Abstract
Introduction: Emerging evidence suggests that Chemotherapy (CT) treated breast cancer survivors (BCS) who have “risk variants” in genes may be more susceptible to cognitive impairment (CI) and/or poor cardiac phenotypes. The objective of this preliminary study was to examine whether there is a relationship between genetic variants and objective/subjective cognitive or cardiac phenotypes. Methods and Analysis: BCS were recruited from Moffitt Cancer Center, Morsani College of Medicine, AdventHealth Tampa and Sarasota Memorial Hospital. Genomic DNA were collected at baseline for genotyping analysis. A total of 16 single nucleotide polymorphisms (SNPs) from 14 genes involved in cognitive or cardiac function were evaluated. Three genetic models (additive, dominant, and recessive) were used to test correlation coefficients between genetic variants and objective/subjective measures of cognitive functioning and cardiac outcomes (heart rate, diastolic blood pressure, systolic blood pressure, respiration rate, and oxygen saturation). Results: BCS (207 participants) with a mean age of 56 enrolled in this study. The majority were non-Hispanic white (73.7%), married (63.1%), and received both CT and radiation treatment (77.3%). Three SNPs in genes related to cognitive functioning (rs429358 in APOE, rs1800497 in ANKK1, rs10119 in TOMM40) emerged with the most consistent significant relationship with cognitive outcomes. Among five candidate SNPs related to cardiac functioning, rs8055236 in CDH13 and rs1801133 in MTHER emerged with potential significant relationships with cardiac phenotype. Conclusions: These preliminary results provide initial targets to further examine whether BCS with specific genetic profiles may preferentially benefit from interventions designed to improve cognitive and cardiac functioning following CT.
Keywords: genetic, cognitive dysfunction, cardiac function, chemotherapy, cancer survivors, breast cancer
An estimated number of 281,550 new cases of invasive breast cancer is predicted among women in the United States in 2021 (Siegel et al., 2021). For these women, although chemotherapy (CT) increases the survival rate (D. Chen et al., 2021), CT-associated cognitive impairment (CI) was found in 13–70% of breast cancer survivors (BCS), (Tao et al., 2017) and continues years beyond treatment (Y. Chen et al., 2022; Ganz, 2012; Pullens et al., 2010; Yamada et al., 2010).
Chemotherapy treatment not only affects cognitive functioning in multiple ways (e.g., memory, processing speed) for BCS, (Deprez et al., 2018; Jim et al., 2012; Pullens et al., 2010; Ren et al., 2019; Wefel et al., 2010), also affecting BCS’s quality of life; (Xuan et al., 2017) and their ability to return to work, resulting in economic burdens and increased psychological stress (Mazanec et al., 2021; Munir et al., 2011; Wefel et al., 2010).
Objective and subjective measures of cognitive functioning were selected for this study. Currently, most research implements use of subjective measures due to the ease of obtaining the patients perception of their cognitive abilities. To be more precise in assessment of cognitive functioning, objective neuropsychological measures provide more validity and reliability in assessment of several cognitive domains, not provided by subjective assessments.
BCS often have ongoing symptoms, such as pain, fatigue, sleep disturbance, and depression, that vary between individuals with current evidence identifying one’s genes may influence one’s symptom experience (E. Boots et al., 2017; S. Doong et al., 2014). Recovery for BCS symptom experience (cognitive, psychological, and physical) may be enhanced if there is increased understanding and evidence explaining the relationships between one’s genetic profile and their symptom experiences. This scientific evidence supports achieving the goals of precision medicine to tailor treatment plans for symptoms based on one’s genetics to promote best health outcomes with minimal consequences (Mirnezami et al., 2012).
CT-induced cardiac dysfunction is a serious side effect of chemotherapy. This notorious side effect occurs ∼10% of BCS with CT (Teske et al., 2018). However, individual variation of CT-induced cardiac dysfunction cannot be fully explained by known clinical risk factors. We hypothesized that genetic variants may provide better explanation. Also, we evaluated a role of genetic risk factors in CT-induced cardiac dysfunction among BCS.
Recent advances in molecular genomic technologies, and extensive genomic research identify risky genomic variations for disease predisposition, progression, and response of treatments. Current knowledge of genomic variation and mutations have been translated into clinical settings for prevention of disease and clinical treatments (Lee et al., 2020; Molina-Moya et al., 2016; Morandi et al., 2016; Percival et al., 2016). For example, BCS with SNPs in CYP2D6 are found to be at an increased risk for adverse effect from the tamoxifen therapy and therefore, BCS with SNPs (rs28371725 and rs16947) in CYP2D6 are to be treated with alternative therapies, such as aromatase inhibitors (Wickramage et al., 2017).
Emerging evidence suggests that genetic factors may influence CT-associated CI among BCS (Ahles et al., 2010; Chan et al., 2019; Park et al., 2019; Stewart et al., 2008; Tan et al., 2019; G. S. Yang et al., 2019). Individual patient genetic profiles may be associated with increased risk for long-term cognitive changes (Carroll et al., 2019; de Frutos-Lucas et al., 2020). Genetic variants in cognitive functioning pathways also may play a role in CT-associated CI. Previous evidence report that CT-treated BCS who have “risk variants” in Apo lipoprotein E (APOE) and catechol-o-methyl transferase (COMT) genes are more susceptible to cognitive dysfunction (Ahles et al., 2010; Buskbjerg et al., 2019; Carroll et al., 2019; Lengacher, Reich, Kip, et al., 2015; Stewart et al., 2008).
After an extensive review of the evidence, 12 SNPs in 11 candidate genes involved in cognitive functioning were selected and investigated for their role among breast cancer survivors. Additionally, we evaluated the potential role of five SNPs from four genes, involved in cardiac symptoms among BCS. Products of these genes considered receptors, carriers, transporters, and metabolic enzymes in various pathways. Gene variations in apolipoprotein E (APOE), brain-derived neurotrophic factor (BDNF), serotonin receptor 2A (5HTR2A), AK091365 (Davies et al., 2016; Rietveld et al., 2014), AKAP6 (Andrews et al., 2017; Davies et al., 2015, 2016), TOMM40 (Davies et al., 2015), BCL11A (Le Hellard et al., 2017), ankyrin repeat and kinase domain containing 1 (ANKK1) (Athanasoulia et al., 2014) and APBA1 (Davies et al., 2016; Sloan et al., 2010) were previously related to cognitive function (Jiang et al., 2016; Koleck et al., 2014; Kurita et al., 2016; Ng et al., 2016; Schillani et al., 2010; Su et al., 2015; Zhang et al., 2015).
Further, SNPs in CDH13 (Angelakopoulou et al., 2012; Bressler et al., 2010), CXCL12 (Ansari et al., 2019; Spiller et al., 2020), CDKN2B (Hu et al., 2019; Trenkwalder et al., 2019), and methylenetetrahydrofolate reductase (MTHFR) (H. L. Yang et al., 2018) have been associated with cardiac function.
The purpose of this sub-analysis was to explore the relationship between SNPs in candidate genes and cognitive and cardiac functioning among BCS with previous CT. If we can identify initial targets for investigation, we can identify whether BCS with specific genetic profiles may preferentially benefit from interventions designed to enhance cognitive and cardiac functioning following CT. An overarching goal of this personalized medicine project is to tailor symptom-relief interventions based on genetic profiles of BCS.
Methods
Participants
Among 212 BCS who consented to participate in our R01 MBSR(BC) Trial, 207 completed the genetic analysis. The overarching purpose of the parent study was to evaluate the efficacy of the Mindfulness-Based Stress Reduction (BC) program compared to the Breast Cancer-Education Support (BCES) program or a usual care (UC) regimen for improving cognitive functioning among BCS, who received adjuvant chemotherapy, or chemotherapy and radiation. Data from this study were obtained at baseline, prior to the randomized intervention.
BCS were recruited from the Moffitt Cancer Center, USF Breast Health Program, AdventHealth Hospital, Tampa, Florida. Sarasota Memorial Hospital, Sarasota Florida, and community groups. Primary inclusion criteria included women age 21 or older diagnosed with stage I, II, or III BC, completed CT or CT and radiation, within 5 years post-treatment, and met the screening criteria for CI with a positive response to 1 of 2 questions: 1) “Have you had difficulty in concentrating on things, like reading a newspaper or watching television; ” and 2) “Have you had difficulty remembering things” (Montazeri et al., 1999). Exclusion criteria included severe psychiatric diagnosis (e.g., bipolar disorder), Stage 0 or Stage IV BC, history of another primary cancer diagnosis with the treatment of CT and/or radiation, metastatic cancer, current neurologic disorder, or a traumatic brain injury.
Study Design and Data Collection
Clinic nurses identified eligible BCS who were then approached by study recruiters. BCS who expressed interest in the study were invited to an orientation session where consent was obtained followed by baseline data collection. The assessment included obtaining 5 mL of blood or buccal (cheek) cell sample for genotyping analysis only at baseline, along with administration of objective neuropsychological assessments and subjective cognitive, clinical treatment, and demographic data. For cardiac data collection, among 212 patients, 28 BCS only participated.
Measurements
Demographics and clinical history. Standard socioeconomic demographic data, including age, gender, ethnicity, highest level of education completed, marital status, income status, and employment status, were collected following orientation. Standard clinical history data on cancer diagnosis, diagnosis date, clinical stage, and treatment were collected as well.
Cognitive performance. Cognitive performance tests were selected based on previous research among cancer patients (Booth-Jones et al., 2005; Jacobs et al., 2007) demonstrating reliability and validity with published normative data for adults (Booth-Jones et al., 2005; Jacobs et al., 2007).
Objective Cognitive Performance. Memory was assessed using tests of visuospatial, verbal, and logical memory: 1) Visuospatial memory was measured by the Brief Visuospatial Memory Test-Revised (BVMT-R) alpha reliability (0.96–0.97) (Benedict, 1997); 2) Verbal memory was measured by the Hopkins Verbal Learning Test-Revised (HVLT-R) alpha reliability (0.76–0.86) (Benedict et al., 1998; Brandt, 1991); and 3) Logical memory was measured by the Logical Memory I and II subscales from the Wechsler Memory Scale-IV (WMS-IV) alpha reliability (0.70–0.90) (D. Wechsler, 2009). Attention/Concentration was measured using the Digit Span subtest of the WAIS-IV, alpha reliability (0.90) (D. Wechsler, 2008), and Part 1 of the Color Trails Test (CTT-1) alpha reliability (0.64) (D’Elia et al., 1996; Maj et al., 1993). Executive functioning was measured by Part 2 of the Color Trails Test (CTT-2) alpha reliability (0.79) (D’Elia et al., 1996; Maj et al., 1993), and Stroop Neuropsychological Screening Test (SNST) alpha reliability (0.73–0.89) (Trenerry et al., 1989). Verbal fluency was measured by the Controlled Oral Word Association Test (COWAT) alpha reliability (0.70) (Benton & Hamsher, 1989) and the Wechsler Test of Adult Reading (WTAR) alpha reliability (0.90–0.97) (Corporation, 2001).
Perceived cognitive performance. The Functional Assessment of Cancer Therapy–Cognitive Function (FACT-Cog) alpha reliability (0.82) was used to assess subjective change of everyday perceived cognitive performance (Wagner, L. I., Sweet, Butt, Lai, & Cella, 2009).
Cardiac measures. Measures of cardiovascular functioning included systolic and diastolic blood pressure, heart rate, respiratory rate, and oxygen saturation, which were collected at the baseline.
Genotyping
Gene and SNP selection. Based on an extensive literature search performed by a molecular epidemiologist (JYP), candidate genes and SNPs were chosen to represent variations of the genes involved in various pathways, including depression, cognitive function, and cardiac phenotype in BC. Among total 208 SNPs from 58 genes identified, 16 SNPs from 14 genes were selected for this study, based on their significant associations with cognitive function and cardiac function (Bal et al., 2016; J. Chen et al., 2004; Fisher et al., 2015; Hirvonen et al., 2004; Koleck et al., 2016; Lengacher, Reich, Kip, et al., 2015; Lengacher, Reich, Paterson, et al., 2015; Martin et al., 2007; Merriman et al., 2014; Ng et al., 2014; Ni et al., 2006; Polesskaya & Sokolov, 2002; Ponce et al., 2009; Prada et al., 2014; Ueland et al., 2001; M. Wagner et al., 2008).
Analysis
Genotyping Analysis
“Genotype” refers to the genetic makeup, thus the alleles or variants of a gene. Each human has two alleles at a gene, with one allele from each parent. A pair of alleles represents the genotype of a specific gene. Because each gene has two alleles, each human can have three possible genotypes at each gene: normal, mixed, and polymorphic (Hughes et al., 2021). A genotype is described in this study as normal/polymorphic if it has two major/minor identical alleles and as mixed if the two alleles are different. Five milliliters of blood were drawn for the genotyping analysis. Genomic DNA was extracted from peripheral blood leukocytes or buccal cells using the Qiagen DNA extraction kit (Qiagen, Valencia, CA) with modifications.
Genetic Measurements
Multiple candidate SNPs involved in cognitive or cardiac function were selected based on previous research and preliminary data. Genotyping methods were used as described for Moffitt’s laboratory (Jim et al., 2012).
The distribution of genotype frequencies of candidate SNPs examined were tested for Hardy-Weinberg equilibrium (HWE) based on categories of race using an asymptotic chi-square test (1 degree of freedom) with a graphical summary or an exact test based on a multinomial distribution if the data are sparse. The HWE is a principle stating that the genetic variation in a population will remain constant from one generation to the next.
Statistical Analysis Plan
Wilcoxon rank sum test or Kruskal Wallis test was used to evaluate whether genetic factors were associated with cognitive or cardiac function. Because we tested multiple genes (each using three genetic models (additive, dominant, and recessive)) and multiple outcomes, we were cognizant of the potential for Type 1 error. Because of our relatively small sample and the anticipated small effect sizes, we did not want to adjust alpha levels in this exploratory study. Rather, we looked for three data patterns to establish consistency: 1) Statistical significance across at least two of the three genetic models on the same measure; 2) Statistical significance across multiple measures within 1 of 5 cognitive domains (i.e., memory (BVMT, HVLT, WMS), attention control (Digit Span, Color Trails 1), executive functioning (Stroop, Color Trails 2), verbal fluency (COWA, WTAR), and perceived cognitive performance (FACT-Cog); and 3) Statistical significance across multiple cognitive domains. All three patterns were necessary to establish consistency. Because the number of participants who were tested for cardiac phenotypes was low, any gene related to any of the 5 cardiac measures was identified with the cognizance that consistency would have to be established in future studies. Statistical significance was established at p < 0.05.
Results
Participant Characteristics
Among 212 BCS consented to participate in the R01 trial, 207 completed this genetic analysis. However, as expected, significant difference of genotype distribution was observed by different racial groups. Due to differential genotype distribution by race, non-White participants were excluded from the analyses, and therefore only participants who self-identified as white were included, resulting in a sample size of 151. Some participants (24.5%) received CT only, while the remainder received both CT and radiotherapy (75%). The majority (65%) of BCS were more than a year since cancer treatment. Selected demographic and clinical characteristics are described in Table 1. Among the 151 BCS included in this analysis, cardiac phenotype data were obtained from 28 BCS (Supplemental Table 1).
Table 1.
Selected Clinical and Demographic Characteristics of Participants (n = 151, White only).
| Variable | Level | N = 151 | % |
|---|---|---|---|
| Age at diagnosis | Mean ± SD | 56 ± 10.5 | |
| Smoking | Never | 96 | 63.6 |
| Former | 46 | 30.5 | |
| Current | 9 | 6.0 | |
| Education status | College | 97 | 64.2 |
| No College | 54 | 35.8 | |
| Marital status | Single, Divorced, Widowed | 46 | 30.5 |
| Married | 102 | 67.5 | |
| Other | 3 | 2 | |
| Employment | Employed < 32 hrs/wk | 23 | 15.2 |
| Employed ≥ 32 hrs/wk | 49 | 32.5 | |
| Othera | 31 | 20.5 | |
| Retired | 42 | 27.8 | |
| Unemployed | 6 | 4 | |
| Income | <$20,000 | 29 | 19.8 |
| $20,000 to < $40,000 | 25 | 17.1 | |
| $40,000 to < $80,000 | 45 | 30.8 | |
| $80,000 to < | 47 | 32.2 | |
| Missing | 5 | –– | |
| Time since completion Category | 1 Year | 52 | 34.4 |
| More than 1 Year | 99 | 65.6 | |
| Cancer treatment Category | Chemo Alone | 37 | 24.5 |
a“Other” responses for employment included “Student”, “Disabled,” “On medical leave,” and a selection of “other” if no available selection fit.
Genetic Analysis Results for Cognition
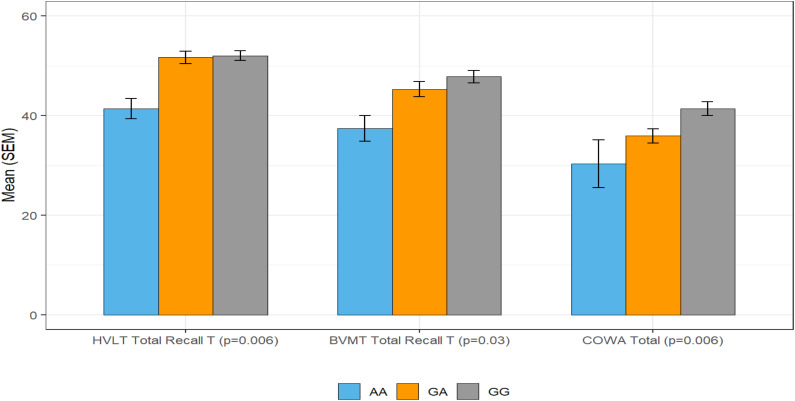
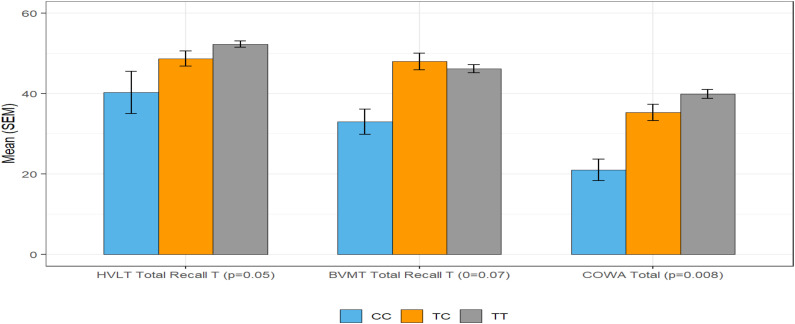
All SNP distributions followed Hardy-Weinberg equilibrium (Table 2). Among 12 SNPs tested, three SNPs (rs429358 in APOE, rs1800497 in ANKK1, and rs10119 in TOMM40) emerged as having the most consistent significant relationships with cognitive outcomes (Memory, Attention control, Executive Functioning, Verbal fluency, and Perceived Cognitive Performance). For both rs429358 in APOE and rs10119 in TOMM40, the same pattern emerged: Statistically significant effects were observed in multiple measures of Memory (HVLT and BVMT), Executive Functioning (Color Trails), and Verbal Fluency (COWA). Effects were observed across all three genetic models (Figures 1 and 2). For rs1800497 in ANKK1, statistically significant effects were observed in multiple measures Memory (HVLT and BVMT), in Attention Control (WAIS Digit Span), Executive Functioning (Stroop), and Perceived Cognitive Performance (FACT-Cog). Effects were observed across all three genetic models.
Table 2.
Characteristics of Candidate Single-nucleotide Polymorphisms (n = 151).
| Gene | SNP ID | Allele | Change | Poly/HT/WT | HWE | Role |
|---|---|---|---|---|---|---|
| APOE | rs429358 | C/T | Cys156Arg | 3/30/118 | 0.91 | Cognitive function (Koleck et al., 2014; Lengacher, Reich, et al., 2015a) |
| BDNF | rs6265 | T/C | Val66Met | 5/44/102 | 1.00 | Cognitive impairment (Ng et al., 2016) |
| HTR2A | rs6313 | A/G | Ser34SER | 26/78/47 | 0.90 | Cognitive function (Kurita et al., 2016) |
| HTR2A | rs6314 | A/G | His452Asn | 0/28/123 | 0.49 | |
| ANKK1 | rs1800497 | A/G | Glu713Lys | 7/45/99 | 0.91 | Cognitive function (Lengacher, Reich, et al., 2015) |
| AK091365 | rs1487441 | G/A | none | 31/81/39 | 0.80 | Cognitive function (Davies et al., 2016; Rietveld et al., 2014) |
| AKAP6 | rs17522122 | G/T | 3’UTR | 39/70/42 | 0.82 | Cognitive function (Andrews et al., 2017; Davies et al., 2015, 2016) |
| TOMM40 | rs10119 | G/A | 3’UTR | 8/60/83 | 0.89 | Cognitive function (Davies et al., 2015) |
| BCL11A | rs7581162 | T/A | Intron | 28/82/41 | 0.71 | Cognitive function (Le Hellard et al., 2017) |
| APBA1 | rs3897757 | A/G | intron | 33/73/45 | 0.97 | Cognitive function (Davies et al., 2016; Sloan et al., 2010) |
| CYP2D6 | rs5758605 | A/G | upstream | 31/70/50 | 0.88 | Cognitive function, depression (Cacabelos, 2007) |
| MTHER | rs1801133 | C/T | Ala222Asp | 19/70/62 | 1.00 | Cognitive function (Jiang et al., 2016; Schiepers et al., 2012) Heart defect (H. L. Yang et al., 2018) |
| Intergenic | rs28714259 | G/A | none | 2/24/125 | 0.91 | Heart failure (Linschoten et al., 2018; Schneider et al., 2017) |
| CHD13 | rs8055236 | G/A | intron | 4/53/94 | 0.72 | Heart disease (Angelakopoulou et al., 2012; Bressler et al., 2010) |
| CXCL12 | rs1746048 | C/T | none | 2/30/119 | 1.00 | Cardiovascular health (Ansari et al., 2019; Spiller et al., 2020) |
| CDKN2B | rs1333049 | G/A | none | 38/75/38 | 1.00 | Coronary artery disease (Hu et al., 2019; Trenkwalder et al., 2019) |
Figure 1.
Cognition and rs10119 Genotype.
Note. Pattern of relationships between genotypes based on rs10119 SNP and phenotype: In rs10119 in TOMM40, statistically significant effects were observed in multiple measures of Memory including Executive Functioning and Verbal Fluency. Effects were observed across all three genetic models with three genotypes (AA, GA, and GG). This figure illustrates how participants that were homozygous for the A variant performed worse on these cognitive measures.
Figure 2.
Cognition and rs429358 Genotype.
Note. Pattern of relationships between genotypes based on rs 429358 SNP and phenotype: In rs429358 in APOE, statistically significant effects were observed in multiple measures of Memory including Executive Functioning and Verbal Fluency. Effects were observed across all three genetic models with three genotypes (CC, CT, and TT). This figure illustrates how participants that were homozygous for the C variant performed worse on these cognitive measures.
Genetic Analysis Results for Cardiac Functioning
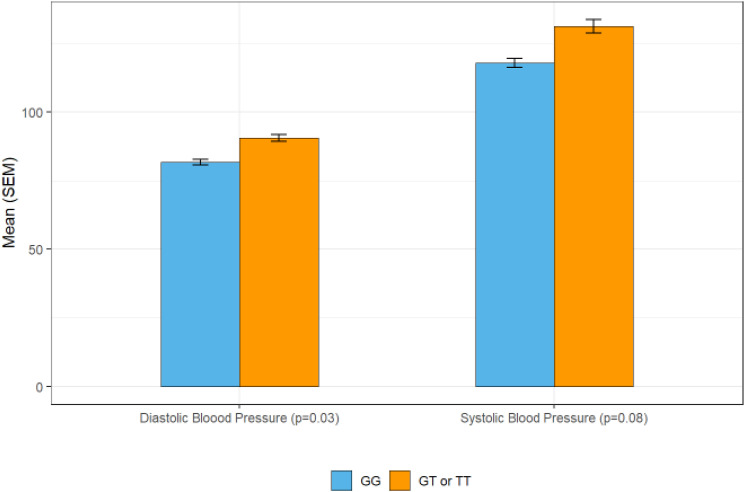
The relationships between SNPs previously associated with cardiac phenotype and cardiac outcomes were exploratory due to the small sample assessed. Two SNPs, rs8055236 in CDH13 and rs1801133 in MTHER emerged as having potential relationships. rs8055236 in CDH13 was associated with diastolic blood pressure in one genetic model (Hetero+poly vs. wild type) (Figure 3). rs1801133 in MTHER was associated with heart rate in the 3-group genetic model.
Figure 3.
Blood Pressure and rs8055236 Genotype.
Note. Pattern of relationships between SNPs and phenotype: In rs8055236 in CHD13, a statistically significant effect was observed in diastolic blood pressure and a similar pattern (but not significant) in systolic blood pressure. This figure illustrates how participants that were homozygous for the G variant tended to have lower blood pressure.
Discussion
Multiple studies have identified CI after CT. However, there is limited research examining the relationship between SNP and cognitive functioning and SNP’s responsible for a substantial risk of CI. Although CI is a major problem for BCS, currently there is no effective pharmacological agent to enhance cognitive functioning in this population.
Our cognitive performance tests have been selected based on previous research on cognitive functioning in cancer patients (Booth-Jones et al., 2005; Jacobs et al., 2007) and have demonstrated reliability and validity and have published normative data for adults and the domains (Bower et al., 2006). Despite the magnitude of the decreased cognitive functioning among chemo-brain BCS, there are gaps in knowledge due to variability in results due to differences in types of assessment, particularly objective and subjective assessments, and design and co-morbidities or the cancer itself (Pullens et al., 2010). Most intervention research studies use subjective measures; however, the objective measures are more precise and identify the real domain deficits in areas of cognitive functioning, such as executive functioning, memory and verbal fluency that include extensive batteries of neuropsychological tests. Although objective tests are not disputed as vital, objective measures require extensive assessment training in the delivery in a 1:1 assessment for each person as compared to assessment by subjective measures that can be performed individually by each patient. Therefore, subjective measures may be deemed as more useful, than neuropsychological tests for research.
CI symptoms of BCS vary for each person and evidence indicates genetic variations are likely to influence frequency and intensity of the symptom (S. H. Doong et al., 2015). Recently, our team and others reported on cognitive functioning in BCS studies with genetic influences (Boots, E. A. et al., 2017; Gonzalez et al., 2015; Lengacher, Reich, Kip, et al., 2015). Based on our previous study (Lengacher, Reich, Kip, et al., 2015), we investigated whether genetic variations may be associated with a multitude of CI or poor cardiac function experienced by BCS. In this study, we observed that three SNPs (rs429358 in APOE, rs1800497 in ANKK1, and rs10119 in TOMM40) were associated with CI.
These SNPs and genes were previously investigated by our team and others. A previous study suggested APOE SNP as a genetic biomarker for increased vulnerability to CT-related CI (Ahles et al., 2003). Our team (Lengacher, Reich, Paterson, et al., 2015) and others previously reported that APOE SNP rs429358 is associated with a decline in cognitive function (Koleck et al., 2014; O'Donoghue et al., 2018; Small et al., 2011). For example, Koleck et al. (2014) performed a longitudinal study to examine the role of APOE SNP in the cognitive function of CT treated BCS (Koleck et al., 2014). They observed that changes in performance on tasks of executive function, attention, verbal learning and memory, and visual learning and memory were influenced by APOE genotype.
In this current analysis, we validated that APOE (rs429358) showed statistically significant association between this SNP and cognitive functioning in BCS. rs1800497 in ANKK1 also was associated with CI in BCS. rs1800497 causes an amino acid change from Glu to Lys at codon 713, reducing D2 dopamine receptor binding and glucose metabolism in the brain (Ponce et al., 2009). Phenotypes affected by this SNP are related to CI, and our finding of an association between this SNP and CI is supported by our previous study (Lengacher, Reich, Paterson, et al., 2015) and others (Berryhill et al., 2013; Noble, 2000; Savitz et al., 2013; Spellicy et al., 2014).
TOMM40 is located at chromosome 19q13.32 and encodes the mitochondrial outer membrane complex (Gottschalk et al., 2014). With, a GWAS study on longitudinal cognitive ability data reported that SNPs in TOMM40 were significantly associated with age-related cognitive decline (Davies et al., 2014). In addition, SNPs in regulatory region of TOMM40 influenced functional effect, suggesting that TOMM40 may be associated with CI (Davies et al., 2014; Lin et al., 2017). Currently, a role of SNPs in TOMM40 among BCS with chemo-treatment was not investigated in CI. Our results suggested that genetic variations in TOMM40 may be related to CI among BCS.
CT-induced cardiac dysfunction is a serious and common side effect of treatment (Norton et al., 2020). Around 30% of patients develop heart failure or mild left ventricular dysfunction within 6 months among CT-treated patients and these cardiac events occur more often in patients with 65 years or older (Necela et al., 2017). However, individual variation of CT-induced cardiac dysfunction cannot be explained by known clinical risk factors. We hypothesized that genetic variants may provide better explanation. We conducted a systematic literature search to identify genetic risk factors for CT-induced cardiac dysfunction. When testing the relationships between 5 SNPs and cardiac outcomes, two SNPs, rs8055236 in CDH13 and rs1801133 in MTHER emerged as having a significant relationship with cardiac phenotype in one genetic model.
Cadherin-13 (CDH13) was known the important role in human diseases such as cancer, and cardiovascular disease. High expression of CDH13 was reported in diseased vascular endothelial during atherosclerosis (Takeuchi et al., 2007). Previous studies reported that SNPs in CDH13 associated with coronary artery disease (Burton et al., 2007; Chotchaeva et al., 2016; Samani et al., 2007). In addition, CDH13 was identified as one of the risk loci for CAD in a meta-analysis (van der Harst & Verweij, 2018). However, the biological function of CDH13 on the cardiovascular system has not been established yet, especially in BCS. In previous genome-wide association studies for coronary heart disease (CHD) with 25,000 subjects and 5794 CHD events,, rs8055236 SNP in CDH13 was associated with CHD risk (Angelakopoulou et al., 2012; Baudhuin, 2009; Bressler et al., 2010; Yan et al., 2009). Our results are consistent with results from previous studies.
Methylenetetrahydrofolate reductase (MTHFR) rs1801133 leads to reduced enzyme activity and causes extracellular matrix remodeling (Wang et al., 2006). This SNP was significantly associated with the risk of obstructive heart defects in previous studies (Weiner et al., 2012; Yin et al., 2012). Further, a meta-analysis of 7697 cases and 13,125 controls reported that MTHFR rs1801133 was associated with risk of congenital heart defects (OR. 1.25, 95% CI: 1.03–1.51) (Mamasoula et al., 2013). The T allele of the MTHFR rs1801133 is associated with reduced activity of MTHFR (Castro et al., 2004) and increased plasma homocysteine level (DeVos et al., 2008). Our results are consistent with a significant association between polymorphisms in MTHFR gene and increased risk of cardiac dysfunction among CT-treated BCS. There are several strengths and limitations of this analysis. The first strength is that objective neuropsychological assessments were used to test objective measures of cognitive functioning, as well as self-reports subjective measures for each participant. As a limitation, first, the biological function of the SNPs identified were not fully established. The impact of SNPs in expression or activity of genes needs to be investigated to assess potential mechanisms. Second, the sample size of this study, especially for cardiac function was small. Therefore, this sub-analysis lacked sufficient power for broad generalization. Third, non-white patients were excluded for statistical analysis. Therefore, these results may not be applied in non-white patients. Fourth we used vital signs as a cardiac outcome. Other cardiac outcomes could be further assessed for their association in the analysis. Finally, results from cardiac functioning analysis were significant only in one genetic model. Therefore, these results presented may be exploratory. This study supports an initial step supporting goals of personalized medicine to tailor symptom-relief interventions based on genetic profiles of BCS. These genetic findings may be used in the future to assist health care providers, nurses, and physicians to identifying women with breast cancer most at risk for cognitive decline and cardiac dysfunction.
In summary, we reported significant associations between genetic variations and CT-induced CI or cardiac function in BCS. These results suggest that the heterogeneity in CI and cardiac function may be influenced by genetic profiles in genes involved in cognitive function or cardiac phenotype in BCS. Further, these preliminary results provide initial targets to further examine whether BCS with specific genetic profiles may preferentially benefit from interventions designed to improve cognitive and cardiac functioning following CT.
Ethics, Knowledge Dissemination, and Impact of Study
This translational research used neuropsychological, genetic, and cognitive impairment (CI). If CI is influenced by genetic profiles, these aspects may impact medical costs of cancer care. Genetic profile identification has the potential to facilitate the development of personalized medicine pathways for optimal treatment response. Evidence may provide for broader dissemination of the intervention as a non-pharmacological treatment in cancer centers among CT-induced CI among BCS.
Supplemental Material
Supplemental Material for Translational Genomic Research: The Association between Genetic Profiles and Cognitive Functioning or Cardiac Function Among Breast Cancer Survivors Completing Chemotherapy by Jong Y. Park, Cecile A. Lengacher, Richard R. Reich, Hyun Y. Park, Junmin Whiting, Anh Thy Nguyen, Carmen Rodríguez, Hongdao Meng, Sara Tinsley, Katterine Chauca, Liliana Gordillo-Casero, Trudy Wittenberg, Anisha Joshi, Katherine Lin, Roohi Ismail-Khan, John V. Kiluk, and Kevin E. Kip in Biological Research For Nursing
Author Contributions: Study design: JYP, CAL, RRR, CR, HM, KEK; data collection: JYP, CAL, ATN, CR, ST, KC, LGC, TW, AJ, KL; analysis and interpretation of data: JYP, CAL, RRR, HYP, JW, ATN, CR, HM, ST, KC, LGC, TW, AJ, KL, RIK, JK, KEK; manuscript preparation: JYP, CAL, RRR, CR, HM, KC, LGC, RIK, JK, KEK.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This research was supported in part from the award Number 1 R01 CA199160-01 from the National Cancer Institute supported the project described. This work has been supported in part by the Biostatistics and Bioinformatics Shared Resource and the Participant Research, Interventions, & Measurement core at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Data Accessibility Statement: Data will be made available to faculty researchers at Universities in the United States through a data sharing agreement and to approved members of the scientific community 3 years after the final grant completion date. This dissemination plan will undergo yearly review. Pending results, effective intervention materials and programs will be shared through a Web site or national repository.
Ethics Declarations: This study protocol was approved by the Institutional Review Board at the University of South Florida to ensure the ethical treatment of participants.
Dissemination: This reported analysis as part of a larger clinical trial, may provide for ideal delivery and broader dissemination of potential interventions based on genetic profile as a non-pharmacological treatment in cancer centers and settings of CT-induced CI among BCS. Results of this study will be disseminated via peer-reviewed publications, scientific meetings, and oncology conferences.
Supplemental Material: Supplemental material for this article is available online.
ORCID iDs
Jong Y. Park https://orcid.org/0000-0002-6384-6447
Sara Tinsley https://orcid.org/0000-0003-3012-7398
References
- Ahles T. A., Saykin A. J., McDonald B. C., Li Y., Furstenberg C. T., Hanscom B. S., Mulrooney T. J., Schwartz G. N., Kaufman P. A. (2010). Longitudinal assessment of cognitive changes associated with adjuvant treatment for breast cancer: Impact of age and cognitive reserve. Journal of Clinical Oncology, 28(29), 4434–4440. 10.1200/JCO.2009.27.0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahles T. A., Saykin A. J., Noll W. W., Furstenberg C. T., Guerin S., Cole B., Mott L. A. (2003). The relationship of APOE genotype to neuropsychological performance in long-term cancer survivors treated with standard dose chemotherapy. Psycho-oncology, 12(6), 612–619. 10.1002/pon.742. [DOI] [PubMed] [Google Scholar]
- Andrews S. J., Das D., Anstey K. J., Easteal S. (2017). Association of AKAP6 and MIR2113 with cognitive performance in a population-based sample of older adults. Genes, Brain and Behavior, 16(4), 472–478. 10.1111/gbb.12368. [DOI] [PubMed] [Google Scholar]
- Angelakopoulou A., Shah T., Sofat R., Shah S., Berry D. J., Cooper J., Palmen J., Tzoulaki I., Wong A., Jefferis B. J., Maniatis N., Drenos F., Gigante B., Hardy R., Laxton R. C., Leander K., Motterle A., Simpson I. A., Smeeth L., Hingorani A. D. (2012). Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: Cardiovascular Biomarker Genetics Collaboration. European Heart Journal, 33(3), 393–407. 10.1093/eurheartj/ehr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari W. M., Humphries S. E., Naveed A. K., Khan O. J., Khan D. A., Khattak E. H. (2019). Effect of coronary artery disease risk SNPs on serum cytokine levels and cytokine imbalance in premature coronary artery disease. Cytokine, 122(5), 154060. 10.1016/j.cyto.2017.05.013. [DOI] [PubMed] [Google Scholar]
- Athanasoulia A. P., Siever C., Uhr M., Ising M., Stalla G. K., Schneider H. J. (2014). The effect of the ANKK1/DRD2 Taq1A polymorphism on weight changes of dopaminergic treatment in prolactinomas. Pituitary, 17(3), 240–245. 10.1007/s11102-013-0496-y. [DOI] [PubMed] [Google Scholar]
- Bal J. K., Beuvier T., Vignaud G., Chebil M. S., Ben-Jabrallah S., Ahmed I., Grohens Y., Gibaud A. (2016). Swelling of poly(n-butyl methacrylate) films exposed to supercritical carbon dioxide: A comparative study with polystyrene. Langmuir, 32(7), 1716–1722. 10.1021/acs.langmuir.5b04436. [DOI] [PubMed] [Google Scholar]
- Baudhuin L. M. (2009). Genetics of coronary artery disease: Focus on genome-wide association studies. American Journal of Translational Research, 1(3), 221–234. [PMC free article] [PubMed] [Google Scholar]
- Benedict R. (1997). Brief visuo-Spatial Memory Test-Revised (BVMT-R). Psychological Assessment Resources. [Google Scholar]
- Benedict R. H. B., Schretlen D., Groninger L., Brandt J. (1998). Hopkins verbal learning test revised: Normative data and analysis of inter-form and test-retest reliability. Clinical Neuropsychologist, 12(1), 43–55. 10.1076/clin.12.1.43.1726. [DOI] [Google Scholar]
- Benton A., Hamsher K. (1989). Multilingual aphasia examination. Retrieved from Spanish version available. http://www4.parinc.com/Default.aspx.
- Berryhill M. E., Wiener M., Stephens J. A., Lohoff F. W., Coslett H. B. (2013). COMT and ANKK1-Taq-Ia genetic polymorphisms influence visual working memory. Plos One, 8(1), Article e55862. 10.1371/journal.pone.0055862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth-Jones M., Jacobsen P. B., Ransom S., Soety E. (2005). Characteristics and correlates of cognitive functioning following bone marrow transplantation. Bone Marrow Transplantation, 36(8), 695–702. 10.1038/sj.bmt.1705108. [DOI] [PubMed] [Google Scholar]
- Boots E., Schultz S., Clark L., Racine A. M., Darst B. F., KoscikOkonkwo R. L.O. C., Carlsson C. M., Gallagher C. L., Hogan K. J., Bendlin B. B., Asthana S., Sager M. A., Hermann B. P., Christian B. T., Dubal D. B., Engelman C. D., Johnson S. C., Okonkwo O. C. (2017. a). BDNF Val66Met predicts cognitive decline in the Wisconsin registry for Alzheimer’s prevention. Neurology, 88(22), 2098–2106. 10.1212/WNL.0000000000003980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boots E. A., Schultz S., Clark L., Racine A. M., Darst B. F., KoscikOkonkwo R. L.O. C., Carlsson C. M., Gallagher C. L., Hogan K. J., Bendlin B. B., Asthana S., Sager M. A., Hermann B. P., Christian B. T., Dubal D. B., Engelman C. D., Johnson S. C., Okonkwo O. C. (2017. b). BDNF Val66Met predicts cognitive decline in the Wisconsin registry for Alzheimer’s prevention. Neurology, 88(22), 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower J. E., Ganz P. A., Desmond K. A., Bernaards C., Rowland J. H., Meyerowitz B. E., Belin T. R. (2006). Fatigue in long-term breast carcinoma survivors: A longitudinal investigation. Cancer, 106(4), 751–758. 10.1002/cncr.21671. [DOI] [PubMed] [Google Scholar]
- Brandt J. (1991). The Hopkins verbal learning test: Development of a new memory test with six equivalent forms. Clinical Neuropsychologist, 5(2), 125–142. 10.1080/13854049108403297. [DOI] [Google Scholar]
- Bressler J., Folsom A. R., Couper D. J., Volcik K. A., Boerwinkle E. (2010). Genetic variants identified in a European genome-wide association study that were found to predict incident coronary heart disease in the atherosclerosis risk in communities study. American Journal of Epidemiology, 171(1), 14–23. 10.1093/aje/kwp377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton P. R., Clayton D. G., Cardon L. R., Craddock N., Deloukas P., Duncanson A., Primary I. (2007). Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature, 447(7145), 661–678. 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskbjerg C. D. R., Amidi A., Demontis D., Nissen E. R., Zachariae R. (2019). Genetic risk factors for cancer-related cognitive impairment: A systematic review. Acta Oncologica, 58(5), 537–547. 10.1080/0284186x.2019.1578410. [DOI] [PubMed] [Google Scholar]
- Cacabelos R. (2007). Molecular pathology and pharmacogenomics in Alzheimer’s disease: Polygenic-related effects of multifactorial treatments on cognition, anxiety and depression. Methods and Findings in Experimental and Clinical Pharmacology, 29(Suppl A), 1–91. [PubMed] [Google Scholar]
- Carroll J. E., Small B. J., Tometich D. B., Zhai W., Zhou X., Luta G., Ahles T. A., Saykin A. J., Nudelman K. N. H., Clapp J. D., Jim H. S., Jacobsen P. B., Hurria A., Graham D., McDonald B. C., Denduluri N., Extermann M., Isaacs C., Dilawari A. A., Living With Cancer Study (2019). Sleep disturbance and neurocognitive outcomes in older patients with breast cancer: Interaction with genotype. Cancer, 125(24), 4516–4524. 10.1002/cncr.32489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro R., Rivera I., Ravasco P., Camilo M. E., Jakobs C., Blom H. J., de Almeida I. T. (2004). 5,10-methylenetetrahydrofolate reductase (MTHFR) 677C-->T and 1298A-->C mutations are associated with DNA hypomethylation. Journal of Medical Genetics, 41(6), 454–458. 10.1136/jmg.2003.017244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A., Yeo A., Shwe M., Tan C. J., Foo K. M., Chu P., Khor C. C., Ho H. K. (2019). An Evaluation of DNA Methyltransferase 1 (DNMT1) single nucleotide polymorphisms and chemotherapy-associated cognitive impairment: A prospective, longitudinal study. Scientfic Reports, 9(1), 14570. 10.1038/s41598-019-51203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Kelly C., Haw T. J., Lombard J. M., Nordman I. I. C., Croft A. J., Ngo D. T. M., Sverdlov A. L. (2021). Heart failure in breast cancer survivors: Focus on early detection and novel biomarkers. Current Heart Failure Reports, 18(6), 362–377. 10.1007/s11897-021-00535-w. [DOI] [PubMed] [Google Scholar]
- Chen J., Lipska B. K., Halim N., Ma Q. D., Matsumoto M., Melhem S., Kolachana B. S., Hyde T. M., Herman M. M., Apud J., Egan M. F., Kleinman J. E., Weinberger D. R. (2004). Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): Effects on mRNA, protein, and enzyme activity in postmortem human brain. American Journal of Human Genetics, 75(5), 807–821. 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sheng J., Tang X., Zhao Y., Zhu S., Liu Q. (2022). Clemastine rescues chemotherapy-induced cognitive impairment by improving White matter integrity. Neuroscience, 484(12), 66–79. 10.1016/j.neuroscience.2022.01.001. [DOI] [PubMed] [Google Scholar]
- Chotchaeva F., Balatskiy A., Samokhodskaya L., Tkachuk V., Sadovnichiy V. (2016). Association between T-cadherin gene (CDH13) variants and severity of coronary heart disease manifestation. International Journal of Clinical and Experimental Medicine, 9(3), 4059–4064. [Google Scholar]
- Corporation T. P. (2001). Wechsler test of adult reading. Harcourt Assessment. [Google Scholar]
- Davies G., Armstrong N., Bis J. C., Bressler J., Chouraki V., Giddaluru S, Hofer E., Ibrahim-Verbaas C. A., Kirin M., Lahti J., van der Lee S. J., Le Hellard S., Liu T., Marioni R. E., Oldmeadow C., Postmus I., Smith A. V., Smith J. A., Thalamuthu A., Deary I. J. (2015). Genetic contributions to variation in general cognitive function: A meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Molecular Psychiatry, 20(2), 183–192. 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G., Harris S. E., Reynolds C. A., Payton A., Knight H. M., Liewald D. C., Lopez L. M., Luciano M., Gow A. J., Corley J., Henderson R., Murray C., Pattie A., Fox H. C., Redmond P., Lutz M. W., Chiba-Falek O., Linnertz C., Saith S., Deary I. J. (2014). A genome-wide association study implicates the APOE locus in nonpathological cognitive ageing. Molecular Psychiatry, 19(1), 76–87. 10.1038/mp.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G., Marioni R. E., Liewald D. C., Hill W. D., Hagenaars S. P., Harris S. E., Ritchie S. J., Luciano M., Fawns-Ritchie C., Lyall D., Cullen B., Cox S. R., Hayward C., Porteous D. J., Evans J., McIntosh A. M., Gallacher J., Craddock N., Pell J. P., Deary I. J. (2016). Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Molecular Psychiatry, 21(6), 758–767. 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Frutos-Lucas J., Cuesta ., López-Sanz D., Peral-Suárez Á., Cuadrado-Soto E., Ramírez-Toraño F., Brown B. M., Serrano J. M., Laws S. M., Rodríguez-Rojo I. C., Verdejo-Román J., Bruña R., Delgado-Losada M. L., Barabash A., López-Sobaler A. M., López-Higes R., Marcos A., Maestú F. (2020). The relationship between physical activity, apolipoprotein E epsilon4 carriage, and brain health. Alzheimer’s Research & Therapy, 12(1), 48. 10.1186/s13195-020-00608-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Elia L. F., Satz P., Uchiyama C.L., White T. (1996). Color trails test. Psychological Assessment Resources, Inc. [Google Scholar]
- Deprez S., Kesler S. R., Saykin A. J., Silverman D. H. S., de Ruiter M. B., McDonald B. C. (2018). International cognition and cancer task force recommendations for neuroimaging methods in the study of cognitive impairment in non-CNS cancer patients. Journal of the National Cancer Institute, 110(3), 223–231. 10.1093/jnci/djx285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVos L., Chanson A., Liu Z., Ciappio E. D., Parnell L. D., Mason J. B., Tucker K. L., Crott J. W. (2008). Associations between single nucleotide polymorphisms in folate uptake and metabolizing genes with blood folate, homocysteine, and DNA uracil concentrations. American Journal of Clinical Nutrition, 88(4), 1149–1158. 10.1093/ajcn/88.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong S., Dhruva A., Dunn L. B., West C., Paul S. M., Cooper B. A., Elboim C., Abrams G., Merriman J. D., Langford D. J., Leutwyler H., Baggott C., Kober K., Aouizerat B. E., Miaskowski C. (2014). Associations between cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression in patients prior to breast cancer surgery. Biological Research for Nursing, 17(3), 237–247. 10.1177/1099800414550394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong S. H., Dhruva A., Dunn L. B., West C., Paul S. M., Cooper B. A., Elboim C., Abrams G., Merriman J. D., Langford D. J., Leutwyler H., Baggott C., Kober K., Aouizerat B. E., Miaskowski C. (2015). Associations between cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression in patients prior to breast cancer surgery. Biological Research for Nursing, 17(3), 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher P. M., Holst K. K., Adamsen D., Klein A. B., Frokjaer V. G., Jensen P. S., Svarer C., Gillings N., Baare W. F., Mikkelsen J. D., Knudsen G. M. (2015). BDNF Val66met and 5-HTTLPR polymorphisms predict a human in vivo marker for brain serotonin levels. Human Brain Mapping, 36(1), 313–323. 10.1002/hbm.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz P. A. (2012). Doctor, will the treatment you are recommending cause chemobrain? Journal of Clinical Oncology, 30(3), 229–231. 10.1200/JCO.2011.39.4288. [DOI] [PubMed] [Google Scholar]
- Gonzalez B. D., Jim H. S., Booth-Jones M., Small B. J., Sutton S. K., Lin H. Y., Park J. Y., Spiess P. E., Fishman M. N., Jacobsen P. B. (2015). Course and predictors of cognitive function in patients with prostate cancer receiving androgen-deprivation therapy: A controlled comparison. Journal of Clinical Oncology, 33(18), 2021–2027. 10.1200/JCO.2014.60.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk W. K., Lutz M. W., He Y. T., Saunders A. M., Burns D. K., Roses A. D., Chiba-Falek O. (2014). The broad impact of TOM40 on neurodegenerative diseases in aging. Journal of Alzheimers Disease and Parkinsonism, 1(1), 12. 10.13188/2376-922X.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen M., Laakso A., Någren K., Rinne J. O., Pohjalainen T., Hietala J. (2004). C957T polymorphism of the dopamine D2 receptor (DRD2) gene affects striatal DRD2 availability in vivo. Molecular Psychiatry, 9(12), 1060–1061. 10.1038/sj.mp.4001561. [DOI] [PubMed] [Google Scholar]
- Hughes E., Tshiaba P., Wagner S., Judkins T., Rosenthal E., Roa B., Gallagher S., Meek S., Dalton K., Hedegard W., Adami C. A., Grear D. F., Domchek S. M., Garber J., Lancaster J. M., Weitzel J. N., Kurian A. W., Lanchbury J. S., Gutin A., Robson M. E. (2021). Integrating clinical and polygenic factors to predict breast cancer risk in women undergoing genetic testing. JCO Precision Oncology, 5(5), 307–316. 10.1200/PO.20.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Su G., Wang X. (2019). The roles of ANRIL polymorphisms in coronary artery disease: A meta-analysis. Bioscience Reports, 39(12), Article BSR20181559. 10.1042/bsr20181559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S. R., Small B. J., Booth-Jones M., Jacobsen P. B., Fields K. K. (2007). Changes in cognitive functioning in the year after hematopoietic stem cell transplantation. Cancer, 110(7), 1560–1567. 10.1002/cncr.22962. [DOI] [PubMed] [Google Scholar]
- Jiang W., Xu J., Lu X. J., Sun Y. (2016). Association between MTHFR C677T polymorphism and depression: A meta-analysis in the Chinese population. Psychology, Health & Medicine, 21(6), 675–685. 10.1080/13548506.2015.1120327. [DOI] [PubMed] [Google Scholar]
- Jim H. S., Park J. Y., Permuth-Wey J., Rincon M. A., Phillips K. M., Small B. J., Jacobsen P. B. (2012). Genetic predictors of fatigue in prostate cancer patients treated with androgen deprivation therapy: Preliminary findings. Brain, Behavior, and Immunity, 26(7), 1030–1036. 10.1016/j.bbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleck T. A., Bender C. M., Sereika S. M., Ahrendt G., Jankowitz R. C., McGuire K. P., Ryan C. M., Conley Y. P. (2014). Apolipoprotein E genotype and cognitive function in postmenopausal women with early-stage breast cancer. Oncology Nursing Forum, 41(6), E313–E325. 10.1188/14.ONF.E313-E325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleck T. A., Bender C. M., Sereika S. M., Brufsky A. M., Lembersky B. C., McAuliffe P. F., Puhalla S. L., Rastogi P., Conley Y. P. (2016). Polymorphisms in DNA repair and oxidative stress genes associated with pre-treatment cognitive function in breast cancer survivors: an exploratory study. Springerplus, 5(1), 422. 10.1186/s40064-016-2061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita G. P., Ekholm O., Kaasa S., Klepstad P., Skorpen F., Sjøgren P. (2016). Genetic variation and cognitive dysfunction in opioid-treated patients with cancer. Brain and Behavior, 6(7), Article e00471. 10.1002/brb3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hellard S., Wang Y., Witoelar A., Zuber V., Bettella F., Hugdahl K., Espeseth T., Steen V. M., Melle I., Desikan R., Schork A. J., Thompson W. K., Dale A. M., Djurovic S., Andreassen O. A. (2017). Identification of gene loci that overlap between schizophrenia and educational attainment. Schizophrenia Bulletin, 43(3), 654–664. 10.1093/schbul/sbw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. M., Oh M. H., Go J. H., Han K., Choi S. Y. (2020). Molecular subtypes of triple-negative breast cancer: Understanding of subtype categories and clinical implication. Journal of Genetics and Genomics, 42(12), 1381–1387. 10.1007/s13258-020-01014-7. [DOI] [PubMed] [Google Scholar]
- Lengacher C. A., Reich R. R., Kip K. E., Paterson C. L., Park H. Y., Ramesar S., Jim H. S., Alinat C. B., Park J. Y. (2015. a). Moderating effects of genetic polymorphisms on improvements in cognitive impairment in breast cancer survivors participating in a 6-week mindfulness-based stress reduction program. Biological Research for Nursing, 17(4), 393–404. 10.1177/1099800415577633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengacher C. A., Reich R. R., Paterson C. L., Jim H. S., Ramesar S., Alinat C. B., Budhrani P. H., Farias J. R., Shelton M. M., Moscoso M. S., Park J. Y., Kip K. E. (2015. b). The effects of mindfulness-based stress reduction on objective and subjective sleep parameters in women with breast cancer: A randomized controlled trial. Psycho-oncology, 24(4), 424–432. 10.1002/pon.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H., Lin E., Lane H. Y. (2017). Genetic biomarkers on age-related cognitive decline. Frontiers in Psychiatry, 8(11), 247. 10.3389/fpsyt.2017.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linschoten M., Teske A. J., Cramer M. J., van der Wall E., Asselbergs F. W. (2018). Chemotherapy-related cardiac dysfunction: A systematic review of genetic variants modulating individual risk. Circulation: Genomic and Precision Medicine, 11(1), Article e001753. 10.1161/circgen.117.001753. [DOI] [PubMed] [Google Scholar]
- Maj M., D'Elia L., Satz P., Janssen R., Zaudig M., Uchiyama C., Starace F., Galderisi S., Chervinsky A., World Health Organization (1993). Evaluation of two new neuropsychological tests designed to minimize cultural bias in the assessment of HIV-1 seropositive persons: A WHO study. Archives of Clinical Neuropsychology, 8(2), 123–135. 10.1093/arclin/8.2.123. [DOI] [PubMed] [Google Scholar]
- Mamasoula C., Prentice R. R., Pierscionek T., Pangilinan F., Mills J. L., Druschel C., Pass K., Russell M. W., Hall D., Töpf A., Brown D. L., Zelenika D., Bentham J., Cosgrove C., Bhattacharya S., Riveron J. G., Setchfield K., Brook J. D., Bu’Lock F. A., Keavney B. D. (2013). Association between C677T polymorphism of methylene tetrahydrofolate reductase and congenital heart disease: Meta-analysis of 7697 cases and 13,125 controls. Circulation: Cardiovascular Genetics, 6(4), 347–353. 10.1161/CIRCGENETICS.113.000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Cleak J., Willis-Owen S. A., Flint J., Shifman S. (2007). Mapping regulatory variants for the serotonin transporter gene based on allelic expression imbalance. Molecular Psychiatry, 12(5), 421–422. 10.1038/sj.mp.4001952. [DOI] [PubMed] [Google Scholar]
- Mazanec S. R., Park S., Connolly M. C., Rosenzweig M. Q. (2021). Factors associated with symptom distress in women with breast cancer prior to initiation of chemotherapy. Applied Nursing Research, 62(3), 151515. 10.1016/j.apnr.2021.151515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriman J. D., Aouizerat B. E., Cataldo J. K., Dunn L., Cooper B. A., West C., Paul S. M., Baggott C. R., Dhruva A., Kober K., Langford D. J., Leutwyler H., Ritchie C. S., Abrams G., Dodd M., Elboim C., Hamolsky D., Melisko M., Miaskowski C. (2014). Association between an interleukin 1 receptor, type I promoter polymorphism and self-reported attentional function in women with breast cancer. Cytokine, 65(2), 192–201. 10.1016/j.cyto.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnezami R., Nicholson J., Darzi A. (2012). Preparing for precision medicine. New England Journal of Medicine, 366(2), 489–491. 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- Molina-Moya B., Kazdaglis G., Lacoma A., Prat C., Gomez A., Villar-Hernandez R., García-García E., Haba L., Maldonado J., Samper S., Ruiz-Manzano J., Domínguez J. (2016). Evaluation of GenoFlow DR-MTB array test for detection of rifampin and isoniazid resistance in mycobacterium tuberculosis. Journal of Clinical Microbiology, 54(4), 1160–1163. 10.1128/JCM.03341-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montazeri A., Harirchi I., Vahdani M., Khaleghi F., Jarvandi S., Ebrahimi M., Haji-Mahmoodi M. (1999). The European Organization for Research and Treatment of Cancer Quality of Life Questionnaire (EORTC QLQ-C30): Translation and validation study of the Iranian version. Supportive Care in Cancer, 7(6), 400–406. 10.1007/s005200050300. [DOI] [PubMed] [Google Scholar]
- Morandi A., Bonnedfond A., Lobbens S., Yengo L., Miraglia Del Giudice E., Grandone A., Lévy-Marchal C., Weill J., Maffeis C., Froguel P. 2016). Associations between type 2 diabetes-related genetic scores and metabolic traits, in obese and normal-weight youths. The Journal of Clinical Endocrinology & Metabolism, 101(11), 4244–4250. 10.1210/jc.2016-2432 [DOI] [PubMed] [Google Scholar]
- Munir F., Kalawsky K., Lawrence C., Yarker J., Haslam C., Ahmed S. (2011). Cognitive intervention for breast cancer patients undergoing adjuvant chemotherapy: A needs analysis. Cancer Nursing, 34(5), 385–392. 10.1097/NCC.0b013e31820254f3. [DOI] [PubMed] [Google Scholar]
- Necela B. M., Axenfeld B. C., Serie D. J., Kachergus J. M., Perez E. A., Thompson E. A., Norton N. (2017). The antineoplastic drug, trastuzumab, dysregulates metabolism in iPSC-derived cardiomyocytes. Clinical and Translational Medicine, 6(1), 5. 10.1186/s40169-016-0133-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T., Chan M., Khor C. C., Ho H. K., Chan A. (2014). The genetic variants underlying breast cancer treatment-induced chronic and late toxicities: A systematic review. Cancer Treatment Reviews, 40(10), 1199–1214. 10.1016/j.ctrv.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Ng T., Teo S. M., Yeo H. L., Shwe M., Gan Y. X., Cheung Y. T., Foo K. M., Cham M. T., Lee J. A., Tan Y. P., Fan G., Yong W. S., Preetha M., Loh W. J., Koo S. L., Jain A., Lee G. E., Wong M., Dent R., Chan A. (2016). Brain-derived neurotrophic factor genetic polymorphism (rs6265) is protective against chemotherapy-associated cognitive impairment in patients with early-stage breast cancer. Neuro-oncology, 18(2), 244–251. 10.1093/neuonc/nov162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X., Bismil R., Chan K., Sicard T., Bulgin N., McMain S., Kennedy J. L. (2006). Serotonin 2A receptor gene is associated with personality traits, but not to disorder, in patients with borderline personality disorder. Neuroscience Letters, 408(3), 214–219. 10.1016/j.neulet.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Noble E. P. (2000). The DRD2 gene in psychiatric and neurological disorders and its phenotypes. Pharmacogenomics, 1(3), 309–333. 10.1517/14622416.1.3.309. [DOI] [PubMed] [Google Scholar]
- Norton N., Crook J. E., Wang L., Olson J. E., Kachergus J. M., Serie D. J., Necela B. M., Borgman P. G., Advani P. P., Ray J. C., Landolfo C., Di Florio D. N., Hill A. R., Bruno K. A., Fairweather D. (2020). Association of genetic variants at TRPC6 with chemotherapy-related heart failure. Frontiers in Cardiovascular Medicine, 7(5), 142. 10.3389/fcvm.2020.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue M. C., Murphy S. E., Zamboni G., Nobre A. C., Mackay C. E. (2018). APOE genotype and cognition in healthy individuals at risk of Alzheimer’s disease: A review. Cortex, 104(5), 103–123. 10.1016/j.cortex.2018.03.025. [DOI] [PubMed] [Google Scholar]
- Park J. Y., Lengacher C. A., Reich R. R., Alinat C. B., Ramesar S., Le A., Paterson C. L., Pleasant M. L., Park H. Y., Kiluk J., Han H., Ismail-Khan R., Kip K. E. (2019). Translational genomic research: The role of genetic polymorphisms in MBSR program among breast cancer survivors (MBSR[BC]). Translational Behavioral Medicine, 9(4), 693–702. 10.1093/tbm/iby061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival N., George A., Gvertson J., Hamill M., Fernandez A., Davies E., Rahman N., Banerjee S. (2016). The integration of BRCA testing into oncology clinics. British Journal of Nursing, 25(12), 690–694. 10.12968/bjon.2016.25.12.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya O. O., Sokolov B. P. (2002). Differential expression of the “C” and “T” alleles of the 5-HT2A receptor gene in the temporal cortex of normal individuals and schizophrenics. Journal of Neuroscience Research, 67(6), 812–822. 10.1002/jnr.10173. [DOI] [PubMed] [Google Scholar]
- Ponce G., Pérez-González R., Aragüés M., Palomo T., Rodríguez-Jiménez R., Jiménez-Arriero M. A., Hoenicka J. (2009). The ANKK1 kinase gene and psychiatric disorders. Neurotoxicity Research, 16(1), 50–59. 10.1007/s12640-009-9046-9. [DOI] [PubMed] [Google Scholar]
- Prada D., Colicino E., Power M. C., Cox D. G., Weisskopf M. G., Hou L., Spiro Iii A., Vokonas P., Zhong J., Sanchez-Guerra M., Herrera L. A., Schwartz J., Baccarelli A. A. (2014). Influence of multiple APOE genetic variants on cognitive function in a cohort of older men - results from the normative aging study. BMC Psychiatry, 14(1), 223. 10.1186/s12888-014-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullens M. J., De Vries J., Roukema J. A. (2010). Subjective cognitive dysfunction in breast cancer patients: A systematic review. Psycho-oncology, 19(11), 1127–1138. 10.1002/pon.1673. [DOI] [PubMed] [Google Scholar]
- Ren X., Boriero D., Chaiswing L., Bondada S., St Clair D. K., Butterfield D. A. (2019). Plausible biochemical mechanisms of chemotherapy-induced cognitive impairment (“chemobrain”), a condition that significantly impairs the quality of life of many cancer survivors. Biochimica et Biophysica Acta- Molecular Basis of Disease, 1865(6), 1088–1097. 10.1016/j.bbadis.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld C. A., Esko T., Davies G., Pers T. H., Turley P., Benyamin B., Chabris C. F., Emilsson V., Johnson A. D., Lee J. J., de Leeuw C., Marioni R. E., Medland S. E., Miller M. B., Rostapshova O., van der Lee S. J., Vinkhuyzen A. A., Amin N., Conley D., Koellinger P. D. (2014). Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proceedings of the National Academy of Sciences of the United States of America, 111(38), 13790–13794. 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani N. J., Erdmann J., Hall A. S., Hengstenberg C., Mangino M., Mayer B., Dixon R. J., Meitinger T., Braund P., Wichmann H. E., Barrett J. H., König I. R., Stevens S. E., Szymczak S., Tregouet D. A., Iles M. M., Pahlke F., Pollard H., Lieb W., the Cardiogenics, C (2007). Genomewide association analysis of coronary artery disease. New England Journal of Medicin, 357(5), 443–453. 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitz J., Hodgkinson C. A., Martin-Soelch C., Shen P. H., Szczepanik J., Nugent A. C., Herscovitch P., Grace A. A., Goldman D., Drevets W. C. (2013). DRD2/ANKK1 Taq1A polymorphism (rs1800497) has opposing effects on D2/3 receptor binding in healthy controls and patients with major depressive disorder. International Journal of Neuropsychopharmacology, 16(9), 2095–2101. 10.1017/S146114571300045X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiepers O. J., Harris S. E., Gow A. J., Pattie A., Brett C. E., Starr J. M., Deary I. J. (2012). APOE E4 status predicts age-related cognitive decline in the ninth decade: Longitudinal follow-up of the Lothian Birth Cohort 1921. Molecular Psychiatry, 17(3), 315–324. 10.1038/mp.2010.137. [DOI] [PubMed] [Google Scholar]
- Schillani G., Martinis E., Capozzo M. A., Era D., Cristante T., Mustacchi G., Conte M. A., DE Vanna M., Grassi L., Giraldi T. (2010). Psychological response to cancer: Role of 5-HTTLPR genetic polymorphism of serotonin transporter. Anticancer Research, 30(9), 3823–3826. [PubMed] [Google Scholar]
- Schneider B. P., Shen F., Gardner L., Radovich M., Li L., Miller K. D., Jiang G., Jr, Lai D., O'Neill A., Sparano J. A., Davidson N. E., Cameron D., Gradus-Pizlo I., Mastouri R. A., Suter T. M., Foroud T., Sledge G. W. (2017). Genome-wide association study for anthracycline-induced congestive heart failure. Clinical Cancer Research, 23(1), 43–51. 10.1158/1078-0432.Ccr-16-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A. (2021). Cancer statistics, 2021. Ca: a Cancer Journal for Clinicians, 71(1), 7–33. 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- Sloan C. D., Shen L., West J. D., Wishart H. A., Flashman L. A., Rabin L. A., Santulli R. B., Guerin S. J., Rhodes C. H., Tsongalis G. J., McAllister T. W., Ahles T. A., Lee S. L., Moore J. H., Saykin A. J. (2010). Genetic pathway-based hierarchical clustering analysis of older adults with cognitive complaints and amnestic mild cognitive impairment using clinical and neuroimaging phenotypes. American Journal of Medical Genetics Neuropsychiatric Genetics, 153b(5), 1060–1069. 10.1002/ajmg.b.31078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small B. J., Rawson K. S., Walsh E., Jim H. S., Hughes T. F., Iser L., Andrykowski M. A., Jacobsen P. B. (2011). Catechol-O-methyltransferase genotype modulates cancer treatment-related cognitive deficits in breast cancer survivors. Cancer, 117(7), 1369–1376. 10.1002/cncr.25685. [DOI] [PubMed] [Google Scholar]
- Spellicy C. J., Harding M. J., Hamon S. C., Mahoney J. J., 3rd, Reyes J. A., Kosten T. R., Newton T. F., De La Garza R., Nielsen D. A. (2014). A variant in ANKK1 modulates acute subjective effects of cocaine: A preliminary study. Genes, Brain and Behavior, 13(6), 559–564. 10.1111/gbb.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller W., Jung K. J., Lee J. Y., Jee S. H. (2020). Precision medicine and cardiovascular health: Insights from Mendelian randomization analyses. Korean Circulation Journal, 50(2), 91–111. 10.4070/kcj.2019.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A., Collins B., Mackenzie J., Tomiak E., Verma S., Bielajew C. (2008). The cognitive effects of adjuvant chemotherapy in early stage breast cancer: A prospective study. Psycho-oncology, 17(2), 122–130. 10.1002/pon.1210. [DOI] [PubMed] [Google Scholar]
- Su Y. Y., Liang X., Schoepf U. J., Varga-Szemes A., West H. C., Qi R., Kong X., Chen H. J., Lu G. M., Zhang L. J. (2015). APOE polymorphism affects brain default mode network in healthy young adults: A STROBE article. Medicine, 94(52), Artcile e1734. 10.1097/MD.0000000000001734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T., Adachi Y., Ohtsuki Y., Furihata M. (2007). Adiponectin receptors, with special focus on the role of the third receptor, T-cadherin, in vascular disease. Medical Molecular Morphology, 40(3), 115–120. 10.1007/s00795-007-0364-9. [DOI] [PubMed] [Google Scholar]
- Tan C. J., Lim S. W. T., Toh Y. L., Ng T., Yeo A., Shwe M., Foo K. M., Chu P., Jain A., Koo S. L., Dent R. A., Ng R. C. H., Yap Y. S., Lim E. H., Loh K. W., Chay W. Y., Lee G. E., Tan T. J. Y., Beh S. Y., Chan A. (2019). Replication and meta-analysis of the association between BDNF Val66Met polymorphism and cognitive impairment in patients receiving chemotherapy. Molecular Neurobiology, 56(7), 4741–4750. 10.1007/s12035-018-1410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., Lin H., Yan Y., Xu X., Wang L., Zhang J., Yu Y. (2017). Impairment of the executive function in breast cancer patients receiving chemotherapy treatment: A functional MRI study. European Journal of Cancer Care, 26(6). 10.1111/ecc.12553. [DOI] [PubMed] [Google Scholar]
- Teske A. J., Linschoten M., Kamphuis J. A. M., Naaktgeboren W. R., Leiner T., van der Wall E., Kuball J., van Rhenen A., Doevendans P. A., Cramer M. J., Asselbergs F. W. (2018). Cardio-oncology: An overview on outpatient management and future developments. Netherlands Heart Journal, 26(11), 521–532. 10.1007/s12471-018-1148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenerry M. R., Crosson B., Deboe J., Leber W. R. (1989). Stroop neuropsychological screening test manual. Psychological Assessment Resources. [Google Scholar]
- Trenkwalder T., Nelson C. P., Musameh M. D., Mordi I. R., Kessler T., Pellegrini C., Debiec R., Rheude T., Lazovic V., Zeng L., Martinsson A., Gustav Smith J., Gådin J. R., Franco-Cereceda A., Eriksson P., Nielsen J. B., Graham S. E., Willer C. J., Samani N. J. (2019). Effects of the coronary artery disease associated LPA and 9p21 loci on risk of aortic valve stenosis. International Journal of Cardiology, 276(4), 212–217. 10.1016/j.ijcard.2018.11.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueland P. M., Hustad S., Schneede J., Refsum H., Vollset S. E. (2001). Biological and clinical implications of the MTHFR C677T polymorphism. Trends in Pharmacological Sciences, 22(4), 195–201. 10.1016/s0165-6147(00)01675-8. [DOI] [PubMed] [Google Scholar]
- van der Harst P., Verweij N. (2018). Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circulation Research, 122(3), 433–443. 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Schuhmacher A., Schwab S., Zobel A., Maier W. (2008). The His452Tyr variant of the gene encoding the 5-HT2A receptor is specifically associated with consolidation of episodic memory in humans. International Journal of Neuropsychopharmacology, 11(8), 1163–1167. 10.1017/S146114570800905X. [DOI] [PubMed] [Google Scholar]
- Wagner L. I., Sweet J., Butt Z., Lai J.-s., Cella D. (2009). Measuring patient self-reported cognitive function: Development of the functional assessment of cancer therapy-cognitive function instrument. Journal of Supportive Oncology, 7(6), W32–W39. [Google Scholar]
- Wang I. J., Chiang T. H., Shih Y. F., Lu S. C., Lin L. L., Shieh J. W., Wang T. H., Samples J. R., Hung P. T. (2006). The association of single nucleotide polymorphisms in the MMP-9 genes with susceptibility to acute primary angle closure glaucoma in Taiwanese patients. Molecular Vision, 12(3), 1223–1232. [PubMed] [Google Scholar]
- Wechsler D. (2008). Wechsler Adult Intelligence Scale-Fourth Edition (WAIS-IV). NCS Pearson. [Google Scholar]
- Wechsler D. (2009). Wechsler memory scale-fourth edition. http://www.pearsonpsychcorp.es/.
- Wefel J. S., Saleeba A. K., Buzdar A. U., Meyers C. A. (2010). Acute and late onset cognitive dysfunction associated with chemotherapy in women with breast cancer. Cancer, 116(14), 3348–3356. 10.1002/cncr.25098. [DOI] [PubMed] [Google Scholar]
- Weiner A. S., Gordeeva L. A., Voronina E. N., Boyarskikh U. A., Shabaldin A. V., Filipenko M. L. (2012). Polymorphisms in folate-metabolizing genes and risk of having an offspring with congenital anomalies in the West Siberian region of Russia: a case-control study. Prenatal Diagnos, 32(11), 1041–1048. 10.1002/pd.3952. [DOI] [PubMed] [Google Scholar]
- Wickramage I., Tennekoon K. H., Ariyaratne M. A. Y., Hewage A. S., Sundralingam T. (2017). CYP2D6 polymorphisms may predict occurrence of adverse effects to tamoxifen: A preliminary retrospective study. Breast Cancer: Targets and Therapy, 9, 111–120. 10.2147/BCTT.S126557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan H., Gan C., Li W., Huang Z., Wang L., Jia Q., Cheng H. (2017). Altered network efficiency of functional brain networks in patients with breast cancer after chemotherapy. Oncotarget, 8(62), 105648–105661. 10.18632/oncotarget.22358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T. H., Denburg N. L., Beglinger L. J., Schultz S. K. (2010). Neuropsychological outcomes of older breast cancer survivors: Cognitive features ten or more years after chemotherapy. Journal of Neuropsychiatry and Clinical Neurosciences, 22(1), 48–54. 10.1176/appi.neuropsych.22.1.4810.1176/jnp.2010.22.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G. S., Kumar S., Dorsey S. G., Starkweather A. R., Kelly D. L., Lyon D. E. (2019). Systematic review of genetic polymorphisms associated with psychoneurological symptoms in breast cancer survivors. Supportive Care in Cancer, 27(2), 351–371. 10.1007/s00520-018-4508-3. [DOI] [PubMed] [Google Scholar]
- Yang H. L., Yang Y. L., Yu C. H., Shiao S. P. K. (2018). Meta-prediction of MTHFR gene polymorphism and air pollution on the risks of congenital heart defects worldwide: A transgenerational analysis. International Journal of Environmental Research and Public Health, 15(8), 1660. 10.3390/ijerph15081660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Hu Y., North K. E., Franceschini N., Lin D. (2009). Evaluation of population impact of candidate polymorphisms for coronary heart disease in the Framingham heart study offspring cohort. BMC Proc, 3(Suppl 7), S118. 10.1186/1753-6561-3-s7-s118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M., Dong L., Zheng J., Zhang H., Liu J., Xu Z. (2012). Meta analysis of the association between MTHFR C677T polymorphism and the risk of congenital heart defects. Annals of Human Genetics, 76(1), 9–16. 10.1111/j.1469-1809.2011.00687.x. [DOI] [PubMed] [Google Scholar]
- Zhang W., Cao Y., Wang M., Ji L., Chen L., Deater-Deckard K. (2015). The Dopamine D2 Receptor Polymorphism (DRD2 TaqIA) interacts with maternal parenting in predicting early adolescent depressive symptoms: Evidence of differential susceptibility and age differences. Journal of Youth and Adolescence, 44(7), 1428–1440. 10.1007/s10964-015-0297-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Translational Genomic Research: The Association between Genetic Profiles and Cognitive Functioning or Cardiac Function Among Breast Cancer Survivors Completing Chemotherapy by Jong Y. Park, Cecile A. Lengacher, Richard R. Reich, Hyun Y. Park, Junmin Whiting, Anh Thy Nguyen, Carmen Rodríguez, Hongdao Meng, Sara Tinsley, Katterine Chauca, Liliana Gordillo-Casero, Trudy Wittenberg, Anisha Joshi, Katherine Lin, Roohi Ismail-Khan, John V. Kiluk, and Kevin E. Kip in Biological Research For Nursing