Abstract
Unipolar localization of IcsA on the surface of Shigella flexneri is required for efficient formation of actin tails and protrusions in infected eucaryotic cells. Lipopolysaccharide (LPS) mutations have been demonstrated to affect either the establishment or the maintenance of IcsA in a unipolar location, although the mechanism is unknown. In order to analyze the contribution of virulence plasmid determinants on the unipolar localization of IcsA, we examined the localization of IcsA expressed from a cloned plasmid copy in two different genetic backgrounds. The localization of IcsA was first examined in a virulence plasmid-cured derivative of the wild-type S. flexneri 2a isolate 2457T. This approach examined the contribution of virulence plasmid-borne factors, including the previously identified virulence plasmid-borne protease that is responsible for cleaving IcsA in the outer membrane and releasing the 95-kDa secreted form from the cell surface. IcsA localization in a related but nonpathogenic Escherichia coli strain expressing LPS of the O8 serotype was also examined. IcsA surface presentation in both of these genetic backgrounds continued to be unipolar, demonstrating that virulence plasmid-borne determinants are not responsible for unipolar localization of IcsA. The unipolar localization of IcsA in the E. coli background suggests that a common pathway that allows IcsA to be spatially restricted to one pole on the bacterial cell surface exists in Shigella and E. coli.
Shigella flexneri is the causative agent of bacillary dysentery. The bacterium is capable of mediating its uptake by colonic epithelial cells. Following uptake, S. flexneri is released from the phagocytic vacuole into the cytoplasm of the host cell (23). S. flexneri can undergo intra- and intercellular movement in which host cell actin is recruited to form a polymerized actin tail that propels the organism through the cytoplasm and into adjacent host cells (4). This characteristic allows the organism to spread without being released back into the intestinal milieu.
The spreading phenotype can be assessed by the formation of actin tails and protrusions in infected host cells, the formation of plaques in confluent tissue culture cell monolayers, and the ability to provoke keratoconjunctivitis in a guinea pig eye (23). The ability to spread intercellularly is dependent upon the activity of one protein, IcsA, borne by the large virulence plasmid of Shigella spp. (4, 12, 14, 16). Although icsA mutants continue to be invasive, they do not form plaques or provoke keratoconjunctivitis in the guinea pig eye and they exhibit reduced virulence in monkey models of infection due to the inability to catalyze intercellular movement (28).
IcsA is a 120-kDa outer membrane protein that can be cleaved at the carboxyl terminus, leaving a carboxyl-terminal membrane anchor embedded in the outer membrane and releasing a 95-kDa form to the exterior of the bacterial cell (9, 11). IcsA is a unique virulence factor because it is asymmetrically localized to the old pole in the outer membrane of the bacterium (11). The establishment and maintenance of this unipolar localization has been shown to be essential for virulence as assessed by plaque formation and the ability to cause keratoconjunctivitis in the guinea pig eye. This requirement for virulence was first demonstrated by the analysis of a group of lipopolysaccharide (LPS) mutants (24–26). It is clear from these studies that the LPS plays a role in either establishment or maintenance of IcsA at one pole of the bacterium.
There have been four published reports that address the role of IcsA cleavage in intercellular spread, but there is no consensus that implicates IcsA proteolysis as an essential step in proper unipolar localization and subsequent formation of actin tails and protrusions in eucaryotic cells. Two groups have recently cloned and identified a protease involved in cleavage of IcsA. This gene, sopA (icsP), is also borne by the large virulence plasmid although it is not linked with the icsA locus. SopA (IcsP) has homology with the OmpT and OmpP outer membrane proteases of Escherichia coli K-12 (7, 29).
The original approach to addressing the importance of IcsA cleavage involved site-directed mutagenesis of the putative Arg-Arg cleavage site (at amino acid sites 758 to 759) in IcsA. Fukuda et al. demonstrated that alteration of this site results in the absence of cleavage and secretion of IcsA. The absence of cleavage, however, does not affect virulence as assessed by invasion, plaque formation, actin polymerization, and IcsA localization (9). Based on these results, Fukuda et al. concluded that cleavage is not essential for IcsA function. In an earlier study, d’Hauteville and Sansonetti introduced similar mutations in this region to analyze its role as a putative phosphorylation site. Mutagenesis of the Arg-Arg region prevented phosphorylation in vitro by protein kinase A and, paradoxically, resulted in an ability to form slightly larger plaques relative to those of wild-type Shigella in tissue culture cells, suggesting a super-intercellular-spreading phenotype (5). In subsequent analysis of these mutants, d’Hauteville et al. demonstrated that cleavage of IcsA was indeed abolished and that the unipolar localization of IcsA was affected in these strains (6). Loss of cleavage resulted in the inability to unipolarly localize IcsA, resulting in the formation of actin clouds around the bacteria in infected monolayers. While these phenotypes are consistent with the inability to form plaques in confluent monolayers, these mutants were not reevaluated for plaque-forming ability in the follow-up study (6).
Recently, Egile et al. cloned and identified sopA from S. flexneri 5 (7). Mutagenesis by insertional inactivation of sopA resulted in lack of cleavage of IcsA. This lack of cleavage also affected the surface presentation of IcsA, causing a delocalization of IcsA from its unipolar position. Due to it aberrant position, IcsA-catalyzed actin polymerization results in an actin cloud surrounding the bacterium rather than an actin tail at one end. This phenotype leads to a reduced ability to form plaques and a reduction in virulence, suggesting that cleavage is essential for proper formation of actin tails (7). In the most recent report, Shere et al. cloned icsP from S. flexneri 2a, which encodes a protein identical to SopA except for the first seven amino acids (29). Mutagenesis of this gene results in a dramatic decrease in the amount of cleavage of IcsA, but in contrast to the observations of Egile et al. (7), a small amount of the cleaved form of IcsA is detected in the supernatant. Reduced cleavage of IcsA from the bacterial surface results in an alteration of IcsA localization. IcsA continues to be unipolarly localized but is also distributed in random patches over the surface of a bacterial cell. Actin tail formation continues, but the tail extends up the sides of the bacteria. However, this alteration in IcsA cleavage and localization does not affect the ability of the mutant to form plaques in confluent tissue culture monolayers (29). These results are consistent with the data reported by Fukuda et al. (9) and suggest that reduction of cleavage results in only minor alteration of the normal functioning and localization of IcsA. In cell extracts, the icsP mutant was able to move faster than wild-type Shigella, suggesting that more surface-associated IcsA may result in faster intracellular movement, consistent with the super-intercellular-spreading phenotype originally reported by d’Hauteville and Sansonetti (5).
Despite these reports, the importance of cleavage of IcsA for establishing unipolar localization and efficient intercellular spread remains unclear. In order to examine the factors involved in the establishment of IcsA at one pole, we have taken two approaches which rely on expression of a wild-type form of IcsA from a plasmid. The first approach examines expression of icsA in a virulence plasmid-cured derivative of S. flexneri 2a. By examining IcsA localization in this background by confocal microscopy, we analyzed the contribution of virulence plasmid-borne determinants, including SopA/IcsP, on the unipolar establishment and maintenance of IcsA in the outer membrane. The second approach involves analyzing IcsA localization in an OmpT-deficient (IcsA-permissive) strain of E. coli. This strategy addresses the contribution of Shigella-specific factors involved in unipolar localization of IcsA. Elimination of OmpT, which cleaves IcsA at the same position as SopA/IcsP, prevents secretion of IcsA in the E. coli background (21). Using these approaches, we demonstrate that IcsA can be efficiently unipolarly localized in the absence of the Shigella virulence plasmid and in the genetically related but nonvirulent background of E. coli. These results demonstrate that unipolar localization of IcsA is either intrinsic to the IcsA protein or involves recognition of a common apparatus or guide that is present both in Shigella and in E. coli.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in these experiments are listed in Table 1. Plasmids were maintained in E. coli DH5α. All bacteria were grown at 37°C in tryptic soy broth (Difco) unless otherwise stated. Antibiotics were supplemented as needed at the following concentrations: 100 μg/ml for ampicillin and 50 μg/ml for kanamycin.
TABLE 1.
Strains used in this study
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | φ80 dlacZΔM15 endA1 recA1 hsdR17 (rK− mK−) supE44 thi-1 λ− gyrA relA1 F− Δ(lacZYA-argF)U169 | Gibco BRL |
| 2443 | thr-1 leuB6 Δ(gpt-proA)66 argE3 thi-1 rfbO8lacY1 ara-14 galK2 xyl-5 mtl-1 mgl-51 rpsL31 kdgK51 | 19 |
| AD202 | CGSC 7297; araD139 Δ(argF-lacZYA)UompT::Kan flhD5301 fruA25 relA1 rpsL150 rbsR22 deoC1 | 1, Coli Genetic Stock Center |
| ATM379 | thr-1 leuB6 Δ(gpt-proA)66 argE3 thi-1 rfbO8lacY1 ara-14 galK2 xyl-5 mtl-1 mgl-51 rpsL31 kdgK51 ompT::Kan | This study |
| ATM402 | ATM379 pHS3192 | This study |
| S. flexneri | ||
| 2457T | Wild-type serotype 2a | 8 |
| BS103 | Virulence plasmid-cured derivative of 2457T | 18 |
| BS489 | BS103/pHS3192 | This study |
| BS585 | BS103/pRCS6 | This study |
| CVD 1203 | 2457T/ΔicsA ΔaroA | 22 |
| BS543 | 2457T/ΔicsA | This study |
| BS583 | BS543/ΔicsA pHS3192 | This study |
| BS584 | BS543/ΔicsA pRCS6 | This study |
Genetic techniques.
IcsA was expressed from pHS3192, which contains icsA on a 6.5-kb EcoRI-SalI fragment from the virulence plasmid of S. flexneri 5 (4). icsA on this plasmid construct is expressed under the control of its native promoter.
A moderate-copy-number plasmid, pK184IQ, was constructed to allow for control of icsA expression from a lac promoter. This cloning vector was constructed by insertion of lacIq at the SspI site in pK148 (13). The lacIq gene was cloned as a 1.2-kb EcoRI fragment from pNK627 (34) and treated with Klenow fragment (New England Biolabs) to generate a blunt-end fragment prior to insertion in pK184. PCR primers were chosen to amplify icsA from the virulence plasmid of 2457T (16). The icsA PCR product was cloned into pK184IQ, generating pRCS6. In pRCS6, the first codon of icsA is downstream of the lac promoter and ribosome binding site. The amount of IcsA produced from pRCS6 could be modulated by the addition of isopropylthiogalactopyranoside (IPTG).
The wild-type aroA allele from 2457T was transduced into CVD 1203, an icsA deletion mutant of 2457T (22). Selection was based on growth on M9 minimal medium supplemented with nicotinic acid, and the ΔicsA aroA+ transductant was named BS543. Transduction and preparation of lysates of the generalized transducing phage P1L4 were performed according to the methods of Miller (20). ATM379 was constructed by transducing ompT::Kan from E. coli AD202 into E. coli 2443 (which carries rfb expressing the O8 LPS serotype [rfbO8]), followed by selection for kanamycin resistance. The introduction of ompT::Kan was confirmed by PCR with ompT-specific primers and by lack of cleavage of IcsA following expression from pHS3192 in ATM402.
Protein analysis.
Whole-cell and supernatant protein extracts were prepared as previously described (26). Briefly, bacteria were grown to mid-exponential phase (optical density at 600 nm, 0.5 to 0.7) and standardized to reflect equivalent cell numbers. We chose to examine bacteria in exponential phase to achieve conditions comparable to those used in other studies that addressed the localization of IcsA (6, 7, 9, 25, 26, 29). The bacteria were collected, the pellet was resuspended in lysing solution and boiled for 10 min, and the supernatant was filtered through a 0.22-μm-pore-size low-level-protein-binding polyvinylidene difluoride filter (Schleicher and Schuell) to remove any bacterial cells. The supernatant proteins were precipitated by the addition of 20% trichloroacetic acid to a final concentration of 10%. The relative amounts of proteins produced by the different strains were estimated from the intensities of the signals produced on Western blot analysis. Western blotting was performed essentially as previously described (26) with a mouse monoclonal antibody to IcsA followed by binding of an alkaline phosphatase-conjugated goat anti-mouse secondary antibody (Gibco BRL). Reactive bands were visualized by the application of the chemiluminescent alkaline phosphatase substrate, CDP-Star (Boehringer Mannheim), and subsequent exposure to X-ray film (X-Omat; Kodak).
Labeling of bacteria.
Labeling of bacteria grown in vitro was performed by growing bacteria to mid-exponential phase, washing them, and fixing them to polylysine-treated coverslips with 3% buffered formalin. IcsA was detected with a mouse monoclonal antibody against IcsA and then with a rhodamine-conjugated goat anti-mouse secondary antibody (Kirkegaard and Perry). Fluorescence microscopy was performed with an Olympus BX50 microscope or with a Bio-Rad MRC-600 confocal microscope. Image processing of samples analyzed on the confocal microscope was performed with confocal microscope operating software (COMOS; Bio-Rad). In order to assess and quantitate the number of bacteria properly localizing IcsA to one pole, at least 300 bacteria were counted and scored for unipolar localization or trailing and circumferential expression of IcsA. This experiment was repeated at least three times.
RESULTS
IcsA protein expression and secretion.
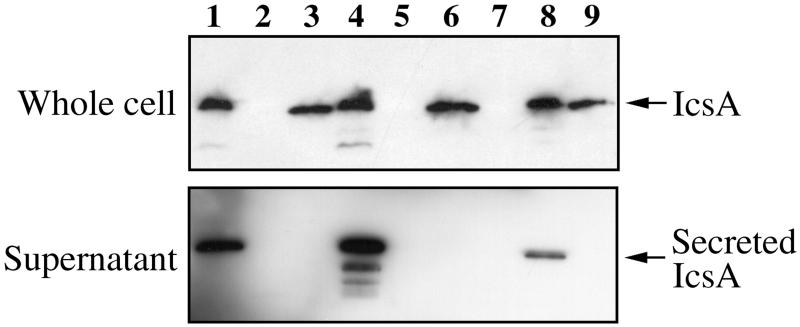
To analyze the contribution of virulence plasmid-borne determinants in IcsA unipolar localization, a cloned copy of icsA (pHS3192) was introduced into BS103, a virulence plasmid-cured derivative of 2457T, to generate BS489. BS489 was examined for expression and secretion of IcsA. As a control, pHS3192 was introduced into BS543, an icsA deletion derivative of 2457T, to generate BS583. This plasmid clone of icsA has been previously described (4) and complements the ΔicsA defect in BS543 as assessed by a plaque assay (data not shown). The production of IcsA was examined in these strains by isolating total proteins from whole-cell lysates and probing with monoclonal antibody specific for IcsA. In Fig. 1, a 120-kDa band is visible in lanes 3 and 4, indicating that the cell-associated levels of IcsA are similar in BS583 and BS489. These levels are also similar to that of 2457T (lane 1), confirming that expression of IcsA from the medium-copy-number plasmid vector pBR325 does not result in dramatic alteration of cell-associated IcsA in the bacteria. No IcsA protein was detected in BS543, the icsA deletion strain (lane 2). Supernatant proteins were examined to detect the secreted form of IcsA, which should be present in strains containing the large virulence plasmid-borne protease. The expected 95-kDa secreted form of IcsA was detected only in strains containing the virulence plasmid, 2457T and BS583. The level of IcsA secretion was higher in BS583, and lower-molecular-weight degradation products were seen. We believe that these degradation products, which are occasionally seen in protein samples from the wild-type, 2457T, are sample-handling artifacts. The cleaved form of IcsA is not involved in actin-based motility, and differences in cleavage patterns among the strains do not affect localization in the outer membrane. No secreted form of IcsA was detected in BS489, indicating that other potential chromosomally encoded proteases did not cleave IcsA from the surface in this strain.
FIG. 1.
Western blot analysis of whole-cell and supernatant protein preparations with anti-IcsA monoclonal antibody. Upper panel, whole-cell lysates; lower panel, supernatant protein preparations. Lane 1, 2457T, wild-type; lane 2, BS543, ΔicsA mutant; lane 3, BS489, virulence plasmid-cured strain expressing IcsA from pHS3192; lane 4, BS583, ΔicsA mutant expressing IcsA from pHS3192; lane 5, BS585, virulence plasmid-cured strain expressing IcsA from pRCS6, no IPTG; lane 6, BS585, virulence plasmid-cured strain expressing IcsA from pRCS6, 0.6 mM IPTG; lane 7, BS584, ΔicsA mutant expressing IcsA from pRCS6, no IPTG; lane 8, BS584, ΔicsA mutant expressing IcsA from pRCS6, 0.6 mM IPTG; lane 9, E. coli ATM402 ompT::Kan rfbO8 expressing IcsA from pHS3192. The samples were prepared from equivalent cell numbers.
The cloned version of icsA on pHS3192 was also introduced into E. coli ATM379 (ompT::Kan rfbO8) to create ATM402 to examine the unipolar localization of IcsA in the absence of Shigella-specific factors. This strain of E. coli was chosen because we had previously demonstrated that expression of the O8 LPS serotype in S. flexneri was permissive for unipolar IcsA localization (26). The ompT gene present in this strain was inactivated by introduction of a kanamycin insertion into ompT by P1 transduction. The OmpT protease has been shown to completely cleave IcsA from the surface of E. coli by cleavage at the same amino acid residue as that cleaved by the Shigella virulence plasmid-borne protease SopA/IcsP (7, 9, 29). Protein profiles from this strain indicated that IcsA was expressed at slightly lower levels than in the Shigella background. However, secretion of IcsA did not occur in this background (Fig. 1, lane 9).
To examine the possible contributions of sequences upstream and downstream of icsA in pHS3192, icsA-specific primers were chosen to PCR amplify the coding region of icsA from 2457T, beginning with the first codon and ending with the stop codon. This PCR product was cloned downstream of the lac promoter and ribosome binding site of pK184IQ to yield pRCS6. Because pK184IQ carries lacIq, expression of icsA could be controlled by the addition of IPTG. pRCS6 was introduced into BS543 (ΔicsA) and BS103 to yield BS584 and BS585, respectively. In order to determine the appropriate amount of induction required to mimic wild-type levels of IcsA, icsA expression from this plasmid was induced over a range of IPTG concentrations (data not shown). A concentration of 0.6 mM IPTG induced IcsA levels comparable to that of the wild type, while the absence of IPTG resulted in no detectable IcsA (Fig. 1, lanes 5 to 8). Analysis of supernatant proteins demonstrated that IcsA was cleaved in BS584 but not cleaved in BS585 (virulence plasmid cured).
Surface labeling with IcsA-specific antiserum.
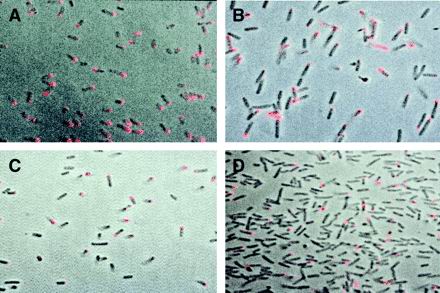
To examine the cell surface localization of IcsA, bacteria were grown to mid-exponential phase and then fixed onto coverslips and probed with monoclonal antiserum specific for IcsA. About 50% of the wild-type parent, 2457T, was labeled with the antiserum. The label was tightly localized to one pole (Fig. 2A). In BS583, which expresses IcsA from a plasmid-borne cloned copy, 44% of the bacteria were labeled and this label appeared predominantly unipolar (Fig. 2C). There were a few variant cells in the population expressing IcsA over their entire surfaces. Although the population of cells expressed levels of IcsA similar to that in the wild type, the aberrantly labeled cells may have been due to the expression of IcsA from a multicopy plasmid. Expression of cloned IcsA from pHS3192 in the virulence plasmid-cured strain BS489 demonstrated that the majority of cells expressed IcsA on the surface. More importantly, the localization of IcsA in this plasmid-cured strain continued to be unipolar, demonstrating that virulence plasmid-borne determinants (e.g., the virulence plasmid-borne protease) were not involved in the establishment of IcsA unipolarity (Fig. 2B). Again, as expression of IcsA was from a plasmid-borne copy, we observed a few bacteria that were labeled over their entire cell surfaces. Interestingly, expression of IcsA from pHS3192 in E. coli ATM402 also resulted in surface expression that was predominantly unipolar (Fig. 2D).
FIG. 2.
Labeling of bacteria grown in vitro with monoclonal antibody to IcsA. Fluorescence images demonstrating antibody binding (shown in red) are overlaid on a phase-contrast image of whole bacterial cell following confocal microscopy and image analysis with the COMOS program. (A) 2457T; (B) BS489, a virulence plasmid-cured strain expressing icsA from pHS3192; (C) BS583, a ΔicsA mutant expressing icsA from pHS3192; (D) E. coli ATM402 ompT::Kan rfbO8 expressing icsA from pHS3192.
In our previous study (26), we noted cell-to-cell variation in IcsA labeling patterns within a population of bacteria. Therefore, it is necessary to examine many individual bacteria to assess the predominant phenotype that a given population of bacteria may exhibit with respect to localization of IcsA. Such a quantitative analysis is necessary to validate the labeling pattern in Fig. 2. These results are presented in Table 2 and confirm that the trend in IcsA labeling observed in Fig. 2 is representative of the bacteria in the sample.
TABLE 2.
Quantitative analysis of IcsA labeling pattern
| Strain | Virulence plasmid | icsAa | % of bacteria with indicated IcsA labeling patternb
|
||
|---|---|---|---|---|---|
| Polar | Halo or trail | No label | |||
| 2457T | + | 49 | 0 | 51 | |
| BS583 ΔicsA | + | pHS3192 | 44 | 6 | 50 |
| BS489 | − | pHS3192 | 48 | 7 | 45 |
| E. coli ATM402 | − | pHS3192 | 32 | 1.5 | 66.5 |
| BS585 | − | pRCS6 | 18 | 2.6 | 79.4 |
Source of icsA provided in trans from a plasmid vector.
Percentages are based on results from at least three different experiments in which at least 300 bacteria were counted following labeling in vitro with anti-IcsA monoclonal antibody and then examined by confocal microscopy to analyze individual bacteria. Those with polar labeling had label at only one pole of the bacterium. Those with a halo or trail had label around the entire organism, with some bacteria demonstrating a unipolar concentration, or label from one pole to the middle of the cell.
Surface-labeling experiments were also done with pRCS6, which contains only the coding sequence of IcsA under the control of the lac promoter. IcsA expression was induced from pRCS6 in both the ΔicsA mutant (BS584) and the virulence plasmid-cured strain (BS585). Labeling with IcsA-specific antiserum demonstrated that IcsA was unipolarly localized in both the presence and absence of the large virulence plasmid (data not shown). Quantitative analysis of the labeling pattern of BS585 demonstrated that fewer bacteria were labeled when icsA was expressed from pRCS6 although the predominant labeling pattern was still unipolar and that the overall percentage of unipolarly labeled bacteria was the same for both pHS3192 and pRCS6 (Table 2). These results confirm the data obtained with pHS3192 and further demonstrate that sequences upstream and downstream of icsA are not required in IcsA localization.
DISCUSSION
A number of genes involved in LPS biosynthesis have been shown to affect IcsA localization due to their effect on the length of the LPS chain. Removal of the O side chain results in aberrant localization of IcsA ranging from circumferential expression over the entire cell surface to labeling of the bacteria from the pole to the midsection. This distribution of IcsA dramatically alters the formation and position of the actin tail and results in an inability to efficiently form protrusions or fireworks to move out of the infected host cell (24–26, 33). Thus, the LPS in the outer membrane is important for the establishment or maintenance of IcsA at the pole. These types of analyses demonstrate the importance of unipolar localization and maintenance of IcsA for the virulence of S. flexneri but do not define a mechanism for the establishment of IcsA at the old pole of the bacterial cell or indicate the role of the LPS in this process.
Compartment-specific localization of proteins in procaryotes is becoming more recognized as new advances in microscopy and the availability of antisera specific for various proteins allows their subcellular localization to be studied. For example, many of the proteins involved in cell division in E. coli have been localized to the septum and analysis of differentiating organisms like Bacillus spp. and Caulobacter crescentus have served as model systems to study compartment-specific protein localization and expression (17). The ActA protein of Listeria monocytogenes is a functional homologue of IcsA, although there is no sequence homology between IcsA and ActA at the amino acid level. ActA is responsible for actin-based motility in cells infected with L. monocytogenes and is also spatially restricted, appearing everywhere on the bacterial cell surface except the new pole (15). This spatial orientation is also critical to proper formation of actin tails by L. monocytogenes (15, 30). Very little is known about the mechanisms that target a protein to a specific site in the bacterial cells in these systems. Models to describe this process must include a mechanism to allow for both the establishment and maintenance of compartment-specific localization.
Specific proteolysis has been proposed as one model to explain compartment-specific protein localization. In this model, the localized protein would be proteolytically cleaved everywhere in the bacteria except the ultimate targeted site. Specific proteolysis has been demonstrated to be important for spatial distribution of methyl-accepting chemotaxis protein A (McpA) in C. crescentus. This proteolysis is important in removing McpA from the stalked cell but is not involved in establishing this protein at the pole of the swarmer cell (2). d’Hauteville et al. proposed specific proteolysis of IcsA as a mechanism to establish and maintain the unipolar localization of IcsA in S. flexneri (6). This hypothesis was based on their observation that strains of S. flexneri that do not cleave IcsA express IcsA over the bacterial surface and form smaller plaques in confluent HeLa cell monolayers. This model proposes that in wild-type Shigella, IcsA is targeted over the entire bacterial cell and is specifically removed everywhere except the old pole (6, 7). In this model, IcsA would not contain information that was important for its own targeting (other than the proteolytic cleavage site) and would not be localized unipolarly in the absence of the protease.
In order to examine the relevance of the virulence plasmid-borne protease that cleaves IcsA and to define bacterial components involved in unipolar localization of IcsA, we chose to examine IcsA localization in a virulence plasmid-cured derivative of S. flexneri. This strategy allowed us to examine the effect of all virulence plasmid-borne determinants, including the SopA/IcsP protease, on IcsA localization. Previous work by Goldberg et al. demonstrated that, unlike secretion of other virulence factors in S. flexneri, IcsA secretion is not dependent on the virulence plasmid-borne type III secretory apparatus (11), which makes it feasible to examine IcsA localization and secretion in the absence of the virulence plasmid. Copy number and expression levels of IcsA are critical factors in this experiment; too little IcsA expressed on the surface does not mimic the wild-type levels, while too much IcsA results in loss of unipolar localization (27, 31). IcsA protein levels expressed from the plasmid-borne versions of icsA and from the wild-type virulence plasmid copy were equivalent as assessed by Western blotting. An additional control was to complement an icsA mutant with the plasmid-borne copy of icsA. This complemented strain was able to form plaques and unipolarly localize IcsA. Analysis of the virulence plasmid-cured strain expressing icsA from either pHS3192 or pRCS6 demonstrated that IcsA localized and was maintained at the old pole, identical to IcsA localization in the wild-type S. flexneri background in the absence of any other virulence plasmid-borne determinants. These results indicate that the virulence plasmid-borne protease is not responsible for establishing or maintaining IcsA at one pole.
Furthermore, expression of icsA from pHS3192 in a OmpT− mutant strain of E. coli expressing a complete O side chain resulted in unipolar localization of IcsA. These results demonstrate that the IcsA protein contains the information required for its own targeting to one pole of the bacterial cell. Alternatively, IcsA may recognize structures or guide proteins present both in Shigella and E. coli. Nevertheless, even if the latter model is correct, our data suggest that virulence-specific guide factors borne by the virulence plasmid or the Shigella chromosome are not required for unipolar localization of IcsA. Our results are in agreement with the analyses of mutants by Fukuda et al. (9) and Shere et al. (29) that demonstrate that IcsA cleavage is not required for unipolar localization. Thus, the ultimate role of the SopA/IcsP protease in IcsA positioning and function remains unresolved.
In C. crescentus, compartment-specific transcription is responsible for asymmetric localization of flagellar gene mRNA that results in protein localization to the swarmer cell. This model of compartment-specific localization involves specific DNA sequences upstream of the transcription start site that act like a “zip code” for localized transcription (10). The polar localization of IcsA expressed from pRCS6, which contains only the coding sequence of icsA, suggests that a similar zip code directing mechanism is not responsible for establishment of IcsA at the pole in Shigella. However, this result does not address the contribution of mRNA sequences within the coding sequence of icsA. Internal mRNA sequences have recently been demonstrated to be important for specific secretion of Yop proteins from Yersinia enterocolitica via the type III secretory apparatus, demonstrating yet another mechanism for targeting proteins to specific areas of the bacterial cell (3).
There has been little work to define IcsA domains important for unipolar localization. Suzuki et al. demonstrated that the transport of IcsA to the outer membrane is sec dependent across the inner membrane and self-catalyzed across the outer membrane, where subsequent cleavage releases the protein to the exterior of the cell (31). Structure-function domain mapping studies have illustrated that although the carboxyl-terminal portion of IcsA is required for insertion and transport across the outer membrane, this domain is not responsible for unipolar localization (32).
In our study, unipolar localization of IcsA in E. coli demonstrates that IcsA by itself contains information that directs it to the pole. This observation predicts that internal signals in IcsA provide the means to establish this protein at the old pole. Because unipolar localization occurs in virulence plasmid-cured Shigella sp. and E. coli, a common pathway must exist in both genetic backgrounds. Since it is not known at which step unipolarity is established, several models can be envisioned. Efficient unipolar localization may require a chaperone or guide protein that functions at the inner or outer membrane or periplasmic space, which allows IcsA to recognize the old pole. Alternatively, there may be a protein or component such as a remnant from the cell division apparatus that marks a site at the old pole for recognition by specific residues in IcsA. Both models predict that IcsA interacts with proteins or structures found in both E. coli and Shigella that allow the protein to be unipolarly localized in both genetic backgrounds. Future efforts will be directed at identifying the domains of IcsA and the putative targets that interact with IcsA.
ACKNOWLEDGMENTS
We thank Marcia B. Goldberg at Albert Einstein College of Medicine of Yeshiva University for the gift of IcsA-specific monoclonal antibodies, Fernando Noriega at the Center for Vaccine Development for the gift of CVD1203, and Keith A. Lampel for the gift of primers. We also thank the Biomedical Instrumentation Center at USUHS for technical assistance with confocal analysis and Reinaldo E. Fernández for technical assistance with protein analysis.
This work was supported by NIAID grant AI24656 (to A.T.M.) and USUHS grant RO7385 (to A.T.M.).
REFERENCES
- 1.Akiyama Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem Biophys Res Commun. 1990;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 2.Alley M R K, Maddock J R, Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D M, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 4.Bernardini M L, Mounier J, d’Hauteville H, Coquis-Rondon M, Sansonetti P J. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci USA. 1989;86:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.d’Hauteville H, Sansonetti P J. Phosphorylation of IcsA by cAMP-dependent protein kinase and its effect on intercellular spread of Shigella flexneri. Mol Microbiol. 1992;6:833–841. doi: 10.1111/j.1365-2958.1992.tb01534.x. [DOI] [PubMed] [Google Scholar]
- 6.d’Hauteville H, Lagelouse R D, Nato F, Sansonetti P J. Lack of cleavage of IcsA in Shigella flexneri causes aberrant movement and allows demonstration of a cross-reactive eukaryotic protein. Infect Immun. 1996;64:511–517. doi: 10.1128/iai.64.2.511-517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egile C, d’Hauteville H, Parsot C, Sansonetti P J. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol Microbiol. 1997;23:1063–1073. doi: 10.1046/j.1365-2958.1997.2871652.x. [DOI] [PubMed] [Google Scholar]
- 8.Formal S B, Dammin G J, LeBrec E H, Schneider H. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol. 1958;75:604–610. doi: 10.1128/jb.75.5.604-610.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda I, Suzuki T, Munakata H, Hayashi N, Katayama E, Yoshikawa M, Sasakawa C. Cleavage of Shigella surface protein VirG occurs at a specific site, but the secretion is not essential for intracellular spreading. J Bacteriol. 1995;177:1719–1726. doi: 10.1128/jb.177.7.1719-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gober J W, Champer R, Reuter S, Shapiro L. Expression of positional information during cell differentiation in Caulobacter. Cell. 1991;64:381–391. doi: 10.1016/0092-8674(91)90646-g. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg M B, Parsot C, Barzu O, Sansonetti P J. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol. 1993;175:2189–2196. doi: 10.1128/jb.175.8.2189-2196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg M B, Theriot J A. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc Natl Acad Sci USA. 1995;92:6572–6576. doi: 10.1073/pnas.92.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jobling M G, Holmes R K. Construction of vectors with p15a replicon, kanamycin resistance, inducible lacZα and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 1990;18:5315–5316. doi: 10.1093/nar/18.17.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kocks C, Marchand J-B, Gouin E, d’Hauteville H, Sansonetti P J, Carlier M-F, Cossart P. The unrelated surface proteins ActA of Listeria monocytogenes and IcsA of Shigella flexneri are sufficient to confer actin-based motility on Listeria innocua and Escherichia coli respectively. Mol Microbiol. 1995;18:413–423. doi: 10.1111/j.1365-2958.1995.mmi_18030413.x. [DOI] [PubMed] [Google Scholar]
- 15.Lasa I, Cossart P. Actin-based bacterial motility: towards a definition of the minimal requirements. Trends Cell Biol. 1996;6:109–114. doi: 10.1016/0962-8924(96)81001-4. [DOI] [PubMed] [Google Scholar]
- 16.Lett M-C, Sasakawa C, Okada N, Sakai T, Makino S, Yamada M, Komatsu K, Yoshikawa M. virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the virG protein and determination of the complete coding sequence. J Bacteriol. 1989;171:353–359. doi: 10.1128/jb.171.1.353-359.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddock J R, Alley M R K, Shapiro L. Polarized cells, polar actions. J Bacteriol. 1993;175:7125–7129. doi: 10.1128/jb.175.22.7125-7129.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurelli A T, Blackmon B, Curtiss R., III Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun. 1984;43:397–401. doi: 10.1128/iai.43.1.397-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier U, Mayer H. Genetic location of genes encoding enterobacterial common antigen. J Bacteriol. 1985;163:756–762. doi: 10.1128/jb.163.2.756-762.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 21.Nakata N, Tobe T, Fukuda I, Suzuki T, Komatsu K, Yoshikawa M, Sasakawa C. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol Microbiol. 1993;9:459–468. doi: 10.1111/j.1365-2958.1993.tb01707.x. [DOI] [PubMed] [Google Scholar]
- 22.Noriega F R, Losonsky G, Wang J Y, Formal S B, Levine M M. Further characterization of ΔaroA ΔvirG Shigella flexneri 2a strain CVD 1203 as a mucosal Shigella vaccine and as a live-vector vaccine for delivering antigens of enterotoxigenic Escherichia coli. Infect Immun. 1996;64:23–27. doi: 10.1128/iai.64.1.23-27.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsot C, Sansonetti P J. Invasion and the pathogenesis of Shigella infections. Curr Top Microbiol Immunol. 1996;209:25–42. doi: 10.1007/978-3-642-85216-9_2. [DOI] [PubMed] [Google Scholar]
- 24.Rajakumar K, Jost B H, Sasakawa C, Okada N, Yoshikawa M, Adler B. Nucleotide sequence of the rhamnose biosynthetic operon of Shigella flexneri 2a and role of lipopolysaccharide in virulence. J Bacteriol. 1994;176:2362–2373. doi: 10.1128/jb.176.8.2362-2373.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandlin R C, Lampel K A, Keasler S P, Goldberg M B, Stolzer A L, Maurelli A T. Avirulence of rough mutants of Shigella flexneri: requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect Immun. 1995;63:229–237. doi: 10.1128/iai.63.1.229-237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandlin R C, Goldberg M B, Maurelli A T. Effect of O side chain length and composition on the virulence of Shigella flexneri 2a. Mol Microbiol. 1996;22:63–73. doi: 10.1111/j.1365-2958.1996.tb02656.x. [DOI] [PubMed] [Google Scholar]
- 27.Sandlin, R. C., and A. T. Maurelli. Unpublished observation.
- 28.Sansonetti P J, Arondel J, Fontaine A, d’Hauteville H, Bernardini M L. OmpB (osmo-regulation) and icsA (cell-to-cell spread) mutants of Shigella flexneri; vaccine candidates and probes to study the pathogenesis of shigellosis. Vaccine. 1991;9:416–422. doi: 10.1016/0264-410x(91)90128-s. [DOI] [PubMed] [Google Scholar]
- 29.Shere K D, Sallustio S, Manessis A, D’Aversa T G, Goldberg M B. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol Microbiol. 1997;25:451–462. doi: 10.1046/j.1365-2958.1997.4681827.x. [DOI] [PubMed] [Google Scholar]
- 30.Smith G A, Portnoy D A, Theriot J A. Listeria monocytogenes ActA protein is sufficient to direct actin-based motility. Mol Microbiol. 1995;17:945–951. doi: 10.1111/j.1365-2958.1995.mmi_17050945.x. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Lett M-C, Sasakawa C. Extracellular transport of VirG protein in Shigella. J Biol Chem. 1995;270:30874–30880. doi: 10.1074/jbc.270.52.30874. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Saga S, Sasakawa C. Functional analysis of Shigella VirG domains essential for interaction with vinculin and actin-based motility. J Biol Chem. 1996;271:21878–21885. doi: 10.1074/jbc.271.36.21878. [DOI] [PubMed] [Google Scholar]
- 33.Van Den Bosch L, Manning P A, Morona R. Regulation of O-antigen chain length is required for Shigella flexneri virulence. Mol Microbiol. 1997;23:765–775. doi: 10.1046/j.1365-2958.1997.2541625.x. [DOI] [PubMed] [Google Scholar]
- 34.Zumstein L, Wang J C. Probing the structural domains and function in vivo of Escherichia coli DNA topoisomerase I by mutagenesis. J Mol Biol. 1986;191:333–340. doi: 10.1016/0022-2836(86)90130-0. [DOI] [PubMed] [Google Scholar]