Abstract
The Actinidia (kiwifruit) is an emerging fruit plant that is severely affected by salt stress in northern China. Plants have evolved several signaling network mechanisms to cope with the detrimental effects of salt stress. To date, no reported work is available on metabolic and molecular mechanisms involved in kiwifruit salt tolerance. Therefore, the present study aims to decipher intricate adaptive responses of two contrasting salt tolerance kiwifruit species Actinidia valvata [ZMH (an important genotype), hereafter referred to as R] and Actinidia deliciosa [‘Hayward’ (an important green-fleshed cultivar), hereafter referred to as H] under 0.4% (w/w) salt stress for time courses of 0, 12, 24, and 72 hours (hereafter refered to as h) by combined transcriptome and metabolome analysis. Data revealed that kiwifruit displayed specific enrichment of differentially expressed genes (DEGs) under salt stress. Interestingly, roots of R plants showed a differential expression pattern for up-regulated genes. The KEGG pathway analysis revealed the enrichment of DEGs related to plant hormone signal transduction, glycine metabolism, serine and threonine metabolism, glutathione metabolism, and pyruvate metabolism in the roots of R under salt stress. The WGCNA resulted in the identification of five candidate genes related to glycine betaine (GB), pyruvate, total soluble sugars (TSS), and glutathione biosynthesis in kiwifruit. An integrated study of transcriptome and metabolome identified several genes encoding metabolites involved in pyruvate metabolism. Furthermore, several genes encoding transcription factors were mainly induced in R under salt stress. Functional validation results for overexpression of a candidate gene betaine aldehyde dehydrogenase (AvBADH, R_transcript_80484) from R showed significantly improved salt tolerance in Arabidopsis thaliana (hereafter referred to as At) and Actinidia chinensis [‘Hongyang’ (an important red-fleshed cultivar), hereafter referred to as Ac] transgenic plants than in WT plants. All in all, salt stress tolerance in kiwifruit roots is an intricate regulatory mechanism that consists of several genes encoding specific metabolites.
Introduction
Fruit plants play a promising role in human lives due to their nutritional benefits for health. Abiotic stresses cause a massive loss in fruit crop production by restricting their growth and development. Most of the fruit plants, due to varying ability of salt tolerance, are severely damaged by salt stress due to being perennials. To withstand the damaging effects of salt stress, plants have evolved several tolerance mechanisms. Many genes have been induced or suppressed in fruit plants under salt stress [1–3]. The decreased cultivable area due to rising sea level, decrease in groundwater recharge, traditional irrigation methods, and increased evapotranspiration is a major concern worldwide [4]. Therefore, it is highly prudent to identify metabolic pathways and salt stress-responsive genes to improve fruit plant genetic resources that can thrive well in salt-affected soils.
Plant survival in adverse environmental conditions is based on integrating stress-adaptive metabolic and structural changes in endogenous developmental programs [5]. The complex processes of regulation of salt tolerance in plants and salt adaptation involve the control of water flux and osmotic adjustments at the cellular level through osmoprotectant biosynthesis [6]. Whereas ionic stress caused by salt copes with controlled influx and efflux of ions at the plasma membrane and sequestering of the ions in the vacuole [5]. Similarly, the imbalanced redox homeostasis and energy supply in plants are regulated by reprogramming the metabolism and impaired cellular structure [7]. Various functional genes and regulatory genes control these regulatory events. Therefore, it is imperative to understand the role of metabolic pathways and complex defense-related signaling networks involved in salt stress tolerance in plants.
Generally, the salt tolerance in plants is associated with the accumulation of specific metabolites [8] and the number of genes involved in the biosynthesis of these metabolites [9, 10]. Some of these genes are BADH [11], NHX1 [12], AISP [13], P5CS [14], CCD1 [15], WRKY8 [16], NAC06 [17], ERF11 [18], and MADS25 [19]. Similarly, some important primary and secondary metabolites for salt stress tolerance are sugars, polyols, amino acids [20, 21], nitrogen-containing compounds, terpenes, and phenolic compounds [22]. Recently, high-throughput omics technologies, i.e. transcriptome and metabolome, have been developed, which are critical in identifying the regulatory networks and metabolic reorganizations for controlling salinity tolerance in plants [23]. To date, several integrated studies of transcriptome and metabolome have been reported in some plants against salt stress [23–26].
The Actinidia, commonly known as kiwifruit, is a recently adopted fruit plant gaining popularity among customers due to its unique flavor and nutritional benefits. It is comprised of 54 species and 21 varieties, a total of 75 taxa [27, 28]. However, most of the available commercial kiwifruit cultivars are salt-sensitive. Additionally, there is no reported work available on transcriptome and metabolome analysis in kiwifruit for salt stress so far. Previously, we screened different kiwifruit genotypes to assess their tolerance ability against salt stress [29, 30]. Based on previous results, the compoarative analysis of transcriptome and metabolome was conducted to decipher the intricate metabolic pathways and their regulatory gene network involved in conferring salt tolerance in kiwifruit. The data obtained from the present study will facilitate researchers to identify salt stress-responsive networks in kiwifruit. Additionally, it will deepen the understanding of the salt resistance mechanism in fruit trees and accelerate the cultivation of salt-tolerant varieties in kiwifruit.
Results
Performance of R and H for salt stress tolerance
In our previous study, we reported the varying tolerance ability of R and H against salt stress [29]. In the present study, we put efforts to assess the salt stress tolerance of R and H at different time courses. Plants were treated with 0.4% NaCl (w/w) for 12 h, 24 h, and 72 h by keeping 0 h (untreated) plants as control. The leaves of the H plants showed noticeable wilting and necrosis compared with R plants (Fig. 1Ai). Similarly, the roots of the H plants turned into brownish color under salt stress while no visible evidence for salt stress damage was found in R roots (Fig. 1Aii).
Figure 1.
Morphological changes of R and H, an overview of RNA-seq data, and validation of DEGs expression pattern in R and H under 0.4% NaCl stress. (A) Morphological changes of R and H in (i) leaves and (ii) roots. (B) Overview of RNA-seq data (i) expression-dependent heatmap of samples obtained from R and H by using data from spearman correlation coefficient analysis. The x-axis and y-axis represents samples taken from Av and H under salt stress, (ii) statistics of commonly up-regulated genes in R and H, (iii) statistics of up-regulated and down-regulated DEGs in roots, and (iv) validation of RNA-seq data by RT-qPCR analysis. (C) GO terms and KEGG enrichment analysis of up-regulated genes in R4 vs H4. (i) GO terms enrichment analysis. The x-axis represents enriched GO terms and y-axis represents the number of up-regulated genes in a particular GO term. The red columns are for GO terms under biological process, the Green color represents GO terms under cellular components and the blue color stands for GO terms in molecular function. (ii) KEGG enrichment analysis. Vertical columns represent enriched pathways and horizontal columns are for annotated genes. R, Actinidia valvata; H, A. deliciosa; h, hours.
Overview of transcriptome profiling and validation of RNA-seq data by qPCR
A global transcriptional profile of roots sampled from R and H treated with or without salt stress was carried out to thoroughly examine the intricate mechanisms involved in enhanced salt stress tolerance. For convenience, R and H were used to represent ZMH from Actinidia valvata and ‘HWD’ from A. deliciosa, while 1, 2, 3, and 4 represent 0 h, 12 h, 24 h, and 72 h, respectively. The statistics for PacBio and Illumine Hi-seq transcriptome sequencing are presented in the online supplementary material (Table S1). The hierarchical clustering of 24 samples by using Spearman correlation coefficient analysis data showed that all samples from H and R were clustered in clade one and clad two, respectively (Fig. 1Bi). It signifies that the gene expression profile for samples was consistent. A pairwise comparison method was adopted to identify DEGs responsive to salt stress. We identified 100 004 DEGs for R1 vs H1, 111 907 DEGs for R2 vs H2, 106 669 DEGs for R3 vs H3, and 108 617 DEGs for R4 vs H4 (Fig. 1Bii, iii, and Fig. S1, see online supplementary material). Based on the phenotype results (Fig. 1A) and the total number of common DEGs (Fig. 1B, ii, iii, and Fig. S1, see online supplementary material), we selected the R4 vs H4 comparison for further analysis. We did RNA-seq data validation by checking the relative expression of 16 randomly selected genes with RT-qPCR (Fig. S2, see online supplementary material). The detail for the pair of primers used for RT-qPCR are given in the supplementary section (Table S2, see online supplementary material). The scatter plot of 16 genes from RT-qPCR and RNA-seq data showed a linear trend (Fig. 1Biv). The higher correlation (R2 = 0.8449) between RT-qPCR and RNA-seq results implied that our RNA-seq data is reliable for further assessment.
Differentially expressed salt stress-responsive genes
To find the salt stress-responsive DEGs for kiwifruit, the GO enrichment analysis and KEGG analysis for DEGs in R4 vs H4 were carried out. The GO enrichment analysis provided additional information about the key processes and pathways that distinguished the response of R and H against salt stress. This analysis resulted in identification of 126 422 GO terms in which 78 750 were statistically significant under biological process, cellular process, and molecular function. The most prominent GO terms that were commonly found in roots included metabolic process, cellular processing, cell, cell parts, catalytic activity, binding, response to stimulus, metabolic process, signaling, detoxification, signal transducer activity, antioxidant activity, etc. (Fig. 1Ci).
According to KEGG analysis, 10 out of the top 50 pathways were specifically identified based on their relevance to salt stress tolerance. Among these 10 pathways, most numbers of annotated DEGs were linked to protein processing (1817, 6.56%), biosynthesis of amino acids (1620, 5.87%), plant hormone signal transduction (1607, 5.82%), starch and sucrose metabolism (101, 3.67%), pyruvate metabolism (711, 2.58%), oxidative phosphorylation (590, 2.14%), glutathione metabolism (533, 1.93%), phenylpropanoid biosynthesis (523, 1.90%), glycine, serine, and threonine metabolism (409, 1.48%), and arginine and proline metabolism (323, 1.17%) (Fig. 1Cii).
Important pathways conferring salt stress tolerance
Salt stress can lead to over-accumulation of various metabolites in plants. The salt tolerance ability of plants is typically assessed by the maintenance of proper levels of metabolic processes and defense responses. We carried out comprehensive study for important metabolic KEGG pathways to find out the reason behind the elevated salt tolerance ability of the R plants. We selected glutathione metabolism pathway [31–33], pyruvate metabolism pathway [34, 35], glycine betaine metabolism pathway [36–38], ABA signaling pathway [39, 40], and MYB transcription factors [41, 42] due to their important role in conferring salt tolerance in plants that is extensively reported by researchers in the past (Fig. 1Cii).
ABA signaling was altered in R under salt stress
In the plant hormone signal transduction pathway, a total of 1607 annotated genes were identified among which 833 were up-regulated DEGs. These up-regulated genes belong to Auxin (176 genes), ABA (112 genes), CTK (113 genes), BR (110 genes), Ethylene (106 genes), GA (39 genes), JA (77 genes), and SA (100 genes) (Fig. S3, see online supplementary material). As the role of ABA in salt stress is well reported previously, we did a comprehensive analysis of ABA signaling in kiwifruit. The expression pattern of the top 20 DEGs for both species under 0.4% salt stress is presented in the heatmap (Fig. 2Ai). The R_transcript_19927 (Abscisic acid receptor PYL9) was found to play role in the tolerance mechanism as its expression was significantly higher in R compared to H. After performing RT-qPCR, we obtained a similar expression pattern for R_transcript_19927 as in RNA-seq data (Fig. 2Aii). As additional evidence, we measured the ABA contents in kiwifruit. Surprisingly, the trend for change in contents was identical to gene expression results. The ABA contents in R significantly increased over time, while no significant change was found in H (Fig. 2Aiii).
Figure 2.
Important pathways conferring salt stress tolerance in kiwifruit. (A) Analysis of the ABA pathway. (i) Heatmap for top 20 up-regulated DEGs. (ii) The expression profile of identified candidate gene R_transcript_19927 (Abscisic acid receptor PYL9). (iii) Contents of abscisic acid (ABA) hormone. (B) Analysis of glycine betaine (GB) pathway. (i) Heatmap of top 20 up-regulated DEGs. (ii) The expression profile analysis of candidate gene R_transcript_80984 (Betaine Aldehyde dehydrogenase). (iii) Contents of glycine betaine (GB). (C) Analysis of glutathione metabolism pathway. (i) Heatmap for top 20 up-regulated DEGs. (ii) The expression profile for candidate gene R_Transcript_55242 (Glutamate cysteine ligase). (iii) Contents of glutathione. (D) Analysis of pyruvate biosynthesis pathway. (i) Heatmap for 20 up-regulated DEGs. (ii) The expression profile for candidate gene R_transcript_92720 (Aldehyde dehydrogenase). (iii) Contents of pyruvate. (E) Analysis of transcription factors (TFs) families. (i) Heatmap for top 20 MYB family genes. (ii) The expression profile of candidate gene R_transcript_ 18 883 (MYB transcription factor). (iii) the Total number of transcription factors (TF) in each annotated family. (iv) Transcriptional activation activity analysis for MYB1 (R_transcript_18876) and MYB2 (R_transcript_18 883) TFs. Each value represents the mean ± SD of three biological replicates. Full names for abbreviations are presented in the online supplementary material (Table S3).
The R showed enhanced accumulation of glycine betaine than H
For the GB pathway, a total of 246 up-regulated DEGs were identified in R4 vs H4 comparison. According to the heatmap of FPKM values, we found that the expression pattern of R_transcript_45466 and R_transcript_80984 was significantly different in R than in H under salt stress (Fig. 2Bi). Both transcripts are involved in the biosynthesis of GB and annotated as BADH (betaine aldehyde dehydrogenase). We validated the gene expression results of R_transcript_80984 (betaine Aldehyde dehydrogenase) from RNA-seq by RT-qPCR and found both results consistent (Fig. 2Bii). It indicated that R possessed a different salt-induced GB pathway than H. To confirm our assumption, we measured GB contents in both species under the same salt stress. The results showed that R accumulated noticeably higher GB than H (Fig. 2Biii).
Glutathione conferred salt stress tolerance in R plants
Elevated salt stress conditions lead to an increase in the accumulation of ROS in plants. The KEGG analysis identified glutathione metabolism among the top 50 differentially expressed KEGG pathways (Fig 4). The expression profile for the top 20 DEGs in both species under salt stress is presented in the heatmap (Fig. 2Ci). Many transcripts were differentially expressed between R and H plants. The expression pattern for R_transcript_55242 (Glutamate cysteine ligase) was analysed based on FPKM values and validated by RT-qPCR (Fig. 2Cii). These results provided clues about the involvement of glutathione in the enhanced tolerance ability of R plants. We measured glutathione contents in both species under salt stress as evidence to prove our assumption. The R showed a significantly enhanced accumulation of glutathione compared with H (Fig. 2Ciii).
Figure 4.
Overview for metabolome data and integrated metabolome and transcriptome analysis for R and H under salt stress. (A) the biplot of PC1 and PC2 for up-regulated DEMs. (B) Heatmap for hierarchical clustering of 12 samples from R and H. (C) Volcano plot representing the regulation pattern of DEMs. (D) Statistics of up-regulated and down-regulated DEMs in roots. (E) The KEGG enrichment analysis of up-regulated DEMs in R4 vs H4. Vertical columns represent enriched pathways and horizontal columns are for annotated metabolites. (F) Integration of transcriptome and metabolome data. Green color represents a particular metabolite, orange color represents the enzyme catalyzing a particular reaction, and yellow color stands for the production or consumption of energy in a chemical reaction. ALDH, aldehyde dehydrogenase; CoA, coenzyme A; DEM, differentially expressed metabolites; H, A. deliciosa; LDH, L-lactate dehydrogenase; PC, principal component; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase; PK, pyruvate kinase; R, Actinidia valvata.
The R accumulated more pyruvate than H
Pyruvate metabolism provides energy backup for plants to efficiently cope with the catastrophic effects of salt stress. In our analysis, pyruvate fell among the top 50 differentially expressed KEGG pathways (Fig. 4). There were in total 357 up-regulated DEGs in this pathway. Expression profiles of the top 20 DEGs were clearly different in R and H under salt stress (Fig. 2Di). The expression profile for R_transcrip_92720 (Aldehyde dehydrogenase) was analysed and validated by FPKM and RT-qPCR, respectively (Fig. 2Dii). By measuring the contents of pyruvate, we found that R accumulated a significantly higher amount of pyruvate compared with H (Fig. 2Diii).
Transcription factors were differentially expressed in R and H under salt stress
Transcription factors are responsible for the regulation of downstream genes, and they function in multiple biological processes. In the case of our study, salt stress induced many TFs with differential expression patterns between R and H. We found 20 TF families that were prominently expressed in kiwifruit under salt stress (Fig. 2Eiii). The MYB family was deliberately picked based on the abundance of gene numbers. The expression pattern of the top 20 DEGs showed a clear difference between R and H (Fig. 2Ei). Furthermore, the expression pattern of candidate gene R_transcript_ 18 883 (MYB transcription factor) by FPKM analysis and qPCR showed that it was specifically expressed in R under salt stress (Fig. 2Eii). The results from transcriptional activation activity showed that AvMYB2 (R_transcript-18 883) had ability to activate expression of downstream reporter gene (Fig. 2Eiv).
Identification of regulatory networks and hub genes
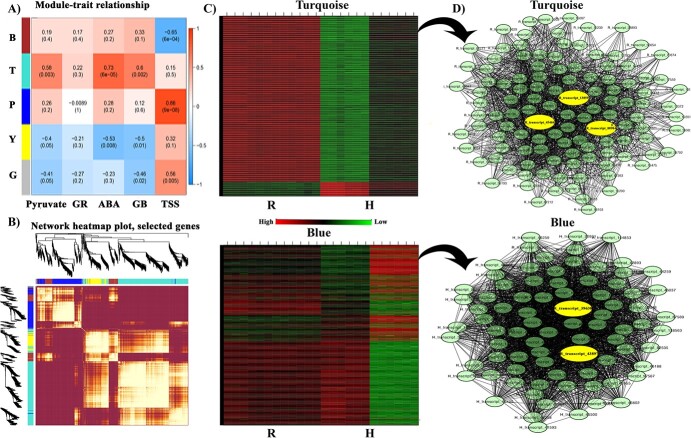
The WGCNA analysis was implied on transcriptome data which identified five distinct color modules based on the co-expression pattern of individual genes presented in the network heatmap plot (Fig. 3B). The clustered dendrogram of DEGs from R and H is presented in the online supplementary material (Fig. S4). The ABA, TSS, glutathione, pyruvate, and GB contents in R and H under salt stress were used as phenotypic input data to carry out module-trait relationship analysis and results are presented in the module-trait relationship plot (Fig. 3A). Out of five color modules, the blue module showed a significant correlation with TSS, while the turquoise model was significantly correlated with pyruvate, GB, and ABA. The turquoise module contained 1057 genes that showed a significant association with pyruvate (R2 = 0.58), ABA (R2 = 0.73), and GB (R2 = 0.6). Similarly, the blue module comprised 354 genes that showed a significant relationship with TSS and GR (R2 = 0.86). The expression profile for all the genes used in turquoise and blue color modules is presented in heatmaps (Fig. 3C). The genes from both modules were selected for their edges and nodes calculation which were later used for gene-network visualization purpose. This analysis resulted in the identification of two genes annotated as betaine aldehyde dehydrogenase (R_transcript_45466 and R_transcript_80984) and one gene annotated as pyruvate kinase (R_transcript_13859) in the turquoise color module (Fig. 3D, upper panel). Similarly, the blue module resulted in the identification of hub genes related to TSS annotated as fructokinase (H_transcript_39638) and another gene related to glutathione annotated as glutathione-s-transferase (H_transcript_43897) (Fig. 3D, lower panel). The changes in contents of TSS are presented in the online supplementary material) (Fig. S5). All identified hub genes are highlighted as yellow color in the network plot.
Figure 3.
Identification of gene networks and key candidate genes involved in enhanced salt tolerance of kiwifruit. (A) Correlation plot representing the trait-modules relationship. (B) Heatmap plot of gene networks. Light colors represent low overlapping and dark color represents high overlapping of gene pairs. (C) Heatmaps for selected color modules. (D) Gene network for the blue and turquoise module. ABA, abscisic acid; B, blue; G, grey; GB, glycine betaine; GR, glutathione reductase; H, A. deliciosa; R, Actinidia valvata; T, turquoise; TSS, total soluble sugars; Y, yellow.
Overview of metabolome profile in R and H under salt stress
Based on GO term results of metabolic processes enrichment, the widely targeted metabolic profiles for R and H were performed under salt stress. Three repetitions for each group were implied to enhance the reliability of experimental data and results. The PCA analysis separated metabolites into two distinct areas of the plot (Fig. 4A). Similarly, the hierarchical clustering for samples resulted in two distinct clades for both species (Fig. 4B). Further analysis revealed that 70 metabolites were up-regulated and 71 metabolites were down-regulated in R4 vs H4 under salt stress (Fig. 4C and 4D upper panel). The Venn diagram results showed the presence of 37 differentially expressed metabolites in R4 vs H4 (Fig. 4D, lower panel).
Differentially expressed metabolites under salt stress
The KEGG pathways analysis for the top 20 metabolic pathways revealed several salt stress-responsive pathways which include flavonoid biosynthesis, pyruvate metabolism, phenylpropanoid biosynthesis, glutathione metabolism, etc. (Fig. 4E). For the identification of key pathways involved in salt stress tolerance, we compared the KEGG pathways for transcriptome and KEGG pathways for metabolome. This comparison resulted in the identification of three overlapping pathways, namely pyruvate metabolism, phenylpropanoid biosynthesis, and glutathione metabolism. These overlapping pathways might play important role in enhanced salt stress tolerance of R.
Integration of transcriptome and metabolome data reveals pyruvate biosynthesis as an important source of energy for R against salt stress
Based on the statistically significant q-value of KEGG pathways and overlapping with RNA-seq KEGG results, we selected the pyruvate metabolism pathway for integration of transcriptome and metabolome analysis results (Figs 4E, 1Cii). The results of pyruvate contents showed its enhanced accumulation under an extended time course of salt stress in kiwifruit (Fig. 2Diii). To determine the association between the pyruvate biosynthesis pathway and its associated genes, we screened out the differentiability expressed genes from R4 vs H4 involved in each step of this pathway. We identified 10 DEGs for enolase among which R_transcript_40556 had a significantly higher expression, 19 genes for pyruvate kinase (PK) among which R_transcript_15014 showed higher expression, three genes for L-lactate dehydrogenase (LDH) and H_transcript_93191 showed higher expression among all, nine genes for pyruvate decarboxylase (PDC) in which R_transcript_13128 showed differential expression, 35 genes for aldehyde dehydrogenase (ALDH) among which R_transcript_9286 gene showed significantly different expression in R. Similarly, there were 10 genes for acetyl-coenzyme A synthetase (CoA) among which R_transcript_48519 was differentially expressed between both species, while 40 genes were found for pyruvate dehydrogenase (PDH) in which the R_transcript_58755 gene showed significant changes in expression for R (Fig. 4F).
Functional validation of AvBADH in at plants
The glycine betaine pathway was deliberately selected due to the two-step conversion of choline (a toxic substance) to betaine (a harmless substance) in the presence of CMO and BADH genes. The second step in this pathway is rate liming. The phylogenetic analysis results showed that AvBADH was clustered with Avicennia marina (Fig. S6, see online supplementary material). The heatmap for 13 AvBADH genes from transcriptome data is given in supplementary files (Fig. S6, see online supplementary material). The sub-cellular localization results suggested that AvBADH gene was localized in peroxysomes and chloroplast of tobacco plant cells (Fig. S6, see online supplementary material). The successful T1 transformants were detected by RT-PCR (Fig. S6, see online supplementary material). We carried out a germination test and pot experiment to assess the performance of T3 generation transgenic lines and WT plants under salt stress. The early-stage and late-stage At plants were treated with 125 mM salt or no salt, and 300 mM salt, respectively. The roots and shoots of transgenic At plants showed higher relative expression of AvBADH gene than WT plants (Fig. S6, see online supplementary material). Under normal conditions, the At seedlings overexpressing AvBADH and WT had similar growth (Figs 5A and 6Ai). In contrast transgenic plants performed better than WT plants under salt stress (Figs 5A and 6Aii, iii). Salt stress negatively impacted growth parameters in WT and overexpressed lines. However, the growth parameters (number of leaves, root length, fresh and dry weight of shoot and root, plant fresh weight, germination and survival rate) of early-stage and late-stage At seedlings overexpressing the AvBADH gene were significantly higher than WT plants (Figs 5B, C, D, 6Aiv, v, vi, vii, and Fig. S6, see online supplementary material).
Figure 5.

Functional validation of AvBADH gene in early-stage At plants under 300 mM NaCl treatment. (A) Phenotype for early-stage transgenic At plants. (B) Plant fresh weight (g), (C) root length (cm), (D) number of leaves, (E) number of roots. Each value represents the mean ± SD of three biological replicates. The ns represents non-significance mean differences while ** denotes significant mean differences at P ≤ 0.01. ns, non-significant; OE, overexpression; WT, wild type.
Figure 6.

Functional validation of AvBADH gene in late-stage At plants under 300 mM NaCl treatment. (A) Phenotypes for transgenic lines and WT A. thaliana. (i) transgenic and WT lines before salt stress treatment, (ii) transgenic and WT lines after 300 mM salt stress for 21 days, (iii) transgenic and WT lines after 300 mM salt stress for 30 days, (iv) fresh and dry weight of shoot (g), (v) fresh and dry weight of root (g), (vi) root length (cm), and (vii) the number of leaves. (B) Histochemical staining of transgenic and WT lines under 300 mM salt stress for 21 days. (i) NBT (first row), DAB (second row), and Trypan blue (third row), (ii) accumulation of hydrogen peroxide (H2O2), (iii) accumulation of malonaldehyde (MDA), and (iv) accumulation of oxygen radicle (O2−). (C) Chlorophyll analysis for transgenic and WT lines under 300 mM salt stress for 21 days. (i) Extracted chlorophyll from leaves, (ii) chlorophyll fluorescence, (iii) SPAD value and Fv/Fm ratio. (D) Assays of (i) BADH enzymes, (ii) APX, (iii) GPX, (iv) SOD, (v) POD, (vi) CAT, (vii) Na+/K+ ratio, (viii) PRO, and (ix) GB. Each value represents the mean ± SD of three biological replicates. The ns represents non-significance mean differences while ** denotes significant mean differences at P ≤ 0.01. ns, non-significant; OE, overexpression; WT, wild type.
The histochemical staining results showed significantly lower production of H2O2, O2−, and MDA in transgenic lines over-expressing AvBADH than WT under 300 mM salt stress for 21 days (Fig. 6B). The At plants over-expressing AvBADH produced significantly higher chlorophyll contents and Fv/Fm compared to WT plants (Fig. 6C). The transgenic lines over-expressing AvBADH showed higher BADH enzyme activity and accumulated higher Na+, K+, Cl−, Na+/K+ ratio, APX, GPX, CAT, Proline, and glycine betaine than WT plants under 300 mM salt stress for 21 days (Fig. 6D and Fig. S12, see online supplementary material).
Functional validation of AvBADH in Ac plants
We constructed an overexpression vector for the AvBADH gene to perform the transformation in Ac (‘Hongyang’) through Agrobacterium-mediated transformation. The positive transgenic lines were detected by using specific pair of primers through RT-PCR (Fig. S13, see online supplementary material). The WT and transgenic plants were grown on an MS medium containing 0.4% (w/v) salt (NaCl). The plant samples were taken at zero-day (control), three days, and five days after salt stress treatment for subsequent analysis. The transgenic plants showed higher relative expression of AvBADH gene than WT plants (Fig. S14, see online supplementary material). The phenotype analysis showed that transgenic lines performed better than WT plants after five days of salt stress (Fig. 7A). The transgenic plants showed improved fresh weight, chlorophyll contents, and BADH enzyme activity than WT plants under salt stress (Fig. 7B, C, and D). On the contrary, the transgenic lines accumulated less H2O2 than WT plants under salt stress, suggesting that overexpression of the AvBADH gene significantly improved ROS scavenging in transgenic kiwifruit plants (Fig. 7E).
Figure 7.

Functional validation of AvBADH gene in kiwifruit under 0.4% salt stress for 0 days, 3 days, and 5 days. (A) Phenotype for transgenic and WT lines. (B) Fresh weight (g) of plants. (C) Chlorophyll contents (SPAD value). (D) BADH enzyme activity. (E) Accumulation of H2O2. Each value represents the mean ± SD of three biological replicates. The ns represents non-significance mean differences while ** denotes significant mean differences at P ≤ 0.01. ns, non-significant; OE, over expression; WT, wild type.
Discussion
Kiwifruit is a recently domesticated and relatively new fruit plant. It is highly popular among consumers due to its remarkably high vitamin C content [27]. Unfortunately, it is highly prone to abiotic stress especially salt stress because of its fleshy and shallow root system. Therefore, the kiwifruit industry is under constant pressure to develop new rootstocks that can assist plants to thrive well in salt-affected areas. In the past, the seedlings from H species have widely been used as rootstock which is sensitive to environmental stresses. Recently, the R species showed high potential for rootstock as ZMH genotype performed very well against different abiotic stress [29, 43]. Keeping in view the above-mentioned results, the integrated transcriptome and metabolome study was designed to underpin the fundamental difference in regulatory pathways between both species under salt stress.
The ABA signal transduction is involved in salt stress tolerance
Plant hormones are signaling molecules that are reported to be actively involved in salt stress tolerance [44–48]. The salt-responsive hormones contain ABA (abscisic acid), JA (jasmonic acid), and SA (salicylic acid) [49]. ABA is the most important salt-responsive phytohormone that, under high levels, activates kinase cascades and regulates gene expression in response to salt stress. Also, increased endogenous levels of ABA under salt stress can regulate osmotic stress by closing stomata [50]. The SnRK2s (sucrose non-fermenting-1 related protein kinase 2) play an important role in osmoregulation under salt stress. In the absence of ABA, the PP2Cs (protein phosphatase 2Cs) inhibit the kinase activities of SnRK2s. Upon osmotic stress, the ABA receptor proteins including PYL (PYR1-like) and RCAR (regulatory component of ABA receptor) can inhibit the activity of PP2Cs by perceiving and binding to accumulated ABA, thus restoring the activity of SnRK2s [51]. In the present study, we observed a higher expression level of R_Transcript_19927 (abscisic acid receptor PYL9) and ABA contents in kiwifruit under salt stress (Fig. 9B, C), indicating that the ABA signaling pathway played an important role in salt tolerance of R by mitigating osmotic stress. Our results were on a par with previous studies where increased ABA gene expression [52] and elevated ABA contents [53] were reported under salt stress.
Glycine betaine and TSS play role in salt stress tolerance
Glycine betaine is one of the major osmoprotectants along with TSS that accumulate in plants under salt stress to protect photosynthetic machinery, the structural integrity of proteins, enzymes, membranes, and scavenge ROS [54]. The soluble sugars play an important role in nutrition, osmoprotection and act as metabolite signaling molecule under salt stress in plants [55]. The overaccumulation of soluble sugars serve as carbon source and osmoprotection, maintain osmotic homeostasis and scavenge ROS to cope with the detrimental effects of salt stress in plants [56]. GB can reduce potassium efflux [57], regulate phosphate homeostasis, and increase the enzymatic activity of H+-ATPase in plasma membrane under salt stress [58], indicating that GB influences the regulation of ion homeostasis in plant organs. The GB is synthesized by two-step oxidation of choline via choline monooxygenase (CMO; EC 1.14.15.7) and betaine aldehyde dehydrogenase (BADH; EC 1.2.1.8) [59]. The multifunctional BADH gene responsible for the second oxidation reaction in the GB biosynthesis pathway is reported for salt stress tolerance, production of fragrance, and selectable marker for transgenic plants [60]. In the present study, the higher expression levels of the AvBADH gene and higher contents of GB and TSS protected plants from ionic and oxidative stress by maintaining ion homeostasis and providing osmo-protection, respectively.
Glutathione metabolism helps in scavenging ROS under salt stress.
Enzymatic and non-enzymatic antioxidants involved in ROS scavenging are key players in alleviating oxidative damage in plants caused by salt [61]. Glutathione is a non-enzymatic antioxidant that is found widely in plant tissues. During ROS scavenging, the glutathione is converted to GSSG (glutathione disulfide) by GPX (glutathione peroxidase) enzyme, while GSSG is recycled again to glutathione by GR (glutathione reductase) enzyme. The GPX and GR enzymes are responsible for maintaining glutathione homeostasis and glutathione/GSSG ratio [62]. In the plant cell, the glutathione reductase (GR) gene is responsible for maintaining the supply of reduced glutathione under salt stress. Therefore, the higher expression level of GR and higher Glutathione in salt-tolerant kiwifruit suggested that glutathione played an important role in ROS savaging and maintaining ion homeostasis under salt stress.
Pyruvate metabolism acts as an energy source for kiwifruit during salt stress
Respiration is an important metabolic process that is present in almost all living organisms on the surface of the earth. The key role of pyruvate, an end product of glycolysis, in plants is to provide energy in the form of ATP [63]. Additionally, pyruvate enters into chloroplast through transporter genes (i.e. BASS2) and serves as a precursor for ABA signaling. During glycolysis, phosphoenol pyruvate is converted to pyruvate in the presence of the pyruvate kinase (PK) enzyme. The pyruvate kinase enzyme needs K+ ion in plants as a co-factor for its activity under salt stress. Previously, pyruvate showed decreased expression on overconsumption of proline in the Krebs cycle in A. lagopoides [64]. Thus, we inferred that pyruvate not only provided energy to plant cells but in the presence of the K+ ion served as a precursor for ABA signaling to confer salt tolerance in kiwifruit plants.
Mechanisms underlying the enhanced salt tolerance in R
The present study revealed the enhanced salt tolerance of R compared to H. The results from transcriptome and metabolome analysis enabled us to propose a model to explain the potential mechanism underlying the enhanced salt tolerance in R. Upon exposure to salt stress, the R roots underwent the activation of the ABA signaling pathway. The plant cells received these signals and activated the biosynthesis of metabolites involved in salt stress tolerance. The overaccumulation of GB and TSS protected plants against osmotic stress. Similarly, the glutathione biosynthesis safeguarded plants from ionic stress through scavenging ROS. On the other hand, over-accumulation of pyruvate biosynthesis provided an adequate amount of energy in the form of ATP to carry out uninterrupted cell functioning (Fig. 8A). During the biosynthesis of GB, choline (a toxic substance) is oxidized into glycine betaine (an osmo-protectant) in the presence of CMO and BADH enzymes (Fig. 8B).
Figure 8.

Proposed model for (A) molecular mechanisms underlying enhanced salt tolerance in R, and (B) The biosynthetic pathway of GB in plants. ABA, abscisic acid; BADH, betaine aldehyde dehydrogenase; CMO, choline monooxygenase; GB, glycine betaine; GR, glutathione reductase; GST, glutathione-s-transferase; ROS, radioactive oxygen species.
Conclusion
In the present study, we comprehensively analysed the global changes in transcriptome and metabolome profiles in two kiwifruit species with contrasting salt tolerance abilities. The KEGG pathway enrichment analysis identified key pathways that are directly involved in salt stress tolerance. The WGCNA analysis identified two co-expression modules that were highly correlated with changes in ABA, glutathione, GB, TSS, and pyruvate contents. Furthermore, five candidates that were weighted as candidate hub genes were identified within these modules. Our results suggested that salt stress tolerance in kiwifruit could not be attributed to a single gene or metabolite, but to an intricate regulatory mechanism of pathways. Additionally, the AvBADH gene from GB pathway enhanced salt tolerance of transgenic A. thaliana and A. chinensis plants. Future research on salt stress tolerance should be directed to the candidate genes and metabolic pathways identified in this study.
Materials and methods
Plant material and salt treatment
We selected two Actinidia species A. valvata Dunn. (ZMH, tolerant) and A. deliciosa Planch. var. deliciosa (cultivar ‘Hayward’, HWD, sensitive) based on our previous genotype screening study for salt stress [29]. Plants for both genotypes were propagated through the tissue culture technique on Murashige and Skoog (MS) medium [65] containing 30 g/L sucrose and 7.2 g/L agar (pH 5.8) to obtain true-to-type plants. The explants for both genotypes were obtained from Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, China (latitude 34°43′N, longitude 113°39′E, and altitude 111 m). After rooting, plantlets were transferred into 6 × 6 cm plastic pots with a 3 L holding capacity containing growing media (compost: perlite: vermiculite = 2:1:1). At the 4–6 leaf stage, plants were subjected to 0.4% NaCl per net weight of the growing medium in the pot. Treatment was repeated thrice and there were five plants in each replication. Samples from plant roots were collected at 12 h, 24 h, and 72 h after treatment, considering 0 h as control.
Physiological measurements
The pyruvate contents were measured according to a previously described method [66]. The ABA (abscisic acid) contents were measured by following a previously described protocol [67]. The GB (glycine betaine) and TSS (total soluble sugar) were measured by using commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), while the glutathione was measured according to the previously described protocol [68]. A bio-mate 3 s spectrophotometer (Thermo Scientific, Waltham, MA, USA) was used to check the optical density (OD).
RNA extraction and RNA sequencing
Samples for RNA-seq analysis were collected from the roots of two contrasting kiwifruit genotypes for salt stress. In total, 24 samples were collected from the control and three time-points for both genotypes (2 cultivars × 4 sample collection times × 3 biological replicates × 5 technical replicates). All the samples were sent to Biomarker Technology Company (Biomarker, Beijing, China) for RNA-seq. Total RNA from the roots was isolated according to the manufacturer’s protocol of the Plant Total RNA purification kit (Waryoung, Beijing, China). We used NanoDrop 2000 (Thermo Scientific, MA, USA) system and the NanoDrop 6000 assay kit (Agilent technologies, Beijing, China) to check the RNA concentration and integrity. The cDNA libraries were synthesized from RNA by using magnetic beads linked with Oligo (dT) to enrich eukaryotic mRNA and their concentration was assessed by Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). RNA-seq libraries were sequenced by an Illumina Hiseq platform X Ten (Illumina, CA, USA). To construct PacBio libraries, cDNA was synthesized by using SMRTer PCR cDNA synthesis kit (Clontech, USA) and the PacBio libraries were constructed according to the PacBio protocol (http://www.pacb.com/wp-content, accessed on November 15, 2020).
RNA-seq Data processing
Paired-end raw reads were filtered by in-house perl script to remove adapters, primer sequences, low-quality reads, and reads containing poly (N) to obtain the clean reads in FASTQ format. At the same time, a base distribution map was used to calculate the Q20, Q30 AT and GC contents, and sequence duplication level of clean reads. The SMART Analysis Software suit was used to analyzed the sequencing results from PacBio (http://www.pacb.com/product-and-services /analytical-software/smrt-analysis, accessed on November 15, 2020). The SMART sequencing results were filterd for high quality reads by removing reads with a quality of < 0.75 and length of < 50bp. The high-quality reads were then used to obtain error-corrected circular consensus sequences (CCSs) with the full passes = 0 and quality > 0.75. The CCSs were considered as full-length (FL) transcripts only if the 5′-primer sequence, 3′ primer sequence and a poly A tail were present. The nonchimeric sequences in the FL sequences were considered to be FL-nonchimeric (FLNC) sequences. The FLNC sequences with high-quality bases were obtained for subsequent analysis.
Functional annotation of transcripts and identification of DEGs
The reference transcriptome was assembled de novo from SMART transcript data due to unavailability of reference genome for hexaploid kiwifruit. The clean reads from RNA-seq data were then mapped to the Pacbio reference genome sequence by using Hisat2 tool [69]. The reads with perfect match or one mismatch were used for subsequent analysis. The uni-gene sequences were annotated against KEGG (Kyoto Encyclopedia of Genes and Genomes), GO (gene ontology), NR (non-redundant protein sequence), Pfam (homologous protein family), KOG (eukaryotic ortholog group), and Swiss-Port databases to obtain their annotation by using local blast programs at a threshold E-value = 1e-5. The plant transcription factors (TFs) and TF family classification was predicted by iTAK [70]. The expression levels of the gene were calculated by fragment per kilobase of transcript per million fragments mapped (FPKM) method. Identification of DEGs (Differentially expressed genes) was done by DESeq2 [71] and edgeR R package [72]. The genes were considered differentially expressed at FDR-adjusted P-value < 0.01 and |log2 (fold change) | 2. The enriched DEGs via GO (gene ontology) [73] and KEGG (Kyoto Encyclopedia of Genes and Genomes [74] were used to identify significant metabolic pathways at an FDR-adjusted P-value < 0.05.
qPCR validation of selected DEGs
For the expression validation, 16 DEGs were randomly selected from RNA-seq data to analyse the relative gene expression levels [75]. Total RNA from the roots was isolated according to the manufacturer’s protocol of the Plant Total RNA purification kit (Waryoung, Beijing, China). The RT-qPCR was performed according to a previously described method [76]. All the experiments were repeated thrice. The relative expression of genes was calculated by 2ΔΔCT method [77]. All the pair of primers used for RT-qPCR are listed in Table S1 (see online supplementary material).
WGCNA for identification of regulatory networks and hub genes
We performed WGCNA by using a R-based WGCNA package (v.1.69) with default parameters [78]. The adjacency matrix was constructed by using the FPKM values of DEGs, which was then converted to TOM (topological overlap matrix). Modules were detected by using a dynamic hybrid tree-cut algorithm (DynamicTreeCut, v.1.63–1) with default parameters [79]. The color modules were identified based on phenotype data, and the correlation matrix between phenotype and gene module was computed by an R-based WGCNA package. The networks for color modules were visualized by Cytoscape software (v.3.8.2) [80].
Metabolomic profiling and statistical analysis
The extraction of metabolites from R roots was performed according to the previously described protocol with slight modifications [81]. Primary metabolites were quantified through GC–MS measurements and LC–MS was implied to measure secondary metabolites. Annotation for LC–MS data was done by matching the extracted data from the chromatogram KEGG compound database and KEGG pathway database. An R-based package TargetSearch was used to annotate GC–MS data [82]. The normalized data were subjected to PCA analysis on R-based software Factoextra (v.1.0.6) with default parameters [83]. The Metaboanalyst (v.4.0) software was used to choose DEMs [84]. The VANTED software was used to construct metabolic pathways [85].
Gene cloning, phylogenetic tree, and subcellular localization analysis
For stable transformation in At and Ac, the open reading frame (ORF) of AvBADH was cloned from cDNA of R roots by using a specific pair of primers (Table S4, see online supplementary material). The phylogenetic tree for BADH proteins from R and other plant species was constructed by GenomeNet software (https://www.genome.jp/) and visualized by iTOOLs software. For subcellular localization analysis, the coding sequences (CDSs) of AvBADH was amplified by using a specific pair of primers lacking stop codon (TAG). The PCR product was transferred into a pBI121-GFP vector driven by CaMV 35S promoter. After sequence confirmation, the recombinant 35S pro:AvBADH::GFP, and 35S pro:GFP (control) plasmids were entered into an Agrobacterium tumefaciens strain GV 3101. The cultures of recombinant plasmids in GV3101 (OD600 = 0.6–1.0) were injected into tobacco plant leaves and kept in a growth chamber for 2–3 days. The transformed leaves were analysed on a confocal laser-guided microscope (FV1000; Olympus, Tokyo, Japan) for signal detection.
Transformation of At and Ac plants
For At transformation, the ORF of AvBADH was cloned into pBI121 vector driven by CaMV 35S promoter. A. tumefaciens strain GV3101 harboring the AvBADH was transformed into WT At (Columbia-0) via floral dip method [86]. The selection of T0 positive seeds was done on MS medium containing kanamycin (100 mg/L) (Sigma, Saint Louis, MO, USA). The detection of positive plants was done by RT-qPCR with F-35S + R-BADH primer pair. The positive plants were raised to T3 generation to ensure the maximum homozygosity. Three positive T3 transgenic lines were selected for subsequent functional validation.
For Ac transformation, the ORF of AvBADH was cloned into pBI121 vector driven by CaMV 35S promoter. The A. tumefaciens strain EHA105 harboring the AvBADH was transformed into Ac leaves by following a previously described protocol [87]. We obtained transgenic plants after ~6 months. The positive transformants were identified by PCR and RT-qPCR analysis. Three positive transformants with higher expression levels were used for subsequent analysis.
Assay of salt stress for transgenic At plants
Early-stage At plants
For germination assay, we germinated eight seeds per plate for WT and each OE line on MS medium plates containing 0 mM or 125 mM NaCl salt concentration. Each treatment was replicated on three plates. The germination rate was calculated seven days after transferring plates into the light. The germination was confirmed on the emergence of plumule from seed coat [88]. The Root length was measured in centimeters (cm). The fresh weight of root and shoot was measured in grams (g) on a weight balance immediately after taking samples. The shoot and root samples were oven-dried at 65°C for 72 hours for taking dry weight in grams (g). Plant survival rate was measured by taking the percentage (%) of alive plants for each line.
Late-stage At plants
The 15-day-old transgenic and WT At potted plants were treated with 300 mM NaCl salt stress. For measuring physiochemical parameters, samples were taken from plants after 21 days of no-salt or 300 mM NaCl salt stress treatments. The total chlorophyll contents were measured according to a previously described protocol [89]. The ion contents for Na+, K+, and Cl− were measured following previous methods [90, 91]. The contents for APX (Ascorbate peroxidase, EC 1.11.1.11) GPX (Glutathione-s-transferase, EC 2.5.1.18), CAT (Catalase, EC 1.11.1.6), POD (Peroxidase, EC 1.11.1.7) and SOD (Superoxide dismutase, EC 1.15.1.1) were measured according to previously described methods [90, 92, 93]. BADH enzyme activity was carried out by enzyme-linked immunosorbent (ELISA) assay method using a specific detection kit (ml036337 for BADH activity, Mlbio, Shanghai, China) and membrane damage (MDA) was measured according to the method described by Chen and his team [94]. Proline and GB were measured by previously described methods [95]. A fluorimeter (IMAGING-PAM) was used to record chlorophyll fluorescence and Fv/Fm was calculated by Imaging WinGegE software. The histochemical staining for O2.--, H2O2, and plant cell death was done with nitro blue tetrazolium (NBT), 3,3′-diaminobenzidine (DAB), and trypan blue, respectively [96, 97]. The H2O2 and O2.-- were measured calorimetrically [98, 99].
Transcriptional activation activity (TA) analysis of AvMYB TF
The transcriptional activation (TA) activity was carried out by following the steps described in a previous study [100]. The coding sequence (CDS) of AvMYB was amplified from cDNA of R and inserted into pGBKT7 vector using Novoprotein recombination kit to form pGBKT7-AvMYB1 and pGBKT7-AvMYB2 vectors (Novoprotein, Suzhou, China). Then, the constructs and empty pGBKT7 vector (negative control) were transferred into AH109 yeast strain. Transformed colonies were screened on SD/−Trp, SD/−Trp-His-Ade, and SD/−Trp-His-Ade with X-a-Gal solid media, which were cultured 2–3 days in an incubator at 30°C.
Statistical analysis
Physiological data were analysed for significant differences by one-way ANOVA and t-test. The mean differences were computed through Duncan’s MRT test. The results were presented as means ± SD. All the experients were repreated thrice and all the analyses were carried out with Mac-based R packages.
Core idea
To the best of our knowledge, this is the first-ever study on kiwifruit to decipher intricate regulatory mechanisms involved in salt tolerance through integrated transcriptome and metabolome analysis.
The hexaploid R (ZMH) is a special plant material that offers enhanced salt tolerance.
Gene co-expression network analysis resulted in the identification of two novel genes pyruvate kinase (R_transcript_13859) and fructokinase (H_transcript_39638) related to pyruvate and sugar metabolism in R under salt stress.
The pyruvate metabolism is rarely reported to be involved in salt stress studies. In the present study, the pyruvate metabolism was also identified among important pathways contributing to elevated salt tolerance in R plants under salt stress.
The At and Ac plants overexpressing the AvBADH gene exhibited elevated salt tolerance compared to WT plants under different salt stress concentrations.
Supplementary Material
Contributor Information
Muhammad Abid, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China; Lushan Botanical Garden, Chinese Academy of Sciences, Jiujiang 332900, China.
Shichao Gu, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Yong-Jie Zhang, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Shihang Sun, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Zhi Li, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Dan-Feng Bai, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Leiming Sun, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Xiu-Juan Qi, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Yun-Peng Zhong, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Jin-Bao Fang, Key Laboratory for Fruit Tree Growth, Development and Quality Control, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, China.
Acknowledgments
We thank all the members of the kiwifruit genetic resources innovation and breeding team, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences (Zhengzhou, Henan, China) for supporting and encouraging us to accomplish this work.
This study was funded by Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Science (CAAS-ASTIP-2021-ZFRI), the China Agriculture Research System of MOF and MARA (CARS-26), Yunnan Science and Technology Program (202205AF150043), Sichuan Science and Technology Program (2021YFN0060), and Yangtze River Kiwifruit Industry Technology Research Center (CJZX20210110).
Author Contributions
M.A., Y-P.Z., J-B.F. conceived and designed the experiments. M.A. and S.G. performed the experiments. S.G., Y-J.Z., and D-F.B. helped in collecting the plant samples. M.A. analysed the data and wrote the main manuscript. S.S., Z.L., and L.S. provided valuable suggestions for experiments. Y-P.Z. and X-J.Q. revised and edited the manuscript. Y-P.Z. and J-B.F. supervised this study and obtained funding. All authors read and approved the final version of the manuscript.
Data availability
The authors declare that the data supporting the findings of this study are presented in the article and supplementary information files are available from the corresponding author upon request. Transcriptome raw data is submitted in NCBI public repository under BioProject (PRJNA726156).
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
References
- 1. Samsatly J, Copley TR, Jabaji SH. Antioxidant genes of plants and fungal pathogens are distinctly regulated during disease development in different Rhizoctonia solani pathosystems. PLoS One. 2018;13:e0192682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao C, Jiang W, Zayed Oet al. The LRXs-RALFs-FER module controls plant growth and salt stress responses by modulating multiple plant hormones. Natl Sci Rev. 2021;8:nwaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao R, Sun H, Mei Cet al. The Arabidopsis Ca2+-dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 2011;192:61–73. [DOI] [PubMed] [Google Scholar]
- 4. Mackay A. Climate change 2007: impacts, adaptation and vulnerability. Contribution of working group II to the fourth assessment report of the intergovernmental panel on climate change. J Environ Qual. 2007;37:2407. [Google Scholar]
- 5. Golldack D, Li C, Mohan Het al. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci. 2014;5:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agarwal PK, Shukla PS, Gupta Ket al. Bioengineering for salinity tolerance in plants: state of the art. Mol Biotechnol. 2013;54:102–23. [DOI] [PubMed] [Google Scholar]
- 7. Miller GAD, Suzuki N, Ciftci-Yilmaz Set al. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–67. [DOI] [PubMed] [Google Scholar]
- 8. Li H-W, Zang B-S, Deng X-Wet al. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta. 2011;234:1007–18. [DOI] [PubMed] [Google Scholar]
- 9. Cotsaftis O, Plett D, Johnson AATet al. Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Mol Plant. 2011;4:25–41. [DOI] [PubMed] [Google Scholar]
- 10. Kumar K, Kumar M, Kim S-Ret al. Insights into genomics of salt stress response in rice. Rice. 2013;6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar S, Dhingra A, Daniell H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol. 2004;136:2843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qiao WH, Zhao XY, Li Wet al. Overexpression of AeNHX1, a root-specific vacuolar Na+/H+ antiporter from Agropyron elongatum, confers salt tolerance to Arabidopsis and Festuca plants. Plant Cell Rep. 2007;26:1663–72. [DOI] [PubMed] [Google Scholar]
- 13. Saad RB, Zouari N, Ben Ramdhan Wet al. Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger “AlSAP” gene isolated from the halophyte grass Aeluropus littoralis. Plant Mol Biol. 2010;72:171–90. [DOI] [PubMed] [Google Scholar]
- 14. Hmida-Sayari A, Gargouri-Bouzid R, Bidani Aet al. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Sci. 2005;169:746–52. [Google Scholar]
- 15. Jing P, Zou J, Kong Let al. OsCCD1, a novel small calcium-binding protein with one EF-hand motif, positively regulates osmotic and salt tolerance in rice. Plant Sci. 2016;247:104–14. [DOI] [PubMed] [Google Scholar]
- 16. Gao Y, Liu J, Yang Fet al. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol Plant. 2020;168:98–117. [DOI] [PubMed] [Google Scholar]
- 17. Li M, Chen R, Jiang Qet al. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol Biol. 2020;105:333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han D, Han J, Yang Get al. An ERF transcription factor gene from Malus baccata (L.) borkh, MbERF11, affects cold and salt stress tolerance in Arabidopsis. Forests. 2020;11:514. [Google Scholar]
- 19. Wu J, Yu C, Hunag Let al. Overexpression of MADS-box transcription factor OsMADS25 enhances salt stress tolerance in Rice and Arabidopsis. Plant Growth Regul. 2020;90:163–71. [Google Scholar]
- 20. Shulaev V, Cortes D, Miller Get al. Metabolomics for plant stress response. Physiol Plant. 2008;132:199–208. [DOI] [PubMed] [Google Scholar]
- 21. Gupta B, Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics. 2014;2014:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patel MK, Kumar M, Li Wet al. Enhancing salt tolerance of plants: from metabolic reprogramming to exogenous chemical treatments and molecular approaches. Cell. 2020;9:2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pan J, Li Z, Dai Set al. Integrative analyses of transcriptomics and metabolomics upon seed germination of foxtail millet in response to salinity. Sci Rep. 2020;10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meng X, Liu S, Dong Tet al. Comparative transcriptome and proteome analysis of salt-tolerant and salt-sensitive sweet potato and overexpression of IbNAC7 confers salt tolerance in Arabidopsis. Front Plant Sci. 2020;11:1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xing JC, Zhao BQ, Dong Jet al. Transcriptome and metabolome profiles revealed molecular mechanisms underlying tolerance of Portulaca oleracea to saline stress. Russ J Plant Physiol. 2020;67:146–52. [Google Scholar]
- 26. Wei T, Wang Y, Liu J-H. Comparative transcriptome analysis reveals synergistic and disparate defense pathways in the leaves and roots of trifoliate orange (Poncirus trifoliata) autotetraploids with enhanced salt tolerance. Hortic Res. 2020;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang S, Ding J, Deng Det al. Draft genome of the kiwifruit Actinidia chinensis. Nat Commun. 2013;4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang T, Gleave AP. Applications of biotechnology in kiwifruit (Actinidia). In: Agbo EC, ed. Innovations in Biotechnology. InTech, 2012, 3–30. [Google Scholar]
- 29. Abid M, Zhang YJ, Li Zet al. Effect of salt stress on growth, physiological and biochemical characters of four kiwifruit genotypes. Sci Hortic. 2020;271:109473. [Google Scholar]
- 30. Zhong YP, Xiu-Juan QI, Chen JYet al. Growth and physiological responses of four kiwifruit genotypes to salt stress and resistance evaluation. J Integr Agric. 2019;18:83–95. [Google Scholar]
- 31. Cimini S, Locato V, Giacinti Vet al. A multifactorial regulation of glutathione metabolism behind salt tolerance in rice. Antioxidants. 2022;11:1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan H, Nie Y, Cui Ket al. Integrative transcriptome and metabolome profiles reveal common and unique pathways involved in seed initial imbibition under artificial and natural salt stresses during germination of Halophyte quinoa. Front Plant Sci. 2022;13:853326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nahar K, Hasanuzzaman M, Alam MMet al. Roles of exogenous glutathione in antioxidant defense system and methylglyoxal detoxification during salt stress in mung bean. Biol Plant. 2015;59:745–56. [Google Scholar]
- 34. Zhao X, Chen Z, Leng Pet al. iTRAQ-based quantitative proteomic analysis of the response of Hylotelephium erythrostictum leaves to salt stress. Sci Hortic. 2020;264:109190. [Google Scholar]
- 35. Liu D, Hu R, Zhang Jet al. Overexpression of an agave phosphoenolpyruvate carboxylase improves plant growth and stress tolerance. Cell. 2021;10:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. De la Torre-González A, Montesinos-pereira D, Blasco Bet al. Influence of the proline metabolism and glycine betaine on tolerance to salt stress in tomato (Solanum lycopersicum L.) commercial genotypes. J Plant Physiol. 2018;231:329–36. [DOI] [PubMed] [Google Scholar]
- 37. Rady MOA, Semida WM, Abd El-Mageed TAet al. Up-regulation of antioxidative defense systems by glycine betaine foliar application in onion plants confer tolerance to salinity stress. Sci Hortic. 2018;240:614–22. [Google Scholar]
- 38. Missihoun TD, Willée E, Guegan J-Pet al. Overexpression of ALDH10A8 and ALDH10A9 genes provides insight into their role in glycine betaine synthesis and affects primary metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2015;56:1798–807. [DOI] [PubMed] [Google Scholar]
- 39. Wang W-R, Liang J-H, Wang G-Fet al. Overexpression of PpSnRK1α in tomato enhanced salt tolerance by regulating ABA signaling pathway and reactive oxygen metabolism. BMC Plant Biol. 2020;20:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J, Yu H, Zhang Yet al. Increased abscisic acid levels in transgenic maize overexpressing AtLOS5 mediated root ion fluxes and leaf water status under salt stress. J Exp Bot. 2016;67:1339–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang Q, Jiang T, Xu Let al. A salt-stress-regulator from the poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis. Plant Physiol Biochem. 2017;114:100–10. [DOI] [PubMed] [Google Scholar]
- 42. Wang T, Tohge T, Ivakov Aet al. Salt-related MYB1 coordinates abscisic acid biosynthesis and signaling during salt stress in Arabidopsis. Plant Physiol. 2015;169:1027–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Z, Zhong Y, Bai Det al. Comparative analysis of physiological traits of three Actinidia valvata Dunn. Genotypes during waterlogging and post-waterlogging recovery. Hortic Environ Biotechnol. 2020;61:825–36. [Google Scholar]
- 44. Raghavendra AS, Gonugunta VK, Christmann Aet al. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. [DOI] [PubMed] [Google Scholar]
- 45. Javid MG, Sorooshzadeh A, Moradi Fet al. The role of phytohormones in alleviating salt stress in crop plants. Aust J Crop Sci. 2011;5:726–34. [Google Scholar]
- 46. Ming R, Zhang Y, Wang Yet al. The JA-responsive MYC2-BADH-like transcriptional regulatory module in Poncirus trifoliata contributes to cold tolerance by modulation of glycine betaine biosynthesis. New Phytol. 2020;229:2730–50. [DOI] [PubMed] [Google Scholar]
- 47. Sharma I, Ching E, Saini Set al. Exogenous application of brassinosteroid offers tolerance to salinity by altering stress responses in rice variety 'Pusa Basmati-1′. Plant Physiol Biochem. 2013;69:17–26. [DOI] [PubMed] [Google Scholar]
- 48. Zhu T, Deng X, Zhou Xet al. Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci Rep. 2016;6:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao S, Zhang Q, Liu Met al. Regulation of plant responses to salt stress. Int J Mol Sci. 2021;22:4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao Y, Zhang Z, Gao Jet al. Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep. 2018;23:3340–3351.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barrero JM, Rodríguez PL, Quesada Vet al. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006;29:2000–8. [DOI] [PubMed] [Google Scholar]
- 53. Hamayun M, Khan SA, Khan ALet al. Effect of salt stress on growth attributes and endogenous growth hormones of soybean cultivar 'Hwangkeumkong'. Pakistan J Bot. 2010;42:3103–12. [Google Scholar]
- 54. Ashraf M, Foolad MR. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot. 2007;59:206–16. [Google Scholar]
- 55. Rosa M, Prado C, Podazza Get al. Soluble sugars: metabolism, sensing, and abiotic stress. Plant Signal Behav. 2009;4:388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sami F, Yusuf M, Faizan Met al. Role of sugars under abiotic stress. Plant Physiol Biochem. 2016;109:54–61. [DOI] [PubMed] [Google Scholar]
- 57. Wei D, Zhang W, Wang Cet al. Genetic engineering of the biosynthesis of glycinebetaine leads to alleviate salt-induced potassium efflux and enhances salt tolerance in tomato plants. Plant Sci. 2017;257:74–83. [DOI] [PubMed] [Google Scholar]
- 58. Li D, Zhang T, Wang Met al. Genetic engineering of the biosynthesis of glycine betaine modulates phosphate homeostasis by regulating phosphate acquisition in tomato. Front Plant Sci. 2019;9:1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen THH, Murata N. Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ. 2011;34:1–20. [DOI] [PubMed] [Google Scholar]
- 60. Niazian M, Sadat-Noori SA, Tohidfar Met al. Betaine aldehyde dehydrogenase (BADH) vs. Flavodoxin (Fld): two important genes for enhancing plants stress tolerance and productivity. Front Plant Sci. 2021;12:650215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Naliwajski MR, Skłodowska M. The oxidative stress and antioxidant systems in cucumber cells during acclimation to salinity. Biol Plant. 2014;58:47–54. [Google Scholar]
- 62. Hasanuzzaman M, Nahar K, Anee TIet al. Glutathione in plants: biosynthesis and physiological role in environmental stress tolerance. Physiol Mol Biol Plants. 2017;23:249–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shi H, Ye T, Chan Z. Comparative proteomic responses of two bermudagrass (Cynodon dactylon (L). Pers.) varieties contrasting in drought stress resistance. Plant Physiol Biochem. 2014;82:218–28. [DOI] [PubMed] [Google Scholar]
- 64. Sobhanian H, Motamed N, Jazii FRet al. Salt stress induced differential proteome and metabolome response in the shoots of Aeluropus lagopoides (Poaceae), a halophyte C4 plant. J Proteome Res. 2010;9:2882–97. [DOI] [PubMed] [Google Scholar]
- 65. Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–97. [Google Scholar]
- 66. He L, Jing Y, Shen Jet al. Mitochondrial pyruvate carriers prevent cadmium toxicity by sustaining the TCA cycle and glutathione synthesis. Plant Physiol. 2019;180:198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu N, Ding Y, Fromm Met al. Endogenous ABA extraction and measurement from Arabidopsis leaves. Bio Protoc. 2014;4:e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Moreno-Galván AE, Cortés-Patiño S, Romero-Perdomo Fet al. Proline accumulation and glutathione reductase activity induced by drought-tolerant rhizobacteria as potential mechanisms to alleviate drought stress in Guinea grass. Appl Soil Ecol. 2020;147:103367. [Google Scholar]
- 69. Kim D, Paggi JM, Park Cet al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zheng Y, Jiao C, Sun Het al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 2016;9:1667–70. [DOI] [PubMed] [Google Scholar]
- 71. Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;7:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Young MD, Wakefield MJ, Smyth GKet al. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mao X, Tao C, Olyarchuk JGet al. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–93. [DOI] [PubMed] [Google Scholar]
- 75. Zhang Y, Wang P, Xia Het al. Comparative transcriptome analysis of basal and zygote-located tip regions of peanut ovaries provides insight into the mechanism of light regulation in peanut embryo and pod development. BMC Genomics. 2016;17:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li Y, Fang J, Qi Xet al. Combined analysis of the fruit metabolome and transcriptome reveals candidate genes involved in flavonoid biosynthesis in Actinidia arguta. Int J Mol Sci. 2018;19:1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 78. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the dynamic tree cut package for R. Bioinformatics. 2008;24:719–20. [DOI] [PubMed] [Google Scholar]
- 80. Shannon P, Markiel A, Ozier Oet al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Giavalisco P, Köhl K, Hummel Jet al. 13C isotope-labeled metabolomes allowing for improved compound annotation and relative quantification in liquid chromatography-mass spectrometry-based metabolomic research. Anal Chem. 2009;81:6546–51. [DOI] [PubMed] [Google Scholar]
- 82. Cuadros-Inostroza A, Lisec J, Redestig H, Hannah MA. The TargetSearch Package. Viewpoints 2013.
- 83. Kassambara A, Mundt F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. 2020. (Retrieved from https://github.com/kassambara/factoextra).
- 84. Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinforma. 2019;68:e86. [DOI] [PubMed] [Google Scholar]
- 85. Junker BH, Klukas C, Schreiber F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Davis AM, Hall A, Millar AJet al. Protocol: streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods. 2009;5:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang T, Atkinson R, Janssen B. Choice of agrobacterium strain for transformation of kiwifruit. Acta Hortic. 2007;753:227–32. [Google Scholar]
- 88. Li X, Pan Y, Chang Bet al. NO promotes seed germination and seedling growth under high salt may depend on EIN3 protein in Arabidopsis. Front Plant Sci. 2016;6:1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ganguly M, Datta K, Roychoudhury Aet al. Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance. Plant Signal Behav. 2012;7:502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang M, Fang Y, Ji Yet al. Effects of salt stress on ion content, antioxidant enzymes and protein profile in different tissues of Broussonetia papyrifera. South African J Bot. 2013;85:1–9. [Google Scholar]
- 91. Nazar R, Iqbal N, Syeed Set al. Salicylic acid alleviates decreases in photosynthesis under salt stress by enhancing nitrogen and sulfur assimilation and antioxidant metabolism differentially in two mungbean cultivars. J Plant Physiol. 2011;168:807–15. [DOI] [PubMed] [Google Scholar]
- 92. Ali MB, Hahn EJ, Paek KY. Effects of light intensities on antioxidant enzymes and malondialdehyde content during short-term acclimatization on micropropagated Phalaenopsis plantlet. Environ Exp Bot. 2005;54:109–20. [Google Scholar]
- 93. Ji W, Zhu Y, Li Yet al. Over-expression of a glutathione S-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol Lett. 2010;32:1173–9. [DOI] [PubMed] [Google Scholar]
- 94. Chen K, Chen L, Fan Jet al. Alleviation of heat damage to photosystem II by nitric oxide in tall fescue. Photosynth Res. 2013;116:21–31. [DOI] [PubMed] [Google Scholar]
- 95. Ja’afar U, Aliero AA, Yahaya TO . A study on proline and glycine betaine contents as salinity and drought tolerant indicators in Solanum lycopersicum L. (cultivar: 'Roma'). Glob J Sci Front Res D Agric Vet. 2018;18:29–32. [Google Scholar]
- 96. Huang XS, Wang W, Zhang Qet al. A basic helix-loop-helix transcription factor PtrbHLH of Poncirus trifoliata confers cold tolerance and modulates POD-mediated scavenging of H2O2. Plant Physiol. 2013;162:1178–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Riechmann J, Heard J, Martin Get al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–10. [DOI] [PubMed] [Google Scholar]
- 98. Mukherjee SP, Choudhuri MA. Implications of water stress-induced changes in the levels of endogenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol Plant. 1983;58:166–70. [Google Scholar]
- 99. Hernandez JA, Ferrer MA, Jimenez Aet al. Antioxidant systems and O2.-/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 2001;127:817–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Qu Y, Nong Q, Jian Set al. An AP2/ERF gene, HuERF1, from pitaya (Hylocereus undatus) positively regulates salt tolerance. Int J Mol Sci. 2020;21:4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are presented in the article and supplementary information files are available from the corresponding author upon request. Transcriptome raw data is submitted in NCBI public repository under BioProject (PRJNA726156).