Abstract
The lipopolysaccharide (LPS) structure of Salmonella typhimurium has been correlated with the virulence of wild-type strain LT2. Mutants of LT2 with truncated polysaccharide portions of LPS are less virulent than strains with a complete LPS structure. Polyclonal T cells and monoclonal T-cell hybridomas were more reactive to heat-killed rough mutants than to heat-killed smooth strains, as measured by interleukin-2 (IL-2) production. Using a large panel of strains with truncated LPS molecules, we found that T-cell reactivity decreased with certain lengths of polysaccharide. The decreased response was not due to differential phagocytic uptake, IL-12 production, or major histocompatibility complex class II surface expression by macrophages. Also, LT2 did not mediate any global suppression since addition of LT2 did not diminish the response of T cells specific for antigens unrelated to Salmonella. In an experiment in which processing times were varied, we found that antigens from rough strains were processed and presented more quickly than those associated with smooth strains. At longer processing times, epitopes from LT2 were presented well. We hypothesize that the slower antigen processing and presentation of wild-type Salmonella may be caused by masking of surface antigens by the longer polysaccharide portion of smooth LPS. This blocking of effective antigen presentation may contribute to the virulence of Salmonella.
Typhoid fever, caused by Salmonella typhi, is still a major health problem in developing nations. Worldwide, sixteen million cases of typhoid fever with 600,000 deaths are estimated annually (26). Salmonella typhimurium, a murine pathogen, serves as a model for typhoid fever. S. typhimurium, follows a course of infection in mice similar to that of S. typhi in humans (10, 50). The outcomes by several routes of infection in mice (intraperitoneal [i.p.], intravenous [i.v.] and oral) are similar (23, 50). It is important to understand the immune response to S. typhimurium not only as a model for human disease but also because of its extensive use in experimental vaccine schemes as a carrier for epitopes from other pathogens (6, 63, 64).
One of the main virulence factors of Salmonella is its lipopolysaccharide (LPS), a component of the outer membrane of gram-negative bacteria (54). The LPS structure consists of three regions: O-specific polysaccharide, core polysaccharide, and lipid A. The O side chain gives the bacterium added virulence. Truncations in the polysaccharide portion of LPS cause a reduction in virulence. Based on colony morphology, mutant strains that have incomplete LPS molecules are termed rough while the wild type is termed smooth. Wild-type strains of S. typhimurium have a 50% lethal dose of <10 organisms in susceptible mice (11), while rough mutants are avirulent (50% lethal dose of ≥109) (30). All strains have a lipid A region and a minimum of two 2-keto-3-deoxyoctulosonic acid moieties (53). It is the lipid A region that mediates most of the biological effects of LPS, including the induction of cytokines (interleukin-1 [IL-1], IL-6, IL-12, and tumor necrosis factor alpha from mononuclear cells (13, 58).
The basis for differential virulence of rough and smooth strains is not known. A significant reason is the difference in serum sensitivities between rough and smooth strains (52). Several other comparisons have been made. Rough forms of Salmonella, in some studies, were found to be more potent cytokine inducers than smooth forms (13, 14, 53). Other studies have indicated that virulence may be due to the superior ability of smooth strains to bind and colonize HeLa cells in vitro (44) or murine epithelial cells in vivo (36). Upon oral challenge, smooth strains outcompeted rough strains during infection of the gut (36). Both rough and smooth strains could block phagosome-lysosome fusion to enhance intracellular survival (5, 25). However, during intracellular infection, rough strains were ultimately more susceptible to killing (41, 51). When used in vaccination studies, smooth and rough strains can both induce protective cell-mediated immunity (45). Currently, it is unclear how these observations relate to the mechanism of virulence as controlled by the LPS structure. Our studies address the influence of LPS on antigen presentation and the development of T-cell-mediated immunity.
While γδ T cells play a significant role in resistance to infection (43), protective immunity primarily involves CD4+ αβ T cells and has been demonstrated by adoptive transfer (22) and subset depletion studies (47). Clearance of infection was impaired in T-cell-receptor-alpha-negative (TCR-α−/−) mice which lack αβ T cells (65). To analyze the relationship between bacterial virulence, LPS structure, and antigen presentation, we studied the ability of αβ T-cell hybridomas to respond to both smooth and rough strains of Salmonella. We report a decreased murine T-cell response to antigens associated with strains expressing a wild-type LPS compared to the T-cell response to the same antigens present in rough mutants. The decreased response does not appear to be caused by differences in uptake by phagocytic cells, IL-12 production, major histocompatibility complex (MHC) class II expression, or a globally suppressive event. While the mechanism underlying the differential virulence of smooth and rough strains remains unknown, the generation of epitopes for a T-cell response appears to play a role. T-cell hybridomas reactive to rough strains are not reactive to smooth strains except at significantly longer antigen processing times. Smooth strains, because of their complete LPS structure, may be more resistant to antigen-processing enzymes and may block or slow the processing or availability of particular epitopes. This may impede the activation of T cells and account, in part, for the virulence of Salmonella.
MATERIALS AND METHODS
Bacterial strains.
The Lipopolysaccharide Mutant Kit (Kit 1), containing strains of Salmonella typhimurium, was obtained from the Salmonella Genetic Stock Centre (University of Calgary, Calgary, Alberta, Canada). Strains LT2 and SL1004 were kindly provided by John Spitznagel (Emory University School of Medicine, Atlanta, Ga.). The strains of S. typhimurium used in this study and their relevant traits are listed in Table 1.
TABLE 1.
LPS chemotype and partial genotype of S. typhimurium strains used in this study
| Strain | LPS chemotype | Genotype | Reference |
|---|---|---|---|
| LT2 | Smooth | rfa+ | 71 |
| SL3770 | Smooth | rfa+ | 55 |
| SA1355 | Smooth | rfa+ | 7 |
| SL3749 | Ra | rfaL446 | 55 |
| SA1627 | Ra | Δhis(rfb-his)642 | 48 |
| SL733 | Rb1 | rfaK953 | 69 |
| SL3750 | Rb2 | rfaJ417 | 55 |
| SL3748 | Rb3 | rfaI432 | 55 |
| SL1306 | Rc | galE503 | 37 |
| SL3769 | Rd1 | rfaG471 | 55 |
| SL1004 | Rd1 | rfaG571 | 46 |
| SL3789 | Rd2 | rfaF511 | 55 |
| SA1377 | Re | rfaC630 | 7 |
| SL1102 | Re | rfaE543 | 55 |
| SL3600 | Re | rfaD657 | 33 |
Mice and injections.
Female C3HeB/FeJ mice obtained from the Jackson Laboratory (Bar Harbor, Maine) were used at 8 to 12 weeks of age. Mice were housed in microisolator cages, with laboratory chow and water available ad libitum. Injections of live S. typhimurium SL1004 or SA1377 were done i.p. (108 bacteria/mouse). Previous studies have shown that the outcome is the same by several routes of infection in mice (i.p., i.v., and oral) (23, 50).
S. typhimurium preparation.
Heat-killed S. typhimurium (HKST) strains were prepared by growing bacteria in 50 ml of brain heart infusion (BHI) broth. The cultures were placed on a shaker and incubated overnight at 37°C. Bacteria were then pelleted by centrifugation, washed twice in cold phosphate-buffered saline (PBS), diluted in cold PBS to a concentration of 109/ml, and heated for 90 min in an 80°C water bath. To confirm heat killing, bacteria were plated on individual BHI plates.
Isolation of PEC.
Eight days after injection with bacteria, peritoneal exudate cells (PEC) were recovered by peritoneal lavage with cold (4°C) Hanks balanced salt solution containing 0.06% bovine serum albumin, 10 mM HEPES buffer, 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 10 U of heparin per ml. Cells were centrifuged and resuspended in culture medium: RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum (FCS), 5 × 10−5 M 2-mercaptoethanol, 0.5 mM sodium pyruvate, 10 mM HEPES buffer, 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 2 mM l-glutamine. The recovered population was approximately 60% lymphocytes and 40% macrophages by morphology.
ConA-elicited macrophages.
C3HeB/FeJ mice were injected i.p. with 100 μg of concanavalin A (ConA). After 3 to 5 days, the PEC were recovered as described above and added to 96-well tissue culture plates (105/well). Nonadherent cells were removed by washes with warm culture medium and aspiration. The remaining adherent population was >98% macrophages based on morphology.
Assay media.
For experiments using PEC as a source of antigen-presenting cells (APC) and T cells (see Fig. 1 and 2), the culture medium was RPMI 1640 supplemented with 10% FCS (heat inactivated), 5 × 10−5 M 2-mercaptoethanol, 0.5 mM sodium pyruvate, 10 mM HEPES buffer, 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 2 mM l-glutatmine. The experiments for which the results are shown in Fig. 6 and Table 2 were also conducted with this medium formulation. For experiments using T-cell hybridomas (see Fig. 3, 4, 5, and 7) the culture medium was Dulbecco’s modification of Eagle’s medium (DMEM) plus 50 U of penicillin per ml, 50 μg of streptomycin per ml, 2 mM l-glutamine, and 10% FCS (heat inactivated).
FIG. 1.
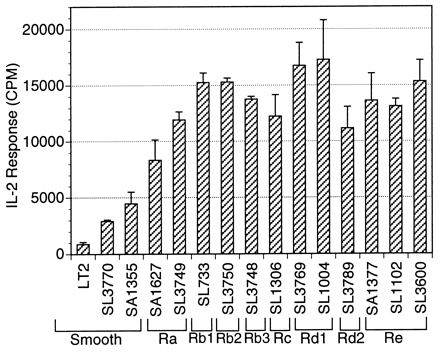
T cells from the PEC population respond to rough strains of heat-killed S. typhimurium better than to smooth strains in a dose-dependent manner. PEC from mice immunized with 108 live SL1004 organisms per mouse were combined with 107 heat-killed bacteria per ml in 96-well microtiter plates at a concentration of 3 × 105/well for 24 h. The levels of IL-2 in the resulting supernatants were measured by using the IL-2-dependent cell line HT-2 as an indicator. The IL-2 response, recorded in counts per minute (CPM), is expressed as the means ± SD (error bars) for triplicate wells.
FIG. 2.
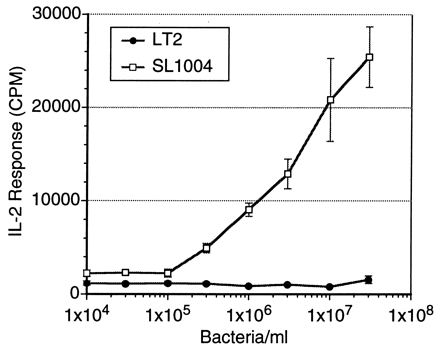
T cells in the PEC population from S. typhimurium SL1004-immunized mice respond to HKST SL1004 (107 organisms/ml), a rough LPS mutant, as the dose increases. The T cells are minimally responsive to wild-type heat-killed LT2. PEC (3 × 105/ml) were combined with 107 heat-killed bacteria per ml in 96-well microtiter plates for 24 h. The level of IL-2 in the resulting supernatants were measured by using the IL-2-dependent cell line HT-2 as an indicator. The IL-2 response, recorded in counts per minute (CPM), is expressed as the means ± SD (error bars) for triplicate wells.
FIG. 6.
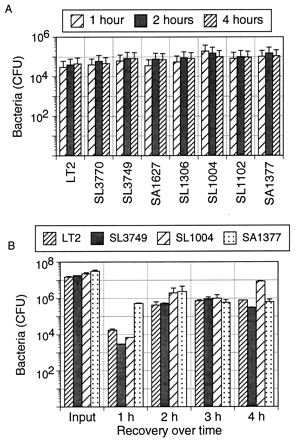
Uptake of the wild-type strain and uptake of LPS mutant strains of S. typhimurium by macrophages are similar. (A) Bacteria were labeled overnight with 20 μCi of [3H]thymidine per ml, washed three times with PBS, and heat killed (80°C for 1 h). The bacteria were added to ConA-elicited macrophages in a 96-well format at a ratio of 10:1. Macrophages were lysed at intervals with 0.05% Triton X-100, and the lysate was dried onto a Spot Plate (Packard). Radioactivity was determined in a Matrix 96 direct beta counter. The number of bacteria were calculated from a standard curve (relating counts per minute and the number of bacterial for each strain) generated from dilutions of heat-killed bacteria dried onto the Spot Plates. (B) Uptake of live S. typhimurium by ConA-elicited macrophages. Macrophages were cultured with live bacteria at a ratio of 1:100 for 1 to 4 h and then lysed with Triton X-100 at each time indicated. Dilutions of input and recovered bacteria were plated on BHI agar for counting.
TABLE 2.
Effect of heat-killed Salmonella on IL-12 production and MHC class II expression by macrophages
| Strain | IL-12 (pg/ml)a | MHC class II (OD405)b |
|---|---|---|
| LT2 | 17,654 ± 893 | 1.57 ± 0.16 |
| SL3749 | 20,824 ± 2,413 | 1.50 ± 0.14 |
| SL1004 | 25,422 ± 2,656 | 1.51 ± 0.15 |
| SA1377 | 19,345 ± 2,629 | 1.53 ± 0.18 |
After 5 days, thioglycolate-elicited peritoneal macrophages were isolated and cultured with 105 heat-killed Salmonella organisms per ml for 24 h. IL-12p40 was detected in the supernatant by a sandwich ELISA. Absorbance was read at 405 nm in a microplate reader. Concentrations were determined with the linear range of a standard curve. All strains tested induced similar levels of IL-12 independent of LPS phenotype.
Thioglycolate-elicited macrophages were isolated and cultured with 107 heat-killed Salmonella organisms per ml for 24 h. The macrophages were then fixed with 0.05% glutaraldehyde. Surface MHC class II molecules were detected with a biotinylated anti-I-Ek antibody. Absorbance was read at 405 nm in a microplate reader. Background was measured with biotinylated mouse IgG. In this experiment the background measured between 0.20 and 0.25. The macrophages all had similar levels of surface MHC class II molecules regardless of incubation with smooth or rough bacteria.
FIG. 3.
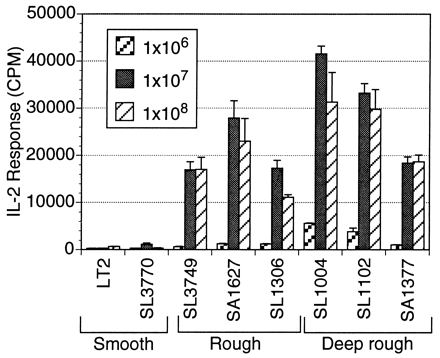
A monoclonal T-cell hybridoma (18-15.18) responds to an antigen(s) from rough strains in a dose-dependent manner. Hybridoma 18-15.18 produces more IL-2 in the presence of LPS mutant strains than in the wild-type LT2 strain. The response increases as LPS length decreases. ConA-elicited macrophages (105/well) were used as APC and combined in a 96-well format with 105 T-cell hybridoma 18-15.18 cells and 107 HKST organisms per well for 24 h. The IL-2 response, recorded in counts per minute (CPM), is expressed as the means ± SD (error bars) for triplicate wells.
FIG. 4.
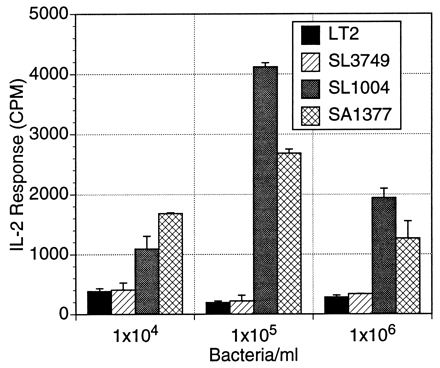
T-cell hybridoma 18-15.18 responds to live rough LPS mutant bacteria better than to live smooth wild-type bacteria in a dose-dependent manner. ConA-elicited macrophages (105/well) were incubated with live, PBS-washed S. typhimurium bacteria in various concentrations for 2 h. The extracellular bacteria were then removed, and the macrophages were fixed with glutaraldehyde. The T-cell hybridoma was added (105 cells/well) and incubated for 24 h. The IL-2 production in the supernatants was measured. The IL-2 response, recorded in counts per minute (CPM), is expressed as the means ± SD (error bars) for triplicate wells.
FIG. 5.
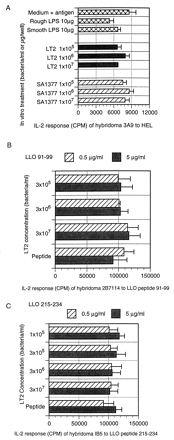
(A) The presence of purified LPS or HKST does not suppress the ability of T-cell hybridoma 3A9 to respond to hen egg lysozyme. (B) Wild-type LT2 does not abrogate the MHC class I-restricted response of T-cell hybridoma 2B7114 to listeriolysin O (LLO) peptide 91–99. (C) LT2 does not diminish the MHC class II-restricted response of T-cell hybridoma IB5 to listeriolysin O (LLO) peptide 215–234. Purified LPS, heat-killed LT2, heat-killed SA1377, and peptide were added together in various concentrations to hybridoma (105 cells/well) and ConA-elicited macrophages (105/well) in a 96-well format and incubated 24 h. The supernatants were evaluated for IL-2 by using the HT-2 cell line as an indicator. The IL-2 response, recorded in counts per minute (CPM), is expressed as the means ± SD (error bars) for triplicate wells. Because hybridomas are specific, cross reactivity did not interfere with the response as shown.
FIG. 7.
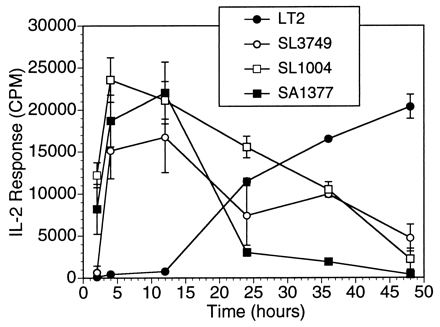
Epitopes for T-cell activation are present in the smooth strain and are revealed over longer processing times. The response of T-cell hybridoma 18-15.18 to heat-killed S. typhimurium LT2 in vitro increases after 24 h of processing. Heat-killed bacteria (107/ml) were incubated with ConA-elicited macrophages (105/well) for various intervals. At each time indicated, macrophages were washed and fixed with 0.05% glutaraldehyde. T cells (hybridoma 18-15.18 [105 cells/well]) were added for 24 h. IL-2 production in supernatants was measured. The IL-2 response, recorded in counts per minute (CPM), is expressed as the means ± SD (error bars) for triplicate wells.
Antigen-specific T-cell hybridomas.
T-cell hybridomas were produced with minor adaptations of previously published techniques (56). Mice were injected with live S. typhimurium SL1004 as described previously. Eleven days later, PEC were harvested as described above. T-cell enrichment was accomplished by the removal of cells adherent to tissue culture dishes and nylon wool. T cells were resuspended in DMEM plus 50 U of penicillin per ml, 50 μg of streptomycin per ml, 2 mM l-glutamine, and 10% FCS and added to 24-well plates containing irradiated spleen cells from syngeneic mice, with heat-killed SL1004 serving as the antigen (107 organisms/ml). After 4 days, T cells were fused to cell line BW1100 (66) by using polyethylene glycol according to standard techniques (31). All hybridomas were subcloned by limiting dilution to ensure monoclonality. The T-cell hybridoma used in experiments presented here, 18-15.18, is I-Ek restricted and expresses the αβ T-cell receptor (unpublished observations). 18-15.18 responds in vitro to antigens from S. typhimurium and is cross reactive with S. dublin, S. minnesota, and Escherichia coli. 18-15.18 does not react either to purified preparations of S. typhimurium LPS, smooth, Ra, Rb, Rc, or Rd (Sigma Chemical Co., St. Louis, Mo.) or to gram-positive organisms (data not shown).
T-cell activation assay.
For experiments using PEC as a source of T cells (see Fig. 1 and 2), the concentration of cells was 3 × 105/well in a 96-well format. Bacteria were added for a period of 24 h. Supernatants were removed and the response was determined by the IL-2 assay described above. In the experiments for which the results are shown in Fig. 3, 4, 5, and 7, hybridomas 18-15.18, 3A9 (I-Ak restricted, hen egg lysozyme specific) (2), 2B7114 (H-2Kd restricted, listeriolysin O [LLO] [peptide 91–99] specific) (21), and IB5 (I-Ek restricted, LLO [peptide 215–234] specific) (56) were used at a concentration of 105 cells/well with ConA-elicited macrophages as APC (105/well). Response to the antigen was determined by IL-2 production. This protocol was modified to monitor the kinetics of antigen processing (see Fig. 7). Various concentrations of HKST were incubated with ConA-elicited macrophages (105/well) for 2 to 48 h. The extracellular bacteria were removed by washing, and the macrophages were fixed with 0.05% glutaraldehyde at each time point. T-cell hybridomas were then added for 24 h, and the response was determined by the IL-2 assay as described below.
Measurement of IL-2.
IL-2 was used as an indicator of T-cell responsiveness as described previously (42). Twenty-four-hour cell-free supernatants from the T-cell activation assay were added to a 96-well plate containing an IL-2-dependent cell line (HT-2, 104 cells/well). The mixture was incubated at 37°C for 24 h. Cells were then pulsed with [3H]thymidine (1 μCi/well) and incubated at 37°C for an additional 18 to 24 h. Cells were harvested onto a glass filter with a Micromate 196 cell harvester (Packard Instrument Co., Inc., Downers Grove, Ill.) and then counted in a Matrix 96 counter (Packard Instrument). The number of counts per minute are directly related to the amount of IL-2 produced based on the linear portion of a standard curve. All assays were performed in triplicate, and results were reported as the average number of counts per minute (± standard deviation) for the triplicate samples.
Measurement of bacterial uptake.
Bacteria were labeled in a manner similar to previously published techniques (9). Bacteria were grown in BHI medium containing 20 μCi of [3H]thymidine for 24 h. The bacteria were pelleted by centrifugation, resuspended in cold PBS, and washed three times. The bacteria were killed by heating in an 80°C water bath for 1 h. The bacteria were plated before and after heat killing on BHI plates to determine the number and inactivation. After the heat-killing procedure, the number of bacteria was determined in a Petroff-Hausser counting chamber. The bacteria were added to ConA-elicited macrophages for various lengths of time. The macrophages were washed three times with warm RPMI to remove extracellular bacteria and then lysed with cold 0.05% Triton X-100. The lysate was dried onto a Spot Plate (Packard Instrument). The level of radioactivity was determined in a Matrix 96 direct beta counter (Packard Instrument). A standard curve was generated by adding several dilutions to the Spot Plate for each bacterial strain. The number of bacteria in each experimental sample was determined by comparing the counts per minute to the counts per minute/number of bacteria for the standard. Uptake of live S. typhimurium was determined by centrifuging, plating, and counting of the bacteria as described above. The bacteria were added to ConA-elicited macrophages for various lengths of time in antibiotic-free RPMI. At each interval, the macrophages were washed three times with RPMI containing gentamicin (50 μg/ml) to remove and kill any extracellular bacteria and then were lysed with 0.05% Triton X-100, and the lysates were diluted in PBS and plated on BHI plates. The plates were incubated overnight at 37°C.
IL-12 ELISA.
Female C3HeB/FeJ mice were injected i.p. with 2.5 ml of thioglycolate. After 5 days, the peritoneal macrophages were isolated and cultured with 105 heat-killed Salmonella organisms per ml for 24 h. IL-12p40 was detected in the supernatant by sandwich enzyme-linked immunosorbent assay (ELISA) (58). C17.8.20 was used as the capture antibody and C15.6.7.6 served as the detection antibody. Both antibodies recognize epitopes on the p40 chain of murine IL-12 and were provided by G. Trinchieri (Wistar Institute, Philadelphia, Pa.). Extravidin alkaline phosphatase was used to bind to the biotinylated detection antibody and p-nitrophenylphosphate served as the substrate. Absorbance was read at 405 nm by using a Microplate Autoreader model EL 311SX (Bio-Tek Instruments, Inc., Winooski, Vt.). The relationship between absorbance and IL-12 concentration was linear between 25 and 5,000 pg/ml, with correlation coefficients consistently >0.990.
MHC class II ELISA.
Female C3HeB/FeJ mice were injected i.p. with 100 μg of ConA. After 5 days, the peritoneal macrophages were isolated and cultured with 107 heat-killed Salmonella organisms per ml for 24 h. The macrophages were then fixed with 0.05% glutaraldehyde. Surface MHC class II molecules were detected by using a biotinylated anti-I-Ek antibody. Extravidin alkaline phosphatase was used to bind to the biotinylated detection antibody and p-nitrophenylphosphate served as the substrate. Absorbance was read at 405 nm with a Microplate Autoreader model EL 311SX (Bio-Tek Instruments). Background was measured by using biotinylated mouse immunoglobulin G (IgG).
RESULTS
Differential reactivity of T cells from Salmonella-immunized mice.
An initial observation showed that T cells isolated from S. typhimurium SL1004-immunized mice were reactive in vitro to heat-killed SL1004 but not to the wild-type LT2 strain. This observation was intriguing since SL1004 differs from LT2 only in LPS structure. This finding suggested that the complete LPS structure was somehow interfering with antigen presentation and/or T-cell recognition.
To study the effects of LPS on T-cell responsiveness, a panel of mutants of LT2 with progressively decreasing lengths of LPS was assembled (Table 1). Most of these mutants are otherwise isogenic. For example, strains LT2 and SA1377 are isogenic, except for the LPS, and generate very different patterns of T-cell reactivity. Figure 1 is representative of the results for several experiments that illustrated the T-cell response to the truncated LPS mutants. In these experiments, peritoneal T cells and macrophages from SL1004-immunized mice reacted in vitro with the heat-killed bacteria. The T-cell response to the strains increased as their LPS lengths decreased in the following ways. A significant increase in response occurred in the LPS transition from smooth to Ra and from Ra to Rb. The T-cell response reached a plateau at this point but in numerous experiments reached a maximum response to the Rd mutant SL1004 as seen from the results presented in later figures. The differential response of the T cells could be seen over a range of bacterial concentrations (105/ml to 108/ml), with 107/ml chosen as a representative dose for future experiments (Fig. 2).
Response of Salmonella-specific hybridomas to live and killed bacteria.
Salmonella-specific T-cell hybridomas were constructed and defined to explore the differential T-cell responses to rough and smooth bacteria in a monoclonal T-cell population. Figure 3 is representative of the data obtained from in vitro assays that tested the reactivity of T-cell hybridoma 18-15.18 to heat-killed Salmonella strains, with ConA-elicited macrophages as APC. At several antigen concentrations, the trend was the same: T cells reacted more strongly to the truncated LPS mutants than to the wild-type strain LT2 at all doses. Again, the T-cell response increased as the LPS length decreased. Increasing the antigen dose by at least 100-fold did not overcome the lack of T-cell activation by the smooth strains. These experiments with T-cell hybridomas indicated that the differential response was not due to variability in polyclonal T-cell populations found in the PEC population. A similar pattern of differential responsiveness was observed with 21 independent T-cell hybridomas including those generated from immunizations with S. typhimurium LT2, SL1004, and SA1377.
For most experiments, heat-killed bacteria were used to avoid complications arising from intracellular survival and replication. However, it is important that the same trend of T-cell reactivity was observed in experiments using live bacteria in vitro. In Fig. 4, the bacteria were allowed to infect adherent ConA-elicited macrophages for 2 h before washing and fixing. T cells responded to the rough strains but not to the smooth strain. At high bacterial loads, there was some cytotoxicity to the macrophages (microscopic inspection). Both rough and smooth strains caused cytotoxicity at high doses. Differential presentation due to LPS structure was also observed for Salmonella inactivated by several mechanisms: phenol, ethanol, formalin, and acetone (data not shown). These results indicated that the differential responses were similar with live and dead bacteria and were not influenced by the method used to kill the bacteria.
Salmonella LT2 does not globally suppress antigen processing or presentation in vitro.
Several investigators have proposed that LT2 or wild-type LPS is suppressive (3, 40). Because of this possibility, we investigated the ability of LT2 to hinder the response of MHC class I- and class II-restricted T-cell hybridomas to antigens not associated with Salmonella. The addition of heat-killed LT2 or purified LPS to hybridoma 3A9 did not suppress its ability to respond to the protein antigen HEL (Fig. 5A). Hybridomas 2B7114 and IB5 recognize LLO peptides 91–99 and 215–234, respectively, from Listeria monocytogenes. LT2 did not globally suppress the response to either LLO peptide (Fig. 5B and C). To determine if the presence of LT2 diminished the T-cell response to SL1004, PEC populations from SL1004-immunized mice were restimulated in vitro with heat-killed SL1004 plus various concentrations of heat-killed LT2. Even at the highest doses of LT2, reactivity to SL1004 was still apparent (data not shown). Thus, LT2 did not influence the ability of several hybridomas to respond to epitopes offered as whole proteins or synthetic peptides nor did it impair the response of peritoneal T cells to bacterial antigens.
Differential T-cell response is not accounted for by bacterial uptake, macrophage IL-12 induction, MHC class II expression, or LPS responsiveness.
To rule out the possibility that the differences in T-cell reactivities were due to differences in the amount of available antigen, two different methods were used to determine the level of uptake of the S. typhimurium strains by macrophages. In Fig. 6A, [3H]thymidine-labeled heat-killed bacteria were added to macrophages for various lengths of time. The macrophages were washed thoroughly and then lysed to obtain only the cell-associated bacteria. There were no differences in uptake among eight strains ranging from smooth to deep rough in LPS character. In Fig. 6B, macrophages were cultured with live bacteria for periods up to 4 h, rinsed with medium containing the antibiotic gentamicin, and then lysed with Triton X-100. The antibiotic kills extracellular bacteria that may be attached to the macrophage surface; therefore, the number of recovered bacteria is a measure of those that were ingested. It appears that the number of both rough and smooth bacteria decreased after 1 h and then increased equally over time. Minor differences in uptake were within the limits of experimental error and could not account for the greatly reduced response to LT2 (Fig. 2 and 3). Uptake does not appear to be the reason epitopes from smooth strains are not presented effectively to T cells.
When macrophages respond to LPS, they produce IL-12 and are activated to upregulate the level of MHC class II surface expression. Both of these factors could influence the T-cell response to a particular strain of S. typhimurium. These macrophage activation parameters were measured in vitro after incubation with smooth and rough bacteria. Table 2 shows results for four representative strains with different LPS structures. The levels of IL-12 induction and MHC class II expression were similar among the rough and smooth strains.
The observations of differential T-cell reactivities to smooth and rough strains were also noted for T cells and macrophages from LPS-nonresponder mice, C3H/HeJ (59) (data not shown). These experiments revealed that bacterium-associated smooth LPS influenced the immune response independently of the ability of the cells to respond to lipid A.
With longer processing times, epitopes from smooth strains are presented to T cells.
The kinetics of processing and presentation of HKST strains were evaluated. In this experiment (Fig. 7), ConA-elicited macrophages were pretreated with HKST for various times and then fixed with glutaraldehyde. The monoclonal T-cell hybridoma (18-15.18) was added after fixation and the IL-2 response was determined. In keeping with other studies of antigen processing (70), there was a rapid increase in effective presentation of SL1004 and SA1377 in 1 to 2 h. In contrast, during the first 12 h, the epitope from LT2 was not presented as effectively as the same epitope carried by SL1004 or SA1377. However, with longer processing times (>24 h), there was a vigorous response to LT2. These results indicated that the relevant epitope was present in LT2, but because of LPS-mediated interference, longer processing times were required.
DISCUSSION
These experiments provide insight into how S. typhimurium interacts with macrophages and T cells to generate an effective immune response. CD4+ T cells have been shown to be protective against Salmonella infection via adoptive transfer or depletion of T cell subsets (17, 47). While antibodies and, to a lesser extent, cytotoxic T lymphocytes can play a role in the immune response to Salmonella, protective immunity against oral or i.p. challenge can be primarily mediated by CD4+ T cells (22, 38, 65). Our studies have focused specifically on the activation of CD4+ T cells by Salmonella antigens. We have observed that this subset of T cells is less responsive to wild-type Salmonella strains than to avirulent strains in vitro (Fig. 1). Our results agree with those of previous studies which showed a diminished response to antigens associated with smooth strains of Salmonella (67). The major difference between the wild-type strains and the mutant strains used in this study is the presence of a full-length LPS. Because the T cells that we used did not respond directly to LPS and because CD4+ T cells generally recognize peptide epitopes (15), it seemed possible that LPS was influencing the generation of peptide epitopes at the level of the macrophage.
We observed an increase in the T-cell response to Salmonella strains with truncated LPS structures. This observation, though initially evaluated with polyclonal T-cell populations (Fig. 1), was also confirmed with a monoclonal T-cell population (Fig. 3) and later with 21 additional independent T-cell hybridomas (data not shown). The hybridoma illustrated here was reactive to an epitope associated with an outer membrane protein preparation in the context of I-Ek.
Our data indicated that LT2 did not adversely affect the presentation of antigens not associated with the LT2 organism. Previously, studies in our laboratory showed that live LT2 organisms did not diminish the response of T-cell hybridoma D011.10 to ovalbumin (8). This finding indicated that LT2 was not globally suppressing processing of other antigens. The addition of heat-killed LT2 bacteria or purified LPS also did not interfere with the processing and presentation of hen egg lysozyme to a specific hybridoma, 3A9 (Fig. 5). Wild-type Salmonella also does not suppress the responses of Listeria-specific hybridomas to their peptide ligand. The hybridomas IB5 and 2B7114 mixed with their peptide ligand and various amounts of LT2 displayed a normal response (Fig. 5B and C). LT2 also did not diminish the T-cell hybridoma response to antigens displayed by SL1004 when the two strains were mixed in vitro with APC and a Salmonella-specific hybridoma (data not shown). While others have noted suppressive mechanisms associated with Salmonella (4, 12, 20), these mechanisms appeared to operate for longer periods of time and by using proliferation and antibody formation as readouts of immune function. Some suppressive mechanisms may operate via cytokines and NO production by macrophages (24, 39). Our use of T-cell hybridomas with minimal activation requirements may represent a system that is not susceptible to the globally suppressive activities of Salmonella as others have noted.
In many instances, rough strains of bacteria have been shown to be taken up preferentially by phagocytic cells (16, 35, 67). Conversely, in other studies, smooth and rough strains have been shown to have no significant differences in the rates of phagocytic uptake (5, 25, 51, 68). We confirmed and extended these observations with eight representative S. typhimurium strains (Fig. 6), using both live and killed bacteria. The effective association of bacteria with macrophages was monitored by uptake of radiolabeled dead bacteria, by the uptake and survival of live bacteria, and also by the ability of bacteria to induce cytokine (IL-12) production (Table 2). In all cases, it is apparent that both smooth and rough strains can associate equally with macrophages as monitored by both physical and functional parameters. Both smooth and rough strains bind LPS binding protein equally, which in turn binds to the CD14 receptor on macrophages (61). The levels of uptake mediated via CD14 should then be similar. While we and others (67) have noted very small differences in uptake levels of rough and smooth strains, it is clear that the small changes in uptake cannot account for the ≥100-fold differences in antigen presentation when smooth and rough strains are compared.
Our studies indicate that Salmonella can alter antigen processing and presentation during the first 12 h of bacterium-macrophage interaction by using T-cell hybridoma activation as our readout. The activation of T-cell hybridomas can occur independently of cytokines and accessory molecules (29). This readout coupled with the use of fixed APC effectively isolates potential regulatory effects at the level of antigen processing and presentation. Allowing additional time for the macrophages to process the heat-killed strains revealed that antigens from LT2 could be presented to T-cell hybridomas (Fig. 7). Thus, rather than the absence of the relevant epitope in LT2, it is apparent that the presence of smooth LPS delays the generation of the epitope during antigen processing. These results are compatible with a physical blocking or masking of epitopes by the polysaccharide portion of LPS. In fact, polysaccharides have been shown to interfere with MHC class II-restricted antigen presentation (18, 34).
It is possible that protein epitopes are protected from proteolysis in the endosome by LPS. Outer membrane proteins have been shown to be closely associated with LPS and may be tightly bound to LPS (1, 19, 32, 60). The phenomenon of LPS masking of outer membrane proteins has been observed for Neisseria gonorrheae in which LPS inhibits binding of cathepsin B to three outer membrane proteins (57). LPS also has been shown to block the binding of bacteriophages and colicins to their outer membrane protein receptors in smooth strains of E. coli (62). S. typhimurium strains with complete LPS structures are insensitive to phage attack, possibly due to steric hindrance by the O side chain (49). Smooth strains are resistant to lysis by the complement pathway membrane attack complex because the complex is prevented by LPS from assembling at the membrane surface (27, 28). It is conceivable that the LPS could disrupt or interfere with processing and presentation of LT2 antigens, especially antigens present in the outer membrane. If the polysaccharide portion of LPS interferes with the processing and availability of certain epitopes, it would follow that the repertoires of T cells generated by smooth and rough strains would be different. In this regard, our preliminary studies do indicate differences in the specificities of such T cells. While the vast majority of T-cell hybridomas generated from mice immunized with rough strains behave as the representative one described here (Fig. 3, rough strains≫smooth strains), we have noted that T-cell hybridomas which recognize epitopes that do not appear to be controlled by the complete polysaccharide portion of LPS exist. Some of these hybridomas, generated from mice immunized with smooth strains, react equally well with both smooth and rough strains. These results indicate that the specificity of the TCR influences the relative response to smooth and rough strains and further indicates that the level of control mediated by LPS is at the level of epitope generation or availability. These results not only have implications for the mechanism of virulence by Salmonella but also relate to the use of Salmonella as a vaccine strain or carrier of passenger epitopes.
In this study, we have attempted to understand the mechanisms by which LT2 is more virulent than strains with truncated LPS by focusing on the initial events in the bacterium-macrophage interaction. Our studies indicate that inefficient antigen processing and presentation are linked to virulence. Thus, we suggest that part of the complex phenomenon of virulence is mediated by altered antigen processing and differential T-cell recognition.
ACKNOWLEDGMENTS
This work was supported in part by National Institute of Allergy and Infectious Diseases grants RO1 AI-35285 and AI-34065.
The advice and technical assistance provided by Marianne Skeen and Mark Miller are greatly appreciated.
REFERENCES
- 1.Alberti S, Rodriquez-Quinones F, Schirmer T, Rummel G, Tomas J M, Rosenbusch J P, Benedi V J. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect Immun. 1995;63:903–910. doi: 10.1128/iai.63.3.903-910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen P M, Unanue E R. Processing and presentation of hen egg-white lysozyme by macrophages. Immunobiology. 1984;168:182–188. doi: 10.1016/S0171-2985(84)80109-6. [DOI] [PubMed] [Google Scholar]
- 3.Al-Ramadi B K, Brodkin M A, Mosser D M, Eisenstein T K. Immunosuppression induced by attenuated Salmonella. Evidence for mediation by macrophage precursors. J Immunol. 1991;146:2737–2746. [PubMed] [Google Scholar]
- 4.Arai T, Matsui K. A purified protein from Salmonella typhimurium inhibits high-affinity interleukin-2 receptor expression on CTLL-2 cells. FEMS Immunol Med Microbiol. 1997;17:155–160. doi: 10.1111/j.1574-695X.1997.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 5.Buchmeier N A, Heffron F. Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect Immun. 1991;59:2232–2238. doi: 10.1128/iai.59.7.2232-2238.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatfield S N, Dougan G, Roberts M. Progress in the development of multivalent oral vaccines based on live attenuated Salmonella. In: Kurstak E, editor. Modern vaccinology. New York, N.Y: Plenum Medical; 1994. pp. 55–86. [Google Scholar]
- 7.Chatterjee A K, Sanderson K E, Ross H. Influence of temperature on growth of lipopolysaccharide-deficient (rough) mutants of Salmonella typhimurium and Salmonella minnesota. Can J Microbiol. 1976;22:1540–1548. doi: 10.1139/m76-226. [DOI] [PubMed] [Google Scholar]
- 8.Cluff C W, Garcia M, Ziegler H K. Intracellular hemolysin-producing Listeria monocytogenes strains inhibit macrophage-mediated antigen processing. Infect Immun. 1990;58:3601–3612. doi: 10.1128/iai.58.11.3601-3612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies W A. Kinetics of killing Listeria monocytogenes by macrophages: correlation of 3H-DNA release from labeled bacteria and changes in numbers of viable organisms by mathematical model. J Reticuloendothel Soc. 1982;32:461–476. [PubMed] [Google Scholar]
- 10.Dunlap N E, Benjamin W, Jr, Briles D E. The intracellular nature of Salmonella infection during the early stages of mouse typhoid. Immunol Ser. 1994;60:303–312. [PubMed] [Google Scholar]
- 11.Eisenstein T K, Killar L M, Sultzer B M. Immunity to infection with Salmonella typhimurium: mouse-strain differences in vaccine- and serum-mediated protection. J Infect Dis. 1984;150:425–435. doi: 10.1093/infdis/150.3.425. [DOI] [PubMed] [Google Scholar]
- 12.Eisenstein T K, Meissler J J, Jr, Miller S I, Stocker B A. Immunosuppression and nitric oxide production induced by parenteral live Salmonella vaccines do not correlate with protective capacity: a phoP::Tn10 mutant does not suppress but does protect. Vaccine. 1998;16:24–32. doi: 10.1016/s0264-410x(97)00160-6. [DOI] [PubMed] [Google Scholar]
- 13.Flad H D, H. L, Rietschel E T, Ulmer A J. Agonists and antagonists for lipopolysaccharide-induced cytokines. Immunobiology. 1993;187:303–316. doi: 10.1016/S0171-2985(11)80346-3. [DOI] [PubMed] [Google Scholar]
- 14.Flebbe L, Vukajlovich S W, Morrison D C. Immunostimulation of C3H/HeJ lymphoid cells by R-chemotype lipopolysaccharide preparations. J Immunol. 1989;142:642–652. [PubMed] [Google Scholar]
- 15.Fremont D H, Hendrickson W A, Marrack P, Kappler J. Structures of an MHC class II molecule with covalently bound single peptides. Science. 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg D, Shilo M. Role of cell wall structure of Salmonella in the interaction with phagocytes. Infect Immun. 1970;2:279–285. doi: 10.1128/iai.2.3.279-285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautreaux M D, Deitch E A, Berg R D. T lymphocytes in host defense against bacterial translocation from the gastrointestinal tract. Infect Immun. 1994;62:2874–2884. doi: 10.1128/iai.62.7.2874-2884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Fernandez M, Carrasco-Marin E, Alvarez-Dominguez C, Outschoorn I M, Leyva-Cobian F. Inhibitory effects of thymus-independent type 2 antigens on MHC class II-restricted antigen presentation: comparative analysis of carbohydrate structures and the antigen presenting cell. Cell Immunol. 1997;176:1–13. doi: 10.1006/cimm.1996.1078. [DOI] [PubMed] [Google Scholar]
- 19.Hancock R E W, Karunaratne D N, Bernegger-Egli C. Molecular organization and structural role of outer membrane macromolecules. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. New York, N.Y: Elsevier Science Publishers; 1994. pp. 263–279. [Google Scholar]
- 20.Hassan J O, Curtiss R., III Virulent Salmonella typhimurium-induced lymphocyte depletion and immunosuppression in chickens. Infect Immun. 1994;62:2027–2036. doi: 10.1128/iai.62.5.2027-2036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiltbold E M, Safley S A, Ziegler H K. The presentation of class I and class II epitopes of listeriolysin O is regulated by intracellular localization and by intercellular spread of Listeria monocytogenes. J Immunol. 1996;157:1163–1175. [PubMed] [Google Scholar]
- 22.Hougen H P, Jensen E T. Experimental Salmonella typhimurium infections in rats. III. Transfer of immunity with primed lymphocyte subpopulations. APMIS. 1990;98:1015–1021. [PubMed] [Google Scholar]
- 23.Hsu H S. Pathogenesis and immunity in murine salmonellosis. Microbiol Rev. 1989;53:390–409. doi: 10.1128/mr.53.4.390-409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang D, Schwacha M G, Eisenstein T K. Attenuated Salmonella vaccine-induced suppression of murine spleen cell responses to mitogen is mediated by macrophage nitric oxide: quantitative aspects. Infect Immun. 1996;64:3786–3792. doi: 10.1128/iai.64.9.3786-3792.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishibashi Y, Arai T. Specific inhibition of phagosome-lysosome fusion in murine macrophages mediated by Salmonella typhimurium infection. FEMS Microbiol Immunol. 1990;2:35–43. doi: 10.1111/j.1574-6968.1990.tb03476.x. [DOI] [PubMed] [Google Scholar]
- 26.Ivanoff B, Levine M M, Lambert P H. Vaccination against typhoid fever: present status. Bull W H O. 1994;72:957–971. [PMC free article] [PubMed] [Google Scholar]
- 27.Joiner K A, Hammer C H, Brown E J, Cole R J, Frank M M. Studies on the mechanism of bacterial resistance to complement-mediated killing. I. Terminal complement components are deposited and released from Salmonella minnesota S218 without causing bacterial death. J Exp Med. 1982;155:797–808. doi: 10.1084/jem.155.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joiner K A, Hammer C H, Brown E J, Frank M M. Studies on the mechanism of bacterial resistance to complement-mediated killing. II. C8 and C9 release C5b67 from the surface of Salmonella minnesota S218 because the terminal complex does not insert into the bacterial outer membrane. J Exp Med. 1982;155:809–819. doi: 10.1084/jem.155.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kappler J W, Skidmore B, White J, Marrack P. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridomas. Lack of independent antigen and H-2 recognition. J Exp Med. 1981;153:1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kita E, Nishikawa F, Kamikaidou N, Oku D, Yasui K, Kashiba S. Mechanism of the protective immunity against murine typhoid: persistence of Salmonella L forms in the liver after immunization with live-cell vaccines. FEMS Microbiol Immunol. 1992;5:191–199. doi: 10.1111/j.1574-6968.1992.tb05901.x. [DOI] [PubMed] [Google Scholar]
- 31.Kruisbeek A M. Production of mouse T cell hybridomas. In: Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Current protocols in immunology. New York, N.Y: Wiley-Interscience; 1991. p. 3.14.1. [DOI] [PubMed] [Google Scholar]
- 32.Kuusi N, M. N, Saxen H, Makela P H. Immunization with major outer membrane protein (porin) preparations in experimental murine salmonellosis: effect of lipopolysaccharide. Infect Immun. 1981;34:328–332. doi: 10.1128/iai.34.2.328-332.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann V, Hammerling G, Nurminen M, Minner I, Ruschmann E, Luderitz O, Kuo T T, Stocker B A. A new class of heptose-defective mutant of Salmonella typhimurium. Eur J Biochem. 1973;32:268–275. doi: 10.1111/j.1432-1033.1973.tb02607.x. [DOI] [PubMed] [Google Scholar]
- 34.Leyva-Cobian F, Unanue E R. Intracellular interference with antigen presentation. J Immunol. 1988;141:1445–1450. [PubMed] [Google Scholar]
- 35.Liang-Takasaki C J, Makela P H, Leive L. Phagocytosis of bacteria by macrophages: changing the carbohydrate of lipopolysaccharide alters interaction with complement and macrophages. J Immunol. 1982;128:1229–1235. [PubMed] [Google Scholar]
- 36.Licht T R, Krogfelt K A, Cohen P S, Poulsen L K, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacLachlan P R, Sanderson K E. Transformation of Salmonella typhimurium with plasmid DNA: differences between rough and smooth strains. J Bacteriol. 1985;161:442–445. doi: 10.1128/jb.161.1.442-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mastroeni P, Villarreal R B, Hormaeche C E. Adoptive transfer of immunity to oral challenge with virulent Salmonellae in innately susceptible BALB/c mice requires both immune serum and T cells. Infect Immun. 1993;61:3981–3984. doi: 10.1128/iai.61.9.3981-3984.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsui K. A purified protein from Salmonella typhimurium inhibits proliferation of murine splenic anti-CD3 antibody-activated T-lymphocytes. FEMS Immunol Med Microbiol. 1996;14:121–127. doi: 10.1111/j.1574-695X.1996.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 40.Matsui K, Arai T. Immunosuppression induced by Salmonella involves inhibition of tyrosine phosphorylation in murine T lymphocytes. FEMS Immunol Med Microbiol. 1993;7:345–354. doi: 10.1111/j.1574-695X.1993.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 41.Mattsby-Baltzer I, Ahlstrom B, Edebo L, de Man P. Susceptibility of lipopolysaccharide-responsive and -hyporesponsive ItyS Mice to infection with rough mutants of Salmonella typhimurium. Infect Immun. 1996;64:1321–1327. doi: 10.1128/iai.64.4.1321-1327.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller M A, Skeen M J, Ziegler H K. Nonviable bacterial antigens administered with IL-12 generate antigen-specific T cell responses and protective immunity against Listeria monocytogenes. J Immunol. 1995;155:4817–4828. [PubMed] [Google Scholar]
- 43.Mixter P F, Camerini V, Stone B J, Miller V L, Kronenberg M. Mouse T lymphocytes that express a γδ T-cell antigen receptor contribute to resistance to Salmonella infection in vivo. Infect Immun. 1994;62:4618–4621. doi: 10.1128/iai.62.10.4618-4621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mroczenski-Wildey M J, Di Fabio J L, Cabello F C. Invasion and lysis of HeLa cell monolayers by Salmonella typhi: the role of lipopolysaccharide. Microb Pathog. 1989;6:143–152. doi: 10.1016/0882-4010(89)90017-x. [DOI] [PubMed] [Google Scholar]
- 45.Muotiala A, Hovi M, Makela P H. Protective immunity in mouse salmonellosis: comparison of smooth and rough live and killed vaccines. Microb Pathog. 1989;6:51–60. doi: 10.1016/0882-4010(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 46.Nakano M, Saito K. Chemical components in the cell wall of Salmonella typhimurium affecting its virulence and immunogenicity in mice. Nature. 1969;222:1085–1086. doi: 10.1038/2221085a0. [DOI] [PubMed] [Google Scholar]
- 47.Nauciel C. Role of CD4+ T cells and T-independent mechanisms in acquired resistance to Salmonella typhimurium infection. J Immunol. 1990;145:1265–1269. [PubMed] [Google Scholar]
- 48.Nikaido H, Levinthal M, Nikaido K, Nakane K. Extended deletions in the histidine-rough-B region of the Salmonella chromosome. Proc Natl Acad Sci USA. 1967;57:1825–1832. doi: 10.1073/pnas.57.6.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nurminen M, Lounatmaa K, Sarvas M, Makela P H, Nakae T. Bacteriophage-resistant mutants of Salmonella typhimurium deficient in two major outer membrane proteins. J Bacteriol. 1976;127:941–955. doi: 10.1128/jb.127.2.941-955.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Brien A D. Influence of host genes on resistance of inbred mice to lethal infection with Salmonella typhimurium. Curr Top Microbiol Immunol. 1986;124:37–48. [PubMed] [Google Scholar]
- 51.Okamura N, Spitznagel J K. Outer membrane mutants of Salmonella typhimurium LT2 have lipopolysaccharide-dependent resistance to the bactericidal activity of anaerobic human neutrophils. Infect Immun. 1982;36:1086–1095. doi: 10.1128/iai.36.3.1086-1095.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ornellas E P, Roantree R J, Steward J P. The specificity and importance of humoral antibody in the protection of mice against intraperitoneal challenge with complement-sensitive and complement-resistant Salmonella. J Infect Dis. 1970;121:113–123. doi: 10.1093/infdis/121.2.113. [DOI] [PubMed] [Google Scholar]
- 53.Raetz C R. Bacterial endotoxins: extraordinary lipids that activate eucaryotic signal transduction. J Bacteriol. 1993;175:5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reeves P. Biosynthesis and assembly of lipopolysaccharide. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. New York, N.Y: Elsevier Science Publishers; 1994. pp. 281–317. [Google Scholar]
- 55.Roantree R J, Kuo T T, MacPhee D G. The effect of defined lipopolysaccharide core defects upon antibiotic resistances of Salmonella typhimurium. J Gen Microbiol. 1977;103:223–234. doi: 10.1099/00221287-103-2-223. [DOI] [PubMed] [Google Scholar]
- 56.Safley S A, Jensen P E, Reay P A, Ziegler H K. Mechanisms of T cell epitope immunodominance analyzed in murine listeriosis. J Immunol. 1995;155:4355–4366. [PubMed] [Google Scholar]
- 57.Shafer W M. Lipopolysaccharide masking of gonococcal outer-membrane proteins modulates binding of bacterial cathepsin G to gonococci. J Gen Microbiol. 1988;134:539–545. doi: 10.1099/00221287-134-3-539. [DOI] [PubMed] [Google Scholar]
- 58.Skeen M J, Miller M A, Shinnick T M, Ziegler H K. Regulation of murine macrophage IL-12 production. J Immunol. 1996;156:1196–1206. [PubMed] [Google Scholar]
- 59.Sultzer B M, Castagna R, Bandekar J, Wong P. Lipopolysaccharide nonresponder cells: the C3H/HeJ defect. Immunobiology. 1993;187:257–271. doi: 10.1016/S0171-2985(11)80343-8. [DOI] [PubMed] [Google Scholar]
- 60.Sultzer B M, Goodman G W. Endotoxin protein: a B-cell mitogen and polyclonal activator of C3H/HeJ lymphocytes. J Exp Med. 1976;144:821–827. doi: 10.1084/jem.144.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ulevitch R J, Tobias P S. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 62.van der Ley P, de Graaff P, Tommassen J. Shielding of Escherichia coli outer membrane proteins as receptors for bacteriophages and colicins by O-antigenic chains of lipopolysaccharide. J Bacteriol. 1986;168:449–451. doi: 10.1128/jb.168.1.449-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verma N K, Ziegler H K, Stocker B A, Schoolnik G K. Induction of a cellular immune response to a defined T-cell epitope as an insert in the flagellin of a live vaccine strain of Salmonella. Vaccine. 1995;13:235–244. doi: 10.1016/0264-410x(95)93308-v. [DOI] [PubMed] [Google Scholar]
- 64.Verma N K, Ziegler H K, Wilson M, Khan M, Safley S, Stocker B A, Schoolnik G K. Delivery of class I and class II MHC-restricted T-cell epitopes of listeriolysin of Listeria monocytogenes by attenuated Salmonella. Vaccine. 1995;13:142–150. doi: 10.1016/0264-410x(95)93127-u. [DOI] [PubMed] [Google Scholar]
- 65.Weintraub B C, Eckmann L, Okamoto S, Hense M, Hedrick S M, Fierer J. Role of αβ and γδ T cells in the host response to Salmonella infection as demonstrated in T-cell-receptor-deficient mice of defined Ity genotypes. Infect Immun. 1997;65:2306–2312. doi: 10.1128/iai.65.6.2306-2312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White J, Blackman M, Bill J, Kappler J, Marrack P, Gold D P, Born W. Two better cell lines for making hybridomas expressing specific T cell receptors. J Immunol. 1989;143:1822–1825. [PubMed] [Google Scholar]
- 67.Wick M J, Harding C V, Normark S J, Pfeifer J D. Parameters that influence the efficiency of processing antigenic epitopes expressed in Salmonella typhimurium. Infect Immun. 1994;62:4542–4548. doi: 10.1128/iai.62.10.4542-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wick M J, Harding C V, Twesten N J, Normark S J, Pfeifer J D. The phoP locus influences processing and presentation of Salmonella typhimurium antigens by activated macrophages. Mol Microbiol. 1995;16:465–476. doi: 10.1111/j.1365-2958.1995.tb02411.x. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto N, Anderson T F. Genomic masking and recombination between serologically unrelated phages P22 and P221. Virology. 1961;14:430–439. doi: 10.1016/0042-6822(61)90334-8. [DOI] [PubMed] [Google Scholar]
- 70.Ziegler H K, Unanue E R. Decrease in macrophage antigen catabolism caused by ammonia and chloroquine is associated with inhibition of antigen presentation to T cells. Proc Natl Acad Sci USA. 1982;79:175–178. doi: 10.1073/pnas.79.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zinder N D, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64:679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]


