Abstract
The pyrrolysyl-tRNA synthetase (PylRS) facilitates the cotranslational installation of the 22nd amino acid pyrrolysine. Owing to its tolerance for diverse amino acid substrates, and its orthogonality in multiple organisms, PylRS has emerged as a major route to install noncanonical amino acids into proteins in living cells. Recently, a novel class of PylRS enzymes was identified in a subset of methanogenic archaea. Enzymes within this class (ΔPylSn) lack the N-terminal tRNA-binding domain that is widely conserved amongst PylRS enzymes, yet remain active and orthogonal in bacteria and eukaryotes. In this study, we use biochemical and in vivo UAG-readthrough assays to characterize the aminoacylation efficiency and substrate spectrum of a ΔPylSn class PylRS from the archaeon Candidatus Methanomethylophilus alvus. We show that, compared with the full-length enzyme from Methanosarcina mazei, the Ca. M. alvus PylRS displays reduced aminoacylation efficiency but an expanded amino acid substrate spectrum. To gain insight into the evolution of ΔPylSn enzymes, we performed molecular phylogeny using 156 PylRS and 105 pyrrolysine tRNA (tRNAPyl) sequences from diverse archaea and bacteria. This analysis suggests that the PylRS•tRNAPyl pair diverged before the evolution of the three domains of life, placing an early limit on the evolution of the Pyl-decoding trait. Furthermore, our results document the coevolutionary history of PylRS and tRNAPyl and reveal the emergence of tRNAPyl sequences with unique A73 and U73 discriminator bases. The orthogonality of these tRNAPyl species with the more common G73-containing tRNAPyl will enable future efforts to engineer PylRS systems for further genetic code expansion.
Keywords: pyrrolysyl-tRNA synthetase, aminoacyl-tRNA synthetase, genetic code expansion, noncanonical amino acids, synthetic biology, amber suppression
Abbreviations: aaRS, aminoacyl-tRNA synthetase; AlloK, NƐ-alloc-l-lysine; Amp, ampicillin; BocK, NƐ-boc-l-lysine; chPylRS, chimera PylRS; HGT, horizontal gene transfer; HMET1, Candidatus Methanohalarchaeum thermophilum; MaPylRS, PylRS enzyme from Candidatus Methanomethylophilus alvus; Ma-tRNAPyl, the pyrrolysine tRNA from Candidatus Methanomethylophilus alvus; MbPylRS, PylRS enzyme from Methanosarcina barkeri; MeH, N-methyl-l-histidine; Mm-tRNAPyl, the pyrrolysine tRNA from Methanosarcina mazei; MOST, Ministry of Science and Technology; MSBL1, Mediterranean Sea Brine Lakes 1 archaeon; ncAA, noncanonical amino acid; PheRS, phenylalanyl-tRNA synthetase; Pyl, pyrrolysine; PylRS, pyrrolysyl-tRNA synthetase; sfGFP, superfolder GFP; Spec, spectinomycin; tRNAPyl, pyrrolysine tRNA
Pyrrolysine (1, Pyl, Fig. 1) is the 22nd naturally occurring proteinogenic amino acid that is encoded in the genomes of certain anaerobic archaea and bacteria (1). In these organisms, Pyl is installed into polypeptides through the combined actions of the pyrrolysyl-tRNA synthetase (PylRS) and pyrrolysine tRNA (tRNAPyl) (2, 3). PylRS specifically recognizes free Pyl and attaches the amino acid to the 3′-hydroxyl of tRNAPyl (4). The product, Pyl-tRNAPyl, then introduces Pyl into proteins in response to in-frame UAG codons during normal ribosomal protein synthesis.
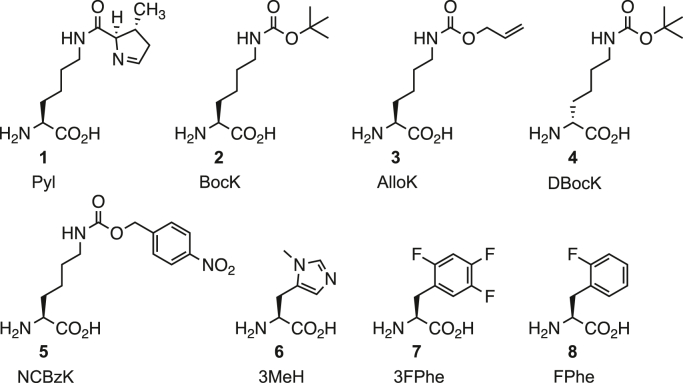
Figure 1.
Structures of relevant noncanonical amino acids used in this study.
Over the past 2 decades, the PylRS•tRNAPyl pair has gained widespread interest for its ability to install noncanonical amino acids (ncAAs) into proteins with site-specific precision in a variety of phylogenetically diverse organisms. Several features of the PylRS•tRNAPyl pair make it an exceptional tool for expanding the genetic code. First, unlike other aminoacyl-tRNA synthetases (aaRSs) that are commonly used for genetic code expansion, the PylRS•tRNAPyl pair does not crossreact with endogenous aaRSs or tRNAs in both bacterial and eukaryotic hosts (5, 6). Owing to this orthogonality, the PylRS•tRNAPyl pair can be used to install ncAAs into proteins in a variety of model organisms. Second, PylRS has a remarkably high tolerance for structurally disparate ncAA substrates, which is attributed to the large size of the amino acid binding pocket within the enzyme’s active site (7). Finally, unlike most aaRSs, PylRS does not interact with the anticodon of its cognate tRNA (8); therefore, the anticodon of tRNAPyl can be mutated to recognize codons other than UAG without impacting tRNA recognition by PylRS (9).
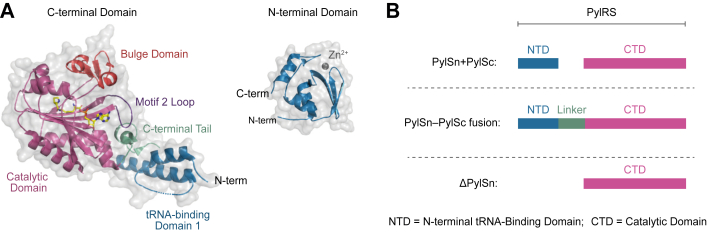
Most PylRS enzymes are comprised of two functional domains including a C-terminal catalytic domain (PylSc) and an N-terminal tRNA-binding domain (PylSn; Fig. 2A) (10). PylSc contains a conserved catalytic core and Rossmann fold, which is typical of class II aaRSs (11, 12). Structure-based analyses have revealed that this domain is likely derived from an ancestral version of the phenylalanyl-tRNA synthetase (PheRS) (11). In contrast, PylSn is a novel RNA-binding protein with no structural or sequence homology to any known RNA-binding proteins (13). A recently determined crystal structure of PylSn in complex with tRNAPyl (14) showed that this domain makes extensive contacts with tRNAPyl, specifically the T and variable loops, confirming earlier biochemical studies (13). The exact physiological function of PylSn remains unknown; however, given its affinity for tRNAPyl, it is hypothesized that this domain serves to recruit tRNAPyl to the catalytic domain. This might allow cells to maintain a low basal level of tRNAPyl, thereby minimizing suppression of UAG codons that are otherwise meant to terminate translation (13).
Figure 2.
Domain organization of the three classes of PylRS enzymes.A, the crystal structures of the PylRS N-terminal tRNA-binding domain (PylSn) (Protein Data Bank [PDB] code:5UD5) and C-terminal catalytic domain (PylSc) (PDB code:2Q7H) from Methanosarcina mazei. The C-terminal domain is shown in complex with adenylated pyrrolysine (yellow). Structural features of each domain are labeled. B, domain organization of the three classes of PylRS enzymes. PylRS, pyrrolysyl-tRNA synthetase.
Based on the arrangement of the PylSn and PylSc domains, PylRS enzymes can be subdivided into three classes, referred to as “PylSn + PylSc,” “PylSn–PylSc fusion,” and “ΔPylSn” (Fig. 2B). In the PylSn + PylSc class, PylSn and PylSc are expressed from two separate genes as distinct polypeptides. Enzymes in this class are most commonly found in bacteria that utilize Pyl (1, 13). In the PylSn–PylSc fusion class, the PylSn and PylSc domains are expressed as a single polypeptide, connected by a variable linker of ∼40 to 155 amino acids. Because of their high activity in various model organisms, enzymes in the PylSn–PylSc fusion class are the most widely used for genetic code expansion (5). The ΔPylSn class is the most recently discovered class of PylRS enzymes (9, 15). Enzymes in this class completely lack the PylSn domain, with PylSc having evolved robust stand-alone activity (16, 17). In recent years, ΔPylSn class enzymes have gained popularity as tools for genetic code expansion, largely because they can be engineered to be mutually orthogonal with PylSn–PylSc fusion enzymes. Because of this mutual orthogonality, ΔPylSn and PylSn–PylSc fusion enzymes can be used, together in the same cell, to simultaneously install two distinct ncAAs (16, 17, 18, 19, 20, 21, 22, 23, 24). However, given the relatively recent discovery of ΔPylSn enzymes, their activity and substrate specificities have not been fully characterized.
In the current study, we use enzyme-kinetic biochemical and in vivo UAG-readthrough translation assays to characterize the aminoacylation activity and substrate specificity of the ΔPylSn class PylRS enzyme from Candidatus Methanomethylophilus alvus (MaPylRS) and the PylSn–PylSc fusion enzyme from Methanosarcina mazei. We also explore the amino acid substrate range of these two PylRS enzymes. Finally, to gain insight into the evolutionary history of the Pyl-decoding trait, we performed molecular phylogenetic studies of 156 PylSc and 105 tRNAPyl sequences from archaeal and bacterial organisms.
Results
Kinetic analysis of PylRS variants with pyrrolysine
Loss of PylSn is normally detrimental to the activity of PylRS (25). Specifically, when PylSn is deleted from PylSn–PylSc fusion enzymes, the truncated enzymes retain aminoacylation activity in vitro but have undetectable activity in vivo (13, 25). Based on these previous studies, we hypothesized that the ΔPylSn enzyme from Ca. M. alvus might have lower aminoacylation activity compared with full-length PylSn–PylSc fusion enzymes. To compare the aminoacylation activity of MaPylRS to PylSn–PylSc fusion enzymes, we performed in vitro activity assays using purified recombinant PylRS with tRNAPyl transcripts. We compared the activity of MaPylRS to the PylSn–PylSc fusion enzymes from M. mazei (MmPylRS), Methanosarcina barkeri (MbPylRS), and an engineered MmPylRS–MbPylRS chimera (chPylRS) with improved solubility (14). Each of these enzymes was previously used to site specifically install diverse ncAAs into proteins in both bacteria and eukaryotes (5, 26), and they are of high interest for synthetic biology and biotechnology applications. Like all aaRSs, the reaction catalyzed by PylRS involves two elementary steps including (i) reaction of the substrate amino acid with ATP to form an aminoacyl adenylate intermediate and (ii) reaction of the aminoacyl adenylate with a terminal hydroxyl of the tRNA to form the aminoacyl-tRNA product (4). Formation of the aminoacyl adenylate intermediate is typically monitored using an ATP–PPi exchange assay; however, poor solubility of the PylSn domain of MmPylRS and MbPylRS requires the use of truncated enzymes for this assay (11, 27, 28). Alternatively, aaRS activity can be assayed by monitoring aminoacyl-tRNA formation, by separating charged and uncharged tRNAs on an acidic-denaturing polyacrylamide gel (29). This assay requires lower enzyme concentrations, within the solubility limit of MmPylRS and MbPylRS, and therefore allows for direct comparison of the activity of ΔPylSn and full-length PylSn–PylSc fusion enzymes. Therefore, we used the latter assay to measure PylRS activity.
To determine kinetic parameters (Km and kcat) for wildtype MaPylRS, we measured aminoacyl-tRNA formation using varying concentrations of the native substrate Pyl (30). Our analysis revealed a higher Km for MaPylRS (35 ± 6 μM), when compared with wildtype MmPylRS (20 ± 4 μM) and MbPylRS (20 ± 2 μM), and the more soluble chPylRS (7.6 ± 0.2 μM). The data indicate weaker Pyl recognition by MaPylRS compared with the PylSn–PylSc fusion enzymes. In addition, kcat was lower for MaPylRS (4.5 ± 0.2 s−1 × 10−3) compared with MmPylRS, chPylRS, and MbPylRS (8.3 ± 0.3, 11 ± 1, and 30 ± 1 s−1 × 10−3, respectively, Table 1 and Fig. S1). These values correspond to an aminoacylation efficiency (kcat/Km) threefold higher for MmPylRS and 11-fold higher for MbPylRS and chPylRS, compared with MaPylRS.
Table 1.
Kinetic properties of different PylRS enzymes with Pyl and lysine analogs
| Entry | Enzyme | tRNAPyl | Amino acid | Km (μM) | kcat (s−1 × 10−3) | kcat/Km (mM−1s−1 × 10−3) | Relative activity |
|---|---|---|---|---|---|---|---|
| 1 | MmPylRSa | Mm | Pyl (1) | 20 ± 4 | 8.3 ± 0.3 | 415 | 100 |
| 2 | MbPylRSa | Mm | Pyl (1) | 20 ± 2 | 30 ± 1 | 1510 | 364 |
| 3 | chPylRSa | Mm | Pyl (1) | 7.6 ± 0.2 | 11 ± 1 | 1447 | 349 |
| 4 | MaPylRS | Ma | Pyl (1) | 35 ± 6 | 4.5 ± 0.2 | 132 | 32 |
| 5 | MmPylRS | Ma | Pyl (1) | 31 ± 11 | 1.2 ± 0.1 | 38.7 | 9.3 |
| 6 | MaPylRS | Mm | Pyl (1) | ND | ND | — | — |
| 7 | MaPylRS | Ma | BocK (2) | 2200 ± 1200 | 2.3 ± 0.2 | 1.0 | 0.003 |
| 8 | MaPylRS | Ma | AlloK (3) | 540 ± 220 | 2.5 ± 0.2 | 4.6 | 0.01 |
| 9 | MmPylRS | Ma | BocK (2) | 1600 ± 900 | 0.9 ± 0.1 | 0.56 | 0.001 |
| 10 | MmPylRS | Ma | AlloK (3) | 720 ± 200 | 1.0 ± 0.1 | 1.4 | 0.003 |
tRNAPyl crossrecognition by different PylRS enzymes
Previous studies have shown that MmPylRS displays significant crossrecognition of heterologous tRNAPyl molecules. Specifically, stop codon readthrough assays both in vitro and in vivo have shown that MmPylRS can efficiently aminoacylate the Ca. M. alvus tRNAPyl (Ma-tRNAPyl), whereas MaPylRS is only active with its homologous tRNAPyl (16, 17, 20). Interestingly, it has been shown that UAG suppression is more efficient when MmPylRS is paired with Ma-tRNAPyl instead of its homologous tRNAPyl (Mm-tRNAPyl) (17). Indeed, we found that MmPylRS displayed twofold greater UAG readthrough with Ma-tRNAPyl compared with Mm-tRNAPyl, using two different ncAA substrates (Fig. S2). The increase in UAG readthrough when MmPylRS is paired with Ma-tRNAPyl might reflect higher activity of MmPylRS with this tRNAPyl; however, it is also possible that the observed increase in UAG readthrough is a result of more efficient UAG decoding by Ma-tRNAPyl in Escherichia coli or better compatibility of Ma-tRNAPyl with the E. coli ribosome. To determine if MmPylRS is more active with Ma-tRNAPyl, we performed additional in vitro aminoacylation assays with MmPylRS and MaPylRS paired with their nonhomologous tRNAPyl. In contrast to UAG readthrough data, we found that, while MmPylRS could aminoacylate Ma-tRNAPyl with Pyl (Km = 31 ± 11 μM, kcat = 1.2 ± 0.1 s−1 × 10−3), the aminoacylation efficiency was 10.7-fold lower than with its homologous tRNA (Table 1). The decrease in aminoacylation efficiency is a result of a sevenfold decrease in kcat; as expected, the Km for Pyl remained unchanged. These data indicate that the observed increase in UAG readthrough when MmPylRS is paired with Ma-tRNAPyl does not result from more efficient tRNA aminoacylation. We were unable to detect aminoacylation of Mm-tRNAPyl by MaPylRS, supporting in vivo data that show that MaPylRS does not recognize the M. mazei tRNAPyl (16, 17, 20) (Fig. S2).
Kinetic analysis of PylRS variants with ncAA substrates
In addition to Pyl, we also determined Km and kcat for two ncAAs that are known substrates of wildtype MmPylRS and MaPylRS, namely NƐ-boc-l-lysine (2, BocK) and NƐ-alloc-l-lysine (3, AlloK; Fig. 1). Despite MaPylRS having a nearly twofold higher Km for Pyl compared with MmPylRS, both enzymes showed similar Km values for the ncAAs BocK and AlloK, albeit 100-fold higher than the Km for Pyl (Table 1). These data indicate that MmPylRS and MaPylRS have a similar tolerance for these lysine-derived ncAAs.
MmPylRS and MaPylRS ncAA substrate range
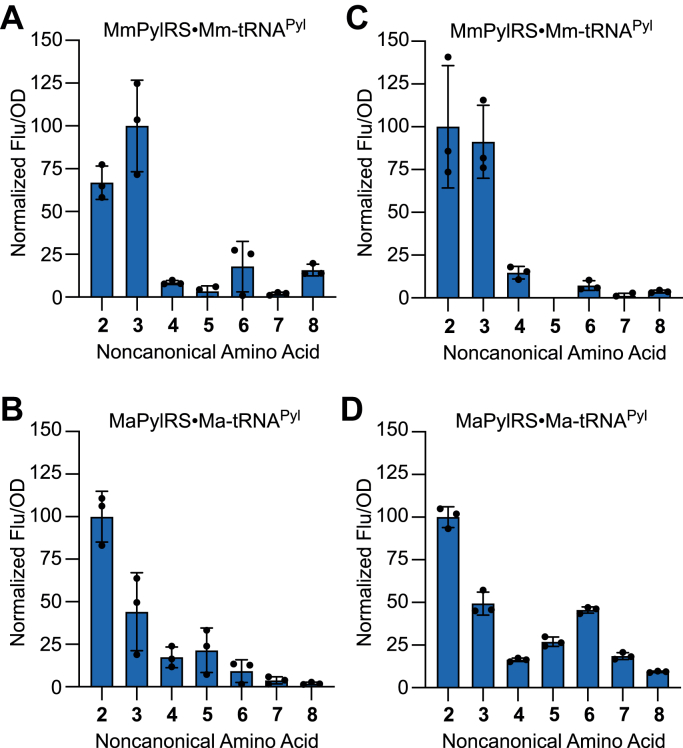
MmPylRS and MaPylRS share highly similar amino acid binding pockets, differing at only two positions: L309 and C348 in MmPylRS are replaced with methionine and valine, respectively, in MaPylRS (Fig. S3). Despite their similarities, studies have shown that MaPylRS and MmPylRS are different in terms of their ability to recognize certain ncAAs (17, 21). To investigate the substrate ranges of MmPylRS and MaPylRS, we performed in vivo UAG-readthrough assays using a library of 359 distinct ncAAs. For these assays, we used superfolder GFP (sfGFP) as a reporter of UAG suppression. We employed two different sfGFP reporters, one containing an in-frame UAG codon at position 2 (sfGFP-2am) and the other containing a UAG codon at position 27 (sfGFP-27am). The sfGFP-2am is an excellent reporter for ncAAs with long polar side chains, whereas sfGFP-27am is better suited for measuring the incorporation of hydrophobic and aromatic ncAAs (31).
We measured sfGFP-2am and sfGFP-27am expression in E. coli that were coexpressing MmPylRS or MaPylRS (along with their homologous tRNAPyl), in the presence of each one of the 359 unique ncAAs. While both wildtype enzymes showed high specificity, rejecting the majority of the ncAAs in our library, differences in substrate recognition of MmPylRS and MaPylRS were evident (Figs. S4 and S5). With the sfGFP-2am reporter, MmPylRS afforded robust sfGFP production with the two lysine analogs BocK and AlloK, as well as N-methyl-l-histidine (6, 3MeH), and ortho-fluoro-l-phenylalanine (8) (Fig. 3A). Similarly, sfGFP production was detected with MaPylRS in the presence of BocK, AlloK, and 3MeH. In addition to these ncAAs, MaPylRS also afforded sfGFP production in the presence of NƐ-boc-d-lysine (4, dBocK) and NƐ-(4-nitrocarbobenzyloxy)-l-lysine (5, NCBzK) (Fig. 3B). With the sfGFP-27am reporter, both enzymes enabled sfGFP synthesis in the presence of BocK, AlloK, ᴅBocK, and 3MeH; however, the sfGFP fluorescence signal in the presence of 3MeH was much higher with MaPylRS than with MmPylRS (Fig. 3, C and D). In addition, MaPylRS afforded significant sfGFP production in the presence of NCBzK and the fluorinated phenylalanine derivative trifluoro-l-phenylalanine (7) (Fig. 3D). Together, these data demonstrate a slightly expanded amino acid substrate spectrum for MaPylRS compared with MmPylRS.
Figure 3.
The substrate range of wildtype PylRS from Methanosarcina mazei and Candidatus Methanomethylophilus alvus. Substrate recognition was determined by measuring in vivo amber suppression using a superfolder GFP (sfGFP) reporter harboring a UAG codon at position 2 (A and B) or position 27 (C and D). sfGFP fluorescence was measured in Escherichia coli cells coexpressing the MmPylRS•Mm-tRNAPyl pair (A and C) or the MaPylRS•Ma-tRNAPyl pair (B and D), in GMML medium supplemented with 1 mM of an ncAA. Numbers correspond to the ncAAs shown in Figure 1. Data are presented as the mean ± SD for three biological replicates. MaPylRS, PylRS enzyme from Candidatus Methanomethylophilus alvus; Ma-tRNAPyl, the pyrrolysine tRNA from Candidatus Methanomethylophilus alvus; MmPylRS, PylRS enzyme from Methanosarcina mazei; Mm-tRNAPyl, the pyrrolysine tRNA from Methanosarcina mazei; ncAA, noncanonical amino acid; PylRS, pyrrolysyl-tRNA synthetase.
To compare the yield of purified proteins that can be obtained using these two PylRS variants, we expressed sfGFP-27am with the ncAA BocK, using either the MmPylRS•Mm-tRNAPyl pair or the MaPylRS•Ma-tRNAPyl pair, and then purified the resultant proteins via immobilized metal ion affinity chromatography. Consistent with an earlier study (18), we found that the MaPylRS•Ma-tRNAPyl pair afforded significantly more pure protein than the MmPylRS•Mm-tRNAPyl pair, with expression yields of 12.2 ± 1.2 and 4.3 ± 0.6 g per liter of culture, respectively (Fig. S6).
The aforementioned experiments, together with our previous data (14, 27), demonstrate that wildtype PylRS from diverse organisms is a catalytically competent aaRS in terms of activity and amino acid substrate specificity. However, to become the synthetic biologist’s workhorse, variants have been created with four or more amino acid substitutions. These engineered PylRS variants “degrade” the enzyme’s affinity for Pyl and extend the substrate range significantly to facilitate incorporation of a large variety of ncAAs into proteins.
Distribution of pyrrolysine encoding in archaea and bacteria
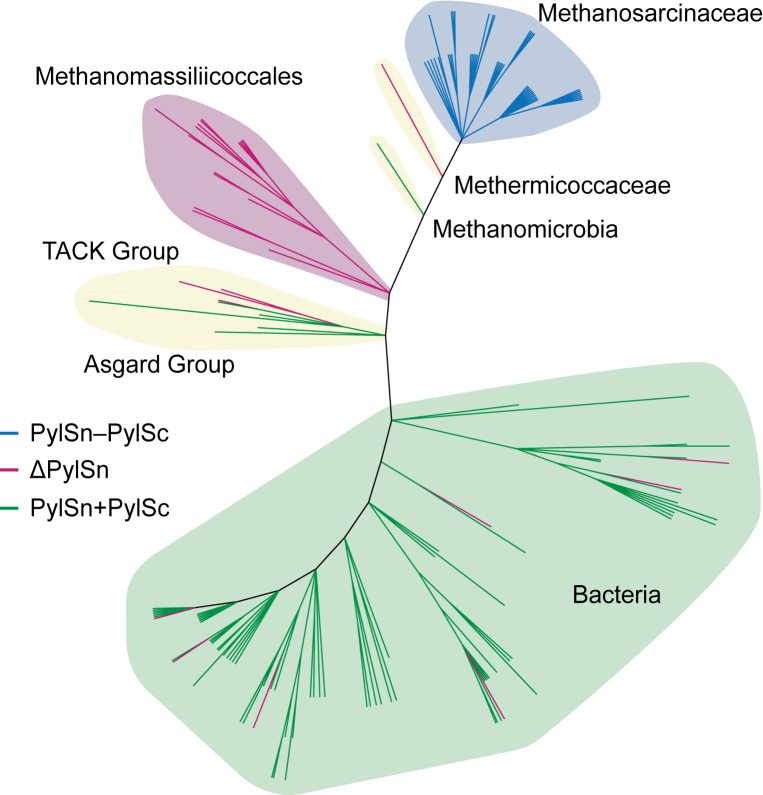
To investigate the phylogenetic distribution of PylRS subclasses in Pyl-encoding organisms, we searched publicly available databases for protein sequences with homology to PylSc. Several additional PylRS sequences were manually curated from recently published archaeal genomes (32). As a result of these searches, we identified PylRS genes in 156 diverse anaerobic bacteria and archaea (Fig. 4).
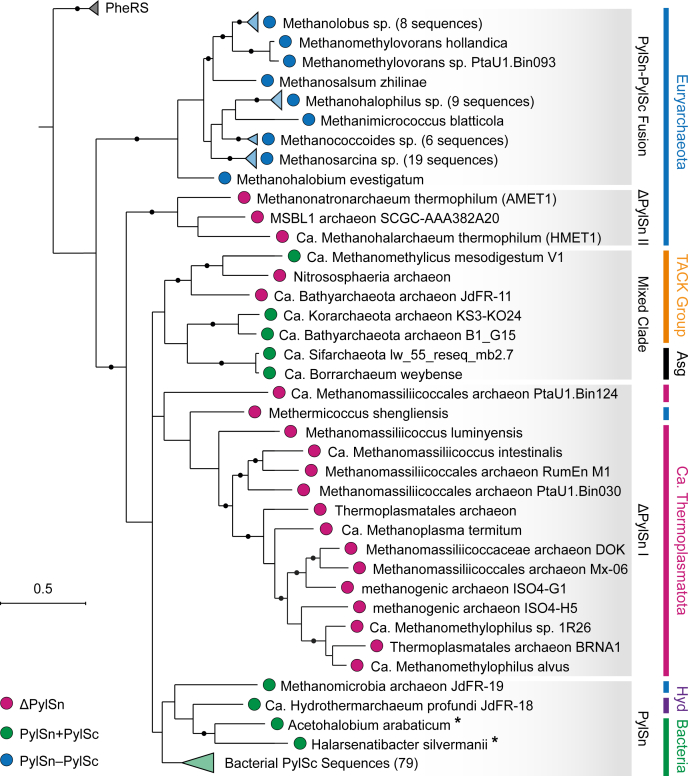
Figure 4.
Unrooted phylogenetic tree depicting the distribution of putative PylRS-encoding organisms. Branches are colored according to which type of PylRS gene (PylSn + PylSc, PylSn–PylSc fusion, or ΔPylSn) is present in the genome. The tree was constructed using phyloT (phylot.biobyte.de) and rendered using iTol. PylRS, pyrrolysyl-tRNA synthetase.
In archaea, we identified PylSc homologs in 75 organisms across eight phyla, including Euryarchaeota, Ca. Thermoplasmatota, Asgardarchaeota, Ca. Hydrothermarchaeota, and the TACK group phyla Thaumarchaeota, Ca. Bathyarchaeota, Ca. Verstraetearchaeota, and Ca. Korarchaeota. As far as we are aware, this is the first time that Pyl-encoding machinery has been identified in the Ca. Korarchaeota phylum. Of the 75 PylRS genes identified in archaea, 47 belong to the PylSn–PylSc fusion class of PylRS enzymes. We found that organisms encoding a PylSn–PylSc fusion enzyme form a monophyletic group comprised entirely of members of the family Methanosarcinaceae, in the order Methanosarcinales (Fig. 4). Within this family, we identified PylRS-encoding genes across eight of nine genera. The only other Pyl-encoding organism that we identified within the order Methanosarcinales is Methermicoccus shengliensis (33, 34). A phylogeny inferred using 122 16S ribosomal RNA sequences from putative Pyl-encoding organisms revealed that M. shengliensis is close relative of the Methanosarcinaceae (Fig. S7). Despite this close taxonomic relationship, M. shengliensis does not encode a PylSn–PylSc fusion enzyme but instead encodes a ΔPylSn class PylRS enzyme. Likewise, another closely related Pyl-encoding relative of the Methanosarcinaceae, Methanomicrobia archaeon JdFR-19, encodes a PylSn + PylSc class enzyme. Together, these observations suggest that fusion of PylSn and PylSc domains likely occurred as a single event in an ancestor of the Methanosarcinaceae.
In total, we identified 21 archaeal genomes encoding homologs of PylSc but not PylSn. For some of these organisms, the PylSn gene might not have been identified because of incomplete genome sequences. For example, the Nitrososphaeria archaeon (isolate SpSt-1131), whose PylRS is assigned to the ΔPylSn class, has an estimated genome completeness of only 68% (35). However, it is reasonable to assume that these organisms do not encode PylSn given that (1) a PylSn gene was not found in the available genome sequence and (2) molecular phylogeny (described later) shows that the PylSc proteins in these organisms are very similar to those from confirmed ΔPylSn organisms. As previously described, the majority of ΔPylSn enzymes (15 of 21 sequences) belong to the Methanomassiliicoccales, an order of methanogenic archaea associated with animal digestive tracts (9, 15, 16, 36). In addition to the Methanomassiliicoccales, which belong to the phylum Ca. Thermoplasmatota, our analysis shows that ΔPylSn enzymes are also present in archaea of the phylum Euryarchaeota, as well as the TACK group phyla Ca. Bathyarchaeota and Thaumarchaeota. Of the 75 PylSc-encoding archaeal genomes that we identified, only seven were found to encode PylSc and PylSn from distinct genes. These PylSn + PylSc class enzymes were found in two members of the phylum Asgardarchaeota, as well as, Euryarchaeota, Ca. Hydrothermarchaeota, and the TACK group phyla Ca. Bathyarchaeota, Ca. Korarchaeota, and Ca. Verstraetearchaeota.
The 81 remaining PylRS-encoding genomes that we identified belong to bacteria originating from four phyla. The majority of these sequences were found in the phylum Firmicutes, with the largest order, Clostridiales, having 41 representative sequences. In addition to Firmicutes, PylRS-encoding genes were found in 14 Deltaproteobacteria, two Actinobacteria, and one Spirochaetes. We identified PylSn homology in all but 11 of the bacterial genomes that encode PylSc. In all cases, PylSn and PylSc were encoded by distinct genes.
Molecular phylogeny of PylSc
To gain insight into the evolutionary history of PylRS, we performed molecular phylogeny using PylSc protein sequences predicted from PylRS-encoding genomes. A phylogenetic tree was inferred using a total of 156 PylSc sequences. PylRS is class II aaRS that shares a most recent common ancestor with PheRS (4, 11, 37). Therefore, we used five representative PheRS sequences from bacteria and archaea as an outgroup to root the tree.
In agreement with previous studies (15, 38), our phylogenetic analysis shows that most PylSc sequences delineate into three distinct clades corresponding to their domain architecture. These include a PylSn–PylSc fusion clade, a ΔPylSn clade, and a PylSn + PylSc clade (Figs. 5 and S8). In a previous analysis, however, it was noted that some sequences do not fit within this grouping. In particular, the Mediterranean Sea Brine Lakes 1 archaeon SCGC-AAA382A20 (MSBL1) PylRS, which is a ΔPylSn class enzyme, does not group within the previously identified ΔPylSn clade (38). Here, we found that the PylSc from MSBL1 instead groups with two unique ΔPylSn class enzymes from the recently identified species Ca. Methanohalarchaeum thermophilum (HMET1) and Methanonatronarchaeum thermophilum (39). Interestingly, all three members of this novel clade (ΔPylSn clade II) are halophiles and were isolated from similar hypersaline environments (39, 40). Given that they occupy similar habitats, horizontal gene transfer (HGT) provides a plausible explanation for the similarities in PylSc among these organisms, despite their relatively distant taxonomic relationships (Fig. S7) (41).
Figure 5.
Maximum-likelihood tree constructed using 156 PylSc protein sequences from bacteria and archaea. The tree was constructed with 100 replicates using MEGA X. Branches with bootstrap support values greater than 75% are indicated with a black circle. Bacterial PylSc sequences are indicated with an asterisk. (Asg = Asgardarchaeota; Hyd = Hydrothermarchaeota).
The archaeon HMET1 (42) harbors two genomic copies of both tRNAPyl and PylRS (43). In this species, one tRNAPyl isoacceptor gene (pylTG) contains the canonical G73 discriminator base, whereas the second tRNAPyl isoacceptor gene (pylTA) contains an unusual A73 at the discriminator position. Experiments in Haloferax volcanii have shown that these tRNAPyl isoacceptors are differentially aminoacylated by the two PylRS isoforms encoded in this organism. The PylRS2 isoform preferentially aminoacylates tRNAPyl with an A73 discriminator and has a motif 2 loop that is shortened by one amino acid compared with the PylRS1 isoform that aminoacylates tRNAPyl with a G73 discriminator (43). We found that the MSBL1 archaeon (40, 44) also harbors a PylRS with a similarly shortened motif 2 loop (40, 43). Furthermore, the tRNAPyl gene in the MSBL1 genome contains the unusual A73 discriminator (Table 2). An earlier study (43) characterized the amino acid length and composition of motif 2 loop that enables HMET PylRS2 to recognize the pylTA gene product, tRNAPylA. In this study, it was shown that when HMET PylRS2 contains a shortened motif 2 loop with the sequence DSKN, a sequence identical to that found in MSBL1, the mutant enzyme can aminoacylate tRNAPylA (lanes 3 and 4, Fig. 6B in Ref. (43)). Thus, HMET1 and MSBL1 of ΔPylSn clade II appear to utilize the same novel mechanism of tRNAPyl recognition, in which a PylRS variant with a shortened motif 2 loop recognizes a tRNAPyl isoacceptor with a unique A73 discriminator base. Notably, the MSBL1 genome is incomplete, and a pylTG and complete PylRS1 gene was not found in the available sequence.
Table 2.
Motif 2 loop sequences and tRNAPyl discriminator base identity of organisms encoding two ΔPylSn enzymes
In a previous study, it was shown that PylRS sequences from Ca. Bathyarchaeota and Ca. Methanomethylicus mesodigestum V1 form a novel clade (38, 45). Our results show that PylSc sequences from TACK group archaea (Nitrososphaeria archaeon, Ca. Bathyarchaeota archaeon JdFR-11, and Ca. Korarchaeota archaeon KS3-KO24) and two newly identified Asgardarchaeota also group within this novel clade. Interestingly, this mixed clade is comprised of PylSn + PylSc class and ΔPylSn class enzymes.
In addition to the possible HGT event described for ΔPylSn clade II, two other possible HGTs are evident in the PylRS tree. First, we found that the M. shengliensis PylSc is most similar to those in ΔPylSn clade I; however, this clade is otherwise entirely comprised of sequences from the Methanomassiliicoccales. This observation suggests possible HGT of the Pyl-encoding operon from the Methanomassiliicoccales to M. shengliensis. The similarities between the M. shengliensis and Methanomassiliicoccales PylSc, as well as other proteins within the Pyl operon, have been noted previously (38, 40). This proposed HGT is further supported by the phylogenetic analysis of tRNAPyl (see “Evolution of tRNAPyl” section). Second, we found that, despite originating from archaea, the PylSc sequences from Methanomicrobia archaeon JdFR-19 and Ca. Hydrothermarchaeum profundi are most similar to bacterial PylSc, in particular those from Acetohalobium arabaticum and Halarsenatibacter silvermanii. Similarities between the Pyl operon in A. arabaticum and archaea have been described previously and are believed to reflect HGT of the Pyl operon from archaea to bacteria (15, 38, 40). Here, our results show that PylSc from the archaea Methanomicrobia archaeon JdFR-19 and Ca. Hydrothermarchaeum profundi and the bacteria H. silvermanii also share these similarities, providing further support for this hypothesis.
Evolution of tRNAPyl
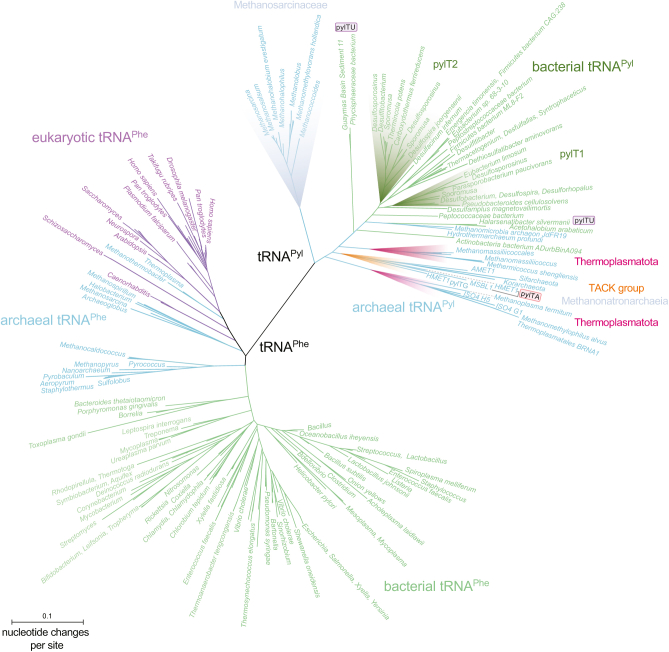
Based on our previous finding that PylRS emerged from duplication of an ancestral PheRS gene (11), we chose tRNAPhe sequences representing all three domains of life as an outgroup for our phylogenetic analysis of tRNAPyl. Indeed, the tRNAPhe sequences served as an ideal outgroup for the tRNAPyl phylogeny as the tRNAPhe and tRNAPyl sequences each clustered into well-supported monophyletic groups (Figs. 6, S9, and S10). Because of the small size of the tRNA, some of the deepest branches in the bacterial side of the tRNAPhe tree are less well supported compared with trees based on the ribosome or the aaRSs (11, 46). Nevertheless, the canonical three-domain phylogenetic pattern is evident in the tRNAPhe sequences with the bacterial sequences forming a grouping distinct and apart from the archaeal and eukaryotic sister lineages. Thus, the phylogeny indicates that the tRNAPyl and tRNAPhe genes diverged before the evolution of the three domains of life, placing an early limit on the evolution of the Pyl-decoding trait.
Figure 6.
Phylogeny of tRNAPheand tRNAPyl. Aligned tRNAPhe sequences were downloaded from the Sprinzl database and serve as an outgroup for a phylogeny of all known tRNAPyl genes (pylT). The distance-based phylogeny was calculated using the PhyML BioNJ algorithm PhyML (68) in SeaView (67). Bootstrap support is indicated in Fig. S9. Branches and species are color coded according to the primary phylogenetic domains: Eukaryota (purple), Archaea (cyan), and Bacteria (green). Subclades among the tRNAPyl sequences are highlighted, including the Methanosarcinaceae (blue), TACK group (orange), Ca. Thermoplasmatota (red), and bacterial pylT1 and pylT2 genes (green). Sequences with A73 (pylTA) or U73 (pylTU) discriminator bases are annotated.
The tRNAPyl phylogeny itself (Figs. S9 and S10) reveals several major subclades that are generally congruent with the clades identified in the PylRS phylogeny (Figs. 5 and S8). The deepest branching lineages in the tRNAPyl tree belong to diverse archaeal species, and the bacterial tRNAPyl sequences do not form a separate clade apart from the archaea. The phylogeny suggests that bacterial tRNAPyl is derived from the archaeal version, consistent with the phylogeny based on PylSc.
The tRNAPyl phylogeny further indicates that the Pyl trait evolved no later than the divergence of the main archaeal lines of descent. The deepest branches in the tRNAPyl tree separate the Methanosarcinaceae from several diverse archaeal groups that have retained the Pyl-decoding trait, including Ca. Korarchaeota and other members of the TACK group in addition to two clades of Ca. Thermoplasmatota. According to the maximum-likelihood phylogeny (Fig. S10), the bacterial tRNAPyl sequences form a well-supported group (bootstrap = 89) that diverged from the TACK and Ca. Thermoplasmatota group after divergence of Thermoplasmatota from the Euryarchaeota. Just as in the PylSc tree (Fig. 5), some euryarchaeal species do not form a clade with the Methanosarcinaceae. The Euryarchaeote Methanomicrobia archaeon JdFR-19 is in a deeply branching lineage closely related to Ca. Hydrothermarchaeum, and the Methanonatronarchaeia are also deeply branching but more similar to the tRNAPyl from Ca. Thermoplasmatota than to that from Methanosarcinaceae.
Several instances of gene duplication are evident in the tRNAPyl tree. As noted previously, the deeply branching HMET1 archaeon contains two tRNAPyl genes, one with the usual G73 (pylTG) discriminator and one with the orthogonal A73 discriminator base (pylTA). The maximum-likelihood tree (Fig. S10) suggests that the pylT duplication occurred after the divergence of Methanonatronarchaeum from Ca. Methanohalarchaeum. MSBL1 is the only other species with the A73 discriminator, and it is also the most closely related to (bootstrap = 85, Fig. S10), and likely derived from, the A73-continaing pylTA from Ca. Methanohalarchaeum.
Duplications of the pylT gene are even more common among the bacterial Pyl-decoding species. There is a relatively deep divergence separating the bacterial tRNAs into two clades (pylT1 and pylT2, green highlights, Fig. 6). Among the pylT2 clade, there is evidence of several additional and independent gene duplication events producing pylT2.1 and pylT2.2 sequences (Fig. S10). The Firmicute of the Negativicutes class, Sporomusa acidovorans, actually encodes three tRNAPyl genes, one of the pylT1 type and two from the pylT2 clade (pylT2.1 and pylT2.2). Finally, we identified three bacterial species with an unusual U73 discriminator base (pylTU). Evolution of the U73 discriminator appears to have occurred twice in the bacterial tRNAPyl. The pylTU-containing Phycisphaeraceae bacterium and Guaymas Basin Sediment 11 tRNAPyl sequences are closely related to each other (bootstrap = 100, Fig. S10) and are deeply branching with respect to other bacterial tRNAPyl species. The H. silvermanii tRNAPyl appears to be an independent change of G73 to U73. Although there are no biochemical data available for pylTU-encoding species, we anticipate that these tRNAs may represent yet another route to a mutually orthogonal tRNAPyl system, as was observed for pylTA (43).
Discussion
We and others have shown that the PylSn–PylSc fusion enzyme from M. mazei and the ΔPylSn enzyme from Ca. M. alvus display robust amber suppression activity in E. coli (16, 17, 21, 47). In this study, we found that, despite its high amber suppression efficiency, the aminoacylation activity of MaPylRS is threefold lower than MmPylRS with the native substrate Pyl. Several factors might account for the apparent discrepancy in aminoacylation efficiency and UAG readthrough of the MaPylRS•Ma-tRNAPyl pair. First, ΔPylSn enzymes have cognate tRNAs that are remarkably distinct from the tRNAs associated with PylSn–PylSc fusion and PylSn + PylSc enzymes (9). In terms of the Ca. M. alvus tRNAPyl, unique features include lack of a base between the acceptor and D stems, a shortened D loop, and an unpaired base in the anticodon stem. These features do not appear to be important for tRNA recognition by MaPylRS (17), thus, their exact functional role (if any) is unclear. It is possible that these features enable Ma-tRNAPyl to suppress amber codons more efficiently than Mm-tRNAPyl, at least in the context of the E. coli ribosome. In support of this hypothesis, our results using MmPylRS show that although Ma-tRNAPyl is aminoacylated less efficiently than Mm-tRNAPyl in vitro, UAG suppression in vivo is twofold greater with Ma-tRNAPyl than with Mm-tRNAPyl. A second factor that might account for the apparent discrepancy in aminoacylation and UAG suppression is post-transcriptional modifications of tRNAPyl. Certain post-transcriptional modifications are globally conserved in each domain of life and, while tRNAPyl is known to be modified in Methanosarcina (2), the modification status of heterologously expressed Mm-tRNAPyl and Ma-tRNAPyl is uncharacterized. The presence or the absence of modifications on Ma-tRNAPyl could improve its ability to suppress UAG codons in E. coli. Finally, a third factor that might compensate for the decreased aminoacylation activity of MaPylRS is the higher solubility of MaPylRS compared with PylSn–PylSc fusion enzymes. The N-terminal domain of PylRS is known to have low solubility, which complicated in vitro characterization and, for many years, precluded structure determination of PylSn (13, 14, 25, 48). Owing to its lack of PylSn, MaPylRS has an expression yield in E. coli ∼20-fold higher and is soluble at concentrations approximately fivefold higher than MmPylRS (18). Thus, while MaPylRS shows reduced aminoacylation activity compared with MmPylRS, this reduction in activity is likely compensated for by an increase in soluble MaPylRS expression and, possibly, increased amber suppression efficiency of Ma-tRNAPyl. In any case, the observation that MaPylRS has lower in vitro activity than MmPylRS suggests that MaPylRS can be optimized to improve its activity in E. coli.
Herein, we also demonstrated that MaPylRS has a greater amino acid substrate range than MmPylRS. Using in vivo UAG-suppression assays, we showed that, compared with MmPylRS, MaPylRS has a greater tolerance for structurally disparate ncAAs, including phenylalanine and histidine derivatives. These observations are in agreement with a previous study demonstrating greater tolerance of MaPylRS for substituted phenylalanine derivatives (21). The distinct substrate specificities of MaPylRS and MmPylRS might reflect subtle differences in the amino acid binding pockets of these enzymes, which differ at positions L309 and C348. To investigate how the substrate binding pocket varies amongst all known PylRS orthologs, we compared the identity of 12 residues that line the amino acid binding pocket and that are generally thought to influence the substrate specificity of PylRS (5). This analysis showed that while most residues in the substrate binding pocket are strictly conserved, there is considerable variability at positions L309, C348, and M350 (residues are numbered according to the MmPylRS sequence, File S1 and Fig. S11). We are currently investigating how the natural variability of PylRS orthologs might influence the substrate specificity of these enzymes. A second factor that might influence substrate recognition is the interaction of PylRS with tRNAPyl. It has been shown that tRNA–aaRS interactions can influence substrate binding (49), and, therefore, it is possible that the lack of the N-terminal tRNA-binding domain contributes to the differences in substrate recognition between MaPylRS and MmPylRS. The broader substrate spectrum of wildtype MaPylRS might prove to be a useful feature of this enzyme for applications in genetic code expansion; however, polyspecificity is not always a desirable feature of orthogonal aaRSs. This is especially true when multiple mutually orthogonal aaRSs are used to simultaneously install distinct ncAAs in the same cell (50). In these cases, overlapping substrates of polyspecific aaRSs can impede accurate translation of a further expanded genetic code.
In this study, we provided an updated molecular phylogenetic analysis of the catalytic domain of PylRS and tRNAPyl. We included several recently identified PylRS and tRNAPyl sequences that enable a better understanding of the evolutionary history of this aaRS•tRNA pair. It is hypothesized that PylRS originated via duplication of the PheRS gene (4, 11); however, when this event occurred is still an open question. Structure-based phylogenetic analysis suggests that PylRS is an ancient enzyme that was present in the microbial community prior to the emergence of the last universal common ancestor of life on earth (11, 51).
Our phylogeny inferred using tRNAPyl sequences agrees with those based on PylSc and points to an ancient origin. Namely, the data suggest that the Phe- and Pyl-decoding traits diverged from an ancestral aaRS•tRNA pair in an event that predated the divergence of bacteria, archaea, and eukaryotes. Despite the small size of the tRNA, tRNAPyl retains a record of its history that is generally congruent with the phylogeny of PylRS sequences. The tRNAPyl and tRNAPhe phylogeny shows that Pyl decoding evolved at the earliest sometime before the divergence of the three domains of life and at the latest before the divergence of the major archaeal phyla. The observation is also evident in the PylRS phylogeny and attests to the ancient origin of Pyl decoding.
The narrow taxonomic distribution of Pyl-decoding organisms, however, and the close linkages of Pyl decoding to methanogenesis, have led to the speculation that PylRS is a more recent archaeal invention, perhaps evolved specifically for methanogenesis and likely originating in an Euryarchaeote (15). The hypothesis that PylRS is a recent archaeal invention was proposed at a time when Pyl decoding was only known to exist in the Methanosarcinaceae, seventh order methanogens, and a few bacteria; however, our data show that Pyl decoding is widespread amongst archaea from diverse lineages representing multiple archaeal phyla. Moreover, the complete Pyl-decoding cassette was recently discovered for the first time in nonmethanogenic archaea, questioning the long-standing assumption that Pyl decoding is strictly tied to methanogenesis (32, 52). Taken together, these results challenge the hypothesis that PylRS emerged recently in archaea, strictly for the purpose of methanogenesis.
The tRNA trees also revealed the evolution of tRNAPyl genes with different and some mutually orthogonal versions that differ at the tRNA discriminator base at position 73 (pylTG, pylTA, and pylTU). In some of these cases, the tRNAPyl duplication events were accompanied by a duplication of PylRS. In other cases, such as in bacterial tRNAPyl sequences (pylT1 and pylT2), we saw evidence of both older and more recent gene duplications without coincident duplication of the PylRS. Thus, our analysis indicates that while tRNAPyl and PylRS normally coevolve, there are instances demonstrating independent evolution of the aaRS and tRNA. These duplications of PylRS and tRNAPyl are doubtless a rich source of aaRS•tRNA pairs for synthetic biology applications, and their existence suggests that microorganisms are capable of yet greater genetic code flexibility in nature.
While the catalytic domain of PylRS is hypothesized to be derived from PheRS, the origins of the N-terminal domain are less clear. Most studies on the evolutionary history of PylRS were conducted at a time when all known archaeal sequences belonged to either the PylSn–PylSc fusion or the ΔPylSn class of PylRS enzymes. This led to the assumption that PylSn + PylSc enzymes were unique to bacteria (1, 13, 14). A more recent comprehensive analysis using genomic and metagenomic data identified several archaeal PylRS sequences that encode PylSn and PylSc as distinct products; however, in most cases, taxonomic classification of these organisms was not possible because of gaps in genome sequences (38). Herein, we have identified additional archaea that encode PylSn + PylSc class PylRS enzymes; several with completely sequenced genomes enabling accurate taxonomic classification. These data show that, unlike PylSn–PylSc fusion and ΔPylSn enzymes, PylSn + PylSc enzymes are widespread in archaea. Given this broad taxonomic distribution, it is conceivable that the split PylRS represents a more ancient form the enzyme (Fig. S12, model 1). Under this model, a single domain fusion event in an ancestor of the Methanosarcinales would account for the monophyletic distribution of PylSn–PylSc fusion enzymes. Interestingly, in all the PylSn + PylSc-encoding archaea that we identified, the PylSn and PylSc genes are in close proximity in the genome, often overlapping or separated by a short stretch of nucleotides. In several cases, a single base pair insertion or deletion is all that is required to convert the split enzyme into a PylSn–PylSc fusion protein.
A second possibility, which is more parsimonious from a structural point of view, is that all extant PylRS enzymes are derived from a ΔPylSn ancestor (Fig. S12, model 2). However, this model is not in line with the currently available data when sequence similarity is considered. Because PylSn + PylSc and ΔPylSn enzymes are more similar to each other than to PylSn–PylSc enzymes, placing ΔPylSn as the ancestral variant implies that PylSn emerged twice during the evolution of PylRS (Fig. S12). We believe that this is much less likely than our proposed model (model 1) in which PylSn + PylSc is the ancestral variant. Model 1 is also more consistent with the widespread phylogenetic distribution of PylSn-encoding organisms. We note that since PylSn is not homologous to any domain of the closest relative of PylRS, PheRS, it is possible that a primordial PylRS existed before the evolution of PylSn. However, we neither have direct evidence of this nor are there known homologs of PylSn to provide further insight into the origin of this domain.
Assuming that extant PylRS enzymes are indeed derived from a PylSn + PylSc ancestor, we were curious as to what factors might have contributed to loss of PylSn in some organisms. Intriguingly, we found that in several archaea that encode a PylSn + PylSc enzyme, the PylSn gene initiates with the noncanonical start codons UUG or GUG. These alternate start codons likely minimize the expression of PylSn with respect to PylSc, which initiates with the canonical AUG (53). Substoichiometric expression of PylSn with respect to PylSc might have provided the original selective pressure for evolution of a PylSc domain with robust stand-alone activity. However, it is likely that additional selective pressures also contributed to loss of PylSn. One possibility is that genome streamlining was a driving force for loss of PylSn. Genome streamlining is selection that favors a reduction in overall genome size and is commonly observed in endosymbiotic organisms living in nutrient-rich environments (54). It has been shown that the process of streamlining can lead to mutations and deletions in the aaRSs of endosymbionts, especially in nonessential domains (55, 56). Consistent with the hypothesis that genome streamlining contributed to loss of PylSn is the fact that the largest monophyletic group of ΔPylSn-encoding archaea, the Methanomassiliicoccales, is comprised of organisms that are primarily endosymbiotic, many of which have been shown to have other hallmarks of genome streamlining, for example, a decrease in overall genome size, increase in gene coding density, and the absence of many common metabolic genes (36, 57, 58, 59). Interestingly, we found that archaea that encode ΔPylSn enzymes have genomes that are on average 1.8-fold smaller than organisms that encode full-length PylRS (File S13), further supporting the hypothesis that genome streamlining might have contributed to loss of PylSn, at least in the case of the Methanomassiliicoccales.
Experimental procedures
Phylogenetic analysis of PylSc
PylRS sequences were retrieved from National Center for Biotechnology Information databases using BlastP. For initial searches, the full-length PylSc sequence from Desulfitobacterium hafniense and the 270 C-terminal residues of M. mazei were used as a query. A subsequent search was performed using the PylSc sequence from Ca. Bathyarchaeota archaeon B1 G15, which identified more disparate PylSc sequences. PylSn protein sequences were retrieved in the same way using the sequence from D. hafniense as a query. For the phylogenetic analysis based on PylSc, protein sequences were aligned using the MUSCLE algorithm (60) and manually trimmed. The phylogenetic tree was constructed in MEGA X (61) using the maximum-likelihood method (100 replicates) with default settings. The PheRS and PylRS sequences used for this analysis are available in File S2. For the phylogenetic analysis based on 16S rRNA sequences, assembled genomes of PylRS-encoding organisms were retrieved from public databases and 16S rRNA sequences were extracted using the ContEst16S webtool (62). The 16S rRNA sequences were aligned using the MUSCLE algorithm (60) and manually trimmed. The phylogenetic tree was constructed in MEGA X using the maximum-likelihood method (100 replicates) with default settings.
Phylogenetic analysis of tRNAPyl
All tRNAPhe sequences (260) were downloaded in aligned format from the Sprinzl database (63). PylRS-encoding genomes were downloaded from National Center for Biotechnology Information nonredundant sequence database (64) and the Joint Genomes Institute integrated microbial genomics (65) database, and tRNAPyl sequences were extracted using the ARAGORN server (66). The tRNAPyl sequences were aligned to the tRNAPhe outgroup by aligning conserved stem and loop segments of the tRNA secondary structure. The program SeaView (67) was used to manually align the tRNA sequences. The complete set of aligned tRNAPhe and tRNAPyl sequences is included in File S3.
Phylogenetic trees were calculated using both distance-based (Figs. 6 and S9) and maximum-likelihood methods (Fig. S10) in the PhyML package (68) inside the SeaView alignment editor (67). The distance-based trees were computed using the BioNJ algorithm in PhyML, and 1000 pseudoreplicate datasets were used to determine bootstrap support values. The maximum-likelihood tree was calculated starting from 100 random trees and using PhyML and a GTR substitution model with eight rate categories, the gamma value and number of invariable sites was based on the maximum-likelihood estimates, and empirical nucleotide frequencies. The tree topology was optimized using the best of nearest neighbor interchanges and the subtree pruning and regrafting algorithms. The following PhyML command was used: phyml -d DNA -m GTR -c 8 -a e -f e -v e -s BEST -o tlr -b -4. Bootstrap supports were calculated based on the Shimodaira–Hasegawa (69) approximate likelihood-ratio test in PhyML (68).
ncAAs
Synthesis of enantiomerically pure l-pyrrolysine for in vitro aminoacylation assays was described previously (30). The preparation and composition of the 359-ncAA library for determining MmPylRS and MaPylRS substrate ranges was also described previously (31). All other ncAAs used in this study were sourced from commercial vendors and used without further purification.
Preparation of tRNA transcripts
tRNA transcripts were prepared from synthetic oligonucleotides using purified recombinant T7 RNA polymerase as described previously (70, 71). Briefly, oligonucleotides containing the various tRNAPyl sequences and a T7 promoter were synthesized by the W.M. Keck Biotechnology Resource Laboratory at Yale University. Synthetic oligonucleotides were designed with 2′-methoxyguanine at the penultimate position of the 5′ end to reduce nontemplated nucleotide addition (72). After in vitro transcription with T7 RNA polymerase, the tRNA transcripts were purified using a 12% polyacrylamide gel containing 7 M urea. Purified tRNA transcripts were dissolved in RNAse-free water and refolded by heating to 80 °C for 10 min, followed by slowly cooling to room temperature over 10 min. The refolded tRNAs were directly used for aminoacylation experiments.
Expression of PylRS variants and in vitro aminoacylation
N-terminally His6-tagged MmPylRS, MbPylRS, chPylRS, and MaPylRS were expressed from pET15b plasmids in E. coli strain BL21(DE3). Protein expression was induced with 1 mM IPTG at 37 °C with shaking. After 3 h, cells were collected by centrifugation at 5000 rpm for 10 min and then lysed by sonication. The lysates were clarified by centrifugation, and PylRS enzymes were purified from the clarified lysate by nickel affinity chromatography using a gravity-flow nickel–nitrilotriacetic acid column, following the manufacturer's protocol. Aminoacylation assays were performed at 37 °C in buffer (100 mM Hepes [pH 7.2], 25 mM MgCl2, 60 mM NaCl, 5 mM ATP, and 1 mM DTT) using 15 μM of tRNAPyl (labeled at the 3′ end with [α-32P]-ATP), 1 μM of purified recombinant enzymes, and amino acid concentrations ranging from 0.25 to eightfold Km, as described previously (27). Aminoacylation was monitored by separating charged from uncharged tRNA exactly as described previously (27).
PylRS tRNA crossrecognition
E. coli strain DH10B was cotransformed with a pBAD plasmid, harboring the sfGFP[2UAG] and Ca. M. alvus or M. mazei pylT genes, and pMW plasmid harboring the wildtype MaPylRS or MmPylRS genes. Freshly transformed colonies were isolated and grown to saturation in 2× YT media supplemented with ampicillin (Amp; 100 μg/ml) and spectinomycin (Spec; 100 μg/ml). Saturated cultures (5 μl) were used to inoculate 150 μl of chemically defined media (47), supplemented with IPTG, arabinose, and 1 mM BocK or AlloK, in a black 96-well plate. Replicate wells with no added ncAA were used to measure background signals. Cultures were incubated at 37 °C in microplate reader (BioTek), and fluorescence intensity (λex = 485 nm, λem = 535 nm) and absorbance at 600 nm were measured every 15 min for 24 h. Data are reported as the fluorescence intensity divided by the absorbance at 600 nm at the 24 h time point after background subtraction.
Substrate range of PylRS variants
For measuring PylRS substrate specificity, E. coli BL21(DE3) were cotransformed with a pET plasmid, harboring the sfGFP[2UAG] or sfGFP[27UAG] and Ca. M. alvus or M. mazei pylT genes and a pCDF plasmid encoding MmPylRS or MaPylRS. Freshly transformed colonies were isolated and cultured in LB media (25 ml) supplemented with Amp (100 μg/ml) and Spec (100 μg/ml) at 37 °C until an absorbance of 0.6 to 0.8 at 600 nm. Cells were harvested by centrifugation, washed twice with M9 salt solution, and then resuspended in GMML medium (M9 salt solution, 1% glycerol, 2 mM MgSO4, and 0.1 mM CaCl2) supplemented with 1 mM IPTG. After washing, aliquots (50 μl) of the cell suspension were loaded into 384-well plates containing 1 mM of each ncAA. Resuspended cell cultures were incubated in a microplate reader (BioTek) at 37 °C, and the fluorescence intensity (λex = 485 nm, λem = 535 nm) and absorbance at 595 nm were monitored continuously for 12 h. Wells A1–2, B1–2, and C1–2 (C0) did not include IPTG or an ncAA. Wells D1–2, E1–2, and F1–2 (C1) did not include IPTG. C1 wells were used as negative controls to subtract the background signal. Data are reported as the fluorescence intensity, divided by the absorbance at 595 nm, at the 12 h time point, following subtraction of the background signal. After an initial screen using the full 359-ncAA library, the aforementioned assay was repeated using only the ncAAs that afforded appreciable sfGFP production (2–8). For the repeat assay, freshly transformed cells were grown overnight in LB containing Amp and Spec (100 μg/ml each), and then overnight cultures (5 μl) were used to inoculate 150 μl of defined media supplemented with Amp and Spec (100 μg/ml each), 1 mM IPTG, and 1 mM of ncAA 2 to 8, in black, clear-bottom, and 96-well plates. Plates were incubated with 12 min of continuous shaking every 15 min, at 37 °C in a BioTek Synergy HT microplate reader. Fluorescence intensity (λex = 485 nm, λem = 528 nm) and absorbance at 600 nm were measured every 15 min for 20 h. All experiments were performed with three biological replicates, and data are reported as the fluorescence intensity, divided by the absorbance at 600 nm at the 20 h time point. Data in Figure 3 were normalized where 0% corresponds to the background fluorescence/absorbance value in the absence of an ncAA, and 100% corresponds to the maximum obtained fluorescence/absorbance value.
Expression and purification of sfGFP containing ncAAs
Chemically competent E. coli BL21(DE3) was cotransformed with a pET plasmid containing sfGFP[27UAG] and the M. alvus or M. mazei pylT gene and a pCDF plasmid carrying MaPylRS or MmPylRS. The cotransformed cells were plated on LB agar supplemented with Amp (100 μg/ml) and Spec (100 μg/ml) and grown overnight at 37 °C. Single colonies were cultured in 10 ml LB media supplemented with appropriate antibiotics and grown overnight. The overnight cultures were used to inoculate 1 l of LB containing antibiotics, and cells were grown at 37 °C with continuous shaking until the absorbance at 600 nm reached 0.6 to 0.8. sfGFP expression was induced with 1 mM IPTG and 1 mM of BocK at 37 °C overnight. The overnight cultures were pelleted by centrifugation (6000g, 20 min), and the pellet was resuspended in lysis buffer (200 mM NaCl, 50 mM Tris, pH 7.5) and lysed by sonication. The lysate was clarified by centrifugation (12,000g, 40 min), and the supernatant was loaded onto a gravity flow column containing pre-equilibrated nickel–nitrilotriacetic acid resin. The resin was washed with 10 column volumes of wash buffer (200 mM NaCl, 50 mM Tris, 20 mM imidazole, pH 7.5), and the sfGFP was eluted using five column volumes of elution buffer (200 mM NaCl, 50 mM Tris, 200 mM imidazole, pH 7.5). The buffer was changed, and protein was concentrated, using Amicon Ultra-4 Centrifugal Filters. The concentration of the purified protein was determined by measuring the absorbance at 280 nm and using a calculated extinction coefficient of 18,910 M−1 cm−1. Yield values for three biological replicates are given in Fig. S6.
Data availability
The raw data for this study are available from the corresponding author upon request.
Supporting information
This article contains supporting information (67, 68).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Profs Oscar Vargas-Rodriguez, Sergey V. Melnikov, and Ilka Heinemann for helpful discussions and Christopher A. Jahn for early experimental contributions. We thank Jiarui Sun and Dr Christian Rinke of the Australian Centre for Ecogenomics at The University of Queensland for providing Asgardarchaeota PylRS sequences.
Author contributions
L.-T. G., K. A., Y.-S. W., D. S., and J. M. T. conceptualization; L.-T. G., K. A., H.-K. J., T. M., X. F., P. O., and J. M. T. investigation; P. O., D. S., and J. M. T. writing–original draft; D.S. supervision.
Funding and additional information
H.-K. J. holds a graduate student fellowship of the Taiwan Academic Talents Overseas Advancement Program of the Ministry of Science and Technology (MOST; grant no.: 110-2917-I-007-006). J. M. T. is a postdoctoral fellow supported by a National Institutes of Health Pathway to Independence Award (grant no.: K99GM141320). This work was supported by grants from the National Institute of General Medical Sciences (grant no.: R35GM122560; to D. S.) and Department of Energy Office of Basic Energy Sciences (grant no.: DE-FG02-98ER20311; to D. S.); the National Natural Science Foundation of China (grant no.: 31901029; to X. F.) and the Natural Science Foundation of Guangdong Province, China (grant no.: 2021A1515010995; to X. F.); Academia Sinica and the Taiwan Ministry of Science and Technology (MOST 107-2113-M-001-025-MY3 and MOST 110–2113-M-001-044; to Y.-S. W.); the Natural Sciences and Engineering Research Council of Canada (grant no.: 04282; to P. O.), Canada Research Chairs (grant no.: 232341; to P. O.), and the Canadian Institutes of Health Research (grant no.: 165985; to P. O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biographies

Li-Tao Guo is currently an associate research scientist in the Department of Molecular, Cellular, and Developmental Biology at Yale University. He is passionate about developing novel biotechnology. After a stint in field of genetic code expansion, he began developing new RNA sequencing technology around an ultra-processive reverse transcriptase. This transcriptase was discovered in the lab of Anna Pyle at Yale University, and is revolutionizing our ability to interrogate the transctriptome.

Kazuaki Amikura is currently a postdoctoral researcher at the Institute of Space and Astronautical Science, Japan Aerospace Exploration Agency. His general interest is in understanding and repurposing the translation machinery for synthetic biology. His current research focuses on understanding the evolution of protein synthesis machinery on earth.

Han-Kai Jiang is a PhD student at Academia Sinica and National Tsing Hua University, Taiwan. He is currently visiting the Department of Molecular Biophysics and Biochemistry at Yale University. His research is focused on chemical and synthetic biology, and he is interested in utilizing non-canonical amino acids as tools to study enzyme biochemistry and protein post-translational modifications. His research at Yale focuses on developing novel biosensors based on aminoacyl-tRNA synthetases.
Edited by Karin Musier-Forsyth
Footnotes
Present address for Jeffery M. Tharp: Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN, USA.
Supporting information
References
- 1.Gaston M.A., Jiang R., Krzycki J.A. Functional context, biosynthesis, and genetic encoding of pyrrolysine. Curr. Opin. Microbiol. 2011;14:342–349. doi: 10.1016/j.mib.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polycarpo C., Ambrogelly A., Bérubé A., Winbush S.M., McCloskey J.A., Crain P.F., et al. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight S.K., Larue R.C., Mahapatra A., Longstaff D.G., Chang E., Zhao G., et al. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 4.Englert M., Moses S., Hohn M., Ling J., O'Donoghue P., Söll D. Aminoacylation of tRNA 2'- or 3'-hydroxyl by phosphoseryl- and pyrrolysyl-tRNA synthetases. FEBS Lett. 2013;587:3360–3364. doi: 10.1016/j.febslet.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan W., Tharp J.M., Liu W.R. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta. 2014;1844:1059–1070. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crnković A., Suzuki T., Söll D., Reynolds N.M. Pyrrolysyl-tRNA synthetase, an aminoacyl-tRNA synthetase for genetic code expansion. Croat. Chem. Acta. 2016;89:163–174. doi: 10.5562/cca2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanagisawa T., Umehara T., Sakamoto K., Yokoyama S. Expanded genetic code technologies for incorporating modified lysine at multiple sites. ChemBioChem. 2014;15:2181–2187. doi: 10.1002/cbic.201402266. [DOI] [PubMed] [Google Scholar]
- 8.Ambrogelly A., Gundllapalli S., Herring S., Polycarpo C., Frauer C., Söll D. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tharp J.M., Ehnbom A., Liu W.R. tRNAPyl: structure, function, and applications. RNA Biol. 2018;15:441–452. doi: 10.1080/15476286.2017.1356561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krahn N., Tharp J.M., Crnković A., Söll D. Engineering aminoacyl-tRNA synthetases for use in synthetic biology. Enzymes. 2020;48:351–395. doi: 10.1016/bs.enz.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kavran J.M., Gundllapalli S., O'Donoghue P., Englert M., Söll D., Steitz T.A. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nozawa K., O'Donoghue P., Gundllapalli S., Araiso Y., Ishitani R., Umehara T., et al. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature. 2009;457:1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang R., Krzycki J.A. PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine. J. Biol. Chem. 2012;287:32738–32746. doi: 10.1074/jbc.M112.396754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T., Miller C., Guo L.-T., Ho J.M.L., Bryson D.I., Wang Y.-S., et al. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 2017;13:1261–1266. doi: 10.1038/nchembio.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borrel G., Gaci N., Peyret P., O'Toole P.W., Gribaldo S., Brugère J.-F. Unique characteristics of the pyrrolysine system in the 7th order of methanogens: implications for the evolution of a genetic code expansion cassette. Archaea. 2014;2014 doi: 10.1155/2014/374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willis J.C.W., Chin J.W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem. 2018;10:831–837. doi: 10.1038/s41557-018-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi A., Iraha F., Ohtake K., Sakamoto K. Pyrrolysyl-tRNA synthetase with a unique architecture enhances the availability of lysine derivatives in synthetic genetic codes. Molecules. 2018;23:2460. doi: 10.3390/molecules23102460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki E., Yanagisawa T., Kuratani M., Sakamoto K., Yokoyama S. Fully productive cell-free genetic code expansion by structure-based engineering of Methanomethylophilus alvus pyrrolysyl-tRNA synthetase. ACS Synth. Biol. 2020;9:718–732. doi: 10.1021/acssynbio.9b00288. [DOI] [PubMed] [Google Scholar]
- 19.Beránek V., Willis J.C.W., Chin J.W. An evolved Methanomethylophilus alvus pyrrolysyl-tRNA synthetase/tRNA pair is highly active and orthogonal in mammalian cells. Biochemistry. 2019;58:387–390. doi: 10.1021/acs.biochem.8b00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meineke B., Heimgärtner J., Lafranchi L., Elsässer S.J. Methanomethylophilus alvus Mx1201 provides basis for mutual orthogonal pyrrolysyl tRNA/aminoacyl-tRNA synthetase pairs in mammalian cells. ACS Chem. Biol. 2018;13:3087–3096. doi: 10.1021/acschembio.8b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tharp J.M., Vargas-Rodriguez O., Schepartz A., Söll D. Genetic encoding of three distinct noncanonical amino acids using reprogrammed initiator and nonsense codons. ACS Chem. Biol. 2021;16:766–774. doi: 10.1021/acschembio.1c00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao L., Liu J., Ghelichkhani F., Rozovsky S., Wang L. Genetic incorporation of ϵ-N-benzoyllysine by engineering Methanomethylophilus alvus pyrrolysyl-tRNA synthetase. ChemBioChem. 2021;22:2530–2534. doi: 10.1002/cbic.202100218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Cao L., Klauser P.C., Cheng R., Berdan V.Y., Sun W., et al. A genetically encoded fluorosulfonyloxybenzoyl-l-lysine for expansive covalent bonding of proteins via SuFEx chemistry. J. Am. Chem. Soc. 2021;143:10341–10351. doi: 10.1021/jacs.1c04259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunkelmann D.L., Willis J.C.W., Beattie A.T., Chin J.W. Engineered triply orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs enable the genetic encoding of three distinct non-canonical amino acids. Nat. Chem. 2020;12:535–544. doi: 10.1038/s41557-020-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herring S., Ambrogelly A., Gundllapalli S., O'Donoghue P., Polycarpo C.R., Söll D. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007;581:3197–3203. doi: 10.1016/j.febslet.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumas A., Lercher L., Spicer C.D., Davis B.G. Designing logical codon reassignment – expanding the chemistry in biology. Chem. Sci. 2015;6:50–69. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo L.-T., Wang Y.-S., Nakamura A., Eiler D., Kavran J.M., Wong M., et al. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16724–16729. doi: 10.1073/pnas.1419737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanagisawa T., Ishii R., Fukunaga R., Kobayashi T., Sakamoto K., Yokoyama S. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J. Mol. Biol. 2008;378:634–652. doi: 10.1016/j.jmb.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 29.Wolfson A.D., Pleiss J.A., Uhlenbeck O.C. A new assay for tRNA aminoacylation kinetics. RNA. 1998;4:1019–1023. doi: 10.1017/s1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong M.L., Guzei I.A., Kiessling L.L. An asymmetric synthesis of l-pyrrolysine. Org. Lett. 2012;14:1378–1381. doi: 10.1021/ol300045c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H.-K., Lee M.-N., Tsou J.-C., Chang K.-W., Tseng H.-W., Chen K.-P., et al. Linker and N-terminal domain engineering of pyrrolysyl-tRNA synthetase for substrate range shifting and activity enhancement. Front. Bioeng. Biotechnol. 2020;8:235. doi: 10.3389/fbioe.2020.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun J., Evans P.N., Gagen E.J., Woodcroft B.J., Hedlund B.P., Woyke T., et al. Recoding of stop codons expands the metabolic potential of two novel Asgardarchaeota lineages. ISME Commun. 2021;1:30. doi: 10.1038/s43705-021-00032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oren A. In: The Prokaryotes: Other Major Lineages of Bacteria and the Archaea. Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. The family Methermicoccaceae; pp. 307–309. [Google Scholar]
- 34.Cheng L., Qiu T.-L., Yin X.-B., Wu X.-L., Hu G.-Q., Deng Y., et al. Methermicoccus shengliensis gen. nov., sp. nov., a thermophilic, methylotrophic methanogen isolated from oil-production water, and proposal of Methermicoccaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2007;57:2964–2969. doi: 10.1099/ijs.0.65049-0. [DOI] [PubMed] [Google Scholar]
- 35.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cozannet M., Borrel G., Roussel E., Moalic Y., Allioux M., Sanvoisin A., et al. New insights into the ecology and physiology of Methanomassiliicoccales from terrestrial and aquatic environments. Microorganisms. 2021;9:30. doi: 10.3390/microorganisms9010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko J.-H., Wang Y.-S., Nakamura A., Guo L.-T., Söll D., Umehara T. Pyrrolysyl-tRNA synthetase variants reveal ancestral aminoacylation function. FEBS Lett. 2013;587:3243–3248. doi: 10.1016/j.febslet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukai T., Crnković A., Umehara T., Ivanova N.N., Kyrpides N.C., Söll D. RNA-dependent cysteine biosynthesis in bacteria and archaea. mBio. 2017;8 doi: 10.1128/mBio.00561-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorokin D.Y., Merkel A.Y., Abbas B., Makarova K.S., Rijpstra W.I.C., Koenen M., et al. Methanonatronarchaeum thermophilum gen. nov., sp. nov. and 'Candidatus Methanohalarchaeum thermophilum', extremely halo(natrono)philic methyl-reducing methanogens from hypersaline lakes comprising a new euryarchaeal class Methanonatronarchaeia classis nov. Int. J. Syst. Evol. Microbiol. 2018;68:2199–2208. doi: 10.1099/ijsem.0.002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan Y., Haroon M.F., Alam I., Ferry J.G., Stingl U. Single-cell genomics reveals pyrrolysine-encoding potential in members of uncultivated archaeal candidate division MSBL1. Environ. Microbiol. Rep. 2017;9:404–410. doi: 10.1111/1758-2229.12545. [DOI] [PubMed] [Google Scholar]
- 41.Spang A., Caceres E.F., Ettema T.J.G. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science. 2017;357:563. doi: 10.1126/science.aaf3883. [DOI] [PubMed] [Google Scholar]
- 42.Sorokin D.Y., Makarova K.S., Abbas B., Ferrer M., Golyshin P.N., Galinski E.A., et al. Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat. Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Gong X., Zhao Q., Mukai T., Vargas-Rodriguez O., Zhang H., et al. The tRNA discriminator base defines the mutual orthogonality of two distinct pyrrolysyl-tRNA synthetase/tRNAPyl pairs in the same organism. Nucl. Acids Res. 2022;50:4601–4615. doi: 10.1093/nar/gkac271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mwirichia R., Alam I., Rashid M., Vinu M., Ba-Alawi W., Kamau A.A., et al. Metabolic traits of an uncultured archaeal lineage -MSBL1- from brine pools of the Red Sea. Sci. Rep. 2016;6 doi: 10.1038/srep19181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanwonterghem I., Evans P.N., Parks D.H., Jensen P.D., Woodcroft B.J., Hugenholtz P., et al. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.170. [DOI] [PubMed] [Google Scholar]
- 46.Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tharp J.M., Ad O., Amikura K., Ward F.R., Garcia E.M., Cate J.H.D., et al. Initiation of protein synthesis with non-canonical amino acids in vivo. Angew. Chem. Int. Ed. Engl. 2020;59:3122–3126. doi: 10.1002/anie.201914671. [DOI] [PubMed] [Google Scholar]
- 48.Yanagisawa T., Ishii R., Fukunaga R., Nureki O., Yokoyama S. Crystallization and preliminary X-ray crystallographic analysis of the catalytic domain of pyrrolysyl-tRNA synthetase from the methanogenic archaeon Methanosarcina mazei. Acta Crystallogr. F. 2006;62:1031–1033. doi: 10.1107/S1744309106036700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ibba M., Hong K.-W., Sherman J.M., Sever S., Söll D. Interactions between tRNA identity nucleotides and their recognition sites in glutaminyl-tRNA synthetase determine the cognate amino acid affinity of the enzyme. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6953–6958. doi: 10.1073/pnas.93.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng Y., Addy P.S., Mukherjee R., Chatterjee A. Defining the current scope and limitations of dual noncanonical amino acid mutagenesis in mammalian cells. Chem. Sci. 2017;8:7211–7217. doi: 10.1039/c7sc02560b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fournier G. Horizontal gene transfer and the evolution of methanogenic pathways. Met. Mol. Biol. 2009;532:163–179. doi: 10.1007/978-1-60327-853-9_9. [DOI] [PubMed] [Google Scholar]
- 52.Brugère J.-F., Atkins J.F., O'Toole P.W., Borrel G. Pyrrolysine in archaea: a 22nd amino acid encoded through a genetic code expansion. Emerg. Top. Life Sci. 2018;2:607–618. doi: 10.1042/ETLS20180094. [DOI] [PubMed] [Google Scholar]
- 53.Tharp J.M., Krahn N., Varshney U., Söll D. Hijacking translation initiation for synthetic biology. ChemBioChem. 2020;21:1387–1396. doi: 10.1002/cbic.202000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wernegreen J.J. In it for the long haul: evolutionary consequences of persistent endosymbiosis. Curr. Opin. Genet. Dev. 2017;47:83–90. doi: 10.1016/j.gde.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Melnikov S.V., van den Elzen A., Stevens D.L., Thoreen C.C., Söll D. Loss of protein synthesis quality control in host-restricted organisms. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11505–E11512. doi: 10.1073/pnas.1815992115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melnikov S.V., Rivera K.D., Ostapenko D., Makarenko A., Sanscrainte N.D., Becnel J.J., et al. Error-prone protein synthesis in parasites with the smallest eukaryotic genome. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E6245–E6253. doi: 10.1073/pnas.1803208115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Söllinger A., Schwab C., Weinmaier T., Loy A., Tveit A.T., Schleper C., et al. Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiol. Ecol. 2016;92:fiv149. doi: 10.1093/femsec/fiv149. [DOI] [PubMed] [Google Scholar]
- 58.Borrel G., Parisot N., Harris H.M.B., Peyretaillade E., Gaci N., Tottey W., et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics. 2014;15:679. doi: 10.1186/1471-2164-15-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borrel G., Harris H.M.B., Parisot N., Gaci N., Tottey W., Mihajlovski A., et al. Genome sequence of "Candidatus Methanomassiliicoccus intestinalis" Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc. 2013;1 doi: 10.1128/genomeA.00453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar R.C. Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee I., Chalita M., Ha S.-M., Na S.-I., Yoon S.-H., Chun J. ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017;67:2053–2057. doi: 10.1099/ijsem.0.001872. [DOI] [PubMed] [Google Scholar]
- 63.Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucl. Acids Res. 2009;37:D159–162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., et al. Database resources of the national center for biotechnology information. Nucl. Acids Res. 2022;50:D20–D26. doi: 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen I.-M.A., Chu K., Palaniappan K., Ratner A., Huang J., Huntemann M., et al. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucl. Acids Res. 2021;49:D751–D763. doi: 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laslett D., Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucl. Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 68.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 69.Shimodaira H., Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- 70.Korencić D., Söll D., Ambrogelly A. A one-step method for in vitro production of tRNA transcripts. Nucl. Acids Res. 2002;30:e105. doi: 10.1093/nar/gnf104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ellinger T., Ehricht R. Single-step purification of T7 RNA polymerase with a 6-histidine tag. Biotechniques. 1998;24:718–720. doi: 10.2144/98245bm03. [DOI] [PubMed] [Google Scholar]
- 72.Kao C., Zheng M., Rüdisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3' terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data for this study are available from the corresponding author upon request.