Abstract
We propose a label-free biosensor based on a porous silicon resonant microcavity and localized surface plasmon resonance. The biosensor detects SARS-CoV-2 antigen based on engineered trimeric angiotensin converting enzyme-2 binding protein, which is conserved across different variants. Robotic arms run the detection process including sample loading, incubation, sensor surface rinsing, and optical measurements using a portable spectrometer. Both the biosensor and the optical measurement system are readily scalable to accommodate testing a wide range of sample numbers. The limit of detection is 100 TCID50/ml. The detection time is 5 min, and the throughput of one single robotic site is up to 384 specimens in 30 min. The measurement interface requires little training, has standard operation, and therefore is suitable for widespread use in rapid and onsite COVID-19 screening or surveillance.
Keywords: Biosensor, Localized surface plasmon resonance (LSPR), SARS-CoV-2, COVID-19, Automatic detection, High throughput
1. Introduction
SARS-CoV-2 detection is critical (Larremore et al., 2021), and there are several techniques available to detect virus in human samples in analytical labs (Table 1 ) (Jayamohan et al., 2021). The nucleic acid amplification test (NAAT) is a mainstream detection technique (Jayamohan et al., 2021) based on the specific amplification of nucleic acid sequences of target analytes such as the ribonucleic acid (RNA) sequence of SARS-CoV-2. NAAT includes polymerase chain reaction (PCR) and isothermal techniques. PCR is currently the most popular technique and is a gold standard due to its high sensitivity and specificity (Zhu et al., 2020). PCR requires thermal cycling in three temperature zones of 95°C, 55°C, and 72°C. PCR typically takes at least 4 h to obtain analytical results due to this thermal cycling process as well as the requirement for sample preparation such as nucleic acid extraction and purification; this can be 24–48 h with logistics included. All NAAT techniques require a clean laboratory environment and trained personnel, thus hindering application for onsite rapid detection of SARS-CoV-2. Isothermal techniques, such as recombinase polymerase amplification (RPA) (Behrmann et al., 2020) and loop-mediated isothermal amplification (LAMP) (Haq et al., 2021) can amplify specific gene targets without thermal cycling, and thus are much faster than PCR (ranging from 20 min to 1 h).
Table 1.
Current SARS-CoV-2 detection techniques in practice (FDA approved) and research, their estimated usage percentage, advantages, and disadvantages. Biosensors include electrochemical (Eissa and Zourob, 2021), electrical (Seo et al., 2020), and optical (Funari et al., 2020a) transduction mechanisms. Examples of “Others” are coughing sound analysis by AI (Laguarta et al., 2020) and waste water analysis to detect population infection before clinical diagnosis (Evangelista and Bryner, 2020).
| Detection techniques | Usage percent | Limit of Detection | Detection time | Advantage | Disadvantage | |
|---|---|---|---|---|---|---|
| NAAT (Jayamohan et al., 2021) | PCR (Zhu et al., 2020) | 40% | N/A | N/A | Sensitive, specific, accurate | Complex procedure, long turn-around time, clean environment and training required |
| Isothermal (Behrmann et al., 2020; Haq et al., 2021) | 6% | 7.74 RNA copies/reaction 95% detection probability | 15–20 min | Fast, simpler than PCR | Sensitivity worse than PCR, limited throughput, need for training | |
| Isothermal (Behrmann et al., 2020; Haq et al., 2021) | N/A | N/A | ||||
| CRISPR (Nouri et al., 2021) | 3% | N/A | N/A | Fast, on-site detection is possible | Complex sample preparation, limited target gene regions, multiplexing challenging | |
| Sequencing (Lu et al., 2020) | 5% | N/A | N/A | Sensitive and specific, can detect virus mutants | Complex procedure, high cost, training | |
| Antigen (Grant et al., 2020; Kyosei et al., 2020; Liu et al., 2021) | 15% | 0.65 ng/ml (95% Confidence Interval of 0.53–0.77 ng/ml) | N/A | Rapid, simple, onsite detection | Limited sensitivity and specificity | |
| Antigen (Grant et al., 2020; Kyosei et al., 2020; Liu et al., 2021) | Approaches PCR assays' sensitivity | ∼10 min | ||||
| Antigen (Grant et al., 2020; Kyosei et al., 2020; Liu et al., 2021) | 360 TCID50/mL | Within 16 min | ||||
| Antibody (Cavalera et al., 2021; Speletas et al., 2020; Tantuoyir and Rezaei, 2021) | 15% | N/A | N/A | Rapid, simple, onsite detection | Limited sensitivity and specificity | |
| Antibody (Cavalera et al., 2021; Speletas et al., 2020; Tantuoyir and Rezaei, 2021) | N/A | N/A | ||||
| Antibody (Cavalera et al., 2021; Speletas et al., 2020; Tantuoyir and Rezaei, 2021) | N/A | N/A | ||||
| CT scan (Lee et al., 2020) | 10% | N/A | N/A | Evaluates impact on organs | Complex procedure, high cost, non-specific, low throughput, training required | |
| Biosensora (Eissa and Zourob 2021; Funari et al., 2020a; Seo et al., 2020) | 3% | 0.8 pg/ml | N/A | Rapid, simple detection | Throughput and onsite operation needs improvement | |
| Biosensora (Eissa and Zourob 2021; Funari et al., 2020a; Seo et al., 2020) | ∼0.08 ng/ml | ∼30min | ||||
| Biosensora (Eissa and Zourob 2021; Funari et al., 2020a; Seo et al., 2020) | Culture medium: 16 pfu/ml; Clinical samples: 242 copies/ml | N/A | ||||
| Other techniquesa | 3% | N/A | N/A | |||
| (Evangelista and Bryner 2020; Laguarta et al., 2020) | N/A | N/A | ||||
FDA: Food and Drug Administration; NAAT: nucleic acid amplification test; PCR: polymerase chain reaction; CRISPR: clustered regularly interspaced short palindromic repeats; CT: computed tomography; N/A: not available.
For research use only.
Antibody detection is faster, simpler, and identifies human immune components specific to viral infection (Tantuoyir and Rezaei, 2021) including in lateral flow assay (LFA) formats (Cavalera et al., 2021). Unfortunately, antibodies lack the specificity of NAAT techniques. Viral antigen detection is also possible with LFA (Grant et al., 2020), ELISA (Kyosei et al., 2020), and chemiluminescence (Liu et al., 2021) techniques, but again suffer sensitivity and detection limit challenges. Indeed, there is still a lack of scalable, high-throughput, accurate, and fully automatic onsite rapid detection techniques for SARS-CoV-2 (Larremore et al., 2021; Mina and Andersen, 2021).
Biosensors are promising for onsite rapid detection of SARS-CoV-2 due to their high sensitivity, ease-of-operation, fast turnaround, and integration with high-throughput applications. Examples include electrochemical (Eissa and Zourob, 2021), electrical (Seo et al., 2020), and optical biosensors (Funari et al., 2020a) for SARS-CoV-2 detection. Of these, optical biosensors provide high sensitivity. In a recent study, biolayer interferometry immunosorbent assay measuring optical signal excited by SARS-CoV-2 antibody-antigen binding reaction has been reported (Dzimianski et al., 2020). This biosensor can be applied to existing BLI platforms for quantitive detection of SARS-CoV-2 antibody.
As a nanostructured material, porous silicon is desirable for high performance optical biosensing due to its high surface area, ease of fabrication and tunable optical properties. It has been used for the sensitive detection of deoxyribonucleic acid (DNA) (Rong and Weiss, 2009), antibody/antigens (Wu et al., 2012a), and viruses (Rossi et al., 2007). However, these biosensors typically require analytical targets to diffuse through the nanopores of porous silicon materials to spatial region of confined field for high sensitivity detecton, leading to longer incubation times and careful selection of pore sizes (Rong and Weiss, 2009). These biosensors typically have detection limits in the range of nM to μM.
Surface Plasmon Polariton (SPP) is a collective oscillation of free electrons in noble metals. It can be excited by photons in visbile spectrum. Many different kinds of plasmonic devices have been designed and studied in detail (Wang et al., 2021), for various applications including biosensing. Surface plasmon resonance (SPR) sensors uses electrical field confined at the interface between metal and dielectric to interact with biomolecules for biosensing applications. SPR sensors solve many of these limitations mentioned before. Here, Au, Ag, and Pt can be used to construct SPP-based biosensors such as SPR (Li et al., 2015) and localized surface plasmon resonance (LSPR) biosensors (Csáki et al., 2018). These SPP-based biosensors have high sensitivity towards molecular binding events near the metal surface because the surface has strong field confinement resulting in strong field-biomolecule interactions (Qu et al., 2020). In contrast to SPR with low nM detection limits, LSPR-based biosensors can be as low as the fM range (Kaye et al., 2017). Furthermore, SPP-based biosensors have biomolecular binding events on the metal surface, thus eliminating the need for diffusion into nanopores. Versus SPR biosensors, LSPR biosensors also have a field confinement much closer to the metal-dielectric interface; thus, they are more suitable for biomolecular detection (Mataji-Kojouri et al., 2020). LSPR has been applied to diagnosing COVID-19 by Funari(Funari et al., 2020b), Behrouzi(Behrouzi and Lin, 2022), and Qiu (Qiu et al., 2020). These LSPR biosensors are mainly based on reflection or transmission interferometry of dielectric structures coated with noble metal thin film, or reflection, transmission and absorption spectra of noble metal nanoparticles.
Here, we describe a new biosensing platform that exploits the advantages of SPP and combines a porous silicon resonant microcavity with the LSPR phenomenon to construct a highly sensitive biosensor for rapid onsite detection. Porous silicon was used to construct a resonant microcavity, and gold thin film was deposited on top of the microcavity to form mode coupling between Tamm Plasmon Polariton (TPP) mode and microcavity resonant mode. The porous silicon resonant microcavity can be constructed relatively easily without complex thin film deposition processes, and its resonant mode has strong field confinement in the central defect layer (Wu et al., 2012a). TPP mode has field confinement at the interface between metal and top Bragg mirror of the microcavity (Kaliteevski et al., 2007). By coupling the resonant cavity mode with TPP mode, the field confined in defect layer of a microcavity can leak out and reinforce TPP field so that the total field in the vicinity of the metal thin film can be enhanced. Moreover, this strong field excites LSPR resonances around the discontinuous gold thin film with nanostructures, which can further enhance the interaction between biomolecules and electrical field. Sensitive detection of biomolecules can be obtained without requiring biomolecules to diffuse through nanopores into the defect layer of microcavity. We propose a SARS-CoV-2 antigen detection system consisting of optical biosensors based on porous silicon, portable fiber spectrometers, and robotic arms performing automatic operation to achieve all of these aims. Robots or autonomous systems have been designed to realize automatic SARS-CoV-2 detection using RT-PCR detection technique. An open-source robot platform has been developed to detect SARS-CoV-2 of 96 samples in about 4 h (Villanueva-Canas et al., 2021). The system involves 6 main steps, and 6 different equipment are needed to complete each step. Another automatic system having better compactness (one single floor-type equipment) and higher throughput has been proposed that can carry out SARS-CoV-2 detection of 192 samples under 180 min at detection limit of 100 copies per reaction (Lu et al., 2022). In this work, we propose a desktop type automatic detection system that can detect SARS-CoV-2 of 384 samples in around 30 min with detection limit of 100 TCID50/ml. The detection system can be extended to include antibody and nuclei acid detection and can thus be considered an all-in-one detection tool.
2. Materials and methods
The proposed biosensor is intended for high-precision measurements by reflection spectroscopy through a portable spectrometer. Using this technique, viral particles do not need to diffuse into the nanopores as is the case for most porous silicon-based biosensors. This proposed method allows one to shorten the detection time. For this new LSPR resonant microcavity biosensor, a salient resonant feature is available for better resolution of small shifts. Thus, the new design can substantially improve the sensitivity and the detection limit versus previously reported LSPR biosensors. For high throughput screening, we fabricated a sensor array in a standard 96-well format on a porous silicon wafer via a fast and low-cost approach. The sensor array is readily scalable to satisfy screening needs (frequency and population size) by simply exploiting silicon wafers of larger diameters in fabricating the porous silicon. The porous silicon wafer can also be cut into different sizes of sensor arrays via a commercially available die saw.
The optical measurement system of the sensor array uses a portable fiber spectrometer to measure the reflection spectrum. A robotic arm automatically aligns each sensor spot with a fiber probe of the spectrometer to save the optical reflection data automatically. Another robotic arm automatically accomplishes the common tasks encountered in biosensing: loading of clinical specimens onto the sensor spot and rinsing the sensor surface after incubation. The automatic optical measurement system can achieve high throughput and scalability corresponding to the sensor array design. Multiple spectrometers can be stacked to achieve simultaneous multiple-spot measurement for a large well array—this will reduce turnaround time. The fully automatic design, from specimen loading to results output, can protect health professionals from biosafety hazards. The biosensor and measurement system proposed here may be a powerful tool to control the COVID-19 pandemic via efficient population screening (Larremore et al., 2021).
2.1. Biosensor fabrication
Previously, researchers have demonstrated optimization of the number of periods in the Bragg mirror for excitation of TPP (Kumari et al., 2018), and for coupling between TPP and resonant mode of a microcavity(Lu et al., 2019). Basically, the Au thin film has an effect on the quality factor of the TPP mode. The number of periods in the top Bragg mirror of the porous silicon microcavity has an effect on quality factor of the resonant microcavity mode, quality factor of the TPP mode, and the coupling between TPP mode and resonant microcavity mode. After considering these factors, we adopted a design of porous silicon resonant microcavity previously reported (Wu et al., 2012b), and used Au thin film with thickness of 30 nm.
Porous silicon was fabricated by electrochemical anodization of silicon wafer (6-inch diameter, boron doped P-type, <100> crystal orientation, 0.01 Ω cm resistivity) in 15% aqueous hydrofluoric (HF) acid (mixing 75 ml 50% aqueous HF with 175 ml 95% ethanol). The etching conditions have been reported previously (Wu et al., 2012b). There are two different porous silicon layers in the resonant microcavity design. The top Bragg mirror consists of six periods of alternating porous silicon layers 1 and 2. The bottom Bragg mirror has the same structure as the top Bragg mirror. A defect layer with the same optical parameter as the second layer is embedded between the two Bragg mirrors. This defect layer introduces a resonance peak in the total reflection band of the Bragg mirror resulting in a resonant microcavity (Wu et al., 2012b). The refractive indices of each porous silicon layer have been estimated from its porosity via Bruggeman effective medium theory (Grigoriev et al., 2020). The thickness of each porous silicon layer was estimated by multiplying etching time with etching speed as characterized by scanning electron microscopy (Rong et al., 2008a). The resulting porous silicon has a circular area with a diameter of 11 cm as shown in Fig. 1 a and b. After anodization, the porous silicon wafer is thermally oxidized in ambient air under 800°C for 30 min. This process converts the silicon hydrogen bond (Si-H) to a silicon oxygen bond (Si-O) and thus stabilizes the porous silicon in ambient air and makes the porous silicon hydrophilic (Rong et al., 2008b).
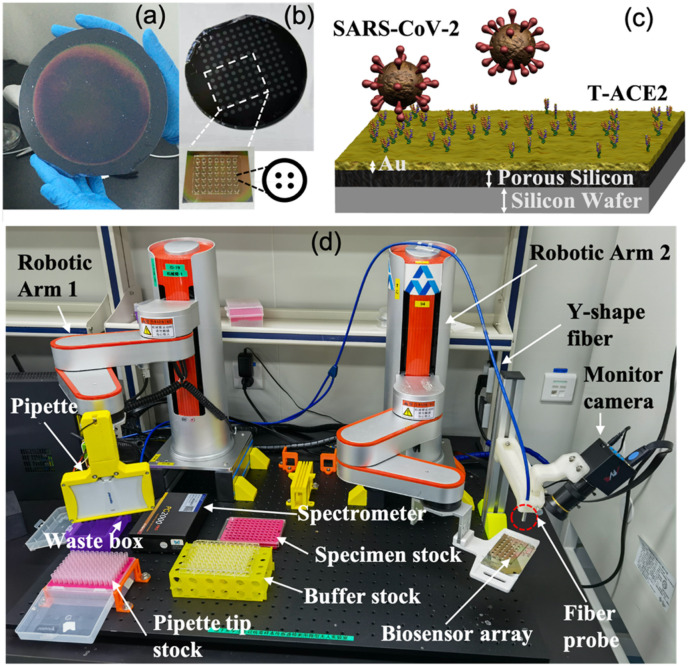
Fig. 1.
(a) Six-inch-diameter wafers; the fabricated porous silicon active area is 11 cm in diameter. (b) Polydimethylsiloxane (PDMS) well-based array on wafer in 96-well format from which a PDMS 6 × 7 well array is cut to form a biosensor chip. The zoom-in view of a single well shows that, within each well, a 2 × 2 spot array is defined through a robotic arm control program with each 0.5-mm diameter spot measured by reflection spectroscopy. (c) Binding of SARS-CoV-2 by engineered trimeric angiotensin converting enzyme-2 (T-ACE2) immobilized on biosensor surface. (d) The proposed sensing platform consists of a biosensor array chip, fiber spectrometer, Y-shaped fiber, 8- or 12-headed pipette, as well as two robotic arms for alignment/measurement and sample loading/rinsing. Note that the halogen white light source is not shown in this figure, and that the thicknesses of the different materials are not to scale.
The 5 nm Ti and 30 nm Au thin films were consecutively deposited (Kim et al., 2007) on the porous silicon wafer via physical vapor deposition (PVD, ZD-400 single chamber high vacuum resistive evaporator; Shenyang Kecheng Vacuum Tech Co. Ltd). The Au thin film is also porous because it conformally covers the porous silicon thin film. The Ti thin film serves as a buffer between the porous silicon and the Au thin film.
The biosensor must be biofunctionalized to specifically detect SARS-CoV-2. First, the gold-coated porous silicon wafer was immersed in a solution of 11-mercaptoundecanoic acid (20 mM) in ethanol (10 ml) and incubated for 24 h (Ahmad and Moore, 2012; Yeom et al., 2011). After the reaction, the surface of the chip was washed with ethanol and dried with air. The functionalized gold-coated porous silicon wafer was then immersed in 0.4 M 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) and a 0.1 M N-hydroxy succinimide (NHS) mixture dissolved in 2-(N-morpholino) ethane sulfonic acid (MES) buffer for 15 min to activate the carboxylic groups of the gold-coated porous silicon wafer. A polydimethylsiloxane (PDMS) well array was fabricated in advance via a mask to form a sensor array on the porous silicon wafer. The PDMS well array has a standard 96-well format with a well diameter of 6 mm and a well depth of 4 mm. The PDMS well array was prepared on the silicon wafer in advance. When ready to use, the sample was cut and peeled off the silicon wafer and placed on a functionalized porous silicon wafer. Sealing between PDMS and the functionalized porous silicon wafer with the activated carboxylic groups is waterproof; thus, there is no leakage between the wells.
To detect SARS-CoV-2, we used engineered trimeric angiotensin converting enzyme-2 (T-ACE2) protein as the probe (Fig. 1c). Engineered T-ACE2 (0.2 mg/ml) has a much higher affinity towards the S-protein of SARS-CoV-2 than monomeric ACE2 (Guo et al., 2021) and was immobilized on the surface for 30 min by covalent binding to the activated carboxylic groups (Torrente-Rodríguez et al., 2020; Zang et al., 2019). This was followed by immersion in blocking buffer (Superblock, Sigma) containing bovine serum albumin (BSA) for 5 min to deactivate the unreacted carboxylic groups. ACE2 also has enhanced affinity for several variants including D614G (Zhou et al., 2021).
2.2. Biosensor measurement setup
The reflection spectrum of the LSPR resonant microcavity biosensor was measured under wet conditions. Fig. 1d shows that the biosensor was measured with a portable fiber spectrometer (PG2000-Pro, Idea Optics, Shanghai, China). A Y-shaped optic fiber was used to guide the incident white light toward the biosensor and collect the reflected light from the biosensor. The Y-shaped fiber has one fiber in the center to collect the reflected light and six fibers around the central fiber to provide the incident light. All seven fibers in the fiber bundle are 100 μm in diameter. The distance from the fiber outer surface to the biosensor surface is 3 mm, and the beam spot on the surface of the biosensor has a diameter of 0.5 mm. The spot size is determined by the diameter of the fiber providing incident light and the distance between the fiber outer surface and the biosensor surface. A halogen white light source was used to provide incident light at a perpendicular angle of incidence. The reflected light was collected and guided to a portable spectrometer for real time spectra data display and saving. The wavelength range of the spectrometer is 200–1100 nm; only data in the 600–1000 nm range is used for analysis. The wavelength sampling rate is 0.48 nm.
2.3. Automatic and high throughput detection
A typical biosensing process consists of the following sequential operation steps:
Step 1, buffer loading; Step 2, biosensor measurement; Step 3, specimen loading; Step 4, biosensor rinsing; Step 5, buffer loading; and Step 6, biosensor measurement.
These six steps can be grouped into three main modules, and these modules can be combined in a mix-and-match way in the graphic programming interface of the robot to fit the needs of various applications. These modules can also be selectively activated or executed ad hoc by clicking the software interface. Two robotic arms (Z-Arm 1632, HITBOT, Shenzhen, China) were custom made and programmed to complete the needed tasks either separately or in synchronization to complete an integrated application. The following main modules were developed to smoothly run the various tasks of the platform via the two robotic arms programmed in the graphical interface. Fig. 1d shows the setup to implement the following three main modules.
2.3.1. Module 1: specimen/buffer loading
This module uses pipettes on Robotic Arm 1 to automatically load either phosphate buffer saline (PBS) buffer or clinical liquid specimens onto the biosensor chip to cover the 6 mm diameter wells. Liquid sample (4 μl) is needed to completely cover the surface. Accurate volume control (±10%) is achieved by controlling the pressure applied on the pipette. After specimen loading, Robotic Arm 2 holds the biosensor chip for 5 min (which is enough for binding of S-protein with T-ACE2); optionally, the robotic arm can keep shaking the biosensor chip for a better binding reaction. Note that the interaction between the virus and the electric field in the vicinity of the metal thin film is instantaneous, and there is no time effect on optical response of the biosensor. Buffer loading does not need incubation or shaking, and its purpose is to make the biosensor surface wet for optical spectral measurements. Simultaneous loading of multiple wells can be achieved with a multi-channel pipette.
2.3.2. Module 2: biosensor measurements
This module uses Robotic Arm 2 for alignment of each spot within each well with the fiber probe to take spectral measurements. The repositioning error of the robotic arm is within 20 μm—this is much smaller than the spot diameter of 0.5 mm. The current application has a 2 × 2 spot array in each well (Fig. 1b), which means four experiments for each specimen (the location and number of spots in each well can also be defined through graphical programming). LabVIEW is used to control the spectrometer and is synchronized with Robotic Arm 2 so that the whole process of aligning a spot and taking its measurement is complete within 1 s. The Robotic Arm 2 moves to the next spot only after the spectral data from the previous spot have been safely saved. Three measurements are typically needed, and the average of the three measurements are used to reduce the impact of alignment error. After all measurements have been completed, the liquid clinical specimen or buffer on the biosensor surface is removed by pipette on Robotic Arm 1 to prepare for the next operation.
2.3.3. Module 3: biosensor rinsing
This module uses a pipette on Robotic Arm 1 for rinsing the surface of the biosensor with PBS. This includes applying PBS buffer on the surface and removing the majority of the buffer from the surface with a pipette. Typically, 20 μl of PBS buffer is used to rinse the surface. The liquid residue has no influence on the result since after rinsing, the biosensor surface will be loaded with PBS buffer before reflection spectral measurement. And shaking the biosensor is optional. Simultaneous rinsing of multiple wells can be achieved by using a multi-channel pipette. Repeated rinsing of the wells can help remove non-specific interferents and can be selected through graphical programming.
2.4. S-ECD viral protein expression and purification
The extracellular domain (1-1208 amino acids) of S protein (S-ECD) of SARS-CoV-2 (Genebank ID: QHD43416.1) was cloned into the pCAG vector (Invitrogen) with a “GSAS” mutation at residues 682 to 685, two proline substitutions at residues 986 and 987, and a C-terminal T4 fibritin trimerization motif followed by one Flag tag and Strep tag. This construct will hereafter be referred to as S-ECD.
The purification process of S-ECD has been described previously (Chi et al., 2020). Briefly, the recombinant protein was overexpressed using HEK 293F mammalian cells (Invitrogen) at 37°C under 5% CO2 in a Multitron-Pro shaker (Infors, 130 rpm). The secreted proteins were purified with anti-FLAG M2 affinity resin (Sigma Aldrich). After loading two times, the anti-FLAG M2 resin was washed with the wash buffer containing 25 mM tris(hydroxymethyl)aminomethane (Tris, pH 8.0) and 150 mM NaCl. The protein was eluted with the wash buffer plus 0.2 mg/ml flag peptide. The eluent of the peptidase domain (PD) was then concentrated and subjected to size-exclusion chromatography (Superdex 200 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM Tris (pH 8.0) and 150 mM NaCl. The peak fractions were collected for further assays.
2.5. Inactivated SARS-CoV-2 virus solution preparation
Clinical specimens (throat swab) obtained from a SARS-CoV-2 infection case were inoculated into Vero cells at 37°C in a 5% CO2 incubator, and the cytopathic effect (CPE) was monitored daily. The positive isolate was further inoculated into Vero cells to generate viral stock. The viral stock was titrated and reached a titer of 106.3 TCID50/ml (tissue culture infectious dose). Beta-propiolactone was added to the suspension of virus stock at 0.05% and kept overnight at 4°C. The inactivated virus solution was incubated at 37°C for 4 h for hydrolyzation of beta-propiolactone. The treated virus was subjected to three consecutive passages on Vero cells to confirm the complete inactivation of SARS-CoV-2. Inactivated SARS-CoV-2 solution was then serially diluted to obtain concentrations from 1 to 106 TCID50/ml. Each dilution was detected by one biosensor well. Each biosensor has four spots of detection; thus, four experiments were done for each dilution.
2.6. Automatic data analysis
The biosensor reflection was measured before and after virus binding on the biosensor surface. The reflection spectrum was analyzed by an in-house developed Matlab program to identify the resonant valley position. Virus binding on the biosensor surface can cause a red shift of the resonant valley because virus binding adds material to the metal surface. The amount of red shift of the resonant valley is the response of the biosensor to virus binding and is proportional to the concentration of the virus in the sample being tested.
Matlab software can automatically identify the resonant valley position for a given reflection spectrum. These tools first smooth the original spectrometer data via a Gaussian window with a pre-selected window size to remove noise and smooth the spectra curve. The Matlab program then finds the peak and valley positions of a given full-width-half-maximum (FWHM) in a given wavelength range. In addition to the resonant valley, five peaks and five valleys were identified, and their respective wavelength positions were recorded. This analysis was done on biosensor spectra before and after virus binding: Either the red shift of the resonant valley alone or the total red shift of the five peaks and five valleys can be used to monitor the response of the biosensor. The latter method can help capture the red shifts due to virus binding in all available wavelength ranges. While the resonant valley can resolve small red shifts, longer wavelength features can provide larger shifts (Wu et al., 2012a).
2.7. Environmental sample treatment
Environmental samples were collected using viral transport medium (VTM) supplied by YOCON Biology Technology Company (Beijing, China). For biosensor detection, samples were applied on the biosensor surface directly without any pre-processing. For RT-PCR detection, viral RNA was extracted using Qiagen RNeasy Mini Kit following the manufacturer's instruction. Then RT-PCR process was carried out.
3. Results and discussion
This section describes biosensor surface characterization, optical reflection spectroscopy, and detection of SARS-CoV-2 are presented.
3.1. Biosensor surface characterization
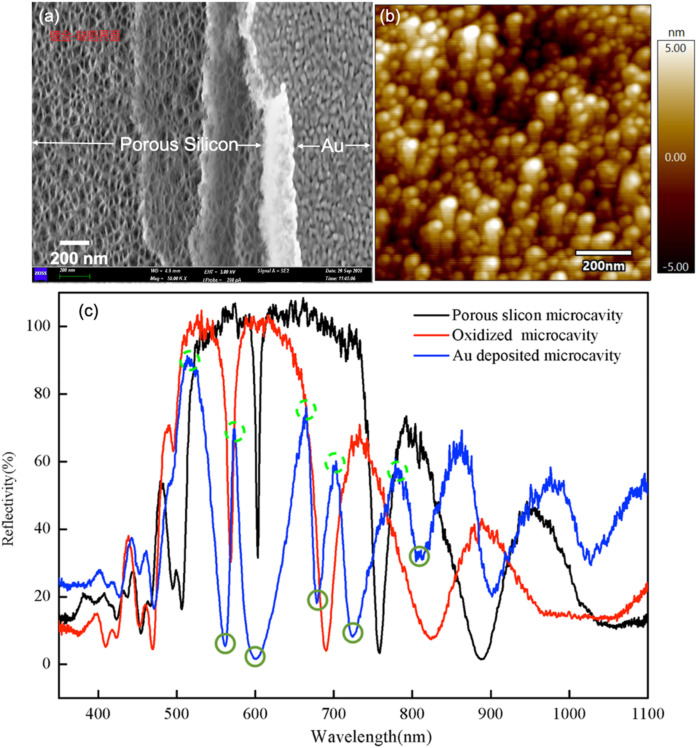
Fig. 2a shows top view scanning electron microscopy (SEM) images of porous silicon coated with gold. Partial removal of the Au thin film from the biosensor surface causes the nanoporous structure of both the porous silicon and gold thin film to be observed. Fig. 2b shows a higher resolution image of the gold surface using an atomic force microscope (AFM). The deposited Au thin film on the porous silicon is formed by clustering gold nanoparticles.
Fig. 2.
(a) Scanning electron microscopy (SEM) image of porous silicon coated with gold thin film. The left half of the SEM shows the porous structure of porous silicon wherein the silicon skeleton and air pores form the nanoscale structures; the right half of the SEM shows the physical vapor deposition (PVD)-deposited gold thin film with nanoscale porous features. (b) Atomic force microscope (AFM) image of the Au-coated porous silicon biosensor surface. The nanoscale features of the gold thin film are obvious. The Au thin film is porous on the nanoscale suggesting that it supports the localized surface plasmon resonance (LSPR) mode. (c) Reflection spectra of porous silicon resonant microcavity in different stages of the fabrication process: after anodization (black), after thermal oxidation (red), and after Au coating (blue). Five peaks (dotted light green circle) and five valleys (solid dark green circle) of the spectrum after Au coating are marked. Either the resonant valley alone (the leftmost valley, as marked in the figure) or all five peaks and five valleys of the porous silicon microcavity can be used for biosensing response calculation.
3.2. Optical reflection spectroscopy
The biosensor is measured by reflection spectroscopy via a portable fiber spectrometer. Fig. 2c shows the reflection spectrum of the porous silicon resonant microcavity after anodization, thermal oxidation, and Au coating. The resonant feature remains obvious after each step of the fabrication process. In addition to the resonant valley, five peaks and five valleys were ordered from the resonant valley towards the longer wavelength direction and were included in the calculation of total red shift as the final response signal of the biosensor. The signal-to-noise ratio of the spectrum is relative to background noise when the white light source is shut off and there is only ambient light; this value is over 100 dB.
3.3. Detection of SARS-CoV-2 related targets
After biofunctionalization with T-ACE2, the biosensor is ready for detection of SARS-CoV-2. We used four kinds of SARS-CoV-2-related targets for detection: (1) S-ECD viral protein; (2) pseudovirus (SinoBiological, Catalog Number PSV001, 1010 copies/ml stock concentration); and (3) inactivated SARS-CoV-2 virus cultured from a positive clinical specimen. (4) environmental samples.
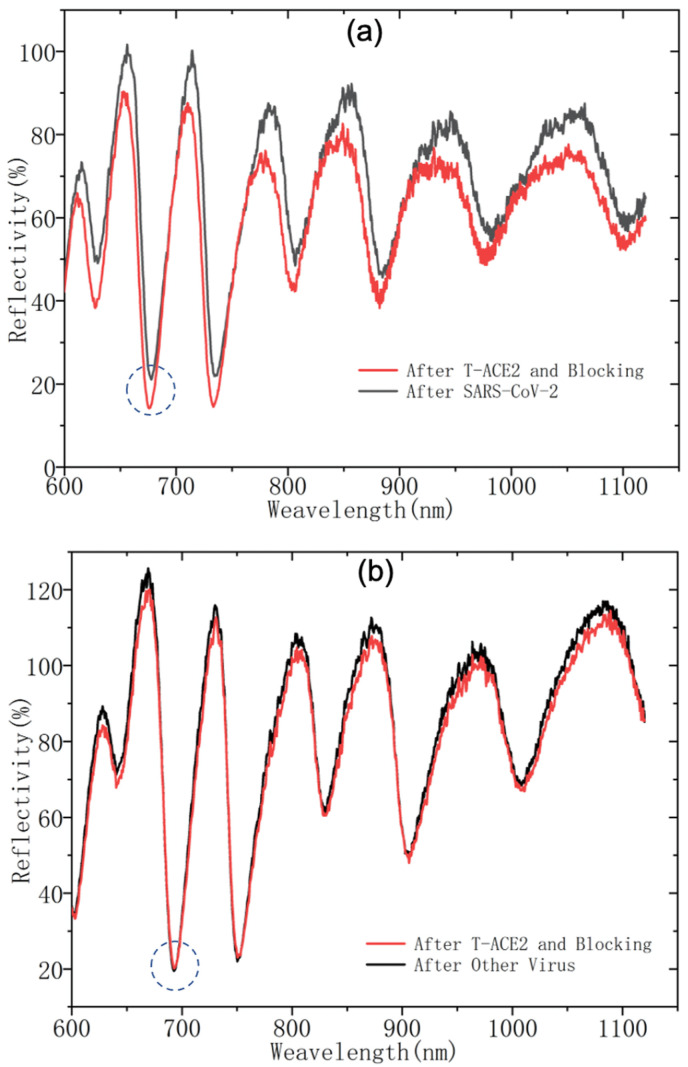
To demonstrate the proof-of-principle operation, Fig. 3 shows representative spectral shifts of the biosensor upon binding reaction with specific (SARS-CoV-2) and non-specific (other viruses) targets. For specific targets, the biosensor generates red shifts for all characteristic peaks and valleys. These shifts are negligible for non-specific targets. In addition, the extent of red shift correlates with the quantity of specific targets present in an unknown specimen.
Fig. 3.
Representative spectral shifts due to binding of (a) specific targets (SARS-CoV-2) and (b) non-specific targets (other viruses). The red shifts of the characteristic spectral peak and valley features are clearly visible for (a) positive results and negligible for (b) negative results. The circled feature is the resonant valley.
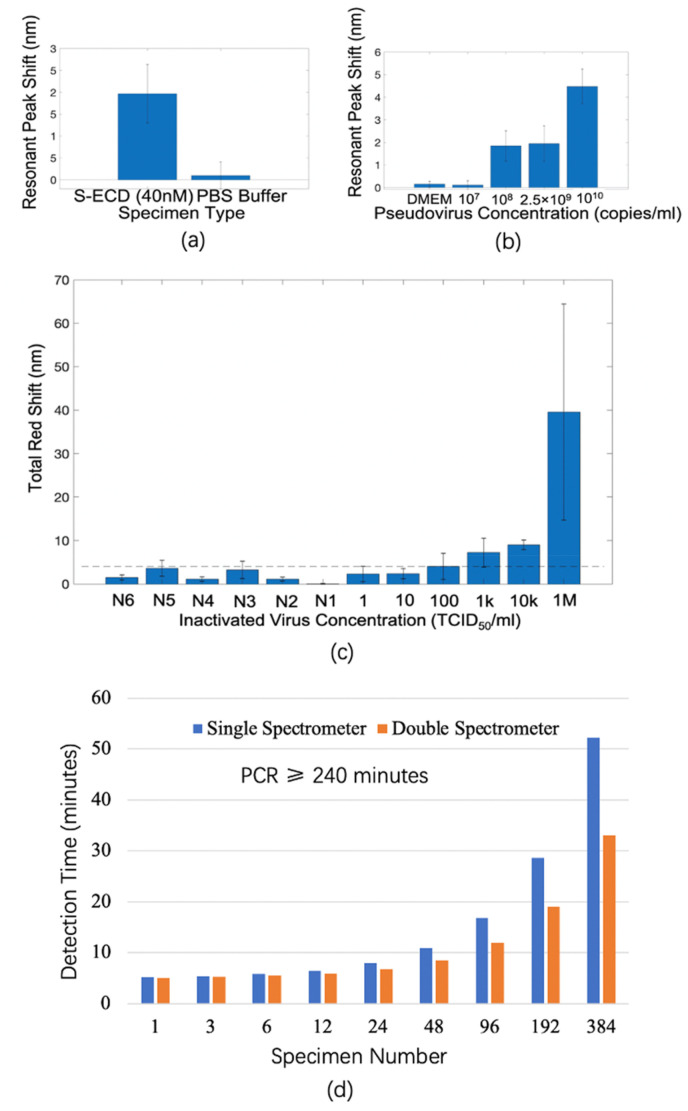
Fig. 4a shows the response of the biosensor (shift of resonant valley only, as marked in Fig. 3) for detection of S-ECD protein (40 nM). The biosensor has a specific response for S-ECD viral protein and a negligible response for PBS buffer only. Fig. 4b shows the response of the biosensor (shift of resonant valley only as marked in Fig. 3) as a function of pseudovirus concentration. The biosensor response increases with increasing pseudovirus concentration, and the limit of detection is more than 107 copies/ml. Three wells with one spot in each well were measured for each kind of specimen. For all experiments shown in Fig. 4a and b; three spectral measurements were taken both before and after binding reaction with T-ACE2. The error bars in Fig. 4a and b shows the standard error of responses in the three wells for the same specimen also demonstrating the well-to-well variation.
Fig. 4.
(a) Detection of S-ECD viral protein [40 nM in phosphate buffer saline (PBS) buffer] with control experiments in PBS buffer only. The error bars show the standard error from three experiments with each experiment carried out in one well; only one spot in each well is measured. The standard error also demonstrates the well-to-well variations. (b) Detection of SARS-CoV-2 pseudovirus at different concentrations with control experiment of dilution medium only (Dulbecco's modified eagle medium (DMEM), Thermo Fisher p/n 10569044). The error bars represent the standard error from three experiments with each experiment carried out in one well; only one spot in each well is measured. That is, the standard error demonstrates the well-to-well variations. (c) Detection of different concentrations of inactivated SARS-CoV-2 virus. The positive samples are a serial dilution of inactivated SARS-CoV-2 from 1 to 106 TCID50/ml. N1 is culture media that contains no virus. N2–N6 are five clinical throat swab specimens from patients infected by human bocavirus, enterovirus, adenovirus, PIV-3, and influenza A, respectively. The height of the vertical bars represents the average total red-shifts of five peaks and five valleys from four experiments with each experiment carried out in one well on four spots. The error bars show standard errors from those four experiments and demonstrate the spot-to-spot variation in the same well. The standard error comes from the non-uniform binding of virus on the biosensor surface reflected by different red shifts for the four spots in a well. The standard errors can be reduced by measuring more spots in a well. The limit of detection serves as a cutoff threshold to differentiate between positive and negative results and is estimated to be 100 TCID50/ml; this is shown as a dashed line. (d) Comparison of detection time between proposed biosensor and polymerase chain reaction (PCR) technique for different numbers of specimens. The time for biosensing includes sample loading, biosensor surface rinsing, and reflection spectroscopy measurement. The time for PCR includes nucleic acid extraction, purification, and amplification. Note that PCR requires at least 240 min regardless of the number of samples.
To optimize the performances of the biosensor for inactivated SARS-CoV-2 detection in BSL-2 lab, we adapted data processing of the biosensor by including five peaks and five valleys as discussed in Section 2.6 and Section 3.2 (Fig. 2c). Fig. 4c shows the total red shift of the biosensor as a function of inactivated SARS-CoV-2 virus concentration. Four spots (2 × 2 array) within each well were measured for each concentration, and three spectral measurements were taken for each spot both before and after virus binding. The height of the red shifts increases as virus concentration increases. The response for negative controls is also shown including the culture medium and five clinical throat swab specimens of patients infected by viruses other than SARS-CoV-2 as diagnosed by PCR: human bocavirus, enterovirus, adenovirus, parainfluenza virus Type 3 (PIV-3), and influenza A. The error bars in Fig. 4c show the standard error of responses of four spots to detect one kind of specimen in the same well, thus demonstrating the spot-to-spot variations. The limit of detection is calculated as three times the standard deviation of the signal levels of all the negative control experiments. As such, the LOD is close to but lower than the response signal level of the biosensor at 100 TCID50/ml. Thus we can estimate the LOD of the biosensor to be 100 TCID50/ml, which is better than the state-of-the-art antigen detection methods with limits of detection of several hundred to thousands of TCID50/ml (Cerutti et al., 2020; Liu et al., 2021; Porte et al., 2020; Scohy et al., 2020). The dynamic range lies in clinically relevant SARS-CoV-2 concentrations in throat swab and saliva specimens (Nouri et al., 2021).
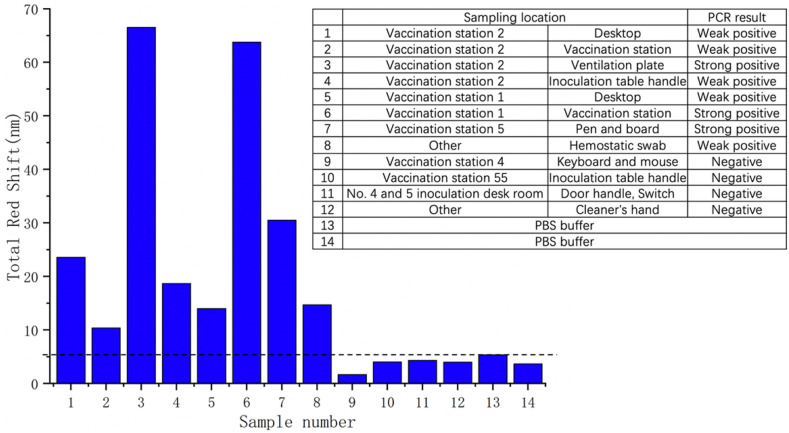
Besides clinical throat swab specimens, we tested the system by environmental samples. The first to twelfth specimens were sampled at different locations in the vaccination hospital, where sample 3, 6 and 7 are strong positive, sample 1, 2, 4, 5 and 8 are weak positive, sample 9, 10, 11 and 12 are negative according to PCR results. The thirteenth and fourteenth specimens are PBS buffer. Similar to the inactivated SARS-CoV-2 virus experiment described in Fig. 4c, we measure a 2 × 2 array (four spots) in each biosensor chip, and do three rounds of measurements to guarantee the detection accuracy. Fig. 5 shows the total reflectance spectrum red shift of each specimen. It is obvious that biosensors with strong positive samples loaded have larger amount of redshift (more than 30 nm), while those loaded with weak negative samples have relatively lower amount of redshift (more than 10 nm). In addition, biosensors with negative samples and PBS buffers loaded are of much lower amount of red shift (less than 5.5 nm) compared to those loaded with positive samples. This experiment illustrates that our biosensor has high accuracy in distinguishing environmentally positive samples from negative samples. Moreover, the signal strength has positive relationship with the viral load in the sample.
Fig. 5.
Detection of different environmental samples. The right table shows the number, sampling location and the PCR test result of each sample. The figure shows the total reflectance spectrum red shift of biosensor.
3.4. Comparison of detection time
We also characterized the time needed to detect different numbers of specimens with the proposed biosensor and its detection system (time including sample loading, biosensor surface rinsing, and reflection spectroscopy measurement) and versus PCR. The shortest turnaround time with our approach is 5 min, and the time increases with the number of specimens (Fig. 4d). Twenty min is needed for a single spectrometer with single well detection and 96 specimens. Less than 1 h is needed for 384 specimens. Roughly 30 min is needed for 384 specimens when using a double spectrometer for simultaneous two-well measurements. In PCR, at least 240 min (4 h) is needed regardless of the number of specimens. The throughput of the biosensor system is much higher than PCR and can accommodate many specimens more easily than PCR for practical applications. The detection time for large specimen numbers can be further reduced, and thus the throughput can be enhanced by integrating more spectrometers in a scalable system for multiple and simultaneous well detection.
The biosensor array is readily scalable. To use smaller number of wells per chip, the silicon wafer can be cut to achieve any array size such as 1 × 6, 4 × 6, or 8 × 6. At the other extreme, scaling can be achieved using silicon wafers with larger diameters such as 18 inches, which is equivalent to 1000 sensor wells per wafer. In our current design of the robotic system, up to four wafers can be mounted on a robotic arm. As a result, up to 4000 clinical specimens can be measured in one batch. The time to measure the optical signal of such a large well array can also be reduced by using multiple fiber spectrometers with multiple fiber probes collecting the optical spectra of multiple wells simultaneously.
A key feature of this work is that both the sensor array and the signal measurement system are readily scalable to accommodate screening of a wide range of sample numbers. We believe that this automatic, high throughput, and scalable detection system could be a useful tool to assist in curbing the SARS-CoV-2 epidemic. Research shows that detection methods that are not as sensitive and specific as RT-PCR—when applied with frequency and good coverage of population—can help prevent epidemic spread without the need for vaccines (Larremore et al., 2021). The system proposed here is well suited for such applications.
4. Conclusions
A localized surface plasmon resonance biosensor based on a porous silicon microcavity has been proposed, fabricated, and demonstrated for highly sensitivity detection of SARS-CoV-2 virus. The biosensor detects the coronavirus via the binding protein T-ACE2. Unlike nucleic acid amplification tests, the biosensor detects virus directly without the need for complex and time-consuming procedures such as nucleic acid extraction, purification, and amplification. As a result, the fastest detection time is 5 min, which is much shorter than PCR tests that can take 4 h. In addition, the biosensor has a sensitivity and limit of detection at least four-fold better than the state-of-the-art antigen detection methods; it is fully automatic and high throughput.
Future work will characterize regeneration and multiplexing: Detection of antibody, nucleic acid, and other viral protein (such as N protein) can all be incorporated in the system. These additional detection targets offer many advantages: Antibody detection can help stratify the stage of disease (i.e., early asymptomatic/pre-symptomatic stage (isolation needed) or in the recovery/clearance stage (isolation not needed)). This can help avoid unnecessary isolation or quarantine measures. Nucleic acid detection and/or other viral protein detection can help confirm positive diagnosis.
This work does have some limitations. First, the use of a protein target rather than a nucleic acid biomarker implies that the sensitivity and specificity will never be as high as PCR-based methods. Less selective methods still have significant value for point-of-care testing and surveillance. Second, the silicon wafer surface could be susceptive to fouling and thus careful longitudinal quality control studies that focus on the surface chemistry are needed. Third, the liquid handling equipment currently makes this a benchtop-sized design. Future integration of microfluidic components can lead to point-of-care designs.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang [grant number 2020R01005]; Westlake University [grant number 10318A992001]; Tencent Foundation [grant number XHTX202003001]; Research Program on COVID-19 of Zhejiang University of Technology, Zhejiang Key R&D Program [grant number 2021C03002]; and the Bright Dream Joint Institute for Intelligent Robotics [grant number 10318H991901].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114861.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- Ahmad A., Moore E. Electrochemical immunosensor modified with self-assembled monolayer of 11-mercaptoundecanoic acid on gold electrodes for detection of benzo a pyrene in water. Analyst. 2012;137(24):5839–5844. doi: 10.1039/c2an35236b. [DOI] [PubMed] [Google Scholar]
- Behrmann O., Bachmann I., Spiegel M., Schramm M., Abd El Wahed A., Dobler G., Dame G., Hufert F.T. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (Exo-IQ) Clin. Chem. 2020;66(8):1047–1054. doi: 10.1093/clinchem/hvaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrouzi K., Lin L.W. Gold nanoparticle based plasmonic sensing for the detection of SARS-CoV-2 nucleocapsid proteins. Biosens. Bioelectron. 2022;195:10. doi: 10.1016/j.bios.2021.113669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalera S., Colitti B., Rosati S., Ferrara G., Bertolotti L., Nogarol C., Guiotto C., Cagnazzo C., Denina M., Fagioli F., Di Nardo F., Chiarello M., Baggiani C., Anfossi L. A multi-target lateral flow immunoassay enabling the specific and sensitive detection of total antibodies to SARS COV-2. Talanta. 2021;223 doi: 10.1016/j.talanta.2020.121737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti F., Burdino E., Milia M.G., Allice T., Gregori G., Bruzzone B., Ghisetti V. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J. Clin. Virol. 2020;132 doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csáki A., Stranik O., Fritzsche W. Localized surface plasmon resonance based biosensing. Expert Rev. Mol. Diagn. 2018;18(3):279–296. doi: 10.1080/14737159.2018.1440208. [DOI] [PubMed] [Google Scholar]
- Dzimianski J.V., Lorig-Roach N., O'Rourke S.M., Alexander D.L., Kimmey J.M., DuBois R.M. Rapid and sensitive detection of SARS-CoV-2 antibodies by biolayer interferometry. Sci. Rep. 2020;10(1):12. doi: 10.1038/s41598-020-78895-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa S., Zourob M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal. Chem. 2021;93(3):1826–1833. doi: 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- Evangelista S., Bryner A. 2020. COVID-19: Using Wastewater to Track the Pandemic. [Google Scholar]
- Funari R., Chu K.-Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169:8. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funari R., Chu K.Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169:8. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92(16):11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- Grigoriev F.V., Sulimov V.B., Tikhonravov A.V. Combined modeling of the optical anisotropy of porous thin films. Coatings. 2020;10(6):517. [Google Scholar]
- Guo L., Bi W.W., Wang X.L., Xu W., Yan R.H., Zhang Y.Y., Zhao K., Li Y.N., Zhang M.F., Cai X., Jiang S.B., Xie Y.H., Zhou Q., Lu L., Dang B.B. Engineered trimeric ACE2 binds viral spike protein and locks it in "Three-up" conformation to potently inhibit SARS-CoV-2 infection. Cell Res. 2021;31(1):98–100. doi: 10.1038/s41422-020-00438-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq F., Sharif S., Khurshid A., Ikram A., Shabbir I., Salman M., Ahad A., Rana M.S., Raja A., Badar N., Tashkandi H., Al Amri T., Azhar E.I., Almuhayawi M.S., Harakeh S., Malik M.F.A. Reverse transcriptase loop-mediated isothermal amplification (RT-LAMP)-based diagnosis: a potential alternative to quantitative real-time PCR based detection of the novel SARS-COV-2 virus. Saudi J. Biol. Sci. 2021;28(1):942–947. doi: 10.1016/j.sjbs.2020.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayamohan H., Lambert C.J., Sant H.J., Jafek A., Patel D., Feng H.D., Beeman M., Mahmood T., Nze U., Gale B.K. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021;413(1):49–71. doi: 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliteevski M., Iorsh I., Brand S., Abram R.A., Chamberlain J.M., Kavokin A.V., Shelykh I.A. Tamm plasmon-polaritons: possible electromagnetic states at the interface of a metal and a dielectric Bragg mirror. Phys. Rev. B. 2007;76(16) [Google Scholar]
- Kaye S., Zeng Z., Sanders M., Chittur K., Koelle P.M., Lindquist R., Manne U., Lin Y.B., Wei J.J. Label-free detection of DNA hybridization with a compact LSPR-based fiber-optic sensor. Analyst. 2017;142(11):1974–1981. doi: 10.1039/c7an00249a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-K., Kerman K., Saito M., Sathuluri R.R., Endo T., Yamamura S., Kwon Y.S., Tamiya E. Label-free DNA biosensor based on localized surface plasmon resonance coupled with interferometry. Anal. Chem. 2007;79(5):1855–1864. doi: 10.1021/ac061909o. [DOI] [PubMed] [Google Scholar]
- Kumari A., Kumar S., Shukla M.K., Kumar G., Maji P.S., Vijaya R., Das R. Coupling to Tamm-plasmon-polaritons: dependence on structural parameters. J. Phys. D Appl. Phys. 2018;51(25) [Google Scholar]
- Kyosei Y., Namba M., Yamura S., Takeuchi R., Aoki N., Nakaishi K., Watabe S., Ito E. Proposal of de novo antigen test for COVID-19: ultrasensitive detection of spike proteins of SARS-CoV-2. Diagnostics. 2020;10(8):594. doi: 10.3390/diagnostics10080594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguarta J., Hueto F., Subirana B. COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J. Eng. Med. Biol. 2020;1(10):275–281. doi: 10.1109/OJEMB.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A., Milind T., Mina M.J., Parker R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021;7(1) doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.Y.P., Ng M.Y., Khong P.L. COVID-19 pneumonia: what has CT taught us? Lancet Infect. Dis. 2020;20(4):384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Cushing S.K., Wu N. Plasmon-enhanced optical sensors: a review. Analyst. 2015;140(2):386–406. doi: 10.1039/c4an01079e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Ju C.H., Han C., Shi R., Chen X.H., Duan D.M., Yan J.H., Yan X.Y. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021;173 doi: 10.1016/j.bios.2020.112817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Li Y.W., Jia H., Li Z.W., Ma D., Zhao J.L. Induced reflection in Tamm plasmon systems. Opt Express. 2019;27(4):5383–5392. doi: 10.1364/OE.27.005383. [DOI] [PubMed] [Google Scholar]
- Lu J., Fan W.H., Huang Z.H., Fan K., Dong J.H., Qin J.S., Luo J.Z., Zhang Z.Z., Sun G.D., Duan C.H., Pan K.Y., Gu W.S., Zhang X. Automatic system for high-throughput and high-sensitivity diagnosis of SARS-CoV-2. Bioproc. Biosyst. Eng. 2022;45(3):503–514. doi: 10.1007/s00449-021-02674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataji-Kojouri A., Ozen M.O., Shahabadi M., Inci F., Demirci U. Entangled nanoplasmonic cavities for estimating thickness of surface-adsorbed layers. ACS Nano. 2020;14(7):8518–8527. doi: 10.1021/acsnano.0c02797. [DOI] [PubMed] [Google Scholar]
- Mina M.J., Andersen K.G. COVID-19 testing: one size does not fit all. Science. 2021;371(6525):126–127. doi: 10.1126/science.abe9187. [DOI] [PubMed] [Google Scholar]
- Nouri R., Tang Z.F., Dong M., Liu T.Y., Kshirsagar A., Guan W.H. CRISPR-based detection of SARS-CoV-2: a review from sample to result. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R., Pizarro G., Vial P., Iruretagoyena M., Dittrich S., Weitzel T. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G.G., Gai Z.B., Tao Y.L., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome Coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Qu J.-H., Dillen A., Saeys W., Lammertyn J., Spasic D. Advancements in SPR biosensing technology: an overview of recent trends in smart layers design, multiplexing concepts, continuous monitoring and in vivo sensing. Anal. Chim. Acta. 2020;1104:10–27. doi: 10.1016/j.aca.2019.12.067. [DOI] [PubMed] [Google Scholar]
- Rong G., Najmaie A., Sipe J.E., Weiss S.M. Nanoscale porous silicon waveguide for label-free DNA sensing. Biosens. Bioelectron. 2008;23(10):1572–1576. doi: 10.1016/j.bios.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Rong G., Weiss S.M. Biomolecule size-dependent sensitivity of porous silicon sensors. Phys. Status Solidi A Appl. Mat. Sci. 2009;206(6):1365–1368. [Google Scholar]
- Rong G.G., Ryckman J.D., Mernaugh R.L., Weiss S.M. Label-free porous silicon membrane waveguide for DNA sensing. Appl. Phys. Lett. 2008;93(16) [Google Scholar]
- Rossi A.M., Wang L., Reipa V., Murphy T.E. Porous silicon biosensor for detection of viruses. Biosens. Bioelectron. 2007;23(5):741–745. doi: 10.1016/j.bios.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Scohy A., Anantharajah A., Bodeus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Correction to rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(9):12257–12258. doi: 10.1021/acsnano.0c06726. [DOI] [PubMed] [Google Scholar]
- Speletas M., Kyritsi M.A., Vontas A., Theodoridou A., Chrysanthidis T., Hatzianastasiou S., Petinaki E., Hadjichristodoulou C., Grp C.S. Evaluation of two chemiluminescent and three ELISA immunoassays for the detection of SARS-CoV-2 IgG antibodies: implications for disease diagnosis and patients' management. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.609242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantuoyir M.M., Rezaei N. Serological tests for COVID-19: potential opportunities. Cell Biol. Int. 2021;45(4):740–748. doi: 10.1002/cbin.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J.B., Min J.H., Yang Y.R., Xu C.H., Rossiter H.B., Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3(6):1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva-Canas J.L., Gonzalez-Roca E., Unanue A.G., Titos E., Yoldi M.J.M., Gomez A.V., Puig-Butille J.A. Implementation of an open-source robotic platform for SARS-CoV-2 testing by real-time RT-PCR. PLoS One. 2021;16(7) doi: 10.1371/journal.pone.0252509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.Q., Yu P., Wang W.H., Zhang X.T., Kuo H.C., Xu H.X., Wang Z.M.M. High-Q plasmonic resonances: fundamentals and applications. Adv. Opt. Mater. 2021;9(7) [Google Scholar]
- Wu B., Rong G.G., Zhao J.W., Zhang S.L., Zhu Y.X., He B.Y. A nanoscale porous silicon microcavity biosensor for novel label-free tuberculosis antigen-antibody detection. Nano. 2012;7(6) [Google Scholar]
- Wu C., Rong G., Xu J., Pan S., Zhu Y. Physical analysis of the response properties of porous silicon microcavity biosensor. Phys. E (Amsterdam, Neth.) 2012;44(7–8):1787–1791. [Google Scholar]
- Yeom S.-H., Kim O.G., Kang B.H., Kim K.J., Yuan H., Kwon D.H., Kim H.R., Kang S.W. Highly sensitive nano-porous lattice biosensor based on localized surface plasmon resonance and interference. Opt Express. 2011;19(23):22882–22891. doi: 10.1364/OE.19.022882. [DOI] [PubMed] [Google Scholar]
- Zang F., Su Z., Zhou L., Konduru K., Kaplan G., Chou S.Y. Ultrasensitive Ebola virus antigen sensing via 3D nanoantenna arrays. Adv. Mater. 2019;31(30) doi: 10.1002/adma.201902331. [DOI] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J.N., Portmann J., Halwe N.J., Ulrich L., Trüeb B.S., Fan X., Hoffmann B., Wang L., Thomann L., Lin X., Stalder H., Pozzi B., de Brot S., Jiang N., Cui D., Hossain J., Wilson M.M., Keller M.W., Stark T.J., Barnes J.R., Dijkman R., Jores J., Benarafa C., Wentworth D.E., Thiel V., Beer M. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592(7852):122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Zhu H.L., Zhang H.Q., Xu Y., Lassakova S., Korabecna M., Neuzil P. PCR past, present and future. Biotechniques. 2020;69(4):317–325. doi: 10.2144/btn-2020-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.