Abstract
The biological impact of ether glycerophospholipids (GP) in peroxisomal disorders and other diseases makes them significant targets as biomarkers for diagnostic assays or deciphering pathology of the disorders. Ether lipids include both plasmanyl and plasmenyl lipids, which each contain an ether or a vinyl ether bond at the sn-1 linkage position, respectively. This linkage, in contrast to traditional diacyl GPs, precludes their detailed characterization by mass spectrometry via traditional collisional-based MS/MS techniques. Additionally, the isomeric nature of plasmanyl and plasmenyl pairs of ether lipids introduces a further level of complexity that impedes analysis of these species. Here, we utilize 213 nm ultraviolet photodissociation mass spectrometry (UVPD-MS) for detailed characterization of phosphatidylethanolamine (PE) and phosphatidylcholine (PC) plasmenyl and plasmanyl lipids in mouse brain tissue. 213 nm UVPD-MS enables the successful differentiation of these four ether lipid subtypes for the first time. We couple this UVPD-MS methodology to reversed-phase liquid chromatography (RPLC) for characterization and relative quantitation of ether lipids from normal and diseased (Pex7 deficiency modeling the peroxisome biogenesis disorder, RCDP) mouse brain tissue, highlighting the ability to pinpoint specific structural features of ether lipids that are important for monitoring aberrant lipid metabolism in peroxisomal disorders.
Graphical Abstract
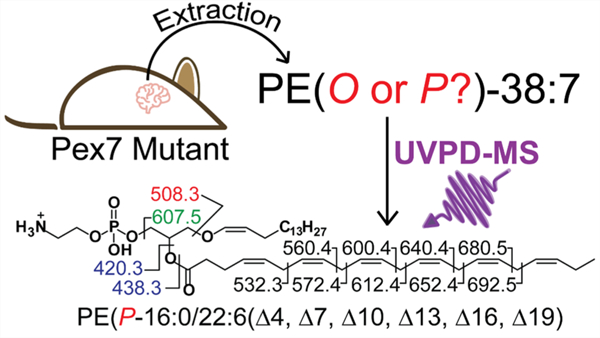
INTRODUCTION
Mammalian glycerophospholipids (GP) exist primarily in diacyl form, i.e. with ester linkages between the sn-1 and sn-2 fatty acids to the glycerol backbone. Ether lipids are a unique type of GP in which the sn-1 chain is connected via an ether or vinyl ether linkage to the glycerol backbone. This seemingly simple structural feature has profound biological implications, and ether lipids have specifically garnered significant interest owing to their mechanistic role in neurological diseases, in particular, peroxisome biogenesis disorders (PBD). There are two PBD groups, Zellweger Spectrum Disorder (ZSD) and Rhizomelic Chondrodysplasia Punctata (RCDP). These are rare but lethal genetically heterogeneous autosomal recessive diseases, characterized by multiple defects in peroxisome functions leading to, among other abnormalities, plasmalogen deficiencies and accumulation of very long chain fatty acids.1–4 In RCDP, plasmalogen deficiency represents the major disease etiology. Peroxisomes are critical to the early, committing steps in ether lipid biosynthesis, and their altered function has clinically devastating outcomes.5–8 Nevertheless, we know very little about the pathophysiological processes. Unraveling the mechanism and underpinnings of PBDs requires more detailed molecular profiles, and this represents a classic case of how the development of new high performance analytical strategies are needed to drive breakthroughs in biological insight. In particular, deciphering the unique lipid signatures associated with PBDs is a key step in this endeavor.
Ether phospholipids in eukarya exist primarily with glycerol-3-phosphate linkages to fatty acids via one ether (sn-1) and one ester (sn-2) bond, while those in archaea are distinctly different, with a glycerol-1-phosphate backbone and two ether-linked isoprenoid alkyl chains.9,10 Ether lipids in eukarya are typically characterized by either an ether or vinyl ether linkage at the sn-1 position, while the sn-2 position is preferentially occupied by polyunsaturated fatty acids such as arachidonic acid (20:4) or docosahexanoic acid (22:6).11,12 In contrast, the sn-1 ether or vinyl ether chain generally contains 16:0, 18:0, or 18:1 compositions.11,12 Plasmalogens, which contain a vinyl ether linkage, are the more-commonly studied class of these two primary ether GP types and are widely distributed, with especially high concentrations in human brain and heart tissues.13–15 In the nervous system, plasmalogens are major components of myelin sheaths and play an essential role in modulating neural synaptic vesicle trafficking.16,17
In addition to a sparse understanding of biosynthetic pathways of ether GP production relative to standard diacyl GPs, structural characterization of ether GPs is more challenging than their diacyl counterparts. Mass spectrometry is the gold standard for identification of GPs, with electrospray ionization (ESI) being the most commonly employed technique.18–20 While single-stage ESI-MS can identify an ether-linked GP via accurate mass measurements,21 this technique does not afford information about individual fatty acids or ether chain composition.22 The use of collisionally activated dissociation methods such as collision-induced dissociation (CID) and higher-energy collisional dissociation (HCD) have been employed for more detailed structural characterization, including fatty acid and ether chain identification, but are generally unable to provide ether linkage or unsaturation element identification. An exception to this is positive ion mode CID or HCD, which enables differentiation of PE-P and PE-O lipids via production of diagnostic PE-P fragments.23 However, PC-P and PC-O ether linkage information is not accessible, nor are acyl or ether chain modifications (such as C═C double bonds), underscoring the inability to decipher these four main classes of ether GPs (PE-P, PE-O, PC-P, and PC-O lipids). Discrimination between all four ether GP subtypes is especially important due to the often isomeric nature of PX-O and PX-P lipids, as any PX-P lipid will have a corresponding isomeric PX-O counterpart which cannot be distinguished using traditional MS/MS methods (for example, PC(P-16:0/18:1) is isomeric with PC(O-16:1/18:1)).
To combat these deficits in traditional MS/MS approaches for the analysis of ether GPs, many research groups have sought to develop more advanced methods. For example, multiple reaction monitoring (MRM) methods coupled with CID in negative and positive ion mode has been used to identify and quantify PE and PC plasmalogens,24 or combined with isobaric mass tagging for analysis of ether lipids at the molecular level/sum composition.25 Other efforts have employed ESI-MS/MS of ether lipids (PE and PC) for characterization at the fatty acid level.23,26 Clever derivatization techniques27–33 and gas-phase reaction methods34,35 have also been developed to facilitate differentiation of ether linkage types and unsaturation location features. For example, shotgun experiments utilizing the Paternò–Büchi (PB) reaction has been utilized for derivatization of the ether lipid alkenyl C═C bond to discriminate isomeric plasmenyl and plasmanyl PC lipids,33 while shotgun ozone-induced dissociation (OzID) combined with MS3 CID can generate ion fragmentation patterns which allow assignment of all structural elements within an ether lipid except for the C═C double bond stereochemistry.34 Additionally, electron-induced dissociation (EID), i.e. electron impact excitation of ions from organics (EIEIO), has also been used for characterization of plasmenyl and plasmanyl lipids.36 At the same time, improved chromatographic methods have enhanced the ability to separate and resolve various plasmalogen phospholipids from biological matrices37–40 However, despite these inroads, the ability to derive headgroup, ether linkage, and unsaturation element information for all four classes of GPs simultaneously (PE-P, PE-O, PC-P, and PC-O) on an LC time scale has remained challenging.
As shown in the present study, ultraviolet photodissociation (UVPD) mass spectrometry overcomes many of these obstacles as it is LC-compatible and commercially available, requires no sample preparation, and can generate both ether linkage C═C double bond and linkage type information independent of phospholipid headgroup. UVPD generates unique and informative fragment ions owing to the higher-energy pathways which are accessed via absorption of UV photons. UVPD has been successfully employed for the characterization of traditionally challenging lipid features, including cyclopropane rings,41 C═C double bonds,42–46 sn-linkage information,44 and hydroxyl groups.47–56 Here, we employ 213 nm UVPD-MS for detailed characterization of plasmenyl and plasmanyl PEs and PCs, including both ether linkage characterization and localization of unsaturation elements, in brain tissue. We focus on delineating the role of ether lipids in neurological diseases, in particular, PBD. We quantify ether GPs in Pex7 mutant mouse models, which genetically mimic the human PBD, rhizomelic chondrodysplasia punctata (RCDP), which is specifically characterized clinically by deficiencies in plasmalogen biosynthesis.1,57–59 Comprehensive characterization of ether GPs sheds new light on the roles and distributions of these biomarkers in peroxisome biogenesis disorders.
EXPERIMENTAL SECTION
Extensive experimental details are provided in the Supporting Information. In brief, the mice used were B6;129S6-Pex7tm2.3Brav, also referred as Pex7null/null, derived from the Pex7 hypomorphic mouse model (B6;129S6-Pex7tm2.0Brav). Cerebral cortex, cerebellum, and hippocampus were collected and snap frozen in liquid nitrogen and stored at 80 °C until lipid extractions were performed, as described in the Supporting Information. All experiments were performed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) equipped with a solid-state 213 nm CryLaS Nd:YAG laser (1.5 uJ/pulse at a repetition rate of 2.5 kHz). MS1 and MS2 spectra were collected at a resolution of 30,000 at m/z 200. MS2 spectra were collected using an AGC target of 1e6 and averaging 5 scans total (with 3 μscans per scan). HCD was performed using normalized collisions energies (NCE) ranging from 18 to 30. 213 nm UVPD was performed in the low-pressure trap of the dual linear ion trap using activation periods of 50 ms (125 laser pulses) and 600 ms (1500 laser pulses) for PE and PC lipids, respectively. Lipids were separated using an Acquity UPLC CSH C18 column (pore size 130 Å, 1.7 μm particle size, 2.1 mm × 100 mm, Waters) on a Dionex Ultimate 3000 UHPLC system (Thermo Fisher Scientific) coupled to the Orbitrap mass spectrometer using a heated ESI source. Targeted LC/UVPD-MS experiments were performed using UVPD with quadrupole isolation, an isolation width of 1 m/z, and a UVPD activation time of 800 ms. For assignment of all fragment ions from direct infusion-mode and LC-mode mass spectra alike, a minimum S/N threshold of 3 along with a ppm error tolerance of 20 ppm was utilized. LipiDex was used for automated analysis of untargeted negative-mode HCD experiments via generation of a custom LipiDex library.60 Quantitative analysis of ether GPs was performed using FreeStyle (Thermo Fisher Scientific). Lipid nomenclature follows the standard LIPID MAPS nomenclature system; in cases of unknown C═C double bond positions, “Δ” is used in place of “E” or “Z”; in cases of unknown sn-1/sn-2 stereochemistry, “_” is used in place of “/”.
RESULTS AND DISCUSSION
Prior to application of the UVPD method for analysis of lipids in murine brain tissue, we first aimed to characterize the HCD and 213 nm UVPD fragmentation patterns of the four ether lipid classes, namely plasmenyl and plasmanyl PE and PC lipids. The structures of the four ether lipid standards (PE(O-16:0/18:1(9Z)), PE(P-18:0/18:1(9Z)), PC(O-16:0/18:1-(9Z)), PC(P-18:0/18:1(9Z))) are displayed in Table S1, along with precursor m/z values and associated ppm error values. PEs and PCs are both detected as protonated species in the positive ion mode, while in the negative ion mode PEs are detected as deprotonated species, and PCs are detected as deprotonated, formate-adducted ions. Positive-mode HCD mass spectra of the four ether lipid standards are shown in Figure S1a–d along with corresponding fragment ion maps (Figure S1e–h). Fragmentation of these four lipids (Figure S1a–d) produces abundant sn-2 acyl chain product ions as well as sn-1 and sn-2 neutral loss ions; however, sn-1 chain product ions (theoretical m/z 267 for P-18:1 and m/z 241 for O-16:0) are notably absent. As has been shown previously, ether-linked GPs do not produce sn-1 chain product ions upon positive-mode CAD.19,23,24 However, PE-P lipids produce characteristic fragment ions that arise only from the rearrangement of PEs containing vinyl ether linkages at sn-1.19,23,24 These fragment ions of m/z 392 and 339 are observed for PE(P-18:0/18:1(9Z)) (Figure S1d). These products are unique to PE-P lipids and can be used to differentiate PE-O and PE-P isomers possessing the same mass because they are absent in MS/MS spectra of PE-O species (Figure S1a). Unique fragment ions of protonated plasmenyl PC (PC(P-18:0/18:1(9Z))) and plasmanyl PC (PC(O-16:0/18:1(9Z) were not observed upon HCD (Figure S1a,c). The absence of any unique fragment ions to identify PCs with ether linkages underscores the need for an alternative MS/MS strategy to allow their identification.
Previous reports of HCD of deprotonated PE-O and PE-P lipids describe the inability to distinguish ether linkage type without the use of multiple-stage MSn methods.19 We examined the HCD spectra of all four deprotonated ether lipids (Figure S2a–d) and created the corresponding fragment ion maps (Figure S2e–h). In fact, fragment ions of low abundance unique to either plasmenyl or plasmanyl PE or PC lipids were found. For PE lipids, plasmanyl lipid PE(O-16:0/18:1(9Z)) generated no fragment ions corresponding to cleavages within the sn-1 chain (Figure S2b), whereas PE(P-18:0/18:1(9Z)) yielded fragment ions of m/z 460 and 267, corresponding to cleavage of the C–O bond of the sn-1 ether linkage (Figure S2d). Similarly, HCD of the PC(P-18:0/18:1(9Z)) formate adduct produced two fragment ions (m/z 488 and 267) consistent with the cleavage of the C–O bond at sn-1 (Figure S2c). These ions are notably absent for the plasmanyl variant of this PC (PC(O-16:0/18:1(9Z)) (Figure S2a). Overall, these results highlight the ability of negative-mode HCD to distinguish PC-O vs PC-P lipids as well as PE-O vs PE-P lipids, whereas positive-mode HCD allows only differentiation of PE-O vs PE-P lipids. Of note, these P-18:0 product ions of m/z 267.2693 may be easily confused with product ions arising from low-abundance 17:1 standard acyl chains of m/z 267.2330 if measured on lower resolution mass spectrometers. Tabulation of all positive- and negative-mode HCD fragment ion assignments for all four ether lipid standards is summarized in Tables S2, S3 along with corresponding ppm error assignments.
To extend the structural characterization of ether GPLs, we employed 213 nm UVPD to allow the determination of alkene regiochemistry. 213 nm UVPD of all four protonated lipids are shown in Figure 1a–d along with corresponding fragment ion maps (Figure 1e–h). The UVPD spectrum of PE-P in Figure 1d exhibits fragments of m/z 339 and 392, the same rearrangement ions which are produced upon HCD of PE-P (Figure S1d). In addition to these two fragment ions which are unique to the PE-P lipid compared to the PE-O lipid, UVPD of the PE-P lipid yields fragment ions originating from cleavage at the C–O bond at the sn-1 ether linkage (ions of m/z 464, 463, 462). In contrast, the PE-O lipid produces no rearrangement ions nor fragment ions corresponding to cleavage within the sn-1 ether chain (Figure 1b). Similarly, plasmenyl PC lipid PC(P-18:0/18:1(9Z)) produces diagnostic ions from the sn-1 ether chain (m/z 506, 505, 504, 490) (Figure 1c), which are not observed for its PC-O counterpart (Figure 1a). These results showcase the ability of positive-mode 213 nm UVPD to distinguish ether linkage types for plasmenyl and plasmanyl PEs and PCs alike, a key asset of this MS/MS method. In addition to ether linkage information, 213 nm UVPD of all four lipids generates diagnostic ions which localize alkene positions in sn-2 acyl chains. For the PC-O and PC-P lipids, ion pairs of m/z 632, 608 or m/z 658, 634, respectively, localize the C═C double bond to the Δ9 position (Figure 1a,c). These ions are spaced apart by exactly 24.00 Da and are consistent with neutral losses indicative of C═C double bond position, as noted in a prior UVPD study of nonether PC lipids.43 213 nm UVPD of protonated PE-O and PE-P also produces diagnostic fragment ion pairs of m/z 590, 576 or m/z 616, 602, respectively. These diagnostic fragment ion pairs differ by 14.01 Da (CH2), in contrast to the PC C═C double bond fragment ions which are spaced 24 Da apart, and are characteristic of neutral losses of C9H20 and C8H18 from the sn-2 acyl chain, allowing confident identification of the alkene position as Δ9 for each PE lipid. These data indicate a difference in the C═C double bond fragmentation pathway in the positive ion mode for PE and PC glycerophospholipids.
Figure 1.
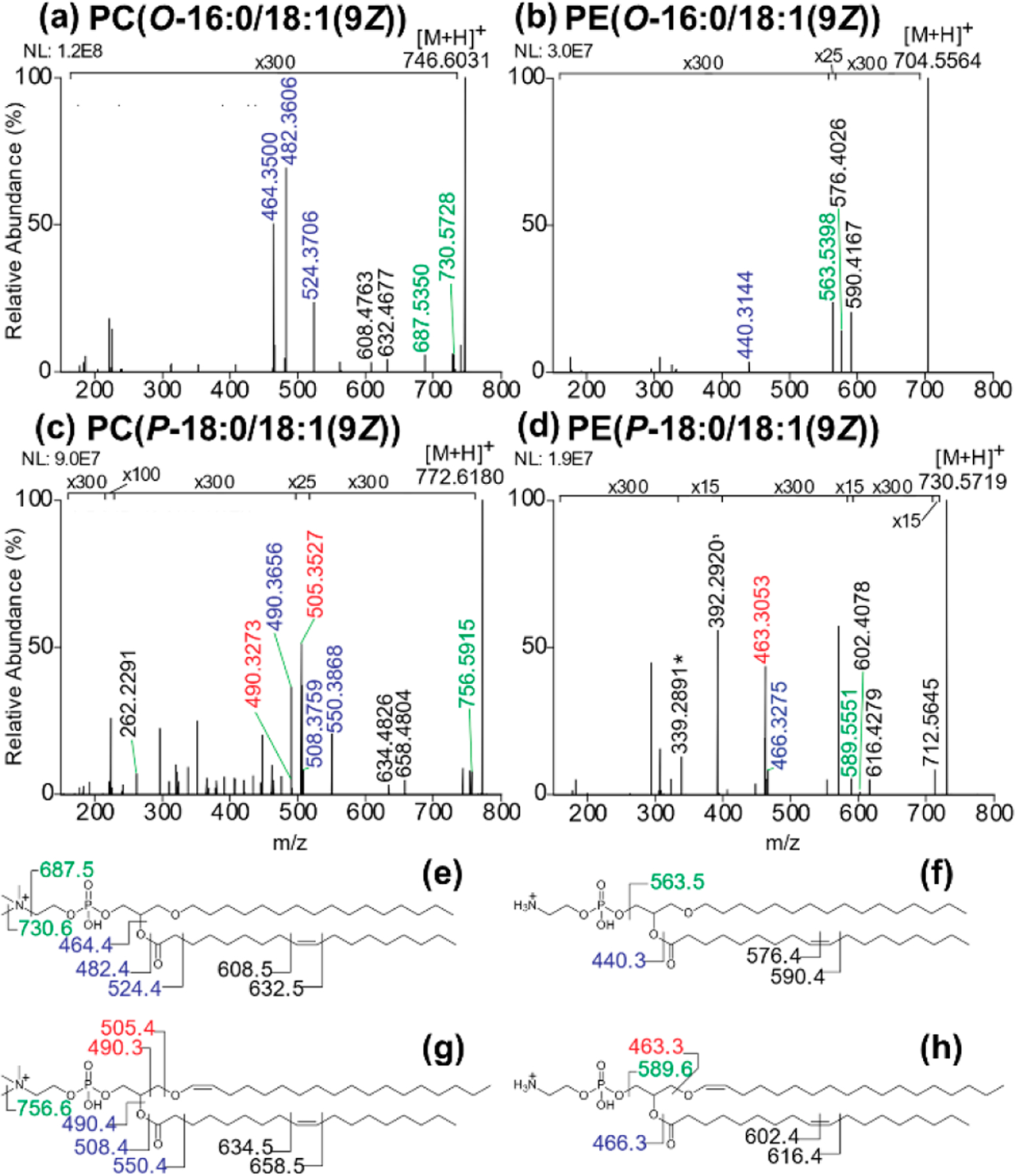
Positive-mode 213 nm UVPD spectra of ether glycerophospholipid standards (a) PC(O-16:0/18:1(9Z)), (b) PE(O-16:0/18:1(9Z)), (c) PC(P-18:0/18:1(9Z)), and (d) PE(P-18:0/18:1(9Z)) with corresponding fragment maps (e-h). The mass accuracy values of the assigned fragment ions are shown in Table S4.
213 nm UVPD was also used for analysis of the four ether lipids in the negative mode (Figure S3a–d and corresponding fragment ion maps in Figure S3e–h). UVPD of deprotonated plasmenyl PE (PE-P) (Figure S3d) shows unique fragments corresponding to cleavage at the sn-1 ether linkage (m/z 460, 461, 462, 267) which are not observed upon fragmentation of the plasmanyl PE (PE-O) (Figure S3b). Similarly, the plasmenyl PC lipid provides unique fragment ions arising from cleavage at the sn-1 ether linkage (m/z 506, 490, 489, 488, 474, 267) (Figure S3c), which are not observed for the plasmanyl PC (Figure S3a). Tabulations of fragment ion assignments with ppm error values for all UVPD data of the four ether lipids are summarized in Tables S4, S5. These results demonstrate the ability of 213 nm UVPD to provide the highest level of characterization for PE and PC ether lipids, differentiating plasmenyl and plasmanyl PE and PC lipids and simultaneously providing C═C double bond localization for all four lipid types.
The UVPD method is readily translated into an LC-MS/MS workflow for analysis of biological extracts. Method optimization and benchmark results obtained using a mixture of six lipids are reported in the Supporting Information (Figure S4, Tables S1, S6). Not only the efficacy of the separation between plasmanyl and plasmenyl lipids but also the separation of these two lipid classes from their standard ester-linked counterparts is crucial for any method aiming to characterize ether lipids in tissue given their often low abundance in comparison to ester-linked lipids. The importance of high accuracy mass measurements is also critical for the workflow, allowing differentiation of isobaric acyl and ether GPs. For example, protonated ester-linked diacyl PC(15:0/18:1) (m/z of 746.5694) differs only by 0.0364 Da compared to its isobaric ether and vinyl ether counterparts, PC(P-16:0/18:0) and PC(O-16:1/18:0) (both m/z 746.6058 Da for the protonated species).
In summary, positive-mode HCD enables distinction of PE-O vs PE-P lipids but does not offer the same level of distinction for PC lipids. In contrast, we have uncovered the ability of negative-mode HCD to differentiate O vs P ether lipids for both deprotonated PE and formate-adducted PC lipids. Negative-mode 213 nm UVPD provides an equivalent level of characterization as negative-mode HCD, similarly distinguishing plasmenyl and plasmanyl PE and PC lipids, with the additional advantage of generating multiple diagnostic fragments for O vs P structural assignment. The highest level of characterization is achieved with positive-mode 213 nm UVPD, enabling distinction of PE-O vs PE-P and PC-O vs PC-P lipids, while additionally generating diagnostic fragment ions for all four of these ether lipid subclasses and allowing double bond localization within the sn-2 acyl chain position.
After optimization of the method, we focused on analysis of ether lipids in brain tissue from Pex7 null mice and their control littermates. Two types of deuterated ether lipids, PE(P-18:0/18:1-d9(9Z)) and PC(P-16:0/16:0-d9) or PC(P-18:0/18:1-d9(9Z)) (structures and mass values in Table S7), were added to each sample prior to extraction to facilitate quantitation based on construction of calibration curves based on chromatographic peak areas over a range of concentrations (0.39–100 pmol) (Figure S5). The resulting calibration curve showed high linearity and a wide dynamic range for both the plasmenyl PE and PC deuterated lipids (Figure S5b). The limit of detection (LOD) and limit of quantitation (LOQ) for the deuterated plasmenyl PE were found to be 5.3 and 16.1 pmol, respectively, and 1.1 and 3.4 pmol, respectively, for the deuterated plasmenyl PC.
The integrated workflow for quantitation and characterization of the ether lipids entailed an untargeted negative mode LC/HCD-MS run prior to a targeted positive mode LC/UVPD-MS run. The untargeted HCD run allowed the initial identification of PE or PC ether lipids via automatic LipiDex analysis, along with manual validation of ether linkage type based on the MS1 and HCD data. The [M – H]– (PE) or [M – H + FA]– (PC) precursor species identified from the negative-mode initial HCD run were then converted to [M + H]+ m/z values to generate a list of precursor masses for the targeted positive-mode LC/UVPD-MS runs, the latter allowing assignment of C═C double bond locations within the sn-2 ether lipid acyl chains.
A list of all mouse brain tissue regions utilized for this study is summarized in Table S8. For this study, we used cerebral cortex samples from five Pex7null/null (HO) mutants and four Pex7WT/WT (WT) littermate controls. Additionally, we utilized samples from the cerebellar and hippocampal area of three Pex7null/null (HO) mutants as well as four Pex7WT/WT (WT) and two Pex7WT/null (HT) controls. The Pex7 null mice are defective in the biosynthesis of the committing alkylglycerol molecule, and thus deficient in generating all ether phospholipid classes. Several mice from each genotype were used to account for the mixed background and expected biological variation.
A representative base peak LC-MS trace of a mouse cerebellum lipid extract (sample C5, genotype HT) is shown in Figure S6a, and 13 distinct ether lipid species are tracked based on 11 unique m/z values corresponding to deprotonated molecular species. The overall low abundances of the various ether lipids relative to the base peak trace, as seen in Figure S6a, underscore the benefits of a two-pronged LC-MS/MS strategy, as even the most abundant ether lipid in this sample (PE(P-18:0/22:6, m/z 774) occurs at only approximately 30% of the base peak chromatogram. Employing HCD with dynamic exclusion provides a fast preliminary screening of ether lipids in the first run prior to the second run which utilizes a targeted UVPD method for deeper structural characterization of the preidentified lipids.
Based on analysis by HCD, only vinyl ether PE lipids were identified across all of the mouse brain tissue samples. Our findings align with previous reports of sum and regional levels of ether GPs in mouse brains, as ether PEs make up only ∼11% of all central nervous system tissue lipids, while ether PCs make up only about 0.48% of all brain tissue lipids.61 The base peak LC traces of all 27 mouse tissue samples (9 cerebellum, 9 hippocampus, 9 cortex) are displayed in Figures S7–S9. Summaries of the identified ether lipids along with their precursor m/z values and the fragment ions used for ether linkage assignment from HCD along with ppm error values are reported in Table S9, along with the m/z values and retention time ranges used for the subsequent LC/UVPD-MS runs.
LC/UVPD-MS runs were then undertaken to characterize the 13 ether lipids identified from the HCD runs. Base-peak LC-MS traces from the positive-mode targeted runs are shown in Figures S10–S12. Peak areas of the targeted ether lipids were normalized to the internal standard (PE(P-18:0/18:1-d9(9Z)) to account for differences in extraction efficiency and were also normalized to tissue mass to account for the total amount of ether GPs extracted from each sample. The quantitative results for the ether lipids identified in the WT and HT sample cerebellum and hippocampus extracts are reported in Figure 2a and Figure 2b, respectively. Interestingly, no statistical differences in normalized peak abundances are observed for the WT and HT genotypes for either tissue type. This is consistent with many autosomal recessive enzyme deficiencies, where reduction in enzyme activity in the heterozygous state does not cause characteristic disease manifestations. In recessive peroxisomal disorders, the affected metabolites remain at normal levels in the heterozygote. Human blood and fibroblast cell lines, as well as tissues from Pex7 deficient mice, show similar levels of plasmalogen subclasses between wild type and heterozygous samples.62–64 Using LC/UVPD-MS it is clear that neither plasmenyl nor plasmanyl PC or PE lipids are affected in the heterozygous state, thus enabling grouping of the WT and HT genotype data.
Figure 2.
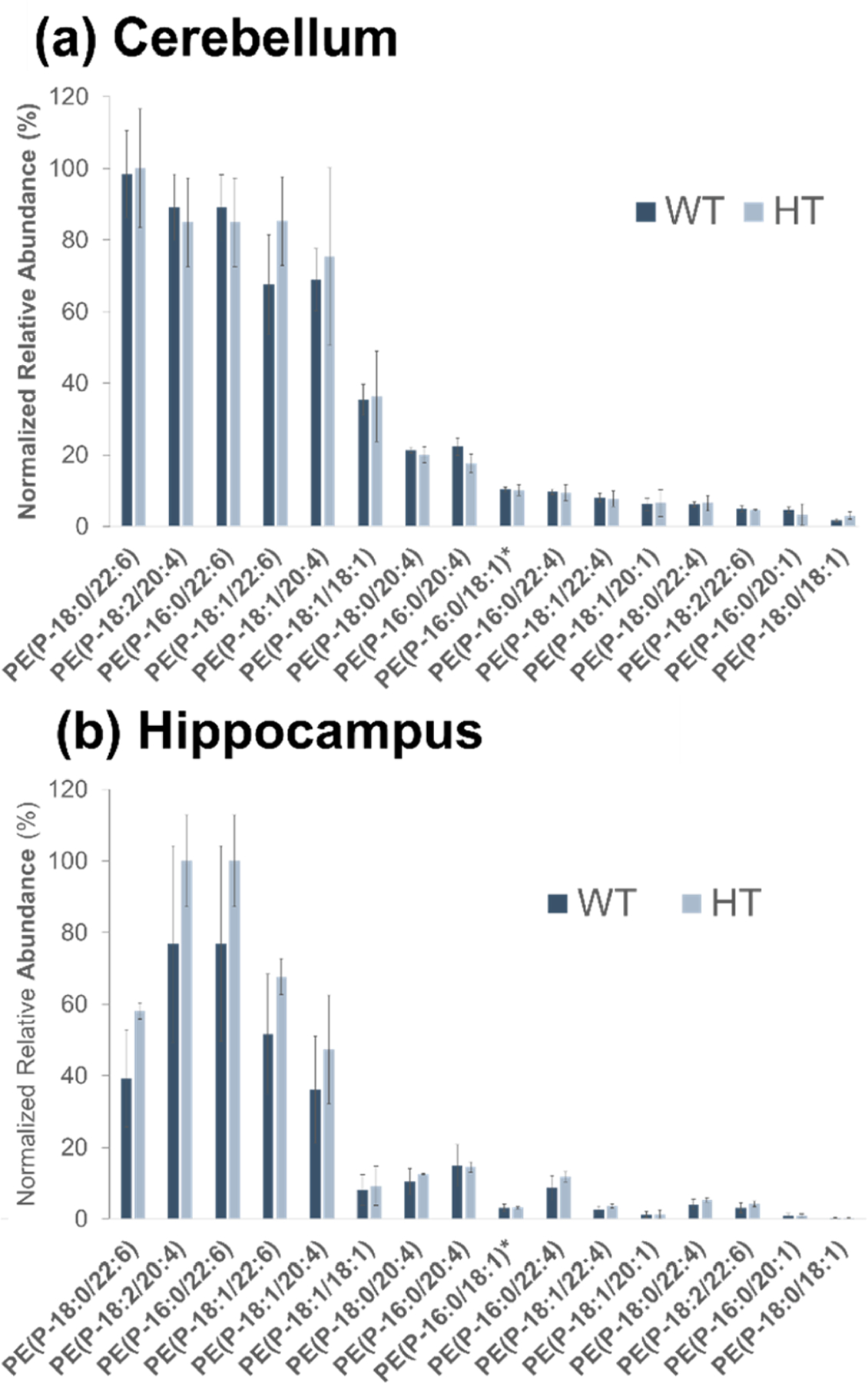
Bar graphs of normalized peak areas of identified ether lipids for (a) cerebellum and (b) hippocampus mouse tissue. Samples are grouped by genotype (WT, HT). No statistically significant differences are observed for WT and HT ether lipid abundances. PE(P-16:0/18:1) and PE(P-18:1/16:0) are isomeric and not distinguishable. The entry on the x-axis is designated as PE(P-16:0/18:1)*.
Pex7 WT and HT genotypes were then grouped together as the “control” group for comparison to the HO (Pex7null/null) samples, as shown in Figure 3. Here, ether lipids are graphed by their normalized relative abundance as a percentage of the most abundant ether lipid in each sample, with notable differences in abundance between the control and disease samples across all three tissue regions (Figure 3). Notably, each of the 16 identified PE-P lipids shows statistically significant differences between the control and disease samples, with the normalized peak area of each Pex7 deficient tissue sample either below the LOD or not detected. While some of the targeted PE-P lipids were below LOQ or below LOD in the normal samples, PE-P lipids of composition 18:0/22:6, 16:0/22:6, 18:1/22:6, and 18:1/20:4 were the most abundant and all above LOD/LOQ in the cerebellum, hippocampus, and cortex tissue samples. Taken in sum, these data show the deficiency of plasmalogen PEs at the individual lipid level, resulting in a general deficiency of all PE-P lipids in the Pex7 disease samples. These results align with previous findings that PE-P lipids are decreased in RCDP and other peroxisomal disorders.59,65,66 Additionally, the LC/HCD-MS data enable discernment of three different pairs of acyl chain isomers such as PE(P-18:0/18:1) and PE(P-16:0/20:1) (both 729.5672 Da) that often go undetected using standard MS-based lipidomics workflows. These findings highlight the ability to discern differences in levels of plasmalogens between control and disease state tissue samples across all ether glycerophospholipid classes. Additional data from the present study focused on evaluating the differences in abundances of standard (nonether) PE and PC glycerophospholipids and are included in the Supporting Information (Figure S14 and companion Additional Results section).
Figure 3.
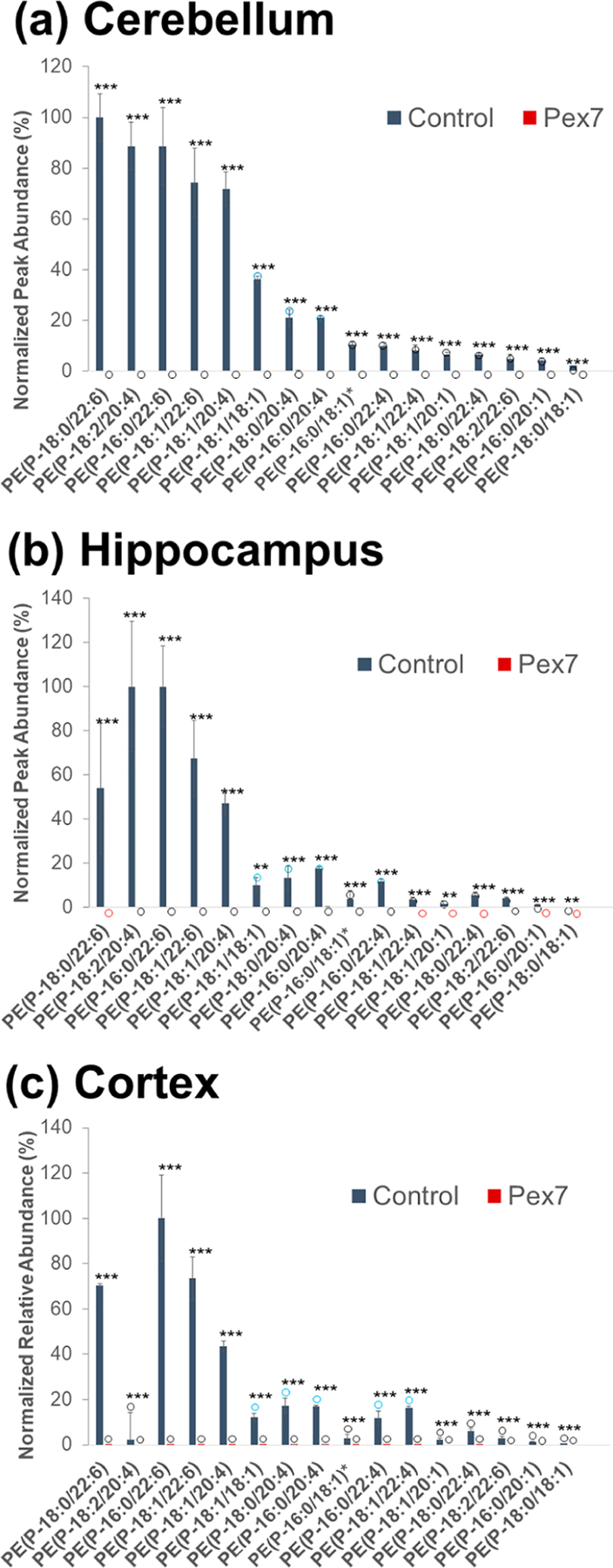
Bar graphs of normalized peak areas of identified ether lipids for (a) cerebellum, (b) hippocampus, and (c) cortex mouse tissue. Samples are grouped by control (genotype WT, HT) and disease (genotype HO, i.e. Pex7 mutation). Statistical significance levels are marked as p < 0.01 (***) and p < 0.05 (**). Blue, black and red circles indicate normalized abundances that are below LOQ, below LOD, and not detected, respectively. PE(P-16:0/18:1) and PE(P-18:1/16:0) are isomeric and not distinguishable. The entry on the x-axis is designated as PE(P-16:0/18:1)*.
Manual curation of the plasmalogen PEs enabled identification of individual species at the C═C double bond location level using the data obtained from the targeted LC/UVPD-MS experiments. An example of this extra level of structural assignment is shown in Figure 4, in which diagnostic fragment ions are detected upon UVPD of the protonated lipid of m/z 748.53 (RT 21.47 min) and mapped onto the structure (Figure 4b). The assignment of these ions enables annotation of this lipid as PE(P-16:0/22:6(Δ4, Δ7, Δ10, Δ13, Δ16, Δ19)). While the regiochemical assignments (sn-1 versus sn-2) of the ether lipids were not explicitly enumerated in this analysis, biologically, ether-linked lipids are known to almost exclusively occur at the sn-1 position for plasmanyl and plasmenyl PEs and PCs, with the sn-2 chain retaining its traditional acyl linkage form.12 Thus, sn-1 and sn-2 positions are inferred from this biological framework; hence, the use of the “/” symbol separating the two sn-designations within the lipid ID nomenclature, indicating assignment of the two sn positions. Identification of C═C double bond position was particularly challenging for PE plasmalogens using 213 nm UVPD owing to the low abundance of diagnostic fragment ions that have been traditionally used for this type of analysis. This is a result of a variety of factors, including the low abundance of ether lipids in these biological samples, ion suppression effects due to coelution of low-abundance ether lipids with high-abundance nonether PE and PC lipids, and the lower fragmentation efficiency of 213 nm UVPD. However, 213 nm UVPD is currently the only commercially available option for UVPD experiments and does generate informative cleavages for the higher-abundance ether lipid species. PE plasmalogens containing multiple unsaturations per acyl chain provided more abundant cleavages at allylic and vinylic positions of C═C bonds that could readily be used to identify the C═C double bond position. For example, the 22:6 acyl chain identified in PE (P-16:0/22:6) (Figure 4) has C═C double bond locations at the 4-, 7-, 10-, 13-, 16-, and 19-Δ positions, which are identified by characteristic aliphatic neutral losses from fragmentation around bis-allylic methylene carbons. Interpretation of UVPD mass spectra via assignment of C═C double bond locations for all of the detected ether lipid species is summarized in Table S10 along with ppm error values that indicate the mass accuracies.
Figure 4.
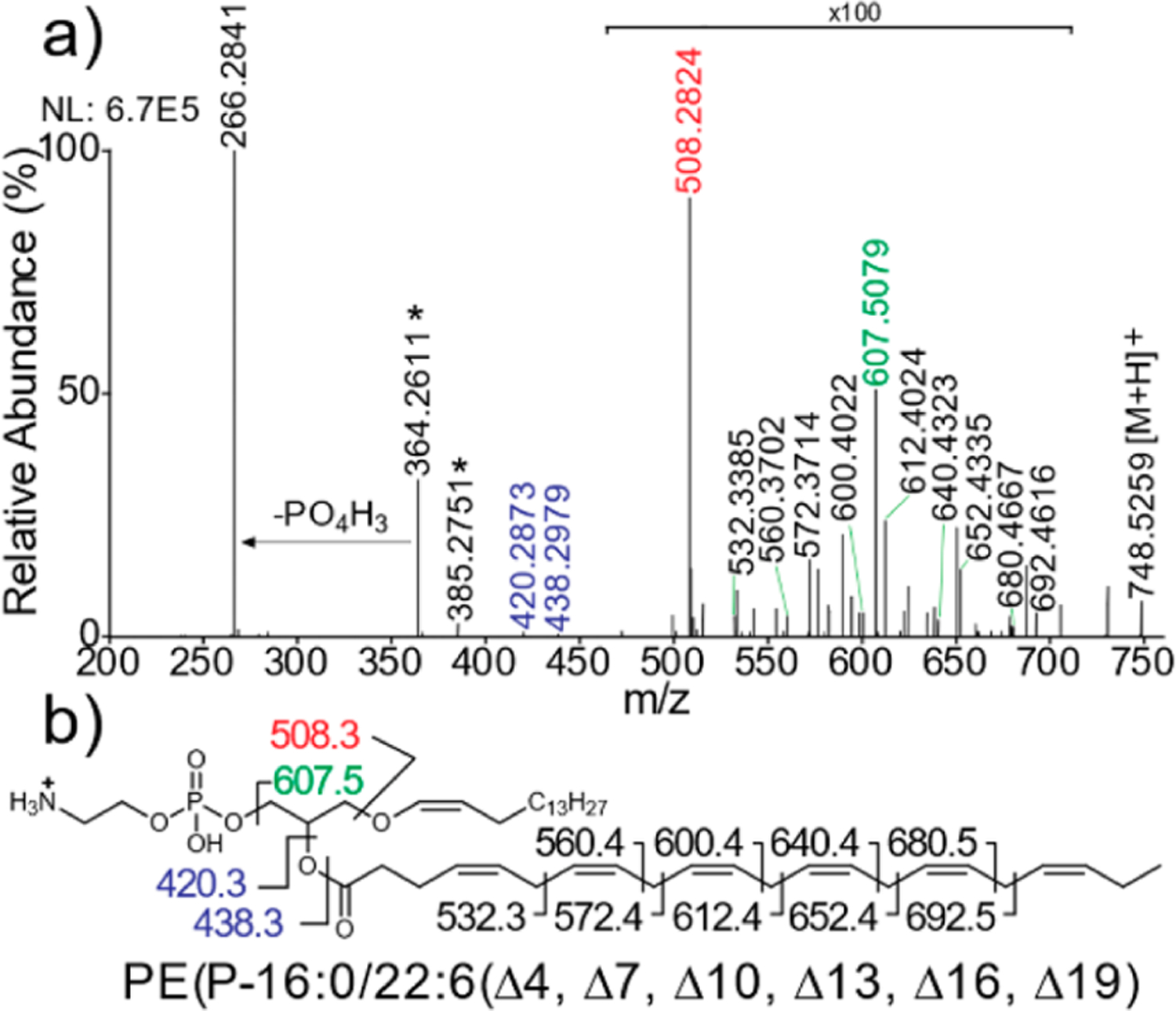
(a) Example of positive-mode targeted 213 nm UVPD spectrum from a mouse cerebellum sample for the precursor ion of m/z 748.53 with (b) fragment map showing identification of the lipid as PE(P-16:0/22:6(Δ4, Δ7, Δ10, Δ13, Δ16, Δ19)). The mass accuracy values of the assigned fragment ions are shown in Table S10.
Overall, the positive-mode LC/UVPD-MS method show-cased in this work enables distinction of all four subclasses of ether lipids (PE-O, PE-P, PC-O, PC-P) along with sn-2 acyl chain double bond assignments on a commercially available platform and without extensive sample preparation. This method is successful for sample-limited applications and for tissue samples which contain inherently low levels of ether lipids. Compared to the new negative-mode HCD approach also spotlighted in this study, UVPD offers the added benefit of double bond localization at the sn-2 position. The greater variety of diagnostic ions produced upon UVPD of ether lipids compared to the more limited array of fragment ions generated upon HCD may facilitate assignment of unique structural moieties in ether lipids in future untargeted biological applications.
While our strategy combining untargeted LC/HCD-MS and targeted LC/UVPD-MS provides the most in-depth characterization of individual ether lipid species for a quantitative workflow, we also aim to further advance the method. In theory, one could forego HCD entirely and rely on 213 nm UVPD for full structural characterization of the ether lipids. However, each 213 nm UVPD scan is slower, meaning that fewer lipids will be sampled in an untargeted manner. For data-dependent acquisition methods, typically the most abundant lipids are subjected to MS/MS, and only a limited number can be analyzed during a chromatographic peak. An inclusion mass list could be applied to target potential ether lipids, but each mass would be subjected to MS/MS prior to cycling back to the top of the list, again slowing analysis and dwelling on features not present in the sample. For these reasons, the untargeted HCD/targeted UVPD approach is expected to offer the best ether lipid coverage and characterization. With respect to sensitivity, while the sensitivity level of our HCD/UVPD method is comparable to other LC/MS-based methods for relative quantitation of lipids, LOD/LOQ values in the pmol range combined with the low abundances of ether lipids in tissue (and the small inherent size of mouse brains) result in many ether lipid species falling below LOD or LOQ. As a result, differences in the abundances of certain lipids between control and disease samples cannot be adequately mapped, and certain diagnostic fragment ions needed for C═C double bond assignments are not observed for the lower-abundance ether lipids. Enrichment of ether lipids and adaptation of the method for nanoLC workflows are potential steps for improvement. Despite these recognized limitations, this targeted LC/UVPD-MS analysis allows the analysis of ether lipid species, including ether linkage type and C═C double bond identification, of low-abundance lipids that would not be accessible with traditional LC-MS/MS methods. We anticipate that this method will enable detailed analysis of the extent of down- and/or upregulation of various ether lipid species in other peroxisomal disorder genotypes as well as other neurological disorders for which ether lipids have been identified as biomarkers.67–69
CONCLUSIONS
We have developed a high-performance mass-spectrometry strategy for analysis of ether lipids in peroxisomal disease (RCDP) and control samples by combining 213 nm UVPD with liquid chromatography. This methodology enables unequivocal mapping of fatty acid, ether chain, ether linkage type (O- vs P-), and C═C double bond localization across PE and PC ether lipid classes. In applying this method to the brain tissue samples from an ether lipid deficient mouse model with Pex7 deficiency which mimic human RCDP, we enable total characterization of the specific ether lipids which are severely downregulated in the Pex7 mutant samples. Additionally, we uncover tissue-specific trends in the abundances of diacyl glycerophospholipids between the normal and disease samples, uncovering novel biological insights into this rare but deadly class of diseases and providing a new framework for elucidation of mechanistic pathways that require molecular level analysis of lipid profiles.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by grants from the National Institutes of Health (National Institute of General Medical Sciences of the National Institutes of Health under award R35GM139658 to JSB), and Welch Foundation (F-1155 to JSB). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Robert A. Welch Foundation or National Institutes of Health. Funding from the UT System for support of the UT System Proteomics Core Facility Network is gratefully acknowledged. Anna Krieger is acknowledged for assistance with lipid extractions.
Footnotes
The authors declare no competing financial interest.
Contributor Information
Molly S. Blevins, Department of Chemistry, University of Texas at Austin, Austin, Texas 78712, United States.
Samuel W. J. Shields, Department of Chemistry, University of Texas at Austin, Austin, Texas 78712, United States.
Wei Cui, Research Institute of the McGill University Health Center, McGill University and the, Montreal, Quebec HA 0C7, Canada.
Wedad Fallatah, Department of Human Genetics, McGill University and the, Montreal, Quebec HA 0C7, Canada; Department of Medical Genetics, King Abdul-Aziz University, Jeddah 21423, Saudi Arabia.
Ann B. Moser, Kennedy Krieger Institute, Baltimore, Maryland 21205, United States; School of Medicine, Johns Hopkins University, Baltimore, Maryland 21205, United States
Nancy E. Braverman, Research Institute of the McGill University Health Center, Department of Human Genetics, and Department of Pediatrics, McGill University and the, Montreal, Quebec HA 0C7, Canada
Jennifer S. Brodbelt, Department of Chemistry, University of Texas at Austin, Austin, Texas 78712, United States.
REFERENCES
- (1).Braverman N; Steel G; Obie C; Moser A; Moser H; Gould SJ; Valle D Nat. Genet 1997, 15, 369–376. [DOI] [PubMed] [Google Scholar]
- (2).Stradomska TJ; Syczewska M; Jamroz E; Pleskaczyńska A; Kruczek P; Ciara E; Tylki-Szymanska A PLoS One 2020, 15, No. e0238796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Steinberg SJ; Dodt G; Raymond GV; Braverman NE; Moser AB; Moser HW Biochim. Biophys. Acta, Mol. Cell Res 2006, 1763, 1733–1748. [DOI] [PubMed] [Google Scholar]
- (4).Moser AB; Kreiter N; Bezman L; Lu S-E; Raymond GV; Naidu S; Moser HW Ann. Neurol 1999, 45, 100–110. [DOI] [PubMed] [Google Scholar]
- (5).Jo DS; Park NY; Cho D-H Exp. Mol. Med 2020, 52, 1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ghosh D; Berg JM J. Am. Chem. Soc 2010, 132, 3973–3979. [DOI] [PubMed] [Google Scholar]
- (7).Zhou YJ; Buijs NA; Zhu Z; Gómez DO; Boonsombuti A; Siewers V; Nielsen J J. Am. Chem. Soc 2016, 138, 15368–15377. [DOI] [PubMed] [Google Scholar]
- (8).Honsho M; Abe Y; Fujiki Y Sci. Rep 2017, 7, 43936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Villanueva L; von Meijenfeldt FAB; Westbye AB; Yadav S; Hopmans EC; Dutilh BE; Damsté JSS ISME J 2021, 15, 168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhou Y; Yu N; Zhao J; Xie Z; Yang Z; Tian B Front. Cell Dev. Biol 2020, 8, 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Mawatari S; Sasuga Y; Morisaki T; Okubo M; Emura T; Fujino T Sci. Rep 2020, 10, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Braverman NE; Moser AB Biochim. Biophys. Acta, Mol. Basis Dis 2012, 1822, 1442–1452. [DOI] [PubMed] [Google Scholar]
- (13).Xu L; Schmitt MV; Ruan H; Jiao Y; Wang X; Wang Y; Yang T; Lienau P; Reichel A; Liu X Int. J. Anal. Chem 2020, 2020, No. e8819437. [Google Scholar]
- (14).Jackson DR; Cassilly CD; Plichta DR; Vlamakis H; Liu H; Melville SB; Xavier RJ; Clardy J ACS Chem. Biol 2021, 16, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gross RW Biochemistry 1985, 24, 1662–1668. [DOI] [PubMed] [Google Scholar]
- (16).Brodde A; Teigler A; Brugger B; Lehmann WD; Wieland F; Berger J; Just WW Hum. Mol. Genet 2012, 21, 2713–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Dorninger F; König T; Scholze P; Berger ML; Zeitler G; Wiesinger C; Gundacker A; Pollak DD; Huck S; Just WW; Forss-Petter S; Pifl C; Berger J Hum. Mol. Genet 2019, 28, 2046–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ivanova PT; Milne SB; Byrne MO; Xiang Y; Brown HA Methods Enzymol 2007, 432, 21–57. [DOI] [PubMed] [Google Scholar]
- (19).Hsu F-F; Turk J J. Chromatogr. B 2009, 877, 2673–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hsu F.-F. u; Turk J J. Am. Soc. Mass Spectrom 2003, 14, 352–363. [DOI] [PubMed] [Google Scholar]
- (21).Rustam YH; Reid GE Anal. Chem 2018, 90, 374–397. [DOI] [PubMed] [Google Scholar]
- (22).Murphy RC; Fiedler J; Hevko J Chem. Rev 2001, 101, 479–526. [DOI] [PubMed] [Google Scholar]
- (23).Zemski Berry KA; Murphy RC J. Am. Soc. Mass Spectrom 2004, 15, 1499–1508. [DOI] [PubMed] [Google Scholar]
- (24).Wu Y; Chen Z; Jia J; Chiba H; Hui S-P Foods 2021, 10, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Tokuoka SM; Kita Y; Shimizu T; Oda Y PLoS One 2019, 14, No. e0226073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Karnati S; Garikapati V; Liebisch G; Van Veldhoven PP; Spengler B; Schmitz G; Baumgart-Vogt E PLoS One 2018, 13, No. e0203464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lydic TA; Townsend S; Adda CG; Collins C; Mathivanan S; Reid GE Methods 2015, 87, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Fhaner CJ; Liu S; Zhou X; Reid GE Mass Spectrom 2013, 2, S0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ryan E; Reid GE Acc. Chem. Res 2016, 49, 1596–1604. [DOI] [PubMed] [Google Scholar]
- (30).Mawatari S; Hazeyama S; Fujino T Lipids 2016, 51, 997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Nie S; Fhaner CJ; Liu S; Peake D; Kiyonami R; Huang Y; Reid GE Int. J. Mass Spectrom 2015, 391, 71–81. [Google Scholar]
- (32).Pradas I; Huynh K; Cabré R; Ayala V; Meikle PJ; Jové M; Pamplona R Front. Physiol 2018, 9, 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lin Q; Zhang D; Xia Y Analyst 2020, 145, 513–522. [DOI] [PubMed] [Google Scholar]
- (34).Deeley JM; Thomas MC; Truscott RJW; Mitchell TW; Blanksby SJ Anal. Chem 2009, 81, 1920–1930. [DOI] [PubMed] [Google Scholar]
- (35).Randolph CE; Shenault DM; Blanksby SJ; McLuckey SA J. Am. Soc. Mass Spectrom 2020, 31, 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Baba T; Campbell JL; Le Blanc JCY; Baker PRS; Ikeda K J. Lipid Res 2018, 59, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Pham TH; Zaeem M; Fillier TA; Nadeem M; Vidal NP; Manful C; Cheema S; Cheema M; Thomas RH Sci. Rep 2019, 9, 5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Rezanka T; Siristova L; Matoulková D; Sigler K Lipids 2011, 46, 765–780. [DOI] [PubMed] [Google Scholar]
- (39).Koch J; Lackner K; Wohlfarter Y; Sailer S; Zschocke J; Werner ER; Watschinger K; Keller MA Anal. Chem 2020, 92, 11268–11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hui S-P; Chiba H; Kurosawa T Anal. Bioanal. Chem 2011, 400, 1923–1931. [DOI] [PubMed] [Google Scholar]
- (41).Blevins MS; Klein DR; Brodbelt JS Anal. Chem 2019, 91, 6820–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Klein DR; Blevins MS; Macias LA; Douglass MV; Trent MS; Brodbelt JS Anal. Chem 2020, 92, 5986–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Klein DR; Brodbelt JS Anal. Chem 2017, 89, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Williams PE; Klein DR; Greer SM; Brodbelt JS J. Am. Chem. Soc 2017, 139, 15681–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Klein DR; Feider CL; Garza KY; Lin JQ; Eberlin LS; Brodbelt JS Anal. Chem 2018, 90, 10100–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).West H; Reid GE Anal. Chim. Acta 2021, 1141, 100–109. [DOI] [PubMed] [Google Scholar]
- (47).Blevins MS; James VK; Herrera CM; Purcell AB; Trent MS; Brodbelt JS Anal. Chem 2020, 92, 9146–9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Buenger EW; Reid GE Eur. J. Mass Spectrom 2020, 26, 311–323. [DOI] [PubMed] [Google Scholar]
- (49).Narreddula VR; McKinnon BI; Marlton SJP; Marshall DL; Boase NRB; Poad BLJ; Trevitt AJ; Mitchell TW; Blanksby SJ Analyst 2021, 146, 156–169. [DOI] [PubMed] [Google Scholar]
- (50).Pham HT; Ly T; Trevitt AJ; Mitchell TW; Blanksby SJ Anal. Chem 2012, 84, 7525–7532. [DOI] [PubMed] [Google Scholar]
- (51).Heiles S Anal. Bioanal. Chem 2021, 413, 5927–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wäldchen F; Becher S; Esch P; Kompauer M; Heiles S Analyst 2017, 142, 4744–4755. [DOI] [PubMed] [Google Scholar]
- (53).Pham HT; Trevitt AJ; Mitchell TW; Blanksby SJ Rapid Commun. Mass Spectrom 2013, 27, 805–815. [DOI] [PubMed] [Google Scholar]
- (54).Pham HT; Julian RR Analyst 2016, 141, 1273–1278. [DOI] [PubMed] [Google Scholar]
- (55).Pham HT; Julian RR Int. J. Mass Spectrom 2014, 370, 58–65. [Google Scholar]
- (56).Pham HT; Julian RR Anal. Chem 2014, 86, 3020–3027. [DOI] [PubMed] [Google Scholar]
- (57).Motley AM; Hettema EH; Hogenhout EM; Brites P; ten Asbroek ALMA; Wijburg FA; Baas F; Heijmans HS; Tabak HF; Wanders RJA; Distel B Nat. Genet 1997, 15, 377–380. [DOI] [PubMed] [Google Scholar]
- (58).Purdue PE; Zhang JW; Skoneczny M; Lazarow PB Nat. Genet 1997, 15, 381–384. [DOI] [PubMed] [Google Scholar]
- (59).Purdue PE; Skoneczny M; Yang X; Zhang J-W; Lazarow PB Neurochem. Res 1999, 24, 581–586. [DOI] [PubMed] [Google Scholar]
- (60).Hutchins PD; Russell JD; Coon JJ Cell Syst 2018, 6, 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Huynh K; Lim WLF; Giles C; Jayawardana KS; Salim A; Mellett NA; Smith AAT; Olshansky G; Drew BG; Chatterjee P; Martins I; Laws SM; Bush AI; Rowe CC; Villemagne VL; Ames D; Masters CL; Arnold M; Nho K; Saykin AJ; Baillie R; Han X; Kaddurah-Daouk R; Martins RN; Meikle PJ Nat. Commun 2020, 11, 5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Fitzner D; Bader JM; Penkert H; Bergner CG; Su M; Weil M-T; Surma MA; Mann M; Klose C; Simons M Cell Rep 2020, 32, 108132. [DOI] [PubMed] [Google Scholar]
- (63).Fallatah W; Smith T; Cui W; Jayasinghe D; Di Pietro E; Ritchie SA; Braverman N Dis. Model. Mech 2020, 13, dmm042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Braverman N; Zhang R; Chen L; Nimmo G; Scheper S; Tran T; Chaudhury R; Moser A; Steinberg S Mol. Genet. Metab 2010, 99, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Brites P; Motley AM; Gressens P; Mooyer PAW; Ploegaert I; Everts V; Evrard P; Carmeliet P; Dewerchin M; Schoonjans L; Duran M; Waterham HR; Wanders RJA; Baes M Hum. Mol. Genet 2003, 12, 2255–2267. [DOI] [PubMed] [Google Scholar]
- (66).Dorninger F; Forss-Petter S; Berger J FEBS Lett 2017, 591, 2761–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Dorninger F; Brodde A; Braverman NE; Moser AB; Just WW; Forss-Petter S; Brügger B; Berger J Biochim. Biophys. Acta 2015, 1851, 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Wood PL; Tippireddy S; Feriante J; Woltjer RL PLoS One 2018, 13, No. e0191815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Miville-Godbout E; Bourque M; Morissette M; Al-Sweidi S; Smith T; Mochizuki A; Senanayake V; Jayasinghe D; Wang L; Goodenowe D; Di Paolo T PLoS One 2016, 11, No. e0151020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


