Abstract

Malic acid derivatives from camu-camu (Myrciaria dubia) fruit exhibited a strong in vitro inhibitory activity toward pancreatic α-amylase and α-glucosidase enzymes. During a bioguided chromatographic fractionation process of the whole fruit (pulp and peelings) polar extract, isomers (S)-4-butoxy-2-hydroxy-4-oxobutanoic acid (1) and (S)-4-butoxy-3-hydroxy-4-oxobutanoic acid (2) (84:16) were isolated and identified as a potent inhibitor of α-amylase (IC50= 11.69 ± 1.75 μg/mL) and α-glucosidase (IC50 = 102.69 ± 4.16 μg/mL). The chemical structures were confirmed by HPLC-ESIMS and 1H and 13C NMR (one- and two-dimensional) analyses. The structure-based virtual screening demonstrated that the aliphatic moiety plays a significant role in the binding mode of the test alkyl malate esters. Compound 1 exhibited the best interaction profile to bind both enzymes, having key structural features to form relevant contacts by involving adequate enzyme–ligand complex stabilization and compactness over time.
Introduction
According to the World Health Organization, 74% of global deaths in 2019 were attributed to lifestyle diseases, such as hypertension, obesity, diabetes mellitus type 2, and the consequent cardiovascular diseases (CVDs).1 The COVID-19 pandemic also showed that lifestyle diseases were the most significant comorbidities causing susceptibility to the SARS-CoV-2 virus and, therefore, the risk of death. The SARS-CoV-2 virus can cause overexpression of angiotensin-converting enzyme II (ACE-2) receptors along the lungs, gastrointestinal tract system, heart, and kidneys, and all whose mechanism of action is binding the ACE-2 receptors located in the host cell’s surface membrane.2 Thus, an alteration of the enzyme signaling and subsequently developing an overactivation of the renin–angiotensin system (RAAS) can be produced as a result of the missed conversion of angiotensin-II into angiotensin (1–7), a vasodilator, such enhancing the risk of injury and death in patients with hypertension and CVDs.3 In this regard, severe clinical conditions in patients with COVID-19 have been related to hypertension (42.3–55.4%), CVDs (30.8%), diabetes (37.3–28.2%), neurological disorders, and cancer. Likewise, recent studies have shown a mortality rate of 17.2–26.0% in patients with COVID-19 and the comorbidities mentioned above.4 This situation was worsened by the sedentary lifestyle resulting from prolonged quarantines as a strategy to contain the SARS-CoV-2 spread worldwide.
Fruit and vegetable are usually rich in vitamin C, phytosterols, and other phytochemicals that act as natural antioxidants decreasing the risk of degenerative and noncommunicable diseases. During the last few years, the ecological importance of the edible fruits belonging to the Myrtaceae family has increased because they are rich in bioactive compounds.5 Among them, camu-camu (Myrciaria dubia) has attracted the attention of researchers because of its high vitamin C content (38.37 ± 2.13 g/100 g dry weight)6 and its potential as a source of phytochemicals to prevent some metabolic diseases7 on the frame of extensive use of the raw material (pulp and peel).8,9
Camu-camu, araca d’agua, cacari, or camo-camo (M. dubia (Kunth) McVaugh) is a tropical fruit belonging to the Myrtaceae family that occurs in flooded areas throughout the Amazon rainforest from Brazil, Peru, Venezuela, and Colombia.10Myciaria dubia fruit (Figure 1) is considered a complex source of phytochemicals such as carotenoids (i.e., lutein, β-carotene, violaxanthin, and luteoxanthin) and polyphenols (i.e., ellagic acid, myricetins, and proanthocyanidins).11 Different in vitro and in vivo studies have shown that M. dubia fruits exhibit health-promoting properties that can potentially prevent noncommunicable chronic diseases, such as hyperglycemia, obesity, and hypertension.7,12,13 For in vitro studies of antihyperglycemic activity, the inhibitors of pancreatic α-amylase and α-glucosidase enzymes have been assessed since their mechanism of action of enzymes is related to the control of postprandial levels of blood glucose, which could significantly impact the treatment of diabetes mellitus type 2.14 The α-amylase and α-glucosidase inhibition values of M. dubia freeze-dried Amazonian fruits have been reported as IC50 = 359 ± 105 and IC50 = 2.98 ± 1.12 μg/mL of reaction,12 respectively, being higher than the value for acarbose (positive control, 3.05 ± 0.25 μg/toward α-amylase and 152 ± 47 μg/mL toward α-glucosidase). Polyphenols casuarictin, ellagic acid, syringic acid, and myricetin showed an excellent statistical correlation with α-amylase and α-glucosidase inhibition values, suggesting their potential as compounds responsible for M. dubia enzyme inhibition.12 However, α-glucosidase inhibition was more potent than α-amylase inhibition since, apart from the fact that they are independent bioassays, the anti-α-amylase activity has been described as structure-dependent instead of total phenolic content-dependent,15 possibly due to several inhibition mechanisms occurring concomitantly during α-glucosidase inhibition; therefore, the activity toward this enzyme is usually higher, depending on the phenolic content.
Figure 1.
Camu-camu (M. dubia) fruits from Colombia.
Malic acid has been reported as responsible for in vitro antihyperglycemic activity in Myrtaceae fruits,16,17 including camu-camu (598.0 mg/100 g fresh weight),18 jaboticaba (1.66 ± 0.01 mg/100 g dry weight),19 pitanga (24.8 ± 0.3 mg/g dry weight),20 and araza (6.7 ± 0.1 mg/g dry weight).20 Computational and kinetics α-glucosidase dynamics simulations confirmed that the l-stereoisomer of malic acid exhibited inhibitory properties on this enzyme (IC50 = 10.68 ± 0.41 mM, with complete enzyme inactivation at 25 mM) and without hydrophobic conditions.21 Likewise, S-malic acid isolated from Flacourtia inermis fruits exhibited inhibitory capacity toward both enzymes (IC50 = 96.40 μg/mL and IC50 = 58.15 μg/mL for α-amylase and α-glucosidase, respectively).16 Also, R-malic acid obtained from Achras sapota L. fruits showed anti-α-amylase activity (IC50 = 25 ± 0.01 μg/mL).17
This work aimed to look for compounds with antihyperglycemic activity through a bioguided fractionation of the polar extract of M. dubia fruits based on α-glucosidase and α-amylase inhibition. The effect of isolated active compounds was extended through structure-based virtual screening using a custom-made library of alkyl malate esters to rank the best-docked compounds and investigate the putative binding mode of these malic acid derivatives.
Results and Discussion
Antihyperglycemic Bioguided Fractionation of the M. dubia Fruit Polar Extract
This work was done to look for sustainable processing strategies for M. dubia fruits found in the Amazonian region. For this reason, the whole fruit (pulp and peelings) was used in all of the experiments, in contrast to the previous works found in the literature.12 The physicochemical characterization of fresh camu-camu showed a pH value of 2.43 ± 0.03, a soluble solid content (SS) of 7.38 ± 0.18° Brix, and a titratable acidity value of 2.20 ± 0.85% citric acid; pH data agreed with previous studies, in which camu-camu was characterized as a highly acidic fruit even in its maturity stage.22,23 The soluble solid content was higher than that is reported in the literature10 because the whole fruit (pulp and peelings) was used in this work.
The acetone/water (7:3, v/v) extract of ca. 4.5 kg of M. dubia fruits was successively partitioned with solvents as follows: pentane, dichloromethane, ethyl acetate, butanol, and water.24 Considering Myrtaceae species exhibited antihyperglycemic activity, the inhibition of α-amylase and α-glucosidase enzymes by the lyophilized fruit and its fractions was measured. Both enzymes are involved in carbohydrate metabolism and, therefore, participate in increasing glucose levels. Thus, the elevated blood glucose level was considered hyperglycemia.25 α-Amylase is responsible for the hydrolysis of complex polysaccharides into small oligosaccharides, and α-glucosidase finally converts all into glucose to be absorbed in the bloodstream.26
Inhibition data of these enzymes and half-maximal inhibitory concentrations (IC50, μg/mL) for the most active fractions are presented in Table 1. Smaller enzyme activity values indicate higher inhibition by the samples. Thus, freeze-dried M. dubia fruits were more active than the positive control (acarbose), in contrast with the data reported by Fujita et al.12 It is noteworthy that Fujita only evaluated the activity of M. dubia frozen pulp, while the whole fruit (i.e., pulp and peelings without seeds) was used in all of the experiments in the present study.
Table 1. In Vitro α-Amylase and α-Glucosidase Inhibitory Activities of M. dubia Fruit and Its Fractions.
| sample | α-amylase activity (mU)a | α-amylase activity IC50 (μg/mL)b | α-glucosidase activity (units/L)a | α-glucosidase activity IC50 (μg/mL)b |
|---|---|---|---|---|
| acarbose | 2.34 ± 0.03a | 139.68 ± 5.37a | 56.35 ± 1.33a | 462.04 ± 4.04 a |
| M. dubia freeze-dried fruit | 1.13 ± 0.14b | 99.07 ± 2.36b | 22.61 ± 0.67b | 267.53 ± 12.67b |
| FAcOEt | 1.53 ± 0.55c | - | 33.76 ± 1.70c | - |
| FBuOH | 0.89 ± 0.64d | 60.74 ± 4.04c | 22.94 ± 0.21b | 237.15 ± 2.39c |
| FWater | 1.00 ± 0.29b | 82.21 ± 3.99d | 41.30 ± 4.09d | 329.71 ± 15.01d |
| F1BuOH | 0.32 ± 0.17a | - | 21.64 ± 3.91a | - |
| F2BuOH | 0.42 ± 0.02b | 23.41 ± 1.99e | 21.31 ± 1.59a | 190.06 ± 7.98e |
| F3BuOH | 1.46 ± 0.81c | - | 20.00 ± 2.50b | - |
| F4BuOH | 0.64 ± 0.51d | - | 7.53 ± 2.25c | - |
| F5BuOH | 2.88 ± 3.05e | - | 33.09 ± 10.16d | - |
| F6BuOH | 0.86 ± 0.25f | - | 20.65 ± 6.55a | - |
| F2.3.2BuOH: (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1) + (S)-4-butoxy-3-hydroxy-4-oxo-butanoic acid (2) (84:16 ratio) | 11.69 ± 1.75f | 102.69 ± 4.16f |
Enzymatic activity at 1000 μg/mL. Activity values are expressed according to the manufacturer’s instructions.
Values are expressed as IC50 ± SD (n = 3) in μg/mL. -, Not determined. Values in the same column/group followed by different superscript, lower-case letters are significantly different by the ANOVA test (p < 0.05). Acarbose was used as the positive control.
Among the fractions shown in Table 1 obtained by partitioning the polar extract, FBuOH was the most active in inhibiting both enzymes, even more active than FAcOEt, which contains more phenolic compounds. This outcome contrasted with the Fujita et al.12 findings, whose antihyperglycemic activity of M. dubia was attributed to the phenolic content. Thus, FBuOH was fractionated by solid-phase extraction (SPE), obtaining six main subfractions (F1BuOH–F6BuOH). The more polar fraction F1BuOH exhibited the highest inhibition of α-amylase activity and was fractionated by preparative HPLC; as a result, a carbohydrate compound was isolated as one of the responsible for this activity (data not shown). Some carbohydrate derivatives, as well as other polymeric compounds such as polysaccharides, peptides, and lipids, have been characterized as natural inhibitors of α-amylase and α-glucosidase activities.27 Then, F2BuOH was subjected to preparative HPLC fractionation, obtaining four main subfractions (F2.1–F2.4). Among them, F2.3BuOH was the most active fraction (α-amylase activity = 0.32 ± 0.01 mU; α-glucosidase activity = 1.98 ± 1.88 units/L). Finally, F2.3BuOH was subsequently fractionated into three subfractions, and F2.3.2BuOH was the most active, with IC50 values of 11.69 ± 1.75 and 102.69 ± 4.16 μg/mL for the inhibition of α-amylase and α-glucosidase, respectively. These values are significantly lower than the positive control acarbose (Table 1), comprising five- to ten-fold higher activities. The inhibitory activities toward two enzymes correlated well, in contrast with de Azevêdo et al.,15 where moderate α-amylase and potent α-glucosidase inhibitory activities were reported for seed and peel extracts of camu-camu from Brazil.
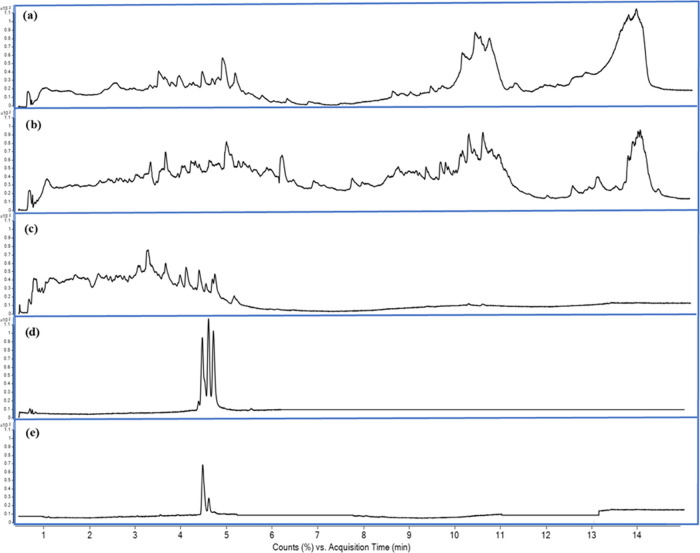
The active F2.3.2BuOH fraction (Figure 2e) was obtained from the parent complex mixture (lyophilized fruit, Figure 2a). This fraction was constituted by two isomeric compounds in an 84:16 ratio, showing the same mass spectrum (Figure 3) with a quasi-molecular ion in ESI negative mode at m/z 189.0769 [M–H]. Water (m/z 171.0665), CO (m/z 160.8422), and C4H9 + H2O (m/z 115.0037) losses were also detected in both mass spectra.
Figure 2.
Total ion chromatogram (TIC) of camu-camu fruit bioactive fractions obtained by UHPLC/MS-QToF analyses (C18 column): (a) lyophilized fruit, (b) FBuOH, (c) F2BuOH, (d) F2.3BuOH, and (e) F2.3.2BuOH.
Figure 3.
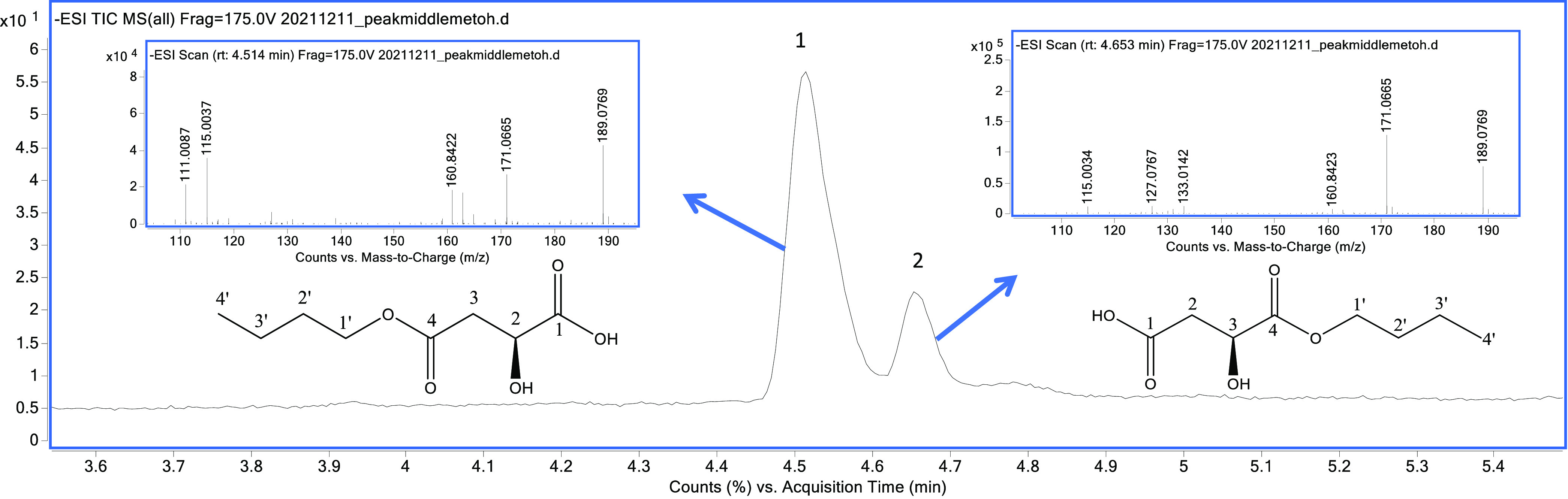
Total ion chromatogram of F2.3.2BuOH by HPLC–MS (C18-column) and HRMS of the isomers: (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1) and (S)-4-butoxy-3-hydroxy-4-oxo-butanoic acid (2).
Then, the NMR spectra were recorded to identify the structure of these isomers. 1H NMR data of F2.3.2BuOH showed 12 aliphatic protons signals corresponding to the major component of the isomeric mixture. Among them, one oxymethine at δH 4.45 and two methylene groups with diasterotopics protons, the first one located at δH 4.13 and 4.10 and the second one resonating at δH 2.60 and 2.52, were detected. The 13C NMR spectrum showed the presence of eight carbons, including one carboxyl group at δC 178.1 (COOH) that correlates in the HMBC spectrum with oxymethine at δH 4.45 and the diasterotopic protons of methylene at δH 2.60 and 2.52. In addition, another carboxyl group located at δC 175.6 (COOR) was detected, showing a correlation in the HMBC experiment with the same methylene group (δH 2.60 and 2.52) and the protons of oxymethylene at δH 4.13 and 4.10. All of these correlations confirmed the presence of a malic acid moiety in the structure. Finally, an n-butyl moiety was evident from 13C NMR data and COSY 1H–1H spectra. The key correlation between the carboxyl group at δC 175.6 (COOR, C4) with the diasterotopic protons of the oxymethylene group at δ H 4.13 and 4.10 confirmed the formation of the ester bond with the carboxyl group at δC 175.6 (COOR, C4). All of the spectroscopic analyses allowed us to conclude the presence of 4-butoxy-2-hydroxy-4-oxo-butanoic acid (Figure 3) as the principal constituent of fraction F2.3.2BuOH. Interestingly, the protons of oxymethylene (H-1′) were diasterotopics, indicating that they are under different magnetic environments, as occurred in prochiral methylene carbons of asymmetric molecules.28
The 1H NMR data of the minor component (F2.3.3BuOH) showed similar signals to those of the compound mentioned above, with the main difference being in the chemical shift signal of the oxymethine proton that appeared at δH 4.03 (H-3/minor isomer) instead of δH 4.45 (H-2/main isomer). The deshielding effect of oxymethine in H-2 (δH 4.45, main isomer) occurs because the oxygen of the carboxylic acid pulls electron density away from the carboxyl carbon (COOH, C1), which inductively pulls electron density away from the adjacent carbon; the same effect occurs in the minor isomer (δH 4.03, H-3) but in less proportion due to the presence of a butyl moiety of ester. Based on this evidence, the presence of the major isomer as a 2-hydroxy-containing monobutyl malate ester and the minor isomer as the 3-hydroxy-containing monobutyl malate ester (i.e., 4-butoxy-3-hydroxy-4-oxo-butanoic acid) (Figure 3) was confirmed. The proportion of two monobutyl malate esters was 84:16. The negative specific rotation ([α]D20= −19.20° ± 2.81) of F2.3.2BuOH suggested an S configuration for the isomeric mixture since it is the same as that reported for the (S)-malic acid commonly found in nature.29
These compounds were not detected in the M. dubia lyophilized fruit after careful screening by HPLC–MS analyses under selecting ion monitoring (SIM) mode, looking for the ion fragment at m/z 189 u ([M–H]−). The fact that malic acid had been reported as a constituent of the M. dubia fruit and that these compounds were detected in the butanolic fraction led us to propose that the monobutyl malate esters were produced as artifacts during the extraction process when the solvent was removed under vacuum. Gronbach et al.30 reported the formation of malic acid esters when alcohols are used for the extraction (sublimation) of sea buckthorn fruits. As mentioned before, in vitro antihyperglycemic activity of malic acid has been reported. This compound has been identified as a constituent of different tropical fruits of the Myrtaceae family, including M. dubia. Additionally, in vivo studies on high-fat-diet-induced mice evidenced antiobesity activity of the M. dubia extract at a 200 mg/kg dose, preventing weight gain and decreasing fasting hyperglycemia.31 Moreover, as the M. dubia pulp improved the biochemical profile (glucose, total cholesterol, and triglycerides contents) in obese rats, the fruit could be considered a functional food ingredient in treating obesity-linked chronic diseases.32 However, this is the first time that monobutyl malate esters are found to be responsible for the inhibition of α-amylase and α-glucosidase, showing better behavior than the positive control (acarbose). Considering that it was not possible to isolate the two pure isomers to measure the individual IC50 biological activity, molecular docking simulations were further utilized to expand the experimental findings through an in silico approach for disclosing the structure-dependent effect of monoalkyl malate esters.
Molecular Docking Studies of Malic Acid and Malate Esters within the Active Site of α-Amylase and α-Glucosidase
Molecular docking was selected as the first-line, structure-based bioactive discrimination method. Although molecular docking can simulate the binding of low-molecular-weight compounds within the active site of target enzymes as a prediction of putative binders and nonbinders,33 the performance of the selected scoring protocol should be assessed before structure-based virtual screening.34 Initially, recently deposited crystal structures of human enzymes were then retrieved from the protein data bank (PDB), such as α-amylase (PDB: 5EMY) and α-glucosidase (PDB: 5NN8), having resolution >2.5 Å andhaving a well-defined active site after optimization with model ligand coordinates35,36 This known information was used to prepare the test enzyme structures for docking studies using AutoDock Vina.37 Therefore, the active site was adopted according to the referenced studies, involving the flexible residues approach (4 Å from the ligand). The cocrystallized ligands were then redocked, and the outcome revealed a suitable behavior of the docking protocol since the superposition of docked and cocrystallized structures of acarbose and the glucosyl epi-cyclophellitol derivative (Figure 4a,b, respectively) led to a root-mean-square deviation (RMSD) of atomic positions <0.8 Å, indicating good convergence. The binder discrimination performance was also evaluated through a benchmarking approach from docking scores of known inhibitors (IC50 < 5 μM), available in the ChEMBL database (i.e., 26 for α-glucosidase and 21 for α-amylase), compared to those of a decoy group (i.e., 1300 for α-glucosidase and 1050 for α-amylase). The area under the curve (AUC) from receiver operating characteristic (ROC) curves was employed to examine the sensitivity and specificity of the docking protocol.38 The AUC and the Boltzmann-enhanced discrimination of ROC (BEDROC) were >0.84 and 0.56, respectively, and accordingly, the validation of the docking protocol was considered successful (Figure 4c,d).
Figure 4.
Docking protocol performance. (a) Cartoon ribbon model of α-glucosidase with the cocrystallized (red) and redocked (blue) acarbose, including the root-mean-square deviation (RMSD) of atomic positions. (b) Cartoon ribbon model of α-amylase with the cocrystallized (red) and redocked (blue) glycosyl glucosyl epi-cyclophellitol derivative, including the RMSD of atomic positions. (c) Receiver operating characteristic (ROC) curve from the benchmarking of the docking protocol using bioactive compounds and decoys within the active site of α-glucosidase. (d) ROC curve from the benchmarking of the docking protocol, using bioactive compounds and decoys within the active site of α-amylase.
To extend the experimental outcome obtained for the monobutyl malate isomeric mixture, structure-based virtual screening, under the validated docking protocol, was performed to understand the binding mode of the studied compounds to rationalize the observed enzymatic activity. Furthermore, an additional set of malate esters having different chain lengths was included in this virtual screening to investigate the putative alkyl-dependent influence, extend the information, and unveil some insights into the enzyme interaction of this compound set. Hence, a small, custom-made library comprising two monobutyl malate esters (1 and 2), malic acid (3), a dibutyl malate ester (4), and a series of seven monoalkyl malate esters increasing the aliphatic chain (C1–C5) (5–11) was initially built and docked using the validated protocol with Autodock Vina using flexible residues. In addition, due to the conformational richness of the test ligands, four additional search algorithms and scoring functions were also employed (i.e., Molegro virtual docker (MVD),39 GOLD,40 LibDock,41 and CDOCKER)42 to be exploited as a consensus docking strategy. The resulting scores from the five docking programs (Table 2) were used to classify the test compounds according to the binding performance by the exponential consensus ranking (ECR) through the previously reported metrics to calculate exponential scores (ESs).43 Results indicated the inhibitor acarbose and the (1 → 6) glucose trimer exhibited the best scoring profile, ranking at the first and second places for both enzymes, respectively, followed by compound (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1) at the third place. Malic acid (3) and methyl malate ester (11) showed the poorest scoring behavior. A previous study using a pseudo-quadratic restraint simulated annealing (PQR-SA)-generated homology model of α-glucosidase reported that compound 3 exhibited a good interacting profile, possibly by competing with the active site residues.21 However, no comparisons with other malic acid derivatives were reported. The results presented here (ECR) revealed an alkyl-dependent scoring trend, with the malic esters with longer alkyl chains being the best-ranked compounds. n-Butyl malate ester 2 and dibutyl malate ester (4) showed a lower rank than 1, and even amyl and isoamyl malic esters (6–7), indicating a plausible important structural factor related to the position of the butyl group substituting the farther carboxyl group from the hydroxyl group. This structure-based virtual screening analysis constitutes the first endeavor to associate the alkyl influence of alkyl malate esters with the putative interaction with these test enzymes.
Table 2. Docking Scores Obtained by Different Programs and Exponential Consensus Ranking for the Test Compounds Docked within the Active Site of α-Glucosidase and α-Amylase.
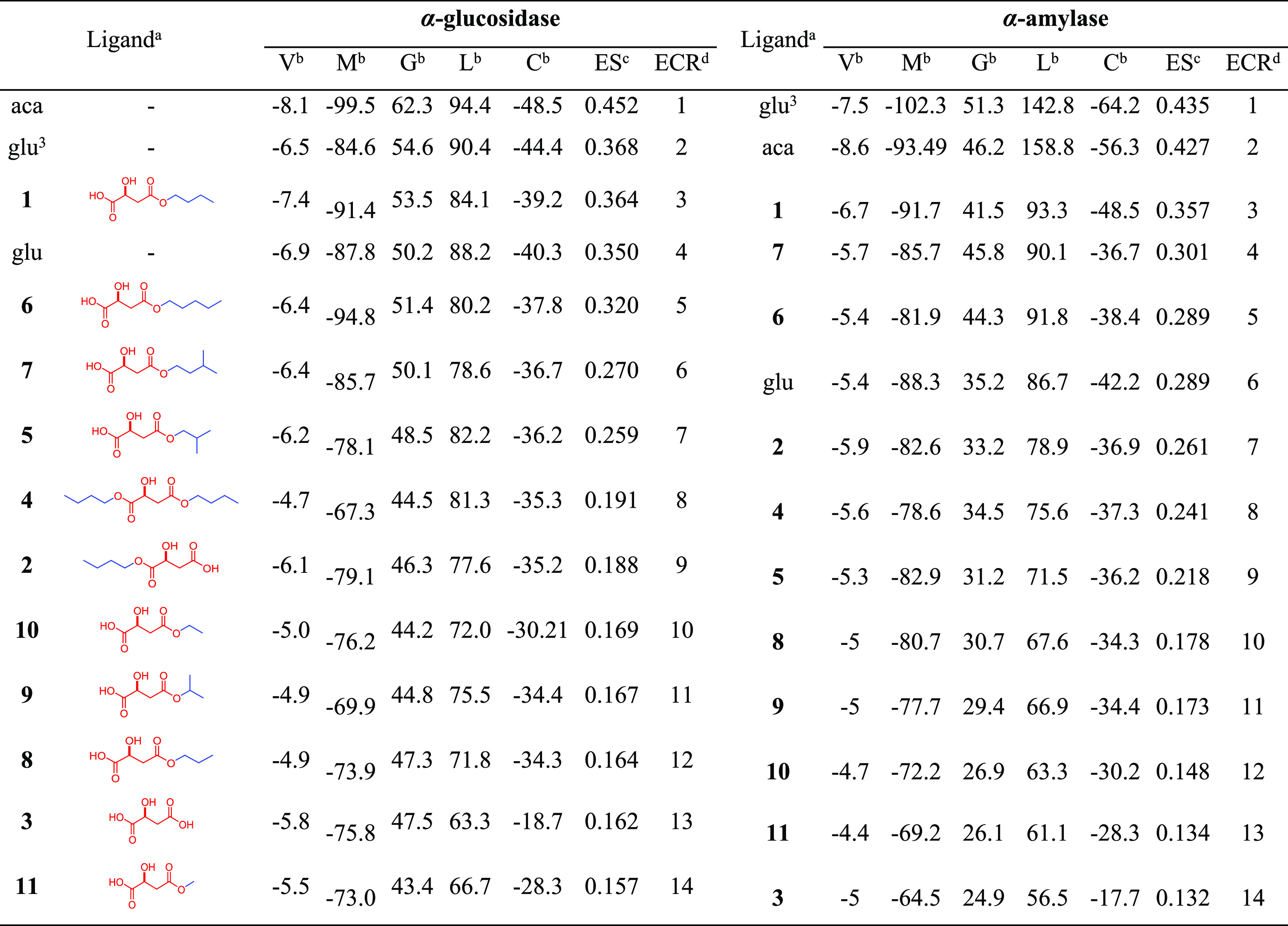
Selected ligands docked within the active site of the two test enzymes: acarbose (aca), glucose (glu), (1 → 6)-glucose trimer (glu3), (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1), (S)-4-butoxy-3-hydroxy-4-oxo-butanoic acid (2), (S)-malic acid (3), dibutyl (S)-malate (4), (S)-4-isobutoxy-2-hydroxy-4-oxo-butanoic acid (5), (S)-4-pentoxy-2-hydroxy-4-oxo-butanoic acid (6), (S)-4-isopentoxy-2-hydroxy-4-oxo-butanoic acid (7), (S)-4-propoxy-2-hydroxy-4-oxo-butanoic acid (8), (S)-4-isopropoxy-2-hydroxy-4-oxo-butanoic acid (9), (S)-4-ethoxy-2-hydroxy-4-oxo-butanoic acid (10), and (S)-4-methoxy-2-hydroxy-4-oxo-butanoic acid (11).
Scores obtained after molecular docking simulations with programs having different search algorithms and scoring functions: V = Vina scores, M = scores MolDock scores, G = GOLD scores, L = LibDock scores, C = CDOCKER scores.
ES = exponential scores obtained from docking scores per program through the metrics previously reported.43
ECR = exponential consensus ranking organized from ES.
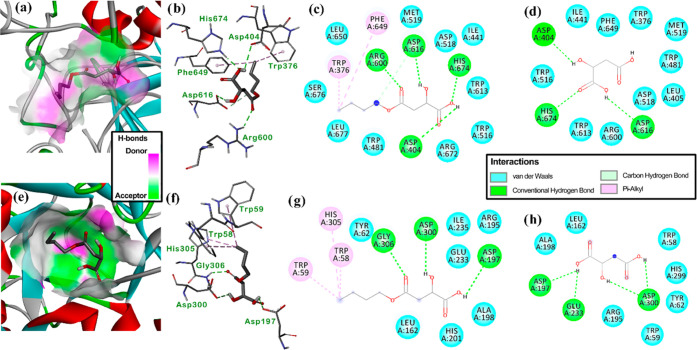
The previous scoring tendency led us to examine the binding mode of (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1) through the individual three- and two-dimensional (3D and 2D, respectively) diagrams to look for relevant interactions between active site residues and ligand moieties (Figure 5). In this regard, the best-docked pose of 1 was fitted very well within the active site of both enzymes (Figure 5a–e). Its binding mode within the active site of both enzymes was found to have a similar profile. It involved H-bonding interactions between the β-hydroxycarboxyl moiety of 1 with Asp404, His674, and Asp616 α-glucosidase residues and with Asp197 and Asp200 α-amylase residues (Figure 5b–f), which were shared with those interaction found for malic acid docking (Figure 5d–h). The better scoring ranking of compound 1 in comparison to that of malic acid can be rationalized by the hydrophobic pi–alkyl interactions of butyl moiety of 1 with Trp376 and Phe649 α-glucosidase residues and with Trp58, Trp59, and His305 α-amylase residues (Figure 5c–g), which might explain plausible stabilization of the α-glucosidase···1 and α-amylase···1 complexes. Indeed, n-butyl substitution appeared to have adequate carbon chain distance to interact with the mentioned residues since the best-docked pose of the other alkyl substitutions (C5–C1) did not favor such multiple hydrophobic interactions. Thus, compound 1 has the acarbose′s polar requirements, particularly in its λ2-azanyl-polyhydroxy-cyclohex-4-ene moiety, involving an additional electrostatic contact favored by the length and hydrophobicity of the n-butyl substituent. These results suggest that these alkyl derivatives of malic acid constitute an interesting series of chemical agents with antihyperglycemic properties to be further explored.
Figure 5.
Representation of the enzyme···ligand complexes: (a) three-dimensional (3D) model of (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1) docked within the active site cavity of α-glucosidase, involving H-bond donor and acceptor zones, (b) 3D residual interaction diagram (RID) of the α-glucosidase···1 complex, (c) 2D RID of the α-glucosidase···1 complex, (d) 2D RID of the α-glucosidase···malic acid (3) complex, (e) 3D model of 1 docked within the active site cavity of α-amylase, involving H-bond donor and acceptor zones, (f) 3D RID of the α-amylase···1 complex, (g) 2D RID of the α-amylase···1 complex, and (h) 2D RID of the α-amylase···3 complex. Residues are differentiated in 2D RIDs by colors according to the interaction type: hydrophobic/van der Waals, aquamarine; polar/conventional hydrogen bond, green; and hydrophobic/pi–alkyl, pink. 3D RIDs depict docked compounds in bold sticks and interaction as dash lines colored according to the aforementioned interaction type. 3D models depict docked compounds in bold sticks and enzymes as secondary structures—colored cartoons.
Molecular dynamics (MD) simulations led us to extend the binding influence of compound 1 on both enzymes to explain the putative ligand···enzyme complex stability through geometric properties over time. Hence, 100 ns MD simulations for the unbound α-glucosidase and α-amylase and docking with compound 1 and acarbose were separately performed. The structural stability of the test molecules was assessed by measuring the time-dependent variations between distinct locations (in Å) of the entire atom set and plotted as root-mean-square deviations (RMSDs) along the simulated time frame (Figure 6a–c). The unbound α-glucosidase and α-amylase evolved normally during the simulation, and good stabilization after 25 and 15 ns was observed with RMSD variations within 1.1–1.6 and 1.3–1.7 Å, respectively. However, the evolution of the enzyme···ligand complexes was found to be similar to that of unbound enzymes, involving structural stabilization as one-step progression. Thus, those complexes between test enzymes and acarbose and compound 1 exhibited a very comparable profile with some particular slight perturbations, reaching an initial stabilization step after 45 ns (0.9–1.3 Å) and 35 ns (0.9–1.4 Å) for α-glucosidase and α-amylase (Figure 6a–c). The evolution of the packing level of the enzyme···ligand complexes during the simulation time, evaluated through the radius of gyration (RoG), was found to be slightly different between both enzymes (Figure 6b–d). In general, unbound α-amylase exhibited lesser overall compactness (typically for α/β-proteins) than unbound α-glucosidase (typically for (α + β)-proteins) (23 vs 24 Å, respectively). However, both enzymes retained their packed levels over time, although α-amylase exhibited slight variations (i.e., 0.2 Å). Complexes between α-amylase and acarbose and compound 1 exhibited a highly similar RoG profile. In the case of the α-glucosidase···acarbose complex, the RoG profile was found to be lower (i.e., 0.1 Å) than that of the unbound enzyme, while the α-glucosidase···compound 1 complex showed a higher RoG profile (i.e., 0.2 Å). Despite the enzyme···ligand interaction affecting the complex 3D packing, a reasonable steady folding reflected its stability after binding between α-glucosidase and compound 1.
Figure 6.
Molecular dynamics (MD) simulations during 100 ns for unbound enzymes (green line) and docked enzymes separately with 1 (red line) and acarbose (blue line). (a) Root-mean-square deviations (RMSDs) along MD-simulated trajectories of α-glucosidase. (b) Radius of gyration along MD-simulated trajectories of α-glucosidase. (c) Root-mean-square deviations (RMSDs) along MD-simulated trajectories of α-amylase. (d) Radius of gyration along MD-simulated trajectories of α-amylase.
Conclusions
Bioguided fractionation of M. dubia fruits (peel and pulp) allowed us to isolate of two monobutyl malate esters, generated during sample work-up, that exhibited significant antihyperglycemic activity by inhibiting α-amylase and α-glucosidase enzymes. Spectroscopic analyses confirmed their structures. The main compound, (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1), showed the best interaction profile among alkyl malate esters to bind both enzymes, resulting in the best ranking by exponential consensus metrics. It exhibited adequate structural features (n-butyl as a hydrophobic moiety and β-hydroxycarboxylic moiety as polar contact favoring H-bonds) to form crucial interactions by involving enzyme–ligand complex stabilization and compactness over time. This knowledge could help to develop new medicines to control hyperglycemia after confirming the viability of using monobutyl malate ester safely in humans. If so, this would be a significant finding for the pharmaceutical industry because it is a compound relatively easy to synthesize.
Material and Methods
General
1H and 13C NMR one- and two-dimensional spectra of compound (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1) were recorded on a Bruker Advance III HD Ascend spectrometer using a 5 mm triple resonance observe TXO cryoprobe with Z-gradients, operating at 700 MHz for the 1H nucleus and 175 MHz for the 13C core (Bruker BioSpin, Rheinstetten, Germany). Data processing was performed using MestReNova 14.2.1-27684. NMR spectra recorded in D2O were referenced to the residual nondeuterated solvent signal at δH 4.79, and the solvent signal at δC 48.84 and formic acid signal at δC 68.45 were set as the internal reference. Analysis by HPLC–MS of 1 was performed on an Agilent 1290 Infinity-II UHPLC system (Agilent, Santa Clara (CA)) equipped with a high-speed binary pump, a sample manager, a multicolumn thermostat, a diode array detector and coupled with an Agilent 6546 Q-ToF mass spectrometer (Agilent Technologies) in negative ESI ionization mode. An LC/MS-QuatroMicro coupled with a 2545 binary gradient manager (Waters Co.) and a fraction collector 2767 (Waters Co.) was used for preparative purposes. The lyophilization process was carried out for 48 h in the Beta 1-8 LDplus lyophilizer equipment (CHRIST, Germany). The main drying phase lasted 40 h at a pressure of 1.0 mbar and a temperature of −20 °C. The final drying phase lasted 8 h at 0.001 mbar at a temperature of −76 °C. The optical rotation of 1 (c = 0.1 mg/100mL in water) was measured on a high-accuracy polarimeter ADP 440 (Bellingham & Stanley, United Kingdom) and calculated with the specific rotation formula.
Plant Material
Fresh camu-camu (M. dubia) fruits were collected at Macagual Amazon Research Center (CIMAZ—Centro de Investigaciones Amazónicas Macagual), Universidad de la Amazonía, geographically located at 1°37′N and 75°36′W, 300 m above sea level, from a farm rural located 20 km from Florencia, Caquetá (Colombia). Ripe fruits were selected according to the peel color (entirely red) and the physicochemical parameters. The pH measurement of M. dubia fruits was performed by a pH meter 370 (Jenway, London, U.K.); the soluble solid content (SS) was determined by an Atago HSR-500 refractometer (Tokyo, Japan), and the results were expressed as °Brix (AOAC protocol 932.12).44 Finally, titratable acidity was determined following the standard AOAC protocol (942.15)44 using a 0.1 N NaOH solution and expressing the result as a percentage of citric acid. All analyses were reported as the average ± standard deviation (n = 5).
Chemicals
For the fractionation of the fruit extract, the following solvents were used: acetone (ITW Reagents, AppliChem GmbH, Darmstadt, Germany), methanol (Applichem GmbH, ITW company, Castellar del Vallés, Barcelona, Spain), butanol (Mallinckrodt, Xalostoe, México), ethyl acetate (Merk–Sigma-Aldrich Co, Billerica, MA), dichloromethane (Merck–Sigma-Aldrich Co, Darmstadt, Germany), and pentane (ITW Reagents, AppliChem GmbH, Darmstadt, Germany). LC/MS optima solvents (formic acid, methanol, and acetonitrile) were acquired from Fisher Scientific (Fair Lawn, NJ). Ultrapure water used for HPLC analysis and enzyme assays was purified by a GenPure Pro UV-TOC/UF water purification system (Thermo Scientific, Atvidaberg, Sweden). Acarbose used as a positive control for the in vitro α-amylase and α-glucosidase inhibition assays was obtained from Sigma-Aldrich Co. (St. Louis, MO). Sodium phosphate dibasic and sodium phosphate monobasic were obtained from Spectrum Chemical MFG, Crop (Gardena, CA).
Extraction
Whole M. dubia fruits (4499 g) were lyophilized, obtaining 807 g of dried fruits. The seeds were manually removed from the dried fruits to get 377 g (peel and pulp), which was successively extracted with three volumes of 500 mL of acetone/water (7:3, v/v). The extracts were combined, and the solvent was removed under vacuum in a rotary evaporator. Then, the aqueous residue was subjected to successive partitioning with pentane (Fpentane), dichloromethane (FDCM), ethyl acetate (FEtOAc), and butanol (FBuOH), 300 mL each; the aqueous residue was denominated as Fwater.24 The solvent from all fractions was removed by distillation under vacuum and lyophilization in the case of aqueous residues, obtaining the following amounts: FPentane, 0.869 g; FDCM, 6.088 g; FEtOAc, 4.600 g; FBuOH, 16.271 g; and Fwater, 114.677 g. Each of these fractions was analyzed by HPLC–MS, and their antihyperglycemic activity was determined by the α-amylase and α-glucosidase inhibition assays.
In Vitro Measurement of Antihyperglycemic Activity
α-Amylase Inhibition Assay
This assay was used to bioguide the fractionation of the M. dubia extract. The α-amylase activity was determined using the MAK009 (Sigma-Aldrich Co, St Louis, MO) enzymatic kit. The assay was carried out according to the protocol provided by the manufacturer.45 For that purpose, the lyophilized M. dubia fruit and its fractions were prepared at 1000 ppm in a pH 7.0 phosphate buffer (50 mM) and subjected to the assay following the kit protocol. Absorbance values were measured at λ 405 nm using a spectrophotometric multiwell plate reader accuSkan GO (Fisher Scientific), and data were analyzed using Thermo Scientific SkanIt Software version 4.0. Pancreatic α-amylase activity values were expressed in mU (milliunits), and calculations were done according to the protocol formula (formula 1)
| 1 |
where B is the amount (nmole) of nitrophenol generated between tinitial and tfinal, the reaction time is tfinal–tinitial, and V is the sample volume (mL) added to the well.
To determine IC50 (μg/mL) values, samples were prepared at different concentrations (500, 250, 100, 50, and 10 ppm), and inhibition values were expressed as a percentage (%) using formula 2
| 2 |
Then, a nonlinear regression method was applied by plotting % α-amylase inhibition vs sample concentration. IC50 is defined as the sample concentration needed to in vitro inhibit the α-amylase activity by 50%.
α-Glucosidase Inhibition Assay
In this case, the α-glucosidase kit (MAK123, Sigma-Aldrich Co., St Louis, MO) was used following the exact protocol of the technical bulletin.46 α-Glucosidase activity of samples was reported as units/L and calculated at a reaction time of 20 min following the protocol formula (formula 3)
| 3 |
IC50 (μg/mL) values were calculated using nonlinear regression curves to determine the half-maximal inhibitory concentration toward α-glucosidase.
Bioguided Fractionation
Based on the results of enzyme inhibition (Table 1), FBuOH (16.271 g) was further fractionated by SPE (solid-phase extraction) using C18 EC cartridges (10 g Chromabond, Macherey-Nagel GmbH & Co., Düren, Germany) and a Manifold (Thermo Scientific HyperSep). Each cartridge was loaded with 1.0 g of sample and eluted successively with MeOH–H2O (0/100, 20/80, 40/60, 60/40, 80/20, 100/0, v/v, 60 mL each) to get six fractions (F1 (271 mg), F2 (784 mg), F3 (1.015 g), F4 (624 mg), F5 (412 mg), and F6 (328 mg)). The solvent was removed from all of the fractions and subjected to the previously described in vitro α-amylase and α-glucosidase inhibition assays.
Ultra-Performance Liquid Chromatography–Mass Spectrometry Chemical Profiling (UHPLC/MS-QToF)
LC–MS analyses of all fractions (100 ppm in methanol) and compound 1 were performed on a Cortecs UPLCr C18 + 1.6 mm column (2.1 mm × 100 mm, 1.6 μm, Waters Co.) with an injection volume of 10 μL. The column temperature was maintained at 40 °C. Acidified water (0.1% formic acid) and acetonitrile (0.1% formic acid) were used as solvents A and B, respectively, with the following gradient: 0–1 min, 5% B; 1–5 min, 5–45%, 5–7 min, 45% B; 10–12 min, 85% B; 14–17 min, 95% B; 17–19 min, 5% B to initial conditions. The column was re-equilibrated for 2 min at initial conditions between injections. The flow rate of the mobile phase was set at 0.5 mL/min. The settings for the mass spectrometer are as follows: ESI ionization = negative, mass scan range = 50–1700 Da, scan time = 0.2 s, capillary voltage = 3500 v, nozzle voltage = 500 v, desolvation temperature = 350 °C, desolvation gas flow rate = 10 L/min, sheath gas temperature = 375 °C, and sheath gas flow rate = 11 L/min. m/z 112.9855 and m/z 966.007 were used as reference masses to check mass accuracy throughout the analysis.
Preparative Liquid Chromatography–Mass Spectrometry (Prep-LC–MS)
The most active fraction, F2BuOH (0.784 g), was dissolved in a mixture of methanol/water (1:4, v/v) at a concentration of 13 mg/mL, filtered through a PTFE filter of 0.45 mm, and injected into the preparative system. An Xbridge BEH C18 OBD 130A prep column (5 μm particle size, 10 mm × 250 mm, Waters Co.) was used. The solvent system was a mixture of water/formic acid (99.9:0.1, v/v, solvent A) and ethanol/formic acid (99.9:0.1, v/v, solvent B), and the flow rate was 7 mL/min. A linear gradient is used as follows: 0–6 min, 5% B; 6–8 min, 5–10% B; 8–11 min, 10–20% B; 11–24 min, 20–90% B; and 26 min, keeping 5% B at initial conditions. Four subfractions were obtained (F2.1BuOH, F2.2BuOH, F2.3BuOH, and F2.4 FBuOH), and F2.3BuOH exhibited the highest inhibition for the two enzymes. This fraction (F2.3BuOH, 22.9 mg) showed the presence of three peaks (Figure 2d); thus, it was subsequently fractionated into three new fractions (F2.3.1BuOH, F2.3.2BuOH, and F2.3.3BuOH) using the above-mentioned conditions. A mixture of two components (3.1 mg) was obtained from F2.3.2BuOH. Their chemical structures were elucidated from UHPLC-ESI–MS and NMR spectra one- and two-dimensional analyses as a mixture of isomeric (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1) and (S)-4-butoxy-3-hydroxy-4-oxo-butanoic acid (2) in an 84:16 ratio, respectively.
Spectroscopic Data of (S)-4-Butoxy-2-hydroxy-4-oxo-butanoic acid (1)
White solid. 1H NMR (700 MHz, D2O): δH 4.45 (dd, J = 7.7, 4.9 Hz, 1H, H-2), 4.13 (td, J = 11.2, 6.3 Hz, 1H, H-1a′), 4.10 (td, J = 11.2, 7.0 Hz, 1H, H-1b′), 2.60 (dd, J = 15.4, 4.9 Hz, 1H, H-3a), 2.52 (dd, J = 15.4, 7.0 Hz, 1H, H-3b), 1.57 (quintet-like, J = 6.3 Hz, 2H, H-2′), 1.29 (sextet, J = 7.7 Hz, 2H, H-3′), 0.83 (t, J = 7.7 Hz, 3H, H-4′) ppm. 13C NMR (125 MHz, D2O): δC 12.9 (C-4′), 18.4 (C-3′), 29.8 (C-2′), 41.5 (C-3), 66.0 (C-1′), 68.5 (C-2), 175.7 (C-4), 178.2 (C-1). [α]D20 = −19.20° ± 2.81 (H2O; c 0.10 g/mL); HR-ESIMS Found 189.0769 [M–H]−, C8H13O5 requires 189.0763; m/z 171.0665 [M–H2O–H]−, 160.8422 [M–CO–H]−. MS data are presented in Figure 3.
Spectroscopic Data of (S)-4-Butoxy-3-hydroxy-4-oxo-butanoic acid (2)
White solid. 1H NMR (700 MHz, D2O): δH 4.13 (td, J = 15.4, 7.7 Hz, 1H, H-1a′), 4.08 (td, J = 15.4, 7.7 Hz, 1H, H-1b′), 4.03 (t, J = 7.7 Hz, 1H, H-3), 2.62 (dd, J = 18.2, 5.6 Hz, 1H, H-2a), 2.56 (dd, J = 18.2, 8.4 Hz, 1H, H-2b), 1.56 (quintet-like, J = 7.7 Hz, 2H, H-2′), 1.30 (sextet-like, J = 7.7 Hz, 2H, H-3′), 0.83 (t, J = 8.4 Hz, 3H, H-4′) ppm. HR-ESIMS Found 189.0769 [M–H]−, C8H13O5 requires 189.0763; m/z 171.0665 [M–H2O–H]−, 160.8423 [M–CO–H]−, 133.0142 [M–C4H9]−. MS data are presented in Figure 3.
Molecular Docking Studies of Mono-n-alkyl Malate Derivatives with α-Amylase and α-Glucosidase Enzymes
A custom-made library comprising 11 malic acid derivatives (i.e., two monobutyl esters (1–2), malic acid (3), a dibutyl ester (4), and seven monoalkyl esters increasing the aliphatic chain (C1–C5) (5–11)) were 2D sketched using Marvin (version 20.11.0, ChemAxon (https://www.chemaxon.com)), and the 3D models were generated through Standardizer (version 20.11.0, ChemAxon (https://www.chemaxon.com)). A random conformational search using the Merck molecular force field (MMFF), without geometric restrictions, included in SPARTAN software (Spartan 14v114 (2013) Wavefunction, Inc., Irvine) was performed with a 500 conformer limit. The energetically most stable conformers were then MMFF-optimized to be used in the docking protocol. The crystal structures of α-glucosidase and α-amylase were imported from the Protein Data Bank RCSB (5NN8 and 5EMY, respectively).35,36 Their sequences and 3D structures were retained without any processing for molecular docking. However, missing hydrogen atoms were added, and ligands, cofactors, and other cocrystallized compounds were removed from the test enzyme molecules. The cocrystallized inhibitors (acarbose for α-glucosidase and glucosyl epi-cyclophellitol derivative for α-amylase) were employed to define the corresponding active site and as a validation criterion of docking calculations (redocking). The active sites for both enzymes were defined in the respective coordinates (−14.5, 32.3, and 95.9 for α-glucosidase and −10.82, −18.20, and 24.5 for α-amylase).
Molecular docking simulations were initially performed with the Autodock/Vina (1.1.2) plug-in for PyMOL (1.3r2) under a Python 2.5.2 environment for Windows.47 Docking simulations were performed between the MMFF-minimized ligand within a cube (dimensions 24 × 24 × 24, grid spacing 1 Å) located at the geometric center of the previously defined active sites of both enzymes. Flexible residues (n = 11) were defined within 4 Å of the test ligand. Additionally, the specificity and sensitivity of the docking protocol were assessed using compounds of diverse chemical nature (i.e., 26 for α-glucosidase and 21 for α-amylase), which have IC50 < 5 μM against each test enzyme, retrieved from the ChEMBL database.48 Furthermore, 50 decoys per active compound were compiled from the Directory of Useful Decoys, Enhanced (DUD-E).49 Thus, decoys and reported bioactive compounds were processed using the same docking protocol with Autodock Vina. The resulting data was then assessed through ROC curves and score enrichment using the screening explorer webserver.50 Once the docking performance was assessed and validated, the docking simulations of test alkyl malate esters were performed. Docking programs Molegro virtual docker (MVD) 6.0,39 GOLD,40 LibDock,41 and CDOCKER,42 having different scoring functions, were additionally used under the same aforementioned docking parameters to assess the docking performance. From the resulting scores from the five programs, the best-docked compounds were therefore top-ranked by a consensus strategy through the exponential score (ES) calculation, following the reported metrics for exponential consensus ranking (ECR) using formula 4.43
| 4 |
where σ = exponential distribution (=10), i = test compound, j = scoring function, and rij = the ranking per program achieved for each test compound.
Finally, 3D models and 2D residual interaction diagrams of the best pose of selected compounds were visualized on Discovery Studio 2016 Visualizer Client (Biovia, San Diego).
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations were run in Gromacs 5.0.5 on a Ubuntu 12.04 server. The best pose of 1 and acarbose obtained from docking and the enzyme crystal structure were employed as input for molecular dynamics simulations of complexes and unbound enzymes. Ligands were prepared by adding hydrogen atoms and the corresponding charges using the AM1-BCC charge scheme in UCSF Chimera. Subsequently, ligand topologies were generated automatically with an ACPYPE script. Protein topologies were obtained in Gromacs using the Amber 99SB force field, and the TIP3P water model was implemented.51 Solvation was performed in a triclinic box using a margin distance of 1.0 nm. The addition of 0.1 M NaCl to complexes and proteins was carried out by randomly replacing water molecules until neutrality. The systems were energy-minimized by 2000 steps of the steepest descent method. NVT equilibration at 310 K for 50 ps, followed by NPT equilibration for 500 ps using the Parrinello–Rahman method at 1 bar as a reference, was conducted on systems using position restraints. Finally, the solute position restraints were released, and a production run for 100 ns was performed. The temperature and pressure were kept constant at 310 K and 1 bar, respectively. The coordinates were recorded with a 1 fs time step. Electrostatic forces were calculated using the particle-mesh Ewald (PME) method. Periodic boundary conditions were used in all simulations, and covalent bond lengths were constrained by the LINear Constraint Solver (LINCS) algorithm.52
Statistical Analysis
Significant differences among samples were determined using Statgraphics Plus 5.1 software (StatPoint, Inc., Herndon, VA), analysis of variance (ANOVA), and Tukey’s tests. Differences at probability level P ≤ 0.05 were considered significant.
Acknowledgments
The authors are thankful to Natalia Cuellar for supplying the fruits. J.M.G.-Ch. thanks FREC members, Diana Paola Forero, Ph.D. and Said Toro, Ph.D., for their guidance in using UHPLC/MS-QToF and the preparative system for the analyses.
This work was supported by grants from the Fondo Nacional de Financiamiento para la Ciencia, la Tecnología y la Innovación, Francisco José de Caldas (contract no. 0459-2013), Red Nacional para la Bioprospección de Frutas Tropicales-RIFRUTBIO, and DIEB (División de Investigación y Extensión de la Universidad Nacional de Colombia-sede Bogotá). J.M.G.-C. thanks financial support for a scholarship from the Ministerio de Ciencia, Tecnología e Innovación of Colombia (Minciencias) grant: Programa de Becas de Excelencia Doctoral del Bicentenario—Corte-I, and Fulbright Visiting Scholar Program 2021.
The authors declare no competing financial interest.
References
- World Health Organization (WHO) . The Top 10 Causes of Death 2020https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
- Ni W.; Yang X.; Yang D.; Bao J.; Li R.; Xiao Y.; Hou C.; Wang H.; Liu J.; Yang D.; Xu Y.; Cao Z.; Gao Z. Role of Angiotensin-Converting Enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 422 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djaharuddin I.; Munawwarah S.; Nurulita A.; Ilyas M.; Tabri N. A.; Lihawa N. Comorbidities and mortality in COVID-19 patients. Gac. Sanit. 2021, 35, S530–S532. 10.1016/j.gaceta.2021.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad Malik J.; Ahmed S.; Shinde M.; Almermesh M. H. S.; Alghamdi S.; Hussain A.; Anwar S. The impact of COVID-19 on comorbidities: A review of recent updates for combating it. Saudi J. Biol. Sci. 2022, 29, 3586–3599. 10.1016/j.sjbs.2022.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri-Numa I. A.; Soriano Sancho R. A.; Pereira A. P. A.; Pastore G. M. Small Brazilian wild fruits: Nutrients, bioactive compounds, health-promotion properties and commercial interest. Food Res. Int. 2018, 103, 345–360. 10.1016/j.foodres.2017.10.053. [DOI] [PubMed] [Google Scholar]
- Cunha-Santos E. C. E.; Viganó J.; Andrade Neves D.; Martínez J.; Teixeira Godoy H. Vitamin C in camu-camu [Myrciaria dubia (H.B.K.) McVaugh]: evaluation of extraction and analytical methods. Food Res. Int. 2019, 115, 160–166. 10.1016/j.foodres.2018.08.031. [DOI] [PubMed] [Google Scholar]
- Azevedo L.; de Araujo Ribeiro P. F.; de Carvalho Oliveira J. A.; Correia M. G.; Ramos F. M.; de Oliveira E. B.; Barros F.; Stringheta P. C. Camu-camu (Myrciaria dubia) from commercial cultivation has higher levels of bioactive compounds than native cultivation (Amazon Forest) and presents antimutagenic effects in vivo. J. Sci. Food Agric. 2019, 99, 624–631. 10.1002/jsfa.9224. [DOI] [PubMed] [Google Scholar]
- Santos I. L.; Freire Miranda L. C.; da Cruz Rodrigues A. M.; Meller da Silva L. H.; Amante E. R. Camu-camu [Myrciaria dubia (HBK) McVaugh]: A review of properties and proposals of products for integral valorization of raw material. Food Chem. 2022, 372, 131290 10.1016/j.foodchem.2021.131290. [DOI] [PubMed] [Google Scholar]
- de Paulo Farias D.; Neri-Numa I. A.; Fernandes de Araújo F.; Pastore G. M. A critical review of some fruit trees from the Myrtaceae family as promising sources for food applications with functional claims. Food Chem. 2020, 306, 125630 10.1016/j.foodchem.2019.125630. [DOI] [PubMed] [Google Scholar]
- Aguirre-Neira J. C.; Reis M. S.; Cardozo M. A.; Raz L.; Clement C. R. Physical and chemical variability of camu-camu fruits in cultivated and uncultivated areas of the Colombian Amazon. Rev. Bras. Frutic. 2020, 42, e-545. 10.1590/0100-29452020545. [DOI] [Google Scholar]
- Fracassetti D.; Costa C.; Moulay L.; Tomás-Barberán F. A. Ellagic acid derivatives, ellagitannins, proanthocyanidins and other phenolics, vitamin C and antioxidant capacity of two powder products from camu-camu fruit (Myrciaria dubia). Food Chem. 2013, 139, 578–588. 10.1016/j.foodchem.2013.01.121. [DOI] [PubMed] [Google Scholar]
- Fujita A.; Sarkar D.; Wu S.; Kennelly E.; Shetty K.; Genovese M. I. Evaluation of phenolic-linked bioactives of camu-camu (Myrciaria dubia McVaugh) for antihyperglycemia, antihypertension, antimicrobial properties and cellular rejuvenation. Food Res. Int. 2015, 77, 194–203. 10.1016/j.foodres.2015.07.009. [DOI] [Google Scholar]
- Fujita A.; Sarkar D.; Genovese M. I.; Shetty K. Improving anti-hyperglycemic and anti-hypertensive properties of camu-camu (Myriciaria dubia McVaugh) using lactic acid bacterial fermentation. Process Biochem. 2017, 59, 133–140. 10.1016/j.procbio.2017.05.017. [DOI] [Google Scholar]
- Karakaya S.; Gözcü S.; Güvenalp Z.; Özbek H.; Yuca H.; Dursunoğlu B.; Kazaz C.; Kiliç C. S. The α-amylase and α-glucosidase inhibitory activities of the dichloromethane extracts and constituents of Ferulago bracteata roots. Pharm Biol. 2018, 56, 18–24. 10.1080/13880209.2017.1414857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevêdo J. C. S.; Fujita A.; de Oliveira E. L.; Genovese M. I.; Pinto Correia R. T. Dried camu- camu (Myrciaria dubia H.B.K. McVaugh) industrial residue: A bioactive-rich Amazonian powder with functional attributes. Food Res. Int. 2014, 62, 934–940. 10.1016/j.foodres.2014.05.018. [DOI] [Google Scholar]
- Alakolanga A. G. A. W.; Savitri Kumar N.; Jayasinghe L.; Fujimoto Y. Antioxidant property and α-glucosidase, α-amylase and lipase inhibiting activities of Flacourtia inermis fruits: characterization of malic acid as an inhibitor of the enzymes. J. Food Sci. Technol. 2015, 52, 8383–8388. 10.1007/s13197-015-1937-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.; Dutta M.; Chaudhury K.; De B. Metabolomic and chemometric study of Achras sapota L. fruit extracts for identification of metabolites contributing to the inhibition of α-amylase and α-glucosidase. Eur. Food Res. Technol. 2016, 242, 733–743. 10.1007/s00217-015-2581-0. [DOI] [Google Scholar]
- Zapata S. M.; Dufour J.-P. Camu-Camu Myrciaria dubia (HBK) McVaugh: Chemical composition of fruit. J. Sci. Food Agric. 1993, 61, 349–351. 10.1002/jsfa.2740610310. [DOI] [Google Scholar]
- Albuquerque B. R.; Pereira C.; Calhelha R. C.; Alves M. J.; Abreu R. M. V.; Barros L.; Oliveira M. B. P. P.; Ferreira I. C. F. R. Jabuticaba residues (Myrciaria jaboticaba (Vell.) Berg) are rich sources of valuable compounds with bioactive properties. Food Chem. 2020, 309, 125735 10.1016/j.foodchem.2019.125735. [DOI] [PubMed] [Google Scholar]
- Sviech F.; Ubbink J.; Prata A. S. Analysis of the effect of sugars and organic acids on the ice melting behavior of pitanga and araza pulp by differential scanning calorimetry (DSC). Thermochim. Acta 2021, 700, 178934 10.1016/j.tca.2021.178934. [DOI] [Google Scholar]
- Gou L.; Zhan Y.; Lee J.; Li X.; Lü Z.-R.; Zhou H.-M.; Lu H.; Wang X.-Y.; Park Y.-D.; Yang J.-M. Effects of L-malic acid on alpha-glucosidase: inhibition kinetics and computational molecular dynamics simulations. Appl. Biochem. Biotechnol. 2015, 175, 2232–2245. 10.1007/s12010-014-1429-6. [DOI] [PubMed] [Google Scholar]
- Akter M. S.; Oh S.; Eun J.; Ahmed M. Nutritional compositions and health promoting phytochemicals of Camu-camu (Myrciaria dubia) fruit: A review. Food Res. Int. 2011, 44, 1728–1732. 10.1016/j.foodres.2011.03.045. [DOI] [Google Scholar]
- Fujita A.; Borges K.; Correia R.; Gombossy de Melo Franco B. D.; Genovese M. I. Impact of spouted bed drying on bioactive compounds, antimicrobial and antioxidant activities of commercial frozen Pulp of camu-camu (Myrciaria dubia Mc. Vaugh). Food Res. Int. 2013, 54, 495–500. 10.1016/j.foodres.2013.07.025. [DOI] [Google Scholar]
- Isaza J. H.; Ito H.; Yoshida T. Oligomeric hidrolizable tannins from Monochaetum multiflorum. Phytochemistry 2004, 65, 359–367. 10.1016/j.phytochem.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Tundis R.; Loizzo M. R.; Menichini F. Natural products as α-amylase and α-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: An update. Mini-Rev. Med. Chem. 2010, 10, 315–331. 10.2174/138955710791331007. [DOI] [PubMed] [Google Scholar]
- Kanwal K. M. K.; Chigurupati S.; Ali F.; Younus M.; Albubayan M.; Wadood A.; Khan H.; Taha M.; Perveen S. Indole-3-acetamides: As potential antihyperglycemic and antioxidant agents; synthesis, in vitro α-amylase inhibitory activity, structure–activity relationship, and in silico studies. ACS Omega 2021, 6, 2264–2275. 10.1021/acsomega.0c05581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Bai Y.; Jin Z.; Svensson B. Food-derived non-phenolic a-amylase and a-glucosidase inhibitors for controlling starch digestion rate and guiding diabetes-friendly recipes. LWT 2022, 153, 112455 10.1016/j.lwt.2021.112455. [DOI] [Google Scholar]
- Jennings W. B. Chemical shift nonequivalence in prochiral groups. Chem. Rev. 1975, 75, 307–322. 10.1021/cr60295a003. [DOI] [Google Scholar]
- Bancroft W. D.; Davis H. L. The optical rotation of malic acid. J. Phys. Chem. A 1930, 34, 897–928. 10.1021/j150311a001. [DOI] [Google Scholar]
- Gronbach M.; Kraußer L.; Broese T.; Oppermann C.; Kragl U. Sublimation for enrichment and identification of marker compounds in fruits. Food Anal. Methods 2021, 14, 1087–1098. 10.1007/s12161-020-01954-6. [DOI] [Google Scholar]
- Abot A.; Brochot A.; Pomié N.; Wemelle E.; Druart C.; Régnier M.; Delzenne N. M.; de Vos W. M.; Knauf C.; Cani P. D. Camu-camu reduces obesity and improves diabetic profiles of obese and diabetic mice: A dose-ranging study. Metabolites 2022, 12, 301. 10.3390/metabo12040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento O. V.; Boleti A.; Yuyama L. K.; Lima E. S. Effects of diet supplementation with camu-camu (Myrciaria dubia HBK McVaugh) fruit in a rat model of diet-induced obesity. An. Acad. Bras. Ciênc. 2013, 85, 355–363. 10.1590/S0001-37652013005000001. [DOI] [PubMed] [Google Scholar]
- Sliwoski G.; Kothiwale S.; Meiler J.; Lowe E. W. Jr. Computational methods in drug discovery. Pharmacol. Rev. 2014, 66, 334–395. 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermaier Y.; Barril X.; Scapozza L. Virtual screening: An in silico tool for interlacing the chemical universe with the proteome. Methods 2015, 71, 44–57. 10.1016/j.ymeth.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Caner S.; Zhang X.; Jiang J.; Chen H.-M.; Nguyen N. T.; Overkleeft H.; Brayer G. D.; Withers S. G. Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic α-amylase. FEBS Lett. 2016, 590, 1143–1151. 10.1002/1873-3468.12143. [DOI] [PubMed] [Google Scholar]
- Roig-Zamboni V.; Cobucci-Ponzano B.; Iacono R.; Ferrara M. C.; Germany S.; Bourne Y.; Parenti G.; Moracci M.; Sulzenbacher G. Structure of human lysosomal acid α-glucosidase–a guide for the treatment of Pompe disease. Nat. Commun., 2017, 8, 1111 10.1038/s41467-017-01263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triballeau N.; Acher F.; Brabet I.; Pin J-P.; Bertrand H-O. Virtual screening workflow development guided by the”receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J. Med. Chem. 2005, 48, 2534–2547. 10.1021/jm049092j. [DOI] [PubMed] [Google Scholar]
- Bitencourt-Ferreira G.; Azevedo W. F. D. Jr.. Molegro Virtual Docker for Docking. In Docking Screens for Drug Discovery; Humana: New York, NY, 2019; pp 149–167. [DOI] [PubMed] [Google Scholar]
- Jones G.; Willett P.; Glen R. C.; Leach A. R.; Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 3, 727–748. 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Rao S. N.; Head M. S.; Kulkarni A.; LaLonde J. M. Validation studies of the site-directed docking program LibDock. J. Chem. Inf. Model. 2007, 47, 2159–2171. 10.1021/ci6004299. [DOI] [PubMed] [Google Scholar]
- Gagnon J. K.; Law S. M.; Brooks C. L. III Flexible CDOCKER: Development and application of a pseudo-explicit structure-based docking method within CHARMM. J. Comput. Chem. 2016, 37, 753–762. 10.1002/jcc.24259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacio-Rodríguez K.; Lans I.; Cavasotto C. N.; Cossio P. Exponential consensus ranking improves the outcome in docking and receptor ensemble docking. Sci. Rep. 2019, 9, 5142 10.1038/s41598-019-41594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AOAC . Association of Official Analytical Chemists. In Official Methods of Analysis of AOAC International, 18th ed.; Association of Official Analytical Chemists: Gaithersburg, USA, 2015http://www.aoac.org/iMIS15_Prod/AOAC. [Google Scholar]
- Sigma-Aldrich Co . Technical Bulletin Amylase Activity Assay Kit. 2016, Catalog number MAK009. https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/377/793/mak009bul.pdf.
- Sigma-Aldrich Co . Technical Bulletin α-Glucosidase Activity Assay Kit. 2016, Catalog number MAK123. https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/286/096/mak123bul.pdf.
- Seeliger D.; de Groot B. L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento A. P.; Gaulton A.; Hersey A.; Bellis L. J.; Chambers J.; Davies M.; Krüger F. A.; Light Y.; Mak L.; McGlinchey S.; Nowotka M.; Papadatos G.; Santos R.; Overington J. P. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014, 42, D1083–D1090. 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysinger M. M.; Carchia M.; Irwin J. J.; Shoichet B. K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empereur-Mot C.; Zagury J-F.; Montes M. Screening explorer-An interactive tool for the analysis of screening results. J. Chem. Inf. Model. 2016, 56, 2281–2286. 10.1021/acs.jcim.6b00283. [DOI] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1-2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J.; Fraaije J. G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem., 1997, 18, 1463–1472. 10.1002/(SICI)1096-987X(199709)18:123.0.CO;2-H. [DOI] [Google Scholar]