Abstract
Introduction
In order to deal with the current pandemic caused by the novel SARS-CoV-2 coronavirus several serological immunoassays have been recently developed with the objective of being used as a complementary diagnostic tool and to support the RT-PCR technique currently considered the “gold-standard” method. However, these new assays need to be evaluated and validated. The purpose of this study was to assess the performance of five immunoassays (two ELISA and three CLIA assays) and one rapid immunochromatographic test for the detection of anti-SARS-CoV-2 antibodies.
Methods
Five semiquantitative immunoassays (MENARINI®, PALEX®, VIRCLIA®, ROCHE® and SIEMENS®) and one lateral flow rapid test (WONDFO®) were performed. A total of 124 samples were studied. Case serum samples (n = 78) were obtained from COVID-19 patients confirmed by real-time RT-PCR/epidemiological-clinical-radiological criteria, and control non-SARS-CoV-2 samples (n = 46) belonged to healthy healthcare workers involved in a seroprevalence study.
Results
Overall, the tests showed sensitivities around 70–90% and specificities greater than 95%, including the immunochromatographic test. In addition, we observed very good agreements among them, being better for the detection of IgG than for IgM antibodies (Cohen's kappa index of 0.95 for VIRCLIA® IgG with ROCHE®), as well as good diagnostic power of the tests as determined by the ROC curves.
Conclusions
This study demonstrates the proper performance of the different immunoassays in order to be applied in the clinical practice as support in the diagnostic approach and in the development of vaccines and seroepidemiological studies of COVID-19.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, coronavirus disease-19; WHO, World Health Organization; real-time RT-PCR, real-time reverse-transcript polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay; CLIA, chemiluminescence immunoassay; PPV, positive predictive value; NPV, negative predictive value; POCT, point-of-care test
Keywords: SARS-CoV-2, Serology, Immunochromatography, ELISA, CLIA, RT-PCR
Abstract
Introducción
Para hacer frente a la pandemia actual causada por el nuevo coronavirus SARS-CoV-2 se han desarrollado recientemente varios inmunoensayos serológicos con el objetivo de ser utilizados como herramienta diagnóstica complementaria y apoyar la técnica de RT-PCR actualmente considerada como el “estándar de oro”. Sin embargo, estos nuevos ensayos deben evaluarse y validarse. El objetivo de este estudio fue evaluar cinco inmunoensayos (dos ELISA y tres ensayos CLIA) y una prueba inmunocromatográfica rápida para la detección de anticuerpos anti-SARS-CoV-2.
Métodos
Se utilizaron cinco inmunoensayos semicuantitativos (MENARINI®, PALEX®, VIRCLIA®, ROCHE® y SIEMENS®) y un test de inmunocromatografía rápida (WONDFO®). Se estudiaron un total de 124 muestras. Las muestras de suero (n = 78) se obtuvieron de pacientes COVID-19 confirmados por RT-PCR en tiempo real/criterios epidemiológicos-clínico-radiológicos. Las muestras control negativas (n = 46) pertenecieron a personal sanitario involucrado en un estudio de seroprevalencia.
Resultados
En general, las pruebas mostraron sensibilidades en torno al 70-90% y especificidades superiores al 95%, incluso la prueba inmunocromatográfica. Además, observamos muy buenas concordancias entre ellas, presentando mayores sensibilidades para la detección de anticuerpos IgG que para IgM (índice kappa de Cohen de 0,95 para VIRCLIA® IgG con ROCHE®), así como un buen poder diagnóstico de las técnicas determinado por las curvas ROC.
Conclusiones
Este estudio demuestra el buen rendimiento de los diferentes inmunoensayos para ser empleados en la práctica clínica como apoyo en el proceso de diagnóstico, en el desarrollo de vacunas y estudios seroepidemiológicos de COVID-19.
Palabras clave: SARS-CoV-2, Serología, Inmunocromatografía, ELISA, CLIA, RT-PCR
Introduction
A novel coronavirus causing severe acute respiratory syndrome emerged in Wuhan (China) in December 2019.1 The virus has spread rapidly all over the world causing a global pandemic. The World Health Organization (WHO) defined the disease as coronavirus disease-19 (COVID-19) and the causative virus as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). On January 2020, the WHO declared it as Public Health Emergency of International Concern. At present, more than 9,296,202 cases have been confirmed as well as 479,133 deaths worldwide.2 COVID-19 has been associated not only with severe morbidity and mortality but also with high transmissibility.3, 4
The development of diagnostic techniques has been increasing since then. Real-time reverse-transcript polymerase chain reaction (real-time RT-PCR) is considered the laboratory method of reference for the diagnosis of acute SARS-CoV-2. However, this method has some limitations. On the one hand, the sensitivity may vary depending on the quality of the sample. On the other hand, the time of infection or the low viral load can generate false negative results.5 Adequate diagnosis of infection is essential, and serological tests may be used as a complementary tool not only to ensure SARS-CoV-2 infected people but also to obtain information about the immunological response of the general population for seroepidemiological studies, to help in vaccine development, to carry out the monitoring of healthcare workers and to study the kinetics of the immunological response, still unknown to date.6
Due to the urgency and demand, a lot of serological tests are being developed on the market with only limited validation on clinical samples. For this reason, many laboratories are evaluating the analytical performance of these assays with promising results.7, 8, 9, 10, 11, 12 Thus, the objective of this study was to evaluate a total of six CE-marked immunoassays, including two enzyme-linked immunosorbent assays (ELISAs), three chemiluminescence immunoassays (CLIAs) and one lateral flow rapid test, for the detection of SARS-CoV-2 IgM, IgG and total antibodies in patients with COVID-19.
Materials and methods
Serum samples
A total of 124 samples were studied. Case serum samples (n = 78) were obtained from COVID-19 patients diagnosed according to the protocols and diagnostic algorithms of our hospital that include epidemiological-clinical-radiological criteria and/or positive real time RT-PCR (LightMix® Modular SARS and Wuhan CoV, TIB MOLBIOL, Berlin, Germany (ROCHE®)). Sample collection time was, on average, 20 days after a positive PCR result/clinical criteria. Control non-SARS-CoV-2 samples (n = 46) belonged to sera from asymptomatic healthcare workers involved in a seroprevalence screening study and with a confirmed negative PCR result. Furthermore, a monitoring of the healthcare personnel was carried out to corroborate their negativity. This retrospective study has been carried out during the epidemic period (April–May 2020).
ELISA assays
Serum samples were run on the Zenit UP A.MENARINI diagnostics instrument and evaluated with the following tests performed according to the manufacturer's instructions: Novel Coronavirus COVID-19 IgG and IgM ELISA (DRG Instruments GmbH, Germany) distributed by A.MENARINI diagnostics and COVID-19 IgG and IgM (DIA.PRO Diagnostic Bioprobes Srl, Milano, Italy) distributed by Palex Medical. These assays provide a semiquantitative in vitro determination of IgG and IgM antibodies based on a double-antigen sandwich principle.
MENARINI® IgG and IgM tests use SARS-CoV-2 recombinant nucleocapsid protein or anti-human IgM specific antibody respectively. This process takes about 2 h.
Palex COVID-19 IgG and IgM enzyme immunoassays use each one nucleocapsid and spike recombinant antigens specific to COVID-19. This process takes about 2 h 30 min.
The cut-off values for the interpretation of the results were calculated according to the manufacturer's instructions.
CLIA assays
COVID-19 VIRCLIA® IgG MONOTEST and COVID-19 VIRCLIA® IgM + IgA MONOTEST (Vircell, S.L., Granada, Spain) are quantitative chemiluminiscent immunoassays based on the reaction of the antibodies with the SARS-CoV-2 antigen coated to the polystyrene surface. This test employs the S1 subunit of the SARS-CoV-2 spike protein and nucleocapsid antigen. It was performed in the VirClia Chemiluminescence analyzer (Vircell microbiologists). The cut-off values for the interpretation of the results were calculated according to the manufacturer's instructions. The process takes about 1 h 30 min.
Elecsys® Anti-SARS-CoV-2 (Roche Diagnostics GmbH, Mannheim, Germany) is an immunoassay for the in vitro semiquantitative detection of antibodies in human serum and plasma. It was carried out on the Cobas® e 411 analyzer (Roche Diagnostics GmbH, Mannheim, Germany). The test is based on a sandwich principle that uses a SARS-CoV-2 recombinant protein representing the nucleocapsid antigen labeled with a ruthenium complex. The process takes 18 min.
SARS-CoV-2 Total (COV2T) is an immunoassay for the in vitro semiquantitative detection of total antibodies (IgG and IgM) in human serum and plasma using the ADVIA Centaur® XP Immunoassay System analyzer (Siemens Healthcare Diagnostics Inc., Tarrytown, USA). The test is based on a sandwich principle using acridinium ester chemiluminescent technology and its duration is about 15 min. The solid phase contains a preformed complex streptavidin-coated microparticles and biotinylated SARS-CoV-2 recombinant antigens. It employs S1 subunit of the SARS-CoV-2 spike protein and the receptor binding domain (RBD).
Rapid immunochromatographic test
WONDFO® SARS-CoV-2 Antibody Test (Lateral Flow Method) (Guangzhou Wondfo Biotech Co., Guangzhou, PR China) is an immunochromatographic assay for rapid, qualitative detection of SARS-CoV-2 IgG/IgM antibody in human whole blood, serum or plasma samples. The test was performed at real time according to the manufacturer's instructions and with serum samples. The result was read visually after 15 min.
Statistical analysis
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated for each serological test. Receiver operator characteristic (ROC) curves were performed using R packages pROC for comparisons of the area under the curve (AUC).13 The Choen Kappa index was calculated for agreement between all analyzed assays and were shown in accompany with 95% confidence interval (CI). A p-value <0.05 was considered statistically significant. Statistical analysis was performed with R Commander.
Results
Sensitivities and specificities of the assays
The sensitivities and specificities as well as the rest of predictive parameters are summarized in Table 1 .
Table 1.
Analytical sensitivities, specificities and predictive parameters for SARS-CoV-2 antibody detection.
| Positive serum samplesa | Negative serum samplesa | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | Accuracy (95% CI) | Diagnostic odds ratio (95% CI) | Youden's index | |
|---|---|---|---|---|---|---|---|---|---|
| MENARINI® IgM | 20/63 | 58/61 | 31.7% (21.6–44.0) | 95.1% (86.5–98.3) | 87.0% (67.9–95.5) | 57.4% (47.7–66.6) | 62.9% (54.1–70.9) | 8.99 (2.51–32.21) | 0.3 |
| MENARINI® IgG | 46/63 | 58/61 | 73.0% (61.0–82.4) | 95.1% (86.5–98.3) | 93.9 (83.5–97.9) | 77.3 (66.7–85.3) | 83.9% (76.4–89.3) | 52.31 (14.44–189.47) | 0.7 |
| MENARINI® Combined IgM or IgG | 49/63 | 57/61 | 77.8% (66.1–86.3) | 93.4% (84.3–97.4) | 92.5% (82.1–97.0) | 80.3% (69.6–87.9) | 85.5% (78.2–90.6) | 49.88 (15.40–161.50) | 0.7 |
| PALEX® IgM | 42/63 | 56/61 | 66.7% (54.4–77.1) | 91.8% (82.2–96.4) | 89.4% (77.4–95.4) | 72.7% (61.9–81.4) | 79.0% (71.0–85.3) | 22.40 (7.81–64.28) | 0.6 |
| PALEX® IgG | 56/63 | 56/61 | 88.9% (78.8–94.5) | 91.8% (82.2–96.4) | 91.8% (82.2–96.4) | 88.9% (78.8–94.5) | 90.3% (83.8–94.4) | 89.60 (26.83–299.26) | 0.8 |
| PALEX® Combined IgM or IgG | 56/63 | 52/61 | 88.9% (78.8–94.5) | 85.2% (74.3–92.0) | 86.2% (75.7–92.5) | 88.1% (77.5–94.1) | 87.1% (80.1–91.9) | 46.22 (16.06–133.07) | 0.7 |
| VIRCLIA® IgM | 38/63 | 58/61 | 60.3% (48.0–71.5) | 95.1% (86.5–98.3) | 92.7% (80.6–97.5) | 69.9% (59.3–78.7) | 77.4% (69.3–83.9) | 29.39 (8.29–104.17) | 0.6 |
| VIRCLIA® IgG | 53/63 | 59/61 | 84.1% (73.2–91.1) | 96.7% (88.8–99.1) | 96.4% (87.7–99.0) | 85.5% (75.3–91.9) | 90.3% (83.8–94.4) | 156.35 (32.76–746.18) | 0.8 |
| VIRCLIA® Combined IgM or IgG | 54/63 | 58/61 | 85.7% (75.0–92.3) | 95.1% (86.5–98.3) | 94.7% (85.6–98.2) | 86.6% (76.4–92.8) | 90.3% (83.8–94.4) | 116.0 (29.83–451.15) | 0.8 |
| SIEMENS® | 49/63 | 59/61 | 77.8% (66.1–86.3) | 96.7% (88.8–99.1) | 96.1% (86.8–98.9) | 80.8% (70.3–88.2) | 87.1% (80.1–91.9) | 103.25 (22.37–476.49) | 0.7 |
| ROCHE® | 50/63 | 59/61 | 79.4% (67.8–87.5) | 96.7% (88.8–99.1) | 96.2% (87.0–98.9) | 81.9% (71.5–89.1) | 87.9% (81.0–92.5) | 113.46 (24.43–526.96) | 0.8 |
| WONDFO® | 46/63 | 59/61 | 73.0% (61.0–82.4) | 96.7% (88.8–99.1) | 95.8% (86.0–98.8) | 77.6% (67.1–85.5) | 84.7% (77.3–90.0) | 79.82 (17.54–363.17) | 0.7 |
Calculated as positive sera among total positive samples and negative sera among total negative samples.
PPV (positive predictive value) and NPV (negative predictive value).
Regarding the sensitivities and specificities, for ELISA assays, PALEX® IgG had a superior sensitivity (88.9%) in comparison with MENARINI®, having both techniques high specificities (91.8% and 95.1% respectively).
Overall, the sensitivities and specificities of CLIA assays were similar showing best results VIRCLIA® IgG (84.1% and 96.1%, respectively). Noteworthy, combining the results of IgM and IgG, i.e. patients with either IgM or IgG positive, would significantly increase the sensitivity of these assays.
In general, specificities were high for all the assays. The best results were obtained for CLIA assays and WONDFO® (96.7%). Moreover, better specificities were obtained for IgM determinations in comparison with their sensitivity values.
The best PPV were observed for CLIA (VIRCLIA® IgG with 96.4%) in comparison with ELISA (MENARINI® IgG with 93.9%). VIRCLIA® IgG, SIEMENS® and ROCHE® had a value of >96%, and only two false positive results were detected in each one. PALEX® IgG and VIRCLIA® Combined IgM or IgG showed the greatest NPV, 88.9% and 86.6%, respectively.
Regarding the accuracies, PALEX® IgG and VIRCLIA® IgG presented the best results (90.3%) whereas for MENARINI® IgM was 62.9%. MENARINI®, PALEX® and VIRCLIA® Combined IgM or IgG obtained 85.5%, 87.1% and 90.3%, respectively.
In general, as shown in Table 1, CLIA assays had the best diagnostic odds ratio (156.35 for VIRCLIA® IgG and 113.46 for ROCHE®) compared to ELISA (best for PALEX® IgG: 89.60) and WONDFO® (79.82) immunoassays.
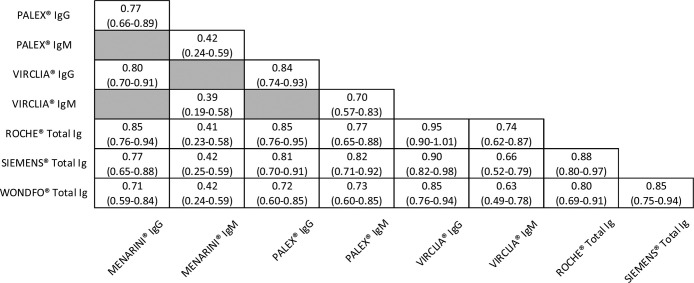
Agreement between serological assays
To determine the agreement between the different techniques, the Choen Kappa index was calculated. The results were summarized in Table 2 and divided according to IgG or IgM antibodies measurement except those for ROCHE®, SIEMENS® and WONDFO® that determine total antibodies.
Table 2.
Agreement between the different serological assays analyzed (κ, (95% CI)).
For IgG, all the assays presented a good or very good concordance, being VIRCLIA® the one that presented the best indexes with the other CLIA assays (0.95 with ROCHE® and 0.90 with SIEMENS®, respectively). WONDFO® had good agreements despite being a rapid and qualitative immunochromatographic test.
However, for IgM, lower correlations were observed in comparison with those for IgG. It is noteworthy to mention the very good agreement, in the case of IgM, between PALEX® (ELISA) and SIEMENS® (CLIA) with a value of 0.82. In contrast, MENARINI® showed only moderate concordances.
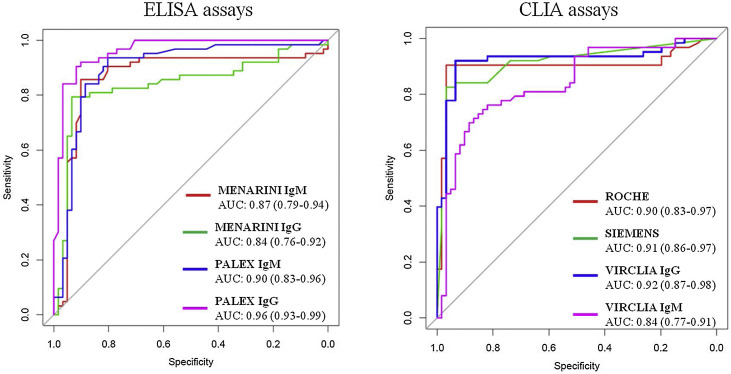
Comparative analysis of ROC curves
ROC curves as well as AUC for ELISA and CLIA assays are shown in Fig. 1 . All the techniques presented areas higher than 0.8 indicating good or very good probability of classification. PALEX® showed the best AUC for IgG determination (0.96) followed by VIRCLIA® IgG (0.92), SIEMENS® (0.91) and ROCHE® and PALEX® IgM with a value of 0.90.
Fig. 1.
Comparative ROC curves for ELISA and CLIA assays.
Youden's index was also used in conjunction with ROC analysis and provided the performance of the diagnostic test. These results are shown in Table 1. PALEX® IgG, VIRCLIA® IgG and ROCHE® got a value of 0.8. A Youden's index of 0.7 was obtained for the combined techniques (IgM or IgG), MENARINI® IgG and WONDFO®. The lowest value was for MENARINI® IgM (0.3).
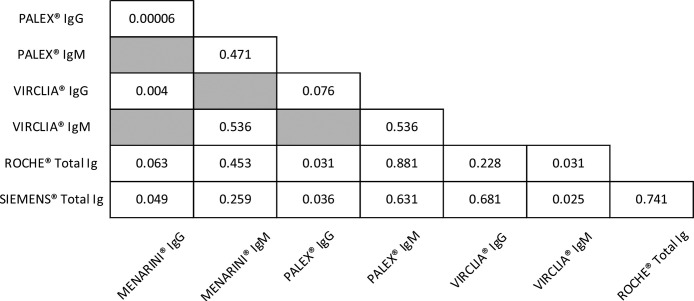
Furthermore, ROC curves for paired comparisons between the serological assays were constructed and the p-values obtained are summarized in Table 3 . For IgM, significant differences (p-value <0.05) were found among CLIA assays. In contrast, for IgG, MENARINI® showed no significant AUC differences only with ROCHE®. PALEX® IgG with SIEMENS® and ROCHE® also presented significant differences (p-value <0.05).
Table 3.
p-Values for the paired comparisons of ROC curves between serological assays.
Discussion
In this study, five automated immunoassays and one rapid lateral flow immunochromatographic test were evaluated using serum samples from COVID-19 patients with PCR-confirmed diagnoses or epidemiological-clinical-radiological criteria for SARS-CoV-2, and sera from healthy hospital personnel, involved in a seroprevalence study, as non-SARS-CoV-2 control samples during the period of April–May 2020.
Regarding the sensitivities and specificities, according to our results, ELISA showed better sensitivities than CLIA (88.9% vs. 85.7%). However, regarding the specificities, CLIA presented better results than ELISA (96.7% vs. 93.4%).
WONDFO® had a sensitivity of 73.0% and 96.7% of specificity. These results are comparable with those obtained in the study carried out by The Health Institute of Carlos III in Madrid in its report of April 2020,14 with values of 77.8% and 95.0%, respectively. Despite being a qualitative test, the good results demonstrate that it could be an adequate method for antibody detection in patients who had overcome the infection, in just 15 min. So, this test has the advantage of being the most rapid and easiest detection method compared to the semiquantitative assays and can be used as point-of-care test (POCT).
In the same way, CLIA presented better PPV in comparison with ELISA.
In general, CLIA showed better results versus ELISA. Differences between the assays may be explained by the SARS-CoV-2 antigen targeted and the format test used. MENARINI® test employs the nucleocapsid (N) protein while PALEX® uses nucleocapsid and spike (S) recombinant antigens. VIRCLIA® employs the S1 subunit of the SARS-CoV-2 spike protein and nucleocapsid antigen, SIEMENS® detects antibodies to the S1 subunit of the SARS-CoV-2 spike protein and the receptor binding domain (RBD), however, ROCHE® uses a recombinant protein representing the nucleocapsid antigen. Okba et al.15 recently reported a study of development of serological assays in which SARS-CoV-2 specific antibody responses were evaluated according to the coronavirus structural proteins in an ELISA format, reaching the conclusion that S1 is more specific than S as an antigen for SARS-CoV-2 serological diagnosis, as well as that RBD and N ELISAs were more sensitive than S1 ELISA in detecting antibodies in mildly infected patients. Besides, they found differences between immunoglobulins, being IgG the most specific.
Another important aspect to mention is the cross-reactivity with other coronaviruses, which could generate false positive results in the serological determinations, as it was also discussed in the Okba et al.15 study. They verified the specificity of the assays in patients with SARS-CoV, MERS-CoV or HCoV-OC43, among others. Results for S1 subunit and RBD results were the best, showing none or little cross-reactivity for SARS-CoV, due to the high degree of similarity between the S1 and RBD of the SARS-CoV and SARS-CoV-2. However, further investigation is needed in order to validate SARS-CoV-2 serological assays that are currently lacking.
In addition, differences found with PCR results may be explained taking into account the viral load of the infection. In mild cases, due to the low viral load and the presence of mild symptoms, these patients may not have produced antibodies which are not detected in the immunoassays, causing discordance with PCR results (false negative results). Yongchen et al.16 reported similar results when evaluating the serological response based on the disease severity, concluding that an immediate antibody response was identified in severe cases compared to non-severe cases, and only one asymptomatic patient showed seroconversion.
Besides, false positive results regarding the PCR were low, in fact, there were 2 symptomatic patients in our study with negative PCR and positive serology. Limitations of the PCR technique have been reported and the majority of errors may occur due to preanalytical factors which interfere in the amount of viral load and/or the ARN stability giving false negative results.17 Assessment of test accuracy in terms of sensitivity and specificity of diagnostic tests are in increasing attention and being studied due to the derived implications.18
The diagnostic odds ratio is a measure of test performance and effectiveness. This statistic parameter is also useful for comparing tests.19 In this study, CLIA assays presented the best results being VIRCLIA® IgG and ROCHE® the most effective techniques (156.35 and 113.46, respectively).
Regarding the agreement between serological assays, for IgG, it was observed good or very good concordances especially between CLIA assays. A Choen Kappa index of 0.95 for VIRCLIA® with ROCHE®, 0.90 for VIRCLIA® with SIEMENS® and 0.88 for ROCHE® with SIEMENS®. Moreover, for IgM, it is important to note the very good agreement between PALEX® and SIEMENS® (0.82). These results based on our data demonstrate that the different techniques are comparable.
Finally, the diagnostic power of these assays was determined by the ROC curves. The high values of AUC mean good or very good classifications. Both, the three CLIA and PALEX® IgG presented the best AUC values, all of them ≥0.9 (except VIRCLIA® IgM, AUC 0.84) as well as a Youden's index of 0.8, but 0.7 for SIEMENS® and 0.6 for VIRCLIA® IgM. In addition, the paired comparisons were constructed in order to evaluate the possibility of exchanging the techniques. Based on our results, no significant differences (p > 0.05) were observed except in the following cases (p < 0.05): VIRCLIA® IgM with SIEMENS® and with ROCHE®; MENARINI® IgG with PALEX® IgG, with VIRCLIA® IgG and with SIEMENS® and PALEX® IgG with SIEMENS® and with ROCHE®.
It is noteworthy to mention the different results obtained for IgG and IgM. Apart from the SARS-CoV-2 antigen target used in each technique, the time that had elapsed between the moment of infection and the sample collection time is an important factor. Some authors have reported the diagnostic value of antibodies’ assays for patients at different times after onset20, 21 concluding that IgM increased during the first week, reached its peak after two weeks and then its level decreased in most patients. Meanwhile, IgG was generated after 1 week and reached its peak level in 3 weeks. In this study, samples were collected on average 20 days after having positive PCR results/epidemiological-clinical-radiological criteria, this fact may explain the better results obtained for IgG than for IgM. Thus, the use of the serological assays tested would need further studies in order to ensure their application as a diagnostic tool in the acute phase of the disease.
In general, our findings demonstrate the good performance of the different immunoassays. In particular, the usefulness of WONDFO® as a rapid complementary diagnostic test; about ELISA, PALEX® IgG showed good results but the best results were found for CLIA highlighting VIRCLIA® IgG, SIEMENS® and ROCHE®. Considering the increasing demand due to the new outbreaks, those which are most profitable would be ROCHE® or SIEMENS® because the determination only takes less than 20 min, they are more economic in comparison with the rest of the assays, and are now available in many laboratories making automation possible according to the logistics organization of each laboratory. Thus, they would be suitable for supplying not only the demand in the clinical context but also the current seroprevalence studies of SARS-CoV-2 infection such as the one carried out in Spain by Pollán et al.22
Several limitations should be mentioned in this study. First, the limited number of samples that may have some impact on the statistical results, so, external validation studies would be needed in a larger number of patients. Second, this comparative study was carried out with the aim of having available as many techniques as possible in order to assist the diagnostic process and taking into account the commercial supply. It is for this reason that the comparison among assays has been carried out independently of the kind of antibodies detected (IgM, IgG or total Ig). Furthermore, false negative and false positive results of antibody detection might affect the analysis, due to different illness severities and sample collection time relative to the onset of infection. Finally, no reliable gold standard for serological tests is currently available for comparative studies, however, RT-PCR is used for this purpose. Thus, some negative PCR values may affect the comparative results due to the performance limitations of this technique.
In conclusion, this study shows the adequate performance as well as the high level of consistency of the investigated serological immunoassays for the detection of SARS-CoV-2 antibodies making them useful for application in the clinical practice as support in the diagnostic approach and in the development of seroepidemiological studies. In this way, further investigations are needed to understand the immunization against SARS-CoV-2 coronavirus and to investigate the generation not only of vaccines but also hyperimmune serum as a therapeutic approach.
Authorship contribution
Silvia Montolio: conceptualization, design of the study, analysis and interpretation of data, writing and review. Carmen Molina: design of the study, analysis and interpretation of data, revision. Frederic Gómez: analysis and interpretation of data, revision and supervision. Ester Picó: analysis and interpretation of data, revision. Núria Serrat: interpretation of data and revision. Inma Palau: data acquisition. Maria Teresa Mestre: analysis of data. Maria Teresa Sans: analysis and interpretation of data, revision, supervision and final approval.
Research funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Conflict of interest
Authors state no conflict of interest.
Acknowledgments
We wish to thank not only the personnel of our laboratory for its technical assistance but also SIEMENS for providing the required reagent in order to carry out this study.
References
- 1.Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2020. Coronavirus disease 2019 (COVID) situation report-157. Available in: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200625-covid-19-sitrep-157.pdf?sfvrsn=423f4a82_2. [Google Scholar]
- 3.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vashist S.K. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics. 2020;10:202. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petherick A. Developing antibody tests for SARS-CoV-2. Lancet. 2020;395:1101–1102. doi: 10.1016/S0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. 2020;81:e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montesinos I., Gruson D., Kabamba B., Dahma H., Van der Wijngaert S., Reza S., et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demey B., Daher N., François C., Lanoix J.-P., Duverlie G., Castelain S., et al. Dynamic profile for the detection of anti-SARS-CoV-2 antibodies using four immunochromatographic assays. J Infect. 2020;21:48. doi: 10.1016/j.jinf.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lassaunière R., Frische A., Harboe Z.B., Nielsen A., Fomsgaard S., Krogfelt K.A., et al. Evaluation of nine commercial SARS-CoV-2 immunoassays. MedRxiv. 2020 doi: 10.1101/2020.04.09.20056325. [DOI] [Google Scholar]
- 11.Bryan A., Pepper G., Wenr M.H., Fink S.L., Morishima C., Chaudhary A., et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise Idaho. J Clin Microbiol. 2020;58:e00941–e1020. doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lippi G., Salvagno G.L., Pegoraro M., Militello V., Caloi C., Peretti A., et al. Assessment of immune response to SARS-CoV-2 with fully automated MAGLUMI 2019-nCoV IgG and IgM chemiluminescence immunoassays. Clin Chem Lab Med. 2020;58:1156–1159. doi: 10.1515/cclm-2020-0473. [DOI] [PubMed] [Google Scholar]
- 13.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuenca Estrella M. Instituto de Salud Carlos III; Madrid: 2020. Informe sobre estrategia de diagnóstico microbiológico del COVID-19 (Actualización) Available in: https://abertis.ccoo.cat/wp-content/uploads/sites/16/2020/05/ANEXO-04-Ensayos-Instituto-Salud-Carlos-III.pdf. [Google Scholar]
- 15.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., et al. SARS-CoV-2 specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yongchen Z., Shenb H., Wanga X., Shia X., Lib Y., Yanc J., et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect. 2020;9:833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne D., Newton D., Evans P., Osman H., Baretto R. Preanalytical issues affecting the diagnosis of COVID-19. J Clin Pathol. 2020;6:1–2. doi: 10.1136/jclinpath-2020-206751. [DOI] [PubMed] [Google Scholar]
- 18.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection. Challenges and implications. N Engl J Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 19.Glas A.S., Lijmer J.G., Prins M.H., Bonsel G.J., Bossuyt P.M.M. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56:1129–1135. doi: 10.1016/S0895-4356(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 20.Hou H., Wang T., Zhang B., Luo Y., Mao L., Wang F., et al. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Translational Immunol. 2020;9:e1136. doi: 10.1002/cti2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M., et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]