Abstract
Iron plays a critical role in the pathophysiology of Mycobacterium tuberculosis. To gain a better understanding of iron regulation by this organism, we have used two-dimensional (2-D) gel electrophoresis, mass spectrometry, and database searching to study protein expression in M. tuberculosis under conditions of high and low iron concentration. Proteins in cellular extracts from M. tuberculosis Erdman strain grown under low-iron (1 μM) and high-iron (70 μM) conditions were separated by 2-D polyacrylamide gel electrophoresis, which allowed high-resolution separation of several hundred proteins, as visualized by Coomassie staining. The expression of at least 15 proteins was induced, and the expression of at least 12 proteins was decreased under low-iron conditions. In-gel trypsin digestion was performed on these differentially expressed proteins, and the digestion mixtures were analyzed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry to determine the molecular masses of the resulting tryptic peptides. Partial sequence data on some of the peptides were obtained by using after source decay and/or collision-induced dissociation. The fragmentation data were used to search computerized peptide mass and protein sequence databases for known proteins. Ten iron-regulated proteins were identified, including Fur and aconitase proteins, both of which are known to be regulated by iron in other bacterial systems. Our study shows that, where large protein sequence databases are available from genomic studies, the combined use of 2-D gel electrophoresis, mass spectrometry, and database searching to analyze proteins expressed under defined environmental conditions is a powerful tool for identifying expressed proteins and their physiologic relevance.
The Mycobacterium tuberculosis genome sequencing project has provided information on sequences of hundreds of newly identified proteins encoded by this pathogen’s DNA. The availability of this information provides new opportunities for increasing our understanding of the pathophysiology of M. tuberculosis in the human host. Toward this end, a major next step is to determine the functions of the proteins revealed by the genome project and their interplay under different physiological conditions in the host.
One physiological condition in the host known to be important in M. tuberculosis infection is the concentration of iron. Iron is an essential nutrient for all pathogens, but this element appears to play an especially critical role in the pathogenesis of tuberculosis. For example, serum containing poorly saturated transferrin, such as human serum, is tuberculostatic, an effect neutralized by the addition of iron (28, 29).
The amount of iron is severely limited in the host at sites of M. tuberculosis replication. A facultative intracellular parasite, M. tuberculosis multiplies within macrophages in the lung and elsewhere. Within the macrophage, iron is limited as a result of the effects of the immunomodulator interferon gamma. This cytokine depletes iron in the labile iron pool of the cell by downregulating transferrin receptor expression and the intracellular concentration of ferritin (6, 7). M. tuberculosis also multiples extracellularly in lung cavities. In the extracellular space, iron is severely limited owing to the high affinity with which it is bound by the host iron-binding proteins transferrin and lactoferrin.
One measure of the importance of iron to M. tuberculosis is the degree to which it goes to obtain this element. The pathogen is known to produce in great abundance at least two high-affinity iron siderophores—exochelins and mycobactins (19, 39, 57). Both exochelins and mycobactins are low-molecular-weight (MW) compounds (MW, ∼700 to 1,000) that are nonribosomally synthesized and contain two fatty acid moieties, salicylic acid, and three modified amino acids per molecule. Exochelins are released extracellularly and may be the most abundant molecule exported by M. tuberculosis. On a molar basis, the concentration of exochelins in M. tuberculosis culture filtrates (∼5 μM) (19) is 150-fold that of the 30-kDa (antigen 85b) major secretory protein of M. tuberculosis (∼30 nM) (22), the most abundant protein exported by this organism. Mycobactins are water-insoluble molecules located in the cell wall of M. tuberculosis cells. In previous studies in our laboratories, we characterized M. tuberculosis exochelins and found that their core structure resembles that of the mycobactins (19). A shorter alkyl side chain on exochelins and a terminal methyl ester or carboxylic acid moiety on this side chain renders exochelins more polar than mycobactins and hence water soluble. It has been proposed by Macham and colleagues that exochelins bind iron in the aqueous extracellular milieu of the mycobacterium and transfer it to mycobactins in the cell wall for subsequent internalization into the bacterial cytoplasm (39, 62). Consistent with this hypothesis, we have demonstrated that exochelins remove iron from human transferrin and lactoferrin and transfer it to mycobactins in the cell wall of live M. tuberculosis cells (18).
To learn more about the role of iron in the physiology of M. tuberculosis, we have been investigating iron-regulated proteins of M. tuberculosis. In this study, we have taken advantage of three major scientific or technological advances to gain a more complete picture of how M. tuberculosis responds to change in the iron concentration in its environment. The first advance, already noted, is the database generated by the M. tuberculosis genome sequencing project (49). The second advance is the development of high-resolution two-dimensional (2-D) gel electrophoresis allowing greatly enhanced separation of proteins. The third advance is the development of mass spectrometric methods for the low-level detection and identification of proteins and peptides. In an effort to learn about cellular components affected by iron levels, we used these three modalities—2-D gel electrophoresis, mass spectrometry (MS), and database searching—to identify proteins expressed under conditions of low or high concentrations of iron. We report the identification of 10 proteins from 11 different gel spots whose expression levels were markedly affected by low- or high-iron concentration conditions. These include two proteins already known to be affected by environmental iron levels in other bacteria, Fur and aconitase, as well as several mycobacterial antigens and enzymes not previously known to be affected by environmental iron levels.
MATERIALS AND METHODS
Bacteria.
M. tuberculosis Erdman strain (ATCC 35801) was obtained from the lungs of guinea pigs infected with the bacteria by aerosol. Frozen bacterial stocks for use in iron studies were prepared from 7H11 agar plates as described (24).
Medium.
Iron-deficient Sauton’s broth was prepared by subjecting the broth to a chelating resin as described (8). Briefly, 5 g of Chelex 100 resin (Bio-Rad, Hercules, Calif.) per liter was added to Sauton’s medium prepared without ferric ammonium citrate and magnesium sulfate (14), and the medium was stirred at 4°C overnight. The Chelex resin-treated medium was passed through a 0.2-μm-pore-size filter into an acid-washed glass flask. Magnesium sulfate (250 mg/liter) and trace amounts of metals including zinc (2 mg of ZnSO4 · 7H2O per liter) and copper (0.5 mg of CuSO4 per liter) were added. This iron-deficient medium was then supplemented with ferric ammonium citrate to the desired iron concentration (1, 15, or 70 μM). The iron concentration in the medium was routinely checked by the ferrozine assay (16).
Cultures.
Bacteria from frozen stocks were cultured on 7H11 agar plates at 37°C in 5% CO2 for 3 weeks, inoculated into Sauton’s medium containing 15 μM Fe at an initial optical density at 540 nm (OD540) of 0.05, and cultured for 3 weeks at 37°C in 5% CO2 without shaking to a final OD540 of approximately 1.0. The bacteria were then subcultured into Sauton’s medium containing either 1 or 70 μM Fe at an initial OD540 of 0.05, grown for 3 weeks to an OD540 of 0.7 to 1.0, harvested, and stored at −20°C until use.
Sample preparation and 2-D gel electrophoresis.
Approximately 20 ml (wet volume) of bacteria was suspended in 80 ml of phosphate buffer containing 100 μM phenylmethylsulfonyl fluoride, 100 μM benzamidine, and 0.5 mM EDTA. The bacterial suspension was sonicated four times for 10 min each time on an ice bath with a 1-cm-diameter probe attached to a sonicator set at 50% duty cycle and a strength of 5 with pulsing (model W-375; Heat System-Ultrasonics, Inc., Farmingdale, N.Y.) and centrifuged at 10,000 × g for 30 min to pellet unbroken bacteria and bacterial cell walls. The supernatant was passed sequentially through a 0.45-μm-pore-size filter and a 0.2-μm-pore-size filter, further clarified by centrifugation at 40,000 × g for 2 h, and fractionated by ammonium sulfate precipitation. Protein concentration was determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.). Two-dimensional gel electrophoresis was performed as described previously (33) with modifications. Protein samples of 300 μg each were dissolved in sample buffer containing 2% sodium dodecyl sulfate, 5% 2-mercaptoethanol, and 10% glycerol and heated at 95°C for 5 min. The samples were centrifuged at 100,000 × g for 10 min to remove any insoluble material and loaded onto 2.4-mm (internal diameter) by 16-cm (length) isoelectric focusing tube gels with a ratio of ampholytes (pH 3 to 10/pH 5 to 7) of 1:4. The samples were focused at 200 V for 2 h, 500 V for 2 h, and 800 V for 16 h. The second-dimension gels were 10 to 20% polyacrylamide linear gradient gels (size, 20 cm by 16 cm by 1.5 mm).
In-gel digestion with trypsin.
The in-gel digestion procedure was similar to the methods of Rosenfeld et al. (53) and Sheer (54) with modifications as described below (11a). Protein spots of interest were cut out of the gel and diced into small pieces with a stainless-steel scalpel or a vortex mixer and placed in siliconized microcentrifuge tubes. The gel was destained and dehydrated by washing three times (∼10 min) with 25 mM NH4HCO3–50% acetonitrile (or until the Coomassie stain was no longer visually detectable). The destained gel particles were then dried under vacuum for 30 min. After rehydration of the particles with a minimal amount of 25 mM NH4HCO3 with 0.1 μg of trypsin per μl, the protein was digested overnight at 37°C. Recovery of the peptides was accomplished by extracting the digestion mixture three times with 50% acetonitrile–5% trifluoroacetic acid. In an effort to reduce the amount of volatile salts (e.g., trifluoroacetic acid and NH4HCO3), the recovered peptides were concentrated in a Speed-Vac vacuum centrifuge (to a final volume of ∼5 μl) and rehydrated at least three times. Control digestions were performed on gel slices that did not contain any protein and revealed trypsin autoproteolysis products and keratin contaminants that were readily identified in the subsequent mass spectrometric analyses (see below).
MALDI-TOF MS of the unseparated digests.
As described in Matsui et al. (41), portions (approximately 1/10th) of the unseparated tryptic digestion mixture were mixed at a 1:1 (vol/vol) ratio with an α-cyano-4-hydroxycinnamic acid matrix (Hewlett-Packard) and analyzed on a ToFSpec SE matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometer (Micromass Inc., Manchester, United Kingdom) with a nitrogen laser, operated in reflectron mode (25). A standard peptide mixture was used to externally calibrate all mass spectra. Postsource decay (PSD) sequencing (26) involved gating a precursor ion to selectively transmit an individual peptide and its metastable fragment ions to the reflectron. The PSD experiments were carried out by varying the reflectron voltage in 9 to 11 steps, with the voltage at each step being reduced to 75% of that at the previous step. The complete PSD spectrum was produced by stitching the segments from individual steps together. Calibration in PSD mode was done by using the fragment ions from a standard peptide, adrenocorticotropic hormone 18-39.
MALDI-CID MS.
Small aliquots of unseparated digestion mixture (each, 1 μl, or approximately 1/10th of the total) were mixed at a 1:1 ratio with the matrix (saturated solution of 2,5-dihydroxybenzoic acid [Aldrich] in acetone). Samples were analyzed by collision-induced dissociation (CID) on a Micromass Autospec orthogonal acceleration TOF mass spectrometer (Micromass Inc.) equipped with an N2 laser (337 nm). After the electric and magnetic sections (MS-1) were tuned manually to transmit the 12C monoisotopic ion of the precursor mass, a two-stage deacceleration electrostatic lens focused the ions into an approximately parallel beam before they entered the gas collision cell (2). The collision cell was filled with Xe gas with a collision energy of 800 eV. Voltage applied periodically from a “push-out” electrode extracted the precursor and product ions into a linear TOF mass analyzer. All spectra were recorded with a microchannel plate detector by using a time-to-digital converter (Precision Instruments, Knoxville, Tenn.) (43).
Database searches for protein identification.
A program available via the internet (http://prospector.ucsf.edu) and developed in the University of California—San Francisco (UCSF) Mass Spectrometry Facility (11b) was used to search genomic databases. The program, MS-Tag, uses fragment ion masses (generated by MALDI-PSD or -CID MS) to search the databases for matches to peptides from known proteins. The following parameters were used in the searches: no errors mode, Mycobacterium species, protein molecular mass range from 1,000 to 120,000 Da, trypsin digest (one missed cleavage allowed), parent ion mass tolerance of ± 1.5 Da, fragment ion mass tolerance of ± 1.5 Da, and allowed fragment ion types a, b, y, a-NH3, b-NH3, y-NH3, b-H2O, and internal. The protein sequences found by using MS-Tag were used to search protein databases for homologous proteins with NCBI’s basic local alignment search tool (BLAST). Basic BLAST searches with the blastp program were performed on the nonredundant database (1). The Wisconsin sequence analysis package, version 8.0 (Genetics Computer Group, Inc., Madison, Wis.), was used to perform sequence alignments (PILEUP) and identity calculations (DISTANCES).
RESULTS AND DISCUSSION
Proteins of M. tuberculosis modulated by iron.
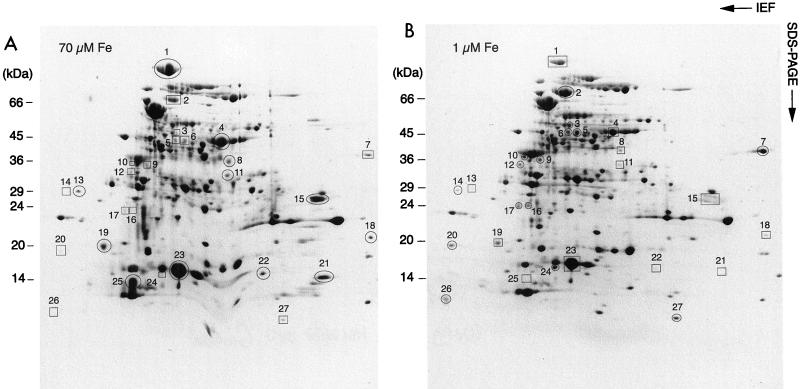
Proteins of M. tuberculosis cultured in Sauton’s medium containing a low concentration (1 μM) or a high concentration (70 μM) of iron were sequentially fractionated by ammonium sulfate precipitation (0 to 20%, 20 to 55%, and 55 to 95%) and analyzed by 2-D gel electrophoresis. With increasing amounts of ammonium sulfate, improved resolution of protein spots was obtained on 2-D gels. Heavy vertical and horizontal streaks were seen on 2-D gels with samples precipitated with 0 to 20% ammonium sulfate, and these streaks severely interfered with analysis of protein spots. (The streak lines are most likely caused by the presence in the sonicates of unusually large amounts of mycobacterial lipids that severely interfere with protein separation on 2-D gels.) Although Triton X-114 extraction improved the resolution of samples precipitated with 20 to 55% ammonium sulfate, the best comparison of protein spots between high-iron-concentration and low-iron-concentration cultures was obtained from samples precipitated with 55 to 95% ammonium sulfate. Of more than 250 protein spots revealed by 2-D gel electrophoresis of samples precipitated with 55 to 95% ammonium sulfate, the expression of at least 15 proteins was induced and the expression of at least 12 proteins was decreased by low iron concentrations (Fig. 1). The protein spots with consistent differential expression from three different batches of low-iron-concentration and high-iron-concentration bacterial cultures were further analyzed by MALDI-MS.
FIG. 1.
2-D gel analysis of proteins of M. tuberculosis cultured in medium containing a (A) high (70 μM) or (B) low (1 μM) concentration of iron. Equal amounts of proteins precipitated by 55 to 95% ammonium sulfate were separated by isoelectric focusing (pH 4 to 7 from left to right) in the first dimension and by linear gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10 to 20% from top to bottom) in the second dimension. The second-dimension gels were stained with Coomassie blue. Open circles and open squares represent protein spots with increased or decreased expression under the indicated condition, respectively. The set of 2-D gels shown here is representative of the results of three independent experiments. The numbers are the protein gel spot numbers, as discussed in the text.
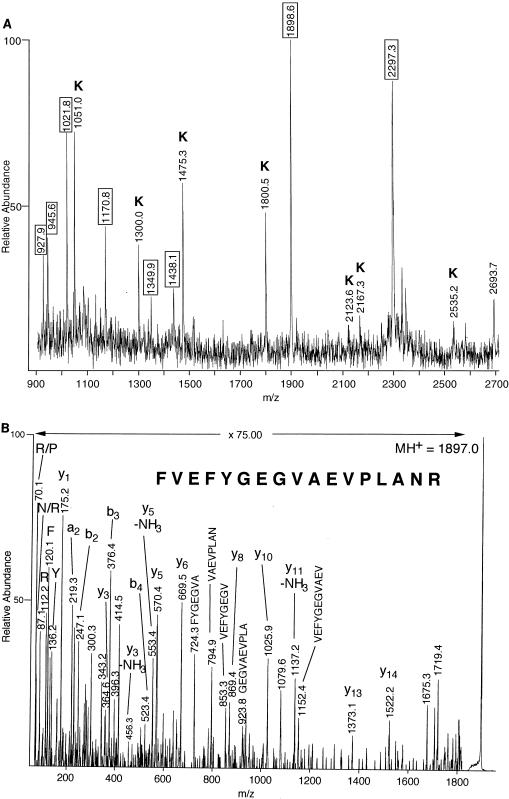
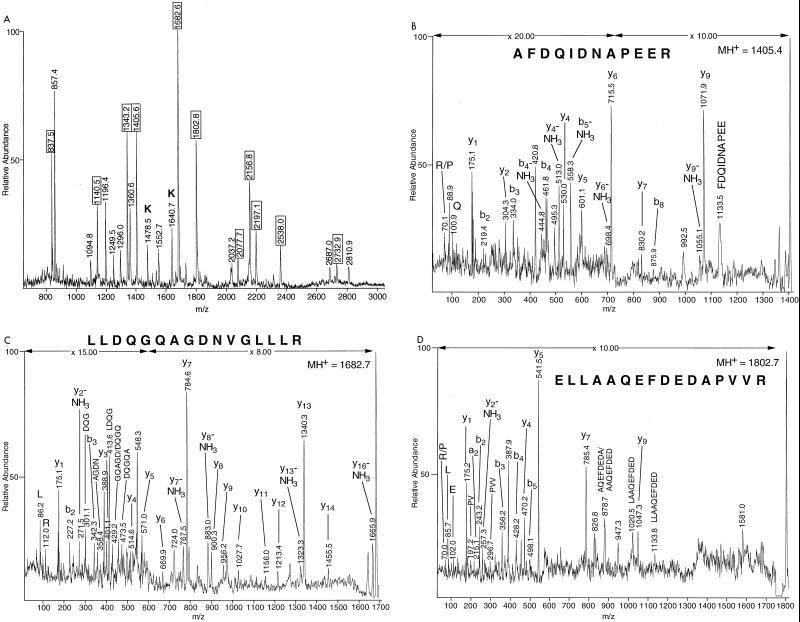
Protein spots from single (spots 1, 2, 15, 18, 21, and 22 to 25) or several (spots 4, 11, and 21) 2-D gels were cut from the gels and digested with trypsin. The resulting digestion mixture was analyzed by MALDI-MS to determine the molecular masses of the tryptic peptides (Fig. 2A and Fig. 3A). To obtain sequence information, masses of the fragment ions of selected tryptic peptides were obtained by using MALDI-PSD (see Fig. 3B to D) or MALDI-CID (see Fig. 2B) MS. MS-Tag fragment ion searches with minimally restricted search parameters (Mycobacterium species and 1,000 to 120,000 Da) of the NCBI protein database offered possible matches to peptide sequences of M. tuberculosis proteins, although in some searches, the peptide fragmentation data also matched peptides in the same protein in an additional mycobacterium species (e.g., aconitase and elongation factor Tu [EF-Tu] [Table 1]). This information, in conjunction with the mass fingerprints of the proteins obtained by MALDI-TOF MS analysis, allowed for the matching of 11 protein spots to M. tuberculosis proteins in the database. BLAST searches of these protein sequences yielded possible identities and functional roles of many of these proteins, as summarized below and in Table 1.
FIG. 2.
Analysis of aconitase homolog by MS. (A) MALDI-TOF MS peptide mass fingerprint spectrum produced by in-gel tryptic digestion of gel spot 1. Masses outlined by boxes indicate tryptic peptides from the M. tuberculosis aconitase protein. Labeling of peaks with K indicates that they are tryptic peptides from contaminating human keratin. (B) MALDI-CID mass spectrum of a tryptic peptide with MH+ at m/z of 1,897.0 (monoisotopic). Fragment ions referred to as a and b ions originate from peptide backbone cleavage with the charge retained on the N terminus, and y ions refer to peptide fragments with the charge retained on the C terminus (5). Multiple bond cleavages internal to the peptide are labeled with the corresponding portion of the peptide sequence. All (or a combination) of these fragment ions were used in the MS-Tag database searches.
FIG. 3.
Analysis of EF-Tu homolog (spot 4) by mass spectrometry. (A) MALDI-TOF MS peptide mass fingerprint spectrum produced by in-gel tryptic digestion of gel spot 4, pooled from four 2-D gels. (B through D) MALDI-PSD mass spectrum of a tryptic peptide with (B) MH+ at m/z of 1,405.4, (C) MH+ at m/z of 1,682.7, and (D) MH+ at m/z of 1,802.7. Fragment ions detected in the PSD analysis were used to search the protein databases with MS-Tag. All three peptides were identified as belonging to EF-Tu proteins from both M. tuberculosis and M. leprae. In the mass fingerprint spectrum (panel A) the peak at m/z of 857.4 was identified as SQRYFR from MALDI-PSD data and was identified as the sole peptide in spot 4 belonging to a minor contaminating protein (NCBI accession no. 1722931) of unknown function. See legend to Fig. 2 for an explanation of peak labels.
TABLE 1.
Summary of proteins identified from 2-D gels of M. tuberculosis cell extracts
| Spot no. | Name (NCBI accession no.), MW | Peptide MH+ (Da) | Δ in mass | Start-end position | Sequenceb |
|---|---|---|---|---|---|
| 1 | Aconitase (NCBI 2791409), 102,449.6 Da | 927.9 | 0.3 | 42–49 | VLAENLLR |
| 845.6 | 0.2 | 747–754 | DYNSFGSR | ||
| 1,021.8 | 0.3 | 839–847 | AVIAESFER | ||
| 1,349.9 | 0.2 | 904–916 | GDGATIEFDAVVR | ||
| 1,438.1 | 0.4 | 773–785 | NQLLDDVSGGYTR | ||
| 1,897.0 | 0.5 | 278–294 | FVEFYGEGVAEVPLANRe | ||
| 2,297.3a | 0.8 | 667–686 | NLPTPSGNTFEWDPNSTYVR | ||
| 2 | PEPCK (NCBI 1871584), 67,253.5 Da | 688.3 | 0 | 524–528 | WIVDR |
| 957.6 | 0.2 | 371–377 | GNDWYFR | ||
| 1,086.7 | 0.1 | 423–432 | TTVPLVTEAR | ||
| 1,114.7 | 0.1 | 496–503 | VFFVNWFR | ||
| 1,163.7 | 0.1 | 203–211 | YITHFPETR | ||
| 1,310.4 | 0.8 | 510–520 | FLWPGFGENSR | ||
| 1,377.8 | 0.2 | 290–301 | AETLGDDIAWMR | ||
| 1,544.1 | 0.4 | 165–178 | AALEKMGDDGFFVKc | ||
| 1,544.1 | 0.3 | 110–121 | SIMKDLYRGCMR | ||
| 4 | EF-Tu (NCBI 399422), 43,593.8 | 837.5 | 0 | 120–126 | EHVLLAR |
| 1,140.5 | 0.07 | 256–265 | TTVTGVEMFR | ||
| 1,343.2 | 1.5 | 365–376 | LIQPVAMDEGLR | ||
| 1,405.6 | 1 | 48–59 | AFDQIDNAPEERf | ||
| 1,682.6 | 0.7 | 267–282 | LLDQGQAGDNVGLLLRf | ||
| 1,802.8a | 0.2 | 158–173 | ELLAAQEFDEDAPVVRf | ||
| 2,077.2a | 0.9 | 140–157 | ADAVDDEELLELVEMEVR | ||
| 2,156.8a | 1.3 | 236–255 | GVINVNEEVEIVGIRPSTTK | ||
| 2,197.1a | 0.6 | 207–225 | ETDKPFLMPVEDVFTITGR | ||
| 2,538.0a | 1.9 | 233–255 | VERGVINVNEEVEIVGIRPSTTR | ||
| 2,732.9a | 1.7 | 93–119 | NMITGAAQMDGAILVVAATDGPMPQTR | ||
| 11 | Oxidoreductase (NCBI 2072661), 29,814.6 | 970.3 | 0.8 | 7–15 | TMFISGASR |
| 1,193.5 | 0.9 | 113–122 | FDLMNGIQVR | ||
| 1,209.5 | 0.9 | 113–122 | FDLMNGIQVRc | ||
| 1,768.7a | 0.7 | 55–71 | ELEEAGGQALPIVGDOR | ||
| 2,037.7a | 1.4 | 194–213 | TMVATAAVQNLLGGDEAMARc | ||
| 2,530.6a | 1.7 | 216–238 | KPEVYADAAYVIVNKPATEYTGK | ||
| 15 | Hypothetical 21.5-kDa protein (NCBI 1731190), 21,534.6 | 866.3 | 0.3 | 75–81 | FTAEELR |
| 922.3 | 0.1 | 95–101 | YNELVER | ||
| 1,239.5 | 0.9 | 83–94 | AAEGYLEAATSR | ||
| 1,584.6a | 0.2 | 60–72 | LQEDLPEQLTELR | ||
| 1,648.2a | 0.6 | 95–108 | YNELVERGEAALER | ||
| 2,538.7a | 0.9 | 122–145 | AEGYVDQAVELTQEALGTVASQTR | ||
| 18 | PPIase (NCBI 1552563), 19,239.5 | 1,602.7 | 0 | 141–154 | HTIFGEVIDAESQR |
| 2,203.8a | 1.4 | 162–182 | TATDGNDRPTDPVVIESITIS | ||
| 2,431.2a | 0.3 | 51–73 | DYSTQNASGGPSGPFYDGAVFHR | ||
| 21 | Lsr2 (NCBI 2113979), 12,098.5 | 770.5 | 0.1 | 50–56 | QWVAAGRd |
| 787.5 | 0.1 | 50–56 | QWVAAGR | ||
| 884.6 | 0.2 | 90–97 | NGHNVSTR | ||
| 1,040.7 | 0.2 | 89–97 | RNGHNVSTR | ||
| 1,286.8 | 0.1 | 73–84 | GAIDREQSAAIR | ||
| 1,570.4 | 0.6 | 98–112 | GRIPADVIDAYHAAT | ||
| 22 | Hypothetical 15.9-kDa protein (NCBI 2113920), 15,312.4 | 1,113.0 | 0.4 | 124–134 | LLGSVPANVSR |
| 1,629.4a | 0.5 | 60–74 | VTGTAPIYEILHDAK | ||
| 1,656.1a | 0.2 | 107–123 | ADLLVVGNVGLSTIAGR | ||
| 1,698.0a | 0.1 | 34–48 | LIIASAYLPQHEDAR | ||
| 2,478.7a | 0.1 | 84–106 | NVEERPIVGAPVDALVNLADEEK | ||
| 23, 24 | HSP 16.3 (α-crystallin) (NCBI 231343), 16,227.4 | 1,163.0 | 0.5 | 91–100 | SEFAYGSFVR |
| 25 | Fur (NCBI 231343), 15,892.0 | 1,410.9 | 1.2 | 1–12 | MSSIPDYAEQLR |
| 1,450.1 | 0.4 | 72–84 | IQPSGSVARYESR | ||
| 1,577.1 | 0.7 | 71–84 | KIQPSGSVARYESR |
MW values indicate average masses (all other masses are monoisotopic).
Sequences in boldface type were determined by MALDI-PSD. Sequences in italic type were determined by MALDI-CID.
Contains one oxidized methionine.
Contains N-terminal pyro-glutamic acid.
MS-Tag search by using this sequence found M. tuberculosis and M. avium aconitase proteins containing this peptide.
MS-Tag searches by using these sequences found M. tuberculosis and M. leprae EF-Tu proteins containing these peptides.
Regulators. (i) Fur.
Protein gel spot 25, whose expression was upregulated under high-iron-concentration conditions and virtually absent under low-iron-concentration conditions, was taken from a single 2-D gel. The gel plug was subjected to in-gel digestion with trypsin and the extracts were analyzed by MALDI-TOF MS to obtain molecular masses of the tryptic peptides. Analysis of the peptide with MH+ at m/z of 1,410.8 by MALDI-CID MS and subsequent searching of the protein sequence databases with MS-Tag identified spot 25 as an M. tuberculosis Fur homolog. Fur (or ferric uptake regulator) proteins are a family of iron-responsive DNA-binding proteins. Subsequent review of the MALDI mass spectrum of the unseparated digestion mixture revealed two other tryptic peptides belonging to the M. tuberculosis Fur protein.
The level of iron in the environment is known to regulate the expression of genes coding for many high-affinity bacterial iron uptake pathways. Under iron-rich conditions, the Fur protein is activated when it binds Fe(II) as a cofactor. This activated repressor is then able to bind the “Fur box,” a consensus sequence located in the promoter region of many bacterial genes. In conditions of iron deprivation, the Fur protein does not bind to the promoter sequence, allowing for the transcription of the genes (47). Homologs of the Fur repressor have been found in many gram-negative bacteria. Their sequences appear to be fairly well conserved, with a high degree of homology with the first-discovered Escherichia coli Fur protein, ranging from 49% homology for Neisseria gonorrhoeae Fur (4) to 76% for Vibrio vulnificus Fur (37). The Fur homolog of M. tuberculosis shows less identity to the Fur proteins of E. coli (22.9%), Legionella pneumophila (28.4%), and N. gonorrhoeae (25.4%). Originally thought of as only a negative repressor, Fur is now known to also positively regulate many genes in E. coli and Salmonella typhimurium (17, 59). Fur may also act as a global regulator affecting gene expression in response to signals besides iron levels. In addition, Fur may be part of a cascade of control elements in which it regulates the expression of secondary regulatory elements (17).
(ii) Aconitase.
Protein gel spot 1, which was expressed at a higher level under conditions of high iron concentration, was excised from a single 2-D gel and subjected to in-gel digestion with trypsin. MALDI-TOF MS analysis of the digestion extracts yielded molecular masses of several tryptic peptides (Fig. 2A). The peptide with MH+ at m/z of 1,897.0 (monoisotopic) was analyzed by MALDI-CID MS to obtain additional sequence information (Fig. 2B). The resultant fragment ion data were used to search the protein databases by using MS-Tag, and the peptide was originally matched to a portion of a Mycobacterium avium protein (MW, 104,025.7 Da). A subsequent BLAST search of the nonredundant protein database with the M. avium peptide sequence revealed the protein to be an aconitase. The tight correlation of the fragmentation data with the M. avium aconitase peptide suggested that spot 1 was an M. tuberculosis aconitase protein whose sequence had yet to be entered into the database. Subsequent to our initial identification of this protein as an aconitase homolog, the sequence for the M. tuberculosis aconitase protein (MW, 102,449.6 Da) was entered; it contains a peptide (FVEFYGEGVAEVPLANR) whose sequence is identical to that of the M. avium peptide that was originally discovered by MS-Tag. A total of eight tryptic peptides from the M. tuberculosis aconitase were detected in the MALDI mass spectrum of the unseparated digest. The M. tuberculosis aconitase protein is highly homologous to the M. avium aconitase (83.1% identity) and to a lesser extent to the aconitases from E. coli (58.3% identity) and the mouse (51.0% identity).
It is not surprising that an aconitase was identified as one of the iron-regulated proteins in these studies. The cytosolic aconitase is a protein with dual roles. It catalyzes the reversible isomerization of citrate and isocitrate via cis-aconitate, as part of the Krebs cycle, and serves as an iron-responsive element (IRE) binding protein. Aconitases are monomeric proteins that contain a single cubane (4Fe-4S) cluster. In the large family of 4Fe-4S proteins, it is unusual, in that only three of the irons are directly ligated to the peptide backbone through cysteines. In aconitase, the fourth iron is labile and coordinates the atoms of the cluster along with a water molecule and the substrate. The instability of the cluster is exploited as a molecular switch, enabling cells to reciprocally regulate the aconitase and RNA binding activity of the protein in response to changes in iron levels. Under conditions of iron deprivation, the apo form (with no 4Fe-4S cluster) of the protein is inactive as an aconitase but active for RNA binding. The IRE-binding function of the protein results in a tight interaction with the IREs contained in the mRNAs of molecules involved in iron storage in mammalian cells, like transferrin receptor and ferritin. Binding of the protein to IREs located in the 5′ untranslated region of mRNAs prevents translation by inhibiting the binding of initiation factors. However, binding of the protein to IREs in the 3′ untranslated region stimulates mRNA translation by protecting the mRNA against degradation (45). When iron levels rise, the protein dissociates from the IRE and is again active as an aconitase. The mechanism underlying the physiological role of the cytosolic aconitase in iron regulation is still unclear. The fact that citrate, the substrate for the aconitase, is capable of binding iron and has been proposed as a possible transporter of intracellular iron in mammals is very curious. In one model, the increased synthesis of iron storage proteins (ferritin and transferrin) and reduced synthesis of iron uptake proteins (transferrin receptors) in iron-replete conditions, in addition to the reduced levels of citrate (conversion to isocitrate by aconitase), eventually lead to reduced intracellular iron levels and the subsequent conversion of the protein back to its iron-binding form (44).
(iii) EF-Tu.
Protein gel spot 4, which is expressed at higher levels when grown in the high-iron-concentration medium, was excised from four 2-D gels and subjected to in-gel tryptic digestion. The resultant digestion mixture was analyzed by MALDI-TOF MS (Fig. 3A). The tryptic peptides with MH+ at m/z of 1,405.6 (Fig. 3B), 1,682.6 (Fig. 3C), and 1,802.8 (Fig. 3D) were further analyzed by MALDI-PSD MS to obtain sequence information. Database searching by using MS-Tag and BLAST revealed spot 4 to be EF-Tu, a helper protein involved in protein synthesis encoded by the tuf gene. A total of 11 tryptic peptides from M. tuberculosis EF-Tu were observed, and additional sequence information was obtained on three of the peptides (see Fig. 3). EF-Tu is a GTPase which promotes the binding of aminoacyl-tRNA to ribosomes (58). The tuf gene of M. tuberculosis was discovered when a λgt11 M. tuberculosis gene library was screened with monoclonal antibodies raised by immunizing rats with live Mycobacterium bovis bacillus Calmette-Guerín. The M. tuberculosis EF-Tu homolog showed high sequence similarity with EF-Tu proteins from several other organisms, including Mycobacterium leprae (95.2% identity), S. typhimurium (75.1% identity), and E. coli (75.1% identity).
Besides its essential role in protein biosynthesis, EF-Tu has been shown by Young and Bernlohr to be methylated and to become membrane associated when E. coli is starved for glucose, galactose, ammonia, glutamate, or phosphate (63). This raises the possibility that EF-Tu may also have roles in the regulation of cell growth and the organism’s response to stress. These investigators propose that EF-Tu’s membrane association allows for its interaction with receptors or proteins that interact with nutrients in the environment that could regulate its methylation. Conditions of nutrient deprivation would result in EF-Tu hypermethylation and subsequent membrane release, allowing for the possibility that the intracellular EF-Tu assumes multiple regulatory roles. In addition to regulating translation through its interaction with tRNA and ribosomes, enabling it to stop the translation of unnecessary proteins and trigger the synthesis of stress-induced proteins, EF-Tu has also been known to act as a transcriptional activator in the presence of RNA polymerase and the appropriate sigma factor (21, 60). Therefore, it may be able to regulate both the translation and transcription of starvation-induced proteins. A preliminary report that patients with tuberculosis but not tuberculin-negative individuals develop an antibody response to the protein suggests a potential role for the protein in serodiagnosis of mycobacterial disease (9).
Antigens. (i) LSR2.
Protein gel spot 21, expressed at a higher level under high-iron-concentration conditions, was excised from four 2-D gels. In-gel tryptic digestion, followed by MALDI-MS analysis, revealed gel spot 21 to be an M. tuberculosis homolog of LSR2, a protein antigen of M. leprae. The digestion mixture was found to contain six tryptic peptides. Additional sequence information was obtained by using MALDI-PSD MS on two of the peptides.
Using polyclonal antibodies from pooled sera of lepromatous patients, Laal et al. (31) screened a λgt11 DNA expression library in an effort to identify genes involved in the immune response to M. leprae infection. These investigators identified LSR2, a dominant T-cell antigen. BLAST searches of this ∼10-kDa protein revealed that M. tuberculosis LSR2 has 92.9% identity with the LSR protein of M. leprae but is not homologous to any other known proteins. Analysis of overlapping peptides spanning the M. leprae LSR sequence showed that two peptides (GVTYEIDLTNKNAA and IDLTNKNAAKLRGD) were recognized by the sera of leprosy patients (56). Single-residue deletions of the peptides enabled the identification of three distinct sequences (GVTY, NAA, and RGD) found to be important for antibody recognition (55). Although nothing is yet known of the M. tuberculosis homolog’s role in the immune response, two of the three sequences important for antibody recognition in leprosy patients, GVTY and RGD, are present in its sequence.
(ii) Hsp16.3 (α-crystallin homolog).
Protein gel spot 23, upregulated under conditions of high iron concentration, was taken from a single 2-D gel and subjected to in-gel digestion and MALDI-MS analysis. Only one M. tuberculosis tryptic peptide was identified in the digestion mixture. MALDI-PSD MS analysis on this peptide (MH+ at m/z of 1,163.0) and database searching revealed gel spot 23 to be a small heat shock protein (Hsp16.3) of the α-crystallin family (61). Mass spectrometric analysis showed gel spot 24 also to have tryptic peptides originating from Hsp16.3, possibly a degraded and/or truncated form of the protein. Members of this family of small heat shock proteins are thought to function as chaperones, acting as molecular surfactants which prevent protein aggregation through nonspecific weak interactions with the properly folded proteins (10, 36). Hsp16.3 is a major M. tuberculosis antigen which can generate a cell-mediated immune response and is thought to be located on the periphery of the cell membrane (27, 32). When Hsp16.3 was overexpressed in wild-type M. tuberculosis, a slower decline in viability after the end of log-phase growth was observed (64). Besides the M. tuberculosis Hsp16.3, an α-crystallin homolog has been detected in M. leprae (46) and M. bovis but not in Mycobacterium smegmatis or the pathogenic species M. avium (64).
In investigations into the regulation of Hsp16.3 expression under various stress conditions, carbon starvation, heat and cold shock, and low pH all failed to induce Hsp16.3 expression (64). The only environmental stress shown to significantly upregulate the expression of Hsp16.3 was growth under microaerobic or anaerobic conditions (12, 64). The environment inside caseous granulomas, where mycobacteria are thought to exist in a dormant state, is unknown. However, many environmental stresses, like oxygen deprivation, may signal mycobacteria to produce proteins like the α-crystallin homolog to aid in the long-term survival of the bacteria. On the other hand, the diversity of this family of small heat shock proteins suggests that the protective capacity of this protein may be general and not necessarily specific to the pathogenic species M. tuberculosis and M. leprae.
Enzymes. (i) PEPCK.
Protein gel spot 2, which was upregulated under low-iron-concentration conditions, was found to be homologous to many GTP-dependent phosphoenolpyruvate (PEP) carboxykinases (PEPCK) from numerous other species, including M. leprae (86.0% identity), Drosophila melanogaster (51.3% identity), and Homo sapiens (52.5% identity). MALDI-MS analysis of the digestion mixture revealed nine tryptic peptides. Further analysis with MALDI-PSD MS was carried out on two of the peptides to obtain sequence information. PEPCK is part of the gluconeogenic pathway, catalyzing the reversible decarboxylation and mononucleotide-dependent phosphorylation of oxaloacetate PEP (42). Most PEPCKs require two metal cations for activity. One of the cations (Mg2+ or Mn2+) must complex with the substrate to form a cation-nucleotide complex. For optimal activity, a second cation (often a transition metal) is required to interact directly with the protein, possibly mediating the interaction of the substrate (oxaloacetate or PEP) with the enzyme to facilitate the formation of the active ternary complex (34). In GTP-dependent PEPCKs, it has been proposed that the second ion may help position the substrate for catalysis by binding the PEP phosphoryl group and the nucleotide β- or γ-phosphate, either directly (23) or through an interaction with water (13, 35, 42).
In early investigations, Fe(II) was found to be the most efficient activator of rat liver cytosolic PEPCK when incubated with the cytosol fraction of the liver homogenate (3). However, studies found a rapid Fe(II)-dependent inactivation after continued incubation with the enzyme in the absence of substrate. This loss of activity is thought to result from PEPCK oxidative damage caused by the reactive oxygen species formed from Fe(II) autoxidation (52). The presence of a ferroactivator, a cytosolic protein factor originally identified as glutathione peroxidase in rat liver and thought to act by removing reactive oxygen species, results in the restoration of enzyme activity (50). It has been proposed that PEPCK may exist in a dynamic equilibrium between an active and inactive form (caused by the Fe(II) autoxidation). If this is the case, the ferroactivator may have a role in shifting the equilibrium towards the active enzyme (52).
(ii) Oxidoreductase.
Protein gel spot 11, which was down-regulated in low-iron-concentration medium, was identified as a homolog of an oxidoreductase. A total of six tryptic peptides was detected by MALDI-MS analysis of the digestion mixture. The large number of microbial alcohol oxidoreductases can be categorized into three major groups: (i) NADP-dependent dehydrogenases, (ii) NADP-independent enzymes which use pyrroloquinoline quinone, heme, or cofactor F420 as the cofactor, and (iii) enzymes that catalyze the essentially irreversible oxidation of alcohols (51). The M. tuberculosis oxidoreductase homolog belongs to a subgroup of the first group, NADP-dependent dehydrogenases, the short-chain alcohol dehydrogenase superfamily. Members of this subgroup are known to act on a large variety of substrates, including sugars, steroids, prostaglandins, aromatic hydrocarbons, antibiotics, and compounds involved in nitrogen metabolism (30). The short-chain alcohol dehydrogenase enzymes do not require any metal ions to function and are typically around 250 amino acids in length. Sequence comparisons between members of the superfamily reveal six conserved domains (30). Among the family members, there is a pattern that 13 residues are largely conserved. Alignments between enzyme pairs typically reveal approximately 25% identity, with the identities for single forms ranging from 14 to 58% (48). The oxidoreductase of M. tuberculosis shows homology to enzymes from Caenorhabditis elegans (48% identity), E. coli (27.6% identity), and H. sapiens (19.5% identity) within this range.
(iii) PPIase.
Gel spot 18, whose expression is reduced in a low-iron-concentration environment, was removed from a single 2-D gel. MALDI-PSD MS analysis of two of the three tryptic peptides found in the digestion mixture (MH+ at m/z of 1,602.7 and 2,203.8) followed by database searching matched them to an M. tuberculosis protein (MW, 19,239.5) with significant homology to peptidyl-prolyl cis-trans isomerases (PPIase).
Immunophilins are housekeeping proteins with many roles, including membrane channeling and protein folding and trafficking (15). Besides exhibiting PPIase activity in the unliganded form, immunophilin-drug (cyclophilin or FK506) complexes inhibit clonal expansion of T cells and have toxic effects on numerous other cellular components. Many intracellular pathogens produce proteins with significant homology to immunophilins and have PPIase activity. The role of these proteins in microbial pathogenicity is as yet unclear; the immunophilins may interact with various partner molecules in mammalian cells or may interact with other components through their PPIase activities by interrupting protein folding or altering protein structure dynamics (20). Many facultative or obligate intracellular pathogens, for example L. pneumophila and Chlamydia trachomatis, produce FK506-binding protein (FKBP)-like immunophilins, such as the Mip (macrophage infectivity potentiator) protein of Legionella spp. These proteins have been proposed to aid in intracellular survival. Mip-protein-negative mutants appear to have a reduced ability to initiate intracellular replication (11). The site of action of Mip protein is not known, nor is it known whether it acts by altering the conformation of other Legionella proteins (20).
The M. tuberculosis PPIase shows homology to immunophilins of the cyclophilin family of several other species including Streptomyces chrysomallus (61.4% identity), C. elegans (48.4% identity), and H. sapiens (46.1% identity). Unlike the Mip proteins of the FKBP family, these bacterial proteins have not been extensively characterized. However, the existence of cyclophilin-like proteins in E. coli, S. typhimurium, and L. pneumophila suggests that a large array of these proteins is produced by bacterial species. There is speculation that these proteins have roles in protein folding and secretion (38).
CONCLUSION
In this study, we have used 2-D gel electrophoresis, MALDI-MS, and database searching to identify M. tuberculosis proteins regulated by extracellular iron levels. Previous studies in our laboratories with 1-D sodium dodecyl sulfate-polyacrylamide gel electrophoresis (unpublished data) did not allow for the direct identification of iron-regulated proteins, since single bands were found to contain multiple comigrating proteins. However, once we obtained the proper conditions for the separation of complex protein mixtures from M. tuberculosis cell lysates by 2-D gel electrophoresis, we were able to analyze and identify single-protein-containing gel spots by MS. The number of M. tuberculosis tryptic peptides detected in digestion mixtures from individual samples varied greatly, ranging from as few as 1 to as many as 11 peptides (Table 1). Therefore, in addition to determining the masses of the tryptic peptides, we chose to generate sequence information. In most cases, useful sequence data could be obtained from only the more abundant signals in the MALDI mass spectra of the digestion mixtures by PSD and/or CID analysis. This sequence data provided a higher level of confidence in the identification of proteins than would have been afforded by tryptic peptide masses alone. This was particularly true in cases where only a few peptide molecular masses could readily be observed over background.
2-D gel electrophoresis identified at least 27 proteins whose expression is modulated by iron concentration—15 proteins are upregulated and 12 proteins are downregulated under low-iron-concentration conditions. Of these proteins, 11 (including two forms of 1 protein) were identified by MALDI-MS and database searching. Two of these proteins (gel spots 15 and 22) were not found to be homologous to any proteins of known function and are listed as hypothetical proteins in Table 1. The identification of two of the proteins as Fur and aconitase homologs suggests the possibility that these proteins function as transcriptional regulators in M. tuberculosis, as in other bacteria, and that they exert control over the expression of many of the other proteins which show differential expression under low- and high-iron-concentration conditions. In future experiments, this may be explored further by comparing the protein expression of Fur or aconitase deletion mutants with wild-type M. tuberculosis or by analyzing genes with upstream Fur or aconitase binding regions. EF-Tu may also play a role in the organism’s response to iron starvation conditions through its ability to regulate protein expression.
Despite the successful identification of 10 proteins in this study, we were unable to obtain sufficient data to identify unambiguously 16 of the proteins that appeared to be under iron regulation. Further refinements in methodology should allow us to identify these latter proteins in addition to other proteins whose expression levels are very low and may be visualized only by silver staining. Alternatively, the application of DNA microarray technologies (for a review, see reference 40) could provide information at the level of RNA message. While such information does not necessarily correlate with changes in protein expression, it may be useful for comparative purposes and/or to suggest additional protein candidates for further investigation by the 2-D gel electrophoresis approach.
Our study demonstrates the feasibility and utility of combining three powerful technologies—2-D gel electrophoresis, MS, and database searching—to study how mycobacteria and other pathogens respond to changes in the environment. Studies of this type should help us to take full advantage of the wealth of new data provided by genomic studies and greatly enhance our understanding of the pathophysiology of M. tuberculosis and other human pathogens.
ACKNOWLEDGMENTS
We thank Karl Clauser for helpful discussions and Michael Tullius for help with the sequence analysis.
This work was supported by the Universitywide AIDS Research Program (R96-SF-1301 and R96-LA-1302) and through the UCSF mass spectrometry facility, which is partially funded by the NCRR (RR01614). Diane Wong was partially supported by a fellowship from the American Foundation for Pharmaceutical Education and by the NIH pharmaceutical sciences training grant (GM07175).
ADDENDUM IN PROOF
Since the submission of this manuscript, the sequence of the complete genome of Mycobacterium tuberculosis has been published (S. T. Cole et al., Nature 393:537–544, 1998.)
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bateman R H, Green M R, Scott G, Clayton E. A combined magnetic sector-time-of-flight mass spectrometer for structural determination studies by tandem mass spectrometry. Rapid Commun Mass Spectrom. 1995;9:1227–1233. [Google Scholar]
- 3.Bentle L A, Snoke R E, Lardy H A. A protein factor required for activation of phosphoenolpyruvate carboxykinase by ferrous ions. J Biol Chem. 1976;251:2922–2928. [PubMed] [Google Scholar]
- 4.Berish S A, Subbarao S, Chen C Y, Trees D L, Morse S A. Identification and cloning of a Fur homolog from Neisseria gonorrhoeae. Infect Immun. 1993;61:4599–4606. doi: 10.1128/iai.61.11.4599-4606.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biemann K. Peptides and proteins: overview and strategy. Methods Enzymol. 1990;193:351–360. doi: 10.1016/0076-6879(90)93426-l. [DOI] [PubMed] [Google Scholar]
- 6.Byrd T F, Horwitz M A. Interferon gamma-activated human monocytes downregulate transferrin receptors and inhibit the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. J Clin Investig. 1989;83:1457–1465. doi: 10.1172/JCI114038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd T F, Horwitz M A. Regulation of transferrin receptor expression and ferritin content in human mononuclear phagocytes. Coordinate upregulation by iron transferrin and downregulation by interferon gamma. J Clin Investig. 1993;91:969–976. doi: 10.1172/JCI116318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calder K M, Horwitz M A. Identification of iron-regulated proteins of Mycobacterium tuberculosis and cloning of tandem genes encoding a low iron-induced protein and a metal transporting ATPase with similarities to two-component metal transport systems. Microb Pathog. 1998;24:133–143. doi: 10.1006/mpat.1997.9999. [DOI] [PubMed] [Google Scholar]
- 9.Carlin N I, Lofdahl S, Magnusson M. Monoclonal antibodies specific for elongation factor Tu and complete nucleotide sequence of the tuf gene in Mycobacterium tuberculosis. Infect Immun. 1992;60:3136–3142. doi: 10.1128/iai.60.8.3136-3142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Z, Primm T P, Jakana J, Lee I H, Serysheva I, Chiu W, Gilbert H F, Quiocho F A. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J Biol Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- 11.Cianciotto N P, Eisenstein B I, Mody C H, Engleberg N C. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J Infect Dis. 1990;162:121–126. doi: 10.1093/infdis/162.1.121. [DOI] [PubMed] [Google Scholar]
- 11a.Clauser, K. Personal communication.
- 11b.Clauser, K., et al. Unpublished data.
- 12.Cunningham A F, Spreadbury C L. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy T H, Nowak T. 1H and 31P relaxation rate studies of the interaction of phosphoenolpyruvate and its analogues with avian phosphoenolpyruvate carboxykinase. Biochemistry. 1985;24:1152–1160. doi: 10.1021/bi00326a014. [DOI] [PubMed] [Google Scholar]
- 14.Eidus L, Hamilton E J. In vitro test with 4,4′-dilsoamyloxythiocarbanilide. Am Rev Respir Dis. 1964;90:258–260. doi: 10.1164/arrd.1964.90.2.258. [DOI] [PubMed] [Google Scholar]
- 15.Fischer G, Schmid F X. The mechanism of protein folding. Implications of in vitro refolding models for de novo protein folding and translocation in the cell. Biochemistry. 1990;29:2205–2212. doi: 10.1021/bi00461a001. [DOI] [PubMed] [Google Scholar]
- 16.Fish W W. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 17.Foster J W, Hall H K. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J Bacteriol. 1992;174:4317–4323. doi: 10.1128/jb.174.13.4317-4323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gobin J, Horwitz M A. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. J Exp Med. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gobin J, Moore C H, Reeve J R, Jr, Wong D K, Gibson B W, Horwitz M A. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci USA. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacker J, Fischer G. Immunophilins: structure-function relationship and possible role in microbial pathogenicity. Mol Microbiol. 1993;10:445–456. doi: 10.1111/j.1365-2958.1993.tb00917.x. [DOI] [PubMed] [Google Scholar]
- 21.Halliday K R. Regional homology in GTP-binding proto-oncogene products and elongation factors. J Cyclic Nucleotide Protein Phosphorylation Res. 1983;9:435–448. [PubMed] [Google Scholar]
- 22.Harth G, Lee B Y, Wang J, Clemens D L, Horwitz M A. Novel insights into the genetics, biochemistry, and immunocytochemistry of the 30-kilodalton major extracellular protein of Mycobacterium tuberculosis. Infect Immun. 1996;64:3038–3047. doi: 10.1128/iai.64.8.3038-3047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebda C A, Nowak T. Phosphoenolpyruvate carboxykinase. Mn2+ and Mn2+ substrate complexes. J Biol Chem. 1982;257:5515–5522. [PubMed] [Google Scholar]
- 24.Horwitz M A, Lee B W, Dillon B J, Harth G. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1995;92:1530–1534. doi: 10.1073/pnas.92.5.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karas M, Bahr U, Giessmann U. Matrix-assisted laser desorption ionization mass spectrometry. Mass Spectrom Rev. 1991;10:335–357. [Google Scholar]
- 26.Kaufmann R, Spengler B, Lutzenkirchen F. Mass spectrometric sequencing of linear peptides by production analysis in a reflectron time-of-flight mass spectrometer using matrix-assisted laser desorption ionization. Rapid Commun Mass Spectrom. 1993;7:902–910. doi: 10.1002/rcm.1290071010. [DOI] [PubMed] [Google Scholar]
- 27.Kingston A E, Salgame P R, Mitchison N A, Colston M J. Immunological activity of a 14-kilodalton recombinant protein of Mycobacterium tuberculosis H37Rv. Infect Immun. 1987;55:3149–3154. doi: 10.1128/iai.55.12.3149-3154.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kochan I. Mechanism of tuberculostasis in mammalian serum. I. Role of transferrin in human serum tuberculosis. J Infect Dis. 1969;119:11–18. doi: 10.1093/infdis/119.1.11. [DOI] [PubMed] [Google Scholar]
- 29.Kochan I, Pellis N R, Golden C A. Mechanism of tuberculostasis in mammalian serum. III. Neutralization of serum tuberculostasis by mycobactin. Infect Immun. 1971;3:553–558. doi: 10.1128/iai.3.4.553-558.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krozowski Z. The short-chain alcohol dehydrogenase superfamily: variations on a common theme. J Steroid Biochem Mol Biol. 1994;51:125–130. doi: 10.1016/0960-0760(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 31.Laal S, Sharma Y D, Prasad H K, Murtaza A, Singh S, Tangri S, Misra R S, Nath I. Recombinant fusion protein identified by lepromatous sera mimics native Mycobacterium leprae in T-cell responses across the leprosy spectrum. Proc Natl Acad Sci USA. 1991;88:1054–1058. doi: 10.1073/pnas.88.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee B Y, Hefta S A, Brennan P J. Characterization of the major membrane protein of virulent Mycobacterium tuberculosis. Infect Immun. 1992;60:2066–2074. doi: 10.1128/iai.60.5.2066-2074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B Y, Horwitz M A. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J Clin Investig. 1995;96:245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee M H, Hebda C A, Nowak T. The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation. J Biol Chem. 1981;256:12793–12801. [PubMed] [Google Scholar]
- 35.Lee M H, Nowak T. Phosphorus-31 nuclear relaxation rate studies of the nucleotides on phosphoenolpyruvate carboxykinase. Biochemistry. 1984;23:6506–6513. doi: 10.1021/bi00321a036. [DOI] [PubMed] [Google Scholar]
- 36.Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 37.Litwin C M, Calderwood S B. Cloning and genetic analysis of the Vibrio vulnificus Fur gene and construction of a Fur mutant by in vivo marker exchange. J Bacteriol. 1993;175:706–715. doi: 10.1128/jb.175.3.706-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Walsh C T. Peptidyl-prolyl cis-trans-isomerase from Escherichia coli: a periplasmic homolog of cyclophilin that is not inhibited by cyclosporin A. Proc Natl Acad Sci USA. 1990;87:4028–4032. doi: 10.1073/pnas.87.11.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macham L P, Ratledge C, Nocton J C. Extracellular iron acquisition by mycobacteria: role of the exochelins and evidence against the participation of mycobactin. Infect Immun. 1975;12:1242–1251. doi: 10.1128/iai.12.6.1242-1251.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall A, Hodgson J. DNA chips: an array of possibilities. Nat Biotechnol. 1998;16:27–31. doi: 10.1038/nbt0198-27. [DOI] [PubMed] [Google Scholar]
- 41.Matsui N M, Smith D M, Clauser K R, Fichmann J, Andrews L E, Sullivan C M, Burlingame A L, Epstein L B. Immobilized pH gradient two-dimensional gel electrophoresis and mass spectrometric identification of cytokine-regulated proteins in ME-180 cervical carcinoma cells. Electrophoresis. 1997;18:409–417. doi: 10.1002/elps.1150180315. [DOI] [PubMed] [Google Scholar]
- 42.Matte A, Tari L W, Goldie H, Delbaere L T. Structure and mechanism of phosphoenolpyruvate carboxykinase. J Biol Chem. 1997;272:8105–8108. doi: 10.1074/jbc.272.13.8105. [DOI] [PubMed] [Google Scholar]
- 43.Medzihradszky K F, Maltby D A, Qiu Y, Yu Z, Hall S C, Chen Y, Burlingame A L. Protein sequence and structural studies employing matrix-assisted laser desorption ionization-high energy collision-induced dissociation. Int J Mass Spectrom Ion Processes. 1997;160:357–369. [Google Scholar]
- 44.Melefors O, Hentze M W. Iron regulatory factor—the conductor of cellular iron regulation. Blood Rev. 1993;7:251–258. doi: 10.1016/0268-960x(93)90012-s. [DOI] [PubMed] [Google Scholar]
- 45.Melefors O, Hentze M W. Translational regulation by messenger RNA protein interactions in eukaryotic cells—ferritin and beyond. Bioessays. 1993;15:85–90. doi: 10.1002/bies.950150203. [DOI] [PubMed] [Google Scholar]
- 46.Nerland A H, Mustafa A S, Sweetser D, Godal T, Young R A. A protein antigen of Mycobacterium leprae is related to a family of small heat shock proteins. J Bacteriol. 1988;170:5919–5921. doi: 10.1128/jb.170.12.5919-5921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ochsner U A, Vasil A I, Vasil M L. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J Bacteriol. 1995;177:7194–7201. doi: 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Persson B, Krook M, Jornvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991;200:537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- 49.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punekar N S, Lardy H A. Phosphoenolpyruvate carboxykinase ferroactivator 1. Mechanism of action and identity with glutathione peroxidase. J Biol Chem. 1987;262:6714–6719. [PubMed] [Google Scholar]
- 51.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 52.Reynolds C H. Activation and inactivation of phosphoenolpyruvate carboxykinase by ferrous ions. Biochem J. 1980;185:451–454. doi: 10.1042/bj1850451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenfeld J, Capdevielle J, Guillemot J C, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 54.Sheer D G. Protein and peptide recovery from polyacrylamide gels. In: Crabb J W, editor. Techniques in protein chemistry. V. San Diego, Calif: Academic Press; 1994. pp. 243–248. [Google Scholar]
- 55.Singh S, Jenner P J, Narayan N P S, Ramu G, Colston M J, Prasad H K, Nath I. Critical residues of the Mycobacterium leprae LSR recombinant protein discriminate clinical activity in erythema nodosum leprosum reactions. Infect Immun. 1994;62:5702–5705. doi: 10.1128/iai.62.12.5702-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh S, Narayan N P, Jenner P J, Ramu G, Colston M J, Prasad H K, Nath I. Sera of leprosy patients with type 2 reactions recognize selective sequences in Mycobacterium leprae recombinant LSR protein. Infect Immun. 1994;62:86–90. doi: 10.1128/iai.62.1.86-90.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snow G A. Isolation and structure of mycobactin T, a growth factor from Mycobacterium tuberculosis. Biochem J. 1965;97:166–175. doi: 10.1042/bj0970166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sprinzl M. Elongation factor Tu: a regulatory GTPase with an integrated effector. Trends Biochem Sci. 1994;19:245–250. doi: 10.1016/0968-0004(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 59.Stojiljkovic I, Hantke K. Functional domains of the Escherichia coli ferric uptake regulator protein (Fur) Mol Gen Genet. 1995;247:199–205. doi: 10.1007/BF00705650. [DOI] [PubMed] [Google Scholar]
- 60.Travers A A, Kamen R I, Schleif R F. Factor necessary for ribosomal RNA synthesis. Nature. 1970;228:748–751. doi: 10.1038/228748a0. [DOI] [PubMed] [Google Scholar]
- 61.Verbon A, Hartskeerl R A, Schuitema A, Kolk A H J, Young D B, Lathigra R. The 14,000-molecular-weight antigen of Mycobacterium tuberculosis is related to the alpha-crystallin family of low-molecular-weight heat shock proteins. J Bacteriol. 1992;174:1352–1359. doi: 10.1128/jb.174.4.1352-1359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wheeler P R, Ratledge C. Metabolism of Mycobacterium tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: ASM Press; 1994. pp. 353–385. [Google Scholar]
- 63.Young C C, Bernlohr R W. Elongation factor Tu is methylated in response to nutrient deprivation in Escherichia coli. J Bacteriol. 1991;173:3096–3100. doi: 10.1128/jb.173.10.3096-3100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan Y, Crane D D, Barry C E R. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial alpha-crystallin homolog. J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]