Abstract
Objective
Reflex cryptococcal antigen (CrAg) screening of blood specimens with a CD4 count of <100 cells/µL was performed at 45 South African CD4 laboratories using a lateral flow assay (LFA). Our objective was to evaluate the reliability of routine LFA results through comparative interlaboratory testing.
Methods
All CrAg-positive and a selected number of CrAg-negative samples from the CD4 laboratories were retested at paired microbiology laboratories using the same LFA. Samples with discordant results were tested at a reference laboratory, using the LFA (with CrAg titers).
Results
During interlaboratory testing, 12,502 samples were retested, with 93 (0.7%) discordant results and a between-laboratory agreement of 99.3% (Cohen’s kappa, 0.98). The proportion of retested samples with discordant results ranged from 0.17% to 5.31% per laboratory pair (median 0.28%), with 3 reporting >3% of results as discordant.
Conclusion
Routine CrAg screening results were reliable, with <1% of samples having discordant results, mainly due to interpretation and transcription errors.
Keywords: cryptococcal antigen, interlaboratory comparison, discordant results, cryptococcal meningitis, lateral flow assay, advanced HIV disease
Cryptococcal antigen (CrAg) screening followed by preemptive antifungal treatment of CrAg-positive persons with advanced HIV disease is recommended by the World Health Organization (WHO) to reduce mortality associated with cryptococcal meningitis.1,2 In October 2016, South Africa’s National Health Laboratory Service (NHLS) initiated reflex CrAg screening of remnant blood specimens with a CD4+ T-cell (CD4) count of <100 cells/µL.3–6 Screening was performed using a CrAg lateral flow assay (LFA) (IMMY) across a national network of 45 CD4 laboratories.5,6 These laboratories are equipped with flow cytometry instruments to perform routine CD4+ T-cell enumeration for the national HIV program. From February 1, 2017, through September 30, 2021, 99% (n = 1,012,493) of eligible patients in the public health sector were screened, with a national prevalence of 6.0%7 (unpublished data, National Institute for Communicable Diseases). Clinician-requested CrAg tests on blood and cerebrospinal fluid (CSF) were performed at NHLS microbiology laboratories using the same LFA method. Patients with a new positive blood CrAg result were recommended to have a lumbar puncture (LP) to exclude cryptococcal meningitis.1 Asymptomatic patients with antigenemia who were CSF CrAg-negative or who declined an LP were recommended to be treated preemptively with fluconazole until antiretroviral treatment-mediated immune reconstitution occurs.
To ensure accurate result reporting through the screening program, all NHLS CD4 laboratories performed verification procedures before patient testing commenced in 2016. This included testing of a verification panel of random CrAg-negative and -positive samples, using the IMMY LFA kit. In addition, an in-house proficiency testing scheme (PTS) was developed in 2017 for CrAg testing of simulated blood samples; NHLS CD4 and microbiology laboratories participated in this PTS.5 However, commonly encountered analytical and postanalytical issues are seldom detected by these quality assurance methods. Most CrAg-positive samples included in the PTS are strongly CrAg-positive or -negative, making assessment of reading faint bands on the test strip a challenge (personal communication, NHLS Quality Assurance Department). This is the most common error noted, where faint results may be interpreted differently by individuals as the results are manually read and verified. Due to manual recording of CrAg test results on a paper worksheet before being transferred manually to the electronic laboratory information system (LIS), transcription errors can also occur.
Routine samples are tested once and authorized results are immediately reported, making it difficult to gauge the reliability of CrAg results in the routine national program. We thus assessed the reliability of routine CrAg LFA results through national interlaboratory comparison, testing during a 7-month period in 2018, with retesting at secondary laboratories and confirmatory testing of samples with discordant results at a reference laboratory. We also conducted a root cause analysis to identify the underlying reasons for discordant results, with recommendations on how these could be rectified in practice.
Methods
Primary CrAg Testing
Remnant-settled ethylenediaminetetraacetic acid (EDTA)-plasma samples with a confirmed CD4 count of <100 cells/µL were routinely tested by 45 CD4 laboratories using the IMMY CrAg LFA, as per the manufacturer’s instructions and an internal standard operating procedure, available through a Q-Pulse quality management system portal. CrAg LFA results were routinely entered by laboratory personnel into the NHLS TrakCare electronic LIS, and authorized results were then immediately accessible to health care providers.
Interlaboratory CrAg Testing
Interlaboratory testing was set up between paired CD4 and microbiology laboratories, most based at the same health care facility, over a 6-month period from April to October 2018. Staff training at each site was provided by the reference laboratory through on-site visits and telephone or video conference calls. The purpose of training was to explain and/or arrange CrAg retesting procedures and logistics at each laboratory pair. Participating laboratory pairs were provided with a supplementary SOP guiding additional requirements. For a period ranging from 3 to 6 months (depending on availability of reagents), each of the 45 CD4 laboratories sent a 2 mL aliquot of settled plasma from all CrAg-positive samples and a minimum of 5 CrAg-negative samples daily to their partner microbiology laboratory. The CrAg LFA results obtained by the CD4 laboratory for the selected samples, CrAg LFA lot numbers, and kit expiry dates were transcribed onto study worksheets and also sent to the microbiology laboratory, where samples were retested using the IMMY CrAg LFA method. The results of these tests, the LFA lot number, and kit expiry date were also recorded.
Confirmation CrAg Testing at the Reference Laboratory
Concordant result lists were sent to the Mycology Reference Laboratory at the National Institute for Communicable Diseases (NICD), Johannesburg, for data capture. For discordant results, plasma samples were shipped on ice to the reference laboratory for testing within 48 hours, that included: (1) IMMY LFA strips from 2 or more lot numbers to test samples (to exclude lot-to-lot variation), with results being independently and manually read by 2 investigators; and (2) serial dilutions were performed in samples with sufficient volumes to determine CrAg titers and to exclude a high-dose hook effect.8
Root Cause Analysis of Discordant Results
Possible reasons for discordant results were investigated according to the confirmatory results. A validity check was performed for quality controls and kit lot number expiry dates recorded on worksheets. For each sample with discordant results, a detailed laboratory report with recommended corrective actions was compiled. Recommendations were based on the type of preanalytical, analytical, and postanalytical errors that frequently occurred (see Supplemental Table 1). The identified issues were resolved through on-site training on CrAg testing, interpretation, and results capture followed by competency recertification. In addition, we delivered a dedicated session on CrAg test troubleshooting within an online series of training seminars to all NHLS end users. Consolidated reports were compiled and shared with each testing laboratory pair at the end of the 6-month test period. Data were captured using REDCap electronic data capture tools hosted at the University of the Witwatersrand9 and Microsoft Excel and analyzed using Stata version 15.0 software (StataCorp). The agreement between CrAg results obtained by the primary and secondary testing laboratories was assessed using the Cohen’s kappa statistic. Ethics approval was obtained from the Human Research Ethics Committee (Medical), University of the Witwatersrand (M140111 of 2014, for 5 years).
Results
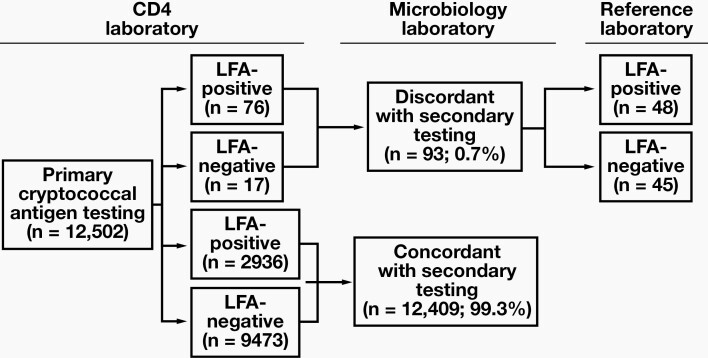
During the interlaboratory comparison period (April through October 2018), 202,345 blood specimens with a CD4 count of <100 cells/µL were eligible for CrAg screening at the 45 CD4 laboratories, of which 200,387 (99%) specimens were tested. The remaining 1958 (1%) samples were either of insufficient volume or otherwise unsuitable for CrAg testing due to hemolysis, an abnormally high viscosity, or high concentration of lipids, with the plasma appearing white or milky in color. Of the 200,387 specimens, 11,576 (5.7%) were CrAg-positive. A total of 12,502 blood samples were included in the retesting program. Of these samples, 2936 CrAg-positive (23.6%) and 9473 CrAg-negative results (76.4%) were concordant between the paired testing laboratories (Figure 1). The overall agreement between paired laboratory CrAg test results was 99.3% (Cohen’s kappa statistic, 0.98). Ninety-three (0.7% of total) samples with discordant results between paired testing laboratories were sent for confirmatory testing to the reference laboratory (see Supplemental Table 2).
Figure 1.
Algorithm to test plasma samples for cryptococcal antigenemia, with primary IMMY lateral flow assay (LFA) testing at CD4 laboratories, retesting of selected samples at secondary microbiology laboratories, and confirmatory testing (IMMY LFA plus titers) of samples with discordant results at a reference laboratory.
Reference Laboratory Results
Of the 93 discordant samples referred to the reference laboratory, 76 (82%) were reported as LFA-positive by the primary testing CD4 laboratory but LFA-negative by the secondary testing microbiology laboratory. Of these 76 samples, the reference laboratory confirmed that 46 (61%) were indeed LFA-negative and thus discordant with the primary testing laboratory’s routine results; 41/46 were also LFA-negative on serial dilutions (5 had insufficient volume for dilutions) (Table 1). Of the remaining 30/76 (39%), 1 had an insufficient volume for confirmatory testing; the other 29 were confirmed as LFA-positive at the reference laboratory (and thus concordant with the primary testing laboratory), and all (except 4 with insufficient volumes for dilutions) had a low CrAg titer of ≤80 (median titer of 5). The remaining 17/93 (18%) samples were originally reported as LFA-negative by the primary testing CD4 laboratory and were LFA-positive on retesting. Fifteen of these 17 samples were confirmed to be LFA-positive at the reference laboratory; 2 had very high LFA titers of ≥2560, 11 had low LFA titers of ≤40, and 2 had insufficient volumes for titer determination. The other 2/17 samples were LFA-negative (on serial dilutions) at the reference laboratory, confirming the result of the primary testing laboratory.
Table 1.
Samples with Discordant Results Between Paired Testing Laboratories Sent for Confirmatory Testing to the Reference Laboratory (n = 93)
| Reference Laboratory Results | Total | |||
|---|---|---|---|---|
| LFA-Positive | LFA-Negative | Insufficient Sample | ||
| Primary laboratory results | ||||
| LFA-positivea | 29 | 46 | 1 | 76 |
| LFA-negativea | 15 | 2 | 0 | 17 |
| Total | 44 | 48 | 1 | 93 |
LFA, lateral flow assay.
aSecondary laboratory results were discordant with those of the primary laboratory.
Discordant Samples Per Testing Laboratory
The total number of samples earmarked for retesting ranged from 46 to 653 per laboratory pair (median of 246), with a range of 0 to 315 positive samples (median of 38) and 46 to 482 negative samples (median of 183) tested per laboratory pair. Twenty-two of the 45 laboratory pairs (49%) reported discordant results, ranging from 0 to 22 per laboratory pair for the 6-month period. Thus 23 of 45 (51%) laboratory pairs reported no discordant results at all during the testing period. The proportion of retested samples with discordant results ranged from 0.17% to 5.31% per laboratory pair (median 0.28%), with 42/45 laboratories reporting a proportion of <1%. Only 3 laboratory pairs, all in the same province, exceeded 3% (laboratory pair A: 22/414 [5.3%], laboratory pair B: 8/243 [3.3%], and laboratory pair C: 10/307 [3.3%]). On closer investigation of the individual data for these 3 laboratory pairs, 14/22 of the primary LFA results from laboratory pair A were confirmed as discordant at the reference laboratory, with 3/14 samples tested on 1 day. Six of 8 primary LFA results for laboratory pair B were confirmed as discordant at the reference laboratory and of these, 5/6 samples were tested at the primary laboratory on the same day. For laboratory pair C, all 10 samples with discordant results were confirmed as such at the reference laboratory and tested separately on nonconsecutive dates.
Discussion
On average, 280,000 patients were screened per annum in the South African national reflex CrAg screening program since it was initiated in 2016.5,10,11 This laboratory screening directly supports South Africa’s large public sector HIV program and, at the time of this study, was aligned to recommendations from both the South African Department of Health and Southern African HIV Clinicians Society to screen HIV-seropositive individuals with a CD4 count of <100 cells/µL.1,2 Clinicians can also request diagnostic CSF CrAg tests from NHLS microbiology laboratories for patients with symptomatic cryptococcal meningitis or order blood screening CrAg tests for asymptomatic patients with CD4 counts of 100 to 200 cells/µL (ie, above the current reflex CrAg testing threshold). The current study was logistically feasible because plasma samples initially tested by a CD4 laboratory could be retested by a paired microbiology laboratory on the same premises using the same standardized method of testing (ie, IMMY LFA). These conditions provided a unique opportunity to evaluate routine laboratory results reported by the national CrAg screening program through interlaboratory analysis.
Accurate result reporting in pathology laboratories is a high priority, and several systems are in place to ensure continuous excellent performance. This includes laboratory accreditation with a national body such as the South African National Accreditation System to ISO 15189:2012, participation in external quality assessment (or proficiency testing) schemes for CD4 and CrAg testing; internal quality control per batch tested (as per manufacturer instructions); and verification of a test method upon implementation using panels of control material.12,13 More than 98% of NHLS laboratories participated in an external quality assessment scheme for CrAg testing (personal communication, NHLS Quality Assurance Department). The limitation of this, and similar PTSs, is that each distributed survey includes a panel of strong-positive and -negative samples with fewer samples with a low antigen concentration to mimic the faint CrAg positivity often seen on LFA strips in the laboratory. The point-of-care nature of the CrAg LFA test may also pose challenges for an external quality assessment scheme, as interpretation and reporting of results are manual. Regular interlaboratory comparison would be an ideal way to assess the true number of samples with discordant results, but is not always feasible in a resource-limited country with staffing shortages and additional reagent and distribution costs.
In this study, we report interlaboratory comparative testing, for a period of 3 to 6 months at each laboratory pair, using both CrAg-negative and -positive samples for parallel testing at CD4 and microbiology laboratories, with confirmatory testing at a reference laboratory, using LFA and LFA-titer methods as standards. Cryptococcus cannot be cultured directly from serum or EDTA-plasma samples. However, published data has confirmed the excellent accuracy of the LFA in serum and plasma to the reference standard of CSF fungal culture among patients with cryptococcal meningitis, with superior performance compared to the older CrAg latex agglutination test.14 In serum, there is good agreement between the enzyme immunoassay (EIA) and LFA for diagnosis of antigenemia.15,16 The manufacturer states in the package insert that the limit of detection for the IMMY EIA is 5.3 ng/mL of antigen, while the IMMY LFA has a lower limit (ie, 1.5 to 3 ng/mL of antigen) and would thus be expected to be more sensitive than the EIA.17,18 There is no equivalent comparison for EDTA-plasma samples since the EIA is not validated for use on this specimen matrix and thus was not used as a reference in this study. This study is not a report on method comparison under ideal laboratory conditions but instead is a reflection of daily CrAg testing results and operational challenges in routine laboratories. In this study, we also did not aim to describe national or local CrAg prevalence or test positivity rates; this information has been reported in earlier publications using a more representative study sample.10,19,20
Overall, only 0.7% of all samples tested in this interlaboratory comparison had discordant results across 45 paired laboratories. The median proportion of tested samples with discordant results across the 45 laboratory pairs was very low (0.28%). Twenty-three laboratory pairs did not report any samples with discordant results, while the 3 laboratories with >3% discordant results tested low daily volumes and routinely reported few CrAg-positive results; the actual number of samples with discordant results equated to <2 per month of testing. Two laboratories had several samples with discordant results reported on the same day. This could indicate a new staff member reporting or verifying results. Additional training was done at the identified 3 laboratories with recertification of staff competency through test witnessing. All other discordant results reported for the 22 testing laboratory pairs were single samples on nonconsecutive days.
The discordant test results noted in this study could be in part caused by very low antigen concentrations, near the limit of detection for the LFA. This is supported by 36 samples with low antigen titers (of 81 total with sufficient volumes for serial dilutions). In addition, 2 of 81 had very high LFA titers of ≥2560, suggesting false-negative results caused by a possible high-dose hook effect. This latter issue is likely to be a far less common problem unless a patient with antigenemia has concurrent symptomatic meningitis or fungemia and thus very high plasma antigen concentrations. On root cause analysis, approximately 95% of discordant samples tested at the reference laboratory were attributed to misinterpretation of LFA bands or transcriptional errors onto the worksheet and/or LIS. Because the LFA results are manually read and interpreted, the visual acuity of the observer or poor ambient lighting could contribute to misclassification of faint-positive bands, for example. The impact of transport to the reference laboratory on sample stability was minimized by shipping samples in cold boxes, with retesting performed within 48 hours. This does not rule out the impact of sample age on retesting, as most samples tested at the primary CD4 laboratories are older than 48 hours (post venesection). Local studies have, however, indicated that CrAg results are stable over time (up to 72 hours post venesection) under controlled laboratory conditions (data not shown) when performed on settled plasma. This is also within the stipulation of the package insert of the IMMY LFA, where storage of specimens is allowed at 2°C to 8°C for up to 72 hours.
Our study highlights the importance of assessing not only strong-positive and -negative samples in PTSs but also samples with concentrations of CrAg close to the limit of detection in a panel to test the ability of laboratories to accurately identify and report on tests with faint lines. A challenge of the CrAg LFA is the manual reading and reporting of results, since this was designed for use at the point of care. The NHLS has conducted a validation of commercially available strip readers (data not shown) in an effort to reduce human error. These readers were either not suitable for multiple strip verification and/or had no LIS connectivity for direct upload of results (most use an Excel-based program for data capture). New-generation readers may address these issues and be validated for introduction into high-volume CrAg testing laboratories. Fully automated CrAg EIA systems as a possible solution to eradicate human error have also been investigated, with high sensitivity and specificity compared to the LFA (>95%). These are, however, expensive and not ideal for low- to medium-volume testing. A limitation of this study was that microbiology laboratories were not blinded to CrAg results from the CD4 laboratory for practical reasons (routinely reported authorized results from the CD4 laboratory were available on the LIS). Samples were collected over a long period to ensure adequate test numbers, yet some laboratories had either low daily test volumes and/or CrAg positivity that may have led to an overestimation of the proportion with discordant results. Not all CrAg-negative samples were retested to save costs. These data are not a reflection of CrAg prevalence because we retested selected CrAg-negative and all CrAg-positive samples at each laboratory pair for a defined period.
Conclusion
CrAg screening results are critically important for clinical decision-making in people with advanced HIV disease, and a positive CrAg test result will prompt invasive investigations and or potentially toxic treatment regimens. Reliable and accurate results are thus essential in a screening program. This is the first study to report the operational discordant results for CrAg testing in a national screening program rather than in the context of method validation or duplicate sample testing. Routine reflex CrAg test results were confirmed to be very reliable. Interlaboratory comparisons are useful to identify errors not detected by PTSs and internal quality control measures and should be conducted more frequently to ensure ongoing excellent performance.
Supplementary Material
Acknowledgments
We thank Adeboye Adelekan from US Centers for Disease Control and Prevention, South Africa; Ivy Rukasha, Molebogeng Kolojane, Phelly Matlapeng, Nikiwe Valashiya, and Siphiwe Kutta from the National Institute for Communicable Diseases; and participating staff at all National Health Laboratory Service CD4 and microbiology laboratories.
Glossary
Abbreviations:
- CrAg
cryptococcal antigen
- LFA
lateral flow assay
- WHO
World Health Organization
- HIV
human immunodeficiency virus
- NHLS
National Health Laboratory Service
- CSF
cerebrospinal fluid
- LP
lumbar puncture
- PTS
proficiency testing scheme
- LIS
laboratory information system
- EDTA
ethylene diamine tetra acetic acid
- NICD
National Institute for Communicable Diseases
- EIA
enzyme immunoassay
Contributor Information
Nozuko P Blasich, Centre for Healthcare-Associated Infections, Antimicrobial Resistance and Mycoses, National Institute for Communicable Diseases, a Division of the National Health Laboratory Service, Johannesburg, South Africa.
Lindi M Coetzee, School of Pathology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa; National Priority Programmes CD4 Unit, National Health Laboratory Service, Johannesburg, South Africa.
Charlotte Sriruttan, Centre for Healthcare-Associated Infections, Antimicrobial Resistance and Mycoses, National Institute for Communicable Diseases, a Division of the National Health Laboratory Service, Johannesburg, South Africa; School of Pathology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa.
Daniel DeSanto, Centre for Healthcare-Associated Infections, Antimicrobial Resistance and Mycoses, National Institute for Communicable Diseases, a Division of the National Health Laboratory Service, Johannesburg, South Africa.
Gregory S Greene, Centre for Healthcare-Associated Infections, Antimicrobial Resistance and Mycoses, National Institute for Communicable Diseases, a Division of the National Health Laboratory Service, Johannesburg, South Africa.
Deborah K Glencross, School of Pathology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa; National Priority Programmes CD4 Unit, National Health Laboratory Service, Johannesburg, South Africa.
Nelesh P Govender, Centre for Healthcare-Associated Infections, Antimicrobial Resistance and Mycoses, National Institute for Communicable Diseases, a Division of the National Health Laboratory Service, Johannesburg, South Africa; School of Pathology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa; Division of Medical Microbiology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa.
Disclosures
CrAg LFA kits for retesting at microbiology laboratories were sourced at no cost to the National Health Laboratory Service from Immuno-Mycologics for the purpose of this project. IMMY was not involved in the design and implementation of the study or analysis of data.
Funding
This work was supported by a Cooperative Agreement between the National Health Laboratory Service and US Centers for Disease Control and Prevention (CDC-RFA-GH15-1575), and a grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (www.niaid.nih.gov) under Award Number R01AI118511. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or US Centers for Disease Control and Prevention.
References
- 1. Govender NP, Meintjes G, Mangena P, et al. Southern African HIV Clinicians Society guideline for the prevention, diagnosis, and management of cryptococcal disease among HIV-infected persons: 2019 update. South Afr J HIV Med. 2019;20(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. Guidelines for the diagnosis, prevention, and management of cryptococcal disease in HIV-infected adults, adolescents, and children; 2018. https://www.who.int/publications/i/item/9789241550277. Accessed May 19, 2022.
- 3. Coetzee L, Cassim N, Moodley K, et al. Roadmap for implementing a national early detection programme for reflexed CrAg screening in national health CD4 laboratories in South Africa. Presented at 7th South African AIDS Conference; Durban, South Africa; June 9–12, 2014. [Google Scholar]
- 4. Coetzee LM, Cassim N, Glencross DK. Rapid scale up of reflexed cryptococcal antigen screening across a CD4 laboratory network in South Africa. Presented at International AIDS Society Conference; Paris, France; July 23-26, 2017. [Google Scholar]
- 5. Govender NP, Glencross DK. National coverage of reflex cryptococcal antigen screening: a milestone achievement in the care of persons with advanced HIV disease. South Afr Med J. 2018;108(7):534–535. [DOI] [PubMed] [Google Scholar]
- 6. Larson BA, Rockers PC, Bonawitz R, et al. Screening HIV-infected patients with low CD4 counts for cryptococcal antigenemia prior to initiation of antiretroviral therapy: cost effectiveness of alternative screening strategies in South Africa. PLoS One. 2016;11(7):e0158986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greene G, Desanto D, Matlapeng P, Govender N. Cryptococcal antigen screening surveillance report, South Africa, February 2017–July 2019. NICD Public Health Surveillance Bulletin, 2019.https://www.nicd.ac.za/wp-content/uploads/2019/12/CRYPTOCOCCAL-ANTIGEN-SCREENING-SURVEILLANCE-NICD-Bulletin-Vol17-Iss3-December2019.pdf. Updated December 1, 2019. Accessed May 19, 2022. [Google Scholar]
- 8. Lourens A, Jarvis JN, Meintjes G, Samuel CM. Rapid diagnosis of cryptococcal meningitis by use of lateral flow assay on cerebrospinal fluid samples: influence of the high-dose “hook” effect. J Clin Microbiol. 2014;52(12):4172–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cassim N, Coetzee LM, Govender NP, et al. District and sub-district analysis of cryptococcal antigenaemia prevalence and specimen positivity in KwaZulu-Natal, South Africa. Afr J Lab Med. 2018;7(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coetzee LM, Cassim N, Glencross DK. Cryptococcal antigen (CrAg) positivity rates reported from a national CD4-reflexed screening programme identify high-risk regions of co-existent HIV/Cryptococcal disease, requiring urgent programmatic focus into care. Presented at 11th Interest Workshop; Lilongwe, Malawi; May 16–19, 2017. [Google Scholar]
- 12. Glencross DK, Aggett HM, Stevens WS, et al. African regional external quality assessment for CD4 T‐cell enumeration: development, outcomes, and performance of laboratories. Cytometry. 2008;74(S1):S69–S79. [DOI] [PubMed] [Google Scholar]
- 13. Glencross DK, Janossy G, Coetzee LM, et al. Large‐scale affordable PanLeucogated CD4+ testing with proactive internal and external quality assessment: in support of the South African national comprehensive care, treatment and management programme for HIV and AIDS. Cytometry. 2008;74(S1):S40–S51. [DOI] [PubMed] [Google Scholar]
- 14. Jarvis JN, Percival A, Bauman S, et al. Evaluation of a novel point-of-care cryptococcal antigen test on serum, plasma, and urine from patients with HIV-associated cryptococcal meningitis. Clin Infect Dis. 2011;53(10):1019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hansen J, Slechta ES, Gates-Hollingsworth MA, et al. Large-scale evaluation of the immuno-mycologics lateral flow and enzyme-linked immunoassays for detection of cryptococcal antigen in serum and cerebrospinal fluid. Clin Vaccine Immunol. 2013;20(1):52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lindsley MD, Mekha N, Baggett HC, et al. Evaluation of a newly developed lateral flow immunoassay for the diagnosis of cryptococcosis. Clin Infect Dis. 2011;53(4):321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Immy Mycologics. ALPHA cryptococcal antigen EIA. https://immy.com/package_inserts/cry101/CRY101%20IFU%20-%20English.pdf. Accessed May 19, 2022.
- 18. Immy Mycologics. Cryptococcal antigen lateral flow assay. 2016. https://www.immy.com/package_inserts/cr2003/CR2003%20IFU%20(Int’l)%20-%20English.pdf. Accessed May 19, 2022.
- 19. Coetzee L-M, Cassim N, Glencross DK. Using laboratory data to categorise CD4 laboratory turn-around-time performance across a national programme. Afr J Lab Med. 2018;7(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coetzee L-M, Cassim N, Sriruttan C, et al. Cryptococcal antigen positivity combined with the percentage of HIV-seropositive samples with CD4 counts <100 cells/μl identifies districts in South Africa with advanced burden of disease. PLoS One. 2018;13(6):e0198993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.