Abstract
Background
Transmitted human immunodeficiency virus (HIV) drug resistance can threaten the efficacy of antiretroviral therapy and pre-exposure prophylaxis (PrEP). Drug-resistance testing is recommended at entry to HIV care in the United States and provides valuable insight for clinical decision making and population-level monitoring.
Methods
We assessed transmitted drug-resistance–associated mutation (TDRM) prevalence and predicted susceptibility to common HIV drugs among US persons with HIV diagnosed during 2014–2018 who had a drug resistance test performed ≤3 months after HIV diagnosis and reported to the National HIV Surveillance System and who resided in 28 jurisdictions where ≥20% of HIV diagnoses had an eligible sequence during this period.
Results
Of 50 747 persons in the analysis, 9616 (18.9%) had ≥1 TDRM. TDRM prevalence was 0.8% for integrase strand transfer inhibitors (INSTIs), 4.2% for protease inhibitors, 6.9% for nucleoside reverse transcriptase inhibitors (NRTIs), and 12.0% for non-NRTIs. Most individual mutations had a prevalence <1.0% including M184V (0.9%) and K65R (0.1%); K103N was most prevalent (8.6%). TDRM prevalence did not increase or decrease significantly during 2014–2018 overall, for individual drug classes, or for key individual mutations except for M184V (12.9% increase per year; 95% confidence interval, 5.6–20.6%).
Conclusions
TDRM prevalence overall and for individual drug classes remained stable during 2014–2018; transmitted INSTI resistance was uncommon. Continued population-level monitoring of INSTI and NRTI mutations, especially M184V and K65R, is warranted amidst expanding use of second-generation INSTIs and PrEP.
Keywords: HIV, drug resistance, surveillance, public health, integrase inhibitor
In this analysis, transmitted human immunodeficiency virus (HIV)-1 drug resistance in 28 US jurisdictions did not increase overall or by drug class during 2014–2018. Continued monitoring is warranted amidst expanding use of HIV pre-exposure prophylaxis and integrase-strand transfer inhibitors.
Human immunodeficiency virus (HIV) antiretroviral therapy (ART) and pre-exposure prophylaxis (PrEP) are cornerstones of HIV treatment and prevention and essential components of the Ending the HIV Epidemic: A Plan for America (EHE) initiative [1]. Viral resistance to ART and PrEP can arise from mutations transmitted at the time of infection or acquired during exposure to suboptimal drug levels; this resistance can reduce the efficacy of ART and PrEP and represents a threat to successful HIV elimination efforts. Transmitted drug-resistance–associated mutations (TDRMs) have been described for all major drug classes and can result in delayed viral suppression or treatment failure for persons initiating ART [2]. For this reason, standard genotypic drug resistance testing is recommended at entry into care for all persons with HIV in the United States to assist with selection of an initial ART regimen [3–5].
Prior estimates of overall TDRM prevalence in the United States have ranged from 11% to 15% [6–9], with non-nucleoside reverse transcriptase inhibitors (NNRTIs) providing the largest contribution (prevalence, 6–10%) followed by nucleoside reverse transcriptase inhibitors (NRTIs; prevalence, 3–8%) and protease inhibitors (PIs; prevalence, 3–5%) [6–9]. Thymidine-analog mutations (TAMs) selected previously by NRTIs no longer in widespread use still contribute substantially to overall NRTI TDRM prevalence [6]. Few studies have assessed population-level transmitted resistance to integrase strand transfer inhibitors (INSTIs) in the United States, and reports of INSTI resistance in treatment-naive individuals have been rare [10–13].
The efficacy of approved PrEP regimens that combine tenofovir disoproxil fumarate (TDF) or tenofovir alafenamide (TAF) with emtricitabine (FTC) can be reduced by drug-resistance mutations. M184V (affecting FTC) and K65R (affecting TDF/TAF and FTC) have been associated with PrEP failure due to infection with drug-resistant virus [14–18]. INSTI resistance is also relevant to PrEP: cabotegravir, an investigational INSTI currently in PrEP efficacy trials, selects for mutations associated with resistance to other INSTIs used for ART [19, 20].
In the United States, all states and territories submit deidentified demographic, risk, clinical, and laboratory data for persons with diagnosed HIV infection to the National HIV Surveillance System (NHSS) at the Centers for Disease Control and Prevention (CDC). Since 2004, HIV-1 nucleotide sequence data from persons with diagnosed HIV residing in select jurisdictions have been incorporated into routine NHSS reporting. The number of participating jurisdictions has grown in recent years; as of 2018, health departments in all US jurisdictions were required to collect and submit HIV-1 nucleotide sequence data to NHSS from drug-resistance tests for all persons with diagnosed HIV in their jurisdiction. This includes the protease (PR), reverse transcriptase (RT), and integrase (IN) segments of the pol region. These data provide valuable insight into population-level drug-resistance patterns and can inform guidelines for clinical management of HIV and PrEP in the United States. We analyzed NHSS data to describe the prevalence of transmitted drug resistance in the United States among persons with HIV diagnosed during 2014–2018.
METHODS
We included persons with HIV infection diagnosed during 2014–2018 with a partial HIV-1 pol nucleotide sequence collected 3 months or earlier of HIV diagnosis and reported to NHSS by 30 June 2019, who were 13 years or older at the time of HIV diagnosis, and who had no evidence of HIV ART use prior to the date of sequence collection. HIV subtype was determined using Context-based Modeling for Expeditious Typing (COMET) version 2.2 [21]; and sequences from subtypes A, B, C, D, F, and G and circulating recombinant forms (CRF) 01 and 02 were included. Sequences were classified by whether they contained the protease and reverse transcriptase (PR-RT) region and/or the integrase (IN) region; sequences less than 500 nucleotides in length were excluded. For persons with multiple PR, RT, or IN sequences meeting the inclusion criteria, the earliest PR, RT, and IN sequence was selected. We included persons whose residence at diagnosis was in 1 of 28 US jurisdictions where 20% or more of HIV diagnoses during 2014–2018 had a sequence meeting the inclusion criteria.
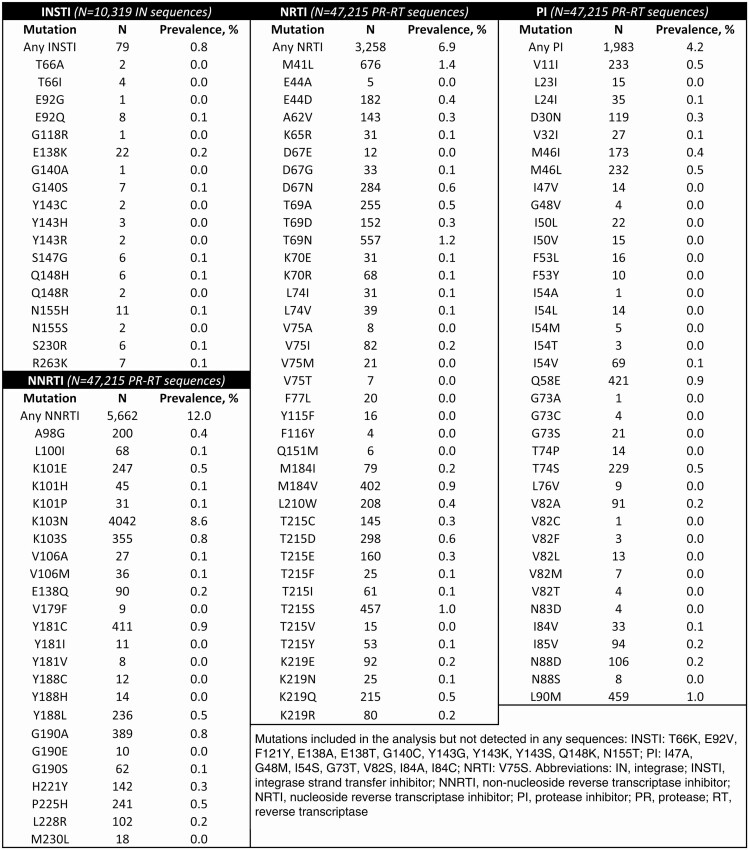
We defined NRTI, NNRTI, and PI TDRMs using the previously published CDC HIV-1 surveillance mutation list [9], which includes RT and PR mutations with a prevalence of 1% or greater in treated persons and omits polymorphic mutations (prevalence ≥0.5% in treatment-naive persons). For INSTI TDRMs, we included 24 non-polymorphic mutations identified by Tzou et al [22] based on published expert lists, conservation in INSTI-naive persons, frequency in INSTI-treated persons, and contribution to reduced in vitro susceptibility: T66A/I/K, E92G/Q, G118R, F121Y, E138A/K/T, G140A/C/S, Y143C/H/R/S, S147G, Q148H/R/K, N155H, S230R, R263K. We included 5 additional rare nonpolymorphic mutations (E92V, Y143G/K, N155S/T) for a total of 29 INSTI TDRMs.
HIV-1 drug-resistance–associated mutations were identified using the Sierra Web Service version 1.1 [23]. Because all sequences were performed 3 months or less after HIV diagnosis among persons with no evidence of prior ART use, all mutations were classified as TDRMs. We reported the number of sequences with each individual mutation, with 1 or more TDRM to each individual drug class (NRTI, NNRTI, PI, and INSTI) and with 1 or more TDRM to any drug class. TDRM prevalence was calculated by dividing the number of sequences with each TDRM by the number of sequences containing the relevant gene (PR, RT, or IN). For each drug class and for select mutations with pronounced effects on commonly used drugs, we calculated the estimated annual percent change during 2014–2018 using log binomial regression.
We used the Stanford HIV Drug Resistance Database (HIVdb) genotypic resistance interpretation system, version 8.2 [24], to predict susceptibility to commonly used antiretroviral drugs. This system assigns a drug penalty score to each mutation and combination of mutations based on prevalence in treated and untreated persons, in vitro phenotypic data, published associations between genotype and virologic suppression, and expert opinion. Scores for individual mutations range from –15 to +60; higher positive numbers indicate higher resistance, 0 indicates no change to susceptibility, and negative numbers indicate increased susceptibility compared with wild-type virus. Individual mutation scores are combined to obtain a final score in 1 of 5 categories: susceptible (0–9), potential low-level resistance (10–14), low-level resistance (15–29), intermediate resistance (30–59), and high-level resistance (≥60). We determined predicted resistance for commonly used antiretroviral drugs assessed by HIVdb using the total number of sequences with 1 or more TDRM affecting the corresponding drug class as the denominator.
RESULTS
A total of 50 747 persons with diagnosed HIV infection from 28 jurisdictions (Alabama, Alaska, Arizona, California, Colorado, Connecticut, District of Columbia, Florida, Idaho, Illinois, Iowa, Louisiana, Maryland, Michigan, Montana, New York, North Dakota, Ohio, Oregon, Pennsylvania, South Carolina, Texas, Utah, Vermont, Virginia, Washington, West Virginia, Wisconsin) had a sequence included in the analysis, representing 35.4% of all HIV diagnoses in these jurisdictions during 2014–2018. Of those with an eligible sequence, 47 215 (93.0%) persons had a PR-RT sequence available and 10 319 (20.3%) had an IN sequence available; 6787 persons (13.4%) had PR-RT and IN sequences available. Characteristics of persons included in the analysis are presented in Table 1; most were male (83.0%) and reported male-to-male sexual contact as a transmission risk factor (69.2%). Demographic and risk characteristics for those with and without a sequence were similar overall, with the exception of stage of HIV infection at diagnosis; a larger proportion of new HIV diagnoses without an eligible sequence had an unknown stage of HIV infection at diagnosis (31.7% vs 12.3%). The most common HIV subtype was subtype B (n = 48, 253, 95.1%), followed by subtype C (n = 606, 1.2%); no other subtype had a prevalence greater than 1.0% (data not shown).
Table 1.
Characteristics of Persons With Diagnosed Human Immunodeficiency Virus (HIV) Infection With and Without an Eligible HIV-1 Nucleotide Sequence: 28 US States, 2014–2018
| New HIV Diagnoses With an Eligible Sequence | New HIV Diagnoses Without an Eligible Sequence | |||
|---|---|---|---|---|
| Characteristics | n | % | n | % |
| Total | 50 747 | 100.0 | 92 662 | 100.0 |
| Sex at birth | ||||
| Female | 8636 | 17.0 | 17 840 | 19.3 |
| Male | 42 111 | 83.0 | 74 822 | 80.7 |
| Age at HIV diagnosis | ||||
| 13–29 years | 21 031 | 41.4 | 37 922 | 40.9 |
| 30–49 years | 21 131 | 41.6 | 39 187 | 42.3 |
| ≥50 years | 8585 | 16.9 | 15 553 | 16.8 |
| Race/ethnicity | ||||
| Black/African American | 19 463 | 38.4 | 37 807 | 40.8 |
| Hispanic/Latino | 14 731 | 29.0 | 26 450 | 28.5 |
| White | 13 392 | 26.4 | 22 351 | 24.1 |
| Other | 3161 | 6.2 | 6054 | 6.5 |
| Transmission category | ||||
| Male-to-male sexual contact | 35 111 | 69.2 | 60 895 | 65.7 |
| IDU | 2617 | 5.2 | 5585 | 6.0 |
| Male-to-male sexual contact and IDU | 1793 | 3.5 | 3321 | 3.6 |
| Heterosexual contact | 11 153 | 22.0 | 22 680 | 24.5 |
| Other | 72 | 0.1 | 181 | 0.2 |
| US Census region | ||||
| Northeast | 8189 | 16.1 | 13 226 | 14.3 |
| Midwest | 6134 | 12.1 | 11 575 | 12.5 |
| South | 23 881 | 47.1 | 45 652 | 49.3 |
| West | 12 543 | 24.7 | 22 209 | 24.0 |
| Year of HIV diagnosis | ||||
| 2014 | 9395 | 18.5 | 20 342 | 22.0 |
| 2015 | 9575 | 18.9 | 19 667 | 21.2 |
| 2016 | 10 923 | 21.5 | 18 215 | 19.7 |
| 2017 | 10 892 | 21.5 | 17 115 | 18.5 |
| 2018 | 9962 | 19.6 | 17 323 | 18.7 |
| Stage of HIV infection at diagnosis | ||||
| Stage 1 | 13 202 | 26.0 | 21 838 | 23.6 |
| Stage 2 | 18 234 | 35.9 | 24 057 | 26.0 |
| Stage 3 (AIDS) | 13 059 | 25.7 | 17 378 | 18.8 |
| Stage unknown | 6252 | 12.3 | 29 389 | 31.7 |
Abbreviation: IDU, injection drug use.
Among the 50 747 persons with a PR-RT and/or IN sequence, 9616 (18.9%) had 1 or more TDRM to any drug class. For individual drug classes, TDRM prevalence was 0.8% (INSTI), 4.2% (PI), 6.9% (NRTI), and 12.0% (NNRTI) (Table 2). The most common individual mutations were K103N (8.6%), M41L (1.4%), and T69N (1.2%); all other mutations had a prevalence 1% or less. M184V prevalence was 0.9%. A total of 31 sequences contained K65R (prevalence, 0.1%); for 19 of 31 (61%) of these sequences, M184V/I was also detected. Among 79 total sequences with INSTI TDRMs, E138K was the most common mutation (n = 22); for 18 of 22 (82%) sequences it was the only TDRM and for 20 of 22 (91%) sequences it was the only INSTI TDRM, indicating minimal impact overall on INSTI susceptibility [25] (1 sequence also contained Q148H and G140S; another contained Q148R). A total of 1180 (2.3%) persons had TDRMs to 2 drug classes, 140 (0.3%) had TDRMs to 3 drug classes, and 1 person had TDRMs to all 4 drug classes.
Table 2.
Prevalence of Transmitted Drug Resistance-Associated Mutations by Drug Class: 28 US States, 2014–2018
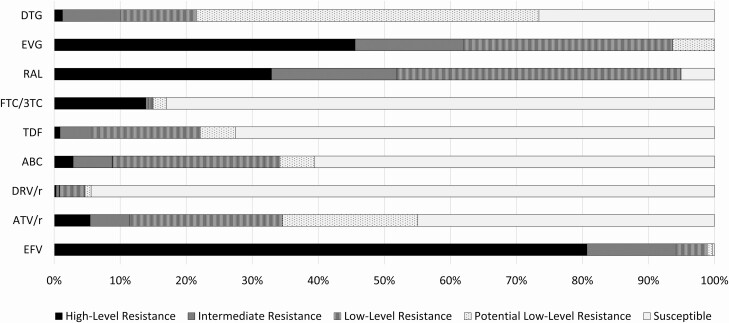
Predicted resistance to common ART drugs among sequences with 1 or more TDRM is shown in Figure 1. The proportion of sequences predicted to maintain susceptibility ranged from 0% (elvitegravir) to 94.4% (darunavir/ritonavir). Predicted high-level or intermediate resistance was uncommon for dolutegravir (8/79 [10.1%]), TDF (174/3258 [5.3%]), emtricitabine/lamivudine (FTC/3TC) (468/3258 [14.4%]), abacavir (ABC) (286/3258 [8.8%]), darunavir/ritonavir (15/1983 [0.8%]), and atazanavir/ritonavir (227/1983 [11.4%]) but much more common for raltegravir (41/79 [51.9%]), elvitegravir (49/79 [62.0%]), and efavirenz (5327/5662 [94.1%]).
Figure 1.
Predicted resistance to commonly used antiretroviral drugs among HIV-1 nucleotide sequences with ≥1 transmitted drug-resistance–associated mutation: 28 US states, 2014–2018. Resistance was predicted by HIVdb genotypic resistance interpretation system, version 8.2. Abbreviations: ABC, abacavir; ATV/r, atazanavir/ritonavir; DRV/r, darunavir/ritonavir; DTG, dolutegravir; EFV, efavirenz; EVG, elvitegravir; FTC/3TC, emtricitabine/lamivudine; RAL, raltegravir; TDF, tenofovir.
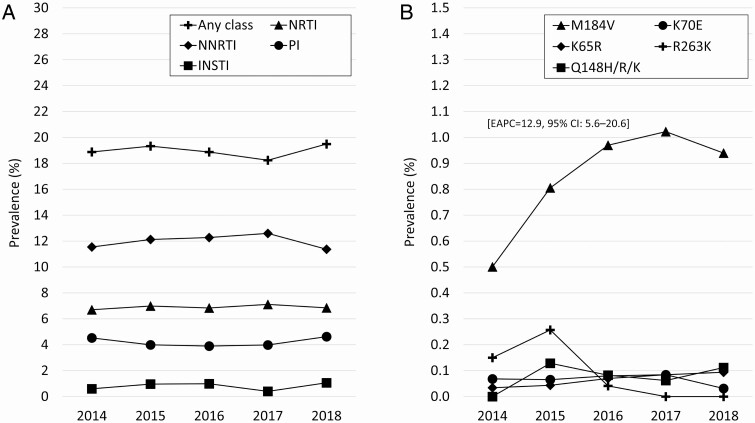
During 2014–2018, TDRM prevalence did not change significantly overall or for individual drug classes (Figure 2A). Among mutations with notable effect on first-line NRTI or INSTI and present in more than 10 sequences in the analysis, only M184V prevalence increased during the time period (estimated 12.9% increase per year; 95% confidence interval, 5.6–20.6), from 0.5% in 2014 to 0.9% in 2018 (Figure 2B).
Figure 2.
Transmitted drug-resistance–associated mutation prevalence by year for (A) individual drug classes and (B) key NRTI and INSTI mutations: 28 US states, 2014–2018. Key mutations include those with frequency N >10 in the analysis, and which substantially decrease susceptibility to first-line NRTIs (M184V, K65R, K70E) or INSTIs (Q148H/R/K, R263K). Abbreviations: CI, confidence interval; EAPC, estimated annual percent change; INSTI, integrase strand transfer inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
DISCUSSION
In this study, we report transmitted HIV-1 drug resistance among more than 50 000 persons with diagnosed HIV, the largest US sample to date. We include data from 28 states where HIV nucleotide sequences from clinical drug-resistance testing were routinely reported during 2014–2018; these 28 states account for more than 70% of all HIV diagnoses in the United States each year [26]. We report for the first time the prevalence of transmitted HIV-1 drug resistance to INSTIs in a large US sample; the more than 10 000 integrase sequences in this analysis were performed during a period of expanding INSTI use in clinical care.
Overall TDRM prevalence in this analysis was 18.9%, the highest prevalence reported to date in a large US study. Direct comparison to prior studies is complicated by differences in surveillance mutation lists used and by our inclusion of INSTI TDRMs. Our analysis used a similar method and mutation list as a prior CDC analysis of 2006 data [9], but with the addition of INSTI mutations that were a small contributor to overall TDRM prevalence. Compared with this previous study, our results suggest that US TDRM prevalence did increase between 2006 and 2014 overall (14.6% to 18.9%), for NNRTI (7.8% to 11.5%), and for NRTI (5.6% to 6.7%), with no change for PI (4.5%) [9]. However, from 2014 to 2018, TDRM prevalence did not increase or decrease overall or for individual drug classes. The low proportion of sequences with TDRMs to multiple drug classes was similar to prior US studies [7, 27] and indicates that an active ART regimen could be constructed for nearly all persons in the analysis.
Transmitted NRTI resistance in this analysis was similar to previous US studies [8, 9, 27] overall and in the prominence of TAMs, which comprised 7 of the 10 most common NRTI mutations (ie, M41L, T215S, T215D, D67N, K219Q, L210W, E44D). These mutations, selected by zidovudine (AZT) and stavudine (d4T), emerged in the 1990s and continue to be transmitted at stable levels despite infrequent use of these drugs in the current era; TAMs have minimal effects on current first-line NRTIs unless combined with other mutations.
Among non-TAM NRTI mutations, M184V, which reduces 3TC and FTC susceptibility, was most common (0.9%), and increased modestly during 2014–2018. However, M184V prevalence overall and by year remains similar to prior analyses [8, 9]. K65R, which significantly reduces susceptibility to tenofovir disoproxil fumarate (TDF) [28], tenofovir alafenamide (TAF) [29], FTC/3TC, and ABC [30], was detected infrequently in this analysis, similar to results from prior studies [8, 9]. Even fewer sequences contained both K65R and M184V/I, a mutation combination noted in prior PrEP failures. This observed prevalence likely underestimates true transmitted K65R and M184V prevalence, as both mutations decay within weeks or months in the absence of drug pressure [31–34] but can persist in minority variant strains not detected by conventional bulk sequencing [35]. Although NHSS data do not capture time since HIV infection, the large proportion of persons with stage 2 or stage 3 infection at diagnosis (Table 1) indicates a majority of people in this study had longstanding HIV infection at the time of diagnosis with ample time for the decay of transmitted resistance. Regardless, continued low prevalence of transmitted M184V and K65R is reassuring and reflects relatively low transmission fitness of these strains in addition to fast reversion dynamics. As PrEP use continues to expand, population-level monitoring of M184V and K65R can provide important insight into the potential impact of drug resistance on future PrEP efficacy.
We report low overall TDRM prevalence for INSTIs with no significant increase or decrease in prevalence during 2014–2018. Most individual INSTI mutations were detected in fewer than 10 sequences; however, high-level or intermediate resistance to the first-generation INSTIs, raltegravir and elvitegravir, was predicted for over half of the 79 sequences with 1 or more INSTI TDRM, which likely reflects their relatively low barrier to resistance and longer period of widespread use. Predicted high-level or intermediate resistance was much less common for dolutegravir. Low INSTI TDRM prevalence throughout 2014–2018 is reassuring, although conclusions about future transmitted INSTI resistance might be premature given that second-generation INSTIs have been widely used for fewer than 5 years and continued expansion of INSTI use is likely for ART and potentially for PrEP.
Transmitted NNRTI resistance prevalence (12.0%) was higher in this study than in prior large US studies [7–9], driven primarily by K103N (8.6%). K103N alone can result in failure of efavirenz-based ART and is responsible for most of the predicted high-level efavirenz resistance in this analysis. Although INSTI-based regimens have largely replaced efavirenz-based ART in the United States, K103N prevalence did not decrease during 2014–2018, reflecting longer intra-host persistence, minimal fitness cost, and more frequent transmission clustering of K103N strains compared with mutations like M184V and K65R [31, 36].
Protease inhibitor TDRM prevalence has changed very little in the past 15 years [6–9], and no significant increase or decrease was seen during 2014–2018. Predicted low, intermediate, and high-level resistance was more common for atazanavir/ritonavir (ATV/r) than for darunavir/r (DRV/r) due primarily to M46I/L, I54V, V82A, and L90M, which have minimal effect on DRV/r. This provides additional support for current ART recommendations favoring DRV/r when a regimen including a PI is needed [3].
This analysis was subject to several limitations. First, this study only includes persons with diagnosed HIV infection with a drug-resistance test performed 3 months or less after HIV diagnosis and reported to public health, and might not reflect prevalence among persons with undiagnosed HIV or those with diagnosed HIV without a drug-resistance test performed or reported. Failure to disclose or report ART use prior to a drug-resistance test could also lead to misclassification of acquired drug resistance. Second, because genotypic resistance testing of the integrase gene is recommended only when INSTI resistance is a concern, it is possible that INSTI TDRM prevalence in this analysis overestimates the true prevalence among all persons with HIV. Third, TDRM prevalence in this analysis is likely underestimated due to reversion of drug-resistant virus to wild-type, which can occur within weeks or months of HIV infection and varies by mutation; most persons in this analysis had longstanding infection at the time of diagnosis. Finally, HIV-1 nucleotide sequence data in this analysis were generated by conventional bulk sequencing that does not characterize minority variants and might underestimate relevant TDRM prevalence.
We document a reservoir of TAMs and TDRMs to PIs and NNRTIs, which continue to be transmitted despite reduced use or discontinuation of the drugs that select for them. This persistent transmission highlights the role that undiagnosed infection and untreated persons play in HIV transmission [36–38], and underscores the importance of early diagnosis and viral suppression in reducing transmitted drug resistance. Efforts to improve diagnosis and treatment in the EHE initiative will be essential for shrinking this reservoir and preventing further transmission of drug resistance. HIV sequence data reported to public health provide valuable insight into trends in population-level drug resistance resulting from the ever-evolving landscape of clinical guidelines, prescribing practices, and public health initiatives to improve testing and treatment programs.
Standard genotypic drug-resistance testing is recommended at entry into care for all persons with HIV in the United States to assist with selection of an initial ART regimen. In a recent modeling analysis, Hyle et al [39] found such testing offered limited clinical benefit and was not cost-effective for people with HIV starting an INSTI-based regimen. These important findings must be balanced by the population-level benefit of drug-resistance monitoring, the meaningful benefit to vulnerable individuals whose resistance profile is consequential, and the miniscule cost of resistance testing relative to other costs associated with HIV treatment. Population-level monitoring of drug resistance also provides public health benefit through the identification of HIV transmission clusters and subsequent efforts to interrupt transmission and improve service delivery to people with HIV or at risk for HIV.
In conclusion, our data confirm transmitted resistance to current first-line ART remains low and support current ART and PrEP recommendations. Drug susceptibility scoring in this analysis indicates that, among sequences with 1 or more TDRM, only a subset is predicted to have intermediate or high-level resistance in practice. Amidst evolving trends in the use of these drugs for ART and PrEP, population-level monitoring remains essential for informing current and future treatment and prevention guidelines.
Notes
Acknowledgments. The authors thank state and local health departments responsible for collecting and reporting HIV surveillance data to the Centers for Disease Control and Prevention (CDC).
Disclaimer. The findings and conclusions in this study are those of the authors and do not necessarily represent the views of the CDC.
Financial support. This work was supported by the Centers for Disease Control and Prevention.
Potential conflicts of interest. W. H. reports receiving royalties on licenses of patents on pre-exposure prophylaxis for HIV prevention and is listed as an inventor on patents by the US government on methods for HIV drug-resistance detection and patents on methods for HIV prevention by chemoprophylaxis, outside the submitted work. J. A. J. and W. H. are named on patents for sensitive assays for HIV-1 drug resistance. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
R Paul McClung, US Public Health Service Commissioned Corps, Atlanta, Georgia, USA; Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Alexandra M Oster, US Public Health Service Commissioned Corps, Atlanta, Georgia, USA; Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
M Cheryl Bañez Ocfemia, Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Neeraja Saduvala, ICF International, Atlanta, Georgia, USA.
Walid Heneine, Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Jeffrey A Johnson, Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Angela L Hernandez, Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
References
- 1. Fauci AS, Redfield RR, Sigounas G, Weahkee MD, Giroir BP. Ending the HIV Epidemic: a plan for the United States. JAMA 2019; 321:844–5. [DOI] [PubMed] [Google Scholar]
- 2. Wittkop L, Günthard HF, de Wolf F, et al. ; EuroCoord-CHAIN Study Group . Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis 2011; 11:363–71. [DOI] [PubMed] [Google Scholar]
- 3. Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in adults and adolescents with HIV. Department of Health and Human Services. Available at: http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. Accessed 23 January 2020.
- 4. Gunthard HF, Calvez V, Paredes R, et al. Human immunodeficiency virus drug resistance: 2018 recommendations of the international antiviral society-USA panel. Clin Infect Dis 2019; 68:177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saag MS, Gandhi RT, Hoy JF, et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2020 recommendations of the International Antiviral Society-USA panel. JAMA 2020; 324:1651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Margot NA, Wong P, Kulkarni R, et al. Commonly transmitted HIV-1 drug resistance mutations in reverse-transcriptase and protease in antiretroviral treatment-naive patients and response to regimens containing tenofovir disoproxil fumarate or tenofovir alafenamide. J Infect Dis 2017; 215:920–7. [DOI] [PubMed] [Google Scholar]
- 7. Ross LL, Shortino D, Shaefer MS. Changes from 2000 to 2009 in the prevalence of HIV-1 containing drug resistance-associated mutations from antiretroviral therapy-naive, HIV-1-infected patients in the United States. AIDS Res Hum Retroviruses 2018; 34:672–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchacz K, Young B, Palella FJ Jr., Armon C, Brooks JT. Trends in use of genotypic resistance testing and frequency of major drug resistance among antiretroviral-naive persons in the HIV Outpatient Study, 1999–2011. J Antimicrob Chemother 2015; 70:2337–46. [DOI] [PubMed] [Google Scholar]
- 9. Wheeler WH, Ziebell RA, Zabina H, et al. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. AIDS 2010; 24:1203–12. [DOI] [PubMed] [Google Scholar]
- 10. De Francesco MA, Izzo I, Properzi M, et al. Prevalence of integrase strand transfer inhibitors resistance mutations in integrase strand transfer inhibitors-naive and -experienced HIV-1 infected patients: a single center experience. AIDS Res Hum Retroviruses 2018; 34:570–4. [DOI] [PubMed] [Google Scholar]
- 11. Menza TW, Billock R, Samoff E, Eron JJ, Dennis AM. Pretreatment integrase strand transfer inhibitor resistance in North Carolina from 2010–2016. AIDS 2017; 31: 2235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen I, Zhang Y, Cummings V, et al. Analysis of HIV integrase resistance in black men who have sex with men in the United States. AIDS Res Hum Retroviruses 2017; 33:745–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aldous AM, Castel AD, Parenti DM. Prevalence and trends in transmitted and acquired antiretroviral drug resistance, Washington, DC, 1999–2014. BMC Res Notes 2017; 10:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Knox DC, Anderson PL, Harrigan PR, Tan DH. Multidrug-resistant HIV-1 infection despite preexposure prophylaxis. N Engl J Med 2017; 376:501–2. [DOI] [PubMed] [Google Scholar]
- 15. Markowitz M, Grossman H, Anderson PL, et al. Newly acquired infection with multidrug-resistant HIV-1 in a patient adherent to preexposure prophylaxis. J Acquir Immune Defic Syndr (1999) 2017; 76:e104–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thaden JT, Gandhi M, Okochi H, Hurt CB, McKellar MS. Seroconversion on preexposure prophylaxis: a case report with segmental hair analysis for timed adherence determination. AIDS 2018; 32:F1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cohen SE, Sachdev D, Lee SA, et al. Acquisition of tenofovir-susceptible, emtricitabine-resistant HIV despite high adherence to daily pre-exposure prophylaxis: a case report. Lancet HIV 2018: S2352-3018(18)30288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Colby DJ, Kroon E, Sacdalan C, et al. Acquisition of multidrug-resistant human immunodeficiency virus type 1 infection in a patient taking preexposure prophylaxis. Clin Infect Dis 2018; 67:962–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoshinaga T, Kobayashi M, Seki T, et al. Antiviral characteristics of GSK1265744, an HIV integrase inhibitor dosed orally or by long-acting injection. Antimicrob Agents Chemother 2015; 59:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Radzio-Basu J, Council O, Cong ME, et al. Drug resistance emergence in macaques administered cabotegravir long-acting for pre-exposure prophylaxis during acute SHIV infection. Nat Commun 2019; 10:2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014; 42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tzou PL, Rhee SY, Descamps D, et al. ; WHO HIVResNet Working Groups . Integrase strand transfer inhibitor (INSTI)-resistance mutations for the surveillance of transmitted HIV-1 drug resistance. J Antimicrob Chemother 2020; 75:170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shafer RW, Jung DR, Betts BJ. Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nat Med 2000; 6:1290–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 2006; 42:1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McColl DJ, Chen X. Strand transfer inhibitors of HIV-1 integrase: bringing IN a new era of antiretroviral therapy. Antiviral Res 2010; 85:101–18. [DOI] [PubMed] [Google Scholar]
- 26. Centers for Disease Control and Prevention. HIV surveillance report, 2018 (updated). Vol. 31. Available at: http://www.cdc.gov/hiv/library/reports/hiv-surveillance.html. Accessed 6 July 2020.
- 27. Rhee SY, Clutter D, Fessel WJ, et al. Trends in the molecular epidemiology and genetic mechanisms of transmitted human immunodeficiency virus type 1 drug resistance in a large US clinic population. Clin Infect Dis 2019; 68:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Squires K, Pozniak AL, Pierone G Jr, et al. ; Study 907 Team . Tenofovir disoproxil fumarate in nucleoside-resistant HIV-1 infection: a randomized trial. Ann Intern Med 2003; 139:313–20. [DOI] [PubMed] [Google Scholar]
- 29. Margot NA, Johnson A, Miller MD, Callebaut C. Characterization of HIV-1 resistance to tenofovir alafenamide in vitro. Antimicrob Agents Chemother 2015; 59:5917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trotta MP, Bonfigli S, Ceccherini-Silberstein F, et al. Clinical and genotypic correlates of mutation K65R in HIV-infected patients failing regimens not including tenofovir. J Med Virol 2006; 78:535–41. [DOI] [PubMed] [Google Scholar]
- 31. Jain V, Sucupira MC, Bacchetti P, et al. Differential persistence of transmitted HIV-1 drug resistance mutation classes. J Infect Dis 2011; 203:1174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Castro H, Pillay D, Cane P, et al. ; UK Collaborative Group on HIV Drug Resistance . Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis 2013; 208:1459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brodard V, Moret H, Beguinot I, et al. Prevalence of detection and dynamics of selection and reversion of K65R mutation in nucleoside reverse transcriptase inhibitor-experienced patients failing an antiretroviral regimen. J Acquir Immune Defic Syndr (1999) 2005; 39:250–3. [PubMed] [Google Scholar]
- 34. Weis JF, Baeten JM, McCoy CO, et al. ; Partners PrEP Study Team . Preexposure prophylaxis-selected drug resistance decays rapidly after drug cessation. AIDS 2016; 30:31–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li JF, Linley L, Kline R, Ziebell R, Heneine W, Johnson JA. Sensitive sentinel mutation screening reveals differential underestimation of transmitted HIV drug resistance among demographic groups. AIDS 2016; 30:1439–45. [DOI] [PubMed] [Google Scholar]
- 36. Wertheim JO, Oster AM, Johnson JA, et al. Transmission fitness of drug-resistant HIV revealed in a surveillance system transmission network. Virus Evol 2017; 3:vex008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drescher SM, von Wyl V, Yang WL, et al. ; Swiss HIV Cohort Study . Treatment-naive individuals are the major source of transmitted HIV-1 drug resistance in men who have sex with men in the Swiss HIV Cohort Study. Clin Infect Dis 2014; 58:285–94. [DOI] [PubMed] [Google Scholar]
- 38. Mourad R, Chevennet F, Dunn DT, et al. ; UK HIV Drug Resistance Database & the Collaborative HIV; Anti-HIV Drug Resistance Network . A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK. AIDS 2015; 29:1917–25. [DOI] [PubMed] [Google Scholar]
- 39. Hyle EP, Scott JA, Sax PE, et al. Clinical impact and cost-effectiveness of genotype testing at human immunodeficiency virus diagnosis in the United States. Clin Infect Dis 2020; 70:1353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]