Abstract
Based on the reactions of different sequences with single ions K+, Na, NH4+, double ions high + low, low + high, and triple ions with different addition orders, the best stable ion combinations of 12 quadruplexes with different DNA sequences were reported. From the fluorescence spectrum, except for HT-V15 and PW17 and AS1411 and HT-V18, the structural stability of G-quadrangle formed basically follows a certain rule. In terms of this experiment, according to circular dichroism, the antiparallel quadrupole structure has the largest proportion among quadrupole structures, and 12 optimal DNA addition schemes and sequences have been obtained through exploration. It is worth mentioning that, on the whole, the best addition scheme of AS1411 and HT-V18 is a three-ion scheme, which provides an effective reference for similar experiments in the future.
1. Introduction
With the deepening of the understanding of DNA structure, the structure of four-stranded DNA has been gradually analyzed and developed, and people found that four-stranded DNA also has important biological functions.1−7 In the presence of univalent alkali metal ions K+ and Na+,8,9 guanine bases on the single strand of DNA form a near-planar g-tetrad10,11 by Hoogsteen hydrogen bonding with guanine on its own chain or other chains. Each G-quad plane consists of four guanines linked by intermolecular hydrogen bonds,12 and the ions reside in the G4 hole. The guanine plane-to-plane-metal ions form simple and stable pyramidal g-tetrahedron through π–π stacking13 of the sugar phosphate skeleton,14−17 but not all of them. The cations in DNA G-tetrahedron are interplanar, subplanar, super-subplanar, and intraplanar. The stability and structure of G4 depend on the type of ions it contains,18 that is to say, G4 is selective for ions19 All these variables contribute to the structural diversity of G-tetrahedron. The link loop (Loop) connecting four G chains in the intramolecular G-tetrahedral plays a key role in its conformation and stability. The shorter the Loop, the easier it is to form a parallel structure and the higher the stability.20,21 The formation sequence of many G-quads in genes and the structure of g-quads with anticancer and antiviral activities remain to be further determined.21 In recent years, the interaction between DNA and single cations22,23 or anions24 has been widely used for the quantitative analysis of disease biomarkers and environmental monitoring. Therefore, it is of great significance to study the factors affecting the structure of G-tetrapod and explore the formation rules of the self-assembly of G-tetrapod.
In this paper, the selectivity of G-tetrachromes to Na+, K+, and NH4+ ions is discussed, and the best fluorescence DNA of each ion is screened out. In addition, the influence of multiple logical arrangements of ions on different G-tetrachromes in the case of double ions and triple ions is discussed. This paper is the first to evaluate the stability of DNA G-quadruplicate in the presence of double and triple ions in different sequences. HT-V15 and PW17 in a dual-ion condition and AS1411 and HT-V18 in a triple-ion condition are more stable than the G-quadruchain structure formed by a single ion.
2. Experimental Section
2.1. Materials and Reagents
Ammonium chloride (NH4Cl, analysis pure), potassium chloride (KCl, analysis pure) from Tianjin Xinbo Chemical Co., LTD., sodium chloride (NaCl, analysis pure) from Tianjin Beilian Fine Chemicals Development Co., LTD., Solarbio pH = 7.8 Tris-HCl, Protoporphyrin IX (PPIX), Sangon Biotech DNA sequence, F97Pro20056 fluorescence spectrophotometer.
2.2. Synthesis of DNA Samples
2.2.1. Pretreatment
The DNA sample was placed in a centrifuge tube for 15 min, and an appropriate amount of Tris buffer was added (pH = 7.8). The DNA sample was heated in a magnetic stirrer with constant temperature heating at 90 °C for 15 min and then cooled naturally to room temperature. Table 1 shows the 12 DNA sequences used in this paper.
Table 1. 12 DNA Sequences Used in the Experiment.
| DNA | DNA sequence |
|---|---|
| AS1411 | GGTGGTGGTGGTTGTGGTGGTGGTGG |
| PS2.M | GTGGGTAGGGCGGGTTGG |
| PS5.M | GTGGGTCATTGTGGGTGGGTGTGG |
| PW17 | GGGTAGGGCGGGTTGGG |
| T30695 | GGGTGGGTGGGTGGGT |
| OT | GGGGTTTTGGGGTTTTGGGGTTTTGGGG |
| AT | GGGTTTAGGGTTTAGGGTTTAGGG |
| BT | GGTTAGGTTAGGTTAGG |
| TT | GGGGTTGGGGTTGGGGTTGGGG |
| HT | GGGTTAGGGTTAGGGTTAGGG |
| HT-V15 | GGGAGGGAGGGAGGG |
| HT-V18 | GGGTAGGGTAGGGTAGGG |
2.2.2. Experimental Steps
Twenty-five microliters of 100 μM DNA was placed into 1, 2, and 3 centrifuge tubes, and 80 μL of the Tris buffer solution and 100 μL of K+, Na+, and NH4+ with a concentration of 100 mM. were added. Then, 400 μL of distilled water was added. The mixture was heated in a magnetic stirrer at 90 °C for 10 min and cooled naturally to room temperature.
2.3. Fluorescent Detection
One hundred microliters of 100 μmol/L PPIX photosensitive reagent was added to annealed 1–3 groups of DNA solution for 1–2 h to form a complex, and the fluorescence intensity of the complex was detected by a fluorescence spectrophotometer and the corresponding relationship was obtained. The fluorescence spectra of the solution were recorded at the excitation wavelength of 405 nm and the emission wavelength was collected at 635 nm. In the same way, the binding relationship between DNA and the dual ions with a large difference in fluorescence and the mixed relationship between the triple ions were detected.
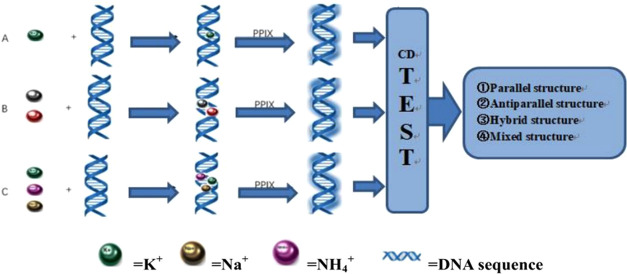
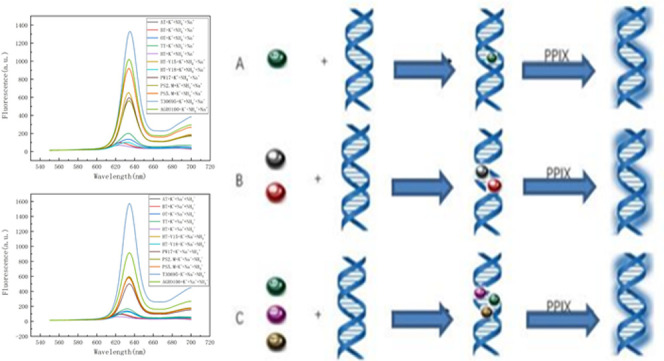
A dual-ion reaction is added after the PPIX reaction, and the last two ions of the triple-ion reaction are added at the same time. The experimental steps are shown in Figure 1.
Figure 1.
Experimental steps of the reaction of different sequences with (A) single ion, (B) double ions, (C) and triple ions.
3. Results and Discussion
3.1. Add a Single Ion
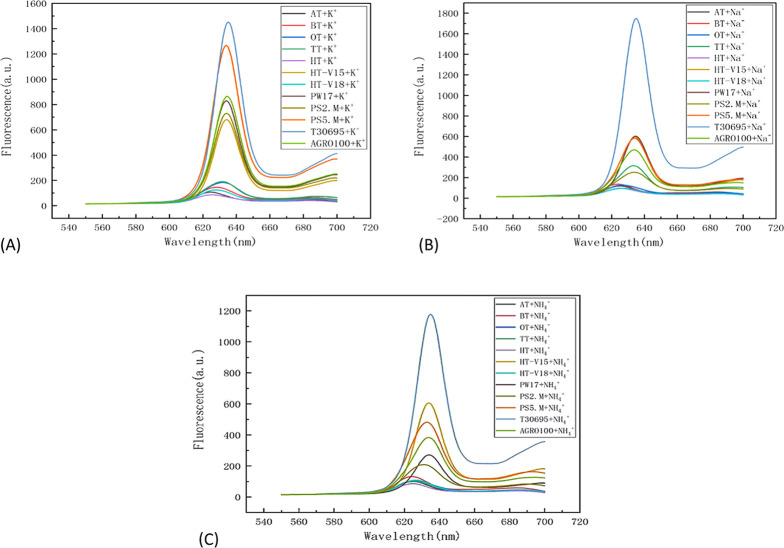
Figure S1 shows the comparison of fluorescence intensities of AS1411, AT, BT, HT, HT-V15, and HT-V18 with K+, Na+, and NH4+ at 635 nm. Figure S2 shows the fluorescence intensity contrast of OT, PS2.M, PS5.M, PW17, T30695, and TT with K+, Na+, and NH4+ at 635 nm. Therefore, under the same conditions, T30595 and PS5.M sequences interact with a univalent ion to produce G-tetrachrosome with stronger stability, followed by PW17, PS2.M, AS1411, and HT-V15 sequence, and the worst stability is the HT sequence.
Table 2 shows the fluorescence intensity summary of reactions of 12 kinds of DNA with K+, Na+, and NH4+ ions and summarizes the fluorescence intensity states of all kinds of DNA under different ions. Therefore, we know that K+ is more conducive to the formation of a stable G-quadruplex structure, and the structure formed by NH4+ and DNA is more unstable, while, in this experiment, the structure formed by Na+ and DNA tends to be more stable.
Table 2. Summary of Fluorescence Intensity.
| fluorescence
intensity |
fluorescence
intensity |
||||||
|---|---|---|---|---|---|---|---|
| DNA | K+ | Na+ | NH4+ | DNA | K+ | Na+ | NH4+ |
| AS1411 | high | low | OT | high | low | ||
| AT | high | low | PS2.M | high | low | ||
| BT | high | low | PS5.M | high | low | ||
| HT | high | low | PW17 | high | low | ||
| HT-V15 | high | low | T30695 | high | low | ||
| HT-V18 | high | low | TT | high | low | ||
Figure 2 compares the fluorescence intensity of 12 kinds of DNA from the perspective of K+, Na+, and NH4+. The most stable sequence of the G-tetrachroid structure was T30695. From the perspective of K+, PS5.M is second only to T30695 in stability. From the perspective of Na+, PW17 and HT-V18 are second only to T30695 in stability. From the perspective of NH4+, HT-V15 is second only to T30695 in stability.
Figure 2.
Comparison of the fluorescence intensities of 12 DNA with (A) K+, (B) Na+, and (C) NH4+.
3.2. Add Double Ions
Figure S3 shows a comparison of the fluorescence intensities of AS1411, AT, BT, HT, HT-V15, and HT-V18 with K+, Na+, and NH4+ as well as ions with poor fluorescence in the high-stability state (high + low) and ions with good fluorescence in the low-stability state (low + high). Figure S4 shows the comparison of fluorescence intensities of OT, PS2.M, PS5.M, PW17, T30695, and TT with K+, Na+, and NH4+ as well as ions with poor fluorescence in the high-stability state and ions with good fluorescence in the low-stability state. On the basis of determining the best single ion, the order of double ion with the best stability was explored.
According to Table 3, in general, the G-quadruchain structure generated by adding ions with poor fluorescence (high + low) in the high-stability state is more stable than the G-quadruchain structure generated by adding ions with good fluorescence (low + high) in the low-stability state. However, there are exceptions where HT-V15 and PW17 can produce satisfactory results when interacting with dual ions under the same conditions. HT-V15 produces a more stable structure with the double ion (low + high) and PW17 produces a more stable structure with the double ion (high + low) than with the single ion, which proves that the double ion can make the structure of the G-quadruplex different.
Table 3. Fluorescence Order of 12 DNA with Single Ions and Specific Dual Ions.
| DNA | fluorescence intensity | DNA | fluorescence intensity |
|---|---|---|---|
| AS1411 | K+ > high + low > low + high > Na+ > NH4+ | OT | K+ > high + low > Na+ > NH4+ > low + high |
| AT | Na+ > K+ > low + high > NH4+ > high + low | PS2.M | K+ > high + low > low + high > Na+ > NH4+ |
| BT | K+ > Na+ > NH4+ > low + high > high + low | PS5.M | K+ > high + low > low + high > Na+ > NH4+ |
| HT | Na+ > NH4+ > K+ > low + high > high + low | PW17 | high + low > K+ > Na+ > low + high > NH4+ |
| HT-V15 | low + high > high + low > K+ > NH4+ > Na+ | T30695 | Na+ > K+ > high + low > NH4+ > low + high |
| HT-V18 | K+ > high + low > NH4+ > Na+ > low + high | TT | Na+ > high + low > low + high > K+ > NH4+ |
Based on the influence of the different effects of double ions on G-quadruplex, the comparison of the fluorescence intensity of different DNA in low + high and high + low states was further explored (Figure 3). The fluorescence images proved that the G-quad structure produced by the high + low state was more stable than that produced by the low + high state. This may be because the more stable individual ions are more influential in the formation of G-quadruplex structures.
Figure 3.
(A, B) Comparison of the fluorescence intensities of 12 kinds of DNA under specific dual ions.
3.3. Add the Triple Ion
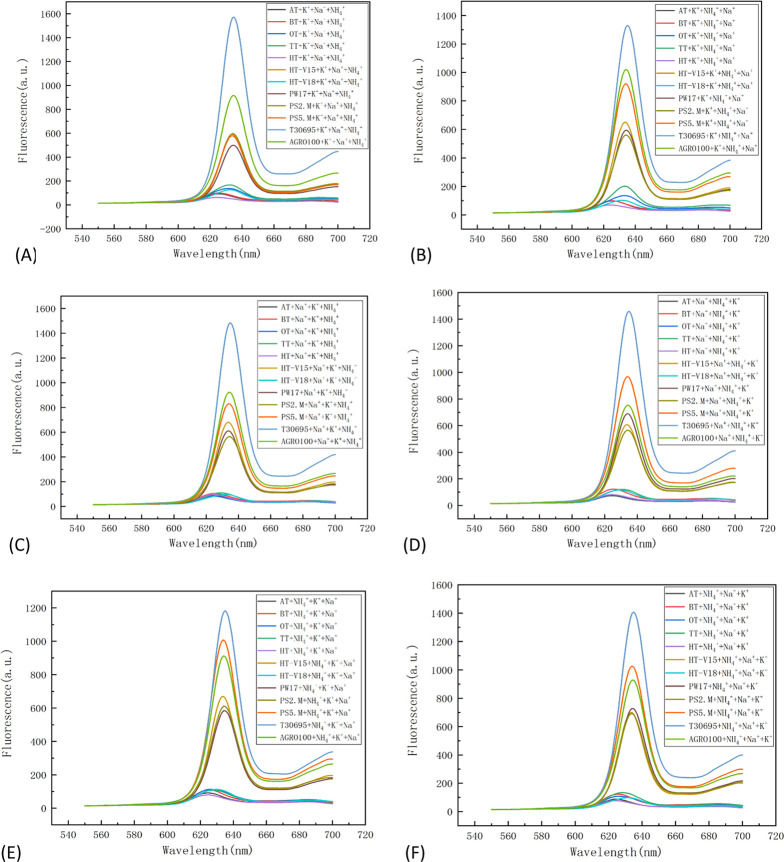
Figure S5 shows the fluorescence intensity contrast of AS1411, AT, BT, HT, HT-V15, and HT-V18 with the three heavy ions in different logical orders at 635 nm. Figure S6 shows the comparison of fluorescence intensities of OT, PS2.M, PS5.M, PW17, T30695, and TT with the triple ions in different logical orders at 635 nm. The analysis results show that the combination of NH4+ + Na+ + K+, followed by the combination of K+ + NH4+ + Na+ and K+ + Na+ + NH4+ can form a more stable G-quadruplex when the triple ion is added. Although K+ is the best ion, it does not appear dominant among the triple ions. In contrast, the G-tetrahedral structure formed by the combination K+ + Na+ + NH4+ is unstable, followed by Na+ + NH4+ + K+ and NH4+ + K+ + Na+, and this large difference requires further analysis.
Table 4 shows a more detailed summary of the structural stability of G-quadruplex formed by the reaction of DNA and triple ions.
Table 4. Summary of Fluorescence Intensity for the Reaction of Triple Ions with DNA.
| fluorescence
intensity |
fluorescence
intensity |
||||
|---|---|---|---|---|---|
| DNA | high | low | DNA | high | low |
| AS1411 | K+ > NH4+ > Na+ | Na+ > NH4+ > K+ | OT | K+ > Na+ > NH4+ | Na+ > NH4+ > K+ |
| AT | K+ > NH4+ > Na+ | Na+ > NH4+ > K+ | PS2.M | NH4+ > Na+ > K+ | K+ > NH4+ > Na+ |
| BT | NH4+ > Na+ > K+ | K+ > NH4+ > Na+ | PS5.M | NH4+ > Na+ > K+ | K+ > Na+ > NH4+ |
| HT | Na+ > K+ > NH4+ | K+ > Na+ > NH4+ | PW17 | NH4+ > Na+ > K+ | K+ > Na+ > NH4+ |
| HT-V15 | NH4+ > Na+ > K+ | K+ > Na+ > NH4+ | T30695 | K+ > Na+ > NH4+ | NH4+ > K+ > Na+ |
| HT-V18 | K+ > Na+ > NH4+ | Na+ > K+ > NH4+ | TT | K+ > NH4+ > Na+ | NH4+ > K+ > Na+ |
Although we reached a general conclusion about the effect of the triple ion on the G-quadruplex, additional findings were found when compared with the 12 ions in different addition orders (Figure 4). Under the same addition order, sequences AS1411 and PS5.M have almost opposite effects. Although the Na+ > K+ > NH4+ combination is an exception, it does not cause a huge difference in the G-quadruplex structure formed by the two sequences under the same conditions.
Figure 4.
(A–F) Comparison of fluorescence intensities of reactions between three ions with different addition orders and 12 DNA.
3.4. Overall Analysis
When we analyze single ions, double ions, and triple ions together (Figure S7), the advantage of single ions is indisputable. However, like double ions, triple ions can also cause surprising changes in the G-quadruplex of some sequences. In sequences AS1411 and HT-V18, G-quadruplex with triple ions is in the most stable position, which makes the triple ion screening experiment meaningful. The different effects of triple ions on G-quadruplex of different sequences are also reflected in the diagram. In sequence TT, the addition of triple ions seems to be only a “burden” on the stability of G-quadruplex, while in sequence AS1411, the addition of triple ions has unexpected effects on the stability of G-quadruplex.
Table 5 further proves that the overall stability order of the G-quadruplex structure formed by 12 kinds of sequences is as follows: single ion > dual ion > triple ion or single ion > triple ion > dual ions such as AT, BT, HT, PS5.M, TT and OT, PS2.M, and T30695. The effects of double and triple ions on the stability of G-quadruplex with different sequences were listed. The most stable G-quadruplex structure formed by HT-V15 and PW17 was the double ion, while the most stable G-quadruplex structure formed by AS1411 and HT-V18 was the triple ion.
Table 5. Summary of Fluorescence Intensity of 12 DNA Reactions with Single Ions, Double Ions, and Triple Ionsa.
| DNA | fluorescence intensity sequence |
|---|---|
| AS1411 | X1X3X2 > X3X2X1 > X2X1X3 > X1X2X3 > X3X1X2 > K+ > X2X3X1 = high + low > low + high > Na+ > NH4+ |
| AT | Na+ > K+ > low + high > NH4+ > X1X3X2 > X1X2X3 > X3X1X2 > high + low > X2X1X3 > X3X2X1 > X2X3X1 |
| BT | K+ > Na+ > NH4+ > low + high > X2X3X1 = X3X2X1 > X3X1X2 > X2X1X3 > X1X3X2 = X1X2X3 > high + low |
| HT | K+ > Na+ > NH4+ > X2X1X3 > X2X3X1 > low + high > X3X1X2 > X3X2X1 > X1X3X2 > high + low > X1X2X3 |
| HT-V15 | low + high > high + low > X3X2X1 > K+ = X2X1X3 > X3X1X2 > X1X3X2 > NH4+ = X2X3X1 > Na+ > X1X2X3 |
| HT-V18 | X1X2X3 > K+ > X2X3X1 > high + low > X3X1X2 > NH4+ > X1X3X2 > X3X2X1 > X2X1X3 > Na+ > low + high |
| OT | K+ > X1X2X3 > X1X3X2 > high + low > Na+ > X3X1X2 > X3X2X1 > NH4+ > low + high > X2X1X3 > X2X3X1 |
| PS2.M | K+ > X3X2X1 > high + low > X3X1X2 > X1X2X3 > X2X3X1 = X2X1X3 = X1X3X2 > low + high > Na+ > NH4+ |
| PS5.M | K+ > high + low > NH4+ > X3X1X2 > X2X3X1 = low + high = X1X3X2 > X2X1X3 > Na+ = X1X2X3 > X3X2X1 |
| PW17 | high + low > K+ > X3X2X1 > X2X3X1 > X2X1X3 > Na+ > X1X3X2 > X3X1X2 > X1X2X3 > low + high > NH4+ |
| T30695 | Na+ > X1X2X3 > X2X1X3 > X2X3X1 > K+ > NH4+ > X1X3X2 > high + low > X3X2X1 = X3X1X2 > low + high |
| TT | Na+ > high + low > X1X3X2 = low + high > K+ > X1X2X3 > X3X2X1 > X2X3X1 > X2X1X3 > X3X1X2 > NH4+ |
Note: X1 = K+, X2 = Na+, X3 = NH4+.
4. CD Spectra
A circular dichroism spectrum is often used to characterize the g-tetrapod structure. According to the intensity and position of the absorption peak, the G-tetrapod obtained by each group of experiments can be roughly divided into four types: (1) parallel quadrupole structure (positive peak at 260 nm, negative peak at 240 nm); (2) antiparallel quadrupole structure (positive peak at 290 nm, negative peak at 260 nm); (3) hybrid structure (positive peak at 290 nm, negative peak at 240 nm, acromial peak at 265 nm); and (4) antiparallel/parallel mixed quadrupole structure (positive peak at 290 nm, positive peak at 260 nm).
4.1. Comparison of the CD Spectra of Single Ions
According to Figures S9 and S10, the quadrupole structures are formed by the reaction of 12 different DNA with strong or weak fluorescence ions (as can be seen from Table 2), among which 9 parallel quadrupole structures are as follows: HT-V15 + K+, PS5.M + K+, PW17 + K+, T30695 + Na+, AS1411 + NH4+, BT + NH4+, HT-V15 + Na+, PW17 + NH4+, and T30695 + NH4+; antiparallel quadrupole substructure has the following 13 species: AS1411 + K+, AT + Na+, BT + K+, HT + Na+, HT-V18 + K+, OT + K+, TT + Na+, AT + NH4+, BT + NH4+, HT + K+, HT-V18 + Na+, OT + NH4+, and TT + NH4+; and mixed quadrupole structure has the following 3 species: PS2.M + K+, PS2.M + NH4+, and PS5.M + NH4+.
In this experiment, the antiparallel quadrupole structure dominates when the 12 types of DNA react with a single ion, and the parallel quadrupole structure begins to dominate when G% gradually increases. Combined with the conclusions in Section 3.1, it can be concluded that the stability is of the order of antiparallel quadrupole structure > parallel quadrupole structure > mixed quadrupole structure. K+ is the most dominant in a single ion, which is further confirmed by the fact that more G-quadruplexes produce antiparallel quadrupole structures when a single K+ acts.
4.2. Comparison of CD Spectra of Dual Ions
According to Figures S11 and S12, there are 12 kinds of quadrupole structures formed by strong and weak fluorescence reactions of different DNA with different addition sequences (as can be seen from Table 3), among which there are 7 kinds of parallel quadrupole structures as follows: AS1411 + L + H, HT-V15 + H + L, HT-V15 + L + H, PS2.M + H + L, PS2.M + L + H, PW17 + H + L, and PW17 + L + H; antiparallel quadrupole substructure has the following 12: AS1411 + H + L, AT + H + L, AT + L + H, BT + H + L, BT + L + H, HT-V18 + H + L, HT-V18 + L + H, OT + H + L, OT + L + H, T30695 + H + L, TT + H + L, and TT + L + H; the hybrid structures are as follows: HT + H + L and HT + L + H; and mixed quadrupole structure has the following 3 species: PS5.M + H + L, PS5.M + L + H, and T30695 + L + H.
The antiparallel quadrupole structure is still dominant in the 12 kinds of DNA reactions with dual ions. Although the parallel quadrupole structure began to appear frequently with the gradual increase of G%, it was not the dominant trend. According to Section 3.2, the dominance was of the order of antiparallel quadrupole structure > parallel quadrupole structure. In the double ion, the combination of high + low has a greater impact on the stability of G-quadruplex, and the antiparallel quadrupole structure is more likely to be generated in this combination, which further confirms that the double ion has an excellent prospect for the stability of G-quadruplex.
4.3. Comparison of CD Spectra of Triple Ions
4.3.1. K+ + Na+ + NH4+
According to Figure S13, there are 12 kinds of quadrupole structures formed by the reaction of DNA with K++Na++NH4+, among which the parallel quadrupole structures are as follows: PW17 + K+ + Na+ + NH4+, T30695 + K+ + Na+ + NH4+, and HT-V15 + K+ + Na+ + NH4+; antiparallel quadrupole substructure has the following 6 kinds: AS1411 + K+ + Na+ + NH4+, BT + K+ + Na+ + NH4+, HT + K+ + Na+ + NH4+, HT-V18 + K+ + Na+ + NH4+, OT + K+ + Na+ + NH4+, and TT + K+ + Na+ + NH4+; hybrid structure has the following kind: AT + K+ + Na+ + NH4+; and mixed structure has the following 2 kinds: PS2.M + K+ + Na+ + NH4+ and PS5.M + K+ + Na+ + NH4+.
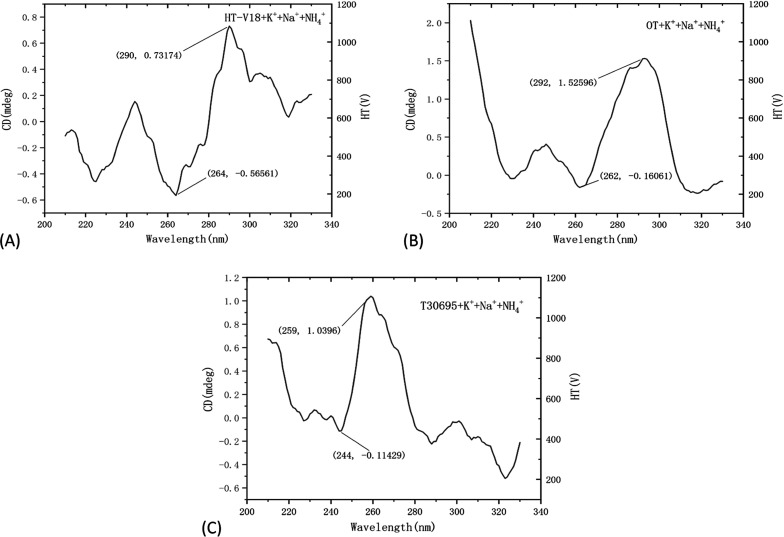
In the 12 DNA combinations of K+ + Na+ + NH4+, the structure of g-tetrad lines is almost antiparallel quadrupole structure. According to Section 3.3, this combination is the best combination of HT-V18, OT, and T30695 sequences, and the obtained G-quadruplex basically exhibits antiparallel quadrupole structure (Figure 5), which confirms the strong repeatability of antiparallel structure in this experiment.
Figure 5.
(A) HT-V18, (B) OT, and (C) T30695 optimal ion addition sequence.
4.3.2. K+ + NH4+ + Na+
According to Figure S14, there are 12 kinds of quadrupole structures formed by the reaction of DNA with K+ + NH4+ + Na+. Among them, the three parallel quadrupole structures are as follows: HT-V15 + K+ + NH4+ + Na+, PW17 + K+ + NH4+ + Na+, and T30695 + K+ + NH4+ + Na+; antiparallel quadrupole substructure has the following six kinds: BT + K+ + NH4+ + Na+, HT + K+ + NH4+ + Na+, HT-V18 + K+ + NH4+ + Na+, OT + K+ + NH4+ + Na+, PS5.M + K+ + NH4+ + Na+, and TT + K+ + NH4+ + Na+; mixed structure has the following kind: PS2.M + K+ + NH4+ + Na+; and hybrid structure has the following two kinds: AS1411 + K+ + NH4+ + Na+ and AT + K+ + NH4+ + Na+.
After the binding of 12 kinds of DNA to K+ + NH4+ + Na+, the G-quadrupole structure gradually increased with G%, and the parallel quadrupole structure began to appear frequently, but most of them were antiparallel quadrupole structures. Combined with Section 3.3, it can be seen that this combination has the most obvious influence on the G-quadruplex structure of AS1411, OT, and TT. According to the comparison, the universality of the antiparallel quadrupole structure can be further proved, and the stability law of the G-quadrupole body is of the order hybrid structure > parallel structure > antiparallel structure. This rule is further verified in the following experiment (Figure 6).
Figure 6.
(A) AS1411, (B) OT, and (C) TT optimal ion addition sequence.
4.3.3. Na+ + K+ + NH4+
According to Figure S15, there are 12 kinds of quadrupole structures formed by the reaction between DNA and Na+ + K+ + NH4+, among which 4 parallel quadrupole structures are as follows: AS1411 + Na+ + K+ + NH4+, PW17 + Na+ + K+ + NH4+, T30695 + Na+ + K+ + NH4+, and PS5.M + Na+ + K+ + NH4+; antiparallel quadrupole substructure has the following 5 kinds: AT + Na+ + K+ + NH4+, HT + Na+ + K+ + NH4+, HT-V18 + Na+ + K+ + NH4+, OT + Na+ + K+ + NH4+, and TT + Na+ + K+ + NH4+; and mixed structure has the following 3 kinds: BT + Na+ + K+ + NH4+, HT-V15 + Na+ + K+ + NH4+, and PS2.M + Na+ + K+ + NH4+.
In the 12 reactions of DNA with Na+ + K+ + NH4+ combinations, the parallel quadrupole structures began to increase with the increase of the G-base content. In this combination, there is no dominant difference between the antiparallel quadrupole structure and the parallel quadrupole structure due to the limited types and the frequent occurrence of the parallel quadrupole structure. According to Section 3.3, this combination is the most stable structure with G-quadruplex generated by sequence AS1411, HT, and T30695, and the phenomenon that parallel quadrupole structure is the most stable structure appears in this combination. However, this does not affect the fact that the antiparallel quadrupole structure is still the most stable (since a large number of data from this experiment show a high level of stability of the generated G-quadrupole) (Figure 7).
Figure 7.
(A) AS1411, (B) HT, and (C) T30695 optimal ion addition sequence.
4.3.4. Na+ + NH4+ + K+
According to Figure S16, there are 12 kinds of quadrupole structures formed by the reaction of DNA with Na+ + NH4+ + K+, among which the parallel quadrupole structure is as follows: T30695 + Na+ + NH4+ + K+; antiparallel quadrupole substructure has the following 4 kinds: BT + Na+ + NH4+ + K+, HT-V18 + Na+ + NH4+ + K+, OT + Na+ + NH4+ + K+, and TT + Na+ + NH4+ + K+; and mixed structure has the following 7 kinds: AS1411 + Na+ + NH4+ + K+, AT + Na+ + NH4+ + K+, PS2.M + Na+ + NH4+ + K+, PS5.M + Na+ + NH4+ + K+, HT-V15 + Na+ + NH4+ + K+, PW17 + Na+ + NH4+ + K+, and HT + Na+ + NH4+ + K+.
Among the 12 combined DNA reactions with Na+ + NH4+ + K+, the parallel/antiparallel quadrupole hybrid structure was unexpectedly dominant. Combined with Section 3.3, it can be seen that this combination can produce a more stable G-quadruplex in HT-V18 (Figure 8A). In this combination, antiparallel quadrupole structure > parallel/antiparallel quadrupole structure can be obtained. The reason may be that the quadrupole structure is not firmly formed in the presence of this combination. The presented hybrid structure is in an inferior position compared with the antiparallel structure, but this conclusion still needs to be verified.
Figure 8.
(A) HT-V18, (B) PS5.M, (C) AS1411, and (D) PS2.M optimal ion addition sequence.
4.3.5. NH4+ + K+ + Na+
According to Figure S17, there are 12 kinds of quadrupole structures formed by the reaction between DNA and NH4+ + K+ + Na+, among which the parallel quadrupole structures are as follows: PW17 + NH4+ + K+ + Na+ and T30695 + NH4+ + K+ + Na+; antiparallel quadrupole substructure has the following 7 kinds: BT + NH4+ + K+ + Na+, HT + NH4+ + K+ + Na+, HT-V18 + NH4+ + K+ + Na+, OT + NH4+ + K+ + Na+, PS2.M + NH4+ + K+ + Na+, PS5.M + NH4+ + K+ + Na+, and TT + NH4+ + K+ + Na+; mixed structure has the following kind: AT + NH4+ + K+ + Na+; and hybrid structure has the following 2 kinds: AS1411 + NH4+ + K+ + Na+ and HT-V15 + NH4+ + K+ + Na+.
In the 12 reactions of DNA with NH4+ + K+ + Na+ combinations, we can see that the antiparallel quadrupole structure is again dominant, which further confirms that the G-quadrupole structure produced in the presence of the 123 combinations is in an unstable state. According to Section 3.3, this combination can produce a stable G-quadruplex structure with PS5.M (Figure 8B).
4.3.6. NH4+ + Na+ + K+
According to Figure S18, there are 12 kinds of quadrupole structures formed by reaction between DNA and NH4+ + Na+ + K+, among which the following 5 kinds of parallel quadrupole structures are found: AS1411 + NH4+ + Na+ + K+, HT-V15 + NH4+ + Na+ + K+, PS2.M + NH4+ + Na+ + K+, PW17 + NH4+ + Na+ + K+, and T30695 + NH4+ + Na+ + K+; antiparallel quadrupole substructure has the following 4 kinds: BT + NH4+ + Na+ + K+, HT + NH4+ + Na+ + K+, OT + NH4+ + Na+ + K+, and TT + NH4+ + Na+ + K+; and mixed structure has the following 3 kinds: AT + NH4+ + Na+ + K+, HT-V18 + NH4+ + Na+ + K+, and PS5.M + NH4+ + Na+ + K+.
In the 12 reactions of DNA with NH4+ + Na+ + K+ in this experiment, the antiparallel quadrupole structure was dominant. As the number of G bases increases, the reactions of this combination with g-quadruplines begin to exhibit frequent parallel quadrupole structures, which is the same phenomenon as the Na+ + K+ + NH4+ combinations. Antiparallel quadrupole structures and parallel quadrupole structures have no obvious advantages in this category. According to Section 3.3, in the presence of this combination, AS1411 and PS2.M have the most stable G-quadrupole structure (Figure 8C,D), and it is concluded that their stability is of the order of regular parallel quadrupole structure > antiparallel quadrupole structure > parallel/antiparallel quadrupole hybrid structure, which corroborates the previous view.
4.4. Overall Analysis
In this part of the experiment, the quadrupole conformation of different DNA sequences in the reaction of single ions, double ions, and triple ions was obtained in detail and summarized in Tables 6–8. It is quite clear that in the course of different combinations of reactions, the antiparallel quadrupole structure produced by G-quadruple has strong repeatability, but it still follows the order of hybrid structure > parallel quadrupole structure > antiparallel quadrupole structure > parallel/antiparallel quadrupole structure in terms of stability, and the hybrid structure usually appears in the sequence with fewer G bases.
Table 6. G-Base Content and the Corresponding Structures of Different DNA Sequencesa.
| G% | parallel structure | antiparallel structure | hybrid structure | mixed structure | |
|---|---|---|---|---|---|
| AS1411 | 65 | low, L + H, X2X1X3, X3X2X1 | H + L, high, X1X2X3 | X1X3X2 | X2X3X1, X3X1X2 |
| AT | 48 | high, low, H + L, L + H, X2X1X3 | X1X3X2, X1X2X3, X3X1X2 | X3X2X1, X2X3X1 | |
| BT | 47 | low | high, H + L, L + H, X1X3X2, X1X2X3, X3X1X2, X3X2X1, X2X3X1 | X2X1X3 | |
| HT | 57 | high, low, X2X1X3, X3X1X2,X3X2X1, X1X2X3, X1X3X2 | H + L, L + H | X2X3X1 | |
| HT-V15 | 80 | H + L, L + H, high, low, X3X2X1, X1X3X2, X1X2X3 | X2X1X3, X3X1X2, X2X3X1 | ||
| HT-V18 | 67 | H + L, L + H, high, low, X1X2X3, X3X1X2, X1X3X2, X2X1X3, X2X3X1 | X3X2X1 | ||
| OT | 57 | H + L, L + H, high, low, X3X2X1, X1X3X2, X1X2X3, X2X1X3, X3X1X2, X2X3X1 | |||
| PS2.M | 67 | H + L, L + H, X3X2X1 | X3X1X2 | high, low, X1X2X3, X2X3X1, X2X1X3, X1X3X2 | |
| PS5.M | 58 | high, X2X1X3 | X3X1X2, X1X3X2 | low, H + L, L + H, X1X2X3, X2X3X1, X3X2X1 | |
| PW17 | 70 | high, low, H + L, L + H, X3X2X1, X1X3X2, X1X2X3, X2X1X3, X3X1X2 | X2X3X1 | ||
| T30695 | 75 | X3X2X1, X1X3X2, X1X2X3, X2X1X3, X3X1X2, X2X3X1 | H + L | L + H | |
| TT | 73 | H + L, L + H, high, low, X3X2X1, X1X3X2, X1X2X3, X2X1X3, X3X1X2, X2X3X1 |
Note: X1 = K+, X2 = Na+, and X3 = NH4+.
Table 8. Query Table of the Ionic Order and Structure of DNA in Different Statesa.
| H/L | high | low | H + L | L + H | X1X2X3 | X1X3X2 | X2X1X3 | X2X3X1 | X3X1X2 | X3X2X1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AS1411 | X1/X3 | A | P | A | P | A | H* | P | M | M | P |
| AT | X2/X3 | A | A | A | A | H | H* | A | M | H | M |
| BT | X1/X3 | A | P | A | A | A | A | H | A* | A | A |
| HT | X2/X3 | A | A | H | H | A | A | A* | M | A | A |
| HT-V15 | X1/X2 | P | P | P | P | P | P | M | M | M | P* |
| HT-V18 | X1/X2 | A | A | A | A | A* | A | A | A | A | M |
| OT | X1/X3 | A | A | A | A | A* | A | A | A | A | A |
| PS2.M | X1/X3 | M | M | P | P | M | M | M | M | A | P* |
| PS5.M | X1/X3 | P | M | M | M | M | A | P | M | A* | M |
| PW17 | X1/X3 | P | P | P | P | P | P | P | M | P | P* |
| T30695 | X2/X3 | P | P | A | M | P* | P | P | P | P | P |
| TT | X2/X3 | A | A | A | A | A | A* | A | A | A | A |
Note: X1 = K+, X2 = Na+, and X3 = NH4+. P = parallel structure; A = antiparallel structure; H = hybrid structure; and M = mixed structure. * is the best order to add to the sequence.
Table 7. DNA Contrast of a Quadrupole Structure Produced in the Presence of a Single Ion, a Double Ion, and a Triple Iona.
| high | low | H + L | L + H | X1X2X3 | X1X3X2 | X2X1X3 | X2X3X1 | X3X1X2 | X3X2X1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| parallel structure | D5, D9, D10, D11 | D1, D3, D5, D10, D11 | D5, D8, D10 | D1, D5, D8, D10 | D5, D10, D11 | D5, D10, D11 | D1, D9, D10, D11 | D11 | D10, D11 | D1, D5, D8,D10, D11 |
| antiparallel structure | D1, D2, D3, D4, D6, D7, D12 | D2,D3, D4,D6, D7, D12 | D1, D2, D3, D6, D7, D11, D12 | D2, D3, D6, D7, D12 | D1, D3, D4, D6, D7, D12 | D3, D4, D6, D7, D9, D12 | D2, D4, D6, D7, D12 | D3, D6, D7, D12 | D3, D4, D6, D7, D8, D9, D12 | D3, D4, D7, D12 |
| hybrid structure | D4 | D4 | D2 | D1, D2 | D2 | |||||
| mixed structure | D8 | D8, D9 | D9 | D9, D11 | D8, D9 | D8 | D3, D5, D8 | D1, D2, D4, D5, D8, D9, D10 | D1, D5 | D2, D6, D9 |
Note: X1 = K+, X2 = Na+, and X3 = NH4+. D1 = AS1411, D2 = AT, D3 = BT, D4 = HT, D5 = HT-V15, D6 = HT-V18, D7 = OT, D8 = PS2.M, D9 = PS5.M, D10 = PW17, D11 = T30695, and D12= TT.
The structure rules of the quadrupoles formed by the reaction are slightly different from those of the single and double ions, depending on the DNA sequence and the different order of addition of the three ions, as shown in Table 5. Although most of the G-quadruplex structures are antiparallel quadrupole structures, it is worth saying that the acromion appears AT 265 nm in the CD images of AS1411 and AT, which proves that the G-quadruplex structure has strong stability, which may be caused by the difference of G%. In HT-V15, PS2.M, and PS5.M, the CD image of M shows the features of both structures, which proves that the structure is parallel/antiparallel mixed quadrupole structure, and the stability of the G-quadrupole structure is low, which may be caused by the influence of other bases (G,T,A) and their arrangement order.
5. Conclusions
-
(1)
For a single ion, K+ can produce a more stable G-quadruplex. When reacting with a single ion, most of the resulting G-quadruplex structures are antiparallel structures.
-
(2)
For double ions, high + low will produce a more stable G-quadruplex structure, and the antiparallel quadrupole structure is dominant. The new rule is antiparallel quadrupole structure > parallel quadrupole structure. In HT-V15 and PW17 sequences, the addition of double ions further promotes the stability of G-quadruplex.
-
(3)
In the presence of triple ions, the G-quadruplex formed by NH4+ + K+ + Na+ combination is more stable. The stability law can be obtained as hybrid structure > parallel quadrupole structure > antiparallel quadrupole structure > parallel/antiparallel quadrupole structure. Among them, in AS1411 and HT-V18, the addition of triple ions makes the stability of G-quadruplex the strongest. Although G-quadruplex has different stabilities under different conditions, the antiparallel quadrupole structure is still dominant, which confirms the repeatability of the antiparallel quadrupole structure in this experiment.
-
(4)
With the increase of the G-base content, parallel quadrupole structure and parallel/antiparallel quadrupole hybrid structure gradually appeared in the G-quadrupole structure, and the hybrid structure usually appeared in AS1411 and AT with a low G content.
-
(5)
Thus, for different sequences, the type and order of the most stable G-quadrustrand are different. The best addition ion for BT, HT, OT, PS2.M, and PS5.M is K+. Na+ was the best addition ion for AT and T30695. The optimal addition of HT-V15 and PW17 were low + high and high + low; the optimal addition order of AS1411 and HT-V18 were K+ + NH4+ + Na+ and K+ + Na+ + NH4+.
-
(6)
Despite the complete and detailed analysis of single ions, double ions, and triple ions in this work, the influence of pH value, temperature, and other conditions is not included. This part of the content will be presented in another comparative experiment of our research group for further discussion. The main objective of this work is to screen out the best stabilizers and order of stability of different kinds of DNA at room temperature.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 21864020 and 51503106), the Natural Science Foundation of Inner Mongolia (Grant Nos. 2018MS02012 and 2019MS02014), the Collaborative Innovation Center for Water Environmental Security of Inner Mongolia Autonomous Region, China (Grant No. XTCX003), and the Inner Mongolia Innovation Guide Project and Research Project of Higher School, Department of Education of Inner Mongolia Autonomous Region (Grant No. NJZC16047).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05243.
Schematic diagram of fluorescence reaction intensity; comparison of fluorescence intensity; and CD spectrogram analysis (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Bates P. J.; Laber D. A.; Miller D. M.; Shelia D. T.; John O. T.; et al. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Jiang R.; He H.; Ma C.; Tang W. Recent advances on G-quadruplex for biosensing, bioimaging and cancer therapy. TrAC, Trends Anal. Chem. 2021, 139, 116257 10.1016/j.trac.2021.116257. [DOI] [Google Scholar]
- Bagheri R.; Karimzadeh F.; Kermanpur A.; Kharaziha M. The novel immobilization of G-quadruplex aptamer on Cu deposited surface using electrochemical method. Mater. Lett. 2021, 282, 128703 10.1016/j.matlet.2020.128703. [DOI] [Google Scholar]
- Sen D.; Walteer G. A sodium-potassium switch in the formation of 4-stranded G-4-DNA. Nature 1990, 344, 410–414. 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- Germann M. W.; Johnson C. N.; Spring A. M. Recognition of damaged DNA: structure and dynamic markers. Med. Res. Rev. 2012, 32, 659–683. 10.1002/med.20226. [DOI] [PubMed] [Google Scholar]
- Park J. Y.; Cho Y. L.; Chae J. R.; Moon S. H.; Cho W. G.; Choi Y. J.; Lee S. J.; Kang W. J. Gemcitabine-Incorporated G-Quadruplex Aptamer for Targeted Drug Delivery into Pancreas Cancer. Mol. Ther. – Nucl. Acids 2018, 12, 543–553. 10.1016/j.omtn.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H.; Greider C. W.; Szostak J. W. Telomeres and telomerase the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 2006, 12, 1133–1138. 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- Schultze P.; Hud N. V.; Smith F. W.; Feigon J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4)). Nucleic Acids Res. 1999, 27, 3018–3028. 10.1093/nar/27.15.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintomé C.; Amrane S.; Mergny J. L.; Alberti P. The exception that confirms the rule: a higher-order telomeric G-quadruplex structure more stable in sodium than in potassium. Nucleic Acids Res. 2016, 44, 2926–2935. 10.1093/nar/gkw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-Y.; Lv Y.; Li S. Molecular dynamics simulation of the stability of (3+1) mixed G-tetraposomes in human telomeres. Acta. Phys.–Chim. Sin. 2009, 25, 783–791. 10.3866/PKU.WHXB200904241. [DOI] [Google Scholar]
- Rizzo A.; Salvati E.; Biroccio A. Methods of studying telomere damage induced by quadruplex-ligand complexes. Methods 2012, 57, 93–99. 10.1016/j.ymeth.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Agrawal P.; Brown R.; Hatzakis E.; Hurley L.; Chen Y. W.; Agra P.; Danzhou Y. The major G-quadruplex formed in the human platelet-derived growth factor receptor beta promoter adopts a novel broken-strand structure in K+ solution. J. Am. Chem. Soc. 2012, 134, 13220–13223. 10.1021/ja305764d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Zhu Y. J.; Wang C.; Pan D.; Liu S.; Yang M.; Xiao Z.; Yang X.; Zhao W.; Zhou X.; et al. Annealing novel nucleobase-lipids with oligonucleotides or plasmid DNA based on H-bonding or pi-pi interaction: Assemblies and transfections. Biomaterials 2018, 178, 147–157. 10.1016/j.biomaterials.2018.06.012. [DOI] [PubMed] [Google Scholar]
- Liu Z.-L.; Tao C.-A.; Wang J.-F. Progress on Applications of G-quadruplex in Biochemical Analysis. Chin. J. Anal. Chem. 2020, 48, 153–163. 10.1016/S1872-2040(19)61212-0. [DOI] [Google Scholar]
- Hänsel-Hertsch R.; Di Antonio M.; Balasubramanian S. DNA G-quadruplexes in the human genome detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachimi A.; Benz A.; Hartig J. S. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009, 17, 6811–6815. 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- Gouci L.; Yang Z. Na+ movement in guanine tetrads. Acta Phys.–Chim. Sin. 2010, 26, 478–486. 10.3866/PKU.WHXB20100219. [DOI] [Google Scholar]
- Chang T.; Li G.; Zhao K.; Ban L.; Pei W.; Bing T.; Shang G. Effects of linking rings, terminal bases and monovalent cations on the structure of G-tetrahydrosome. Chem. J. Chin. Univ. 2014, 35, 2556–2562. [Google Scholar]
- Nakano S. I.; Ayusawa T.; Tanino Y.; Sugimoto N. Stabilization of DNA Loop Structures by Large Cations. J. Phys. Chem. B 2019, 123, 7687–7694. 10.1021/acs.jpcb.9b06074. [DOI] [PubMed] [Google Scholar]
- Han T.; Wang S.; Sheng F.; Wang S.; Dai T.; Zhang X.; Wang G. Target triggered ultrasensitive electrochemical polychlorinated biphenyl aptasensor based on DNA microcapsules and nonlinear hybridization chain reaction. Analyst 2020, 145, 3598–3604. 10.1039/D0AN00065E. [DOI] [PubMed] [Google Scholar]
- Dai T.; Wan Y.; Tian R.; Wang S.; Han T.; Wang G. In Situ Cation Exchange Generated ZnS–Ag2S Nanoparticles for Photothermal Detection of Transcription Factor. ACS Appl. Bio Mater. 2020, 3, 3260–3267. 10.1021/acsabm.0c00232. [DOI] [PubMed] [Google Scholar]
- Liu C.; Wang B.; Han T.; Shi D.; Wang G. Fe Foil-Guided Fabrication of Uniform Ag@AgX Nanowires for Sensitive Detection of Leukemia DNA. ACS Appl. Mater. Interfaces 2019, 11, 4820–4825. 10.1021/acsami.8b18700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.