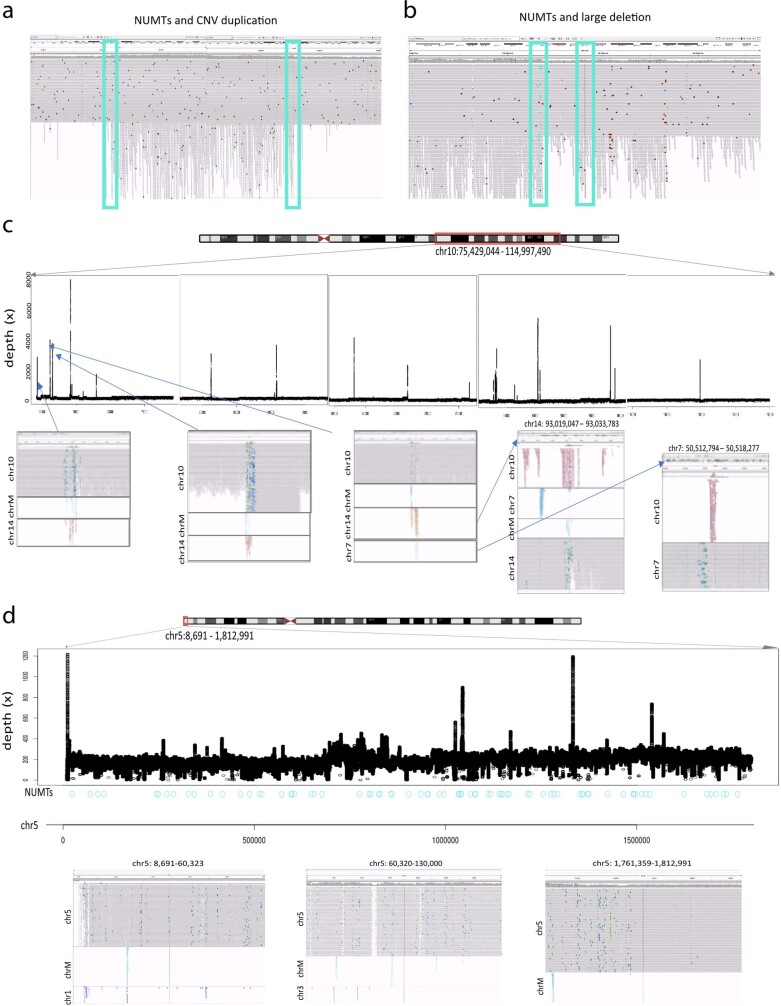
Extended Data Fig. 9. IGV alignments of NUMTs and nuclear chromosomal structure variations.
a. An example of mtDNA fragment inserted into two edges of a CNV duplication. b. An example of mtDNA fragment inserted into two edges of a large deletion. Teal bars indicate the aligned reads which mapped to the nuclear DNA where their mates mapped to the mtDNA, and were highlighted in the teal. c. d. Two examples of cancer genomes carrying mito-chromothripsis observed in this study. c. The sequencing depth of nuclear genome is shown at the top panel. Examples of the read alignment of NUMTs from IGV are shown at the bottom. Reads are coloured by the pair orientation and the chromosome on which their mates can be found. d. The sequencing depth of nuclear genome is shown at the top panel. Teal dots are the locations of NUMT insertions. Examples of the read alignment of NUMTs from IGV are shown at the bottom. Reads are coloured by the pair orientation and the chromosome on which their mates can be found.