Abstract
B7-H3 (CD276) is an immune checkpoint overexpressed in prostate cancer with minimal expression in normal tissues and associated with poor prognosis, making it an excellent therapy target. We interrogated B7-H3 expression and its regulation in metastatic castration-resistant prostate cancer (mCRPC). We found greater expression of B7-H3 transcript relative to other immunotherapy targets (CTLA-4, PD-L1/2), including in tumors that lacked expression of prostate-specific membrane antigen (PSMA). Enzalutamide-resistant mCRPC cells demonstrated increased amounts of B7-H3, and this was associated with resistance signaling pathways. Using a machine-learning algorithm, the gene network of B7-H3 was strongly correlated with androgen receptor (AR) and AR co-factor (HOXB13, FOXA1) networks. In mCRPC samples, the B7-H3 promoter and distal enhancer regions exhibited enhanced transcriptional activity and were directly bound by AR and its co-factors. Altogether, our study characterizes molecular profiles and epigenetic regulation of B7-H3-expressing mCRPC tumors, which informs optimal precision-oncology approaches for mCRPC patients.
Subject terms: Prostate cancer, Cancer genomics
Main
Localized prostate cancer (PC) is curable, but options are limited for recurrent or metastatic tumors developing resistance to androgen-deprivation therapy (ADT) or AR targeted therapy (ART), known as metastatic castration-resistant prostate cancer (mCRPC). Overexpressed tumor antigens, such as PSMA, are targets of novel PET imaging approaches1 as well as precision therapeutics (177Lu-PSMA-617) in mCRPC (https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/215833s000lbl.pdf). Identifying additional mCRPC tumor antigens contributes to new strategies to develop precision antibody-drug conjugates that permit immuno- or cellular therapies2,3.
B7-H3 is a transmembrane glycoprotein in the B7 immune checkpoint superfamily4. Other well-known members, such as PD-L1 and CTLA-4, are targets in various hematologic and solid tumors5–7. B7-H3 is overexpressed in several cancers including prostate cancer, with minimal expression in normal prostatic tissue8–12. Higher expression of B7-H3 correlates with poor cancer prognosis8. B7-H3 has implications for cancer cell transformation and metastasis, and is thought to have a significant effect on the tumor microenvironment and immune suppression11,13. New strategies have been developed to target B7-H3 through antibody-dependent cell-mediated cytotoxicity14, antibody-drug conjugates15 and linking with immunotherapy such as chimeric antigen receptor-T cell16 or NK cell therapies17. However, little is known about the molecular features and regulatory mechanisms of B7-H3 in mCRPC, which prevents the optimal design of such targeted interventions and precludes rational patient selection. To address these barriers, we characterized the genomic, transcriptomic and epigenomic features of B7-H3 expression in mCRPC.
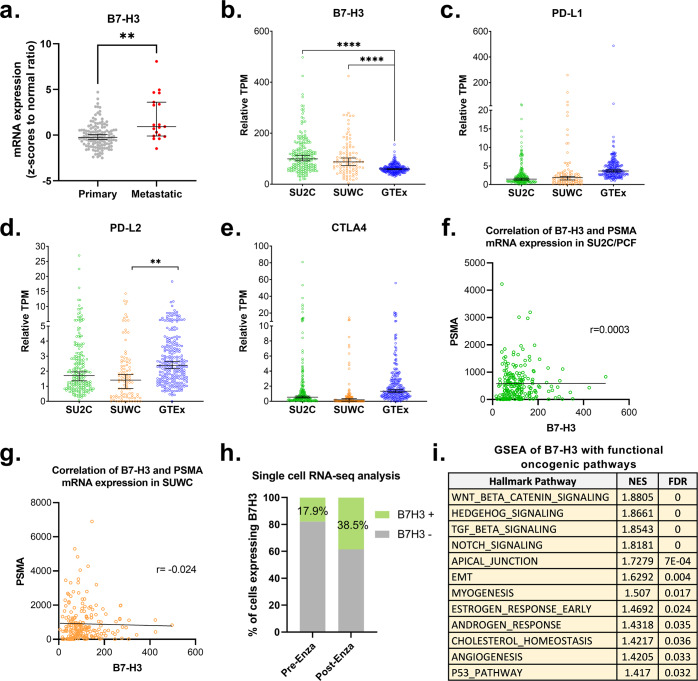
We first evaluated transcript expression profiles of B7-H3 (CD276) and other immune-regulatory genes in mCRPC. We conducted bioinformatic interrogations on whole-exome (WES) and whole-transcriptome sequencing (WTS) data from the datasets including MSKCC 2010 (n = 131, primary PC; n = 19, CRPC)18, SU2C/PCF (n = 208, mCRPC)19, SUWC (n = 101, mCRPC)20, and GTEx (n = 245, benign prostate tissue) datasets21. The MSKCC samples included both primary and metastatic tumors processed through the same platform. mRNA expression of B7-H3 was significantly increased in metastatic PC compared to primary PC (p = 0.004) (Fig. 1a). In other mCRPC datasets, expression of B7-H3 was significantly elevated in both mCRPC datasets compared to benign prostate tissues (median TPM 115, 87 vs. 60, p < 0.0001) (Fig. 1b). We also evaluated the association of the expression of B7-H3 mRNA with protein levels and found significant association in 369 cancer cell lines (p = 1.03E-72) (Supplementary Fig. 1a). In 10 patient-derived xenograft (PDX) models of castration-resistant prostate cancer (LuCaP PDX series)22, we detected B7-H3 expression in each PDX tumor pair and found a positive trend between mRNA and protein expression (n = 10, r = 0.52, p = 0.06) (Supplementary Figs. 1b–c and 4). Based on TPM, other B7 family members including PD-L1 (CD274), PD-L2 (PDCD1LG2), and CTLA-4, exhibited reduced expression in both mCRPC datasets and had limited overall transcript abundance compared to B7-H3 (Fig. 1c–e). Other immunological markers exhibited low abundance or were not overexpressed in mCRPC (Supplementary Fig. 2). The expression of B7-H3 in mCRPC was independent of that of PSMA, and exhibited robust expression even in PSMA-low tumors (Fig. 1f–g). This suggests that targeting B7-H3 could be an attractive alternative for PSMA-negative/low mCRPC patients.
Fig. 1. B7-H3 is selectively overexpressed in mCRPC.
a mRNA expression of B7-H3 in primary and metastatic prostate cancer (PC). Whole-transcriptome sequencing (WTS) data were obtained and analyzed from MSKCC 2010 (primary PC in gray, n = 131; metastatic PC in red, n = 19). All data are median with 95% CI. Statistical significance was using student t-test. **P < 0.01. b–e mRNA expression of B7-H3, PD-L1, PD-L2, and CTLA-4, respectively, in mCRPC (SU2C, green; SUWC, orange) and normal prostate tissue (GTEX, blue). Whole-transcriptome sequencing (WTS) data were obtained and analyzed from SU2C/PCF (n = 208, mCRPC), SUWC (n = 101, mCRPC), and GTEx (n = 245, benign prostate tissue). Data of other B7 family genes are shown in Supplementary Fig. 2. All data are median with 95% CI. Statistical significance was assessed using one-way ANOVA for multiple comparisons. ****P < 0.0001. f, g Lack of correlation of mRNA expression of B7-H3 and PSMA in mCRPC datasets SU2C/PCF (n = 208, r = 0.0003) and SUWC (n = 101, r = −0.024), respectively. Associations were determined by Pearson correlations. h Percentage of cells expressing B7-H3 (B7-H3 positive, green; B7-H3 negative, gray) before (17.9%) and after (38.5%) enzalutamide treatment. ScRNA-Seq analysis was performed on cells from patient before (n = 112) and after enzalutamide (n = 83) treatment. i GSEA of B7-H3 with functional oncogenic pathways. WTS data of mCRPC datasets SU2C/PCF (n = 208) and SUWC (n = 101) mCRPC were combined for GSEA. NES cutoff value 1.4. NES normalized enrichment score. FDR false discovery rate.
To examine how mCRPC tumor cells regulate B7-H3 expression in response to the ART, enzalutamide, we analyzed single-cell mRNA sequencing (scRNA-seq) data of paired biopsy samples from one patient (pre- and post-enzalutamide). We found an increased proportion of B7-H3-expressing tumor cells post-enzalutamide (38.5%) relative to pre-enzalutamide (17.9%) (Fig. 1h). Based on the genomic alterations in mCRPC with high B7-H3 expression, B7-H3 was associated with several known resistance markers including PTEN inactivation and AR-V7 detection19,20,23 (Supplementary Fig. 3). We conducted Gene Set Enrichment Analysis (GSEA)24 on mCRPC datasets (SU2C/PCF, n = 208 and SUWC, n = 101) and identified B7-H3 was enriched of TGF-beta, WNT, and Epithelial-to-Mesenchymal Transition (EMT) signaling pathways (Fig. 1i); each has been associated with resistance to enzalutamide25–27. Altogether, we found robust B7-H3 expression in mCRPC patients with existing molecular or signaling features that promote resistance to ADT and/or ART.
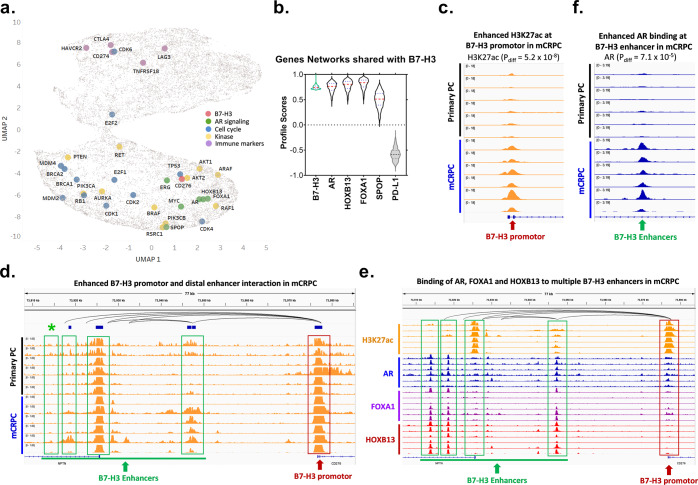
To enhance our mechanistic understanding of B7-H3 expression in mCRPC, we developed a machine-learning algorithm that quantitatively measures the degree of all gene-to-gene interactions to construct gene networks for all detectable genes. We used this algorithm to compare the degree of gene-network interactions between B7-H3 and all other detectable gene networks in the 208 mCRPC patients from the SU2C/PCF study. The overarching degree of gene-network association was visualized on UMAP, depicted through distances on an x-y plane. Remarkably, B7-H3 networks were closely clustered with those of AR, as well as with FOXA1, HOXB13, SPOP, MYC, and ERG (Fig. 2a). CTLA-4, PD-L1, PD-L2, and other immune markers were in distinct clusters (Fig. 2a), which indicated a lack of association with AR-signaling genes. We further examined the similarities of gene networks of B7-H3 and key regulators of AR signaling on a violin plot, in which the degree of overlap represents similarity. We observed that the B7-H3 gene network overlapped with those of AR, HOXB13, and FOXA1, and to a lesser degree with SPOP, but exhibited no intersection with PD-L1 (Fig. 2b). Altogether, these analyses suggest a robust convergence between B7-H3 and multiple genes with known functions in AR signaling.
Fig. 2. B7-H3 is significantly associated with and regulated by AR-signaling pathways.
a Machine-learning (ML)-based UMAP analysis of the association between B7-H3 and key PC pathways. Each dot in UMAP represents one gene. The spatial distance between two genes represents the similarity of their gene networks. Key PC pathways are visualized including AR signaling (green), Cell cycle (blue), Kinases (yellow), and Immune markers (purple) along with B7-H3 (pink). b ML-based analysis of the gene-network association between B7-H3 and key AR-signaling pathway genes in mCRPC patients (SU2C/PCF, n = 208). Data are shown in violin plots, in which red lines represent median and blue lines represent first quartile (lower) and third quartile (upper). The boundary of the violin represents the range of all data points. The degree of overlap of the plots represents the similarity of the networks they are associated with. PD-L1 was used as a negative control. c Comparison of H3K27ac enhancement at B7-H3 promoter in mCRPC and primary PC from representative patient-derived xenografts (PDXs)22. Pdiff indicates the FDR-adjusted P-value for comparison between primary prostate cancer and mCRPC using DESeq2. d Enhanced interaction between B7-H3 distal enhancers (green box) and its promoter (red box) in mCRPC as compared to primary PC. Top track indicates H3K27ac HiChIP data from LNCaP, which reflects long range chromosomal interactions in LNCaP cells. e Binding of AR, FOXA1, and HOXB13 to multiple putative B7-H3 enhancer sequences (green box) in mCRPC. f Increased AR binding in mCRPC at one of the putative CD276 enhancers indicated in d, labeled with green *. Six representative AR ChIP-seq profiles of primary prostate cancer and mCRPC are shown32.
B7-H3 is regulated epigenetically in nasopharyngeal carcinoma28 and glioblastoma29, via histone acetylation and DNA methylation at the promotor, respectively. The convergence between B7-H3 and AR signaling that we identified agreed with prior studies30. We thus sought to interrogate this mechanism of regulation through CHIP-seq data from both primary prostate cancer and mCRPC xenograft samples31,32. Remarkably, we observed enhanced histone-3-lysine-27 acetylation (H3K27ac) marks at the B7-H3 promoter and at putative B7-H3 distal enhancers in mCRPC as compared to primary prostate cancer (Fig. 2c, d), which reflected molecular mechanisms that increased transcription of B7-H3 in mCRPC. Further, we found that AR (and its co-regulators HOXB13 and FOXA1) were directly bound to B7-H3 enhancers (Fig. 2e). Notably, we found that AR exhibited selective binding to one of the B7-H3 putative enhancers in mCRPC as opposed to primary tumors (Fig. 2f). Although AR signaling is active at all stages of prostate cancer, our ChIP-seq analysis illustrated differential epigenetic regulation of B7-H3 transcripts in mCRPC compared to primary prostate cancer.
Our findings provide support that B7-H3-targeting therapies can fulfill an unmet medical need for ADT/ART-resistant mCRPC patients. Strategies to target B7-H3 with checkpoint inhibitors (NCT03729596), monoclonal antibodies (NCT02923180), antibody-drug conjugates (NCT03729596, NCT04145622), or tri-specific killer engager (TriKE) agents, are currently under investigation33,34. These therapeutics could be rationally designed for mCRPC patients that harbor ADT/ART-resistant biomarkers (e.g., PTEN loss, AR-V7 or ERG fusion) or other oncogenic signaling pathways (WNT, EMT, TGF-Beta). Further, targeting B7-H3 may be relevant in mCRPC patients with limited expression of PSMA, although our analysis did not address the status of B7-H3 in neuroendocrine/ small-cell prostate cancers. Our findings established a mechanistic connection between B7-H3 expression and AR-related signaling in mCRPC. This may also hold true in high-risk localized prostate tumors, since B7-H3 immunostaining is reduced after intense neoadjuvant ADT given before radical prostatectomy30. Finally, the epigenetic modifications we found may act as surrogates to measure B7-H3 levels from noninvasive liquid biopsies that include circulating-tumor DNA from mCRPC patients27.
Methods
Gene expression analysis
We normalized the expression of all commonly detected genes across four clinical cohorts (GTEx21, TCGA prostate cancer, SU2C/PCF19, SUWC20). These were originally mapped with distinct platforms and expression units. For each dataset, expression measurements were first converted into transcripts per million (TPM). The SU2C 2019 dataset was converted from fragments per kilobase of exon per million mapped fragments (FPKM) to TPM by scaling each sample such that the sum of expression came to 1 million for each sample35. The TCGA dataset was converted from RNA-seq by Expectation Maximization (RSEM) estimated transcript fraction to TPM by multiplying each value by 1 million36. The GTEx and SUWC datasets were already in TPM units. The TPM expression data for the total samples (n = 1102) in each dataset was merged by gene symbols that were common in all datasets. This merged dataset included 17,044 genes based on scaling each sample by a constant such that the total expression values in each sample sums to 1 million to generate pseudo-TPM values. The final matrix was then used to compare or correlate normalized expression of B7-H3 and all test genes in prostate tissue or metastatic prostate cancer samples. Statistical significance was determined by one-way ANOVA and correlation coefficients based on p-values < 0.05. The scripts and normalized data used to generate the data are shared at: git@github.com:bergo015/GeneNetworkingB7H3.git.
Protein and RNA expression analysis
To analyze the correlation between RNA and protein levels of B7-H3 (CD276), we utilized the Data Explorer tool from the DepMap Portal (https://depmap.org/portal/interactive/)37. On this portal, we elected to examine the public RNA (22Q2) and Proteomics data. This interface generated Pearson or Spearman correlations on CD276 expression levels and provided the statistical significance based on the linear regression. We exported the data points to generate Supplemental Fig. 1a.
Single-cell RNA sequencing analysis
The study from He et al.25 included one patient with paired biopsies prior to enzalutamide treatment and post resistance. Samples were collected with informed consent and ethics approval by the Dana-Farber/Harvard Cancer Center Institutional Review Board under protocol nos. 09–171, 11–104, 13–301, and 01–045. In these paired samples, we examined the tumor cells based on established mechanism to infer tumor cells25. This yielded 112 cells prior to enzalutamide treatment and 83 cells post resistance. To analyze these cells, we determined the positivity of B7-H3 expression based on absolute count values. We assigned B7-H3-positive or -negative cells based on the detection of B7-H3 transcripts in each tumor cells. We determined the proportion of cells with B7-H3 expression by dividing the number of cells expressing B7-H3 by the total cell numbers before enzalutamide treatment and post resistance.
Gene-network construction and analyses
We devised an algorithm that computes gene networks based on serial correlations that measure relative association between all gene pairs. Here we utilized whole-transcriptome sequencing data based on SU2C/PCF19. The input and computing were performed on the servers at the Minnesota Supercomputing Institute. This yielded the gene networks for all 19,135 genes in the format of a square matrix with the rows and columns representing gene IDs. Each element in the 19,135 × 19,135 matrix represents a similarity score of the two genes and their networks. The degree of similarity is measured by numerical values between −1 (highly discordant networks) to 1 (highly similar networks). To generate a B7-H3 gene-network signature, we included all gene networks based on a similarity score of ≥0.7 in reference to B7-H3. We then compared how all elements in this B7-H3 gene-network signature were associated with all other test genes. The degree of similarity to B7-H3 is assessed by the degree of overlap of the B7-H3 gene-network signature and all test genes in a violin plot. To visualize the similarity of the B7-H3 gene network in reference to all other genes, we reduced the 19,135 × 19,135 output matrix using Uniform Manifold Approximation and Projection (UMAP) to 2 dimensions (19,135 × 2 matrix). Plotting the two dimensions on a scatter plot, each point represents a gene, and the distance between each gene and B7-H3 represents the relative gene-network similarity. The scripts, input, and resulting data are provided at git@github.com:bergo015/GeneNetworkingB7H3.git.
Gene set enrichment analysis (GSEA)
Gene profiles were generated by associating B7-H3 expression to all other detected genes in all samples. The mRNA expression profiles were obtained by averaging the mRNA expression of each gene based on whole-transcriptome sequencing data from datasets SU2C/PCF (n = 208, mCRPC) and SUWC (n = 101, mCRPC)19,20. GSEA24 pre-ranked analyses was then performed on this profile to identify enrichment of gene signatures from the 189 C6 oncogenic signatures in MSigDB38. Based on the output files, we tabulated the normalized enrichment scores that were significant based on False Discovery Rates (FDR).
ChIP-seq analysis
ChIP-seq data were previously reported32 and are available under GEO accession number GSE130408. Primary tumors were obtained from prostatectomy specimens; mCRPC samples were obtained from LuCaP series PDXs22. Differential ChIP-seq analysis of AR binding and H3K27ac were performed in Pomerantz et al.32 using DESeq2 and included 23 primary cancer and 15 mCRPC-derived PDXs. The epigenetic regulation at specific chromosomal coordinates was visualized in each figure using IGV viewer.
Immunoblots
Immunoblots was performed following the Licor-system western blot detection protocol (licor.com/bio/support). 20ug of whole cell lysate from LNCaP and HL60 cells were ran in 4–20% SDS-page gel (Biorad, Cat# 4568094) and transferred to Immobilon-FL Transfer Membrane PVDF 0.45 uM pore size (Immobilon cat# IPFL00010). Protein was probed with antibodies against B7-H3 (Invitrogen, Cat # MAS-15693; R&D Systems, Cat # AF1027; Bethyl Laboratories, Cat # A700-026) and GAPDH (Santa Cruz, Cat # sc-32233). Precision plus was used as the protein standard (Bio-Rad, Cat# 161-0363). Primary antibodies were diluted 1:500 and incubated in 5% BSA, PBX and 0.2% tween-20 blocking solution. Secondary antibodies (IRDye 800 CW Goat-anti Mouse, Licor, Cat # 925-32210; IRDye 800 CW Goat-anti Rabbit, Licor, Cat # 925-32211; HRP-Conjugated Mouse-anti Goat, Santa Cruz, Cat # SC2354) were diluted 1:2000 and incubated in 5% BSA, 1× PBX, and 0.2% tween-20 and 0.01% SDS. Bio-rad Chemi-Doc MP Imaging system was used to scan the PVDF membrane for HRP activity detection. The protein expression of B7-H3 was normalized to GAPDH (loading control). All blots were derived from the same experiment and they were processed in parallel. The original scans of the blots are shown in Supplemental Fig. 4.
Statistics
The statistical significance of the mRNA expression of B7-H3 among the three datasets was assessed using one-way ANOVA. The associations between B7-H3 and PSMA were determined through Pearson correlations. The statistical significance was assessed using Chi-square and Fisher’s exact tests for categorical comparison between B7-H3 and genetic alterations. Student t-test was applied for AR-V7 and mRNA expression comparison. Graphs were generated using Prism Graphpad.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank our funding source Ray of Light Foundation (University of Minnesota) and Randy Shaver Cancer Research and Community Fund to support this work. Ray of Light Foundation award from Division of Hematology, Oncology, and Transplantation, University of Minnesota was allocated to the research efforts of J.H. and H.E.B. Randy Shaver Cancer Research and Community Fund from University of Minnesota was allocated to the research efforts of J.H., E.S.A., and N.Z. Some of the figures shown here were created with https://biorender.com/. We acknowledge TCGA for data generated by TCGA Research Network: https://www.cancer.gov/tcga.
Author contributions
X.S. and J.H. conceived the project, designed, and interpreted data. X.S. analyzed data with A.D., H.E.B., and S.T. S.C.B. provided the CHIP-seq analysis. Z.E.S., G.L., S.T., and J.M.D. performed and analyzed the Immunoblots. S.T., A.B., G.L., Z.E.S., and J.M.D. managed the PDX tissue lysates and the acquisition of laboratory reagents. J.W. participated in the machine-learning algorism design. E.S.A., C.J.R., and N.Z. participated in the study design and data interpretation. X.S. and J.H. wrote the manuscript. All authors edited and helped revise the manuscript.
Data availability
Datasets derived from public resources and these resources are provided within the article and below. (1) The MSKCC 201018 data are deposited in at NCBI GEO under accession GSE21032. The analyzed data can also be accessed and explored through the MSKCC Prostate Cancer Genomics Data Portal: http://cbio.mskcc.org/prostate-portal/#x201D. 10.1016/j.ccr.2010.05.026 (2010). (2) The SU2C/PCF19 data are available in Dataset S1 and at www.cbiportal.org, and have been deposited in GitHub, https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019. 10.1073/pnas.1902651116 (2019). (3) For the SUWC dataset20, the accession number for the raw sequencing data reported in the paper is dbGAP: phs001648.v1.p1. 10.1016/j.cell.2018.06.039 (2018). (4) The GTEx21 data are available through the GTEx portal (www.gtexportal.org). 10.3390/jpm5010022 (2015). (5) For the single-cell sequencing (scRNA-seq) dataset25, scRNA-seq expression and clustering data generated in this study are available at https://singlecell.broadinstitute.org/single_cell/study/SCP1244/transcriptional-mediators-of-treatment-resistance-in-lethal-prostate-cancer. Raw sequence data generated in this study are being deposited in dbGaP (accession phs001988.v1.p1). 10.1038/s41591-021-01244-6 (2021). (6) The proteomics data of the 369 cell lines used in Supplementary Fig. 1a were obtained from DepMap Portal using the Data Explorer feature. https://depmap.org/portal/interactive/.
Code availability
The basic machine-learning operation source code, input datasets, and output matrices are provided in the following link: git@github.com:bergo015/GeneNetworkingB7H3.git
Competing interests
E.S.A. is a paid consultant/advisor to Janssen, Astellas, Sanofi, Dendreon, Pfizer, Amgen, Eli Lilly, Bayer, AstraZeneca, Bristol Myers Squibb, ESSA, Clovis, Merck, Curium, Blue Earth Diagnostics, Foundation Medicine, Exact Sciences and Invitae; has received research funding to his institution from Janssen, Johnson & Johnson, Sanofi, Dendreon, Genentech, Novartis, Tokai, Bristol Myers Squibb, Constellation, Bayer, AstraZeneca, Clovis and Merck; and is the coinventor of a patented AR-V7 biomarker technology that has been licensed to Qiagen. J.M.D. has no conflicts relevant to this work. However, he holds equity in and serves as Chief Scientific Officer of Astrin Biosciences. This interest has been reviewed and managed by the University of Minnesota in accordance with its Conflict of Interest policies. None of these companies contributed to or directed any of the research reported in this article. N.Z. is an advisor/consultant for Caris. J.H. is a consultant/advisor to Caris Life Sciences and Astrin Biosciences. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Emmanuel S. Antonarakis, Justin Hwang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-022-00323-2.
References
- 1.Carlucci G, et al. (68)Ga-PSMA-11 NDA approval: a novel and successful academic partnership. J. Nucl. Med. 2021;62:149–155. doi: 10.2967/jnumed.120.260455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrylak DP, et al. PSMA ADC monotherapy in patients with progressive metastatic castration-resistant prostate cancer following abiraterone and/or enzalutamide: efficacy and safety in open-label single-arm phase 2 study. Prostate. 2020;80:99–108. doi: 10.1002/pros.23922. [DOI] [PubMed] [Google Scholar]
- 3.Danila DC, et al. Phase I study of DSTP3086S, an antibody-drug conjugate targeting six-transmembrane epithelial antigen of prostate 1, in metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2019;37:3518–3527. doi: 10.1200/JCO.19.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janakiram M, et al. The third group of the B7-CD28 immune checkpoint family: HHLA2, TMIGD2, B7x, and B7-H3. Immunol. Rev. 2017;276:26–39. doi: 10.1111/imr.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topalian SL. Targeting immune checkpoints in cancer therapy. JAMA. 2017;318:1647–1648. doi: 10.1001/jama.2017.14155. [DOI] [PubMed] [Google Scholar]
- 6.Pandey, P. et al. Revolutionization in cancer therapeutics via targeting major immune checkpoints PD-1, PD-L1 and CTLA-4. Pharmaceuticals (Basel)10.3390/ph15030335 (2022). [DOI] [PMC free article] [PubMed]
- 7.Salik B, Smyth MJ, Nakamura K. Targeting immune checkpoints in hematological malignancies. J. Hematol. Oncol. 2020;13:111. doi: 10.1186/s13045-020-00947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benzon B, et al. Correlation of B7-H3 with androgen receptor, immune pathways and poor outcome in prostate cancer: an expression-based analysis. Prostate Cancer Prostatic Dis. 2017;20:28–35. doi: 10.1038/pcan.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou WT, Jin WL. B7-H3/CD276: an emerging cancer immunotherapy. Front Immunol. 2021;12:701006. doi: 10.3389/fimmu.2021.701006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong F, Yu H, Gao X. Expression of CD24 and B7-H3 in breast cancer and the clinical significance. Oncol. Lett. 2017;14:7185–7190. doi: 10.3892/ol.2017.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yonesaka K, et al. B7-H3 negatively modulates CTL-mediated cancer immunity. Clin. Cancer Res. 2018;24:2653–2664. doi: 10.1158/1078-0432.CCR-17-2852. [DOI] [PubMed] [Google Scholar]
- 12.Zang X, et al. Tumor associated endothelial expression of B7-H3 predicts survival in ovarian carcinomas. Mod. Pathol. 2010;23:1104–1112. doi: 10.1038/modpathol.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapoval AI, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat. Immunol. 2001;2:269–274. doi: 10.1038/85339. [DOI] [PubMed] [Google Scholar]
- 14.Loo D, et al. Development of an Fc-enhanced anti-B7-H3 monoclonal antibody with potent antitumor activity. Clin. Cancer Res. 2012;18:3834–3845. doi: 10.1158/1078-0432.CCR-12-0715. [DOI] [PubMed] [Google Scholar]
- 15.Scribner JA, et al. Preclinical development of MGC018, a duocarmycin-based antibody-drug conjugate targeting B7-H3 for solid cancer. Mol. Cancer Ther. 2020;19:2235–2244. doi: 10.1158/1535-7163.MCT-20-0116. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, et al. Targeting B7-H3 via chimeric antigen receptor T cells and bispecific killer cell engagers augments antitumor response of cytotoxic lymphocytes. J. Hematol. Oncol. 2021;14:21. doi: 10.1186/s13045-020-01024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang S, et al. Targeting B7-H3 immune checkpoint with chimeric antigen receptor-engineered natural killer cells exhibits potent cytotoxicity against non-small cell lung cancer. Front. Pharmacol. 2020;11:1089. doi: 10.3389/fphar.2020.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abida W, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl Acad. Sci. USA. 2019;116:11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quigley DA, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174:758–769 e759. doi: 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keen JC, Moore HM. The Genotype-Tissue Expression (GTEx) Project: linking clinical data with molecular analysis to advance personalized medicine. J. Pers. Med. 2015;5:22–29. doi: 10.3390/jpm5010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen HM, et al. LuCaP prostate cancer patient-derived xenografts reflect the molecular heterogeneity of advanced disease an–d serve as models for evaluating cancer therapeutics. Prostate. 2017;77:654–671. doi: 10.1002/pros.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He MX, et al. Transcriptional mediators of treatment resistance in lethal prostate cancer. Nat. Med. 2021;27:426–433. doi: 10.1038/s41591-021-01244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isaacsson Velho P, et al. Wnt-pathway activating mutations are associated with resistance to first-line abiraterone and enzalutamide in castration-resistant prostate cancer. Eur. Urol. 2020;77:14–21. doi: 10.1016/j.eururo.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyatt AW, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1598–1606. doi: 10.1001/jamaoncol.2016.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang MY, et al. PBK phosphorylates MSL1 to elicit epigenetic modulation of CD276 in nasopharyngeal carcinoma. Oncogenesis. 2021;10:9. doi: 10.1038/s41389-020-00293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, et al. Genetic and clinical characterization of B7-H3 (CD276) expression and epigenetic regulation in diffuse brain glioma. Cancer Sci. 2018;109:2697–2705. doi: 10.1111/cas.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendes AA, et al. Association of B7-H3 expression with racial ancestry, immune cell density, and androgen receptor activation in prostate cancer. Cancer. 2022;128:2269–2280. doi: 10.1002/cncr.34190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pomerantz MM, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 2015;47:1346–1351. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomerantz MM, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 2020;52:790–799. doi: 10.1038/s41588-020-0664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallera, D. A. et al. NK-cell-mediated targeting of various solid tumors using a B7-H3 tri-specific killer engager in vitro and in vivo. Cancers (Basel)10.3390/cancers12092659 (2020). [DOI] [PMC free article] [PubMed]
- 34.Miller J, Z. N. et al. Targeting pan-tumor associated antigen B7H3 via combination of tri-specific killer engager and off-the-shelf NK cell therapy enhances specificity and function against a broad range of solid tumors. J. Immuno Ther. Cancer10.1136/jitc-2020-SITC2020.0470 (2020).
- 35.Zhao Y, et al. TPM, FPKM, or normalized counts? A comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Transl. Med. 2021;19:269. doi: 10.1186/s12967-021-02936-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsherniak A, et al. Defining a cancer dependency map. Cell. 2017;170:564–576 e516. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets derived from public resources and these resources are provided within the article and below. (1) The MSKCC 201018 data are deposited in at NCBI GEO under accession GSE21032. The analyzed data can also be accessed and explored through the MSKCC Prostate Cancer Genomics Data Portal: http://cbio.mskcc.org/prostate-portal/#x201D. 10.1016/j.ccr.2010.05.026 (2010). (2) The SU2C/PCF19 data are available in Dataset S1 and at www.cbiportal.org, and have been deposited in GitHub, https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019. 10.1073/pnas.1902651116 (2019). (3) For the SUWC dataset20, the accession number for the raw sequencing data reported in the paper is dbGAP: phs001648.v1.p1. 10.1016/j.cell.2018.06.039 (2018). (4) The GTEx21 data are available through the GTEx portal (www.gtexportal.org). 10.3390/jpm5010022 (2015). (5) For the single-cell sequencing (scRNA-seq) dataset25, scRNA-seq expression and clustering data generated in this study are available at https://singlecell.broadinstitute.org/single_cell/study/SCP1244/transcriptional-mediators-of-treatment-resistance-in-lethal-prostate-cancer. Raw sequence data generated in this study are being deposited in dbGaP (accession phs001988.v1.p1). 10.1038/s41591-021-01244-6 (2021). (6) The proteomics data of the 369 cell lines used in Supplementary Fig. 1a were obtained from DepMap Portal using the Data Explorer feature. https://depmap.org/portal/interactive/.
The basic machine-learning operation source code, input datasets, and output matrices are provided in the following link: git@github.com:bergo015/GeneNetworkingB7H3.git