Abstract
Biallelic pathogenic variants in SZT2 result in a neurodevelopmental disorder with shared features, including early-onset epilepsy, developmental delay, macrocephaly, and corpus callosum abnormalities. SZT2 is as a critical scaffolding protein in the amino acid sensing arm of the mTORC1 signalling pathway. Due to its large size (3432 amino acids), lack of crystal structure, and absence of functional domains, it is difficult to determine the pathogenicity of SZT2 missense and in-frame deletions, but these variants are increasingly detected and reported by clinical genetic testing in individuals with epilepsy. To exemplify this latter point, here we describe a cohort of 12 individuals with biallelic SZT2 variants and phenotypic overlap with SZT2-related neurodevelopmental disorders. However, the majority of individuals carried one or more SZT2 variants of uncertain significance (VUS), highlighting the need for functional characterization to determine, which, if any, of these VUS were pathogenic. Thus, we developed a novel individualized platform to identify SZT2 loss-of-function variants in the context of mTORC1 signalling and reclassify VUS. Using this platform, we identified a recurrent in-frame deletion (SZT2 p.Val1984del) which was determined to be a loss-of-function variant and therefore likely pathogenic. Haplotype analysis revealed that this single in-frame deletion is a founder variant in those of Ashkenazi Jewish ancestry. Moreover, this approach allowed us to tentatively reclassify all of the VUS in our cohort of 12 individuals, identifying five individuals with biallelic pathogenic or likely pathogenic variants. Clinical features of these five individuals consisted of early-onset seizures (median 24 months), focal seizures, developmental delay and macrocephaly similar to previous reports. However, we also show a widening of the phenotypic spectrum, as none of the five individuals had corpus callosum abnormalities, in contrast to previous reports. Overall, we present a rapid assay to resolve VUS in SZT2, identify a founder variant in individuals of Ashkenazi Jewish ancestry, and demonstrate that corpus callosum abnormalities is not a hallmark feature of this condition. Our approach is widely applicable to other mTORopathies including the most common causes of the focal genetic epilepsies, DEPDC5, TSC1/2, MTOR and NPRL2/3.
Keywords: epilepsy, mTOR, SZT2, variant, genetics
Biallelic variants in SZT2 are associated with early-onset epilepsy, but identifying pathogenic missense variants has proven challenging. Calhoun et al. develop an assay for resolution of variants of unknown significance in SZT2, identify a potential founder variant, and refine the known phenotypic spectrum of SZT2-associated epilepsy.
Introduction
SZT2 (Seizure Threshold 2) encodes a large (>350 kDa) protein with ubiquitous tissue expression and no homology to known functional domains.1 The first association of SZT2 with seizure susceptibility emerged from a mutagenesis screen in mice.1 In this study, homozygous Szt2 knockout mutant mice seized at a lower electrical input relative to wild-type littermates in an acute electroconvulsive model.1 Years later, biallelic SZT2 pathogenic variants were identified in a small cohort of patients with infantile-onset epilepsy and dysgenesis of the corpus callosum.2 Since then, several small case studies report biallelic SZT2 variants in association with a neurodevelopmental disorder characterized primarily by early-onset focal epilepsy, developmental delay, macrocephaly and corpus callosum abnormalities.2–16
For years the function of SZT2 remained elusive until 2017 when SZT2 was identified as a part of the KICSTOR complex, a required component of the amino acid sensing arm of the mTORC1 pathway.17,18 The KICSTOR complex, including SZT2, localizes to the lysosomes only in the presence of amino acids in the extracellular environment.17 In genome-edited cells lacking endogenous expression of SZT2, mTORC1 activity no longer depended on amino acids, i.e. mTORC1 was constitutively active.17 Moreover, cells lacking SZT2 exhibited amino acid-insensitive localization of MTOR to the lysosomal surface, suggesting that SZT2 is a key scaffolding protein.17 Based on these two pivotal studies, the current consensus is that biallelic SZT2 loss-of-function variants produce a mTORopathy due to constitutive mTORC1 activity.17,18
The majority of pathogenic SZT2 variants described to date in the literature are truncations, resulting in complete SZT2 loss-of-function (null). However, there are now also many cases of both missense variants and in-frame deletions that are more difficult to classify, driven primarily by our inability to determine the effect of single amino acid alterations on SZT2 function. In silico measurements of functional constraint based on the number of observed versus expected missense variants (Z-score) shows SZT2 is among the top 15% most constrained genes in the human genome.19 This observation suggests some level of selection, and that some missense variants could impact SZT2 function. Data from clinical diagnostic genetic testing reports 332 variants of uncertain significance (VUS) from 5652 tests; translating to a rate of 5.8% of gene panel tests identifying a SZT2 VUS.20 We focus primarily on SZT2 missense VUS and devise a functional assay to identify loss-of-function SZT2 alleles. Moreover, we use this approach to demonstrate that a recurrent single amino acid deletion is both likely pathogenic and a founder variant in individuals with Jewish ancestry.
Materials and methods
Study participants
Individuals with candidate causative SZT2 variants (accession ID: NM_015284.4 and NP_056099.3) were identified by clinical exome sequencing (n = 11) or gene panel (n = 1). Specifically, inclusion criteria were individuals with biallelic SZT2 variants with an American College of Medical Genetics (ACMG) classification of VUS, likely pathogenic, or pathogenic21 and individuals presenting with either recurrent seizures, macrocephaly, and/or developmental delay. SZT2 variants that were present in homozygous state in individuals within the general population (gnomAD) would have been excluded based on ACMG criteria. The individuals at Northwestern Memorial Hospital and Lurie Children’s Hospital were consented to research under an IRB approved study. We obtained de-identified genetic and clinical data from external colleagues for cases identified through Genematcher.22 These individuals were consented for research under IRB approved studies at their local institutions. SZT2 variant classification was performed according to the ACMG criteria.21
Haplotype analysis in individuals with the recurrent p.Val1984del variant
Exome sequencing data from probands and parents with the SZT2 p.Val1984del variant were obtained. Variant call files (vcfs) were generated with standard GATK best practices. Briefly, sequencing reads were aligned to the human genome (hg38) with BWA-MEM followed by calling and genotyping alleles with GATK HaplotypeCaller and GATK GenotypeGVCFs. Then, GATK SelectVariants was used to subset a vcf file containing variants within 20 Mb of SZT2 p.Val1984del in the individual homozygous due to uniparental disomy (UPD) of all of chromosome 1 (Proband 3). Single nucleotide variants and indels in this region were filtered by minor allele frequencies in gnomAD (see ‘Web resources' section) to generate a list of candidate variants for haplotype analysis. Segregation of alleles in independent trios was used to define haplotype boundaries.
Generation of gene-edited cell lines
pSpCas9(BB)-2A-Puro (PX459) V2.0 was a gift from Feng Zhang (Addgene plasmid 62988; http://n2t.net/addgene:62988; RRID:Addgene_62988). gRNAs targeting SZT2 exons (Supplementary Table 1) were cloned into PX459 as previously described.23 HEK 293T (ATCC® CRL-3216™) cells were seeded into 24-well plates and transfected with pX459 plasmid with cloned gRNAs (500–1000 ng) and ssODN repair oligo (1–2 µl of 10 µM stock) with Lipofectamine 3000 (Invitrogen L3000001) according to manufacturer’s instructions. Cells were treated with puromycin (2.5 µg/ml) for 48 h beginning the day after transfection. Cells were replica plated for cryopreservation and genomic DNA isolation with PureLink™ Genomic DNA Mini Kit (Invitrogen K182002) according to manufacturer’s instructions. gRNA targeting of SZT2 exons were confirmed by T7 endonuclease I (NEB# M0302) according to manufacturer’s instructions. Homology-directed repair was confirmed by amplicon sequencing. Individual clones were collected by limited dilution cloning in 96-well plates followed by similar replica plating as described above. Genomic DNA from each clone was screened by Sanger sequencing as well as amplicon sequencing to confirm genotype as either (i) homozygous for the homology-directed repair (HDR) allele; or (ii) compound heterozygous for the HDR allele and an out-of-frame indel predicted to lead to SZT2 loss-of-function. All primer sequences are provided in Supplementary Table 1.
Amplicon sequencing
Amplicons with Illumina adaptors were generated by two rounds of PCR including the introduction of unique barcodes and standard Illumina primers. Amplicons were sequenced to a depth of at least 1000× on an Illumina Miniseq according to manufacturer’s recommendation. Alleles were analysed with CRISPResso2 using the web interface or command line workflow.24
Immunoblot analysis of mTORC1 activity
Amino acid starvation was performed as previously described.17 Briefly, cells plated in poly-l-lysine coated plates were rinsed in Dulbecco’s phosphate-buffered saline (DPBS) (Gibco 14190250) twice before the addition of amino acid-free Dulbecco’s minimal essential media (DMEM) containing 10% dialysed foetal bovine serum (FBS). For the amino acid starved condition, cells were starved of amino acids for 60 min at 37°C. For cells treated with amino acids, cells were starved of amino acids for 50 min at 37°C followed by incubation with amino acid containing DMEM for 10 min at 37°C. Cells were briefly rinsed with ice-cold DPBS twice. Cells were scraped in ice-cold DPBS and pelleted at 300g for 5 min. The cells were lysed in RIPA Lysis and Extraction Buffer (Thermo Scientific 89900) supplemented with EDTA-free protease inhibitors (Roche Complete PI EDTA-free; Sigma 11836170001) and PhosSTOP™ phosphatase inhibitors (Sigma 4906845001) for 30 min at 4°C. Insoluble material was removed by pelleting at 12 000g for 20 min. Protein concentration was determined by BCA assay (Pierce™ BCA Protein Assay Kit; Cat# 23225) and equal amounts (10–50 µg) of protein were loaded for SDS-PAGE (4–12% gradient gel). Proteins were transferred to PVDF membrane and blocked for 60 min at room temperature with 5% bovine serum albumin (BSA) in PBS-T (PBS + 0.05% Tween20). Primary antibodies (Supplementary Table 2) were incubated overnight at 4°C in blocking buffer. Membranes were washed 3 × 5 min in PBS-T prior to incubation with secondary antibodies (Supplementary Table 2) for 60 min at room temperature. After washing 4 × 5 min in PBS-T, membranes were incubated in Amersham ECL Prime (GE Healthcare) according to manufacturer’s instructions. Membranes were imaged on a Licor Odyssey Fc for 30 s to 60 min.
Fluorescence-activated cell sorting analysis of mTORC1 activity and calculation of CMAS
Amino acid starvation was performed as described above. After starvation, cells were washed twice with DPBS supplemented with 1% v/v Phosphatase Inhibitor Cocktail 3 (Sigma; P0044). Cell were trypsinized using trypLE (Gibco) supplemented with 1% v/v Phosphatase Inhibitor Cocktail 3. Cells were pelleted (300g for 5 min at room temperature) and resuspended in BD Fixation/Permeabilization solution (BD Biosciences 554714). After incubation on ice for 20 min, cells were pelleted and washed twice with BD permeabilization/wash buffer. Phosphorylated S6 (p-S6) was labelled with Alexa488 conjugated antibody (Supplementary Table 2) for 30 min on ice followed by two washes with BD permeabilization/wash buffer. Cells were flow sorted on the BD FACSMelody 3-laser cell sorter. At least 200 000 cells were collected for cells with dim Alexa488-signal (P-S6LOW) and those with bright Alexa488-signal (P-S6HIGH). Genomic DNA was prepared by manufacturer’s recommendation using PureLink™ Genomic DNA Mini Kit (Invitrogen K182002) with slight modification to include a 40 min incubation at 90°C to reverse crosslinks.
Amplicon sequencing and Sanger sequencing were performed on unsorted and sorted cells to determine the constitutive mTORC1 activity score (CMAS). CMAS is calculated for each allele present in the amplicon sequencing dataset. CMAS = % alleles in P-S6HIGH/ % alleles in unsorted cells. CMAS represents an enrichment score to determine whether cells with high mTORC1 activity are enriched for proband-derived SZT2 missense alleles (HDR-mediated). Allele percentage derived from amplicon sequencing and used to calculate CMAS are reported in Supplementary Table 3. Statistical analysis was performed using one-way ANOVA with Tukey’s post hoc test and P < 0.05 was considered significant.
Data availability
The raw data for generation of CMAS scores are provided for applicable SZT2 variants in the Supplementary material. Other data will be provided upon reasonable request.
Results
Genetic characterization of individuals with biallelic SZT2 variants
We describe 12 individuals with biallelic SZT2 variants and of these 24 alleles, truncating variants were identified in a quarter (6/24) of the cohort (Table 1 and Supplementary Fig. 1). This included one individual (Individual 2) who had biallelic SZT2 truncating variants,4 and five individuals carrying a truncating variant (or presumed truncating in the instance of the splice variant) on one allele and a single amino acid change on the other (Individuals 1, 5, 6, 7 and 8). Most individuals carried at least one SZT2 VUS, and these variants accounted for the majority of the variants present in the cohort (missense: 12/24, 50%; in-frame del: 5/24, 21%). One variant (p.Val1984del) was identified in multiple individuals in our cohort and has been previously reported in a single individual.11
Table 1.
Phenotypic details of cohort and genetic details of biallelic SZT2 variants
| Individual | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | DEE | DEE | DEE | SNSz | DEE | IE | DEE | IE | IE | SNSz | FE | DEE |
| Age Sz onset | 2 y | 2 m | 4 y | 3 y | 2 DOL | 2 y | No Sz | 9 m | 3 y | 3 DOL | 20 y | 6 y |
| Current Sz control | Intractable | Intractable | Well controlled | Resolved w/out meds | Intractable | Well controlled | N/A | Well controlled | Well controlled | Resolved w/out meds | Partial | Intractable |
| Development | DD | DD | DD | DD | DD | DD | DD | DD | DD | DD | Normal | Regression |
| Head circ | Macro | Macro | Macro | Micro | Macro | Macro | Micro | Normal | Normal | Macro | Normal | Normal |
| CCA | No | No | No | No | No | Yes | No | No | No | Yes | Yes | No |
| Inheritance | Cmp Het | Cmp Het | Hmz | Hmz | Cmp Het | Cmp Het | Cmp Het | Cmp Heta | Cmp Het | Hmz | Cmp Het | Hmzb |
| Maternally inherited allele | ||||||||||||
| Variant (NM_015284, NP_056099) | c.9407_9408d upTG p.Val3137 Trpfs*48 | c.9703C > T p.Arg3235* | Nonec UPD of all of chr 1 | c.5949_5951 delTGT p.Val1984del | c.2384_5680 del p.His795_His 1893deld | c.841delC p.Gln281Serfs*32 | c.1173_1174 del p.Lys393Glyfs*47 | c.1091-1G > Aa | c.7346G > A p.Arg2449Gln | c.7448C > T p.Ser2483Leu | c.5843G > A p.Arg1948Gln | c.7765C > T p.Arg2589Trp |
| GnomAD AF | 5.0 × 10−6 | 6.6 × 10−6 | Unk | 4.8 × 10−5 | NP | NP | NP | NP | 3.8 × 10−5 | 9.6 × 10−6 | NP | 1.4 × 10−5 |
| CADD | 50 | 50 | N/A | N/A | 50 | 50 | 50 | 50 | 15.37 | 18.74 | 16.936 | 19.83 |
| Polyphen | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 0.897 | 1 | 0.97 | 1 |
| ACMG Class | P | P | N/A | VUS | P | P | P | P | VUS | VUS | VUS | VUS |
| Reclassifiede | P (NT) | P (NT) | N/A | LP | P (NT) | P (NT) | P (NT) | P (NT) | LB | LB | LB | LB |
| Paternally inherited allele | ||||||||||||
| Variant (NM_015284, NP_056099) | c.5949_5951 delTGT p.Val1984del | c.3509_3512 delCAGA p.Thr1170 Argfs*22 | c.5949_5951 delTGT p.Val1984del | c.5949_5951 delTGT p.Val1984del | c.1678G > T p.Ala560Ser | c.6553C > T p.Arg2185Trp | c.4040G > A p.Arg1347His | c.7588A > C p.Ile2530Leua | c.3757C > T p.Arg1253Cys | c.7448C > T p.Ser2483Leu | c.4340A > C p.Glu1447Ala | c.7765C > T p.Arg2589Trp |
| GnomAD AF | 4.8 × 10−5 | NP | 4.8 × 10−5 | 4.8 × 10−5 | NP | 2.4 × 10−5 | 1.2 × 10−4 | 9.6 × 10−6 | 9.6 × 10−6 | 9.6 × 10−6 | NP | 1.4 × 10−5 |
| CADD | N/A | 50 | N/A | N/A | 15.9 | 16.55 | 16.7 | 8.82 | 16.72 | 18.74 | 17.62 | 19.83 |
| PolyPhen | N/A | N/A | N/A | N/A | 0.007 | 1 | 0.004 | 0 | 1 | 1 | 1 | 1 |
| ACMG Class | VUS | P | VUS | VUS | VUS | VUS | VUS | VUS | VUS | VUS | VUS | VUS |
| Reclassifiede | LP | P (NT) | LP | LP | LP | LB | LB | LB | LP | LB | LB | LB |
CADD = combined annotation dependent depletion; CCA = corpus collosum abnormalities; Cmp het = compound heterozygous; DEE = developmental and epileptic encephalopathy; FE = focal epilepsy; gnomAD AF = gnomAD minor allele frequency; Head circ = head circumference; Hmz = homozygous; IE = infantile epilepsy; LB = likely benign; LP = likely pathogenic; Macro = macrocephaly; Micro = microcpehaly; NP = not present; NT = not tested; P = pathogenic; SNSz = suspected neonatal seizure; Sz = seizure; Unk = unknown; w/out meds = without medication.
DNA was only available from one biological parent, but we were unable to ascertain which parent submitted a sample, thus the maternally and paternally inherited variants are randomly assigned in the table for clarity only.
Regions of homozygosity detected suggesting consanguinity.
Uniparental disomy (UPD) of all of chromosome 1.
Exon16-40 deletion.
Variant reclassification after functional testing.
Effect of patient-specific SZT2 variants on mTORC1 activity
We designed a functional assay to measure the effect of these SZT2 VUSs. We utilized CRISPR/Cas9 to edit HEK293T cells at the endogenous SZT2 locus and then quantified mTORC1 signalling in cells starved, or starved and subsequently treated with amino acids (Fig. 1A). Notably HEK293T are diploid for chromosome 1 on which SZT2 is located. First, we created a HEK293T SZT2 null cell line (SZT2KO/KO) and recapitulated the constitutive mTORC1 activity and insensitivity to the presence of amino acids previously observed in SZT2 null cell lines (Supplementary Fig. 2).17,18
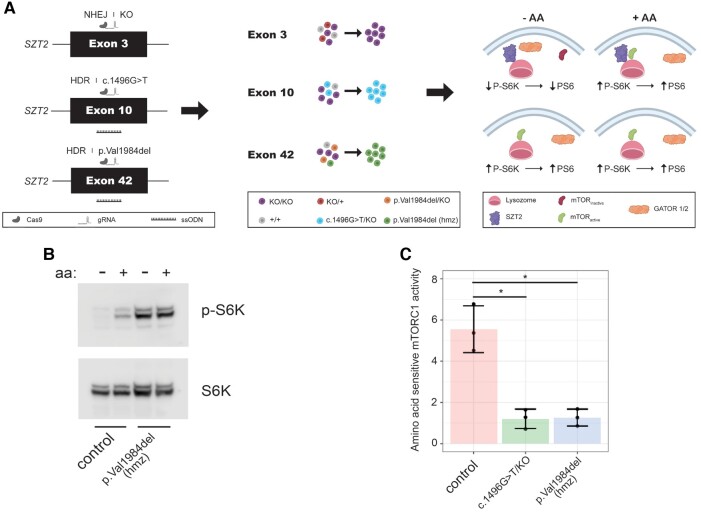
Figure 1.
Development of gene editing approach for functional characterization of SZT2 VUSs. (A) Individual cell clones either homozygous or heterozygous (+ loss-of-function on other allele) for an individual SZT2 variant were generated by gene editing followed by limiting dilution cloning. Immunoblot for amino acid sensitive mTORC1 activity by P-S6K levels was used to determine whether individual variants caused SZT2 loss-of-function. Homology-directed repair rate of cells generated by transfection of px459 encoding targeting gRNAs was analysed using amplicon sequencing and varied between 12% and 25%. (B) Immunoblot of mTORC1 activity in control HEK cells (i.e. unedited) and homozygous HEK SZT2p.Val1984del/p.Val1984del clone. (C) Densitometric quantification of (B). At least three individual replicates were performed. *P < 0.05 (t-test).
We examined one previously published likely pathogenic SZT2 variant (c.1496G > T) and the recurrent p.Val1984del VUS identified in three patients in our cohort (Probands 1, 3 and 4) along with one individual described previously.2,11 c.1496G is the last nucleotide of exon 10 and c.1496G > T yields two possible transcripts: (i) exon skipping resulting in SZT2 p.(Gly412Alafs∗86); or (ii) missense variant SZT2 p.Ser499Ile. RT-PCR from patient fibroblasts suggested exon 10 skipping, though it was unclear whether any of the predicted missense allele p.Ser499Ile transcript was generated, and if so, whether this missense VUS impacted SZT2 function.2 We generated a compound heterozygous SZT2 cell line (HEK.SZT2c.1496G>T/KO) and by analysis of RNA transcripts determined that exon 10 is skipped, producing a truncated protein [p.(Gly412Alafs∗86)] (Supplementary Fig. 3). Furthermore, HEK.SZT2c.1496G>T/KO cells displayed constitutive mTORC1 activity (Fig. 1C and Supplementary Fig. 4). We generated homozygous SZT2 p.Val1984del HEK cells (HEK.SZT2p.Val1984del/p.Val1984del) and determined that this VUS also resulted in constitutively active mTORC1 (Fig. 1B and C). Collectively, these results demonstrate that complete SZT2 loss-of-function and both the previously published c.1496G > T and the recurrent p.Val1984del lead to constitutive mTORC1 signalling also suggesting SZT2 loss-of-function.
A medium-throughput assay for functional characterization of SZT2 VUS
The time-consuming process of limited dilution cloning and identification of clones with desired genotypes led us to investigate methods to increase throughput of our approach. Based on high rates of indel formation (60+%) due to non-homologous end-joining (NHEJ) as well as high HDR efficiency (up to 30%) in HEK293T cells, we considered the possibility of performing functional testing directly after gene editing. Though immunoblot analysis from a pool of cells with different SZT2 alleles would be challenging to interpret, we hypothesized that immunolabelling for phospho-S6 (P-S6) followed by flow cytometry would be a viable alternative. Amino acid starved SZT2 null cells were shown to have both elevated P-S6K and P-S6 levels due to constitutive mTORC1 activity.18 The conceptual framework is as follows: if a variant is loss-of-function variant, then cells homozygous for that variant or compound heterozygous for the variant and a truncating variant induced by NHEJ would lack any functional SZT2 and therefore exhibit constitutive mTORC1 activation. Alternatively, if a variant did not cause SZT2 loss-of-function, cells with one or two copies would express sufficient levels of functional SZT2 for physiological regulation of mTORC1 by amino acid deprivation (Fig. 2A). Subsequent genotyping of unsorted cells as well as the sorted pools of high and low phosphorylated-S6 (i.e. P-S6HIGH and P-S6LOW) allowed us to determine the CMAS for each allele.
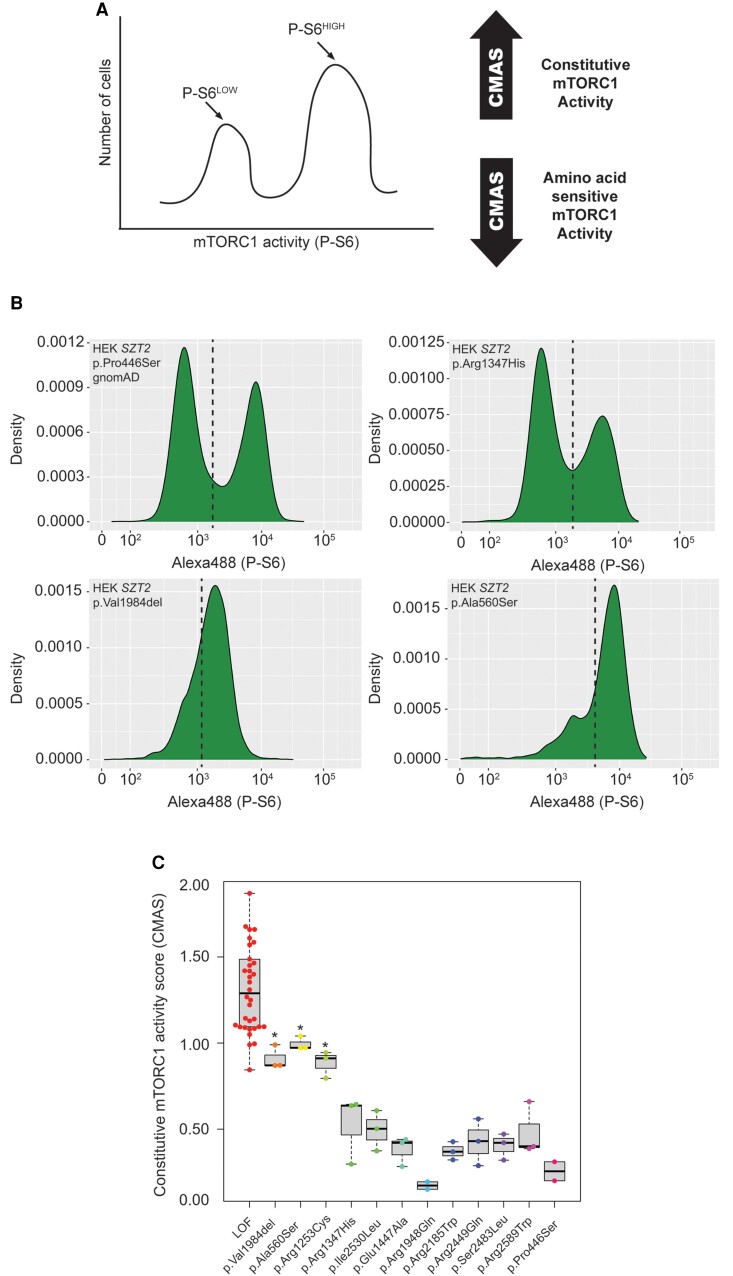
Figure 2.
Development of medium throughput assay for functional characterization of SZT2 VUSs. (A) HEK293T cells were co-transfected with px459 encoding targeting gRNAs and repair oligonucleotides, followed by puromycin selection. Cells were starved of amino acids and then fixed for immunolabelling of phosphorylated S6 and FACS sorting. gDNA was isolated from unsorted and sorted cells followed by amplicon sequencing to confirm CRISPR/Cas9 targeting and to calculate CMAS. (B) Representative FACS plots. Dashed line represents the boundary for sorting P-S6HIGH and P-S6LOW cell populations. (C) CMAS scores derived from amplicon sequencing of sorted and unsorted cells. For all but p.Arg1948Gln (n = 2) and p.Pro466Ser (n = 2), at least three individual replicates were performed. *P < 0.05 (one-way ANOVA with Tukey’s post hoc test).
As a proof of principle, we first performed FACS sorting based on phospho-S6 signal on amino acid starved wild-type HEK and HEK SZT2KO/KO cells. Wild-type HEK cells exhibited a single left-shifted peak (P-S6LOW), while the SZT2KO/KO cells exhibited a single right-shifted peak (P-S6HIGH), indicating constitutive mTORC1 activity (Supplementary Fig. 5). We then used the assay to investigate all of the SZT2 VUSs present in the cohort (Fig. 2B). As an additional negative control, we assayed a common variant in the general population, SZT2 p.Pro446Ser [minor allele frequency (MAF) = 0.3 in gnomAD], and present in a homozygous state in multiple individuals (n = 15 475). All loss-of-function alleles had high CMAS scores (mean CMAS = 1.35 ± 0.36; n = 34) consistent with constitutively active mTORC1, while CMAS for SZT2 p.Pro446Ser was low (0.26 ± 0.07; n = 2), consistent with the protein retaining physiological scaffolding function and amino acid sensitive mTORC1 activity (Fig. 2C and Supplementary Fig. 6). As with the western blot analysis above, p.Val1984del was significantly enriched in the P-S6HIGH (mean CMAS = 0.91 ± 0.06; n = 3) pool, indicative of constitutive mTORC1 activity (Fig. 2B and C). Two missense variants, p.Ala560Ser (mean CMAS = 0.99 ± 0.04; n = 3) and p.Arg1253Cys (mean CMAS = 0.88 ± 0.08; n = 3), were similarly found to be significantly enriched in cells with constitutive mTORC1 activity (Fig. 2B and C). Conversely, the remaining missense variants, p.Glu1447Ala (mean CMAS = 0.38 ± 0.08; n = 3), p.Arg1948Gln (mean CMAS = 0.17 ± 0.03; n = 2), p.Arg2589Trp (mean CMAS = 0.48 ± 0.15; n = 3), p.Arg1347His (mean CMAS = 0.52 ± 0.2; n = 3), p.Arg2185Trp (mean CMAS = 0.37 ± 0.05; n = 3), p.Arg2449Gln (mean CMAS = 0.42 ± 0.13; n = 3), p.Ile2530Leu (mean CMAS = 0.49 ± 0.11; n = 3), and p.Ser2483Leu (mean CMAS = 0.4 ± 0.08; n = 3) were not enriched in the P-S6HIGH pool (Fig. 2B and C) and CMAS were not significantly different to p.Pro446Ser, but rather were consistent with the CMAS of the benign variant, suggesting these variants do not cause SZT2 loss-of-function.
Haplotype analysis of recurrent SZT2 in-frame deletion p.Val1984del
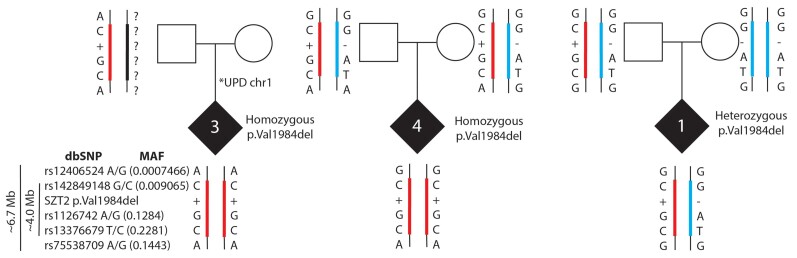
Based on our observation of p.Val1984del in multiple individuals, we hypothesized this variant may be derived from a common ancestor. In support of this observation, SZT2 p.Val1984del is observed at low allele frequency in two populations in gnomAD, specifically in non-Finnish Europeans (MAF = 0.00005420) and Ashkenazi Jewish (MAF = 0.0008679), though no homozygous individuals are reported. Using exome sequencing data we identified a shared haplotype spanning ∼4 Mb including the SZT2 p.Val1984del in all three individuals in our cohort (Fig. 3), suggesting this variant is a founder variant in individuals with Jewish ancestry.
Figure 3.
Shared haplotype suggests SZT2 p.Val1984del is a founder variant in those of Ashkenazi Jewish ancestry. Using exome sequencing data from three study trios in combination with allelic frequencies in the gnomAD population database, all individuals carry the same rare (MAF ranging from 0.00007–0.2) variants spanning a 4 Mb interval. Ref|Alt alleles for SNVs (1 and 5 flanking; 2-4 in haplotype block): (1) rs12406524 = G|A; (2) rs142849148 = G|C; (3) rs1126742 = A|G; (4) rs13376679 = T|C; (5) rs75538709 = G|A.
Clinical characterization of individuals with biallelic SZT2 variants
The functional assays allowed us to tentatively reclassify all of the VUSs in our cohort of 12 individuals. We acknowledge that reclassification of variants based on functional data is relevant in a clinical setting when classification, in particular following ACMG criteria, is based on a ‘well-established assay’, which our approach has not yet achieved.21 Thus, reclassification of the variants as P/LP or B/LB is based on the functional readout of our assay and should not be considered diagnostic reclassification according to ACMG criteria. However, for simplicity sake in this research study we reclassified VUS with CMAS similar to p.Pro446Ser (GnomAD MAF = 0.3, numerous homozygotes) as likely benign (LB) and VUS with CMAS consistent with SZT2 loss-of-function as likely pathogenic (LP). We then grouped these individuals into three groups (P or LP/P or LP, P or LP/LB, LB/LB) and assessed the prevalence of the most common clinical features associated with pathogenic SZT2 variants i.e. early-onset epilepsy, developmental delay, intractable focal seizures, macrocephaly and corpus callosum abnormalities (Tables 1 and 2 and Supplementary Tables 4 and 5).2–16 Individual 2 with biallelic SZT2 truncating variants did not require reclassification but was included in the cohort as a representative of the P/P group for the purpose of phenotypic comparison. While subgrouping the cohort in this manner meant we were unable to perform statistical analysis due to small group sizes, we could make a number of observations. The median seizure onset was 24 months and relatively consistent, with the exception of the LB/LB group where seizure onset was much later (median 13 years). Most individuals with P or LP/P or LP presented with focal seizures, developmental delay and macrocephaly. These core features were less common in the P or LP/LB group, with the exception of developmental delay. Sixty per cent of the P or LP/P or LP group failed to achieve seizure control, while only one individual in either the P or LP/LB or LB/LB group had seizures that were refractory to treatment. Moreover, very few individuals, except Individuals 6, 10 and 11, had corpus callosum abnormalities (Supplementary Table 5 and Supplementary Fig. 7).
Table 2.
Summary of clinical features stratified by ACMG criteria and functional classification
| Category | P or LP/P or LP | P or LP/LB | LB/LB | Cohort |
|---|---|---|---|---|
| Affected individuals | 1–5 (n = 5) | 6–9 (n = 4) | 10–12 (n = 3) | n = 12 |
| Median seizure onset (range) | 24 m (2DOL–4 y) | 24 m (9 m–3 y) | 6y (3DOL–20 y) | 24 m (2DOL–20 y) |
| Focal seizure | 4/5 (80%) | 1/3a (25%) | 1/3 (33%) | 6/11 (50%) |
| Seizures intractable | 3/5 (60%) | 0/3 (0%) | 1/3b (33%) | 4/11 (36%) |
| Developmental delays | 5/5 (100%) | 4/4 (100%) | 2/3 (66%) | 11/12 (92%) |
| Macrocephaly | 4/5c (75%) | 1/4d (25%) | 1/3 (33%) | 7/12 (58%) |
| Corpus callosum abnormalities | 0/5 (0%) | 1/4 (25%) | 2/3 (66%) | 3/12 (25%) |
DOL = day of life; m = months; y = years.
Individual 7 had no report of seizures.
Individual 12 achieved only partial seizure control.
Individual 4 had microcephaly.
Individual 7 had microcephaly.
Discussion
The identification of VUS is one of the most challenging bottlenecks in clinical genetic diagnostics of the modern era. We developed a novel individualized platform that allowed us to recategorize all of the SZT2 VUSs in our cohort of 12 individuals. Of note, we identify a recurrent in-frame deletion (p.Val1984del) in four individuals, including two in a homozygous state (one instance of chromosome 1 uniparental disomy), and one heterozygous in trans with a truncating variant. Two of the individuals (1 and 3) were identified at Lurie Children’s hospital over 2 years. We determined that this variant is a founder variant in individuals with Jewish ancestry. A carrier frequency of 1:576 is estimated based on gnomAD data, but may differ based on local ancestry, and inclusion of this variant in genetic testing panels for individuals with Jewish ancestry requires further investigation.
Based on the necessity for a functional assay to reclassify SZT2 VUSs, we developed a strategy to functionally characterize SZT2 VUSs by knock-in of patient-specific SZT2 variants into HEK 293T cells. The molecular function of SZT2 as a critical regulatory scaffolding protein in the amino acid sensing arm of the mTORC1 signalling pathway had previously been elucidated in HEK 293T.17,18 Advantages of using HEK 293T cells are the high transfection and gene editing efficiencies, both for knockout (NHEJ) and knock-in (HDR) strategies. We successfully developed this strategy as a medium-throughput assay that revealed a recurrent in-frame deletion SZT2 p.Val1984del to be a loss-of-function variant (Figs 2B and C and 3B and C). Using this fluorescence-activated cell sorting (FACS)-based assay we also functionally characterized an additional set of two SZT2 VUSs (p.Ala560Ser and p.Arg1253Cys) as likely loss-of-function variants. The remaining missense VUSs were found to be unlikely to result in loss-of-function as these variants retained amino acid-sensitive mTORC1 activity as observed for a common population variant (p.Pro446Ser) and suggest these variants may be benign. However, while our approach can robustly detect complete loss-of-function alleles, it may not detect more subtle effects on protein function, including partial loss-of-function. For instance, while not significant, with the exception of p.Arg1948Gln, the other missense VUS did have slightly higher CMAS than the gnomAD homozygous missense, this may indicate partial impact on SZT2 function, though this requires further study. Moreover, we know that the clinical features of individuals with pathogenic SZT2 variants are neuronally restricted, even though SZT2 is ubiquitously expressed, thus perturbation of the mTORC1 pathway is more likely to have a detrimental impact during neuronal development and/or function. For instance, it has recently been shown that mTOR regulation is essential for outer radial glia migration during human neuronal development.25 In the future, adaption of this assay in cells of a neuronal lineage, for instance, immortalized ReNcells, may improve on the discriminatory power of the novel assay we present here.
Nakamura et al.26 recently examined mTORC1 activity in lymphoblastoid cell lines (LCLs) from affected individuals. They report elevated mTORC1 activity in cell lines derived from individuals with biallelic SZT2 variants relative to cell lines generated from healthy individuals. There are a few important caveats to their study. First, the developed assay requires generating LCLs from individuals, which is not always possible and decreases throughput. Further, both cell lines generated from individuals with biallelic SZT2 loss-of-function variants show significant response to amino acid treatment after starvation. One of the key findings from the initial studies, and our studies here in HEK293T cells, was that SZT2 gene knockout rendered cells completely insensitive to amino acids.17,18 Although difficult to explain, the finding could be a technical issue with the LCL model.
Similar to previously published biallelic SZT2 cases, most individuals carrying biallelic P or LP variants in this cohort presented with paediatric-onset epilepsy and expressed common features including focal seizures, developmental delay and macrocephaly. However, none of the individuals in this group had corpus callosum abnormalities, suggesting that this is not a cardinal feature of SZT2-associated epilepsy. Moreover, a genotype-phenotype correlation has been suggested; individuals bearing truncating variants may be more likely to have intractable seizures than individuals with missense variants.12 In this cohort, we observed more variability, with Individual 2 (biallelic truncations) having seizure onset at 2 months with intractable seizures, while the other individual with intractable seizures (with seizure onset at two days of life) carried a multi-exon in-frame deletion and a reclassified missense loss-of-function variant (Supplementary Tables 4 and 5). Moreover, the individuals with homozygous p.Val1984del (Individuals 3 and 4) had divergent presentations, with Individual 3 presenting with developmental and epileptic encephalopathy (DEE) and seizures controlled by medication while Individual 4 had a suspected neonatal seizure that resolved without medication. Despite these differing seizure patterns, both individuals have developmental delay and cognitive impairment. These different presentations may be influenced by a variety of factors such as genetic modifiers; in particular Individual 3 has uniparental disomy of chromosome 1 which may contribute to the more severe presentation. Collectively, these results suggest a straightforward genotype-phenotype relationship is unlikely and further studies are needed to characterize the phenotypic spectrum and the impact of genetic variation on protein function.
The majority of individuals with biallelic P/LP variants presented with macrocephaly, this is in keeping with a previous report that just over half of individuals with SZT2 pathogenic variants have macrocephaly.5 Moreover, a previous study found 50% of children with macrocephaly and comorbid developmental delay were found to harbour genetic variants in the PI3K-AKT-mTOR signalling pathway; thus macrocephaly as a cardinal feature of biallelic pathogenic SZT2 variants is a fit with constitutive activation of mTORC1.27 Interestingly, Individual 4 (homozygous SZT2 p.Val1984del) instead is microcephalic, though MRI in this individual also revealed ischemic changes associated with preterm delivery and haemorrhage (Supplementary Table 5) that may contribute to this phenotype. Two additional individuals with SZT2 variants and microcephaly have been reported. The first is Individual 7 in whom our functional analysis suggested that one of the missense variants may not be pathogenic. The second individual is described with compound heterozygous missense variants that have not been functionally tested. Collectively, this suggests that microcephaly is unlikely a consistent feature of SZT2 pathogenic variants, in keeping with other mTORopathies that are generally associated with enhanced cell growth, and thus macrocephaly/megalencephaly.
The remainder of the cohort consisted of seven individuals with one or two alleles not causing clear loss-of-function (n = 7). Several of the core clinical features of intractable focal seizures and macrocephaly were less frequent in these two groups compared to the biallelic P/LP group. As noted above, our functional assay requires additional development and validation to definitively rule out SZT2 as harbouring the causative variant(s) in these individuals, but the more variable phenotype supports the position that these may not be causative. Moreover, SZT2 is a large gene, with many missense variants, indeed there are over 1800 unique missense variants in GnomAD. However, the lack of individual level data in this database precludes an analysis of how many individuals in the general population carry biallelic variants. This study points to an overall larger challenge in the field, where the presence of one P/LP variant in a recessive gene leads to uncertainty in terms of causality i.e. is there an unknown/inconclusive pathogenic variant on the other allele, or is this a spurious finding given loss-of-function allele can be present in the general population for recessive conditions. In the future, large studies from clinical genetic testing data, and individual level genetic information from the general population (e.g. TopMED, UKBiobank) are necessary to address this larger question. However, ultimately for these seven individuals we would recommend further genetic testing or reanalysis of their previous genetic testing, as the SZT2 variants may not be contributory in these individuals.
In summary, here we demonstrate the utility of an individualized platform to recharacterize SZT2 VUS. Importantly, this included a p.Val1984del variant that has a carrier allele frequency of at least 1:576 in those of Jewish ancestry and is a founder variant in this population. While additional modifications are still required to increase throughput, perhaps using saturation mutagenesis or multiplex assays of variant effect (MAVE),28,29 our approach can be applied to characterize VUS in other mTORopathies, including TSC1, TSC2, DEPDC5, NPRL2 and NPRL3, which are the most common causes of focal epilepsies. As the mTORopathies are the targets of multiple new clinical trials for mTOR inhibitors, resolution of VUSs could qualify more individuals with intractable epilepsies for inclusion in these studies.30
Web resources
gnomAD v2.1.1 (https://gnomad.broadinstitute.org); last accessed 23 October 2020.
SeattleSeq (https://snp.gs.washington.edu/SeattleSeqAnnotation138/); last accessed 22 October 2020.
Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/); last accessed 22 October 2020.
Some figure panels were created with BioRender.com.
Supplementary Material
Acknowledgements
This work was supported by the Northwestern University – Flow Cytometry Core Facility supported by Cancer Center Support Grant (NCI CA060553). Flow Cytometry Cell Sorting was performed on a BD FACSAria SORP system and BD FACSymphony S6 SORP system, purchased through the support of NIH 1S10OD011996-01 and 1S10OD026814-01. We would like to thank Andrew Tidball for assistance with pS6 FACs protocol and Greg Calhoun for assistance with graphic design.
Abbreviations
- ACMG
American College of Medical Genetics
- B/LB
benign or likely benign
- CMAS
constitutive mTORC1 activity score
- gnomAD
Genome Aggregation Database
- HDR
homology directed repair
- mTOR
mammalian target of rapamycin
- P/LP
pathogenic or likely pathogenic
- VUS
variant of uncertain significance
Contributor Information
Jeffrey D Calhoun, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Miriam C Aziz, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Hannah C Happ, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Jonathan Gunti, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Colleen Gleason, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Najma Mohamed, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Kristy Zeng, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Meredith Hiller, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Emily Bryant, Ann and Robert H. Lurie Children’s Hospital of Chicago Epilepsy Center and Division of Neurology, Chicago, IL 60610, USA.
Divakar S Mithal, Ann and Robert H. Lurie Children’s Hospital of Chicago Epilepsy Center and Division of Neurology, Chicago, IL 60610, USA.
Irena Bellinski, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Lisa Kinsley, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Mona Grimmel, Ann & Robert H. Lurie Children's Hospital of Chicago, Department of Pediatrics, Epilepsy Center and Division of Neurology, Chicago, IL 60610, USA.
Eva M C Schwaibold, Institute of Human Genetics, Heidelberg University, Heidelberg 69120, Germany.
Constance Smith-Hicks, Department of Neurogenetics, Kennedy Krieger Institute, Baltimore, MD 21205, USA; Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Anna Chassevent, Department of Neurogenetics, Kennedy Krieger Institute, Baltimore, MD 21205, USA.
Marcello Scala, Giannina Gaslini Children’s Hospital, Genova, GE 16147, Italy; Medical Genetic Unit, IRCCS Istituto G.Gaslini, 16147 Genoa, Italy.
Andrea Accogli, Division of Medical Genetics, Department of Specialized Medicine, Montreal Children's Hospital, McGill University Health Centre (MUHC), Montreal, QC, H4A 3J1, Canada; Department of Human Genetics, McGill University, Montreal, QC, Canada.
Annalaura Torella, Division of Medical Genetics, Department of Specialized Medicine, Montreal Children's Hospital, McGill University Health Centre (MUHC), Montreal, QC, H4A 3J1, Canada.
Pasquale Striano, Giannina Gaslini Children’s Hospital, Genova, GE 16147, Italy; Medical Genetic Unit, IRCCS Istituto G.Gaslini, 16147 Genoa, Italy.
Valeria Capra, Giannina Gaslini Children’s Hospital, Genova, GE 16147, Italy; Medical Genetic Unit, IRCCS Istituto G.Gaslini, 16147 Genoa, Italy.
Lynne M Bird, Department of Human Genetics, McGill University, Montreal, QC, Canada.
Issam Ben-Sahra, Telethon Institute of Genetics and Medicine (TIGEM), Pozzuoli, NA 80078, Italy.
Nina Ekhilevich, San Diego Department of Pediatrics and Rady Children’s Hospital Division of Dysmorphology/Genetics, University of California, San Diego, CA 92161, USA.
Tova Hershkovitz, San Diego Department of Pediatrics and Rady Children’s Hospital Division of Dysmorphology/Genetics, University of California, San Diego, CA 92161, USA; Department of Biochemistry and Molecular Genetics, Northwestern Feinberg School of Medicine, Chicago, IL 60610, USA.
Karin Weiss, San Diego Department of Pediatrics and Rady Children’s Hospital Division of Dysmorphology/Genetics, University of California, San Diego, CA 92161, USA; Department of Biochemistry and Molecular Genetics, Northwestern Feinberg School of Medicine, Chicago, IL 60610, USA.
John Millichap, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA; Ann and Robert H. Lurie Children’s Hospital of Chicago Epilepsy Center and Division of Neurology, Chicago, IL 60610, USA.
Elizabeth E Gerard, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA.
Gemma L Carvill, Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60610, USA; Genetics Institute, Rambam Medical Center, Ruth and Bruce Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Haifa, Israel; Ruth and Bruce Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Haifa 3200003, Israel.
Funding
This work was sponsored by National Institutes of Health NINDS R00NS089858 (G.L.C.). We thank the National Institutes of Health, Clinical and Translational Sciences Institute (NUCATS), Northwestern University TL1 award for support (TR001423 to J.D.C.).
Competing interests
G.L.C. holds a collaborative research grant with Stoke Therapeutics for research unrelated to this manuscript, all other authors have nothing to declare.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Frankel WN, Yang Y, Mahaffey CL, Beyer BJ, O’Brien TP. Szt2, a novel gene for seizure threshold in mice. Genes Brain Behav. 2009;8:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basel-Vanagaite L, Hershkovitz T, Heyman E, et al. Biallelic SZT2 mutations cause infantile encephalopathy with epilepsy and dysmorphic corpus callosum. Am J Hum Genet. 2013;93:524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Falcone M, Yariz KO, Ross DB, Foster J II, Menendez I, Tekin M. An amino acid deletion in SZT2 in a family with non-syndromic intellectual disability. PLoS One. 2013;8:e82810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Venkatesan C, Angle B, Millichap JJ. Early-life epileptic encephalopathy secondary to SZT2 pathogenic recessive variants. Epileptic Disord. 2016;18:195–200. [DOI] [PubMed] [Google Scholar]
- 5. Tsuchida N, Nakashima M, Miyauchi A, et al. Novel biallelic SZT2 mutations in 3 cases of early-onset epileptic encephalopathy. Clin Genet. 2018;93:266–274. [DOI] [PubMed] [Google Scholar]
- 6. Nakamura Y, Togawa Y, Okuno Y, et al. Biallelic mutations in SZT2 cause a discernible clinical entity with epilepsy, developmental delay, macrocephaly and a dysmorphic corpus callosum. Brain Dev. 2018;40:134–139. [DOI] [PubMed] [Google Scholar]
- 7. Pizzino A, Whitehead M, Sabet Rasekh P, et al. Mutations in SZT2 result in early-onset epileptic encephalopathy and leukoencephalopathy. Am J Med Genet A. 2018;176:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naseer MI, Alwasiyah MK, Abdulkareem AA, et al. A novel homozygous mutation in SZT2 gene in Saudi family with developmental delay, macrocephaly and epilepsy. Genes Genomics. 2018;40:1149–1155. [DOI] [PubMed] [Google Scholar]
- 9. Kariminejad A, Yazdan H, Rahimian E, et al. SZT2 mutation in a boy with intellectual disability, seizures and autistic features. Eur J Med Genet. 2019;62:103556. [DOI] [PubMed] [Google Scholar]
- 10. Imaizumi T, Kumakura A, Yamamoto-Shimojima K, Ondo Y, Yamamoto T. Identification of a rare homozygous SZT2 variant due to uniparental disomy in a patient with a neurodevelopmental disorder. Intractable Rare Dis Res. 2018;7:245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uittenbogaard M, Gropman A, Brantner CA, Chiaramello A. Novel metabolic signatures of compound heterozygous Szt2 variants in a case of early-onset of epileptic encephalopathy. Clin Case Rep. 2018;6:2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Domingues FS, Konig E, Schwienbacher C, et al. Compound heterozygous SZT2 mutations in two siblings with early-onset epilepsy, intellectual disability and macrocephaly. Seizure. 2019;66:81–85. [DOI] [PubMed] [Google Scholar]
- 13. Iodice A, Spagnoli C, Frattini D, Salerno GG, Rizzi S, Fusco C. Biallelic SZT2 mutation with early onset of focal status epilepticus: Useful diagnostic clues other than epilepsy, intellectual disability and macrocephaly. Seizure. 2019;69:296–297. [DOI] [PubMed] [Google Scholar]
- 14. Sun X, Zhong X, Li T. Novel SZT2 mutations in three patients with developmental and epileptic encephalopathies. Mol Genet Genomic Med. 2019;7:e926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trivisano M, Rivera M, Terracciano A, et al. Developmental and epileptic encephalopathy due to SZT2 genomic variants: Emerging features of a syndromic condition. Epilepsy Behav. 2020;108:107097. [DOI] [PubMed] [Google Scholar]
- 16. Tanaka R, Takahashi S, Kuroda M, et al. Biallelic SZT2 variants in a child with developmental and epileptic encephalopathy. Epileptic Disord. 2020;22:501–505. [DOI] [PubMed] [Google Scholar]
- 17. Wolfson RL, Chantranupong L, Wyant GA, et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng M, Yin N, Li MO. SZT2 dictates GATOR control of mTORC1 signalling. Nature. 2017;543:433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Truty R, Patil N, Sankar R, et al. Possible precision medicine implications from genetic testing using combined detection of sequence and intragenic copy number variants in a large cohort with childhood epilepsy. Epilepsia Open. 2019;4:397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clement K, Rees H, Canver MC, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andrews MG, Subramanian L, Kriegstein AR. mTOR signaling regulates the morphology and migration of outer radial glia in developing human cortex. Elife. 2020;9:e58737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakamura Y, Kato K, Tsuchida N, Matsumoto N, Takahashi Y, Saitoh S. Constitutive activation of mTORC1 signaling induced by biallelic loss-of-function mutations in SZT2 underlies a discernible neurodevelopmental disease. PLoS One. 2019;14:e0221482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yeung KS, Tso WWY, Ip JJK, et al. Identification of mutations in the PI3K-AKT-mTOR signalling pathway in patients with macrocephaly and developmental delay and/or autism. Mol Autism. 2017;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Starita LM, Ahituv N, Dunham MJ, et al. Variant interpretation: Functional assays to the rescue. Am J Hum Genet. 2017;101:315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Theilmann W, Gericke B, Schidlitzki A, et al. Novel brain permeant mTORC1/2 inhibitors are as efficacious as rapamycin or everolimus in mouse models of acquired partial epilepsy and tuberous sclerosis complex. Neuropharmacology. 2020;180:108297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data for generation of CMAS scores are provided for applicable SZT2 variants in the Supplementary material. Other data will be provided upon reasonable request.