Abstract
Focal malformations of cortical development including focal cortical dysplasia, hemimegalencephaly and megalencephaly, are a spectrum of neurodevelopmental disorders associated with brain overgrowth, cellular and architectural dysplasia, intractable epilepsy, autism and intellectual disability. Importantly, focal cortical dysplasia is the most common cause of focal intractable paediatric epilepsy. Gain and loss of function variants in the PI3K-AKT-MTOR pathway have been identified in this spectrum, with variable levels of mosaicism and tissue distribution. In this study, we performed deep molecular profiling of common PI3K-AKT-MTOR pathway variants in surgically resected tissues using droplet digital polymerase chain reaction (ddPCR), combined with analysis of key phenotype data. A total of 159 samples, including 124 brain tissue samples, were collected from 58 children with focal malformations of cortical development. We designed an ultra-sensitive and highly targeted molecular diagnostic panel using ddPCR for six mutational hotspots in three PI3K-AKT-MTOR pathway genes, namely PIK3CA (p.E542K, p.E545K, p.H1047R), AKT3 (p.E17K) and MTOR (p.S2215F, p.S2215Y). We quantified the level of mosaicism across all samples and correlated genotypes with key clinical, neuroimaging and histopathological data.
Pathogenic variants were identified in 17 individuals, with an overall molecular solve rate of 29.31%. Variant allele fractions ranged from 0.14 to 22.67% across all mutation-positive samples. Our data show that pathogenic MTOR variants are mostly associated with focal cortical dysplasia, whereas pathogenic PIK3CA variants are more frequent in hemimegalencephaly. Further, the presence of one of these hotspot mutations correlated with earlier onset of epilepsy. However, levels of mosaicism did not correlate with the severity of the cortical malformation by neuroimaging or histopathology. Importantly, we could not identify these mutational hotspots in other types of surgically resected epileptic lesions (e.g. polymicrogyria or mesial temporal sclerosis) suggesting that PI3K-AKT-MTOR mutations are specifically causal in the focal cortical dysplasia-hemimegalencephaly spectrum. Finally, our data suggest that ultra-sensitive molecular profiling of the most common PI3K-AKT-MTOR mutations by targeted sequencing droplet digital polymerase chain reaction is an effective molecular approach for these disorders with a good diagnostic yield when paired with neuroimaging and histopathology.
Keywords: focal cortical dysplasia, hemimegalencephaly, ddPCR, epilepsy, mosaicism
Pirozzi et al. demonstrate that mosaic hotspot mutations in the PI3K-AKT-MTOR pathway can be detected in individuals with focal cortical dysplasia and hemimegalencephaly via droplet digital PCR. The presence of PI3K-AKT-MTOR hotspot variants is associated with earlier age at seizure onset.
Introduction
Focal malformations of cortical development (FMCD) including focal cortical dysplasia (FCD, defined as localized area of abnormal cortical organization and development), hemimegalencephaly (HMEG, defined as overgrowth of one cerebral hemisphere), megalencephaly (MEG, defined as diffuse overgrowth of both cerebral hemispheres) and dysplastic MEG (DMEG, defined as bilateral cerebral hemisphere overgrowth with extensive bilateral malformations of cortical development), constitute a spectrum of developmental brain disorders associated with brain overgrowth, cellular dysplasia and significant co-morbidities such as intractable epilepsy, autism and intellectual disability.1-7
Mosaic gain of function mutations in the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of the rapamycin (MTOR) signalling pathway have been identified in this spectrum.2,8–14 Identifying a molecular aetiology for these disorders has significant benefits for affected families including understanding the natural history and recurrence risk, as well as more personalized therapeutic interventions for epilepsy. While traditional molecular diagnostic approaches, such as Sanger sequencing or standard-depth multi-gene panels or exome sequencing using peripheral blood-derived DNA, reliably detect inherited or germline genetic variants, these methods lack the sensitivity to detect low-level mosaic (or post-zygotic) variants. Further, the high genomic input requirement for these tests renders them impractical in these disorders due to paucity of affected (or lesional) tissues such as surgically resected epileptic brain tissue samples. To address these challenges as well as improve the molecular diagnostic rate, and better understand genotype–phenotype relationships in these disorders, we designed an ultra-sensitive targeted sequencing panel using droplet digital PCR (ddPCR) that includes six of the most common activating PI3K-AKT-MTOR pathway mutations and tested a large cohort of individuals with FMCD using this panel.4,15
ddPCR is an allele-specific molecular diagnostic method where each PCR reaction is partitioned into ∼20 000 separate emulsion droplets. This method has been optimized to quantify extremely low-frequency single nucleotide variants and copy number abnormalities [down to 0.01% variant allele fraction (VAF), and less than eight molecules of input DNA], and is thus defined as an ultra-sensitive molecular diagnostic technique.16–20 Given its high sensitivity, ddPCR has been increasingly used as a first-tier clinical diagnostic tool to efficiently and reliably detect and quantify common cancer-associated variants [such as the common PIK3CA hotspot mutations—p.C420R, p.E542K, p.E545K, p.H1047L and p.H1047R—in PIK3CA-related overgrowth disorders (PROS), the BRAF p.V600E mutation in brain and thyroid tumours, among others]. It has also been recently used in other paediatric overgrowth, vascular and lymphatic malformation phentypes.21–24 Developmental brain disorders associated with mutations in the PI3K-AKT-MTOR pathway, including FCD and HMEG, have a narrow mutational spectrum with a few ‘hotspot’ mutations representing a sizeable portion of molecularly solved cases.4,11,12,15 We therefore tested these mutational hotspots as a first-tier molecular method in a large cohort of affected individuals to efficiently identify mutations and study the relationships between levels of mosaicism and key neuroimaging and histopathological features.
Materials and methods
Human participants
Children diagnosed with focal intractable epilepsy requiring epilepsy surgery were prospectively enrolled in the Developmental Brain Disorders Research Program under an IRB approved protocol at Seattle Children’s Hospital (IRB no. 13291). All patients signed an informed consent according to the Declaration of Helsinki or were de-identified and included in the study under an IRB approved ‘waiver of consent’. Brain tissue samples were collected during clinically indicated epilepsy surgery. Additional samples (such as saliva, peripheral blood or skin fibroblasts) were also collected from affected individuals when possible.
Inclusion and exclusion criteria
Our study included individuals with epilepsy who underwent epilepsy surgery and were confirmed by neuroimaging and/or histopathologic examination to have one of the following diagnoses: FCD, HMEG, DMEG, other malformations of cortical development or other developmental lesions. Individuals with brain tumours were excluded from this study. Individuals with predominantly somatic overgrowth phenotypes who did not have any brain involvement (i.e. cortical malformations or megalencephaly) were excluded from this study.
Neuroimaging
All patients evaluated at the Pediatric Epilepsy Surgery Center at Seattle Children’s Hospital had brain magnetic resonance imaging (MRI) using the Siemens 1.5 T (Symphony or Avanto) or 3 T (TrioTim or PRISMA) systems, or external site data of equivalent quality. Common preoperative structural data included sagittal T1 MPRAGE (isotropic 1 mm), axial and coronal T2 (2–4 mm), and axial fluid-attenuated inversion recovery (FLAIR) images (2–4 mm). For image analysis, we performed diffusion weighted skull stripping with the brain extraction tool in FSL on T1-data then made three-dimensional renderings with BioImage Suite.25 Where indicated, and as helpful for lesion evaluation, the same skull-stripped mask was used for the T2- or FLAIR-data. Coordinates for brain tissue samples were created using an image guided surgery system (Metronic Stealth) with intra-operative xyz coordinates downloaded and then transformed into MRI space and overlaid using BioImage Suite. Brain MRIs were examined and assessed for the type, distribution and severity of the cortical dysplasia, as well as the presence of any other notable brain MRI abnormalities. All brain images were assessed for laterality of findings (i.e. unilateral or bilateral cortical malformations), symmetry, gradient (anterior versus posterior), lobes or brain regions affected, white matter abnormalities and any mass effect.
Histopathology
Histology sections of all resected brain tissues underwent comprehensive examination to establish a neuropathologic diagnosis. Haematoxylin and eosin stained sections were examined from formalin-fixed, paraffin-embedded (FFPE) from each site of brain tissue resection. In addition, immunohistochemistry was performed on sections from at least one, and often multiple, FFPE tissue block by established methods using mouse monoclonal anti-NeuN (Millipore, MAB 377) and mouse monoclonal anti-GFAP (Dako, Mo761) antibodies to assess neuronal and glial cell populations. FCD was histopathologically diagnosed and subtypes were determined on the basis of criteria established by the International League Against Epilepsy (ILAE) Task Force and described in detail by Blumcke and colleagues.7,26 For patients with histologically confirmed FCD types 2a or 2b, tissue sections from multiple brain regions, including areas with no histopathologic dysplasia, underwent additional immunohistochemical staining using the rabbit polyclonal anti-phospho-S6 (pS6) ribosomal protein (Cell Signaling, 2211). The percentage of pS6-positive cortical grey matter in each site was estimated on the basis of the fraction of cortical neurons with moderate-to-markedly intense cytoplasmic pS6 immunoreactivity. All the neuropathological assessments, including pS6 immunolabelling assessment, were performed by a board-certified paediatric pathologist (R.P.K.), who was blinded with respect to the neuroimaging and ddPCR data.
Sample processing and DNA extraction
We collected 159 specimens from a cohort of 58 children diagnosed with FCD, MEG, HMEG, DMEG and other malformations of cortical development (MCD) including polymicrogyria and multifocal cortical dysplasia. Samples included fresh-frozen (n = 96) and FFPE (n = 28) brain tissues from epilepsy surgery (total brain samples n = 124), as well as peripheral samples including saliva (n = 26), peripheral blood (n = 3), skin (cultured or uncultured fibroblasts) (n = 4), and buccal swab samples (n = 2). Brain samples were either snap-frozen tissues located immediately adjacent to tissues fixed and embedded for neuropathologic examination or were sections from FFPE tissue blocks used for histopathology. Genomic DNA extraction was performed using standard protocols with PureLink™ Genomic DNA Mini Kit (Invitrogen, catalogue number K1820-01). FFPE samples were processed using the GeneRead DNA FFPE Kit (Qiagen, catalogue number 180134) following the manufacturer’s protocol. DNA quality and concentration were quantified using the NanoDrop 3300 (ThermoFisher Scientific) and the Qubit 4 Fluorometer (ThermoFisher Scientific). If DNA quality was suboptimal (defined by 260/280 ratios below 1.8 and/or 260/230 ratio below 2.0), samples were cleaned and concentrated using a column-based kit (Zymo Research Genomic clean and concentrator, catalogue number D4011) and requantified before proceeding with ddPCR.
Droplet digital PCR testing
We designed a custom ddPCR-based panel targeting six mutations in three PI3K-AKT-MTOR pathway genes, namely PIK3CA (p.E542K, p.E545K, p.H1047R), AKT3 (p.E17K) and MTOR (p.S2215Y, p.S2215F). We selected these specific variants as they are the most common variants seen in FCD-HMEG,3,15,27 as well as the most common oncogenic mutations in these genes in somatic tissues in cancer overall (COSMIC, Catalogue of Somatic Mutations in Cancer). All ddPCR probes were validated by Bio-Rad (see Supplementary Table 1). As FFPE samples can have high levels of DNA degradation and nonreproducible sequence artefacts, especially C:G > T:A transitions that can increase the chance of false positive results, all FFPE-derived-DNA samples were treated with uracil DNA glycosylase to minimize PCR amplification errors before ddPCR testing. This method has been successfully used to increase the accuracy of detecting mosaic cancer-associated mutations in FFPE samples.28,29 Here, ∼8–10 ng of DNA was used per ddPCR reaction, and all samples were tested in technical quadruplicates using previously published methods.21,30 Briefly, the ddPCR reaction mix was assembled from 10 μl of ddPCR Supermix for Probes (Bio-Rad, 1863024), 1 μl of ddPCR validated mutant assay (Bio-Rad, see Supplementary Table 1) and 12 μl of water per reaction. A restriction enzyme was not included due to the low DNA input. Then 20 μl of the ddPCR mastermix along with 70 μl of Droplet Generation Oil were loaded into a disposable droplet generator cartridge and placed into a QX200 Droplet Generator (Bio-Rad). Generated droplets were transferred to a 96-well PCR plate and placed in a thermal cycler for amplification (see Supplementary Table 1 for probe-specific annealing temperatures). Amplification cycles were set as follows: denaturing phase at 95°C for 10 min, followed by 40 cycles of 94°C for 30 s and annealing/extension temperature for 1 min, with a final denaturation at 98°C for 10 min followed by holding at 4°C. ddPCR plates were then placed into a QX200 Droplet Reader (Bio-Rad) for fluorescent quantification. Data analysis was performed using the Quantasoft AP software. Positive fluorescence thresholds for mutant (FAM) and wild-type (HEX) associated probes were established using three controls (in quadruplicates) in each run: a no template control (NTC), a wild-type (non-mutant) control and a mutation-positive control (with VAF range 3–30%). Data from quadruplicate wells were pooled for each sample and VAF% was calculated. To account for rare non-specific mutant-positive (i.e. false positive) droplets, the calculated VAF for each sample was compared to the wild-type control run in the same plate. Delta confidence intervals (DCI) were calculated by subtracting the Poisson-normalized maximum fractional abundance (95% CI) for the wild-type control from the Poisson-normalized minimum fractional abundance (95% CI) for each sample of interest. Due to suboptimal DNA quality and amounts from some of the samples, such as FFPE-derived brain tissues, we used more stringent customized cut-offs to minimize false positive results, thus samples were deemed positive for a given mutant allele when their DCI exceeded 0.045 (rather than the DCI cut-off of 0 per the Bio-Rad analysis protocol) and the average number of wild-type-positive droplets for the pooled set of quadruplicate wells exceeded 3000 droplets. If the number of wild-type droplets was between 500 and 3000, samples were retained in the analysis only if the DCI was above one and/or other samples from the same individual showed a clearly positive result. These cut-offs were empirically established by analysing borderline-positive samples (with a DCI between 0 and 0.05) that were assayed in multiple independent experiments with different DNA concentrations. Quantasoft 2D plots of representative examples of mutation-negative and -positive samples are shown in Supplementary Fig. 1A. When samples were analysed in more than one experiment, the ddPCR run producing the highest wild-type positive droplet count and highest DCI was reported here.
Validation of variants via orthogonal probe-based and sequence-based methods
Orthogonal methods of variant identification were used to confirm, where possible, results obtained via ddPCR. We performed smMIPs Amplicon Sequencing and a capture-based custom designed SureSelect NGS (next generation sequencing) panel as previously described.31–36
Statistical analysis
Statistical analysis and relative graph generation was performed in GraphPad Prism v.9.0.2. The appropriate statistical test was chosen on the basis of the number of replicates and type of comparison of interest. Details on specific statistical tests and relative results are provided with the associated data.
Data availability
All generated data are included in this article and associated Supplementary material. The raw data generated and/or analysed during the study are available from the corresponding author on request.
Results
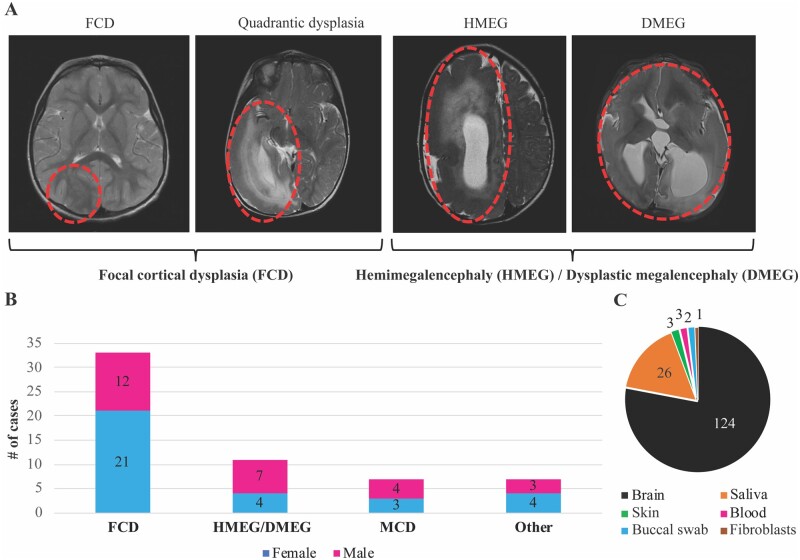
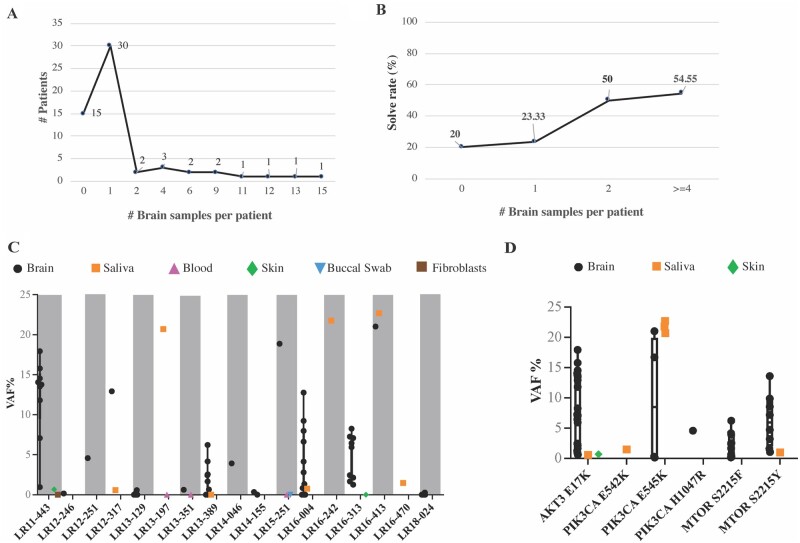
We performed deep molecular analysis of a cohort of 58 children who underwent epilepsy surgery and subgrouped them on the basis of their neuroimaging and histopathologic diagnoses into four groups: FCD (n = 33), HMEG-DMEG (n = 11), other MCD (n = 7) and other cortical lesions (n = 7). The last category includes gliosis (n = 3), hypoxic-ischaemic encephalopathy (HIE, n = 1), meningoangiomatosis (n = 1), mesial temporal sclerosis (MTS, n = 1) and stroke (n = 1). Notably, one individual with HMEG (LR16-242) had contralateral hemimicrencephaly. HMEG with contralateral hemimicrencephaly has been previously reported suggesting that the non-HMEG hemisphere may have developmental abnormalities as well, although molecular studies on the contralateral hemisphere have been limited.37,38 A summary of the clinical subgroups and samples obtained from this cohort, as well as representative brain MR images, are shown in Fig. 1. Of the 58 children tested, we obtained at least one brain tissue from 43 individuals, and were able to obtain two or more brain regions from 13 individuals, including six individuals with nine or more brain tissue samples (Fig. 2A). Following mutation detection (as determined by the cut-off criteria stated previously), any available additional samples from the affected individuals were serially tested to further quantify the levels of mosaicism (VAF) across all available tissues. All in all, the six hotspot mutations (PIK3CA p.E542K, p.E545K, p.H1047R; MTOR p.S2215F and p.S2215Y; AKT3 p.E17K) were detected in 51 samples (n = 44 brain, n = 6 saliva, n = 1 skin) from 17 individuals, with VAFs ranging from 0.14 to 22.67% (Table 1). The overall yield for molecular diagnostic testing, or the molecular solve rate, was 29.31% in this cohort. We identified one of these six hotspots in the brain in 15 of 43 (34.88%) individuals, as well as in peripheral tissues from 2 of 15 (13.33%) individuals for whom surgically resected brain tissues were not available. The molecular diagnostic yield correlated with the number of samples analysed from all individuals. However, the molecular yield increased to 54.5% for individuals with four or more resected brain tissue samples (Fig. 2B and Supplementary Fig. 1A). Most of the tested hotspot mutations were detected in brain samples, although we were able to detect PIK3CA mutations in a small subset of saliva samples (6 of 26 samples, 23.07%), and the AKT3 p.E17K variant was also detected in a skin sample (LR11-443), while MTOR variants were exclusively found in brain samples, albeit we had few non-brain samples in this cohort (Fig. 1C and Supplementary Fig. 1B).
Figure 1.
Overview of the cohort and samples. (A) Representative brain MRIs of patients with focal cortical dysplasia (FCD), hemimegalencephaly (HMEG) and dysplastic megalencephaly (DMEG). Representative affected brain areas are highlighted by red ovals. Patients diagnosed with FCD and quadrantic dysplasia were clustered in the FCD category, while patients diagnosed with HMEG or DMEG were clustered in the HMEG/DMEG category. (B) Bar graph representing the cohort distribution stratified by the four clinical diagnostic categories (FCD, HMEG/DMEG, and malformations of cortical development, MCD, as well as other diagnoses) and sex. The ‘other’ diagnoses include gliosis (n = 3), hypoxic-ischaemic encephalopathy (HIE, n = 1), meningoangiomatosis (n = 1), mesial temporal sclerosis (MTS, n = 1) and stroke (n = 1). (C) Pie chart representing types and numbers of samples collected from the 58 individuals in this series. The vast majority of samples were brain (n = 124, fresh frozen or FFPE).
Figure 2.
Molecular results. (A) Number of brain samples per patient in our cohort. Most of the patients had at least one brain sample (n = 30). (B) The molecular yield (or solve rate) stratified by number of brain samples. The solve rate was calculated as number of positive cases with a given number of brain samples over the total number of cases with the same number of brain samples, and expressed as a percentage. As expected, our solve rate was increased with increasing numbers of brain samples, indicating that the number and type of samples per patient is crucial to identify a causal mosaic mutation. (C) VAF% for the 17 mutation-positive patients. The y-axis represents the VAF%, while patients are listed on the x-axis. All samples belonging to the 17 patients are represented, including negative ones (crossing the zero on the y-axis). Grey shaded boxes are used to represent every other patient to delineate samples belonging to each individual. Different tissues are represented as indicated in the graph legend. Only brain and saliva samples were mutation-positive, whereas blood, buccal swab and skin fibroblast-derived samples were negative. (D) Positive samples stratified by the PI3K-AKT-MTOR hotspot mutation to visualize the range of VAF for each mutation across individuals. Notably, the PIK3CA, p.E545K, variant had the widest VAF% range and was the only hotspot to be present at higher VAF% in saliva rather than brain samples.
Table 1.
Clinical and molecular data of individuals with pathogenic PI3K-AKT-MTOR variants
| Patient ID | Sex | DX (MRI) | DX (NP) | Seizure: age at onset | No. of epilepsy surgeries | Other clinical features | Pathogenic variant | VAF% range | VAF% average | No. of samples | No. of brain samples | No. of positive samples |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LR16-470 | M | HMEG | N/A | N/A | N/A | Hyperpigmentation on several areas of the body | PIK3CA (p.E542K) | 1.47 | 1.47 | 1 | 0 | 1 |
| LR13-197 | F | DMEG | FCD 1 b | 29 days | N/A | Right lower extremity hemihypertrophy with left-sided facial asymmetry | PIK3CA (p.E545K) | 20.68 | 10.36 | 2 | 0 | 1 |
| LR15-251 | M | HMEG | N/A | 1 day | 3 | Flat feet, translucent skin, sebaceous nevus on left cheek, congenital recurrent nocturnal apnoea, adenoidectomy | 18.86 | 6.33 | 3 | 1 | 1 | |
| LR16-242 | F | HMEG | N/A | 1 day | No | Enlarged R cheek and eye (ipsilateral to HMEG), swirled brown macule on R cheek extending to neck | 21.76 | 21.76 | 1 | 0 | 1 | |
| LR16-413 | M | HMEG | FCD | 2 days | 1 | Hypertrophic left lip/cheek | 21.01–22.67 | 21.84 | 4 | 1 | 2 | |
| LR18-024 | F | FCD | FCD 2a | 6 years | 2 | – | 0.28 | 0.11 | 14 | 13 | 2 | |
| LR12-251 | F | FCD | FCD 2a | 3 years | 2 | – | PIK3CA (p.H1047R) | 4.58 | 4.58 | 1 | 1 | 1 |
| LR11-443 | F | HMEG | FCD 2a | 3 days | 2 | Vascular malformations of the skin, organomegaly, scoliosis, visual loss in left eye | AKT3 (p.E17K) | 13.51–13.76 | 13.64 | 11 | 9 | 10 |
| LR12-317 | M | HMEG | N/A | 7 days | 1 | Vascular abnormalities of lower extremities; PWS on the legs, two small lesions on chest and arms | 0.58–12.93 | 6.76 | 2 | 1 | 2 | |
| LR13-351 | F | FCD | FCD 2a | ∼1 month | – | 0.62 | 0.32 | 2 | 1 | 1 | ||
| LR16-313 | F | FCD | FCD 2a | 4 days | 1 | – | 1.26–8.25 | 4.25 | 10 | 9 | 9 | |
| LR12-246 | M | HMEG | FCD 2a | 3 days | 2 | Sacral lipoma | MTOR (p.S2215F) | 0.16 | 0.16 | 1 | 1 | 1 |
| LR13-129 | M | FCD | FCD 2 b | 11 months | 4 | – | 0.24–0.57 | 0.13 | 9 | 9 | 2 | |
| LR13-389 | F | FCD | FCD 2a | 5 weeks | 2 | Amblyopia of right eye, dissociated vertical deviation, anisometropia | 0.65–6.22 | 1.14 | 16 | 15 | 6 | |
| LR14-046 | M | FCD | FCD 2 b | 7 months | 2 | – | 3.91 | 3.91 | 1 | 1 | 1 | |
| LR14-155 | F | FCD | FCD 2 b | 5 years | 1 | – | 0.34 | 0.2 | 3 | 2 | 1 | |
| LR16-004 | M | HMEG | FCD 2a | 3 weeks | 1 | – | MTOR (p.S2215Y) | 0.96–13.59 | 4.23 | 12 | 11 | 9 |
List of the diagnoses, clinical features and molecular results for the 17 mutation-positive cases. When a dash (–) is present in the ‘other clinical features’ cells, it refers to absence of other clinical features. Abbreviations: DX = diagnosis; NP = neuropathology; N/A = not available.
Molecular results
Using our ultra-sensitive and highly targeted ddPCR PI3K-AKT-MTOR panel, we were able to detect and quantify levels of mosaicism in 17 individuals including six with the PIK3CA p.E545K mutation; five with the MTOR p.S2215F mutation and three with AKT3 p.E17K, and one for each of the remaining mutations: PIK3CA p.E542K, PIK3CA p.H1047R and MTOR p.S2215Y. The levels of mosaicism (or VAF ranges) are graphically shown in Fig. 2C, including all positive and negative samples from these 17 individuals. Individual LR16-413 had the highest overall VAF of the cohort, with two samples collected and testing positive for PIK3CA p.E545K at 21.01% in frozen brain tissue and 22.67% in saliva. Notably, blood and fibroblast-derived DNA from this patient were previously tested by a single molecular inversion probe (smMIPs) targeted multi-gene panel of PI3K-AKT-MTOR pathway genes with no mutation detected.11 In contrast, individual LR18-024 had the lowest level of mosaicism (PIK3CA p.E545K at a VAF of 0.14%) of the entire cohort in one of the available FFPE brain samples. When both brain and saliva-derived DNA samples (i.e. paired samples) were available for a given individual, the levels of mosaicism were consistently higher in the brain, with the notable exception of the PIK3CA p.E545K mutation that had a higher VAF in saliva compared to brain (individual LR16-413) (Fig. 2D). The distribution of mutations and VAFs per each sample for all mutation-positive individuals are shown in Supplementary Table 2. Children who tested negative for all six hotspot mutations, as defined by the previously stated cut-off criteria, are listed in Supplementary Table 3 (n = 41).
Genotype–phenotype correlation
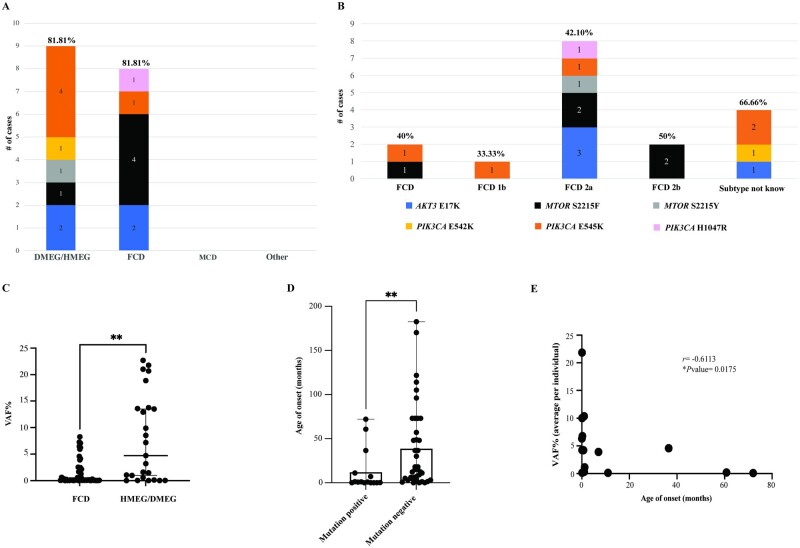
Following mutation detection, we aimed to characterize and better define genotype–phenotype relationships among individuals with these common mutations, correlating the molecular data with key neuroimaging and histopathology features. We found that about 82% of individuals diagnosed with HMEG-MEG by neuroimaging were positive for these six mutational hotspots, compared to only 24.24% of individuals with FCD (Fig. 3A). While the number of mutation-positive individuals within each category is not sufficient to establish a statistically significant correlation, we observed a possible association between FCD (by neuroimaging) and MTOR mutations (p.S2215F, four of the seven positive individuals), and between HMEG-MEG and PIK3CA mutations (p.E542K and p.E545K, six out of nine positive individuals), followed by AKT3 p.E17K present in both cohorts (two of the nine positive HMEG cases, and two of the eight positive FCD cases) (Fig. 3A and B). None of the individuals diagnosed with MCD or other cortical lesions (n = 14) were positive for the PI3K-AKT-MTOR mutations tested. We further stratified the cohort on the basis of the histopathological diagnosis when available, and confirmed that mutations were enriched in FCD type 2, with a molecular solve rate of 42.1% (Fig. 3B), a finding that has been corroborated by other studies.39,40 For five individuals, we could only obtain histopathology reports confirming FCD with no further specification of the FCD subtype (40% solve rate, Fig. 3B and Supplementary Table 5), while no histopathology data were available in six individuals (67% solve rate, Fig. 3B). Interestingly, only 1 of 10 patients with FCD type 1 was positive for a hotspot mutation (patient LR13-197, PIK3CA p.E545K, 33% of FCD 1b cohort, Fig. 3B). Further, no mutation was detected in the single patient with FCD type 3. Overall, we were able to establish that individuals with FCD had a VAF% range that is significantly lower (eight individuals, 55 samples, VAF% range 0–8.25; mean = 1.32; median = 0.12) than individuals presenting with HMEG/DMEG (nine individuals, 33 samples, VAF% range 0–22.67; mean = 8.51; median = 7.18) (Fig. 3C). Detailed molecular and clinical data for all mutation-positive individuals are provided in Supplementary Tables 2 and 3, and detailed ddPCR results for individuals who tested negative for the six hotspots (n = 41) are provided in Supplementary Table 4.
Figure 3.
Genotype–phenotype correlations of PI3K-AKT-MTOR hotspot mutations. (A) Hotspot mutations detected in our cohort are stratified according to the four diagnostic categories and represented as stacked bar graphs. Notably, only FCD and HMEG/DMEG cases were positive for the six positive hotspots tested, while none of the MCD or ‘other’ diagnoses were positive for these variants. The frequency of PIK3CA mutations was higher in DMEG/HMEG with a solve rate of 81.81%. In contrast, FCD had higher frequency of MTOR mutations (9/11 cases) with an overall solve rate of 24.24%. (B) The same data from graph A was stratified by the neuropathology classification/diagnosis and represented via stacked bar graph (number of cases in our cohort n = 58). Solve rate for each category is shown as a percentage and calculated as number of positive cases/number of total cases within the same neuropathology subtype. FCD type 2a had the highest overall molecular yield in our series. (C) VAF% range in the cohort stratified on the basis of the neuroimaging diagnosis. The FCD cohort presents a VAF range significantly lower than the HMEG/DMEG cohort. Mann–Whitney test, two tailed, P < 0.0001, Mann–Whitney U = 411.5. (D) Bar graph showing the age of onset of seizures in the hotspot mutation-positive (n = 15) versus hotspot mutation-negative (n = 39) cohorts. Each dot represents one individual, the top of the bar indicating the mean age of onset (12.79 months in mutation-positive, 39.46 months in mutation-negative) and error bars represent the full range. Kolmogorov–Smirnov test revealed a significant difference with **P = 0.0037. (E) Correlation analysis of seizure age of onset and VAF% in hotspot mutation-positive patients (n = 15). Spearman correlation analysis demonstrated a significant negative correlation with an r = −0.6113 and *P = 0.0175.
To determine whether these highly activating mutations were associated with more severe epilepsy, we compared the age of onset of epilepsy between mutation-positive and -negative individuals and identified that individuals who tested positive had a significantly earlier age of onset of epilepsy (average 12.79 months, ±23.81, n = 15) compared to mutation-negative individuals (average 39.46 months, ±47.72, n = 39) using the Kolmogorov–Smirnov test (P-value = 0.0037, Fig. 3D). Further, we correlated the age of onset of epilepsy with the average VAF% for 15 of the mutation-positive individuals and identified a negative correlation between these variables with an earlier age of onset of epilepsy as the VAF% increases (r = 0.6273, P = 0.0142) using the Spearman correlation test (Fig. 3E, see Table 1 for a complete list of seizure age of onset and VAF%).
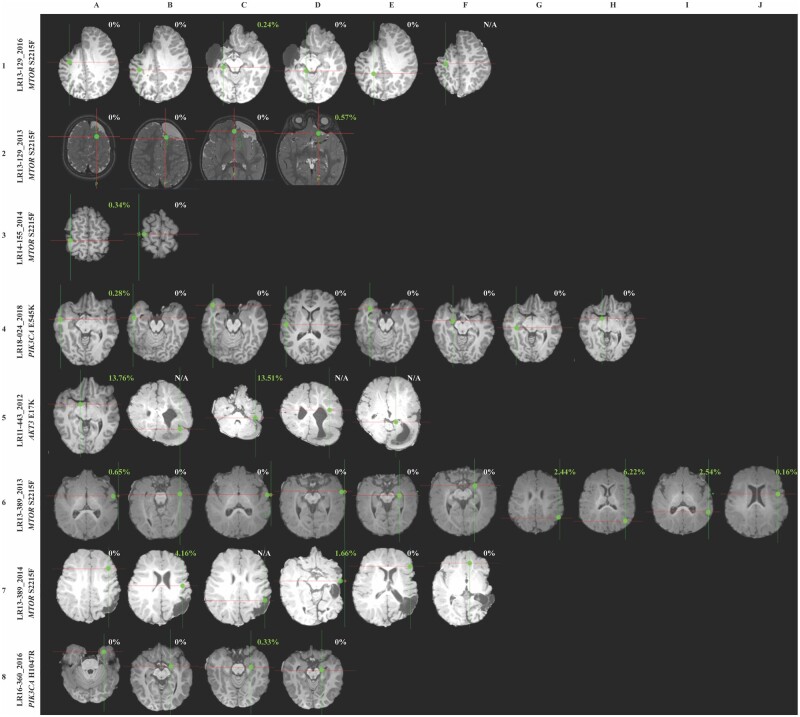
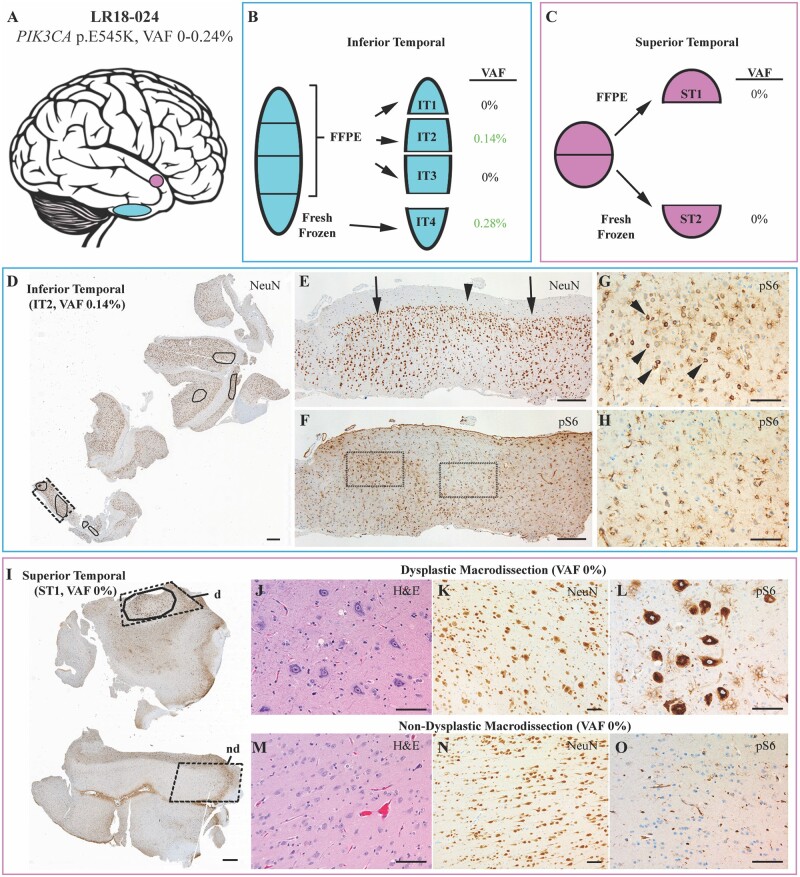
To further delineate a correlation between mutation burden and severity of the phenotype in individuals with mutations, we analysed their neuroimaging and histopathology features and paired them with VAF% when matched samples were available. For six of the positive individuals, we were able to overlay brain MR imaging data of the resection sites obtained during epilepsy surgery with their relative mutation burden (VAF%, Fig. 4). Notably, in several individuals, the severity and distribution of the malformation by neuroimaging was extensive despite low VAF% in those regions. For example, patient LR13-129 (row 1 in Fig. 4) had extensive involvement of the frontal, temporal and parietal regions, and required a total of four epilepsy surgeries, yet harboured the MTOR, p.S2215F, variant at an extremely low VAF (0.24–0.57%) in these brain tissues. Similarly, patient LR18-024 had an extremely low VAF for PIK3CA, p.E545K, in surgically resected tissues despite more extensive involvement by neuroimaging and histopathology (row 4 in Figs 4 and 5). As a comparison, we assessed brain MR images from individuals who tested negative for all six mutational hotspots (data shown in Supplementary Fig. 2). However, we cannot rule out the possibility that these individuals could have other variants in the same genes or other FMCD-associated genes, limiting further correlations.
Figure 4.
Neuroimaging findings and correlations with genotype. Representative brain MRIs showing brain biopsy or tissue resection locations alongside identified VAF of PI3K-AKT-MTOR hotspot mutations. For each image, cross-hairs reflect the site from which a tissue sample was obtained for molecular analysis, with letters at the top reflecting the multiple samples taken. At each location, ddPCR results are shown for the respective region. Green dots represent the exact location of resection, and ddPCR results are indicated as VAF% for positive samples. The year in which the surgery was performed is indicated after the underscore, as some patients underwent multiple brain surgeries (LR13-129, LR13-389). The reported VAF% did not directly correlate with the severity and distribution of visible cortical lesions.
Figure 5.
Histopathological findings and correlations with genotype in tissue samples from patient LR18-024. (A) Separate resected pieces of brain tissue (shown schematically) were received from the inferior (blue) and superior (pink) temporal lobe. (B) The inferior temporal piece was divided into four portions: three were formalin-fixed and embedded in paraffin (FFPE, IT-1, -2, -3) and one (IT4) was fresh frozen, as shown in the scheme. VAF% results obtained via ddPCR for the four portions are shown next to each subresection. (C) The superior temporal piece was divided in two halves: one half submitted as FFPE (ST1) and one half as fresh frozen (ST2). (D) A NeuN immunostained section at low magnification shows the entire block face from IT-2 with areas of cortical dysplasia (outlined). (E) Higher magnification of the region indicated by dashed rectangle in D, which shows abnormal clustering of neurons in two areas of histological dysplasia (arrows) with intervening histologically normal cortex (arrowhead). (F) pS6 immunolabelling of a tissue section adjacent to (E), including dysplastic (G) and non-dysplastic (H) foci. pS6-immunoreactive neurons (arrowheads in F) are present in the dysplastic focus, whereas only glia cells show immunoreactivity in the non-dysplastic zone (G). (I) A pS6 immunostained section showing the entire block face from the superior temporal piece (ST1) with the dysplastic focus encircled. The dashed outlines indicate areas that were macrodissected from the block to enrich for histologically dysplastic (d) and non-dysplastic (nd) cortex. Images from the macrodissected regions highlight the cellular enlargement, disorganization, and strong pS6 immunoreactivity that differentiate the dysplastic (J–L) and non-dysplastic (M–O) cortex. Scale bars = 1 mm in D and H; 300 μm in E and F; and 100 μm in G, H and J–O.
Next, we aimed to identify correlations between genotypes and histopathological features for all 58 individuals in our cohort (as summarized in Supplementary Table 5). For three of the mutation-positive children, we performed a matched analysis comparing histopathological findings with the gene and VAF% for brain regions that were independently analysed by ddPCR as well as studied by haematoxylin and eosin staining and immunostaining for the neuronal marker NeuN and for the MTOR pathway hyperactivation marker pS6 (Fig. 5, Supplementary Fig. 3 and Supplementary Table 6). Notably, in patient LR18-024 we detected the PIK3CA p.E545K mutation at a very low level (VAF 0.28%) in only one (inferior temporal 4, IT4) of nine fresh-frozen brain samples, despite lack of clear dysplastic neuropathology in this region (Fig. 5A and Supplementary Table 6). Conversely, the superior temporal region (fresh-frozen sample ST2) that tested negative for the PIK3CA mutation by ddPCR demonstrated strongly positive pS6 staining and a dysplastic focus with clear FCD2a histopathology. Due to clinical and procedural workflows, the brain samples that were analysed for histopathology were FFPE blocks, while the DNA samples analysed initially for ddPCR were extracted from fresh-frozen sections from the same or contiguous areas, thus the initial molecular analysis might represent perilesional sublocations within the same resected brain sample. Therefore, to further confirm these ultra-low VAF levels and correlate them with histopathology even more accurately, we obtained FFPE scrolls from these two regions (inferior and superior temporal) using the same blocks that were analysed for histopathology. For the inferior temporal region, three tissue blocks (IT 1, 2 and 3) were available with 20 sections each collected and analysed. DNA from these three regions were tested for all six mutational hotspots, and only one of the three blocks (block IT2) tested positive for PIK3CA p.E545K (VAF 0.14%), confirming that this area was indeed positive albeit at a very low VAF%. For the superior temporal specimen, we were also able to perform manual dissection of the dysplastic and non-dysplastic areas (block ST1), comparing these with the full scrolls of the same block. Despite enriching for the affected regions in this sample, we confirmed the absence of the PIK3CA p.E545K mutation, validating the results obtained from the fresh-frozen sample (Fig. 5). A list of ddPCR results for the fresh-frozen and FFPE samples for the inferior temporal and superior temporal specimens for individual LR18-024 is listed in Supplementary Table 6. We observed the same phenomenon in other individuals, such as LR13-129 and LR16-313, where the severity of cortical dysplasia did not correlate with VAFs (i.e. dysplastic and non-dysplastic areas were present at different levels in the same brain regions that tested positive via ddPCR for MTOR p.S2215F VAF 0–0.57% in LR13-129 and AKT3 p.E17K VAF 1.26–8.25% in LR16-313) (Supplementary Fig. 3 and Supplementary Table 6).
Finally, we compared the number of epilepsy surgeries with the number of samples obtained per individual between mutation-positive (n = 17) and mutation-negative individuals (n = 41). We found no statistically significant differences in the number of surgeries in these two cohorts. However, we found that the number of samples per individual obtained from mutation-positive children was significantly higher, with an average of 4.74 samples versus 1.65 (P < 0.01), compared to mutation-negative individuals (Supplementary Fig. 3).
Comparison of droplet digital PCR with other molecular diagnostic methods
To inform our molecular diagnostic approaches for this spectrum, we compared ddPCR data with several ultra-deep sequencing methods that have been used by us and others to detect mosaic mutations and accurately quantify VAF% in different tissues, including single molecule molecular inversion probes (smMIPs), amplicon sequencing and a capture-based custom designed SureSelect NGS panel.31–35 We compared results obtained with these methods for selected mutation-positive individuals on the basis of sample availability (Supplementary Table 2). Most variants detected by ddPCR were confirmed via at least one of these orthogonal approaches when testing was performed on the same region or sample, except for two ultra-low VAF variants: the PIK3CA p.E5454K variant in LR18-024 (VAF 0.28%) and the MTOR p.S2215F variant in LR12-246 (VAF 0.16%). Further, while the level of concordance in the detection rates between the other targeted methods (smMIPs, ultra-deep NGS and amplicon sequencing) was high, the concordance level decreased when comparing ddPCR and smMIPs (13/16, 81.25%) or ddPCR and amplicon sequencing (6/9, 66.67%), due to presence of false positives (one sample for case LR13-389, in bold in Supplementary Table 2) and false negatives (three samples for case LR13-389 in italic in Supplementary Table 2) in both smMIPs and amplicon sequencing data. Importantly, these other sequencing methods failed to detect several of the mutations detected by ddPCR due to suboptimal depth of coverage compared to ddPCR, which has the highest sensitivity. Exome sequencing was performed on two patients (LR14-155, saliva sample; and LR16-004, saliva and one brain region), and while this method was able to detect the MTOR variant in patient LR16-004 at similar VAF% found via ddPCR, it did not detect the low MTOR VAF variant in patient LR14-155, as expected.
Discussion
In this study, we performed ultra-sensitive targeted molecular profiling to delineate the molecular spectrum of FMCD phenotypes caused by common or hotspot PI3K-AKT-MTOR pathway mutations with analysis of complementary clinical, neuroimaging and histopathology data. As recently reported by us and other groups, we identified a key trend for an association between FCD and MTOR mutations, and between HMEG-MEG and PIK3CA mutations.1,4,14,15,27,41 Furthermore, we confirmed the association between VAF% and phenotype, with FCD brain lesions having on average lower VAF% than HMEG/DMEG.27,41 These finding support the hypothesis that the time window during which these mutations occur in the developing human brain might have a larger impact than the type of variant itself, with HMEG-associated genetic variants potentially arising earlier in foetal development and thus involving a larger fraction of the developing brain than FCD-associated variants.41 Within the FCD cohort, the two MTOR hotspot mutations were present in five individuals (four with p.S2215F and one with p.S2215Y). In contrast, five individuals from our HMEG-MEG cohort had the PIK3CA p.E545K hotspot mutation, and one had PIK3CA p.E542K. The AKT3 p.E17K mutation was detected in both HMEG-DMEG and FCD cohorts (2/9 HMEG-MEG; 2/8 FCD). Notably, we found differences in the molecular diagnostic yield between FCD subtypes, with only 1/10 individuals with FCD 1 positive for one of the hotspot mutations (10% of FCD 1 cohort, 33% of FCD 1b cohort); while 41.66% of the FCD 2 cohort had one of the hotspot mutations (10/24 cases), including specifically 42.10% of FCD 2a (8/19 individuals) and 50% of the FCD 2b (2/4). However, it is important to note that FCD 2a is the most common form of FCD overall and was present at a higher frequency in our cohort (19 of the 40 individuals diagnosed with FCD by neuropathology, Fig. 3B). These differences suggest that FCD type 1 is probably more causally heterogeneous and may result from other genetic factors, including other PI3K-AKT-MTOR pathway mutations that could be peri-lesional (or at the periphery of the lesion) or non-genetic factors such as hypoxic-ischaemic insults.3,27,42 Our study further supports the evidence that the highly recurrent AKT3 E17K variant is causal of HMEG and FCD, with three mutation-positive children in this series (LR11-443, LR13-351, LR16-313) who all had FCD2a histopathology. Unfortunately, neuropathologic evaluation of the fourth individual with the AKT3 variant (LR12-317) was not available.
We further aimed to address the key question of whether there is a relationship between mutation burden in the brain and severity of the cortical dysplasia, as several reports have suggested no direct correlation between levels of mosaicism (or VAF%) and phenotype severity.3,27,43 We therefore assessed whether there are any correlations between age of onset of epilepsy and the VAF%, with the hypothesis that children with a higher mutation burden have earlier seizure onset, as a higher VAF might indicate an earlier developmental time point for the occurrence of the mutation in the developing brain with more severe functional dysregulation of neural networks, as recently published by other groups.27 In our cohort, all children with an average VAF > 5% had very early-onset epilepsy (between days of life 0 and 30), whereas children with VAFs <1% had a later age of onset of epilepsy (between 3 and 6 years of age). These data support the finding that these oncogenic PI3K-AKT-MTOR pathway mutations are strong drivers of epilepsy in children and might have direct correlations with the underlying mechanisms, with probable independent effects from neuroimaging or histopathology, supporting similar observations in animal models.44
A similar genotype–phenotype correlation in PIK3CA-related vascular abnormalities was recently reported in a cohort of individuals.22 These findings may have a direct impact on prognosis and possibly therapies including future eligibility for treatment using PI3K-AKT-MTOR pathway inhibitors. However, our data also demonstrate no correlation between VAF and the severity of the brain malformation or the structural lesions at the macroscopic (MRI) or microscopic (histopathology) levels, as the VAF% across all individuals did not correlate directly with the severity of the cortical malformation or degree of dysplasia by imaging, confirming our findings and those of other groups.4,14,15,27,43,45–47 On retrospective analysis of brain MR data, we could not identify specific MRI features that were exclusively present in the mutation-positive cohort and that would distinguish mutation-positive individuals from mutation-negative individuals. When comparing mutation-positive and -negative subgroups, the number of samples obtained per individual were higher from mutation-positive individuals supporting the hypothesis that more extensive surgical resection have been performed in this cohort (Supplementary Fig. 4).
In our study, five individuals (LR12-317, LR13-129, LR15-251, LR16-313 and LR18-024) had a hotspot mutation detected in FFPE brain samples (Supplementary Table 2). In individual LR18-024, we were able to directly compare ddPCR results from fresh-frozen and FFPE samples from the same brain regions obtained during the same surgical procedure (Fig. 5 and Supplementary Table 6). Not surprisingly, the VAF% was lower in the FFPE sample than the fresh-frozen brain sample, possibly due to increased DNA degradation and lower yield in these samples. However, another explanation for this variance could be an actual difference in the number of mutant cells in the two samples as they were from slightly different subregions, as discussed previously. When examining VAF% in the same brain tissue blocks that were characterized histopathologically, we were able to confirm that the brain region with normal histoarchitecture (the inferior temporal lobe) was positive for the PIK3CA p.E454K via ddPCR, while the brain region with a focus of FCD2a histopathology (superior temporal lobe) was negative (Fig. 5 and Supplementary Table 6). These observations further support the notion that levels of mosaicism do not always correlate with histopathology or, alternatively, that mutations may have non-cell autonomous consequences in the developing human brain.
Our study also demonstrated the utility of ultra-sensitive and highly targeted molecular diagnostic screening by ddPCR for mosaic mutations in suspected PI3K-AKT-MTOR related brain malformations (FCD, HMEG and MEG). Due to the nature of this spectrum and the implications of establishing a molecular diagnosis for successfully treating these disorders, we believe that a first-tier ddPCR-based molecular screen holds promise for improving the accuracy and efficiency of detection of mutations. In highly mosaic disorders such as FCD and HMEG, pathogenic mutations can be present at extremely low levels, even in severely affected tissues.2,8–12,14,15,21,27,45 For this reason, it is crucial to use a molecular diagnostic method with sufficient sensitivity to detect low-level mosaicism, and our study shows that ddPCR provides an optimal approach for this purpose.21–24 When considering sample availability and selection, it is imperative to select the relevant tissue to efficiently identify mutations. Our data clearly shows a relationship between the number and types of sample obtained per individual with the molecular solve rate. Our solve rate was enhanced, in fact, when testing at least one brain tissue sample per individual (solve rate 26.86 versus 13.33% in non-brain samples), with the highest yield when individuals had six or more brain specimens tested (solve rate 75%, Fig. 2). Given that brain samples might not be available for children who are not eligible for epilepsy surgery, an alternative tissue source could be saliva-derived DNA, as we were able to detect hotspots mutations in six out of seven individuals on whom we had at least one saliva sample, with only one positive individual (LR13-389) having a negative result in saliva (with VAFs of 1.66–6.22% in the brain) (Supplementary Fig. 1A). The ability to identify mosaic mutations in saliva samples in our cohort could be explained by the embryonic origin of the cell types that are usually found in these samples. Saliva specimens are mostly constituted by water with a small cellular fraction: the latter generally containing leucocytes and squamous epithelial cells.48 Leucocytes, being of mesodermal origin, are the same cells from which DNA is derived from peripheral blood samples. On the other hand, squamous epithelial cells are derived from the ectoderm; the germlayer that gives rise to the neuroectoderm and subsequently to neuronal cells.49,50 Therefore, we hypothesize that the ability to detect FMCD-associated mosaic variants in saliva samples depends on its content of squamous epithelial cells, with a higher yield than peripheral blood due to the shared embryonic origin with neuronal cells. Furthermore, we were able to detect a variant in one skin sample out of three (AKT3 p.E17K, LR14-155), adding to the evidence that tissues that share their embryonic origin (i.e. ectoderm) should be prioritized for molecular diagnostics. As expected, we were not able to detect mutations in peripheral blood, although these samples were a minority in our cohort (n = 5), These findings have important implications for sample validation in clinical laboratories offering diagnostic testing for these and other related developmental brain disorders.
We molecularly solved 29.31% of individuals in this series. Importantly, we were able to solve 9 of 11 individuals with HMEG-DMEG (81.81%) and 10 of 24 individuals with FCD type 2a, highlighting the contribution of PIK3CA and AKT3 hotspot mutations in HMEG-DMEG and MTOR mutations in FCD2a. Overall, these data support that screening these common mutations in lesional brain tissues comprises an efficient first-tier molecular diagnostic approach in affected children. The additional advantage of using a ddPCR-based approach is the minimal DNA input requirement. Our assay used only used 8 ng of genomic DNA per well, with four replicate wells per hotspot. This amount of DNA is generally available from most clinically obtained samples. This specific advantage with ddPCR compared to other deep sequencing methods such as smMIPs, exome and genome sequencing, which have a much higher genomic input requirement with lower sensitivity for mosaicism detection makes ddPCR optimal for use as a molecular diagnostic tool in this spectrum. Our results also show that ddPCR has deeper coverage when compared to these other methods and can discriminate type 1 and type 2 errors (false positives and negatives, respectively), demonstrating higher specificity and sensitivity. Last, ddPCR is optimal for low-quality or degraded DNA, such as FFPE samples (e.g. archival surgical tissues)16 and samples containing traces of substances known to inhibit regular PCR reactions.51
While ddPCR provides targeted and ultra-high depth coverage, it is notably a low-throughput method with probes designed to detect specific mutational sites. It is therefore probably an inefficient molecular diagnostic tool in disorders with a very wide mutational spectrum. We acknowledge that the molecular spectrum of FCD, HMEG and MEG is constantly expanding, and while most genetic variants are mosaic, a fraction of individuals has germline or constitutional variants in other genes such as DEPDC5 or SLC35A2, among others.27,52,53
In conclusion, our study combining deep molecular profiling of the most common oncogenic PI3K-AKT-MTOR pathway mutations in FMCD with detailed analysis of clinical, neuroimaging and histopathology features shows several key and novel findings including (i) confirmation of an association between PIK3CA mutations and HMEG-MEG and MTOR mutations and FCD; (ii) a strong correlation between the age of onset of epilepsy in FCD and HMEG-MEG with mutational burden (mosaicism level); and (iii) weak or no correlation between levels of mosaicism of mutations and histopathological abnormalities. Last, our study supports the utility of ddPCR in a clinical setting for targeted testing of this under-characterized spectrum to efficiently detect low-level mutations, allowing early molecular diagnosis and facilitating molecularly targeted therapies (i.e. PI3K-AKT-MTOR pathway inhibitors).
Supplementary Material
Acknowledgements
We thank the families and their provider for their participation in this study.
Funding
This work was supported by funding from: the Brotman-Baty Institute (to G.M.M., J.H., J.O., E.N., R.S. and R.K.), Jordan’s Guardian Angels and the Sunderland Foundation (to F.P. and G.M.M.); the Tuscany Region—DECODEE project (to R.G.) and grant no. NIH 1R01NS092772 (to W.B.D.).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Abbreviations
- ddPCR
droplet digital PCR
- DMEG
dysplastic megalencephaly
- FCD
focal cortical dysplasia
- FFPE
formalin-fixed-paraffin embedded
- FMCD
focal malformations of cortical development
- HMEG
hemimegalencephaly
- MEG
megalencephaly
- smMIPs
single molecule molecular inversion probes
- VAF
variant allele fraction
Contributor Information
Filomena Pirozzi, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA.
Matthew Berkseth, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA.
Rylee Shear, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA.
Lorenzo Gonzalez, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Andrew E Timms, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA.
Josef Sulc, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA.
Emily Pao, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA.
Nora Oyama, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA.
Francesca Forzano, Department of Clinical Genetics, Guy's and St Thomas NHS Foundation Trust and King's College London, London, UK.
Valerio Conti, Pediatric Neurology, Neurogenetics and Neurobiology Unit and Laboratories, Children's Hospital A. Meyer-University of Florence, Italy.
Renzo Guerrini, Pediatric Neurology, Neurogenetics and Neurobiology Unit and Laboratories, Children's Hospital A. Meyer-University of Florence, Italy.
Emily S Doherty, Section of Clinical Genetics, Carilion Clinic Children's Hospital, Roanoke, VA, USA.
Sulagna C Saitta, Division of Medical Genetics, Department of Obstetrics and Gynecology, David Geffen School of Medicine at the University of California Los Angeles, Los Angeles, CA, USA.
Christina M Lockwood, Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA, USA; Brotman-Baty Institute for Precision Medicine, University of Minnesota, Seattle, WA, USA.
Colin C Pritchard, Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA, USA; Brotman-Baty Institute for Precision Medicine, University of Minnesota, Seattle, WA, USA.
William B Dobyns, Division of Genetics and Metabolism, Department of Pediatrics, University of Minnesota, Minneapolis, MN, USA.
Edward Novotny, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA; Division of Pediatric Neurology, Department of Neurology, Seattle Children’s Hospital, Seattle, WA, USA; Department of Neurology, University of Washington, Seattle, WA, USA.
Jason N N Wright, Department of Radiology, Seattle Children’s Hospital, Seattle, WA, USA.
Russell P Saneto, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA; Division of Pediatric Neurology, Department of Neurology, Seattle Children’s Hospital, Seattle, WA, USA.
Seth Friedman, Center for Clinical and Translational Research, Seattle Children’s Hospital, Seattle, WA, USA.
Jason Hauptman, Department of Neurological Surgery, University of Washington, Seattle, WA, USA.
Jeffrey Ojemann, Department of Neurological Surgery, University of Washington, Seattle, WA, USA.
Raj P Kapur, Department of Laboratories, Seattle Children’s Hospital, Seattle, WA, USA.
Ghayda M Mirzaa, Center for Integrative Brain Research, Seattle Children’s Research Institute, Seattle, WA, USA; Brotman-Baty Institute for Precision Medicine, University of Minnesota, Seattle, WA, USA; Division of Genetic Medicine, Department of Pediatrics, University of Washington, Seattle, WA, USA; Institute for Stem Cell and Regenerative Medicine, University of Washington, Seattle, WA, USA.
References
- 1. Oegema R, Barakat TS, Wilke M, et al. International consensus recommendations on the diagnostic work-up for malformations of cortical development. Nat Rev Neurol. 2020;16(11):618–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mirzaa GM, Campbell CD, Solovieff N, et al. Association of MTOR mutations with developmental brain disorders, including megalencephaly, focal cortical dysplasia, and pigmentary mosaicism. JAMA Neurol. 2016;73(7):836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guerrini R, Barba C. Focal cortical dysplasia: An update on diagnosis and treatment. Expert Rev Neurother. 2021;21(11):1213–1224. [DOI] [PubMed] [Google Scholar]
- 4. Iffland PH2nd, Crino PB. Focal cortical dysplasia: Gene mutations, cell signaling, and therapeutic implications. Annu Rev Pathol. 2017;12:547–571. [DOI] [PubMed] [Google Scholar]
- 5. Fauser S, Sisodiya SM, Martinian L, et al. Multi-focal occurrence of cortical dysplasia in epilepsy patients. Brain. 2009;132(Pt 8):2079–2090. [DOI] [PubMed] [Google Scholar]
- 6. Najm IM, Tilelli CQ, Oghlakian R. Pathophysiological mechanisms of focal cortical dysplasia: A critical review of human tissue studies and animal models. Epilepsia. 2007;48 (Suppl 2):21–32. [DOI] [PubMed] [Google Scholar]
- 7. Najm IM, Sarnat HB, Blumcke I. Review: The international consensus classification of Focal Cortical Dysplasia—a critical update 2018. Neuropathol Appl Neurobiol. 2018;44(1):18–31. [DOI] [PubMed] [Google Scholar]
- 8. Riviere JB, Mirzaa GM, O'Roak BJ, et al. ; Finding of Rare Disease Genes (FORGE) Canada Consortium . De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44(8):934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mirzaa GM, Poduri A. Megalencephaly and hemimegalencephaly: Breakthroughs in molecular etiology. Am J Med Genet Part C Semin Med Genet. 2014;166c(2):156–172. [DOI] [PubMed] [Google Scholar]
- 10. Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138(Pt 6):1613–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mirzaa G, Timms AE, Conti V, et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight. 2016;1(9):e87623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alcantara D, Timms AE, Gripp K, et al. Mutations of AKT3 are associated with a wide spectrum of developmental disorders including extreme megalencephaly. Brain. 2017;140(10):2610–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stutterd C, McGillivray G, Stark Z, et al. ; Melbourne Genomics Health Alliance . Polymicrogyria in association with hypoglycemia points to mutation in the mTOR pathway. Eur J Med Genet. 2018;61(12):738–740. [DOI] [PubMed] [Google Scholar]
- 14. Dobyns WB, Mirzaa GM. Megalencephaly syndromes associated with mutations of core components of the PI3K-AKT-MTOR pathway: PIK3CA, PIK3R2, AKT3, and MTOR. Am J Med Genet Part C Semin Med Genet. 2019;181(4):582–590. [DOI] [PubMed] [Google Scholar]
- 15. Jesus-Ribeiro J, Pires LM, Melo JD, et al. Genomic and epigenetic advances in focal cortical dysplasia types I and II: A scoping review. Front Neurosci. 2020;14:580357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arvia R, Sollai M, Pierucci F, Urso C, Massi D, Zakrzewska K. Droplet digital PCR (ddPCR) vs quantitative real-time PCR (qPCR) approach for detection and quantification of Merkel cell polyomavirus (MCPyV) DNA in formalin fixed paraffin embedded (FFPE) cutaneous biopsies. J Virol Methods. 2017;246:15–20. [DOI] [PubMed] [Google Scholar]
- 17. Uchiyama Y, Nakashima M, Watanabe S, et al. Ultra-sensitive droplet digital PCR for detecting a low-prevalence somatic GNAQ mutation in Sturge-Weber syndrome. Sci Rep. 2016;6:22985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kliman D, Castellano-Gonzalez G, Withers B, et al. Ultra-sensitive droplet digital PCR for the assessment of microchimerism in cellular therapies. Biol Blood Marrow Transplant. 2018;24(5):1069–1078. [DOI] [PubMed] [Google Scholar]
- 19. Zhu G, Ye X, Dong Z, et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn. 2015;17(3):265–272. [DOI] [PubMed] [Google Scholar]
- 20. Falzone L, Musso N, Gattuso G, et al. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med. 2020;46(3):957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piacitelli AM, Jensen DM, Brandling-Bennett H, et al. Characterization of a severe case of PIK3CA-related overgrowth at autopsy by droplet digital polymerase chain reaction and report of PIK3CA sequencing in 22 patients. Am J Med Genet A. 2018;176(11):2301–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zenner K, Cheng CV, Jensen DM, et al. Genotype correlates with clinical severity in PIK3CA-associated lymphatic malformations. JCI Insight. 2019;4(21):e129884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng YY, Yuen ML, Rath EM, et al. CDKN2A and MTAP are useful biomarkers detectable by droplet digital PCR in malignant pleural mesothelioma: A potential alternative method in diagnosis compared to fluorescence in situ hybridisation. Front Oncol. 2020;10:579327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Paolis E, De Bonis M, Concolino P, et al. Droplet digital PCR for large genomic rearrangements detection: A promising strategy in tissue BRCA1 testing. Clin Chim Acta. 2020;513:17–24. [DOI] [PubMed] [Google Scholar]
- 25. Poliachik SL, Poliakov AV, Jansen LA, et al. Tissue localization during resective epilepsy surgery. Neurosurg Focus. 2013;34(6):E8. [DOI] [PubMed] [Google Scholar]
- 26. Blumcke I, Thom M, Aronica E, et al. The clinicopathologic spectrum of focal cortical dysplasias: A consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52(1):158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baldassari S, Ribierre T, Marsan E, et al. Dissecting the genetic basis of focal cortical dysplasia: A large cohort study. Acta Neuropathol. 2019;138(6):885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Do H, Dobrovic A. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase. Oncotarget. 2012;3(5):546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berra CM, Torrezan GT, de Paula CA, Hsieh R, Lourenço SV, Carraro DM. Use of uracil-DNA glycosylase enzyme to reduce DNA-related artifacts from formalin-fixed and paraffin-embedded tissues in diagnostic routine. Appl Cancer Res. 2019;39(1). [Google Scholar]
- 30. Luks VL, Kamitaki N, Vivero MP, et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J Pediatr. 2015;166(4):1048–1054.e1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338(6114):1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hardenbol P, Baner J, Jain M, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21(6):673–678. [DOI] [PubMed] [Google Scholar]
- 33. Cantsilieris S, Stessman HA, Shendure J, Eichler EE. Targeted capture and high-throughput sequencing using molecular inversion probes (MIPs). Methods Mol Biol. 2017;1492:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eijkelenboom A, Kamping EJ, Kastner-van Raaij AW, et al. Reliable next-generation sequencing of formalin-fixed, paraffin-embedded tissue using single molecule tags. J Mol Diagn. 2016;18(6):851–863. [DOI] [PubMed] [Google Scholar]
- 35. Csernak E, Molnar J, Tusnady GE, Toth E. Application of targeted next-generation sequencing, TruSeq custom amplicon assay for molecular pathology diagnostics on formalin-fixed and paraffin-embedded samples. Appl Immunohistochem Mol Morphol. 2017;25(7):460–466. [DOI] [PubMed] [Google Scholar]
- 36. Kuo AJ, Paulson VA, Hempelmann JA, et al. Validation and implementation of a modular targeted capture assay for the detection of clinically significant molecular oncology alterations. Pract Lab Med. 2020;19:e00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salamon N, Andres M, Chute DJ, et al. Contralateral hemimicrencephaly and clinical-pathological correlations in children with hemimegalencephaly. Brain. 2006;129(Pt 2):352–365. [DOI] [PubMed] [Google Scholar]
- 38. Sener RN. Hemimegalencephaly associated with contralateral hemispheral volume loss. Pediatr Radiol. 1995;25(5):387–388. [DOI] [PubMed] [Google Scholar]
- 39. Lee JH, Huynh M, Silhavy JL, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44(8):941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. D'Gama AM, Geng Y, Couto JA, et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann Neurol. 2015;77(4):720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marsan E, Baulac S. Review: Mechanistic target of rapamycin (mTOR) pathway, focal cortical dysplasia and epilepsy. Neuropathol Appl Neurobiol. 2018;44(1):6–17. [DOI] [PubMed] [Google Scholar]
- 42. Blumcke I, Coras R, Busch RM, et al. Toward a better definition of focal cortical dysplasia: An iterative histopathological and genetic agreement trial. Epilepsia. 2021;62(6):1416–1428. [DOI] [PubMed] [Google Scholar]
- 43. Guerrini R, Cavallin M, Pippucci T, et al. Is focal cortical dysplasia/epilepsy caused by somatic MTOR mutations always a unilateral disorder? Neurol Genet. 2021;7(1):e540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roy A, Skibo J, Kalume F, et al. Mouse models of human PIK3CA-related brain overgrowth have acutely treatable epilepsy. eLife. 2015;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumari K, Sharma MC, Kakkar A, et al. mTOR pathway activation in focal cortical dysplasia. Ann Diagn Pathol. 2020;46:151523. [DOI] [PubMed] [Google Scholar]
- 46. Lin YX, Lin K, Kang DZ, et al. Similar PDK1-AKT-mTOR pathway activation in balloon cells and dysmorphic neurons of type II focal cortical dysplasia with refractory epilepsy. Epilepsy Res. 2015;112:137–149. [DOI] [PubMed] [Google Scholar]
- 47. Dobyns WB. Primary microcephaly: New approaches for an old disorder. Am J Med Genet. 2002;112(4):315–317. [DOI] [PubMed] [Google Scholar]
- 48. Theda C, Hwang SH, Czajko A, Loke YJ, Leong P, Craig JM. Quantitation of the cellular content of saliva and buccal swab samples. Sci Rep. 2018;8(1):6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Costa G, Kouskoff V, Lacaud G. Origin of blood cells and HSC production in the embryo. Trends Immunol. 2012;33(5):215–223. [DOI] [PubMed] [Google Scholar]
- 50. Jones KB, Klein OD. Oral epithelial stem cells in tissue maintenance and disease: The first steps in a long journey. Int J Oral Sci. 2013;5(3):121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dingle TC, Sedlak RH, Cook L, Jerome KR. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem. 2013;59(11):1670–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Picard F, Makrythanasis P, Navarro V, et al. DEPDC5 mutations in families presenting as autosomal dominant nocturnal frontal lobe epilepsy. Neurology. 2014;82(23):2101–2106. [DOI] [PubMed] [Google Scholar]
- 53. Ribierre T, Deleuze C, Bacq A, et al. Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J Clin Invest. 2018;128(6):2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All generated data are included in this article and associated Supplementary material. The raw data generated and/or analysed during the study are available from the corresponding author on request.