meijer M, et al. A single-cell model for synaptic transmission and plasticity in Human iPSC-derived neurons. Cell Rep. 2019;27:2199–2211 e2196. doi: 10.1016/j.celrep.2019.04.058. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S, et al. Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science. 2020;369:561–565. doi: 10.1126/science.aay3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, et al. A deleterious Nav1.1 mutation selectively impairs telencephalic inhibitory neurons derived from Dravet syndrome patients. eLife. 2016;5:e13073. doi: 10.7554/eLife.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman GE, et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun. 2017;8:2225. doi: 10.1038/s41467-017-02330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoue M, Kuriyama H. Glucocorticoids inhibit acetylcholine-induced current in chromaffin cells. Am. J. Physiol. 1989;257:C906–C912. doi: 10.1152/ajpcell.1989.257.5.C906. [DOI] [PubMed] [Google Scholar]
- 42.Schoepe S, Schacke H, May E, Asadullah K. Glucocorticoid therapy-induced skin atrophy. Exp. Dermatol. 2006;15:406–420. doi: 10.1111/j.0906-6705.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez-Resendis O, et al. Glucocorticoid-cholinergic interactions in the dorsal striatum in memory consolidation of inhibitory avoidance training. Front Behav. Neurosci. 2012;6:33. doi: 10.3389/fnbeh.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang YY, Dong H, Yang BB, Macdonald JF, Lu WY. Interaction of acetylcholinesterase with neurexin-1beta regulates glutamatergic synaptic stability in hippocampal neurons. Mol. Brain. 2014;7:15. doi: 10.1186/1756-6606-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthews JG, Ito K, Barnes PJ, Adcock IM. Defective glucocorticoid receptor nuclear translocation and altered histone acetylation patterns in glucocorticoid-resistant patients. J. Allergy Clin. Immunol. 2004;113:1100–1108. doi: 10.1016/j.jaci.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Liston C, Gan WB. Glucocorticoids are critical regulators of dendritic spine development and plasticity in vivo. Proc. Natl Acad. Sci. USA. 2011;108:16074–16079. doi: 10.1073/pnas.1110444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breen MS, et al. PTSD blood transcriptome mega-analysis: Shared inflammatory pathways across biological sex and modes of trauma. Neuropsychopharmacology. 2018;43:469–481. doi: 10.1038/npp.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosso IM, et al. Insula and anterior cingulate GABA levels in posttraumatic stress disorder: preliminary findings using magnetic resonance spectroscopy. Depress. Anxiety. 2014;31:115–123. doi: 10.1002/da.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moller AT, Backstrom T, Nyberg S, Sondergaard HP, Helstrom L. Women with PTSD have a changed sensitivity to GABA-A receptor active substances. Psychopharmacology. 2016;233:2025–2033. doi: 10.1007/s00213-014-3776-y. [DOI] [PubMed] [Google Scholar]
- 50.Geuze E, et al. Reduced GABAA benzodiazepine receptor binding in veterans with post-traumatic stress disorder. Mol. Psychiatry. 2008;13:74–83, 73. doi: 10.1038/sj.mp.4002054. [DOI] [PubMed] [Google Scholar]
- 51.Jaffe AE, et al. Decoding shared versus divergent transcriptomic signatures across cortico-amygdala circuitry in PTSD and depressive disorders. Am. J. Psychiatry. 2022;179:673–686. doi: 10.1176/appi.ajp.21020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huckins, L. M. et al. Polygenic regulation of PTSD severity and outcomes among World Trade Center responders. Preprint at medRxiv10.1101/2020.12.06.20244772 (2020).
- 53.Brennand K, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mariani J, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl Acad. Sci. USA. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qian X, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicholas CR, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell. Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Powell, S. K. et al. Induction of dopaminergic neurons for neuronal subtype-specific modeling of psychiatric disease risk. Mol. Psychiatry, 10.1038/s41380-021-01273-0 (2021). [DOI] [PMC free article] [PubMed]
- 59.Ninomiya E, et al. Glucocorticoids promote neural progenitor cell proliferation derived from human induced pluripotent stem cells. Springerplus. 2014;3:527. doi: 10.1186/2193-1801-3-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raciti M, et al. Glucocorticoids alter neuronal differentiation of human neuroepithelial-like cells by inducing long-lasting changes in the reactive oxygen species balance. Neuropharmacology. 2016;107:422–431. doi: 10.1016/j.neuropharm.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 61.Provencal N, et al. Glucocorticoid exposure during hippocampal neurogenesis primes future stress response by inducing changes in DNA methylation. Proc. Natl Acad. Sci. USA. 2020;117:23280–23285. doi: 10.1073/pnas.1820842116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buss C, et al. Intergenerational transmission of maternal childhood maltreatment exposure: implications for fetal brain development. J. Am. Acad. Child Adolesc. Psychiatry. 2017;56:373–382. doi: 10.1016/j.jaac.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carson R, Monaghan-Nichols AP, DeFranco DB, Rudine AC. Effects of antenatal glucocorticoids on the developing brain. Steroids. 2016;114:25–32. doi: 10.1016/j.steroids.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daskalakis NP, Rijal CM, King C, Huckins LM, Ressler KJ. Recent genetics and epigenetics approaches to PTSD. Curr. Psychiatry Rep. 2018;20:30. doi: 10.1007/s11920-018-0898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yehuda R, et al. Putative biological mechanisms for the association between early life adversity and the subsequent development of PTSD. Psychopharmacology (Berl.) 2010;212:405–17. doi: 10.1007/s00213-010-1969-6. [DOI] [PubMed] [Google Scholar]
- 66.Stevens JS, Jovanovic T. Role of social cognition in post-traumatic stress disorder: a review and meta-analysis. Genes Brain Behav. 2019;18:e12518. doi: 10.1111/gbb.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Velikonja T, Fett AK, Velthorst E. Patterns of nonsocial and social cognitive functioning in adults with autism spectrum disorder: a systematic review and meta-analysis. JAMA Psychiatry. 2019;76:135–151. doi: 10.1001/jamapsychiatry.2018.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Kloet CS, et al. Enhanced cortisol suppression in response to dexamethasone administration in traumatized veterans with and without posttraumatic stress disorder. Psychoneuroendocrinology. 2007;32:215–226. doi: 10.1016/j.psyneuen.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 69.OMIM. Online Mendelian Inheritance in Man: An Online Catalog of Human Genes and Genetic Disorders, https://omim.org/ (2021).
- 70.Firth HV, et al. DECIPHER: Database of chromosomal imbalance and phenotype in Humans using ensembl resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buxbaum JD, et al. The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron. 2012;76:1052–1056. doi: 10.1016/j.neuron.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 74.Abrahams BS, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol. Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tylee DS, et al. Blood transcriptomic comparison of individuals with and without autism spectrum disorder: a combined-samples mega-analysis. Am. J. Med Genet B Neuropsychiatr. Genet. 2017;174:181–201. doi: 10.1002/ajmg.b.32511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, C. et al. AutismKB 2.0: a knowledgebase for the genetic evidence of autism spectrum disorder. Database (Oxford)2018, 10.1093/database/bay106 (2018). [DOI] [PMC free article] [PubMed]
- 77.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hess JL, et al. Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia. Schizophr. Res. 2016;176:114–124. doi: 10.1016/j.schres.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cocchi E, Drago A, Serretti A. Hippocampal pruning as a new theory of schizophrenia etiopathogenesis. Mol. Neurobiol. 2016;53:2065–2081. doi: 10.1007/s12035-015-9174-6. [DOI] [PubMed] [Google Scholar]
- 81.Clifton NE, et al. Genetic association of FMRP targets with psychiatric disorders. Mol. Psychiatry. 2021;26:2977–2990. doi: 10.1038/s41380-020-00912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4th ed (DSM-IV) (American Psychiatric Press, 1994).
- 83.First, M. B., Spitzer, R. L., Gibbon, M. & Williams, J. B. Structured Clinical Interview for DSM-5—Research Version (SCID-5 for DSM-5, Research Version; SCID-5-RV, v.1.0.0). (American Psychiatric Publishing, 2015).
- 84.Blake DD, et al. The development of a clinician-administered PTSD scale. J. Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- 85.Kahler DJ, et al. Improved methods for reprogramming human dermal fibroblasts using fluorescence activated cell sorting. PLoS ONE. 2013;8:e59867. doi: 10.1371/journal.pone.0059867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paull D, et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat. Methods. 2015;12:885–892. doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- 87.Breen, M. S. et al. Modeling gene x environment interactions in PTSD using glucocorticoid-induced transcriptomics in human neurons. Preprint at bioRxiv10.1101/2021.03.01.433391 (2021).
- 88.Piechota M, et al. Transcriptional signatures of steroid hormones in the striatal neurons and astrocytes. BMC Neurosci. 2017;18:37. doi: 10.1186/s12868-017-0352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salvador E, Shityakov S, Forster C. Glucocorticoids and endothelial cell barrier function. Cell Tissue Res. 2014;355:597–605. doi: 10.1007/s00441-013-1762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schaffter N, et al. Serum cortisol as a predictor for posttraumatic stress disorder symptoms in post-myocardial infarction patients. J. Affect. Disord. 2021;292:687–694. doi: 10.1016/j.jad.2021.05.065. [DOI] [PubMed] [Google Scholar]
- 91.Reddy TE, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 95.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoffman GE, Schadt EE. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinf. 2016;17:483. doi: 10.1186/s12859-016-1323-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 99.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Piñero J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wittenberg GM, et al. Major depressive disorder is associated with differential expression of innate immune and neutrophil-related gene networks in peripheral blood: a quantitative review of whole-genome transcriptional data from case-control studies. Biol. Psychiatry. 2020;88:625–637. doi: 10.1016/j.biopsych.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 104.Satterstrom FK, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Rubeis S, et al. Synaptic transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pantazatos SP, et al. Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: evidence for altered glial endothelial and ATPase activity. Mol. Psychiatry. 2017;22:760–773. doi: 10.1038/mp.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boyle EI, et al. GO::TermFinder–open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang, J. & Liao, Y. WebGestaltR: Gene Set Analysis Toolkit WebGestaltR. R package version 0.4.3. https://CRAN.R-project.org/package=WebGestaltR (2020).
- 111.Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap (2019).
- 112.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marchese, S. et al. Altered gene expression and PTSD symptom dimensions in World Trade Center responders. Preprint at medRxiv10.1101/2021.03.05.21252989 (2021). [DOI] [PubMed]
- 116.Mertens J, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mariani J, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Comparison of clinical characteristics of veterans with and without PTSD. Supplementary Table 2. a, Demographic information and PTSD severity of individuals included within this dataset. b, Data dictionary of terms used in demographic feature table. Supplementary Table 3. Curated disorder-associated gene sets. Supplementary Table 4. Results from meta-analysis of differential expression by batch for PBMCs and NGN2-neurons treated with HCort.-based meta-analysis was conducted using METAL with a Cochran’s Q test for heterogeneity. a, Meta-analysis results for PBMCs treated with 2.5 nM DEX. b, Meta-analysis results for PBMCs treated with 5 nM DEX. c, Meta-analysis results for PBMCs treated with 50 nM DEX. d, Meta-analysis results for NGN2-neurons treated with 100 nM HCort. e, Meta-analysis results for NGN2-neurons treated with 1,000 nM HCort. Supplementary Table 5. a,b, GO enrichment results of coregulated expression modules that vary significantly in PBMCs treated with DEX (a), and NGN2-neurons treated with HCort (b). Supplementary Table 6. a,b, Results from meta-analysis of PTSD-effect differential expression by batch for NGN2-neurons treated with 100 nM (a) and 1,000 nM of HCort (b). Sample size-based meta-analysis was conducted using METAL with a Cochran’s Q-test for heterogeneity.
Data Availability Statement
Owing to constraints reflecting the original patient consents, the hiPSCs will be made available by the authors upon reasonable request and IRB approval. RNA-seq files will be uploaded to the gene expression omnibus and will release following manuscript publication.
All computational code is available at GitHub: https://github.com/BreenMS.